Abstract
It is well recognized that flowering plants maintain a particularly broad spectrum of factors to support gene expression in mitochondria. Many of these factors are pentatricopeptide repeat (PPR) proteins that participate in virtually all processes dealing with RNA. One of these processes is the post-transcriptional generation of mature 5′ termini of RNA. Several PPR proteins are required for efficient 5′ maturation of mitochondrial mRNA and rRNA. These so-called RNA PROCESSING FACTORs (RPF) exclusively represent P-class PPR proteins, mainly composed of canonical PPR motifs without any extra domains. Applying the recent PPR-nucleotide recognition code, binding sites of RPF are predicted on the 5′ leader sequences. The sequence-specific interaction of an RPF with one or a few RNA substrates probably directly or indirectly recruits an as-yet-unidentified endonuclease to the processing site(s). The identification and characterization of RPF is a major step toward the understanding of the role of 5′ end maturation in flowering plant mitochondria.
Keywords: Arabidopsis thaliana, RESTORER OF FERTILITY-like proteins, RNA PROCCESSING FACTORS, endonucleolytic processing, pentatricopeptide repeat proteins, post-transcriptional 5′ maturation
Introduction
In mitochondria and chloroplasts, pentatricopeptide repeat (PPR) proteins are the major contributors to the wealth of factors dedicated to post-transcriptional processes.1 With the help of the overwhelming fundamental knowledge about the model plant species Arabidopsis thaliana (Arabidopsis) and its well-developed genomic and genetic tools, it was possible to assign a large number of PPR proteins to specific post-transcriptional processes.1 PPR proteins specify cytidines to be edited, are involved in splicing, and also function as RNA PROCESSING FACTORs (RPF) required for the formation of distinct 5′ ends of mitochondrial transcripts.
In this review, we will summarize the recent progress made in the analysis of 5′ processing of mitochondrial RNAs. In this context, we will focus on the recently identified RPF. These PPR proteins, exclusively P-class members,2 specify the post-transcriptional formation of distinct 5′ termini of one or several mitochondrial transcripts. Despite the considerable progress made in the identification and characterization of the proteins involved in 5′ mRNA maturation, we can still only speculate about the biological importance of post-transcriptional 5′ end processing.
5′ termini of most mitochondrial-encoded RNAs derive from post-transcriptional processing
Mitochondrial transcripts exhibit a surprising complexity often with multiple transcripts of disparate lengths for a given gene.3-5 In recent years, it became clear that often different 5′ termini account for this transcript complexity.6-8 Generally, there are two different ways how 5′ ends can be created. Transcription initiation directly generates primary 5′ ends. Theoretically, these termini contain 5′ triphosphate groups, but these seem to be unstable so that also single phosphates can be found at primary 5′ termini. Nevertheless, the presence of two or three phosphates assigns these 5′ ends to transcription initiation and discriminates them from post-transcriptionally generated 5′ ends also called secondary ends. These termini never contain more than a single phosphate group. The discrimination between both types of ends is still difficult and often ambiguous since techniques to differentiate between both types of ends depend on the presence or absence of the tri- or diphosphate group.7,9 Therefore, new techniques that allow an unambiguous discrimination between both types of 5′ termini would be highly appreciated.
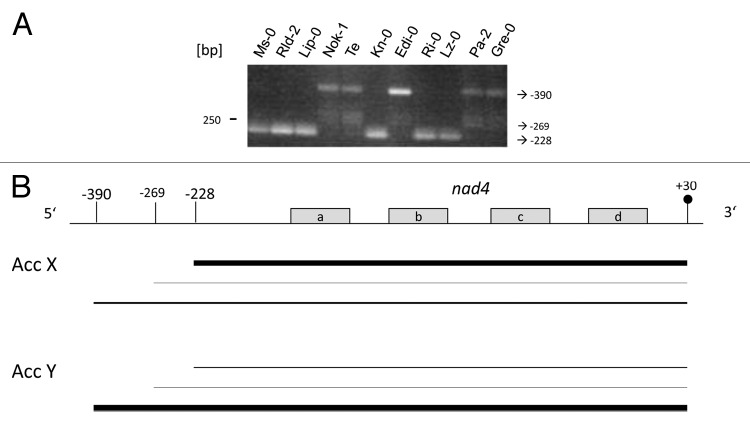
Despite these difficulties, several comprehensive analyses revealed that both transcription initiation as well as post-transcriptional 5′ processing contribute to the complex transcript pattern.6-8 In many cases there are several promoters but also multiple processing events that give rise to RNAs with many different 5′ ends (Fig. 1). These ends can be assigned to one or several major RNAs, which can easily be detected by northern blot analysis, or to low abundant transcripts that represent precursor RNAs or by-products of the post-transcriptional 5′ end formation process. The 5′ extremities of these extremely low abundant RNAs are detectable only by PCR, as for instance, the -269 5′ end of a nad4 transcript in Arabidopsis (Fig. 2).
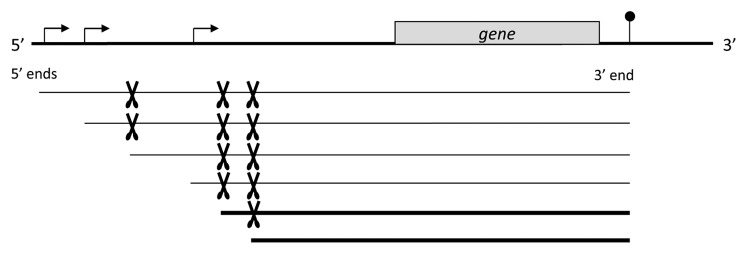
Figure 1. In plant mitochondria complex transcript pattern comprise multiple RNAs with identical 3′ ends (pinhead) but different 5′ termini. The latter originate directly from transcription initiation at multiple promoters (given by bent arrows) and/or from several post-transcriptional processing events (indicated by scissors). For many mitochondrial genes, low abundant RNAs (thin lines) are detectable. These might represent processing intermediates or by-products, or might be primary precursor molecules generated directly by transcription. The maturation process may result into one or several mature RNA species (thick lines), which contain predominantly post-transcriptionally generated 5′ termini.
Figure 2. Natural genetic variation generating nad4 transcript length polymorphisms in Arabidopsis. (A) CR-RT-PCR analysis was used to examine nad4 transcript termini in different Arabidopsis accessions. The PCR products of different sizes are generated from RNA molecules with distinct 5′ ends, whose positions are given in respect to the translation initiation codon on the right hand side of the image. These ends and the 3′ terminus (+30 in respect to the translation termination codon) have been mapped previously in other accessions.7,24 (B) Schema depicting the different nad4 mRNA species. Thick lines indicate major RNAs and thin lines represent low abundant transcript species. Grey boxes represent to nad4 exons a to d. Positions of 5′ and 3′ ends are indicated. Acc X represents ecotypes Ms-0, Rld-2, Lip-0, Kn-0, Ri-0, Lz-0 containing the short version of the mature nad4 mRNA (“short” thick line) and Acc Y represents ecotypes Nok-1, Te, Edi-0, Pa-2, and Gre-0 containing the long version of the major nad4 mRNA (“long” thick line). Schema not drawn to scale.
Even if there remained some doubts about the origin of one or the other 5′ end it seems clear that the 5′ termini of most major RNAs derive from post-transcriptional processing events and not directly from transcription initiation. In Arabidopsis, these post-transcriptionally generated 5′ termini were found in distances up to 645 nucleotides upstream of the translation initiation codon.8
There is no general sequence or secondary structure conservation at mitochondrial transcript termini
A comparison of the nucleotide sequences from positions -20 to +20 relative to the 5′ ends of all major mitochondrial transcripts failed to reveal conserved sequence motifs apart from about 24 nucleotides that are found more or less identical at the 5′ extremities of nad6 and atp9 mRNAs as well as of 26S rRNA.8 Otherwise, sequence conservation was found only at those few major ends that are located within conserved promoter motifs. Similarly, there is also only very little secondary structure conservation at the 5′ ends. Beside some small simple stem loops, a few tRNA-like structures (t-elements) were found either directly upstream or downstream of some 5′ termini. Some of these t-elements, which evolved from true tRNAs, were found to be processed either by the mitochondrial RNase P-like enzyme PROP (5′ end) or RNase Z (3′ end) in vitro, strongly suggesting that these enzymes are also involved in cleavage at the 5′ ends of t-elements in vivo. Likewise, such t-elements are also found at 3′ termini. Thus, depending on the location of these tRNA-like structures relative to the co-transcribed mRNA, processing of the t-elements contributes to 5′ and/or 3′ end formation of mitochondrial mRNAs in vivo, which was confirmed for 3′ end formation of the nad6 mRNA by PROP.10-12 Up to now, no evidence has been found for the presence of a 5′ to 3′ exonucleolytic activity in plant mitochondria, suggesting that an involvement of such an activity in 5′ processing is rather unlikely.4
In contrast to the variability at the 5′ ends, more or less no variation has been observed at the 3′ termini of plant mitochondrial transcripts. For almost all genes investigated, transcripts have single 3′ ends slightly scattering over a few nucleotides.8 Less heterogeneity of 3′ termini was also observed when mitochondrial untranslated regions (UTR) from wheat were compared with Arabidopsis.13 While there are eight 3′ UTR conserved between both species, only one such case is observed among 5′ UTR. Interestingly, some of these identical 3′ ends coincide with breakpoints in homology between the 3′ UTR sequences in wheat and in Arabidopsis.
Two 3′ to 5′ exoribonucleases, the dual targeted RNase R homolog 1 (RNR1) and the mitochondrial polynucleotide phosphorylase (PNPase), are found to be involved in 3′ end formation of atp9 transcripts. The latter enzyme is crucial also for the degradation of 18S rRNA processing by-products and for the turnover of the rRNA itself. Unfortunately, the general role of these enzymes in transcript maturation and RNA turnover has not yet been addressed but it seems that they act in concert with PPR proteins in the post-transcriptional generation of mRNA 3′ termini.14,15 Similar to a mechanism found in chloroplasts,16,17 a P-class PPR protein binds to the 3′ terminal region of a mitochondrial RNA (mtRNA), thereby protecting the transcript from degradation by the mitochondrial PNPase and concomitantly defining the 3′ end of the mRNA. This mechanism has very recently been described for MTSF1, a PPR protein required for 3′ end formation of the nad4 mRNA in mitochondria of Arabidopsis.18
Natural genetic variation affecting 5′ termini of RNAs
Studies of mitochondrial transcripts in different Arabidopsis accessions revealed that RNA polymorphisms are a frequent phenomenon.19-21 In general, deviating transcript lengths are either connected to differences in the mitochondrial DNA (mtDNA) or they originate from variations in nuclear genes required for 5′ processing. So far three RNA polymorphisms have been linked to mtDNA. In three accessions, the generation of larger cox2 mRNAs is connected to an accumulation of a specific mtDNA recombination product, indicating that this transcript length polymorphism is actually the result of a polymorphic mtDNA configuration stoichiometry. Likewise, alternative mtDNA arrangements located far upstream of the cox3 gene influence the lengths of the corresponding mRNAs in Landsberg erecta (Ler). In contrast, a polymorphism observed for ccmC mRNAs is linked to a sequence stretch of about 60 base pairs located a few nucleotides upstream of the distinct 5′ ends of the 1200-nucleotide, long ccmC transcript in Columbia (Col-type), and of the 1100-nucleotide, short ccmC mRNA in C24 (C24-type). These deviating sequences define the C24- and the Col-type ccmC mtDNA configurations, respectively, and it is reasonable to assume that they contain crucial parts of the cis-elements required for 5′ end formation of the different ccmC transcripts (see below).22
In the different Arabidopsis accessions, far more transcript length polymorphisms are linked to nuclear-encoded genes and these polymorphisms exhibit an amazing variability. The nad4 major transcript is found in two lengths differing at their 5′ ends. A long version was initially identified in Ler, while a short type of this transcript was found in Col, but this polymorphism is prevalent in many different accessions (Fig. 2).23 A different situation was found for the nad9 gene, for which either one or two transcripts are detected in the different accessions. Similar to the nad4 mRNA polymorphism, the nad9 transcript variation is frequently observed but the distribution of both polymorphisms among different accessions does not exhibit any correlation. Another form of a nuclear DNA-associated mtRNA polymorphism was observed for the nad3-rps12 transcripts. In different accessions the sizes of these dicistronic RNAs vary between 900 and 1000 nucleotides. Like other transcript-length polymorphisms, the distinct sizes of the nad3-rps12 transcripts result from different 5′ termini.21 Yet, another frequent polymorphism was found also among ccmB transcripts. Here an additional 5′ end is formed in many accessions.19
In contrast to the above mentioned frequent mRNA polymorphisms, a deviating nad2 mRNA pattern was detected exclusively in Can-0. In this accession, the “standard” nad2 transcript present in all other investigated ecotypes was found only in minor amounts. Instead, additional larger nad2 RNAs accumulate to comparatively high levels in Can-0, whereas these transcript species are more or less absent from the other accessions.21
Besides the clear polymorphisms of major transcripts, accession-specific differences were observed in the abundance of different RNA species or were found among minor abundant RNAs.19,21 Such cases are well documented for rpl5 and cox1 transcript species, respectively. It seems that PPR proteins are also involved in the generation of these minor RNA polymorphisms; however, further studies are required to identify the nuclear genes related to the distinct polymorphisms.
RESTORER of FERTILITY-like PPR proteins are required for 5′ end processing of mitochondrial RNAs in Arabidopsis
From the studies described above the following features of 5′ ends of plant mitochondrial transcripts emerged: First, often a number of transcripts with different 5′ termini can be detected for a given gene. Second, a remarkable variation of 5′ termini is seen among different Arabidopsis accessions. Third, these variations often affect main transcript species. Fourth, these polymorphisms are exclusively observed at post-transcriptionally generated 5′ termini.
The 5′ end polymorphisms of mitochondrial transcripts are per se a very interesting phenomenon, but they also paved the way for the further analyses of 5′ processing events. The transcript polymorphisms are based on natural genetic variation, which provides the basis for the map-based identification of nuclear-encoded genes required for 5′ processing. This experimental approach led to the identification of genes encoding RNA PROCESSING FACTOR1 (RPF1) and RNA PROCESSING FACTOR2 (RPF2) in Arabidopsis.23,24
RPF1 (At1g12700) was identified on the upper arm of chromosome 1, where it is located within a gene cluster encoding several P-class PPR proteins.23 This gene encodes a PPR protein with high similarity to RESTORERs of FERTILITY (RF) identified in cytoplasmic male sterility (CMS)/RF systems in several plant species.25-32 CMS is a maternally inherited deficiency to produce or release fertile pollen.33-36 This trait, which is caused by chimeric, CMS type-specific mitochondrial genes, is used for the generation of F1 hybrid seed from various crop species. In the presence of an RF gene, male fertility is reconstituted in the F1 plants by reducing the steady-state level of the deleterious CMS-associated mitochondrial protein either by decreasing the stability of the corresponding RNA or by reducing translation. In Arabidopsis, up to 26 RF-like PRR genes have been identified predominantly in two clusters on chromosome 1.27,32 In this model species, CMS has recently been observed in offspring of a crossing between the distantly related accessions Shahdara (Sha) and Monte-Rosso (Mr-0). Male sterility seems to be caused by the interaction of a novel open reading frame (orf117) present in Sha with two nuclear genome regions on chromosomes 1 and 3 in Mr-0. Since the nuclear genes in these regions have not yet been identified it remains unclear whether the CMS in this autogamous model plant species is associated with one of the nuclear genes encoding an RF-like PPR protein.37
RPF1 is required for the efficient generation of the -228 5′ end of the nad4 mRNA (Table 1). In Ler and many other accessions, the RPF1 gene has lost its function by the presence of a premature translation stop codon, which leads to the accumulation of a 5′ extended larger nad4 mRNA with a 5′ end at position -390 (Fig. 2).
Table 1.
| Protein | AGI number | Number of PPRs | RF-like | Target RNA | 5′end affected# |
|---|---|---|---|---|---|
|
RPF1 |
At1g12700 |
14 |
yes |
nad4 |
-228 in Col |
|
RPF2 |
At1g62670 |
15 |
yes |
nad9 |
-202 in Col |
|
cox3 |
-380 in Col |
||||
|
RPF3 |
At1g62930 |
15 |
yes |
ccmC |
-484/-482 in Col |
|
RPF4* |
|
|
yes |
ccmB |
-231 and -200 in Ler |
|
RPF5 |
At4g28010 |
17 |
no |
nad6 |
-179 in Col |
|
atp9 |
-84 in Col |
||||
| 26S |
+1 in Col |
||||
|
RPF6* |
|
|
yes |
ccmC |
-391/-390 in C24 |
| RPF8* | no | nad2 | -122 in Col |
Analysis in progress. (#) Positions given with respect to the translation initiation codon or to 5′ end of the mature 26S rRNA.
RPF2 (At1g62670) was mapped to the lower arm of chromosome 1, where it is located in the second cluster containing most of the genes encoding RF-like PPR proteins in Arabidopsis.24 RPF2 is required for the generation of a nad9 mRNA with a 5′ end at position -202, which is absent from C24 (Table 1). Likewise, this nad9 transcript species does not accumulate in the rpf2-1 mutant, in which the RPF2 gene is knocked out. Further studies of the rpf2-1 line showed RPF2 to be also involved in 5′ maturation of cox3 RNAs. Unlike RPF1, where the absence of 5′ processing at position -228 of the nad4 transcript is strictly correlated with the presence of a premature translation stop codon in all accessions investigated, the RPF2 gene function is inactivated in different ways in the different ecotypes, either by distinct recombination events or by nucleotide substitutions.24
Since both RPF1 and RPF2 encode RF-like PPR proteins, it was speculated that this group of proteins plays a particular role in 5′ processing of mitochondrial transcripts. Therefore, several mutants with T-DNA insertions in different RF-like PPR genes were screened for alterations of mitochondrial mRNAs. This identified RPF3, encoded by At1g62930, which is required for the accumulation of the ccmC mRNA in accession Col (Table 1).22 Interestingly, the function of RPF3 is restricted to Col-type ccmC transcripts, whereas this protein seems to have little effect on the generation of ccmC mRNAs derived from the C24-type ccmC mtDNA configuration (see above). In contrast to RPF1 and RPF2, whose inactivation has no consequences for the steady-state levels of the Nad4, Nad9, and Cox3 polypeptides, the knockout of the RPF3 gene leads to a substantial reduction of the CcmC polypeptide. Nevertheless, the knockout plants did not reveal any macroscopic phenotype and the activities of respiratory Complexes I, III, and IV are also indistinguishable from wild-type. Thus, it was speculated that the CcmC protein is dispensable for the function of flowering plant mitochondria.
Apart from RPF1 to 3 other RF-like PPR proteins are involved in 5′ processing of mtRNA. Although still under investigation in our laboratory, linkage analyses demonstrate that RPF4 and RPF6 encode RF-like PPR proteins confirming the important role of this group of proteins in the mitochondrial 5′ maturation process (Table 1). RPF4 is involved in the generation of additional ccmB transcripts in Ler and many other accessions (K Stoll, C Jonietz, A Hölzle, and S Binder, unpublished results), while RPF6 is required for the 5′ end formation of ccmC RNAs transcribed from the C24-type ccmC mtDNA configuration (K Stoll, C Jonietz, C Stutzer, and S Binder, unpublished results).8
Taken together, these studies demonstrate the important role of RF-like PPR proteins in 5′ processing of mtRNA. A comparative analysis of the genes encoding RF-like PPR proteins from angiosperms showed that this group of PPR genes undergoes an extraordinary rapid evolution with high rates of non-synonymous to synonymous substitutions.32 The enhanced diversity of RPF2 alleles found in Arabidopsis accessions suggests that RF-like proteins show an extreme variability also within a species, which explains the frequent appearance of 5′ end polymorphisms.
But what is the function of the real RF proteins? Up to now, the exact molecular function of most of these proteins is unknown. Well described is the role of RF1A from the rice Boro II CMS/fertility restoration system. Analogous to RF-like RPF, this protein is involved in an endonucleolytic cleavage reaction. Only the presence of RF1A allows an endonucleolytic cut within the dicistronic, CMS-specific B-nad6-orf79 transcript, which releases a stable orf79 mRNA. In contrast to the dicistronic mRNA, the orf79 transcript is translated with very low efficiency, which results into fertility restoration.31,38
An influence of both mtDNA sequence and nuclear-encoded genes on mitochondrial transcripts has also been observed in Silene vulgaris. In this species, sequences as well as arrangements of mtDNA exhibit extreme intraspecific variations.39 As exemplarily demonstrated by the analysis of the atp1 gene, variability of both mtDNA sequences and nuclear-encoded genes influences the corresponding transcript pattern.40
P-class PPR proteins are involved in efficient 5′ end formation of plant mitochondrial transcripts
Beside the RF-like RPF, PPR proteins not classified as RF-like proteins have a strong impact on 5′ termini of mitochondrial transcripts as well. Very recently, RPF5 (At4g28010) was found to be required for 5′ maturation of three different RNA species sharing conserved sequences at the processing sites (Table 1).41 While the knockout of RPF5 has a moderate effect on processing of atp9 mRNAs, it provokes severe defects in processing of the nad6 mRNA and the 26S rRNA. Thus, this protein is involved in processing of mRNAs and rRNA indicating that there is no fundamental difference between 5′ processing of both types of RNA in plant mitochondria.
Despite the important role of RPF5 in nad6 mRNA processing and the severe reduction of corresponding mature mRNA, the steady-state level of the Nad6 polypeptide is unaltered in the rpf5-1 mutant. Interestingly, also the strongly reduced processing of the 26S rRNA seems to have no effect on the accumulation of mitochondrial-encoded proteins, although the majority of the 26S rRNA in the mutant contains the premature -60 5′ end. This observation suggests that ribosomes containing the aberrant 5′ elongated large rRNA are functional.41
The RPF5 gene was identified by the analysis of several Arabidopsis lines with T-DNAs in genes encoding regular P-class PPR proteins. Altered transcript patterns as they were found in the rpf5-1 mutant have never been observed in any of the Arabidopsis accessions, strongly suggesting that “natural mutants” of this gene do not exist or are extremely rare. A rare polymorphism is also observed for the nad2 mRNA, which is found to deviate from its normal size only in a single accession. Our preliminary analyses indicate that the corresponding RPF, which we designate RPF7, is encoded by a “normal” P-class PPR gene and not by an RF-like gene (Table 1B, Stoll and S Binder, unpublished results).
Taken together, these studies show that all of the factors involved in 5′ processing belong to the P-class PPR proteins. High variability of 5′ ends in different Arabidopsis accessions seems to be restricted to those 5′ termini that require an RF-like PPR protein for processing. The presently observed overrepresentation of the RF-like proteins among the RPF might just be related to the experimental approach that has been used for their identification. This approach is based on natural genetic variation, which is extraordinarily high among the fast-evolving RF-like genes and this feature led to a preferential identification of such genes. Most likely, a systematic analysis of knockout mutants affecting regular P-class PPR proteins will identify more non-RF-like processing factors.
Apart from the RPF described in Arabidopsis, another P-class PPR protein influencing mitochondrial 5′ termini was identified in maize. MPPR6, which is conserved in other species including Arabidopsis, is required for correct 5′ end formation of the rps3 mRNA and proper translation of this transcript. A knockout of this gene interferes with a normal development of maize kernels (further details see below).42
Mitochondrial RPF bind upstream of the processing sites
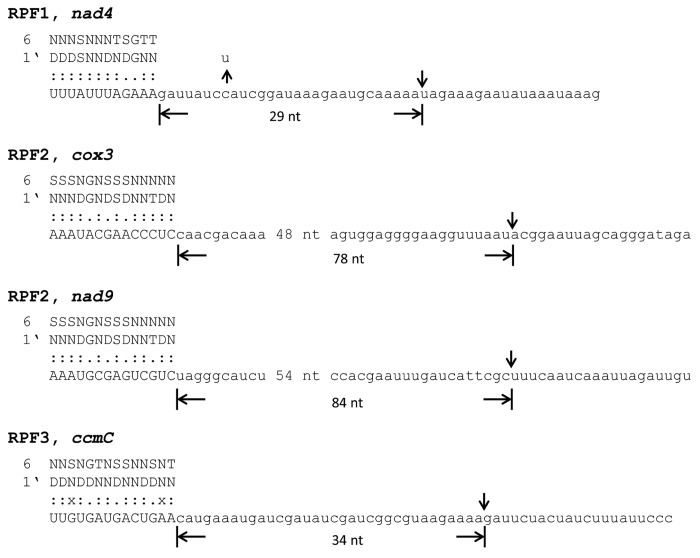
As mentioned above the recent elucidation of the combinatorial amino acid code for RNA recognition by PPR motifs now allows the prediction of binding sites for PPR proteins.43,44 These putative target sites of the above described RPF are summarized in Figure 3. The lengths of the potential binding sites vary between 11 and 14 nucleotides. This comparatively small variation correlates with the very similar number of canonical PPR motifs found within RPF, which ranges between 14 and 17 (Table 1). However, considering the small number of RPF identified these characteristics must be regarded as preliminary and variation of binding site features might increase with the identification and analysis of additional RPF. In addition, the predictions now need to be experimentally confirmed. This can be done either directly by electrophoretic mobility shift assays or indirectly by detection of RNA footprints, which represent small RNA that were protected from exonucleolytic degradation by the bound PPR proteins. Such RNA footprints from chloroplasts have recently been identified in deep-sequence data.45
Figure 3. Binding site predictions of RNA PROCESSING FACTORs 1, 2, 3, and 5. Following the recently suggested amino acid code for the interaction of PPRs with distinct nucleotide identities, binding sites were predicted on different RNA substrates. Amino acid identities of positions 6 and 1’ (position 1 of the following repeat) are given in the single letter code. Perfect matches (:), poor matches (.), and mismatches (x) are indicated. All potential binding sites were identified on 5′ leader sequences. The distances between the 3′ terminal nucleotides of the putative target sites and the 5′ ends of the mature RNA are given in nucleotides (nt). An RNA editing site identified in the 5′ UTR of the nad4 mRNA is indicated (U above a vertical arrow).
All of the predicted binding sites are located in the leader sequences upstream of the 5′ end of the mature RNAs (Fig. 3). The distances between the C-terminal motifs of the proteins and the cleavage sites range between 29 and 84 nucleotides. The positioning of the binding sites 5′ to the processing sites is similar to the potential attachment sites of the RNA editing factors. The latter bind with their last C-terminal motif in identical distances of five nucleotides upstream of the edited cytidine.43 Thus, the editing factors seem to bind much closer to processing sites than presumably PPR proteins involved in 5′ end maturation do.
Particularly interesting binding predictions were obtained for the three target RNAs of RPF5.43,44 While the recognition patterns between RPF5 and nad6 as well as RPF5 and 26S rRNA are found to be identical, a different interaction is predicted between RPF5 and the atp9 precursor RNA (Fig. 3). The distinct interaction patterns correspond well with the differing importance of this protein in 5′ processing of the three RNAs. While RPF5 is crucial for 5′ end formation of the nad6 mRNA and 26S rRNA, this protein has just a supporting role in 5′ end formation of the atp9 mRNA.
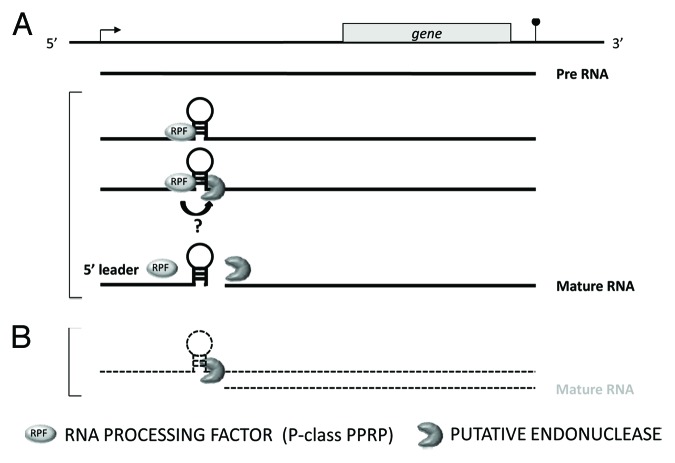
In contrast to the mitochondrial 5′ mRNA processing, 5′ end formation of plastid-located mRNAs follows a completely different mechanism. Proteins like PPR10 from maize attach to the RNA, thereby protecting the bound RNA molecule from degradation by 5′ exonucleolytic activity. As consequence, the binding sites of these proteins correspond to about 20 nucleotides at the 5′ terminus of the processed mRNA.16 The predicted binding of the mitochondrial RPF upstream of the processing sites and not directly to the 5′ terminal sequences of the mRNA indicates that 5′ maturation in mitochondria follows a basically different mechanism. The distant binding and the variability of the distances between the putative RPF binding and the cleavage sites implies that folding of the RNA might be important for processing (Fig. 4), which seems to be irrelevant for 5′ end formation in plastids. To the contrary, 3′ processing of mitochondrial mRNAs is very similar to the formation of 3′ termini in chloroplasts. In both organelles, PPR proteins act as protein caps protecting bound RNAs from degradation by 3′ exonucleases.16,18
Figure 4. Schema depicting a model for the endonucleolytic generation of a mature 5′ end of a mitochondrial transcript. A promoter (bent arrow) and the 3′ end (pinhead) are indicated. (A) Potentially two different proteins are involved in this process. An RNA PROCESSING FACTOR (RPF) interacts sequence-specifically with the target RNA. This interaction might stabilize a secondary structure (here indicated by a stem loop structure) that directs a potential, yet unidentified, endonuclease to the cleavage site. The endonucleolytic cut releases a 5′ leader molecule and a 5′ mature mRNA. Transcript species in this pathway are given as bold lines. Whether the recruitment process requires a direct contact between an RPF and the putative endonucleolytic protein is unclear. Possibly these proteins collaborate indirectly via the RNA substrate. (B) The secondary structure might also be spontaneously formed in the absence of the RPF, which might allow an inefficient generation of low amounts of mature mRNA (transcripts given as dashed lines). Such background levels of 5′ matured mRNA are frequently observed in plant lines lacking the different RPFs.
Plant mitochondrial 5′ end processing is achieved by endonucleolytic cleavage
How the plant mitochondrial 5′ ends are generated is not yet finally clear. As mentioned in the previous sections, there is evidence for endonucleolytic processing of several 5′ ends. This evidence was obtained indirectly by the analysis of the cleavage products. An endonucleolytic cut generates two RNA molecules. A 5′ matured mRNA and a 5′ leader molecule. Thus, the detection of a 5′ leader molecule with a 3′ end that coincides with the nucleotide just upstream of a 5′ end of the mature mRNA is a very strong indirect evidence for an endonucleolytic cleavage reaction. But unfortunately the 5′ leaders are rather unstable, which hampers the straight forward detection of such molecules. Nevertheless, it was possible to detect such 5′ leader molecules.
Up to now nothing is known about the enzyme that performs the endonucleolytic cleavage. It is highly unlikely that the RPF themselves exhibit endonucleolytic activity, although a recombinant DYW-type PPR protein showed such an activity in vitro.46 Rather, one or more endonucleases are involved in the scission reaction. In contrast to plastids, where a whole bunch of endonucleases has been described,47 only two such enzymes have been identified in mitochondria. These proteins, PROP and RNase Z, are predominantly involved in tRNA processing.10-12 When tRNAs or t-elements are co-transcribed with mRNAs, these enzymes contribute also to the post-transcriptional generation of 5′ or 3′ ends of mitochondrial mRNAs. Considering the requirement of tRNA secondary structure for processing by the tRNA processing enzymes one might speculate that other secondary structures, potentially stabilized by RPF, might be recognized by these endonucleolytic enzymes suggesting important roles of PROP and RNase Z also in mRNA as well as rRNA processing (Fig. 4A). But up to now, this model is rather speculative and requires experimental support. In addition, it is also possible that other yet unidentified endonucleases are involved in the cleavage reaction. Nevertheless, a mechanism involving secondary structures could also explain the residual generation of mature 5′ ends observed in the absence of an RPF in the corresponding knockout mutants (Fig. 4B). This “background” processing might be the result of a spontaneous formation of higher order structures, which allows a cleavage by the processing enzyme alone. Such a scenario might indicate that the cooperation of a potential endonuclease with an RPF does not necessarily require a direct contact between these proteins. Instead, the collaboration might be mediated by a distinct secondary structure of the RNA substrate that undergoes cleavage. This scenario is also supported by the fact that all RPF are P-class PPR proteins that lack additional domains required for protein-protein interactions. However, a direct interaction of RPF with a putative endonuclease cannot be completely ruled out.
Influence of 5′ end processing on RNA stability or protein accumulation
Up to now the analyses of the different PPR proteins required for processing did not reveal a homogenous picture about the influence of 5′ end maturation on the stability of RNAs. In the ecotype Ler, where RPF1 is inactivated, a defined nad4 precursor molecule accumulates to amounts very similar or even identical to the level of the mature mRNA in Col demonstrating more or less identical stabilities of both transcript species.23 To contrary, in the rpf5–1 mutant a defined 5′ extended 26S rRNA precursor RNA accumulates only to less than 50% in comparison to the mature 26S rRNA in wild type. This is accompanied by the detection of a 3′ truncated 26S rRNA degradation product, which contains the immature -60 5′ end. These observations demonstrate that the 5′ immature rRNA species is less stable than the completely processed mature rRNA suggesting that in the case of 26S rRNA 5′ maturation and degradation or even 3′ processing might be somehow interconnected. How the stability of an RNA species is increased by 5′ processing is unknown at present.
For all other processing events that have been studied in detail a relationship between 5′ maturation and RNA stability is less obvious since no distinct precursor molecules accumulate in the absence of processing. Thus a targeted approach to detect and analyze RNA degradation products is required to elucidate a detailed picture of a potential link between 5′ processing and the stability of an RNA.
Likewise, the impact of 5′ processing on the accumulation of mitochondrial-encoded proteins does not follow a consistent blueprint. This might be linked to transcript-specific features that could have different consequences for translation. But a consistent schema might be also blurred by other factors. First, in both natural and T-DNA mutants, there are still background amounts of 5′ matured mRNAs that can be translated. Second, often larger precursor RNAs accumulate in the absence of the RPF and it is unclear whether such 5′ extended RNAs can be also correctly translated or not. Third, low turn-over rates of mitochondrial-encoded proteins might allow protein accumulation indistinguishable from wild-type even if translation proceeds only with low efficiency. This could conceal a potential effect of 5′ processing in translation initiation. Thus, for most 5′ processing events it remains unknown whether there is any influence on translation. Up to date, only two instances have been described, in which impaired 5′ processing or the formation of alternative 5′ termini affects mitochondrial protein accumulation. First, the knockout of RPF3, which comes along with the lack of ccmC mRNA accumulation, leads to a severe decrease of the CcmC protein level.22 Second, the absence of MPPR6 in maize provokes the accumulation of 5′ extended rps3 mRNAs and also strongly reduces the accumulation of the ribosomal protein S3 (Rps3), which, in addition, is truncated at its N terminus. In contrast to RPF1, 2, 3, and 5, which are predicted to bind to sites within the cleaved 5′ leaders (Fig. 3), MPPR6 binds to the 5′ terminal region of the rps3 mRNA. Thus, it appears that this PPR protein is predominantly required for correct translation, which is corroborated by the generation of an N-terminally truncated Rps3 protein. This aberrant polypeptide seems to be non-functional, indicated by the substantially impaired mitochondrial translation in mppr6 mutants. The formation of 5′ extended rps3 mRNA species in the corresponding mutant might be an indirect outcome of the MPPR6 function in translation initiation. Probably, this PPR protein influences secondary structure formation, which, in turn, affects 5′ processing of the rps3 transcript.42 To get more information about the functional relationship between 5′ processing and translation, we need to understand how protein biosynthesis works in plant mitochondria. In this context, a mitochondrial in vitro translation system would be an extremely valuable tool to study a potential impact of transcript maturation on translation initiation.
Conclusion
Although substantial progress has been made in the characterization of 5′ processing of mitochondrial transcripts in plants, still many important questions about this frequently observed process remain open. Future studies will be focused on the identification of the endonucleolytic enzyme(s) required for cleavage and on the exact molecular function of the RPF in processing. But most importantly, studies should concentrate on the functional relationship between 5′ and 3′ processing as well as RNA stability and of course the impact of 5′ end maturation on translation. These studies will finally bring down general information about the importance of 5′ processing in plant mitochondrial gene expression.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Benjamin Stupfler for his transcript studies in different Arabidopsis accessions. We are deeply grateful to Ian Small for binding site predictions. Work on 5′ processing of mitochondrial RNA is supported by the Deutsche Forschungsgemeinschaft (Bi 590/10-2).
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/26129
References
- 1.Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008;13:663–70. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binder S, Brennicke A. Gene expression in plant mitochondria: transcriptional and post-transcriptional control. Philos Trans R Soc Lond B Biol Sci. 2003;358:181–8, discussion 188-9. doi: 10.1098/rstb.2002.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gagliardi D, Binder S. Expression of the plant mitochondrial genome. In: Logan D, ed. Plant mitochondria. Ames, IA, USA: Blackwell Publishing, 2007:50-95. [Google Scholar]
- 5.Binder S, Hölzle A, Jonietz C. RNA processing and RNA stability in plant mitochondria. In: Kempken F, ed. Plant Mitochondria. New York, NY, USA: Springer Science and Business Media, 2010:pp. 107-30. [Google Scholar]
- 6.Kühn K, Richter U, Meyer EH, Delannoy E, de Longevialle AF, O’Toole N, Börner T, Millar AH, Small ID, Whelan J. Phage-type RNA polymerase RPOTmp performs gene-specific transcription in mitochondria of Arabidopsis thaliana. Plant Cell. 2009;21:2762–79. doi: 10.1105/tpc.109.068536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kühn K, Weihe A, Börner T. Multiple promoters are a common feature of mitochondrial genes in Arabidopsis. Nucleic Acids Res. 2005;33:337–46. doi: 10.1093/nar/gki179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forner J, Weber B, Thuss S, Wildum S, Binder S. Mapping of mitochondrial mRNA termini in Arabidopsis thaliana: t-elements contribute to 5′ and 3′ end formation. Nucleic Acids Res. 2007;35:3676–92. doi: 10.1093/nar/gkm270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kempken F, Bolle N, Forner J, Binder S. Transcript end mapping and analysis of RNA editing in plant mitochondria. In: Leister D, Herrmann JM, eds. Mitochondria; Practical Protocols. Totowa, NJ, USA: Humana Press Inc., 2007:177-92. [DOI] [PubMed] [Google Scholar]
- 10.Canino G, Bocian E, Barbezier N, Echeverría M, Forner J, Binder S, Marchfelder A, Marchfelder A. Arabidopsis encodes four tRNase Z enzymes. Plant Physiol. 2009;150:1494–502. doi: 10.1104/pp.109.137950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutmann B, Gobert A, Giegé P. PRORP proteins support RNase P activity in both organelles and the nucleus in Arabidopsis. Genes Dev. 2012;26:1022–7. doi: 10.1101/gad.189514.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gobert A, Gutmann B, Taschner A, Gössringer M, Holzmann J, Hartmann RK, Rossmanith W, Giegé P, Rossmanith W, Giegé P. A single Arabidopsis organellar protein has RNase P activity. Nat Struct Mol Biol. 2010;17:740–4. doi: 10.1038/nsmb.1812. [DOI] [PubMed] [Google Scholar]
- 13.Choi B, Acero MM, Bonen L. Mapping of wheat mitochondrial mRNA termini and comparison with breakpoints in DNA homology among plants. Plant Mol Biol. 2012;80:539–52. doi: 10.1007/s11103-012-9966-2. [DOI] [PubMed] [Google Scholar]
- 14.Perrin R, Lange H, Grienenberger JM, Gagliardi D. AtmtPNPase is required for multiple aspects of the 18S rRNA metabolism in Arabidopsis thaliana mitochondria. Nucleic Acids Res. 2004;32:5174–82. doi: 10.1093/nar/gkh852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perrin R, Meyer EH, Zaepfel M, Kim YJ, Mache R, Grienenberger JM, Gualberto JM, Gagliardi D, Gualberto JM, Gagliardi D. Two exoribonucleases act sequentially to process mature 3′-ends of atp9 mRNAs in Arabidopsis mitochondria. J Biol Chem. 2004;279:25440–6. doi: 10.1074/jbc.M401182200. [DOI] [PubMed] [Google Scholar]
- 16.Pfalz J, Bayraktar OA, Prikryl J, Barkan A. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 2009;28:2042–52. doi: 10.1038/emboj.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prikryl J, Rojas M, Schuster G, Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc Natl Acad Sci U S A. 2011;108:415–20. doi: 10.1073/pnas.1012076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haïli N, Arnal N, Quadrado M, Amiar S, Tcherkez G, Dahan J, Briozzo P, Colas des Francs-Small C, Vrielynck N, Mireau H, Briozzo P, Colas des Francs-Small C, Vrielynck N, Mireau H. The pentatricopeptide repeat MTSF1 protein stabilizes the nad4 mRNA in Arabidopsis mitochondria. Nucleic Acids Res. 2013;41:6650–63. doi: 10.1093/nar/gkt337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forner J, Hölzle A, Jonietz C, Thuss S, Schwarzländer M, Weber B, Meyer RC, Binder S, Weber B, Meyer RC, Binder S. Mitochondrial mRNA polymorphisms in different Arabidopsis accessions. Plant Physiol. 2008;148:1106–16. doi: 10.1104/pp.108.126201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forner J, Weber B, Wiethölter C, Meyer RC, Binder S. Distant sequences determine 5′ end formation of cox3 transcripts in Arabidopsis thaliana ecotype C24. Nucleic Acids Res. 2005;33:4673–82. doi: 10.1093/nar/gki774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoll B, Stoll K, Steinhilber J, Jonietz C, Binder S. Mitochondrial transcript length polymorphisms are a widespread phenomenon in Arabidopsis thaliana. Plant Mol Biol. 2013;81:221–33. doi: 10.1007/s11103-012-9993-z. [DOI] [PubMed] [Google Scholar]
- 22.Jonietz C, Forner J, Hildebrandt T, Binder S. RNA PROCESSING FACTOR3 is crucial for the accumulation of mature ccmC transcripts in mitochondria of Arabidopsis accession Columbia. Plant Physiol. 2011;157:1430–9. doi: 10.1104/pp.111.181552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hölzle A, Jonietz C, Törjek O, Altmann T, Binder S, Forner J. A RESTORER OF FERTILITY-like PPR gene is required for 5′-end processing of the nad4 mRNA in mitochondria of Arabidopsis thaliana. Plant J. 2011;65:737–44. doi: 10.1111/j.1365-313X.2010.04460.x. [DOI] [PubMed] [Google Scholar]
- 24.Jonietz C, Forner J, Hölzle A, Thuss S, Binder S. RNA PROCESSING FACTOR2 is required for 5′ end processing of nad9 and cox3 mRNAs in mitochondria of Arabidopsis thaliana. Plant Cell. 2010;22:443–53. doi: 10.1105/tpc.109.066944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bentolila S, Alfonso AA, Hanson MR. A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc Natl Acad Sci U S A. 2002;99:10887–92. doi: 10.1073/pnas.102301599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown GG, Formanová N, Jin H, Wargachuk R, Dendy C, Patil P, Laforest M, Zhang J, Cheung WY, Landry BS, Laforest M, Zhang J, Cheung WY, Landry BS. The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. Plant J. 2003;35:262–72. doi: 10.1046/j.1365-313X.2003.01799.x. [DOI] [PubMed] [Google Scholar]
- 27.Desloire S, Gherbi H, Laloui W, Marhadour S, Clouet V, Cattolico L, Falentin C, Giancola S, Renard M, Budar F, et al. Identification of the fertility restoration locus, Rfo, in radish, as a member of the pentatricopeptide-repeat protein family. EMBO Rep. 2003;4:588–94. doi: 10.1038/sj.embor.embor848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu J, Wang K, Huang W, Liu G, Gao Y, Wang J, Huang Q, Ji Y, Qin X, Wan L, et al. The rice pentatricopeptide repeat protein RF5 restores fertility in Hong-Lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162. Plant Cell. 2012;24:109–22. doi: 10.1105/tpc.111.093211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kazama T, Toriyama K. A pentatricopeptide repeat-containing gene that promotes the processing of aberrant atp6 RNA of cytoplasmic male-sterile rice. FEBS Lett. 2003;544:99–102. doi: 10.1016/S0014-5793(03)00480-0. [DOI] [PubMed] [Google Scholar]
- 30.Koizuka N, Imai R, Fujimoto H, Hayakawa T, Kimura Y, Kohno-Murase J, Sakai T, Kawasaki S, Imamura J, Sakai T, Kawasaki S, Imamura J. Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. Plant J. 2003;34:407–15. doi: 10.1046/j.1365-313X.2003.01735.x. [DOI] [PubMed] [Google Scholar]
- 31.Wang Z, Zou Y, Li X, Zhang Q, Chen L, Wu H, Su D, Chen Y, Guo J, Luo D, et al. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell. 2006;18:676–87. doi: 10.1105/tpc.105.038240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujii S, Bond CS, Small ID. Selection patterns on restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proc Natl Acad Sci U S A. 2011;108:1723–8. doi: 10.1073/pnas.1007667108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Budar F, Barthome R. cytoplasmic male sterilities and mitochondrial gene mutations in plants. In: Logan D, ed. Plant mitochondria. Ames, IA, USA: Blackwell Publishing, 2007:278-307. [Google Scholar]
- 34.Fujii S, Toriyama K. Genome barriers between nuclei and mitochondria exemplified by cytoplasmic male sterility. Plant Cell Physiol. 2008;49:1484–94. doi: 10.1093/pcp/pcn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chase CD. Cytoplasmic male sterility: a window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 2007;23:81–90. doi: 10.1016/j.tig.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 36.Hanson MR, Bentolila S. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell. 2004;16(Suppl):S154–69. doi: 10.1105/tpc.015966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gobron N, Waszczak C, Simon M, Hiard S, Boivin S, Charif D, Ducamp A, Wenes E, Budar F, Ducamp A, Wenes E, Budar F. A cryptic cytoplasmic male sterility unveils a possible gynodioecious past for Arabidopsis thaliana. PLoS One. 2013;8:e62450. doi: 10.1371/journal.pone.0062450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kazama T, Nakamura T, Watanabe M, Sugita M, Toriyama K. Suppression mechanism of mitochondrial ORF79 accumulation by Rf1 protein in BT-type cytoplasmic male sterile rice. Plant J. 2008;55:619–28. doi: 10.1111/j.1365-313X.2008.03529.x. [DOI] [PubMed] [Google Scholar]
- 39.Sloan DB, Müller K, McCauley DE, Taylor DR, Storchová H. Intraspecific variation in mitochondrial genome sequence, structure, and gene content in Silene vulgaris, an angiosperm with pervasive cytoplasmic male sterility. New Phytol. 2012;196:1228–39. doi: 10.1111/j.1469-8137.2012.04340.x. [DOI] [PubMed] [Google Scholar]
- 40.Müller K, Storchova H. Transcription of atp1 is influenced by both genomic configuration and nuclear background in the highly rearranged mitochondrial genomes of Silene vulgaris. Plant Mol Biol. 2013;81:495–505. doi: 10.1007/s11103-013-0018-3. [DOI] [PubMed] [Google Scholar]
- 41.Hauler A, Jonietz C, Stoll B, Stoll K, Braun H-P, Binder S. RNA Processing Factor 5 is required for efficient 5′ cleavage at a processing site conserved in RNAs of three different mitochondrial genes in Arabidopsis thaliana. Plant J. 2013;74:593–604. doi: 10.1111/tpj.12143. [DOI] [PubMed] [Google Scholar]
- 42.Manavski N, Guyon V, Meurer J, Wienand U, Brettschneider R. An essential pentatricopeptide repeat protein facilitates 5′ maturation and translation initiation of rps3 mRNA in maize mitochondria. Plant Cell. 2012;24:3087–105. doi: 10.1105/tpc.112.099051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond CS, Small I. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012;8:e1002910. doi: 10.1371/journal.pgen.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yagi Y, Hayashi S, Kobayashi K, Hirayama T, Nakamura T. Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS One. 2013;8:e57286. doi: 10.1371/journal.pone.0057286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruwe H, Schmitz-Linneweber C. Short non-coding RNA fragments accumulating in chloroplasts: footprints of RNA binding proteins? Nucleic Acids Res. 2012;40:3106–16. doi: 10.1093/nar/gkr1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakamura T, Sugita M. A conserved DYW domain of the pentatricopeptide repeat protein possesses a novel endoribonuclease activity. FEBS Lett. 2008;582:4163–8. doi: 10.1016/j.febslet.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 47.Stoppel R, Meurer J. The cutting crew - ribonucleases are key players in the control of plastid gene expression. J Exp Bot. 2012;63:1663–73. doi: 10.1093/jxb/err401. [DOI] [PubMed] [Google Scholar]