Abstract
Mitochondrial RNA editing factor 12 (MEF12) was identified in a screen for editing defects of a chemically mutated plant population in Arabidopsis thaliana. The MEF12 editing protein is required for the C to U change of nucleotide nad5-374. The MEF12 polypeptide is characterized by an exceptionally long stretch of 25 pentatricopeptide repeats (PPR) and a C-terminal extension domain. Editing is lost in mutant plants with a stop codon in the extending element. A T-DNA insertion substituting the 10 C-terminal amino acids of the extension domain reduces RNA editing to about 20% at the target site in a mutant plant. These results support the importance of the full-length extension module for functional RNA editing in plant mitochondria.
Keywords: RNA editing factor, plant mitochondria, PPR protein, MEF12, E-domain
Introduction
RNA editing in plastids and mitochondria of plants alters more than 500 specific cytidines to uridines. These RNA editing sites are identified by specific trans-acting proteins, which recognize sequence motifs just upstream of the editing site target in the mRNA population.1-6 All presently identified protein factors required for single or few sites, mostly in Arabidopsis thaliana, belong to a subfamily of the pentatricopeptide repeat proteins (PPR proteins).7-11 In this subgroup, all proteins are extended beyond the PPR elements by at least one C-terminal extension, the E domain.
Several RNA editing proteins terminate with this E domain in plastids12-14 and in mitochondria.15,16 Several other of these specific RNA editing PPR proteins are further extended beyond the E domain by a region usually terminating with the three amino acids DYW.17-23
To identify further of these factors, we have developed a forward genetic screen to analyze mitochondrial RNA editing sites in a mutant population of Arabidopsis thaliana plants.24,25 The procedure searches for deviations in the RNA editing efficiency at one or more sites in pools of several mutant plants. With this approach, we have identified several site-specific PPR proteins, and in addition, found a group of novel RNA editing proteins, the MORF protein family.26 These MORF proteins are involved in editing at multiple sites distinct from the PPR proteins, which directly target specific RNA sequences.26,27
In this report, we identify mitochondrial editing factor 12 (MEF12) as a site-specific mitochondrial RNA editing factor by mapping of an EMS-induced mutation in the MEF12 gene and a T-DNA insertion mutant. MEF12 is the largest PPR protein extended by an E domain in Arabidopsis and targets the nad5-374 RNA editing site.
Results
A mutant plant disturbed in editing at site nad5-374
Our SNaPshot screen for RNA editing mutants in a population of chemically (EMS) mutagenized Arabidopsis thaliana plants24,25 identified an individual, which is not edited at nucleotide nad5-374 in the mRNA for subunit 5 of complex I of the respiratory chain in plant mitochondria (Fig. 1A, lower panels). Wild-type (wt) plants alter the genomically encoded C very efficiently to U with no C detectable in sequence tracings of total mitochondrial cDNA (Fig. 1A, upper panels). The mutation in the now named mef12-1 plant line abolishes RNA editing at the nad5-374 site but does not disturb editing of other nearby sites in the nad5 mRNA, for example, site nad5-358 (Fig. 1A, right panels). The gene mutated in mef12-1 thus codes for the editing factor MEF12, which is required specifically for RNA editing at site nad5-374. The other 380 mitochondrial editing sites analyzed in the SNaPshot screen are fully processed in the mef12-1 mutant.
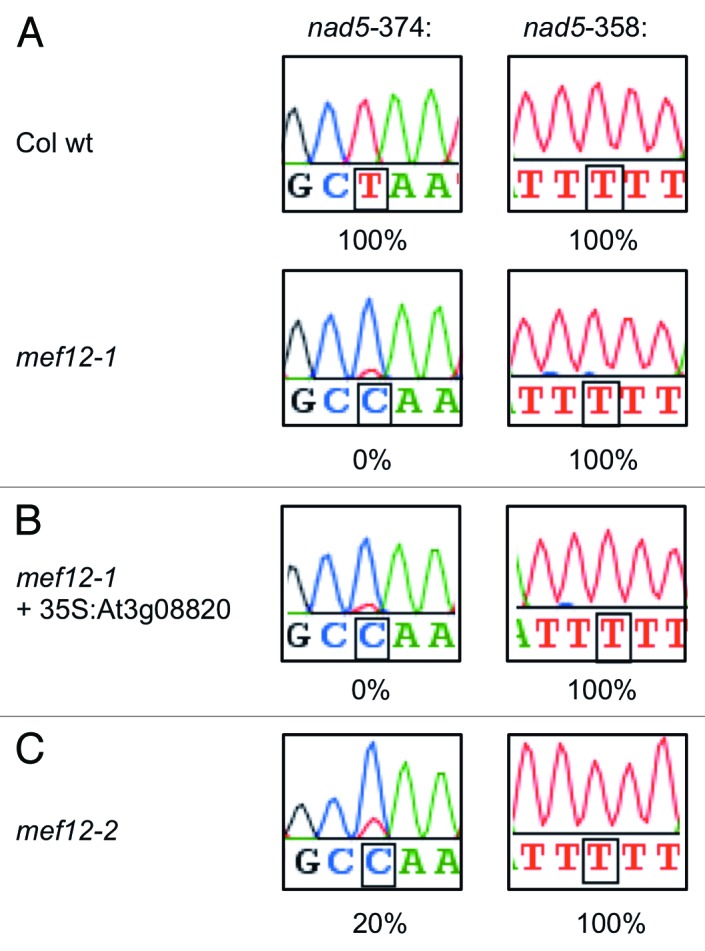
Figure 1. In the mef12-1 mutant plants no RNA editing is observed at site nad5-374. (A) In the EMS mutated plants of line mef12-1 RNA editing is absent at only one site in the mitochondrial mRNA coding for subunit 5 of respiratory chain complex I. The nearest editing site 16 nucleotides upstream at nad5-358 is not affected in mef12-1 and is edited independently. The small T peak visible in mef12-1 at site nad5-374 is less than 10% of the C peak and, thus, attributed to background. Color traces are: G-black, A-green, T-red, C-blue. (B) Transfection of protoplasts from the mef12-1 mutant plants with the Col wild type gene coding for a DYW PPR protein at locus At3g08820 (mef12-1 + 35S:At3g08820) does not restore the ability for RNA editing at site nad5-374. The editing site 16 nucleotides upstream in the nad5 mRNA, site nad5-358, is not affected. (C) A second mutant identifies the MEF12 nuclear gene. For the second candidate locus for the MEF12 gene, locus At3g09040, a mutant plant with a T-DNA insertion was analyzed. The insertion in the E-domain of the encoded protein reduces editing at the target site nad5-374 to 20% and, thus, identifies this locus to code for the MEF12 PPR protein. This mutant was therefore renamed mef12–2. Color traces are: G-black, A-green, T-red, C-blue.
Genomic mapping of the MEF12 gene
The MEF12 gene was mapped by crossing the mef12-1 mutant line of the Arabidopsis thaliana Columbia ecotype (Col) with wild-type plants of ecotype Landsberg erecta (Ler). As in wild-type Col, this site is fully edited also in wild-type ecotype Ler plants.28 The F2 generation yielded in 400 individual plants the about 25% homozygous mutant plants expected with no editing at site nad5-374. SNPs and other markers differentiating between the Ler and Col sequences were employed to map the crossovers closest to the mutated gene. This analysis defined the genomic interval for the mef12-1 mutation to about 1.3 Mb on chromosome three.
In this interval of 1.3 Mb in the Arabidopsis thaliana nuclear genome, six genes annotated for PPR proteins are encoded. Two of these genes encode P type PPRs, one specifies an E class PPR, and three code for PPR proteins with DYW domains. One of the genes for DYW PPR proteins encodes MEF22,16 a second has been identified recently to code for MEF10.17 Since so far no P type PPR proteins have been assigned specific RNA editing functions, we focused on the most likely candidate genes, At3g08820 coding for a DYW PPR protein and At3g09040 coding for an E class PPR protein. Both genes were sequenced in the mutant and compared with their wild-type counterparts. Both open reading frames in the mutant contain single G to A nucleotide transitions typical for EMS-induced mutations. In the DYW PPR protein coded by At3g08820, the altered nucleotide at position 1470 changes a serine codon to an asparagine codon and in the E class PPR protein of At3g09040 a stop codon is introduced for the tryptophane codon (amino acid position 975) by the nucleotide alteration at nucleotide 2925.
Target prediction analysis of the intracellular location of the two proteins suggests a location in the plastid for the DYW PPR protein of At3g08820 with Predotar and “other” with TargetP. Both algorithms predict the putative presequence structure of the E class PPR protein of At3g09040 to target the attached protein to the mitochondrion.
RNA editing is not recovered by the wild-type DYW PPR protein gene coded by At3g08820 in the mef12-1 mutant
To identify which of the two candidate genes codes for the MEF12 RNA editing factor, we initiated cloning of both these genes. However, we succeeded only in cloning the gene encoded at At3g08820, we could not clone the gene of At3g09040. We could thus test only locus At3g08820 for complementation by transient transformation. The Col wild-type At3g08820 gene under control of the 35S-CaMV promoter was introduced into protoplasts of the mef12-1 mutant. In these protoplasts, RNA editing at site nad5-374 is not recovered, no editing is observed in the mitochondrial nad5 cDNA sequence analysis (Fig. 1B, mef12-1 + 35S:At3g08820). This result suggests but does not prove that not the gene at At3g08820, but the one at At3g09040 encodes the MEF12 RNA editing factor.
A second mutant identifies the MEF12 gene
To investigate whether At3g09040 does encode the MEF12 RNA editing factor, we analyzed the homozygous T-DNA insertion line available for this locus from the FLAG resource. In this line FLAG_394H02, sequence analysis confirmed that the T-DNA is inserted in the E domain region (Fig. 2). In the mutant plants, RNA editing at site nad5-374 is reduced by 80% to a residual editing level of about 20% (Fig. 1C). The nearby site nad5-358 is fully edited in this mutant plant. This second mutant line thus confirms that locus At3g09040 codes for the MEF12 RNA editing factor and the T-DNA mutant line was renamed accordingly mef12-2.

Figure 2. Mutations in the E domain disturb the RNA editing function of the MEF12 PPR protein. (A) The MEF12 PPR protein encoded at locus At3g09040 contains 25 PPR repeats and a C-terminal E domain. The N-terminal mitochondrial target sequence is predicted to be about 30 amino acids, the first S repeat begins about 70 amino acids further downstream. The P, L, and S type repeats are shaded in different colors. The G to A transition at nucleotide 2925 (arrow) causes the change of a UGG trytophan codon to a UGA translational stop in the mef12-1 mutant plant line. The T-DNA insertion in the mef12-2 mutant (triangle) is located 54 amino acids further toward the C terminus. The open reading frame in mef12-2 continues into the T-DNA insertion. (B) Comparison of the E-domains in mutants of MEF12 and in PPR proteins with short E-domains narrows the functional E-domain border to few amino acids. Alignment of the predicted protein sequences of MEF12 with those of the non-functional mef12-1 mutation, the partially functional mef12-2 T-DNA insertion and the termini of several short E-class PPR proteins from Arabidopsis thaliana. This type of short PPR editing factors is found in mitochondria but not in plastids. The protein encoded by the mef12-1 mutant is only few amino acids shorter than other functional mitochondrial editing factors, suggesting that a region of very few amino acids is crucial for a functional E-domain. If transcribed and translated, the protein synthesized in the mef12-2 mutant contains all but ten amino acids of the full length MEF12 E-domain and continues into the T-DNA insertion sequence. The observed 20% editing in the mef12-2 mutant suggest that this fusion protein can principally perform the RNA editing function. Conserved amino acids are inversely shaded. Amino acid sequences are shown N to C terminus from left to right for the E-domain of the respective proteins.
Lack of editing at site nad5-374 shows no phenotypic defect
Both mutant plants of mef12-1 and mef12-2 show overall growth phenotypes indistinguishable from wild-type Col plants under normal growth chamber conditions.
The effect of the proline amino acid introduced in the NAD5 protein instead of the leucine derived from this RNA editing event may be tolerated by the NAD5 protein to some extent. The NADH dehydrogenase, complex I of the respiratory chain, may still retain sufficient activity although the proline encoded by the unedited mRNA is expected to introduce a kink in the NAD5 protein. On the other hand, a functional importance of this amino acid alteration in the NAD5 protein is suggested by the conservation of this editing event and the change to leucine in other higher plant species as for example Brassica napus and Beta vulgaris. In the dicot Vitis vinifera and in the monocots rice (Oryza sativa), maize (Zea mays) and wheat (Triticum aestivum) this amino acid is already genomically encoded as a T nucleotide containing triplet.
In Arabidopsis thaliana another editing event occurs at nad5-358 just 16 nucleotides upstream of site nad5-374 within the presumed specificity recognition region of the site. Generally, the RNA editing PPR proteins recognize the sequence further upstream from nucleotide -4 relative to the respective editing event for about as many nucleotides as there are PPR elements. The 25 such repeats in MEF12 (Fig. 2A) could thus cover this upstream editing site. It would be interesting to determine whether this upstream event affects editing at site nad5-374 (Fig. 3 and discussion).
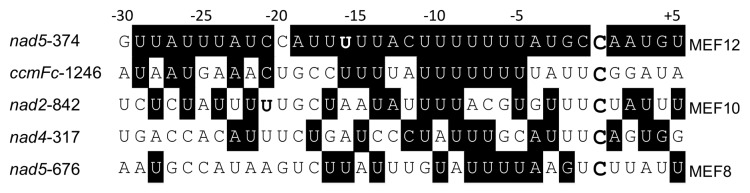
Figure 3. Alignment of the nucleotide sequence of the nad5-374 target editing site with other non-target editing sites. The nucleotides around the nad5-374 editing site (large bold C) show a very high content of U nucleotides especially in the presumed target window of PPR recognition between the nucleotides -4 to -25 or -30. Comparison with the most similar sequences preceding other editing sites (large bold C) accordingly selects for sites with U-rich upstream regions in Arabidopsis thaliana. Of the four most similar sequences aligned here, editing site nad2-842 is fully edited in the mef12-1 mutant and is a target of MEF10.17 Site nad5-676 is also not recognized by MEF12, but requires the MEF8 protein. Vice versa, MEF10 and MEF8 do not influence the MEF12 target site at nucleotide nad5-374.43 Differentiation between the different target sites should thus involve those nucleotides, which differ between the cis-recognition sequences. Nucleotides identical to the nad5-374 MEF12-target site are inversely shaded. Sequences are shown 5′ to 3′ from left to right.
The RNA editing target of MEF12 differs from non-target editing sites
For RNA editing in mitochondria and plastids, the specific C is identified by cis-elements located upstream (5′) of the edited C. The RNA sequence -4 to -20 or -25 nucleotides upstream of the edited nucleotide contains the binding site of a specific PPR protein.5,6,29-33 This sequence at editing site nad5-374 is characterized by an abundance of U nucleotides. Consequently, other editing sites most similar in their recognition sequence are likewise U-rich (Fig. 3). All of these similar sites are edited normally in the mef12-1 mutant. Site nad2-842 is the target site of MEF10,17 confirming that MEF10 cannot compensate for MEF12 and vice versa. This implies that nucleotides differing between the two target sites are the discriminatory identities and have to be identified by the MEF12 and the MEF10 proteins, respectively. Analogously should the differing nucleotides be crucial to differentiate nad2-842 against all other similar sites.
Proteins similar to MEF12 in other plant species
BLAST searches of the databases reveal proteins similar to MEF12 in other plant species (not shown). These are more similar in structure and sequence to the MEF12 protein than any other RNA editing PPR protein, suggesting that they are orthologs of the Arabidopsis thaliana MEF12 protein (At3g09040). Also, in the monocot Oryza sativa (rice), a very similar and apparently functional protein is found at OS04G43430. This is not expected, since in the rice mitochondrial genome the MEF12 target site is already encoded as a T and accordingly does not need to be edited by the MEF12 ortholog. This observation suggests that the MHEF12 protein has another function in addition to the RNA editing at site nad5-374. This additional function should also not be crucial under greenhouse conditions and/or may be at least partially compensated by another protein. Alternatively, the additional function requires only the PPR elements and not the full-length E-domain and can be maintained by the truncated E-domains of mef12-1 and mef12-2. On the other hand, if the genomic reversion of the C to a T nucleotide at site nad5-374 occurred rather recently in rice, the apparent ortholog of MEF12 may not yet have drifted in its sequence structure.
Discussion
MEF12 is the longest RNA editing PPR protein in Arabidopsis thaliana
The 25 PPR elements in MEF12 are the largest number of repeats so far identified in any RNA editing factor in Arabidopsis thaliana and other higher plants. Only one P-type PPR protein is longer with 26 repeats (At5g55840) and two other P-type proteins also contain 25 PPRs (At3g18110 and At4g31850). The size distribution of the RNA editing E-class PPR proteins in Arabidopsis thaliana shows that the next largest such protein contains only 23 PPR repeats. On average, 13.6 PPR elements are present in these RNA editing proteins in Arabidopsis thaliana. In the moss Physcomitrella patens, the average E-domain RNA editing PPR proteins are larger than in Arabidopsis thaliana, the number of repeats averages to 20.2 and the two largest E(DYW)-class PPR proteins have 26 PPR elements. One of these, PpPPR77, has been identified as a bona fide RNA editing factor.34
In other flowering plant species, PPR proteins similar to MEF12 in Arabidopsis thaliana are found, which likewise contain this large number of repeats and, thus, may be true orthologs. An apparently functional ortholog is encoded even in rice, where the single MEF12 target site identified in Arabidopsis thaliana is already a genomic T and does not require RNA editing. This finding suggests that MEF12 may have another function beyond RNA editing of the here identified target site. Since there is no detectable phenotype of the loss of editing or this potential other function, the latter may involve only the PPR elements and is not compromised by the lack of the E domain in mutants mef12-1 and mef12-2. In this scenario the MEF12 protein may be involved also in any other RNA processing step similar to the variety of functions supported by the P-type PPR proteins in Arabidopsis thaliana and in other plants.35,36 Both mutants of MEF12 analyzed here may then be defect in the editing activity which requires the E domain, but may represent the wild type in the second function, which thus cannot be distinguished in these plants.
Prediction of the target sequence and nucleotide contacts
Presuming that each of these amino acid motifs binds to one nucleotide at the MEF12 target site,5,6 an extremely high specificity could be achieved with the 25 repeat elements. On the other hand, it is possible that not all of these elements actually contact a specific nucleotide but that some of them function as spacers and are looped out from the contact with the RNA.5,6,35,36 In terms of still achieving sufficient specificity in the bulk of the unedited mitochondrial mRNA, only about eight nucleotide identifications are required. This would leave up to 19 PPRs free to act as spacers. In the MEF12 instance such spacers between the bound nucleotides may be part of the strings of uridines since such U-rich stretches are also found in other editing sites (Fig. 3).
To identify the nucleotides likely to be contacted, we analyzed the nucleotide identities predicted to be targeted by the criteria derived from coincidences between matching amino acid and nucleotide identities in previously analyzed PPR proteins.5,6 This prediction does not include L, L2, and S2 repeat elements, which therefore do not show any coincidences in the analysis. Our recently developed improved prediction program, which includes the L, L2, and S2 repeat elements,37 predicts the bona fide target sequence of MEF12 at nad5-374 only at position 14 of all known editing sites in mitochondria. This low ranking may be explained if not all of the PPR repeats actually contact the RNA and therefore would not have matching signature amino acids. Since all of the PPR elements are included in the in silico analysis, such spacer elements will distort and dilute the predicted specificity. Furthermore, such spacers may shift some of the PPR elements against the RNA and, thus, preclude a one-on-one alignment and lead to a lower rank by matching.
Interestingly, the first major stretch of seven consecutive uridines coming from the edited C between nucleotides -5 and -11 does not seem to be fully contacted, but the second five uridines motif between nucleotides -14 and -18 seems to be targeted as judged from the matches observed. The actual contact site may then be located from nucleotides -11 or -12 up to nucleotide -21. This would yield 10 or 11 consecutive nucleotides, sufficient for a specific identification of the editing site. Particularly intriguing could be nucleotides -13 and -12, which are AC predicted to be contacted in the nad5-374 target sequence but are not present in this combination in any of the other similar motifs (Fig. 3). The upstream editing site at nad5-358 (nucleotide position -16) better matches the prediction as an edited U, but the unedited C could probably also be tolerated and accommodated by the MEF12 PPR moiety at this position.
Provided that some or all of the L elements act as spacers as previously suggested,5,6 the binding domain may be interrupted. This would dramatically decrease the affinity of the MEF12 protein to the RNA. A similar effect could be achieved by only some of the repeats actually binding to RNA nucleotides. Such a lower affinity may actually be an important condition for the RNA editing PPR proteins since they will have to be removed rapidly from the RNA after editing is completed. This will be most crucial in closely spaced editing sites and their respective overlapping recognition domains as observed here for the upstream editing site at nad5-358. If indeed attachment of MEF12 is better after editing of the upstream site, the PPR protein involved in the latter will have to be detached before the MEF12 editing site at nad5-374 can be addressed by the MEF12 protein. Further refinement of the prediction code and/or experimental evidence will be required to evaluate these options.
The MEF12 target site is also influenced by the MORF3 protein
In a more general function, individual representatives of the small family of MORF proteins are required for most if not all RNA editing sites in mitochondria as well as in plastids.26,27 Of these, the MEF12 editing site nad5-374 depends on the presence of MORF3 for full editing. In the apparent knockout mutant morf3-1, RNA editing is reduced to 60%, suggesting that MORF3 is a prominent MORF protein for this site while one or more of the other MORF proteins can at least partially substitute for MORF3.26 It remains to be investigated whether the MORF3 protein directly interacts with the MEF12 protein. This has been found for other MEFs as, for example, MEF10, which interacts with the mitochondrially and plastid located MORF8.17,26
The C terminus of the E-domain is essential for MEF12 function in RNA editing
The translational stop codon introduced by the mef12-1 EMS mutation and the ensuing loss of the N terminus of the E domain completely blocks the function of MEF12 in editing the target site identified at nad5-374. To get an impression of the functional relevance of this premature end of the protein, we compared the positions of C-termini in other similar RNA editing PPR proteins with rather short E domains (Fig. 2B). The alignment of these E-class PPR proteins with the location of the mef12-1 mutation induced stop codon, reveals that only few C-terminal amino acids may be crucial for function. For example, the MEF20 protein is only two amino acids longer than the truncated mutant MEF12-1 protein (Fig. 2B). MEF18 has only three more C-terminal amino acids than MEF12-1, MEF3 five, and MEF9 and MEF19 six. Provided the E domains in these E-domain PPR proteins function similarly, the few missing amino acids must be crucial. They may be important for interaction with the target RNA and/or for essential interaction with other editing proteins such as one of the MORF proteins, possibly MORF3.26 The mef12-1 mutation highlights one of the requirements of the E domain for functional competence and provides another clue for the functional analysis of the E domain in the RNA editing process.2
This importance of the C-terminal border of a minimal E-domain length is also supported by the second mutant line mef12-2. The T-DNA insertion in mef12-2 is located downstream of the mef12-1 mutation and of the functional termini of the wild-type MEF3, MEF9, MEF18, and MEF20 proteins (Fig. 2B). Both mRNA and protein should be synthesized from mef12-2, since 20% editing is detected at the nad5-374 target site in comparison to the less than 10% editing in mef12-1 (Fig. 1). Either the expression level of the fusion protein is compromised in mef12-2 or this longer protein is only partially functional. In any case, the basic functionality of this MEF12-2 protein also suggests that the amino acids just downstream of the termination codon in mef12-1 are decisive for function of the E-domain PPR protein in RNA editing in plant mitochondria.
Materials and Methods
Plant growth and preparation of DNA and RNA
Wild type and mutant Arabidopsis thaliana plants were grown in chambers as described previously.37 DNA or RNA were prepared from young fully developed leaves by standard protocols.37 The EMS mutant population of ecotype Col of Arabidopsis thaliana had been purchased from Lehle Seeds. The T-DNA insertion line was kindly provided by the FLAG resources. T-DNA insertion sites and the presence of homozygous mutant plants were analyzed by PCR with appropriate primers.
SNaPshot screening and identification of mutants
To analyze the EMS mutated Arabidopsis thaliana plants for deviations in RNA editing, leaves from 10 individual mutant plants were pooled in respective batches and the total RNA was isolated. About 400 sites in mitochondrial mRNAs were analyzed for deviations in any individual by multiplexed single base extension.24,25 Plant lines with reduced or absent RNA editing due to an EMS mutation were selected from these lines as described.24,25 In each mutant line, altered RNA editing was verified by cDNA sequence analysis. Sequences were determined commercially by LGC genomics, 4base lab, or Macrogen.
RNA editing site analysis
Specific cDNA fragments were generated from the relevant region of the nad5 mRNA containing the investigated nad5-374 editing site by RT-PCR with the respective primers used in the SNaPshot analysis.24,25,38 These sequences obtained from wild type and mutant plants were analyzed for the percentages of C or T by comparing the respective areas under the peaks at the RNA editing sites as described.39
Protoplast transformation of mutant plant cells
Transformation of protoplasts40 from EMS mutant plants of mef12-1 was done with the wt Col reading frame encoded at At3g08820 under control of the 35S promoter in vector pMDC12341 with a GFP cassette from psMGFP442 and a multiple cloning site from pET43 (Merck Millipore Novagen®).
Acknowledgments
We thank Dagmar Pruchner and Angelika Müller for excellent experimental help. We are very grateful to the Department of Human Genetics at the Universität Ulm for generously letting us use their sequencing machine for the SNaPshot analyses.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from the Deutsche Forschungsgemeinschaft. MT is a Heisenberg fellow.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/25484
References
- 1.Bock R, Hermann M, Kössel H. In vivo dissection of cis-acting determinants for plastid RNA editing. EMBO J. 1996;15:5052–9. [PMC free article] [PubMed] [Google Scholar]
- 2.Chateigner-Boutin AL, Small I. Plant RNA editing. RNA Biol. 2010;7:213–9. doi: 10.4161/rna.7.2.11343. [DOI] [PubMed] [Google Scholar]
- 3.Fujii S, Bond CS, Small ID. Selection patterns on restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proc Natl Acad Sci USA. 2011;108:1723–8. doi: 10.1073/pnas.1007667108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujii S, Small I. The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 2011;191:37–47. doi: 10.1111/j.1469-8137.2011.03746.x. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi K, Kawabata M, Hisano K, Kazama T, Matsuoka K, Sugita M, et al. Identification and characterization of the RNA binding surface of the pentatricopeptide repeat protein. Nucleic Acids Res. 2012;40:2712–23. doi: 10.1093/nar/gkr1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond CS, et al. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012;8:e1002910. doi: 10.1371/journal.pgen.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Small ID, Peeters N. The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci. 2000;25:46–7. doi: 10.1016/S0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- 8.Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008;13:663–70. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 10.O’Toole N, Hattori M, Andres C, Iida K, Lurin C, Schmitz-Linneweber C, et al. On the expansion of the pentatricopeptide repeat gene family in plants. Mol Biol Evol. 2008;25:1120–8. doi: 10.1093/molbev/msn057. [DOI] [PubMed] [Google Scholar]
- 11.Rivals E, Bruyère C, Toffano-Nioche C, Lecharny A. Formation of the Arabidopsis pentatricopeptide repeat family. Plant Physiol. 2006;141:825–39. doi: 10.1104/pp.106.077826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotera E, Tasaka M, Shikanai T. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature. 2005;433:326–30. doi: 10.1038/nature03229. [DOI] [PubMed] [Google Scholar]
- 13.Okuda K, Myouga F, Motohashi R, Shinozaki K, Shikanai T. Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc Natl Acad Sci USA. 2007;104:8178–83. doi: 10.1073/pnas.0700865104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chateigner-Boutin A-L, Ramos-Vega M, Guevara-García A, Andrés C, de la Luz Gutiérrez-Nava M, Cantero A, et al. CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 2008;56:590–602. doi: 10.1111/j.1365-313X.2008.03634.x. [DOI] [PubMed] [Google Scholar]
- 15.Takenaka M. MEF9, an E subclass PPR protein, is required for an RNA editing event in the nad7 transcript in mitochondria of Arabidopsis thaliana. Plant Physiol. 2010;152:939–47. doi: 10.1104/pp.109.151175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takenaka M, Verbitskiy D, Zehrmann A, Brennicke A. Reverse genetic screening identifies five E-class PPR proteins involved in RNA editing in mitochondria of Arabidopsis thaliana. J Biol Chem. 2010;285:27122–9. doi: 10.1074/jbc.M110.128611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Härtel B, Zehrmann A, Verbitskiy D, van der Merwe JA, Brennicke A, Takenaka M. MEF10 is required for RNA editing at nad2-842 in mitochondria of Arabidopsis thaliana and interacts with MORF8. Plant Mol Biol. 2013;81:337–46. doi: 10.1007/s11103-012-0003-2. [DOI] [PubMed] [Google Scholar]
- 18.Kim S-R, Yang J-I, Moon S, Ryu C-H, An K, Kim KM, et al. Rice OGR1 encodes a pentatricopeptide repeat-DYW protein and is essential for RNA editing in mitochondria. Plant J. 2009;59:738–49. doi: 10.1111/j.1365-313X.2009.03909.x. [DOI] [PubMed] [Google Scholar]
- 19.Okuda K, Chateigner-Boutin A-L, Nakamura T, Delannoy E, Sugita M, Myouga F, et al. Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell. 2009;21:146–56. doi: 10.1105/tpc.108.064667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Q-B, Jiang Y, Chong K, Yang Z-N. AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana. Plant J. 2009;59:1011–23. doi: 10.1111/j.1365-313X.2009.03930.x. [DOI] [PubMed] [Google Scholar]
- 21.Zehrmann A, Verbitskiy D, Härtel B, Brennicke A, Takenaka M. RNA editing competence of trans-factor MEF1 is modulated by ecotype-specific differences but requires the DYW domain. FEBS Lett. 2010;584:4181–6. doi: 10.1016/j.febslet.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 22.Zehrmann A, van der Merwe JA, Verbitskiy D, Härtel B, Brennicke A, Takenaka M. The DYW-class PPR protein MEF7 is required for RNA editing at four sites in mitochondria of Arabidopsis thaliana. RNA Biol. 2012;9:155–61. doi: 10.4161/rna.18644. [DOI] [PubMed] [Google Scholar]
- 23.Verbitskiy D, Zehrmann A, van der Merwe JA, Brennicke A, Takenaka M. The PPR protein encoded by the LOVASTATIN INSENSITIVE 1 gene is involved in RNA editing at three sites in mitochondria of Arabidopsis thaliana. Plant J. 2010;61:446–55. doi: 10.1111/j.1365-313X.2009.04076.x. [DOI] [PubMed] [Google Scholar]
- 24.Takenaka M, Brennicke A. Multiplex single-base extension typing to identify nuclear genes required for RNA editing in plant organelles. Nucleic Acids Res. 2009;37:e13. doi: 10.1093/nar/gkn975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takenaka M, Brennicke A. Using multiplex single-base extension typing to screen for mutants defective in RNA editing. Nat Protoc. 2012;7:1931–45. doi: 10.1038/nprot.2012.117. [DOI] [PubMed] [Google Scholar]
- 26.Takenaka M, Zehrmann A, Verbitskiy D, Kugelmann M, Härtel B, Brennicke A. Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc Natl Acad Sci USA. 2012;109:5104–9. doi: 10.1073/pnas.1202452109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bentolila S, Heller WP, Sun T, Babina AM, Friso G, van Wijk KJ, et al. RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing. Proc Natl Acad Sci USA. 2012;109:E1453–61. doi: 10.1073/pnas.1121465109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zehrmann A, van der Merwe JA, Verbitskiy D, Brennicke A, Takenaka M. Seven large variations in the extent of RNA editing in plant mitochondria between three ecotypes of Arabidopsis thaliana. Mitochondrion. 2008;8:319–27. doi: 10.1016/j.mito.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Chaudhuri S, Maliga P. Sequences directing C to U editing of the plastid psbL mRNA are located within a 22 nucleotide segment spanning the editing site. EMBO J. 1996;15:5958–64. [PMC free article] [PubMed] [Google Scholar]
- 30.Farré J-C, Leon G, Jordana X, Araya A. cis Recognition elements in plant mitochondrion RNA editing. Mol Cell Biol. 2001;21:6731–7. doi: 10.1128/MCB.21.20.6731-6737.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyamoto T, Obokata J, Sugiura M. Recognition of RNA editing sites is directed by unique proteins in chloroplasts: biochemical identification of cis-acting elements and trans-acting factors involved in RNA editing in tobacco and pea chloroplasts. Mol Cell Biol. 2002;22:6726–34. doi: 10.1128/MCB.22.19.6726-6734.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verbitskiy D, van der Merwe JA, Zehrmann A, Brennicke A, Takenaka M. Multiple specificity-recognition motifs enhance RNA editing in plant mitochondria. J Biol Chem. 2008;283:24374–81. doi: 10.1074/jbc.M803292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kempken F, Bolle N, Bruhs A. Higher plant in organello systems as a model for RNA editing. Endocytobiosis Cell Res. 2009;19:1–10. [Google Scholar]
- 34.Sugita M, Ichinose M, Ide M, Sugita C. Architecture of the PPR gene family in the moss Physcomitrella patens. RNA Biol. 2013;10 doi: 10.4161/rna.24772. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitz-Linneweber C, Williams-Carrier R, Barkan A. RNA immunoprecipitation and microarray analysis show a chloroplast Pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell. 2005;17:2791–804. doi: 10.1105/tpc.105.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams-Carrier R, Kroeger T, Barkan A. Sequence-specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA. 2008;14:1930–41. doi: 10.1261/rna.1077708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takenaka M, Zehrmann A, Brennicke A, Graichen K. Improved computational target site prediction for pentatricopeptide repeat RNA editing factors. PLoS One. 2013;8:e65343. doi: 10.1371/journal.pone.0065343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takenaka M, Brennicke A. RNA editing in plant mitochondria: assays and biochemical approaches. Methods Enzymol. 2007;424:439–58. doi: 10.1016/S0076-6879(07)24020-0. [DOI] [PubMed] [Google Scholar]
- 39.Zehrmann A, van der Merwe JA, Verbitskiy D, Brennicke A, Takenaka M. A DYW-domain containing PPR-protein is required for RNA editing at multiple sites in mitochondria of Arabidopsis thaliana. Plant Cell. 2009;21:558–67. doi: 10.1105/tpc.108.064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–72. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 41.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–9. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forner J, Binder S. The red fluorescent protein eqFP611: application in subcellular localization studies in higher plants. BMC Plant Biol. 2007;7:28. doi: 10.1186/1471-2229-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verbitskiy D, Zehrmann A, Härtel B, Brennicke A, Takenaka M. Two related RNA-editing proteins target the same sites in mitochondria of Arabidopsis thaliana. J Biol Chem. 2012;287:38064–72. doi: 10.1074/jbc.M112.397992. [DOI] [PMC free article] [PubMed] [Google Scholar]


