SUMMARY
Antigen-presenting dendritic cells (DCs) must survive bacterial infection in order to present antigen information to naive T-cells. The greater ability of DC's host defense is evident from the report that DCs are more resistant to Listeria monocytogenes than macrophages. However, the molecular mechanism of this resistance is unclear. We found that Listeria replicate more slowly in wild type DCs compared to fascin1 knockout DCs. This finding is significant because fascin1, an actin bundling protein, is specifically and greatly induced upon maturation of dendritic cells, but not other blood cells including macrophages and neutrophils. Infection by Listeria makes phagosomes more acidic in wild type DCs than in fascin1 knockout DCs, suggesting that fascin1 facilitates phagolysosomal fusion for killing of phagocytosed Listeria. We further found that fascin1 binds to LC3, an autophagosome marker, both in vivo and in vitro. Listeria are associated with LC3 to a greater extent in wild type DCs than in fascin1 KO DCs, suggesting that fascin1 facilitates autophagy for eradication of cytoplasmic Listeria. Taken together, our results suggest that fascin1 plays critical roles in the survival of DCs during Listeria infection, allowing DCs to function in innate and adaptive immunity.
Keywords: Fascin, Dendritic cells, Listeria monocytogenes, phagosomes, autophagy, LC3, pH, OmniBank
INTRODUCTION
Dendritic cells (DCs) play central roles in innate and adaptive immunity (1-3). DCs phagocytose antigens such as bacterial pathogens and change their functions from antigen sampling to antigen presentation. Phagosome-engulfed bacteria are degraded by lysosomal fusion for presentation of pathogen information to naive CD4+ T-cells via the MHC-II pathway. DCs also process intracellular bacteria that have escaped from phagosomes into the cytoplasm and present antigen information to naïve CD8+ T-cells via the MHC-I pathway.
Listeria monocytogenes (Lm) is a pathogen that can cause serious infections in immunocompromised individuals and pregnant women. Lm enters cells via phagocytosis (4) and for epithelial cells, via a clathrin-dependent pathway (5). Lm lyses phagosomes with listeriolysin O (LLO), a pore-forming cytolysin, and escapes into the cytoplasm to proliferate. Intracellular Lm polymerizes actin to form comet tails for rapid intracellular movement, and transmigrates directly into adjacent cells to promote new infections (4, 6, 7).
DCs have been demonstrated to play a crucial role in eradication of Lm in mice: DCs, but not macrophages, are essential for priming naive cytotoxic T-lymphocytes that are specific for Lm antigens (8). In vitro, Lm infection of bone marrow-derived DCs (BM-DCs) enhances their maturation, as well as their ability to stimulate T-cell differentiation (9, 10). Importantly, BM-DCs have been shown to be more resistant to Lm than are macrophages (11, 12). This resistance must be critical for DCs’ primary function of antigen presentation because DCs have to survive infection and present antigen information to naive T-cells. While limited escape of Lm from phagosomes into the cytosol has been suggested to be a reason for the resistance (11, 12), the molecular mechanism(s) and molecule(s) for the higher killing activity shown by DCs are not clear. Such molecule(s) must be either specifically expressed or activated in DCs, but not in macrophages.
Fascin1 is an unique actin-bundling protein that is very highly and specifically induced upon maturation of DCs, but not expressed in other blood cells including macrophages, neutrophils, T- and B-cells (13). By characterizing DCs from fascin1 knockout (KO) mice, we have demonstrated that fascin1 causes large changes in the organization of the peripheral actin cytoskeleton of DCs: Fascin1 makes veil-like dorsal membrane ruffling more vigorous, and promotes in vitro chemotactic motility, as well as in vivo migration into draining lymph nodes (14). Consistent with the above result, genome-wide, expression profile analyses of mouse DCs isolated in vivo have revealed that fascin1 is abundantly expressed, in particular, in migratory DCs (15).
The fascin1-mediated, massive changes in the peripheral actin cytoskeleton could affect bactericidal activity of DCs because the actin cytoskeleton is involved in several aspects of host defense mechanisms: For example, bacterial entry via phagocytosis is controlled by the peripheral actin cytoskeleton (16-18). Assembly and bundling of actin filaments have been reported to regulate phagolysosomal fusion (19, 20). Autophagy, which is able to kill cytoplasmic bacteria that have escaped from phagosomes, requires actin and actin binding proteins including Arp2/3 and cortactin (21-23). Fascin1 might increase cell-to-cell transmission of Lm, because actin bundling by fascin1 has been shown to increase Lm motility in vitro in an Arp2/3-independent way (24).
We found, using BM-DCs, that fascin1 KO DCs are more susceptible to Lm infection than wild type DCs. DCs expressing high levels of fascin1 are free from Lm infection, suggesting critical roles of fascin1 in bacterial eradication.
MATERIALS AND METHODS
Antibodies and reagents
The following antibodies were used: FITC-conjugated hamster anti-mouse CD11c monoclonal, FITC-conjugated rat anti-mouse CD86 (B7-2) monoclonal, FITC-conjugated rat anti-mouse I-A/I-E (MHC-II) monoclonal, rabbit anti-Lm polyclonal (BD Biosciences, San Jose, CA), rabbit anti-LC3 polyclonal, and mouse anti-LC3 monoclonal (MBL international, Woburn, MA), mouse anti-fascin monoclonal (55k-2) (25), rabbit anti-fascin monoclonal and rabbit anti-actin monoclonal antibody (Cell Signaling Technology, Danvers, MA). GM-CSF was purchased from Invitrogen (Camarillo, CA). A recombinant, non-fusion, fascin1 protein was prepared as described previously (26). A recombinant GST-LC3 fusion protein was expressed in bacteria as described (27).
Preparation of bone marrow-derived dendritic cells (BM-DCs)
Fascin1 KO heterozygous mice were generated by Lexicon Pharmaceuticals, Inc. (Woodlands, TX) from an ES cell line (OST124903) (Lexicon's OmniBank® library of gene KO ES cell clones), and backcrossed with C57/BL6 female mice for more than 14 generations (28). DCs were prepared from bone marrow isolated from fascin1 KO homozygous mice and their wild-type littermates, as described previously (14). All experimental procedures and protocols for mice are approved by the Animal Care and Facilities Committee at Rutgers. Mice were housed in an AAALAC-accredited animal facility at Rutgers.
Lm Infection and measurements of colony-forming units (CFU)
Infection of DCs with Lm was performed essentially as described in Westcott et al (11, 12). Briefly, DCs were infected with a wild type strain of Lm (10403S, a gift from Dr. D. A. Portnoy) at a moi of 0.5 except specified otherwise. At 30min post infection, DCs were gently washed with fresh culture media (DMEM containing 10ng/ml GM-CSF), and gentamycin (final, 10μg/ml) was added to kill extracellular bacteria (gentamycin is present throughout the entire experiments). At varying times after infection, DCs were harvested, washed 3 times with PBS, and 10% of cells were used for counting viable cell numbers with a hemocytometer, following staining with trypan blue. The rest of DCs were homogenized in PBS containing 0.1% Triton X-100. Cell disruption was confirmed by phase contrast microscopy. The homogenates were serially diluted, and plated on LB agar plates overnight 37°C to determine colony-forming units (CFU).
Measurements of bacterial uptake
To determine whether fascin1 affects bacterial uptake, wild type and fascin1 KO DCs were infected with Lm at a moi of 20. At 30min post infection, gentamycin (10μg/ml) was added to kill extracellular bacteria. DCs were washed three times with PBS, and homogenized in PBS with 0.1% Triton X-100. Bacteria incorporated into DCs were measured by determining CFU as described above.
Immunofluorescence of Lm-infected DCs
Infection with Lm caused rapid maturation of DCs, as judged from the expression of fascin1, CD86 and MHC-II: fascin1 expression can be clearly observed as early as 30-60min after Lm infection. This is in contrast to DC maturation by LPS, which takes as long as 7hr after the addition of LPS (14).
Lm infection changed the morphology of DCs to a more rounded form that tended to detach from the substrate. To keep rounded DCs on coverslips for immunofluorescence staining, DCs were cytospun (500×g for 5min), fixed with methanol and processed for immunofluorescence as described previously (14). In some cases, DCs were fixed with 3.7% formaldehyde, permeabilized with 0.2% Triton x-100, and processed for immunofluorescence as described (14). Images were taken as Z-stacks (0.2μm spacing) with a DeltaVision Image Restoration Microscope system (Applied Precision Instrument, LLC Issaquah, WA), deconvolved either with the softWoRx software (Applied Precision Instruments) or the Huygens software (Scientific Volume Imaging, Hilversum, Netherlands). Projected images were generated with SoftWoRx or ImageJ (http://rsb.info.nih.gov/ij/). Exposure times for imaging and settings for deconvolution were constant for all samples to be compared within any given experiment. In some experiments, images were taken on a Nikon TE300 microscope with a 60x objective lens (NA 1.4). For presentation, image contrast and brightness were adjusted with Photoshop (Adobe, San Jose, CA).
Analyses of Phagosomal and vacuolar pH
A mutant strain Lm (DP L-2319, δhly δplcA δplcB, a gift from D. A. Portnoy) was used for measurements of pH of Lm-engulfed phagosomes. As this strain lacks LLO and two phospholipases, it is defective in vacuolar lysis and is not able to escape from phagosomes (29). Lm were double labeled with a pH-sensitive dye 5-chloromethyll fluorescein diacetate (CMFDA) and a pH-insensitive dye tetramethyl-rhodamine, and phagosomal pH was determined by ratiometric imaging of phagosomal Lm essentially as described (12). Briefly, exponentially growing Lm were washed three times with PBS, and then labeled with CMFDA (100μM) for 10min at 37°C. The labeling reaction was stopped by addition of PBS containing 10% fetal calf serum (FCS). CMFDA-labeled Lm were washed with PBS three times and labeled with 0.2mg/ml of tetramethylrhodamine at 37°C for 45min. The reaction was stopped with PBS containing 10% FCS.
One hour after infection with double-labeled Lm at a moi of 3, DCs were imaged in the FITC and Rhodamine channels with a DeltaVision microscopy system and ratio images of FITC to Rhodamine were generated with the softWoRx software (Applied Precision Instruments). For conversion from the ratio to the pH, double-labeled Lm were incubated in PBS at varying pH from 3 to 7.5 (adjusted with HCl), and the ratio of an FITC fluorescence intensity divided by a corresponding Rhodamine fluorescence intensity was plotted against the pH of each buffer.
For a vacuolar pH measurement, a LysoSensor yellow/blue dye (DN-160, Molecular Probes) was used. This dye is incorporated into acidic organelles including phagosomes and lysosomes, and ratio imaging of 525nm/470nm gives pH values. DCs were labeled with 20μM LysoSensor dye for 2 min and washed with DMEM containing 10% FCS. Images were taken with DeltaVision microscopy with an excitation wavelength of 360±40nm and two emission wavelengths of 457±30nm and 528±38nm, and ratio images of 528nm emission divided by 457nm emission were generated with the SoftWoRx software. Live-cell ratiometric imaging was made before infection, and 30min, 1hr and 3hr after infection (moi of 3). To convert the ratio to pH, LysoSensor-labeled DCs were permeabilized with 20mM HEPES-buffered DMEM at varying pH (3 to7.5) containing 0.1% TritonX-100 or 1μM nigericin and valinomycin (Sigma Chemical Co.), and the ratios were determined as described above.
Autophagy analyses
At varying time post infection, DCs were cytospun and fixed with 3.7% formaldehyde, permeabilized with 0.2% Triton X-100, and double stained with an anti-LC3 and anti-Lm antibody. DCs were imaged using a Deltavision microscope system as described above, and Lm associated with LC3 were quantitated and expressed as percentages of association.
Fascin1-LC3 binding
Fascin1-LC3 interaction was examined by co-immunoprecipitation and a GST-pull down assay. For immunoprecipitation, HeLa cells were homogenized in an immunoprecipitation buffer containing 50mM Tris-HCl, 50mM NaCl, 0.05% Triton X-100, 0.2mM DTT, 0.1mM EDTA. 0.1mM EGTA, 5mM MgCl2 and 10% glycerol (pH 7.5), and the homogenates were centrifuged at 16,000xg for 10min without a brake. Anti- LC3 antibody was added to the supernatants. An anti-LAMP1 antibody was used as a control. The immune complexes were collected by addition of Protein A Sepharose beads (GE Healthcare, Piscataway, NJ), separated on SDS-PAGE, immunoblotted with anti-LC3, anti-LAMP1 and anti-fascin1 antibodies.
A GST-pull down assay was performed as described previously (30). Briefly, GST-LC3 that had been bound to glutathione-Sepharose 4B beads (GE Healthcare) was incubated with GFP-fascin1 (final 0.5mg/ml) for 45 min in the immunoprecipitation buffer described above. After centrifugation at 600xg for 5 min, glutathione-beads were washed extensively with the immunoprecipitation buffer, lysed in SDS-PAGE sample buffer, and analyzed by Western blotting to detect binding between LC3 and GFP-fascin1. As a control, GST alone was bound to glutathione-Sepharose beads, and incubated with GFP-fascin1 in the same way as above. The bead-bound fraction was processed in the same way as above.
Proximity Ligation Assay (PLA)
PLA was performed according to the manufacturer's instruction (O-link Bioscience, Uppsala, Sweden). Briefly, DCs were fixed with methanol at −20°C for 30sec, then further fixed with 3.7% formaldehyde for 5min, and permeabilized with 0.2% Triton X-100. This fixation method that was previously used to simultaneously visualize fascin1 and actin (25), was found to be suitable for simultaneous visualization of fascin1 and LC3. Fixed DCs were incubated with anti-fascin1 mouse monoclonal and anti-LC3 rabbit polyclonal antibodies. The mouse and rabbit antibodies were labeled with anti-mouse and anti-rabbit PLA probes, each of which was conjugated with oligonucleotides. If these two oligonucleotides were located at a close proximity (within ~40nm), they were ligated to form a closed circle, which was amplified with DNA polymerase and detected with fluorescently labeled oligonucleotides. As a positive control, DCs were labeled with anti-fascin1 and anti-actin antibodies, the antigens of which are known to interact each other (25, 31).
Electron microscopy
Thin-section electron microscopy was performed according to protocols by the electron microscopy facility at Rutgers University. Briefly, DCs were infected with wild type strain Lm at a moi of 1, and gentamycin was added at 30min post infection. At 50min post infection, DCs were fixed with 2% glutaraldehyde/2% paraformaldehyde in 0.1M cacodylate buffer for 5min, scrape-harvested into an Eppendorf tube, further fixed in the same solution for 55min, washed with PBS and post-fixed with 1% OsO4 for 1hr. After washing with PBS, samples were dehydrated with graded solutions of ethanol, cells were embedded with a low viscosity embedding media kit (Electron Microscopy Sciences, Hartford, PA). Thin sections were observed with a JEOL 100CX microscope (Peabody, MA) at 80kV, and about 200 bacteria were imaged for each experiment.
Statistical analysis
Statistical analyses were performed using unpaired, two-tailed, Student t test (http://www.physics.csbsju.edu/stats/t-test.html).
RESULTS
Fascin1 KO DCs are more susceptible to Lm infection than wild type DCs
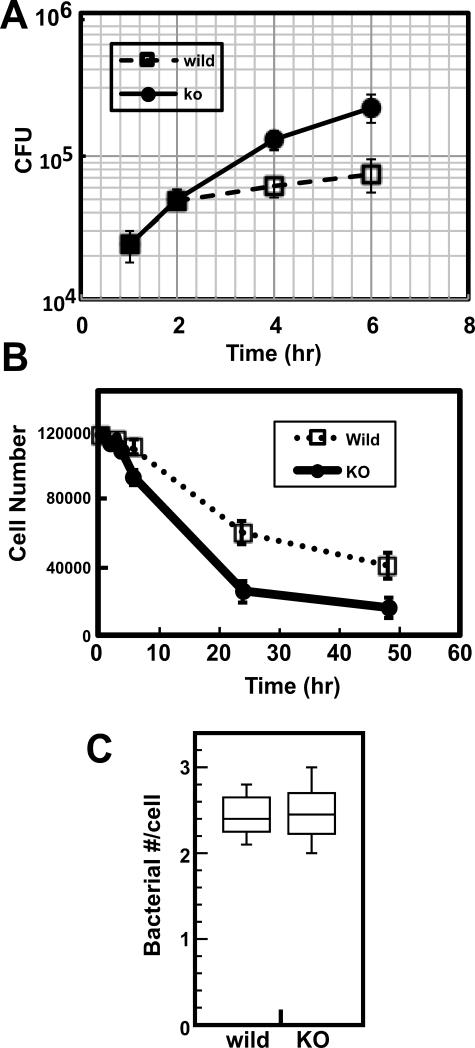
We examined, by infecting DCs at a moi of 0.5, whether fascin1 affects replication rates of intracellular bacteria in wild and fascin1 KO DCs. At this moi, both wild and fascin1 KO DCs were mostly viable during the initial 6hr incubation, allowing us to determine bacterial replication rates. Measurements of intracellular bacterial numbers at 1hr, 2hr, 4hr and 6hr (Fig. 1A) revealed that Lm replicated faster in fascin1 KO DCs than in wild type DCs. During the initial 2hr, Lm grew in a similar rate in both wild type and fascin1 KO DCs. However, Lm growth became much slower at 4hr and 6hr in wild type DCs, whereas Lm grew more consistently in KO DCs. The doubling times (estimated from CFU. between 1 and 4hr) were 45min in fascin1 KO DCs and 90min in wild type DCs. The longer doubling time of Lm in wild type DCs, as well as the growth inhibition at around 4hr, is consistent with the previous report for wild type DCs by Westcott et al (11). The susceptibility to Lm infection in fascin1 deficient DCs is further confirmed by tracking viability of DCs for longer incubation: As Fig. 1B shows, cell number for fascin1 null DCs decreased much more rapidly at 24hr and 48hr than that of wild-type DCs. At 24hr post infection the bacterial number (9.4×105) of fascin1 KO DCs was twice as high as that (4.6x105) of wild type DCs. However, this difference in bacterial number is an underestimate because the cell number of KO DCs was twice as low as that of wild type DCs (B).
Figure 1.
Higher susceptibility of fascin1 KO DCs to Lm infection. A. Lm replication during the initial 6hr of infection. Wild type (wild) and fascin1 KO (KO) DCs were infected with Lm at a moi of 0.5 for 30min. After killing extracellular Lm with gentamycin, growth of intracellular Lm was measured as described in the Materials and Methods section. B. Viability of wild type (wild) and fascin1 KO (KO) DCs for 48hr after infection. More wild type DCs can survive than fascin1 KO DCs. C. Bacterial uptake after 30min post infection. No statistically significant difference in phagocytic activities between wild type and fascin1 KO DCs was observed. Representative results are shown from at least three independent experiments. Error bars, standard deviation.
The bacterial numbers at 1hr post infection were similar between wild type and fascin1 KO DCs, suggesting that the resistance of wild type DCs to Lm infection is not due to a difference in phagocytic activity. To confirm this, we measured bacterial uptake at 30min post infection. To increase an accuracy of measurements of bacterial uptakes, DCs were infected at a moi of 20. As Fig. 1C shows, both wild type and fascin1 KO DCs ingested similar number of bacteria during 25 min of phagocytosis. Student t-test revealed no statistical significance (p=0.57).
DCs with fascin1 expression are resistant to Lm infection
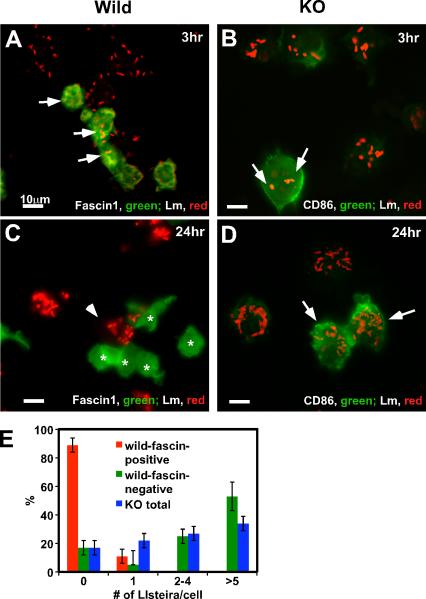
Immunofluorescence revealed that wild type DCs with high fascin1 expression are very resistant to Lm infection (Fig. 2). DCs were infected with Lm at a moi of 3 and at 3hr and 24hr post infection with Lm, wild type (A & C) and fascin1 KO (B & D) DCs were stained with an anti-Lm antibody and counterstained with an anti-fascin1 (for wild type DCs) or anti-CD86 antibody (for KO DCs). At 3hr post infection, wild-type DCs with low levels of fascin1 expression (labeled green) were infected with several Lm (arrows in Fig. 2A; Lm labeled red). At 24hr post infection (C), some DCs expressed high levels of fascin1 (indicated by asterisks). Remarkably, these DCs with high fascin1 expression had no Lm infection whereas adjacent fascin1-negative DCs (indicated by an arrowhead) were heavily infected. The presence of fascin1-positive and fascin1-negative DCs in wild type DC preparations is consistent with our previous report that LPS-matured, CD11c-positive DCs from wild type mice have two distinct populations in terms of fascin1 expression: One population (about 40% of total DCs) shows two orders of magnitude higher fascin1 expression than the other (14).
Figure 2.
Fascin1-expressing DCs are resistant to Lm infection. A-D, Immunofluorescence images of wild type (A & C) and fascin1 KO (B & D) DCs infected with Lm for 3hr (A & B) and 24hr (C & D). DCs were infected with Lm at a moi of 3.0. At indicated time, wild type DCs were stained with anti-Lm (red) and anti-fascin1 (green) antibodies and fascin1 KO DCs were stained with anti-Lm (red) and anti-CD86 (green) antibodies. Bar, 10μm. E, Quantitative analyses of Lm infection in fascin1-positive, wild type DC (red bars), fascin1-negative, wild type DCs (green), and fascin1 KO DCs (blue). Twenty four hours after infection, wild type, fascin1-positive DCs (n=49), as well as wild type, fascin1-negative DCs (n=94), and fascin1 KO (n=100) DCs, were analyzed by immunofluorescence, and the number of intracellular Lm per DC was counted. For each type of DC, non-infected cells and cells infected with one, two to four, or more than 5 Lm were expressed as percentages of total cells. P values of Lm infection between fascin1-positive, wild type DCs and fascin1-negative wild type DCs and between fascin1-positive, wild type DCs and KO DCs are less than 0.0001. On the other hand, the difference between fascin1-negative, wild type DCs and fascin1 KO DCs is statistically insignificant. Error bars, standard deviation. Representative results from three independent experiments.
Fascin1 KO DCs infected with Lm at 3hr post infection started to express CD86 (B), a maturation marker, indicating that KO DCs also became matured by Lm infection. Unlike wild type DCs, however, fascin1 KO DCs were heavily and uniformly infected with Lm at 24hr (D). Quantitative analyses (E) confirmed the above observations: no Lm was detected in 90% of DCs with high fascin1 expression at 24hr post infection (red bars). In contrast, more than 80% of fascin1 KO DCs were heavily infected (blue bars). Fascin1-negative, wild type DCs (green bars) were infected with Lm as heavily as fascin1 null DCs, suggesting that fascin1 expression is well correlated with the resistance to Lm infection in DCs.
Wild type DCs exhibit phagosomal acidification to a higher extent than fascin1 KO DCs
DCs have to control proliferation of Lm in two major steps. The first one is to eradicate phagosome-engulfed Lm and the second is to kill cytoplasmic Lm that have escaped from phagosomes. Phagolysosomal fusion is one of the primary host defense systems that can kill phagosome-entrapped bacteria. Because phagolysosomal fusion is controlled by the actin cytoskeleton (19, 20), fascin1 may alter this fusion process. Because the fusion efficiency depends on the phagosomal pH (32), we measured phagosomal pH.
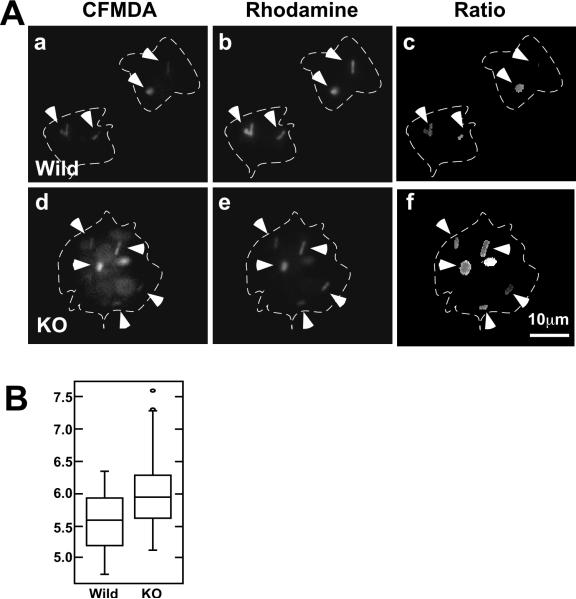
We found that wild type DCs showed lower phagosomal pH than fascin1 KO DCs (Fig. 3). To determine phagosomal pH, we utilized fluorescently labeled Lm as a pH indicator (Lm double-labeled with pH-sensitive (CMFDA) and pH-insensitive (Rhodamine) dyes), and performed ratiometric imaging. This is based on the observation that CMFDA fluorescence is lower in an acidic environment whereas Rhodamine fluorescence is independent of pH. An Lm mutant (DP L-2319) that is unable to escape from phagosomes was used for infection to confirm that Lm is kept contained within phagosomes during the experimental time window. Fig. 3A shows representative images of wild type (a-c) and fascin1 KO (d-f) DCs at 1.5hr post infection with fluorescently labeled Lm (a & d, 529nm; b & e; 617nm; c & f; ratio image). Ratio images (c & f) indicate that Lm in wild type DCs show lower ratios than Lm in fascin1 KO DCs. This observation indicates that phagosomes of wild type DCs are in a more acidic environment. Quantitative ratiometric analyses with box plots confirmed this (B): The median phagosomal pH was 5.6 (n=44) for wild type DCs and 6.0 (n=87) for fascin1 KO DCs (p<0.0001). The pH value (pH 5.6) of wild type DCs is similar to what was recently reported by Westcott et al (12).
Figure 3.
Phagosomal pH is lower in wild type DCs than in fascin1 KO DCs at 1hr post infection. A, Fluorescent images at 529nm (CFMDA), 617nm (Rhodamine), and corresponding ratiometric (529nm/617nm) images of CFMDA/Rhodamine-double-labeled Lm phagocytosed in DCs. A Lm mutant DP L-2319 (Δhly ΔplcA ΔplcB) was used for infection (moi of 3) to keep Lm entrapped in phagosomes. Ratio (529/617) images revealed that phagosomal pH in wild type DCs is lower than that in fascin1 KO DCs. Arrowheads, phagocytosed Lm. Cell peripheries are indicated by dashed lines. B, Box plot of phagosomal pH determined by ratiometric analyses of CFMDA/Rhodamine-double-labeled Lm as described in Materials and Methods. Representative results are shown from three independent experiments.
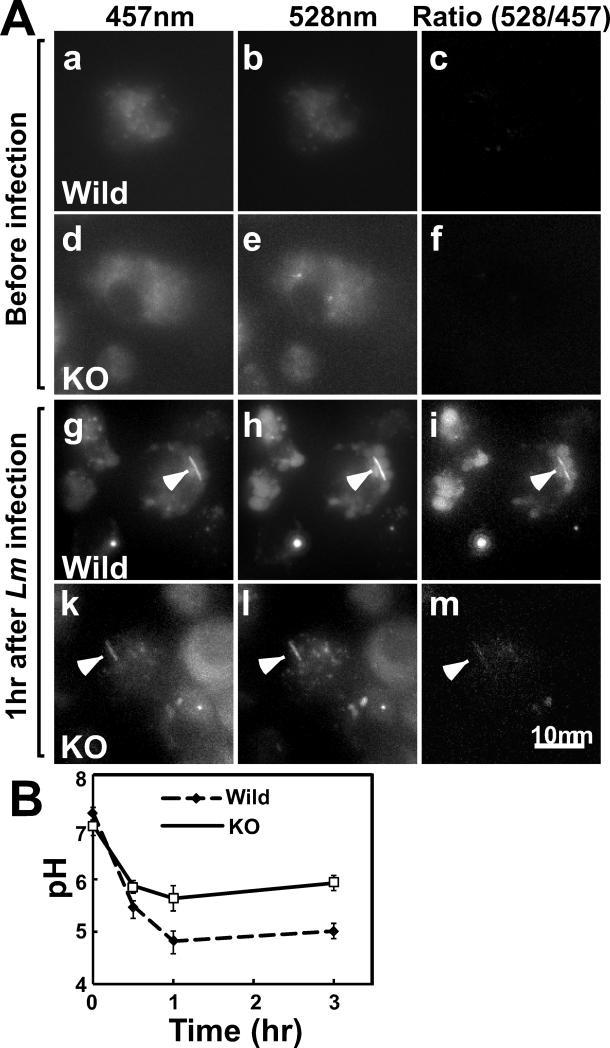
To expand the above analyses, we used a LysoSensor yellow/blue dye (DN-160, Molecular Probes) and examined whether fascin1 affects the pH of intracellular acidic vacuoles. This dye is incorporated into acidic organelles including phagosomes, late endosomes and lysosomes; and ratio imaging of 528nm/457nm gives pH values. After incorporation of the dye, DCs were infected with wild type Lm, and ratiometric imaging was performed at different time points. Fig. 4A shows representative images at 457nm and 528nm emission, as well as their corresponding ratiometric images. Before infection (a-f), the ratiometric images of both wild type (a-c) and fascin1 KO (d-f) DCs were dark, indicating that vacuolar pH was neutral for both wild type and fascin1 KO DCs. After Lm infection (g-m), however, ratiometric images showed many brighter vacuoles in wild type DCs (g-i) than in fascin1 KO DCs (k-m). In addition, infected Lm was also labeled with this dye (indicated by arrowheads) and exhibited brighter ratiometric values in wild type DCs than in fascin1 KO DCs.
Figure 4.
Vacuolar pH is lower in wild type DCs than in fascin1 KO DCs. A, Ratiometric images of wild type (a-c, g-i) and fascin1 KO (d-f, k-m) DCs. Representative images at 457nm and 528nm emission, as well as corresponding ratio images before (a-f) and after (g-m) infection are shown. Arrowheads indicate Lm. B, Time course of vacuolar pH changes of wild type and fascin1 KO DCs. Error bars, standard deviation. Representative results from three independent experiments.
Fig. 4B shows the time course of vacuolar pH change determined by the above ratiometric analyses. Prior to infection, the vacuolar pH was neutral to alkaline (7.0-7.2) in both wild type and fascin1 KO DCs. Lm infection induced rapid acidification of the vacuolar pH. Importantly, wild type DCs exhibited a greater extent of acidification than did fascin1 KO DCs. At 1-3hr after infection the vacuolar pH of wild type DCs became acidic to the range between 4.6 and 5.0. On the other hand, the vacuolar pH of fascin1 KO DCs was lowered to a lesser extent, ranging between 5.4 and 5.8 during the same time window (p<0.0001). These results suggest that fascin1 is critical for acidification of not only phagosomes, but also other acidic vacuoles, including late endosomes, phagolysosomes, and lysosomes.
Fascin1 binds to LC3, a component of autophagosomes
Autophagy is known to eradicate Lm that have escaped into the cytoplasm from phagosomes. Because autophagy is controlled by the actin cytoskeleton (21, 22, 33), fascin1 could also affect an autophagy process. Inspection of the fascin1 sequence revealed that fascin1 contains two conserved sequence of Y152AHL155 and Y314WTL317 (mouse fascin1 sequence, NP_032010), which corresponds to a motif for the LC3-interacting region (W/F/Y-X-X-L/I/V) (34, 35). Indeed, we found that fascin1 binds to LC3 in vivo, as well as in vitro. As Fig. 5A shows, fascin1 was detected in LC3 immunoprecipitates from HeLa cells (lane 2), suggesting that LC3 and fascin1 form a complex in vivo. As a control, we examined whether fascin1 is present in LAMP1 immunoprecipitation and found that fascin1 was not detected in LAMP1 immunoprecipitates (lane 1). The association between LC3 and fascin1 appears to be direct as measured with in vitro binding assays. GST-pull down assays (B) showed that GST-LC3 bound to fascin1 (lane 3), but not GST alone (lane 2).
Figure 5.
In vivo and in vitro association of fascin1 with LC3. A, Co-immunoprecipitation of LC3 and fascin1. Lane1, LAMP1 immunoprecipitate as a control; lane 2, LC3 immunoprecipitate. Immunoprecipitates were analyzed by Western blotting with anti-LAMP1, anti-fascin1 and anti-LC3 antibodies. An asterisk in lane 2 is a cross-reactive IgG band. B, In vitro GST-pull down assay to show direct binding between LC3 and fascin1. GST alone (lane 2) and GST-LC3 (lane 3) bound to glutathione Sepharose beads were incubated with non-fusion, recombinant fascin1 and bound fractions were analyzed by Western blotting with anti-fascin1 and anti-LC3 antibodies. Lane 1, input; lane 4, GST-LC3 without fascin1 incubation showing no fascin1 is present in the original GST-LC3 preparation. C, Proximity Ligation Assay (PLA). a-c, wild type DCs; d-f, fascin1 KO DCs. a & d, phase-contrast image; b & e, fluorescent signals showing close association between fascin1 and LC3 by PLA (see Materials and Methods for detail); c & f, merged images. Cell peripheries were marked by dashed lines. Note that fascin1 KO DCs show no PLA signals, confirming that fluorescent signals detected in wild type DCs indeed indicate the association between fascin1 and LC3.
A proximity ligation assay (PLA) revealed that both fascin1 and LC3 in wild type DCs were co-localized as a complex inside DCs. In a PLA system, two proteins are probed with antibodies conjugated with short nucleotide stretches. If they are within ~40nm, these nucleotides can be ligated into circular DNAs, which are amplified and detected as a fluorescent spot with complementary fluorescent oligonucleotides. As Fig. 5C shows, speckle-like fluorescent spots were detected in the perinuclear region of Lm-infected, wild type DCs (panels a-c). In contrast, no fluorescent signals were detected in fascin1 KO DCs (panels d-f), confirming that the fluorescent signals in wild type DCs indeed represent complex formation between fascin1 and LC3. To validate the authenticity of the PLA, we examined whether this assay can detect the known association between fascin1 and actin (31). Unlike the perinuclear localization of fascin1-LC3 complexes, a PLA assay to detect fascin1-actin association revealed many speckle-like fluorescent spots in membrane ruffles, as well as filopodia, where both proteins are known to be co-localized (supplemental Fig. 1).
Fascin1 appears to promote Lm-LC3 association
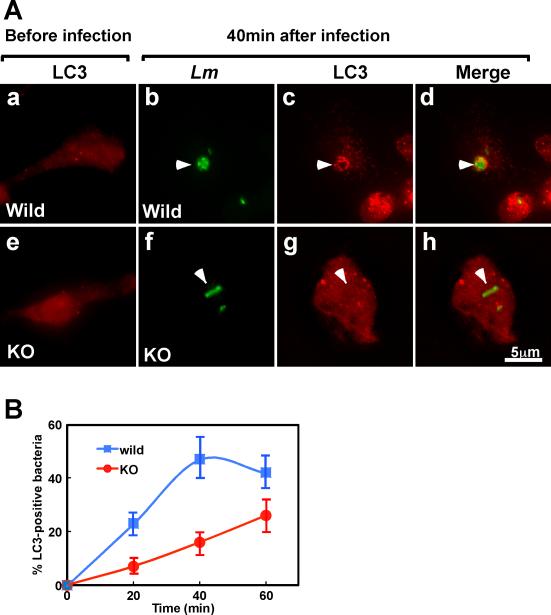
The LC3-fascin interaction may facilitate autophagy of cytoplasmic bacteria. We thus examined whether fascin1 alters the extent of LC3-Lm association, an indication of autophagy-mediated bacterial trapping. Before infection, both wild type and KO DCs show very few LC3 puncta (Fig. 6A, panels a and e). At 40min post infection, Lm in wild type DCs (indicated by arrowheads in panel b-d) were found to be much more frequently associated with LC3 than were Lm in fascin1 KO DCs (indicated by arrowheads in panel f-h). Quantitative measurements of LC3-positive Lm at 20min, 40min and 60min revealed that wild type DCs exhibited much faster kinetics of LC3-Lm association than did fascin1 KO DCs (B). The fraction of LC3-positive bacteria in wild type DCs increased from 23 to 47% from 20min to 40min and leveled off at 60min. On the other hand, the fraction of LC3-positive Lm in fascin1 KO DCs was just increased from 7% at 20min to 16% at 40min and 25% at 60min. These results suggest a possibility that Lm is processed via autophagy with a faster kinetics in wild type DCs than in fascin1 KO DCs.
Figure 6.
Fascin1 facilitate Lm-LC3 association. A, Wild type (a-d) and fascin1 KO (e-h) DCs were labeled with anti-LC3 and anti-Lm antibodies. a & e, before infection; b-d & f-h, 40min after infection with a wild type strain of Lm. Arrowheads in b-d indicates that Lm is surrounded with LC3 in wild type DCs whereas arrowheads in f-h show Lm that is not associated with LC3. B, Time-course analysis of Lm-LC3 association. Wild type DCs show Lm-LC3 association to a greater extent and with faster kinetics. Error bars, standard deviation. Representative result from three independent experiments.
The faster kinetics of LC3 association could occur if Lm in wild type DCs entered more quickly into the cytoplasm than those in fascin1 KO DCs. This was possible because fascin1 reduced phagosomal pH from 6.0 to 5.5 and because the activity of LLO, a pore-forming toxin protein required for cytoplasmic entry, becomes maximum at a phagosomal pH of 5.5 (36, 37). It is also possible that the LC3-Lm association described above may not be derived from cytoplasmic Lm but from vacuolar Lm in a compartment known as SLAPs (spacious Listeria-associated phagosomes): SLAPs has been shown to be associated with LC3 (38) though the SLAP assembly appears to occur at a later time point of 4hr in macrophages. To test these possibilities, we measured, by electron microscopy, what fractions of Lm were cytoplasmic and whether SLAP-like vacuoles were observed. Wild and fascin1 KO DCs were infected with wild type Lm, and fixed at 50min post infection, at which time wild type and fascin1 KO DCs show difference in the LC3 and Lm association.
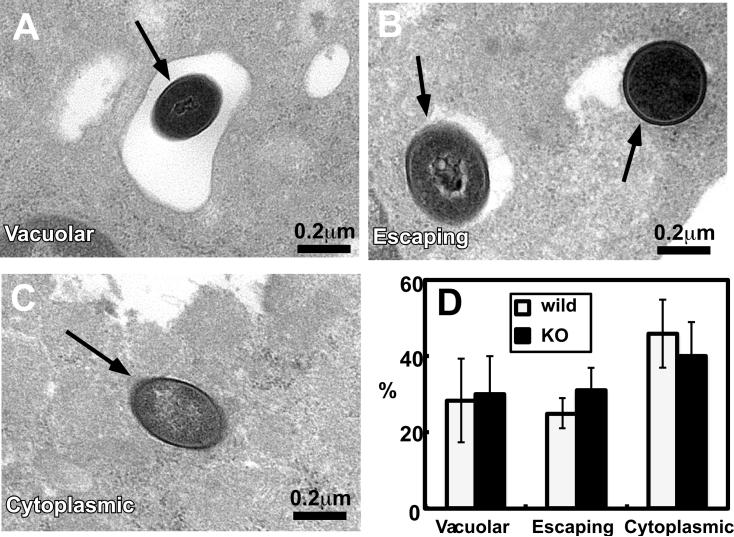
Fig. 7 shows three typical images of wild type Lm: They represent Lm in vacuoles (A), Lm apparently escaping from vacuoles (B), and Lm in the cytoplasm (C). Quantitative analyses to determine the fractions of these three types of Lm in DCs (D) revealed that Lm rapidly escaped from phagosomes in DCs and that there is no significant difference in cytoplasmic penetration between wild type and fascin1 KO DCs: At 50min post infection, the fractions of cytoplasmic Lm were about 46±9% for wild type DCs and 40±9% for fascin1 KO DCs. The fractions of Lm escaping from phagosomes were 25±4% for wild type and 31±6% for fascin1 KO DCs. Electron microscopy also revealed few SLAP-like vacuoles, which is defined by the presence of more than three Lm in a spacious vacuoles. This is consistent with the previous report that only 10% of macrophages exhibited SLAPs and the majority of Lm is cytoplasmic in macrophages at 1hr post infection (38).
Figure 7.
Electron microscopic analyses revealed that cytoplasmic penetration of Lm is similar between wild type and fascin1 KO DCs. DCs were infected with wild type strain Lm (moi of 1) and fixed at 50min post infection. A-C, representative images of Lm in DCs. A, Lm in vacuoles; B, Lm apparently escaping from phagosomes; C, Lm in the cytoplasm. D, quantitative analyses of cytoplasmic penetration of wild strain Lm in wild type and fscin1 KO DCs. Error bars, standard deviation. Representative results from three independent experiments.
Cytoplasmic penetration of Lm was also examined by immunofluorescence analyses. DCs infected with Lm for 1hr were labeled with an antibody against Arp2/3, a marker of cytoplasmic Lm (Supplemental Fig. 2). We found that the fractions of Arp2/3-positive Lm were similar between wild type (43±14%) and fascin1 KO DCs (44±24%). In contrast, mutant strain Lm (DP L-2319) that cannot escape from phagosomes were not labeled with the Arp2/3 antibody, confirming that Lm labeled with the Arp2/3 antibody represent cytoplasmic Lm. These observations are consistent with the electron microscopic analyses and suggest that the difference in phagosomal pH between wild and fascin1 KO DCs did not largely affect the cytoplasmic entry. Taken together, the faster kinetics of LC3-Lm association suggests that wild type DCs show higher autophagy activity than fascin1 KO DCs.
Fascin1 is involved in promoting autophagy flux
To monitor autophagy dynamics, we examined whether wild type DCs have a higher autophagy flux (a fusion process from autophagosomes to autophagolysosomes) than do fascin1 KO DCs. By expressing tfLC3 (LC3 tandemly tagged with GFP and RFP) (39), we measured whether fascin1 alters autophagy flux in LPS-matured DCs. As Fig. 8A and B shows wild type DCs displayed more red LC3 puncta than did fascin1 KO DCs. Quantitative analyses (C) revealed that 70% of wild type DCs showed red LC3 while most fascin1 KO DCs (70%) exhibited green puncta. These results indicate that tfLC3 is in the acidic compartments in wild type DCs, suggesting that fascin1 may promotes autophagosome-lysosome fusion by increasing autophagic flux.
Figure 8.
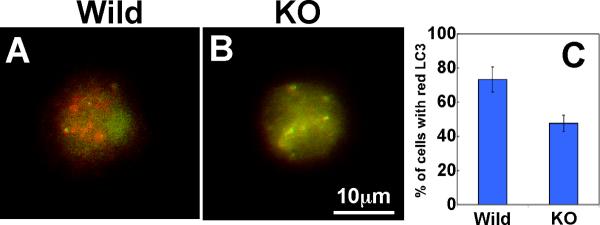
Wild type DCs show higher autophagy flux than fascin1 KO DCs. Wild type (A) and fascin1 KO (B) DCs were transfected with tfLC3 (LC3 tandemly tagged with GFP and RFP), and matured by the addition of LPS. Red signals in wild type DCs indicate autophagosomal fusion of tfLC3. C, quantitative analyses showing percentages of DCs with red tfLC3. Error bars, standard deviation.
DISCUSSION
Our results suggest that fascin1 plays a role in innate immunity by facilitating eradication of Lm in DCs. This novel role of fascin1 would also be critical for adaptive immunity because DCs have to survive infection to present pathogen information to T-cells. The ability of fascin1 to protect DCs from Lm infection is significant: Fascin1 reduces a Lm replication rate by 2 times during the 1-4hr period after infection (Fig. 1). This reduction is comparable to that reported for ATG5, a major constituent of autophagy, on Shigella replication in MEFs (40). Because macrophages express no fascin1, our results would explain the previous report that macrophages are more susceptible to Lm infection than DCs (11, 12).
Role of fascin1 in phagosomal acidification
Before infection, both wild type and fascin1 KO DCs show a similar vacuolar pH from neutral to alkaline, which is consistent with the lack of fascin1 expression in immature wild type DCs (13). While Lm infection acidifies phagosomal pH of both wild type and fascin1 KO DCs, fascin1 expression resulted in further acidification (Fig. 3). It has been demonstrated that maturation activates the vacuolar proton pump (V-ATPase) by increasing the association between the V0 and V1 sectors of V-ATPase (41). Because the V1 sector of V-ATPase binds to actin filaments (42-45), it is possible that fascin1, via bundling actin filaments, could promote the V0- V1 association of V-ATPase on phagosomes, promoting V-ATPase activity, thus lowering phagosomal pH more efficiently and accelerating phagolysosomal fusion of phagosome-entrapped bacteria. We are in the process of testing this possibility.
Our observation of acidic phagosomal pH is in agreement with a recent report by Westcott et al., that Lm-engulfed phagosomes of DCs are acidic (12). There are, however, conflicting reports regarding phagosomal pH in DCs (46, 47). One group has reported that phagosomal pH in DCs is kept neutral to alkaline due to ROS production by the recruitment of NOX2 to phagosomes. The authors claimed that alkaline pH is critical to prevent excessive degradation of possible antigen peptides and thus preserve antigen information for presentation to T-cells (46). On the other hand, a recent study has shown that phagosomal pH is acidified with a variety of phagocytosed particle including latex beads, and that limited proteolysis occurs by NOX2-mediated modification of cathepsins in a pH-independent manner (47).
Regardless of the origin of the conflict about pH of the latex bead-engulfed phagosomes, Lm-engulfed phagosomes must be acidic. Unlike latex beads used in the studies by Savina et al. (46), Lm escapes into the cytoplasm from phagosomes. This escape necessitates acidic phagosomal pH because phagosomal escape requires the activity of Listeriolysin O (LLO), a pore forming toxin protein that is active only in an acidic environment (48). Perhaps, DCs’ responses to Lm are quite different from those to opsonized latex beads. We recognized, for example, that Lm infection rapidly induced fascin1 expression to a maximum within 1hr whereas LPS activation took as long as 7hr to induce fascin1 expression to a similar level (data not shown). This observation indicates that Lm infection rapidly induces DC maturation by pattern recognition receptor engagement whereas opsonized latex beads may keep DCs in an immature state for a prolonged time. It is worthy of note that the timing of cytoplasmic penetration of Lm coincides with the timing of fascin1 expression, as well as that of acidification (Figs 3 & 4).
Role of fascin1 in autophagy
Our results suggest that fascin1 may promote autophagy. How does fascin1 play such a role? Fascin1-LC3 binding is likely to be involved. Intracellular cytoplasmic Lm has been shown to evade autophagy-mediated killing in many types of cells including fibroblasts, macrophages and MDCK cells (49-52). Lm uses the virulence factor, ActA, for the evasion (51-54), and the binding of ActA to Arp2/3 or VASP is essential for evading autophagy (52). It has been suggested that the binding of ActA with Arp2/3 or VASP results in “coating’ of the surfaces of Lm with cytoplasmic host proteins. This “coating” would disguise intracellular Lm as a cytoplasmic structure, thereby evading the autophagy machinery. It is possible that fascin1 could negate this disguise. As an ActA/Arp2/3/VASP complex polymerizes F-actin on the surface of Lm, fascin1 would recruit LC3 via its binding to F-actin. Such LC3 recruitment could facilitate autophagic recognition, blocking the autophagy evasion. We are in the process of testing this hypothesis.
Fascin1 has been reported to promote Lm motility in vitro (24), which could increase cell-to-cell transmission of Lm. However, we observed that wild type DCs with high fascin1 expression has no Lm infection, suggesting that increased Lm motility, if it ever happens in DCs, does not facilitate cell-to-cell transmission of Lm. Indeed, we found that cell-to-cell transmission (judged from the size of Lm-infected DC colonies) is lower in wild type DCs than in fascin1 KO DCs (supplemental Figure 3).
Conclusion
We have demonstrated that fascin1 contributes to survival of DCs against Lm infection. Judging from the proposed roles of fascin1 in lowering phagosomal pH and facilitating autophagy, it is possible that fascin1 protects DCs from infection by other bacterial pathogens. Fascin is abundantly expressed in both Drosophila hemocytes and sea urchin coelomocytes, both of which are phagocytic defense cells, involved in the innate immune system of these organisms (55-58). These results suggest an evolutionarily conserved role of fascin in innate immunity, and it would be interesting to see whether fascin1 also protects these non-mammalian phagocytic defense cells from bacterial infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. M. Dustin (New York University) for his suggestion to examine effects of fascin1 on Lm infection. We thank Dr. M. A. Conti (NIH/NHLBI) and Dr. M. Mitsuyama (Kyoto University, Japan) for their comments and critical reading of this manuscript. We appreciate the gifts of Listeria monocytogenes strains from Dr. D. A. Portnoy (University of California at Berkeley), and GST-LC3 constructs from Drs J. Feng (State University of New York at Buffalo) and T. Kamata (Shinshu University, Japan).
Abbreviations
- KO
knockout
- DCs
Dendritic cells
- LLO
listeriolysin O
- FCS
fetal calf serum
Footnotes
This work was supported by R03AI099315 (to SY) and Johnson & Johnson Proof of Concept Fund (to FM).
References
- 1.Steinman RM, Hemmi H. Dendritic cells: translating innate to adaptive immunity. Curr Top Microbiol Immunol. 2006;311:17–58. doi: 10.1007/3-540-32636-7_2. [DOI] [PubMed] [Google Scholar]
- 2.Mellman I, Steinman RM. Dendritic cells: specialized and regulated antigen processing machines. Cell. 2001;106:255–258. doi: 10.1016/s0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- 3.Trombetta ES, Mellman I. Cell biology of antigen processing in vitro and in vivo. Annu Rev Immunol. 2005;23:975–1028. doi: 10.1146/annurev.immunol.22.012703.104538. [DOI] [PubMed] [Google Scholar]
- 4.Portnoy DA, Auerbuch V, Glomski IJ. The cell biology of Listeria monocytogenes infection: the intersection of bacterial pathogenesis and cell-mediated immunity. J Cell Biol. 2002;158:409–414. doi: 10.1083/jcb.200205009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veiga E, Cossart P. Listeria hijacks the clathrin-dependent endocytic machinery to invade mammalian cells. Nat Cell Biol. 2005;7:894–900. doi: 10.1038/ncb1292. [DOI] [PubMed] [Google Scholar]
- 6.Boujemaa-Paterski R, Gouin E, Hansen G, Samarin S, Le Clainche C, Didry D, Dehoux P, Cossart P, Kocks C, Carlier MF, Pantaloni D. Listeria protein ActA mimics WASp family proteins: it activates filament barbed end branching by Arp2/3 complex. Biochemistry. 2001;40:11390–11404. doi: 10.1021/bi010486b. [DOI] [PubMed] [Google Scholar]
- 7.Welch MD, Rosenblatt J, Skoble J, Portnoy DA, Mitchison TJ. Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science. 1998;281:105–108. doi: 10.1126/science.281.5373.105. [DOI] [PubMed] [Google Scholar]
- 8.Jung S, Unutmaz D, Wong P, Sano G, De los Santos K, Sparwasser T, Wu S, Vuthoori S, Ko K, Zavala F, Pamer EG, Littman DR, Lang RA. In vivo depletion of CD11c(+) dendritic cells abrogates priming of CD8(+) T cells by exogenous cell-associated antigens. Immunity. 2002;17:211–220. doi: 10.1016/s1074-7613(02)00365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brzoza KL, Rockel AB, Hiltbold EM. Cytoplasmic entry of Listeria monocytogenes enhances dendritic cell maturation and T cell differentiation and function. J Immunol. 2004;173:2641–2651. doi: 10.4049/jimmunol.173.4.2641. [DOI] [PubMed] [Google Scholar]
- 10.Paschen A, Dittmar KE, Grenningloh R, Rohde M, Schadendorf D, Domann E, Chakraborty T, Weiss S. Human dendritic cells infected by Listeria monocytogenes: induction of maturation, requirements for phagolysosomal escape and antigen presentation capacity. Eur J Immunol. 2000;30:3447–3456. doi: 10.1002/1521-4141(2000012)30:12<3447::AID-IMMU3447>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 11.Westcott MM, Henry CJ, Cook AS, Grant KW, Hiltbold EM. Differential susceptibility of bone marrow-derived dendritic cells and macrophages to productive infection with Listeria monocytogenes. Cell Microbiol. 2007;9:1397–1411. doi: 10.1111/j.1462-5822.2006.00880.x. [DOI] [PubMed] [Google Scholar]
- 12.Westcott MM, Henry CJ, Amis JE, Hiltbold EM. Dendritic cells inhibit the progression of Listeria monocytogenes intracellular infection by retaining bacteria in major histocompatibility complex class II-rich phagosomes and by limiting cytosolic growth. Infect Immun. 2010;78:2956–2965. doi: 10.1128/IAI.01027-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mosialos G, Birkenbach M, Ayehunie S, Matsumura F, Pinkus GS, Kieff E, Langhoff E. Circulating human dendritic cells differentially express high levels of a 55-kd actin-bundling protein. Am J Pathol. 1996;148:593–600. [PMC free article] [PubMed] [Google Scholar]
- 14.Yamakita Y, Matsumura F, Lipscomb MW, Chou PC, Werlen G, Burkhardt JK, Yamashiro S. Fascin1 promotes cell migration of mature dendritic cells. J Immunol. 2011;186:2850–2859. doi: 10.4049/jimmunol.1001667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller JC, Brown BD, Shay T, Gautier EL, Jojic V, Cohain A, Pandey G, Leboeuf M, Elpek KG, Helft J, Hashimoto D, Chow A, Price J, Greter M, Bogunovic M, Bellemare-Pelletier A, Frenette PS, Randolph GJ, Turley SJ, Merad M. Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol. 2012;13:888–899. doi: 10.1038/ni.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haglund CM, Welch MD. Pathogens and polymers: microbe-host interactions illuminate the cytoskeleton. J Cell Biol. 2011;195:7–17. doi: 10.1083/jcb.201103148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pizarro-Cerda J, Sousa S, Cossart P. Exploitation of host cell cytoskeleton and signalling during Listeria monocytogenes entry into mammalian cells. Comptes rendus biologies. 2004;327:523–531. doi: 10.1016/j.crvi.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Rottner K, Stradal TE, Wehland J. Bacteria-host-cell interactions at the plasma membrane: stories on actin cytoskeleton subversion. Dev Cell. 2005;9:3–17. doi: 10.1016/j.devcel.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Girao H, Geli MI, Idrissi FZ. Actin in the endocytic pathway: from yeast to mammals. FEBS Lett. 2008;582:2112–2119. doi: 10.1016/j.febslet.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 20.Kjeken R, Egeberg M, Habermann A, Kuehnel M, Peyron P, Floetenmeyer M, Walther P, Jahraus A, Defacque H, Kuznetsov SA, Griffiths G. Fusion between phagosomes, early and late endosomes: a role for actin in fusion between late, but not early endocytic organelles. Mol. Biol. Cell. 2004;15:345–358. doi: 10.1091/mbc.E03-05-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monastyrska I, He C, Geng J, Hoppe AD, Li Z, Klionsky DJ. Arp2 links autophagic machinery with the actin cytoskeleton. Mol. Biol. Cell. 2008;19:1962–1975. doi: 10.1091/mbc.E07-09-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, Pandey UB, Kaushik S, Tresse E, Lu J, Taylor JP, Cuervo AM, Yao TP. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010 doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yano T, Kurata S. Induction of autophagy via innate bacterial recognition. Autophagy. 2008;4:958–960. doi: 10.4161/auto.6802. [DOI] [PubMed] [Google Scholar]
- 24.Brieher WM, Coughlin M, Mitchison TJ. Fascin-mediated propulsion of Listeria monocytogenes independent of frequent nucleation by the Arp2/3 complex. J Cell Biol. 2004;165:233–242. doi: 10.1083/jcb.200311040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamashiro-Matsumura S, Matsumura F. Intracellular localization of the 55-kD actin-bundling protein in cultured cells: spatial relationships with actin, alpha-actinin, tropomyosin, and fimbrin. J Cell Biol. 1986;103:631–640. doi: 10.1083/jcb.103.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ono S, Yamakita Y, Yamashiro S, Matsudaira PT, Gnarra JR, Obinata T, Matsumura F. Identification of an actin binding region and a protein kinase C phosphorylation site on human fascin. J Biol Chem. 1997;272:2527–2533. doi: 10.1074/jbc.272.4.2527. [DOI] [PubMed] [Google Scholar]
- 27.Jiang H, Cheng D, Liu W, Peng J, Feng J. Protein kinase C inhibits autophagy and phosphorylates LC3. Biochem Biophys Res Commun. 2010;395:471–476. doi: 10.1016/j.bbrc.2010.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamakita Y, Matsumura F, Yamashiro S. Fascin1 is dispensable for mouse development but is favorable for neonatal survival. Cell Motil Cytoskeleton. 2009;66:524–534. doi: 10.1002/cm.20356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gedde MM, Higgins DE, Tilney LG, Portnoy DA. Role of listeriolysin O in cell-to-cell spread of Listeria monocytogenes. Infect Immun. 2000;68:999–1003. doi: 10.1128/iai.68.2.999-1003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamashiro S, Yamakita Y, Totsukawa G, Goto H, Kaibuchi K, Ito M, Hartshorne DJ, Matsumura F. Myosin phosphatase-targeting subunit 1 regulates mitosis by antagonizing polo-like kinase 1. Dev Cell. 2008;14:787–797. doi: 10.1016/j.devcel.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamashiro-Matsumura S, Matsumura F. Purification and characterization of an F-actin-bundling 55-kilodalton protein from HeLa cells. J Biol Chem. 1985;260:5087–5097. [PubMed] [Google Scholar]
- 32.Kinchen JM, Ravichandran KS. Phagosome maturation: going through the acid test. Nat Rev Mol Cell Biol. 2008;9:781–795. doi: 10.1038/nrm2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monastyrska I, Rieter E, Klionsky DJ, Reggiori F. Multiple roles of the cytoskeleton in autophagy. Biol Rev Camb Philos Soc. 2009;84:431–448. doi: 10.1111/j.1469-185X.2009.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 2010;584:1379–1385. doi: 10.1016/j.febslet.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 36.Geoffroy C, Gaillard JL, Alouf JE, Berche P. Purification, characterization, and toxicity of the sulfhydryl-activated hemolysin listeriolysin O from Listeria monocytogenes. Infect Immun. 1987;55:1641–1646. doi: 10.1128/iai.55.7.1641-1646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beauregard KE, Lee KD, Collier RJ, Swanson JA. pH-dependent perforation of macrophage phagosomes by listeriolysin O from Listeria monocytogenes. J Exp Med. 1997;186:1159–1163. doi: 10.1084/jem.186.7.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Birmingham CL, Canadien V, Kaniuk NA, Steinberg BE, Higgins DE, Brumell JH. Listeriolysin O allows Listeria monocytogenes replication in macrophage vacuoles. Nature. 2008;451:350–354. doi: 10.1038/nature06479. [DOI] [PubMed] [Google Scholar]
- 39.Kimura S, Fujita N, Noda T, Yoshimori T. Monitoring autophagy in mammalian cultured cells through the dynamics of LC3. Methods Enzymol. 2009;452:1–12. doi: 10.1016/S0076-6879(08)03601-X. [DOI] [PubMed] [Google Scholar]
- 40.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 41.Trombetta ES, Ebersold M, Garrett W, Pypaert M, Mellman I. Activation of lysosomal function during dendritic cell maturation. Science. 2003;299:1400–1403. doi: 10.1126/science.1080106. [DOI] [PubMed] [Google Scholar]
- 42.Vitavska O, Merzendorfer H, Wieczorek H. The V-ATPase subunit C binds to polymeric F-actin as well as to monomeric G-actin and induces cross-linking of actin filaments. J Biol Chem. 2005;280:1070–1076. doi: 10.1074/jbc.M406797200. [DOI] [PubMed] [Google Scholar]
- 43.Wieczorek H, Huss M, Merzendorfer H, Reineke S, Vitavska O, Zeiske W. The insect plasma membrane H+ V-ATPase: intra-, inter-, and supramolecular aspects. J Bioenerg Biomembr. 2003;35:359–366. doi: 10.1023/a:1025733016473. [DOI] [PubMed] [Google Scholar]
- 44.Vitavska O, Wieczorek H, Merzendorfer H. A novel role for subunit C in mediating binding of the H+-V-ATPase to the actin cytoskeleton. J Biol Chem. 2003;278:18499–18505. doi: 10.1074/jbc.M212844200. [DOI] [PubMed] [Google Scholar]
- 45.Holliday LS, Lu M, Lee BS, Nelson RD, Solivan S, Zhang L, Gluck SL. The amino-terminal domain of the B subunit of vacuolar H+-ATPase contains a filamentous actin binding site. J Biol Chem. 2000;275:32331–32337. doi: 10.1074/jbc.M004795200. [DOI] [PubMed] [Google Scholar]
- 46.Savina A, Jancic C, Hugues S, Guermonprez P, Vargas P, Moura IC, Lennon-Dumenil AM, Seabra MC, Raposo G, Amigorena S. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell. 2006;126:205–218. doi: 10.1016/j.cell.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 47.Rybicka JM, Balce DR, Chaudhuri S, Allan ER, Yates RM. Phagosomal proteolysis in dendritic cells is modulated by NADPH oxidase in a pH-independent manner. EMBO J. 2012;31:932–944. doi: 10.1038/emboj.2011.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glomski IJ, Gedde MM, Tsang AW, Swanson JA, Portnoy DA. The Listeria monocytogenes hemolysin has an acidic pH optimum to compartmentalize activity and prevent damage to infected host cells. J Cell Biol. 2002;156:1029–1038. doi: 10.1083/jcb.200201081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dortet L, Mostowy S, Cossart P. Listeria and autophagy escape: involvement of InlK, an internalin-like protein. Autophagy. 2012;8:132–134. doi: 10.4161/auto.8.1.18218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meyer-Morse N, Robbins JR, Rae CS, Mochegova SN, Swanson MS, Zhao Z, Virgin HW, Portnoy D. Listeriolysin O is necessary and sufficient to induce autophagy during Listeria monocytogenes infection. PLoS One. 2010;5:e8610. doi: 10.1371/journal.pone.0008610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogawa M, Yoshikawa Y, Mimuro H, Hain T, Chakraborty T, Sasakawa C. Autophagy targeting of Listeria monocytogenes and the bacterial countermeasure. Autophagy. 2011;7:310–314. doi: 10.4161/auto.7.3.14581. [DOI] [PubMed] [Google Scholar]
- 52.Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, Mimuro H, Nakagawa I, Yanagawa T, Ishii T, Kakizuka A, Sztul E, Chakraborty T, Sasakawa C. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol. 2009;11:1233–1240. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- 53.Birmingham CL, Canadien V, Gouin E, Troy EB, Yoshimori T, Cossart P, Higgins DE, Brumell JH. Listeria monocytogenes evades killing by autophagy during colonization of host cells. Autophagy. 2007;3:442–451. doi: 10.4161/auto.4450. [DOI] [PubMed] [Google Scholar]
- 54.Lam GY, Czuczman MA, Higgins DE, Brumell JH. Interactions of Listeria monocytogenes with the autophagy system of host cells. Advances in immunology. 2012;113:7–18. doi: 10.1016/B978-0-12-394590-7.00008-7. [DOI] [PubMed] [Google Scholar]
- 55.Shim J, Lee SM, Lee MS, Yoon J, Kweon HS, Kim YJ. Rab35 mediates transport of Cdc42 and Rac1 to the plasma membrane during phagocytosis. Mol Cell Biol. 2010;30:1421–1433. doi: 10.1128/MCB.01463-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tirouvanziam R, Davidson CJ, Lipsick JS, Herzenberg LA. Fluorescence-activated cell sorting (FACS) of Drosophila hemocytes reveals important functional similarities to mammalian leukocytes. Proc. Natl. Acad. Sci. USA. 2004;101:2912–2917. doi: 10.1073/pnas.0308734101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edwards RA, Bryan J. Fascins, a family of actin bundling proteins. Cell Motil Cytoskeleton. 1995;32:1–9. doi: 10.1002/cm.970320102. [DOI] [PubMed] [Google Scholar]
- 58.Otto JJ, Kane RE, Bryan J. Formation of filopodia in coelomocytes: localization of fascin, a 58,000 dalton actin cross-linking protein. Cell. 1979;17:285–293. doi: 10.1016/0092-8674(79)90154-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.