Abstract
Background
Sympathetic nerve wiring in the mammalian heart has remained largely unexplored. Resolving the wiring diagram of the cardiac sympathetic network would help establish the structural underpinnings of neurocardiac coupling.
New Method
We used two-photon excitation fluorescence microscopy, combined with a computer-assisted 3-D tracking algorithm, to map the local sympathetic circuits in living hearts from adult transgenic mice expressing enhanced green fluorescent protein (EGFP) in peripheral adrenergic neurons.
Results
Quantitative co-localization analyses confirmed that the intramyocardial EGFP distribution recapitulated the anatomy of the sympathetic arbor. In the left ventricular subepicardium of the uninjured heart, the sympathetic network was composed of multiple subarbors, exhibiting variable branching and looping topology. Axonal branches did not overlap with each other within their respective parental subarbor nor with neurites of annexed subarbors. The sympathetic network in the border zone of a 2-week-old myocardial infarction was characterized by substantive rewiring, which included spatially heterogeneous loss and gain of sympathetic fibers and formation of multiple, predominately nested, axon loops of widely variable circumference and geometry.
Comparison with Existing Methods
In contrast to mechanical tissue sectioning methods that may involve deformation of tissue and uncertainty in registration across sections, our approach preserves continuity of structure, which allows tracing of neurites over distances, and thus enables derivation of the three-dimensional and topological morphology of cardiac sympathetic nerves.
Conclusions
Our assay should be of general utility to unravel the mechanisms governing sympathetic axon spacing during development and disease.
Keywords: sympathetic nervous system, transgenic mice, enhanced green fluorescent protein, two-photon fluorescence microscopy, heart, 3-D reconstruction
1. Introduction
The cardiac sympathetic nervous system serves a key physiological role in orchestrating the heart’s response to physical or emotional stress, contributing to the maintenance of circulatory homeostasis. The heart receives its sympathetic input via fibers originating in neurons of cervical and paravertebral ganglia (Hassankhani et al., 1995; Kimura et al., 2012). Sympathetic fibers run in multi-nerve bundles across the epicardial surface before they eventually branch off to innervate their target territories inside the myocardial wall (Ieda et al., 2007; Kimura et al., 2012). A number of previous studies utilizing immunohistochemical approaches have quantitated sympathetic axon density in healthy myocardium as well as its changes in the diseased heart (Clarke et al., 2010; Ieda et al., 2007; Mühlfeld et al., 2010; Zhou et al., 2004). However, the spatial relationship of individual axons with other axons within the cardiac sympathetic network, i.e., the circuit wiring, has remained largely unexplored. Yet this information is critical to an understanding of neural network function in normal and diseased states (Ramon y Cajal, 1995; Swadlow et al., 1980). Thus, resolving the wiring diagram of the cardiac sympathetic network would help elucidate the structural underpinnings of neurocardiac coupling. Further, the ability to reconstruct the sympathetic circuit would provide a valuable tool to study developmental cues that determine network geometry and connectivity, as well as to gauge the efficacy of therapeutic interventions in correcting sympathetic miswiring in the diseased heart. Finally, imaging the sympathetic arbor in living heart raises the possibility to directly interrogate the impact of changes in axon density and/or connectivity on functional properties of the underlying tissue.
As an attempt at solving the cardiac sympathetic circuit, we sought to develop an assay that would allow detailed reconstruction of local network architecture in the intact heart. Here, we used two-photon excitation fluorescence microscopy, combined with computer-assisted image analyses, to map the local sympathetic circuits in intact, living hearts from adult transgenic mice expressing a fluorescent reporter protein in all peripheral adrenergic neurons. We demonstrate that our technique enables imaging of axonal projections within finite volumes of Langendorff-perfused mouse hearts. It also enables delineation of axonal geometry and spacing as well as identification of organizational principles of local sympathetic circuits. Application of this intact-system approach to chronically infarcted myocardium revealed several distinct alterations in axonal microanatomy and arborization in the peri-infarct border zone. Although we imaged only small volumes within the normal and injured mouse heart, this approach provides, to the best of our knowledge, the first glimpses at the 3-D micro-patterning of the sympathetic tree in intact mammalian myocardium. The results thus constitute a starting point to unravel the structural organization of the cardiac sympathetic network. Deciphering the connectional information in well-defined sample volumes will aid in understanding the structural underpinnings of sympathetic nervous function in the healthy and diseased heart.
2. Materials and methods
2.1. Generation and identification of transgenic mice expressing enhanced green fluorescent protein in peripheral sympathetic neurons
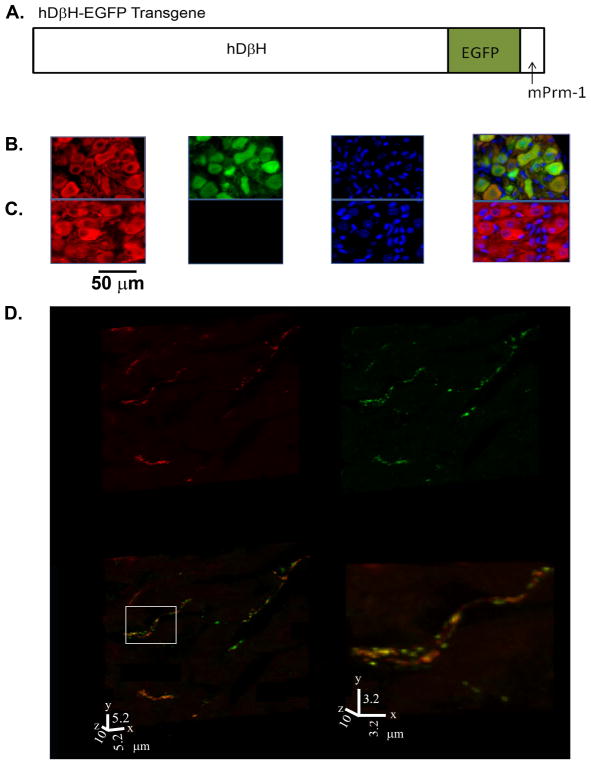
A transgenic mouse model was generated to quantitate cardiac sympathetic innervation density and patterning in living heart. The mouse model utilized the 5.9-kb human dopamine β-hydroxylase (hDβH) gene promoter sequence (provided by Dr. Palmiter, University of Washington, Seattle, Washington; (Mercer et al., 1991)) fused to the EGFP-encoding cDNA sequence. A 530-base pair fragment of the murine protamine-1 gene (mPrm1) including the poly(A) adenylation site and signal was inserted downstream of the EGFP sequence (Figure 1A). The hDβH-EGFP fusion gene was linearized and microinjected into the male pronucleus of fertilized C3HeB/FeJ embryos using standard techniques (Hogan et al., 1986; Steinhelper et al., 1990). The microinjected embryos were cultured to the two-cell stage, implanted into the oviducts of pseudopregnant females and allowed to develop to term. The resulting pups were screened for the presence of the transgene by a modified polymerase chain reaction (PCR) protocol (Rubart et al., 2003; Steinhelper et al., 1990). The transgene-specific oligonucleotide primers were (5′ ATCGAGCTGAAGGGCATCGACTTCAAGGAG3′) and (5′CTCCAGCAGGACCATGTGATCGCGCTTCTC3′). A segment of an endogenous single copy mouse gene provided an internal control for the reaction.
Figure 1.
Generation of hDβH-EGFP transgenic mice. A, Schematic diagram of the hDβH-EGFP transgene. mPrm-1 denotes murine protamine-1. B, Portion of a midsection through a stellate ganglion from an adult hDβH-EGFP transgenic mouse (line 4) stained for expression of tyrosine hydroxylase (TH; red) and EGFP (green), and counterstained with nuclear Hoechst (blue). The three-channel merged image is shown in the right panel. TH-expressing cells exhibit both cytoplasmic and nuclear anti-EGFP immunofluorescence. C, Section through a control stellate ganglion stained as in the transgenic ganglion, showing TH-expressing and non-expressing cells. D, Volume renderings of a 30-slice confocal z-stack obtained from a 10-μm-thick section from the left ventricular free wall of an adult hDβH-EGFP heart (line 4) dually stained with an antiTH and an anti-GFP antibody, followed by exposure to Alexa546- (red) and Alexa633- (green) conjugated secondary antibodies. The yellow color in the merged image indicates co-localization of TH and EGFP. The right panel is a magnified view of the boxed region in the merged image.
2.2. Immunolabeling
Mice of either sex and genotype between 12 and 38 weeks of age received intraperitoneal injections of heparin (125 I.U. / kg bodyweight) and were sacrificed via cervical dislocation in accordance with animal usage protocols. The heart was quickly excised, the ascending aorta was cannulated with an 18-gauge cannula, and the heart was perfused retrogradely at constant pressure with oxygenated Tyrode’s solution, containing (in mmol/L) 140 NaCl, 5 KCl, 1 MgCl2, 2 CaCl2, 10 HEPES and 10 glucose, pH of 7.4 (adjusted with 1 M NaOH). Once the coronary effluent became clear of blood, the perfusate was switched to a fixation solution containing (in mmol/L) 50 cacodylic acid, 333 paraformaldehyde, and 114 NaCl; pH 7.4. After 10 minutes of continuous perfusion, the heart was submerged in fixation solution at ~5 °C overnight. Cervical and stellate ganglia were removed bilaterally and submersion fixed overnight. Post fixation, the tissues were cryoprotected via overnight incubation in an ice-cold 30%-sucrose-PBS solution, embedded in O.C.T. (Tissue-Tek) tissue freezing medium, stored at −80 °C for 1 hour, sectioned at 10 μm thickness, and mounted on frosted slides.
The following primary antibodies were used: a rabbit polyclonal antibody directed against tyrosine hydroxylase (Millipore, Inc; AB 152); a chicken polyclonal antibody directed against tyrosine hydroxylase (Abcam; AB76442); a rabbit polyclonal antibody directed against GFP (Millipore, Inc.; AB3080); a rabbit polyclonal antibody directed against connexin43 (Millipore; AB1728); a FITC-preconjugated, goat polyclonal antibody directed against GFP (Novus Biologicals, NB100-1771). All antibodies had been affinity purified. Secondary antibodies were either goat polyclonal anti-rabbit IgG or goat polyclonal anti-chicken IgY conjugated to one of the fluorophores, Alexa546, Alexa555, Alexa633 (all Life Technologies, Grand Island, NY), or DyLight405 (Thermo Scientific, Waltham, MA). These antibodies had been affinity-purified and had been highly adsorbed to minimize species cross-reactivity.
Sections were incubated with 0.2% Triton X-100 (Sigma-Aldrich, St. Louis, MI) in PBS for 1 hour, followed by 30 minutes of blocking with 2% BSA. Sections were then incubated for 12 hours with primary antibodies in PBS supplemented with 2% BSA and 10% normal goat serum, and subsequently reacted with appropriate secondary antibodies for 1.5 h, followed by a 5-min incubation in PBS containing Hoechst (1:1000). All incubation steps were performed at room temperature, and between all incubation steps the slides were thoroughly washed with PBS three times for 5 minutes each. Sections were mounted in Vectashield (H-1000; Vectorlabs, Burlingame, CA) or ProLong Gold solution (A21103; Life Technologies, Inc, Grand Island, NY). Coverslips were fixed to the slides with nail polish. Sections that had been incubated with secondary antibody without having been labeled with primary antibody served as control.
To determine the empirical limit of co-localization (see section 2.4.), sections from an adult non-transgenic heart were first incubated with a rabbit polyclonal antibody against TH, followed by dual labeling with Alexa546- and Alexa633-conjugated goat polyclonal anti-rabbit IgG. The control experiments skipped incubation with the primary antibodies.
2.3. Immunofluorescence-based assessment of hDβH-EGFP transgene expression in sympathetic ganglia
Ten-micron-thick midsections from cervical and stellate ganglia of transgenic and control animals between 12 and 38 weeks of age were assayed for dual EGFP and TH expression by examining the distributions of anti-GFP and anti-TH immunoreactivities. Tissue sections were double stained with a FITC-preconjugated rabbit polyclonal antibody directed against GFP and a rabbit polyclonal antibody directed against TH as outlined above. A polyclonal goat anti-rabbit IgG conjugated to Alexa555 was used to visualize anti-TH immunoreactivity. Stained sections were imaged using a Leica fluorescence microscope equipped with a 20x 0.5 NA objective and the following filter cubes (excitation / emission): 340–380 / 425 LP (Hoechst), 450–490 / 515 LP (FITC or EGFP) and 515–560 / 590 LP (Alexa555). We acquired 440 × 340 μm2 non-overlapping images from each of the 3 sections per ganglion per animal. Exposure times for each excitation wavelength were the same for sections with and without primary antibody incubation. FITC and Alexa555 fluorescence images from hDβH-EGFP ganglia were thresholded against images obtained from wild-type ganglia stained with anti-GFP antibodies and from transgenic ganglia incubated only with the anti-rabbit IgG secondary antibody, respectively. Transgenic neurons were identified by virtue of co-localization of cytoplasmic anti-TH and anti-GFP immunofluorescence. Penetrance of transgene expression was defined as the percentage of TH-positive neurons expressing EGFP. Metamorph software (Molecular Devices, Sunnyvale, CA) was used for image processing.
2.4. Quantitative co-localization studies
Four-chamber longitudinal sections were obtained from hearts of transgenic and control mice between 12 and 38 weeks of age and dually stained with a rabbit polyclonal antibody directed against GFP and a chicken polyclonal antibody directed against TH as outlined above. Alexa555- and Alexa633-conjugated goat polyclonal antibodies directed against rabbit IgG and chicken IgY, respectively, were used to visualize tissue distribution of anti-GFP and anti-TH immunereactivities. Tissue samples were imaged using a confocal laser scanning microscope (Olympus FV1000). We used a 63x 1.42 NA oil-immersion (RI = 1.515) objective and optically zoomed-in by a factor of 4. Alexa555 fluorescence was excited with a 559-nm laser and detected through a band-pass emission filter of 570–625 nm. Alexa633 fluorescence was excited with a 635-nm laser and collected through a band-pass filter of 655–755 nm. Images were acquired at the Nyquist frequency (i.e., twice the maximal spatial frequency) in the x-y direction (pixel size = 0.106 μm) and oversampled by a factor of ~3 in the z direction (z-step size = 0.3 μm), with a 12-bit range. In our imaging system, the maximal spatial frequency is determined by the resolution of the microscope, and for the particular imaging conditions that we used the resolution is ~200 nm in x-y plane, and ~850 nm along the z axis (Scriven et al., 2008). We thus used the zoom-in feature of the microscope to obtain a pixel size that corresponded to the Nyquist frequency. Images for each detection channel were acquired sequentially using a pixel dwell time of 4 μs and a Kalman integration of 4. Laser power, pixel time, and detector sensitivity were adjusted for both channels individually to obtain relatively balanced signal intensity distributions for each channel.
Confocal image stacks were deconvolved using Huygen’s Professional software 32-bit version 4.1.1p2. Point spread functions used for deconvolution were calculated based on lens specifications and pinhole diameters of 1 airy unit. Post-deconvolution, image stacks were thresholded in each channel to remove background noise. To eliminate single pixel noise, a median filter with a 3 × 3 Kernel was applied. The deconvoluted and filtered image stacks were then segmented into their three orthogonal stacks and maximum projections in the XY dimensions were obtained using Metamorph software (Molecular Devices, Sunnyvale, CA). Maximum projections were used for co-localization analyses. Pearson’s coefficients were derived from these projections using the ImageJ plugin “Colocalisation Threshold” (Costes et al., 2004).
2.5. Two-photon laser scanning microscopy (TPLSM) of Langendorff-perfused mouse heart
2.5.1. Heart preparation
Five minutes after intraperitoneal injection of heparin (125 I.U. / kg bodyweight), the mouse was sacrificed by dislocation of the neck, the heart rapidly removed, connected to a Langendorff apparatus, and the coronary arteries were continuously perfused at 60 cm H2O via a customized, 18G cannula in the aortic root with oxygenated, HEPES-buffered Tyrode’s solution (room temperature). The solution consisted of the following (in mmol/L): 140 NaCl, 5 KCl, 1 MgCl2, 2 CaCl2, 10 HEPES, 10 D-glucose at a pH of 7.4 adjusted with 1 M NaOH. The heart was then transferred to a custom-built holding apparatus (designed and manufactured by M. H. Soonpaa) on the stage of the laser scanning microscope. Once the heart was in a fixed position, the buffer was switched to Tyrode’s solution supplemented with 1 μm ryanodine (R-500; Alomone Labs, Jerusalem, Israel) and 50 μM cytochalasin D (C8273; Sigma-Aldrich, St. Louis, MO) to uncouple contraction from excitation.
2.5.2 TPLSM imaging
TPLSM was performed on an Olympus FV1000 confocal scanning microscope (Olympus, Center Valley, PA) modified for two-photon excitation. Illumination for 2-photon excitation was provided by a mode-locked Titanium:Sapphire laser (Spectra-physics, Santa Clara, CA), which generates trains of 100-fs pulses at a repetition rate of 80 MHz. Hearts were imaged through a 60X 1.2 NA water-immersion objective with a working distance of 280 μm. Illumination wavelength was 940 nm, close to the previously measured peak of the two-photon excitation cross section for EGFP (Herz et al., 2010). Emitted light was split by two dichroic mirrors (DM485 and DM570) in series, passed through narrow-band pass filters (420–460 nm [blue], 495–540 nm [green], and 575–630 nm [red]) and collected with three independent, external photomultiplers. Images were taken at a zoom of 2 or 4 in the x-y dimension (pixel size = 0.207 μm and 0.103 μm, respectively) and at 0.5-μm steps in the z-direction, with a 12-bit dynamic range. Because the axial resolution of the imaging system has been measured previously to be ≥ 1 μm under the imaging conditions employed (Rubart, 2004; Young et al., 2011), a z-step size of 0.5 μm was in accordance with the Nyquist criteria. A Kalman filter of 2 was used. Adjacent stacks had ~10% overlap and were obtained along a nominal vertical distance of 50 μm starting from the epicardial surface. Pixel dwell time was set to 8 μs, resulting in a ~7-min acquisition time per stack composed of 100 individual 512 × 512 pixel images. We obtained a total of 9 (3 × 3) adjacent z-stacks per heart.
2.6. 3-D tracking of EGFP-labeled axons in two-photon image stacks
To reconstruct the 3-D trajectory of the sympathetic arbor from the two-photon imaging data, we adapted a previously developed semi-automated tracing algorithm (Evers et al., 2005; Schmitt et al., 2004). Following the removal of pixels with autofluorescence (see Supplementary Data section 4.), images were thresholded assuming that pixels with intensities greater than the mean background +3 SD represented axonal segments. The reconstruction is based on a geometric model wherein neurite segments are approximated as discrete series of circular cylinders (termed ‘snaxels’) which are generated by moving a two-dimensional circular cross section along an axis in three-dimensional space. The direction of the axis can change with location along a neurite segment, and the cross section is always perpendicular to the tangent of the axis. To achieve maximal congruency of the reconstructed neurite segment with the two-photon image stacks, the snaxels’ axes and diameters are fitted into the image data using the active contour model by Kass et al. (1988). To outline the connections among reconstructed neurite segments, their respective centerlines are connected to each other at branching points and all information about axonal signal intensity was removed by replacing axonal segments with a thin line of uniform thickness, giving rise to the 3-D topological skeleton of the axonal arbor. The examination of axons involved multiple viewing of z-stacks at different brightness and contrast settings to verify their relationship with adjacent axon segments. The tracing algorithm was implemented in Amira version 4.1 and generously provided by Felix Evers, University of Cambridge, Cambridge, United Kingdom.
In order to verify reconstruction accuracy, two different persons independently traced the same portions of an image s tack. Supplementary Figure 9 shows the resulting two 3-D topological skeletons, revealing no large-scale discrepancies that would have lead to different interpretations of the connectedness of axonal branches.
2.7. Permanent coronary artery occlusion
Coronary artery occlusion was performed as described previously (Murry et al., 2004). Animals were intubated and ventilated (2% isoflurane with supplemental oxygen), and the left coronary artery was occluded at the inferior border of the left auricle with an 8-0 Prolene ligature. All procedures were in accordance with institutional guidelines. All animal procedures were performed in compliance with the institutional guidelines at the Indiana University School of Medicine and are in accordance with the recommendations of the Panel on Euthanasia of the American Veterinary Medical Association and the National Institutes of Health publication Guide for the Care and Use of Laboratory Animals.
2.8. Statistical Analyses
The t-test or Mann-Whitney test was used for comparison between two groups. The Kruskal–Wallis test (followed by Dunn’s post-hoc test) was used for comparison between multiple groups. Values are presented as mean ± SEM. SigmaPlot (Systat Software, Inc., San Jose. CA) was used for all statistical analyses.
3. Results
3.1. Generation and analysis of hDβ H-EGFP transgenic mice
A fluorescence-based reporter transgene was generated to enable sympathetic nerve tracking in intact, living heart. The reporter transgene used the human dopamine β-hydroxylase gene promoter to target expression of EGFP to sympathetic neurons (Figure 1A) (Mercer et al., 1991). A total of 6 independent mouse lines bearing the hDβH-EGFP transgene were generated. Dual immunofluorescence staining for EGFP and the sympathetic neuron-specific marker tryrosine hydroxylase (TH) in sections from superior cervical and stellate ganglia, which together provide the vast majority of postganglionic sympathetic neurons innervating the heart (Hassankhani et al., 1995; Nam et al., 2013), revealed a mosaic pattern of transgene expression in the founder lines, with anywhere from ~10% to ~80% of TH-positive neurons exhibiting anti-GFP immunoreactivity. After crossing the mosaic expressing mice which were generated in C3HeB/FeJ inbred mice, into a DBA/2J background, one out of the six lines (line 4) was found to exhibit nearly full transgene penetrance, with ≥ 95% of sympathetic ganglion neurons staining for EGFP (8 superior cervical and stellate ganglia distributed among 4 hDβH-EGFP mice between 12 and 38 weeks of age). Figures 1B and C show representative examples of double TH and EGFP immunofluorescence stainings in midsections of stellate ganglia dissected from a transgenic mouse (line 4) and a non-transgenic littermate, respectively. Transgenic sympathetic neurons expressed EGFP in both cyto- and nucleoplasm, whereas anti-EGFP immunoreactivity was not detectable in non-neuronal cells of transgenic ganglia, or in any cell of non-transgenic ganglia. Size and morphology of transgenic neurons appeared to be unaltered compared with their non-transgenic counterparts. To examine whether EGFP also labeled intracardiac axons of postganglionic sympathetic neurons, a histological section from an adult hDβH-EGFP transgenic heart (line 4) was double stained for TH and EGFP, and subjected to confocal microscopy. Volume rendering of the confocal z-stack (Figure 1D) revealed that voxels containing EGFP coincided with voxels containing TH, as indicated by the abundance of yellow voxels in the merged image (lower panels in Figure 1D), suggesting that EGFP molecules had diffused or had been transported along axons to stain postganglionic sympathetic neurons in their entirety.
Transgenic hearts appeared macroscopically and microscopically normal (Supplemental Figure 1A and B), and average heart weight / body weight ratio of mice was similar between genotypes (Supplemental Figure 1C). Mean densities of sympathetic neuron somae in peripheral ganglia were not significantly different between transgenic and non-transgenic mice (Supplemental Figure 2A), and transmembrane action potentials recorded from the somae of isolated wild-type and transgenic postganglionic sympathetic neurons were superimposable (Supplemental Figure 2B). Finally, sympathetic axon density in the myocardium was similar between genotypes (Supplemental Figure 3).
Overall, these results demonstrated that EGFP was selectively expressed in sympathetic ganglion neurons of adult hDβH-EGFP transgenic mice. EGFP labeled postganglionic sympathetic neurons in their entirety; including nuclei, cytoplasm and axons, and its prolonged expression had no discernible adverse effects on sympathetic nerve and cardiac structure, or on electrophysiological properties of sympathetic neurons. Only hDβH-EGFP transgenic mice from line 4 which had been backcrossed into the DBA/2J background for at least 5 generations were used in subsequent experiments.
3.2. Cardiac EGFP distribution tracks sympathetic nerves - quantitative co-localization analyses
Our initial observations (Figure 1D) suggested that intramyocardial EGFP distributes along sympathetic axons in adult hDβH-EGFP transgenic hearts. To quantitatively assess the extent of co-localization between the TH enzyme and the EGFP reporter in myocardial tissue, we used indirect immunofluorescence in combination with confocal microscopy and digital image analysis. Ten-micron-thick sections from adult hDβH-EGFP hearts were reacted with primary antibodies directed against EGFP and tyrosine hydroxylase, followed by co-incubation with Alexa546- and Alexa633-conjugated secondary antibodies. Preliminary experiments confirmed that the combinations and concentrations of primary antibodies used for co-localization analyses enabled specific antigen recognition in the tissue of interest (Supplemental Figure 4). Further, no signal bleed through between the two fluorophores was observed under the optical conditions employed (Supplemental Figure 5), supporting the utility of this combination for co-localization analyses.
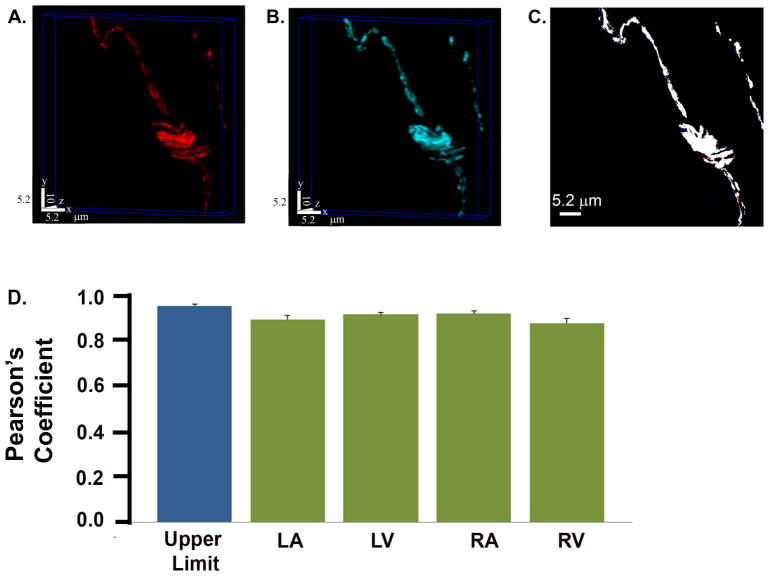
Figures 2A and B display a pair of volume-rendered confocal image stacks of a cardiac section labeled with antibodies specific for TH and EGFP. The abundance of white pixels in the maximum projection of the merged z-stack (Figure 2C) implies that most of the EGFP was co-incident with TH. Numerical analyses confirmed this visual impression: the mean of the Pearson’s correlation coefficient, averaged across a total of 15 pairs of identically processed image stacks, was 91 ± 1 % for the LV. This value was not significantly different from the empirical limit of co-localization (95 ± 1%; P > 0.05) which was measured in sections incubated with the TH antibody twice and reacted with Alexa546-conjugated secondary after the first and an Alexa633 secondary after the second incubation (Supplemental Figure 6). Average Pearson’s coefficients from the other anatomic regions for which quantitative co-localization analyses were performed, ranged between 87 ± 2 % in the right ventricle to 92 ± 1 % in the right atrium (Figure 2D). The magnitudes of region-specific coefficients were not significantly different from the empirical maximum of co-localization (P > 0.05) that we can record under our experimental conditions. We did not detect EGFP-expressing nerves that were tyrosine hydroxylase negative or vice versa, confirming specificity and full penetrance, respectively, of transgene expression at the level of individual intracardiac axons. Overall, these results supported the notion that the intracardiac EGFP distribution faithfully recapitulates the micro-anatomy of the sympathetic network and further suggested the suitability of the hDβH-EGFP transgenic mouse model to image sympathetic nerves in living heart.
Figure 2.
Intracardiac distribution of EGFP tracks sympathetic axons. A and B, Pair of volume-rendered confocal z-stacks obained from a cardiac section (52.7 × 52.7 × 10.0 μm3) co-stained with tyrosine hydroxylase (A; Alexa546 label) and EGFP (B; Alexa633 label) antibodies. C, Co-localization map of the maximum intensity xy projection of the merged stack. If the pixel is white, both proteins occupy that pixel. D, Bar graphs of mean ± SEM Pearson’s co-localization coefficients for the right ventricle (RV), right atrium (RA), left atrium (LA), and left ventricle (LV). Values are from 15 image stacks each per region equally distributed among 3 hearts. There was no significant difference between each region and the empirical limit of co-localization (P > 0.05 by ANOVA).
3.3. Live morphology of the local sympathetic network in left ventricular subepicardium as reconstructed from two-photon imaging data
We next sought to examine the utility of the hDβH-EGFP mouse model in conjunction with TPLSM imaging to three-dimensionally reconstruct local sympathetic arbors in living heart. We choose to study innervation in subepicardial layers of the left ventricular free wall for two reasons. First, this anatomical region is readily accessible to two-photon excitation fluorescence imaging in the living mouse heart (Rubart et al., 2003; Rubart, 2004). Second, previous studies by others have demonstrated marked structural remodeling of sympathetic innervation in the epicardial border zone of chronic myocardial infarctions (Clarke et al., 2010; Zhou et al., 2004). Thus, direct comparisons of 3-D reconstructions obtained from uninjured versus infarcted epicardial layers would provide insights into disease-related variations in local sympathetic wiring.
Pilot experiments in Langendorff-perfused non-transgenic hearts revealed the presence of two-photon excited tissue autofluorescence which spectrally overlapped with EGFP signals and thus could be mistaken as representing sympathetic axon segments in transgenic hearts (Supplemental Figure 7). Because the emission profile of tissue autofluorescence was found to be readily distinguishable from that of EGFP under our vital imaging conditions (Supplemental Figure 7), we were able to dissect the two signal components in TPLSM image stacks taken from Langendorff-perfused hDβH-EGFP hearts as demonstrated in Supplemental Figures 7 and 8.
A representative example of a three-dimensional reconstruction of EGFP-expressing axons from two-photon imaging data acquired in a living heart is shown in Figure 3A (see also Online Movie 1). All EGFP-labeled axons in a 230 μm by 280 μm by 50 μm volume within the outermost left ventricular epicardial layer in an adult hDβH-EGFP heart were imaged by TPLSM and processed images were reconstructed in 3-D. The data set consisted of 9 image stacks, each stack containing 100 images (12 bit, 512 × 512 pixels).
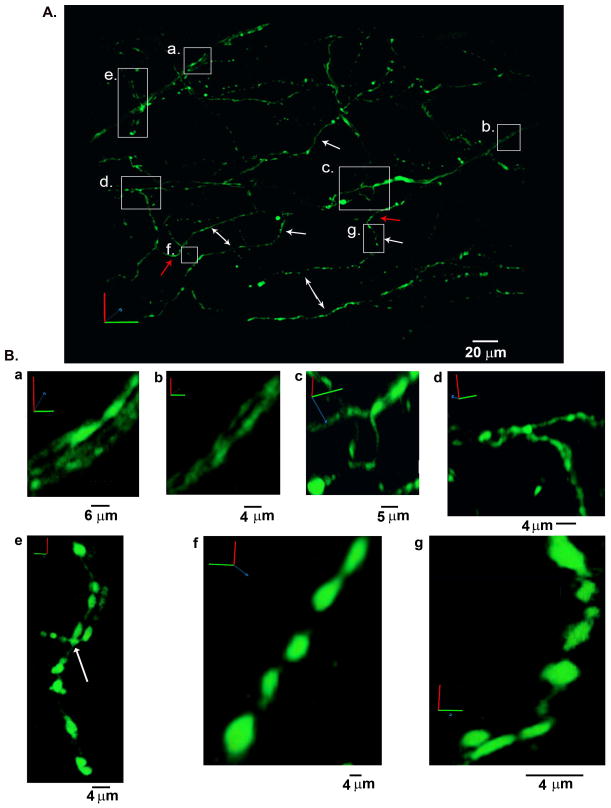
Figure 3.
Three-dimensional reconstruction of the sympathetic network within a portion of the left ventricular subepicardium in an adult, Langendorff-perfused hDβH-EGFP heart. A, 3-D rendering of the sympathetic arbor. All sympathetic axons (labeled with EGFP) in a 230 μm by 280 μm by 50 μm segment were imaged by TPLSM. A montage of 9 (3 × 3) overlapping image stacks provided the dataset for reconstruction. Images (512 × 512) were collected at a voxel size of 0.2 μm × 0.2 μm × 0.5 μm with 12-bit resolution. B, Magnified views of the boxed regions labeled a through g in A. Ba and Bb, three- (Ba) and two-axon bundle (Bb) on the epicardial surface; Bc, branching of an axon while its constituent two axons did not branch; Bd and Be, binary (Bd) and tertiary (Be; arrow) axonal branching; Bf and Bg, intramyocardial axonal segments with boutons. Small letters refer to the correspondingly labeled boxed regions in A.
Previous studies by others have demonstrated that postganglionic sympathetic nerves invade the base of the heart and then run over the epicardial surface predominately in multi-nerve bundles (Kanazawa et al., 2010). In agreement with this finding, the volume rendered two-photon image stack revealed the presence of two nerve bundles on the epicardial surface (boxes labeled a. and b. in Figure 3A; magnified views of portions of each epicardial bundle are shown in panels Ba and Bb of Figure 3). No axon branches off the epicardial nerve in the left upper corner. In contrast, the other nerve bundle executes a sharp, nearly orthogonal, inward turn before invading the myocardial wall and eventually splitting into its constituent two axons (Figure 3Bc).
Several intramural axons (three representative examples are denoted by single-headed white arrows in Figure 3A) were found to follow a zig-zag trajectory, wherein segments of the same axon running tangentially to, but at different depths from, the epicardial surface are connected by axonal portions that project orthogonally, i.e. parallel to the transmural axis. In addition, some of the tangentially projecting axon segments run parallel to each other over distances of several tens of microns with more or less regular spacing between them (marked by double-headed arrows in Figure 3A). Such ordered arrangement may reflect the alignment of axons in parallel to the long axes of their apposed cardiomyocytes, because myocytes in subepicardial layers have been shown previously to preferentially orient their longitudinal axes to the surface lining (Lunkenheimer et al., 2006).
Transmurally projecting portions of sympathetic axons (denoted by white single-headed arrows in Figure 3A) span distances ranging from 10 to 30 μm. These values correspond to once or twice the average depth of adult rodent ventricular cardiomyocytes (~13 μm; Satoh et al., 1996) suggesting that those axonal segments extend over one or two layers of myocytes. Other axons (denoted by red arrows in Figure 3A) made sharp turns without much change along the transmural axis. Thus, the staircase-like 3-D trajectories of axons enable a synchronized signal transfer to multiple cells lying at different and/or similar tissue depths within laterally and/or longitudinally adjoined muscle regions, indicating that axonal sub-arbors supply target territories of variable geometry.
Axonal branching in the target zone enables innervation of multiple cells by the same arbor, promoting a coordinated spread of neural signal transmission among target cells. In the two-photon imaging data shown in Figure 3A, we found multiple bifurcations (magnified views of bifurcation examples are displayed in panels Bc and Bd) and also one example of a trifurcation (Figure 3Be, arrow).
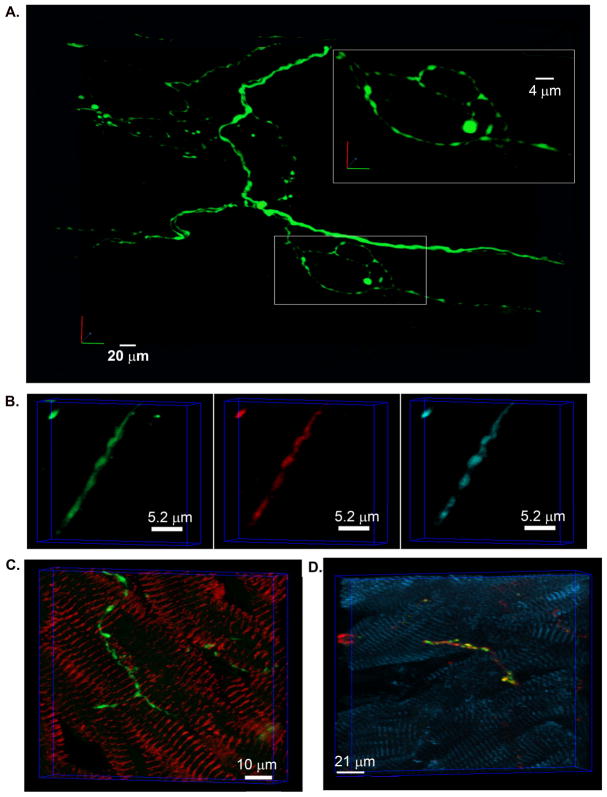
Most intramural neurites typically exhibited high constrast in EGFP fluorescence, giving rise to their bead-on-a-string morphology as shown in Figure 3Be–g. Intramural portions of sympathetic axons also formed multiple loops of varying length that were arranged both in series and in parallel with each other (Figures 3Be and 4A).
An identical pattern of periodic, intensely fluorescent axonal swellings (termed boutons or varicosities) that were connected by thin, weakly fluorescent segments, was also seen in confocal image stacks obtained from fixed cardiac sections triple immunostained for PGP9.5 (a panneuronal marker), tyrosine hydroxylase and EGFP (Figure 4B), or stained for tyrosine hydroxylase and sarcomeric α-actinin (Figure 4C). This result indicates that the punctuated fluorescence pattern of EGFP-labeled neurites mirrors the normal microanatomy of intramural sympathetic fibers. Additional immunofluorescence analyses of cardiac sections supported the notion that boutons constitute functional neurotransmitter release sites, i.e., neuroeffector junctions, as indicated by their co-localization with anti-synapsin I immunoreactivity, a synaptic protein necessary for neurotransmitter exocytosis (Figure 4D) (Shcherbakova et al., 2007). Such en passant alignment of synaptic boutons along much of the length of axon-cell contacts enables axially propagating action potentials to successively trigger transmitter release from all sites along that axon, causing rapid dispersion of the signal among the target cell population.
Figure 4.
Similar morphology of intramural sympathetic axons in living and fixed cardiac tissue. A, 3-D reconstruction of a branching sympathetic axon in the left ventricular subepicardium of a Langendorff-perfused hDβH-EGFP heart. All EGFP-expressing axons in a 130 μm by 165 μm by 40 μm segment were imaged by TPLSM. The montage of 7 (512 × 512) overlapping image stacks provided the data set for reconstruction. B, Volume rendering of a confocal image stack that was acquired from a 10-μm thick section of an hDβH-EGFP heart triple immunostained for PGP9.5 (green), EGFP (red), and tyrosine hydroxylase (cyan). C, Volume rendering of a confocal image stack obtained from a 10-μm-thick cardiac section dually stained for tyrosine hydroxylase and sarcomeric α-actinin (left panel). D, Volume rendering of a confocal image stack obtained from 10-μm-thick cardiac sections triple immunostained for tyrosine hydroxylase (red), synapsin I (green), and sarcomeric α-actinin (cyan). Yellow fluorescent regions, indicating co-localization of tyrosine hydroxylase and synapsin I, superimpose on axonal boutons, suggesting they constitute functional neurotransmitter release sites.
Overall, these findings demonstrate that levels of transgenic reporter protein expression were adequate to three-dimensionally resolve the sympathetic arbor within a finite volume of an intact, living mouse heart by two-photon molecular excitation imaging. Moreover, the 3-D reconstruction of two-photon image sets revealed several structural characteristics of the sympathetic arbor that synergize to effect coordinated signal transmission within input territories. These include varicosity-laden axons, axonal branching and looping, and omni-directional projections of axon branches.
3.4. 3-D rendering of the centerline skeleton
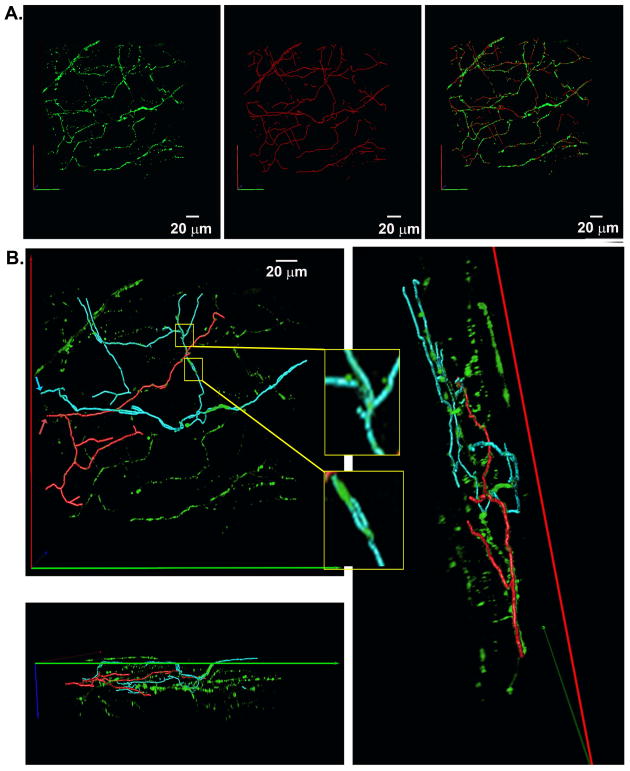
We adapted a previously developed computer-assisted tracing algorithm to reconstruct the 3-D trajectory of the sympathetic arbor from the two-photon imaging data and to subsequently derive the axonal branching topology from it (see Supplemental Figure 9) (Evers et al., 2005; Schmitt et al., 2004). The complete topological skeleton superimposed onto the 3-D two-photon image data set from Figure 3A is displayed in Figure 5A (see also Online Movie 2). Visual inspection of the 3-D rendered skeleton suggested that the local network is partitioned into multipe subarbors. Accordingly, to further examine the spatial extension, orientation, and branching topology of individual sub-arbors within the confines of the imaged volume, we mapped 3-D trajectories of two sub-arbors by selectively highlighting all axonal segments that were physically linked to subarbor-specific reference points. The two reference points were chosen to be the entry points of the sub-arbors’ intial segment into the volume (arrows in Figure 5B). Overlays of the 3-D subtree skeletons and the 3-D imaging data are displayed in Figure 5B (see also Online Movie 3), revealing repetitive bifurcations in either subtree. In addition, the cyan trajectory contained a tertiary branching node and multiple small-diameter loops (zoom-in views). Proximal segments of child axons were oriented at sharp angles with each other and/or with the distal segment of their parent branch at their bi- or trifurcation points, yet child axons were non-recurrent, i.e., they did not turn back in the opposite direction of the parent branch. Branches were of variable lengths and changed, often abruptly, their spatial orientation, giving rise to meandering trajectories extending along all three anatomical axes. Child branches within their respective subarbor did not overlap with one another. Cumulative path lengths of the red and cyan arbor in Figure 5 were 431 μm and 510 μm, respectively. Together, these findings suggest that differences in geometrical properties (i.e., length and angle of branches) between subtrees combine with those in branching and looping topology to give rise to non-stereotypical geometries of the innervation domains of an axon arbor on a microscopic scale.
Figure 5.
3-D topological skeleton of the sympathetic arbor within a finite volume of the left ventricular subepicardium. A, A complete 3-D skeleton of all green fluorescent axons was generated using the semi-automated tracing algorithm and rendered in 3-D. The left, middle and right panel show the 3-D original imaging data, the 3-D skeleton, and the overlay of the 3-D original data and the 3-D skeleton, respectively. B, Two examples of sympathetic subtree trajectories. The subtrees’ trajectories (cyan and red traces) were obtained by highlighting all axonal segments that were physically linked to their respective upstream reference points. The two reference points were chosen to be the entry points of the subtrees’ initial segments into the imaged volume (red and cyan arrows). Panels show the overlay of the two colored subtree skeletons and the 3-D EGFP data set at three different view angles. Inserts display small-diameter loops denoted by the yellow boxes.
Because the locations of end-branches of the traced subtrees typically coincided with the image borders, they most likely exceeded the boundaries of the imaged volume, and it thus seems improbable that they represent axon terminals. Also, because the size of the imaged volume was too small to backtrack each subtree’s proximal trajectory, we were unable to determine whether they originated from the same or different parent axons, or whether they directly connected with axons from other neurons outside the imaged volume.
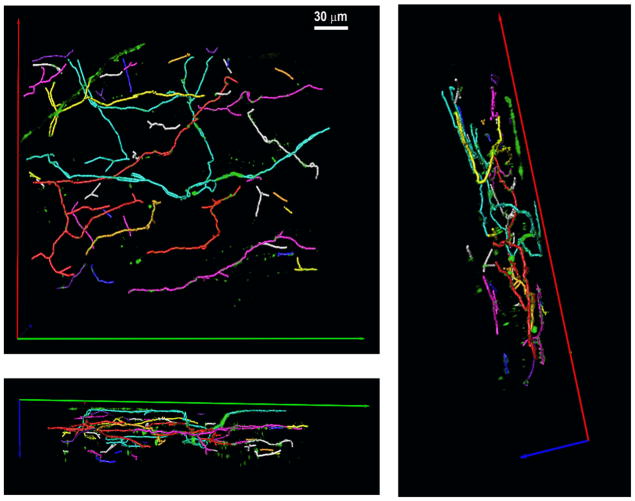
The analyses above define the arrangement of branches within individual subarbors. To further delineate the relative spacing of neighboring subarbors with respect to one another, we obtained their 3-D trajectories and assigned them distinct colors. Figure 6 shows the resulting 3-D color map of the trajectory ensemble (see also Online Movie 4) revealing no, or only minimal, overlap of individually traced subarbors. Such tiled arrangement ensures that the arbors achieve comprehensive coverage of input territories while minimizing the redundancy with which each territory is sympathetically innervated. The total axon length, defined as the sum of lengths of all branches, was found to be 4271 μm, which when normalized to the total volume imaged (3.22 nL), yielded an axon density of 0.0013 μm/μm3. Taking previsouly published average values for the dimensions of an adult rodent cardiomyocyte (Satoh et al., 1996), the image volume would contain about 50 cardiomyocytes, giving rise to an axon length of ~85 μm per individual cardiomyocyte.
Figure 6.
Tiled arrangement of sympathetic subarbors in the left ventricular subepicardium. Volume-rendered trajectories were obtained for each invidually identified subarbor using the 3-D tracing algorithm and assigned contrasting colors. The 3-D color map is shown from three different view angels.
Overall, the three-dimensionally reconstructed wiring map of the local sympathetic arbor unveils organizational principles that synergistically combine with the morphological characteristics described above to achieve coordinated dispersion of sympathetic signals to the subepicardium. From the availabe data set it cannot be determined whether axons in the scanned volume emanate from the same and/or different postganglionic sympathetic neurons.
3.5. Live morphology of remodeled sympathetic innervation in the peri-infarct border zone
Previous studies employing immunohistochemical assays have demonstrated pronounced structural remodeling of the cardiac sympathetic nervous system in chronically infarcted hearts of large and small mammals. The remodeling process entails both loss and gain of sympathetic axons in the peri-infarct zone (Clarke et al., 2010; Li et al., 2004; Zhou et al., 2004). Here, we sought to probe the utility of our reporter mouse model for assessment of changes in sympathetic innervation patterning occurring in the infarct border zone of living hearts. Accordingly, adult hDβH-EGFP mice underwent permanent occlusion of the distal left anterior descending coronary artery. Two weeks later, the hearts were excised and processed for immunohistology or subjected to TPLSM imaging. Coronary artery ligation caused left ventricular infarction with regional wall thinning and interstitial fibrosis (Supplemental Figure 10). Co-staining of 10-μm sections from the infarcted heart for TH and sarcomeric α-actinin to label sympathetic nerves and cardiomyocytes, respectively, revealed a 50% increase in axon density in the peri-infacrt zone (0.18%) compared with the non-infarcted basal portion of the left ventricular free wall (0.12%; Supplemental Figure 10D). Quantitative co-localization analyses demonstrated that the EGFP distribution reproduces the anatomy of sympathetic axons in the border zone of chronic myocardial infarctions, confirming the suitability of our reporter model to track remodeling of the cardiac sympathetic network in previously infarcted, living heart (Supplemental Figure 11).
We next obtained a three-dimensional reconstruction of EGFP-expressing axons from two-photon imaging data acquired in a living, previously infarcted heart. All EGFP-labeled axons in a 230 μm by 250 μm by 50 μm volume within the epicardial border zone in an adult hDβH-EGFP heart at 2 weeks after ligation of the distal left anterior descending coronary artery were imaged by TPLSM. Volume renderings of the two-photon image data are displayed in Figure 7 (see also Online Movie 5).
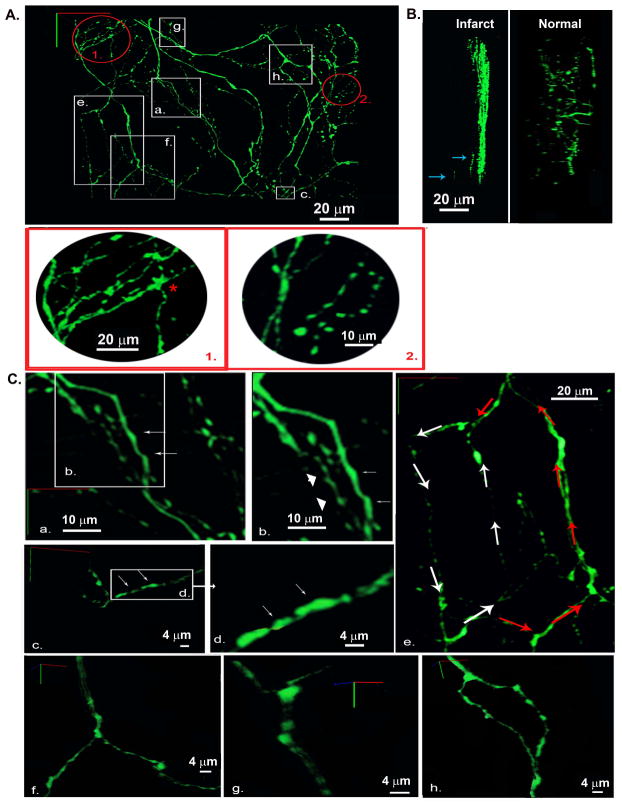
Figure 7.
Three-dimensional reconstruction of the local sympathetic neurite network within the epicardial border zone of a 2-week-old myocardial infarction in a living, adult hDβH-EGFP heart. A, 3-D rendering of the sympathetic arbor. All sympathetic axons (labeled with EGFP) in a 230 μm by 250 μm by 50 μm segment were imaged by TPLSM. A montage of 9 (3 × 3) overlapping image stacks provided the dataset for reconstruction. Images (512 × 512) were collected at a voxel size of 0.2 μm × 0.2 μm × 0.5 μm with 12-bit resolution. Red-boxed regions 1 and 2 are magnified views of the circled regions 1 and 2 in A demonstrating locally increased axon density. B, 3-D renderings at a 90°-rotation about the y axis of 3-D data sets obtained from an infarcted and a non-infarcted hDβH-EGFP heart. Arrows denote axonal fragments at depth. C, Magnified views of the boxed regions labeled a through h in A. a and b, axons with smooth contours (arrows) and with periodic swellings (arrowheads); c and d, eccentric boutons (arrows); e, two axonal loops with different circumferential length; f and g, branching of dual-axon bundles while their constituent axons do not branch; h, bifurcation where the parent axon branches into childs.
A +90°-rotation of the 3-D rendered image data about the y axis (Figure 7B, left panel) reveals nearly complete absence of EGFP-expressing axons at tissue depths between 10 and 50 μm below the epicardial surface. Only a few axon fragments were detected at depths exceeding 10 μm (denoted by arrows in Figure 7B; see also Online Movie 5), giving rise to a steep, epi- to endocardially oriented innervation gradient within the image volume. This pattern sharply contrasts with the spread of EGFP-expressing axons across the entire epicardial-endocardial axis in 3-D image data sets obtained from the left ventricular subepicardium of uninjured hDβH-EGFP hearts (right panel of Figure 7B). Also, sympathetic nerve density appeared to largely vary within the anterior portion of the imaged volume. Close inspection of the three-dimensionally reconstructed image data revealed that axons in small subvolumes (denoted by the circles labeled 1 and 2 in Figure 7A; see also zoom-in views of both regions) were observed to form multiple loops. Similarly sized axon loops were also observed in normal cardiac tissue (see Figure 4A) and most likley represent sympathetic fibers on vascular processes (Long et al., 2009). Overall, these findings suggest that both loss and gain of axons occur simultaneously within the epicardial peri-infarct border zone, profoundly altering sympathetic innervation micro-patterning. The non-uniform changes in sympathetic nerve density assessed by 3-D rendering of tissue EGFP fluorescence are in agreement with those seen in the accompanying immunohistochemical analyses (Supplemental Figure 10).
Inspection of the 3-D image data unveiled largely variable EGFP fluroescence patterns both within and among individual axons. Some axonal segments exhibited relatively smooth contours with little variation in diameter (arrows in Figure 7Ba and Bb), whereas others displayed periodic swellings alternating with thin interconnecting segments. Occasionally, these axonal swellings were not symmetrical to the shafts’ axes (Figure 7Bc and Bd). Further, we detected the presence of larger-diameter axon loops (Figure 7Be), suggesting that multiple axon loops with largely variable circumference and geometry co-exist within the anterior compartment of the imaged volume.
Sympathetic nerves in the imaged cuboid followed a predominantly tangential trajectory and were observed to run in bundles or as solitary nerve fibers. Representative examples of nerve bundles splitting into their respective two constituent axons are shown in Figure 7Bf and Bg. Binary or tertiary branching of solitary fibers was frequently observed throughout the imaged volume. Representative examples of a bi- and trifurcation are displayed in Figures 7Bh and 7 1. (asterisk), respectively.
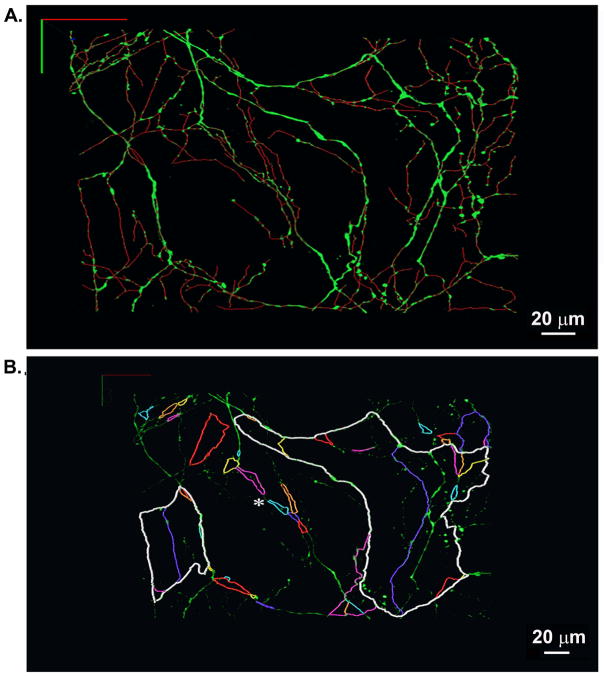
We subsequently used the semi-automatic tracing algorithm to generate the 3-D wiring diagram of the imaged network. Overlays of the 3-D topological skeleton and the 3-D image data are presented in Figure 8A (see also animated Online Movie 6). Visual inspection of the 3-D skeleton suggested that multiple axonal loops with variable diameter and geometry interconnect to form plexuses. To further explore plexus topology, we sequentially traced the 3-D trajectories of all identified loops that were physically linked with each other within a selected subvolume. Trajectories of individually identified loops were assigned contrasting colors and superimposed onto the 3-D display of the two-photon image data set to generate the loop map shown in Figure 8B. The map unveils several distinct structural properties of the axonal plexuses. First, trajectories of juxtaposed loops were oberved to partially overlap, giving rise to a nested-loop configuration of the local network. Second, portions of individual loops could be heterogeneous with regard to fiber morphology and axonal branching topology. Third, single loops followed meandering trajectories of largely varying lengths. Loop length ranged from 23 μm to 872 μm. Fourth, a subset of juxtaposed loops were coupled in series via short axonal segments (see asterisk in Figure 8B).
Figure 8.
3-D rendering of the skeletonized sympathetic arbor within a finite volume of the epicardial border zone of a 2-week-old myocardial infarction in a living hDβH-EGFP heart. A, A complete 3-D skeleton of all green fluorescent neurites was generated using the semi-automated tracing algorithm (upper panel) and superimposed onto the 3-D rendering of the two-photon image data. B, Multiple-loop architecture of the sympathetic circuitry in the epicardial infarct border zone. Individual loops that were found to be physically linked with each other were identified and tracked sequentially in 3-D. Their colored trajectories were superimposed onto the volume-rendered two-photon image data to generate a loop map. Asterisk denotes juxtaposed loops linked in series via short axonal segments.
Overall, these results demonstrate that the hDbH-EGFP reporter mouse, combined with two-photon fluorescence microscopy and quantitative image processing, enables high-fidelity 3-D reconstructions of myocardial infarction - induced sympathetic remodeling on a microscopic scale in the living heart.
4. Discussion
Here, we demonstrate the utility of the hDβH-EGFP reporter mouse model, combined with two-photon excitation microscopy and computer-assisted image analysis, to quantitate sympathetic axon density and to three-dimensionally resolve the sympathetic arbor in a finite volume of circuitry within the Langendorff-perfused heart. Levels of reporter protein expression in intramyocardial segments of sympathetic axons in our study were adequate for in situ imaging by two-photon molecular excitation, without evoking discernible toxicity. Further, generation of the 3-D topological skeleton from the two-photon data set provides information on short-range connectivity among sympathetic axons within the left ventricular epcardium of a normal and injured heart.
Cardiac sympathetic nerve density is an important determinant of the functional response of the myocardium to sympathetic nerve activity. Conventional methods to quantitate sympathetic innervation in heart muscle often involve embedding and mechanical sectioning (Ieda et al., 2007; Li et al., 2004) or dehydration and heating (Cao et al., 2000; Mühlfeld et al., 2010). Such techniques can markedly perturb tissue micro-morphology compared to what is manifested in the intact system. The method we present here allows quantitation of cumulative axon length per unit volume of tissue within intact, living heart, i.e., under conditions of preserved three-dimensional structure. Density or total length of sympathetic axons are widely used surrogates for the degree of sympathetic activation of target tissue (Cao et al., 2000; Ieda et al., 2007; Li et al., 2004; Mühlfeld et al., 2010), although the correlation between anatomical innervation density and neuronal function was not routinely established in these studies. Accordingly, the extent to which total axon length correlates with amount of sympathetic neurotramsitter release in the taregt tissue needs to be determined for the experimental condition being investigated, if it is to be used a a read-out of sympathetic activity.
Using three-dimensional reconstructions of the network’s topology, our method enables identification of organizational features that synergisticaly contribute to a coordinated and rapid dispersion of sympathetic signals within the target tissue. These include the formation of en passant neuroeffector junctions along much of the axon-cell contacts, as well as branching and looping of axons while they meander, both radially and tangentially, over distances of several hundreds of microns through small tissue volumes, giving rise to multiple annexed innervation domains of variable geometry. Compartmentalization of the target tissue into multiple, mutually exclusive input territories (tiling) optimizes axonal spacing, providing a structural foundation for minimizing transmitter diffusion distances between the synapses and postsynaptic receptor sites while balancing neurotransmitter overflow to the target tissue.
Volume-renderings of two-photon image stacks further revealed that portions of intramural axons projecting tangentially to the epicardial surface run parallel to each other with more or less regular spacing between them (see Figure 3A). Intriguingly, previous studies by others have demonstrated that the majority of left ventricular epicardial cardiomyocytes are oriented with their longitudinal axes approximately tangential to the epicardial lining Lunkenheimer et al., 2006). Further, myocardial capillaries have been shown to preferentially run parallel to the long axes of their juxtaposed cardiomycoytes (Bruce et al., 2008). It is therefore likely that sympathetic fibers extend predominately longside their apposed cardiomyocytes and capillaries. Such triple co-linearity may serve to optimize the frequency of sympathetic synapses per axon / myocyte, while reducing effective neurotransmitter diffusion distances to nearby capillary endothelial cells.
Our finding of simultaneous loss and gain of sympathetic axons in the border zone of a chronic myocardial infarction is in agreement with previous studies by others employing mechanical sectioning methods combined with immunohistology for monitoring sympathetic axon density in infarcted heart tissue (Clarke et al., 2010; Zhou et al., 2004; Li et al., 2004; Cao et al., 2000). In contrast to these existing techniques, our approach preserves continuity of structure, which allows tracing of neurites over distances, and thus enables not only quantitation of sympathetic nerve density but also derivation of changes in the three-dimensional and topological morphology of cardiac sympathetic nerves, i. e., the sympathetic circuit wiring, in the injured heart muscle. Indeed, our results reveal pronounced re-organization of sympathetic arborization in the epicardial border zone, manifested in formation of multiple nested and/or serial closed loops of widely variable length and geometry. Sympathetic axons in the uninjured left ventricular epicardium were also found to loop, suggesting axonal looping occurs normally. A previous immunohistological study by the Segal laboratory revealed a mesh-like arrangement of sympathetic nerves in the walls of excised, intact arteries (Long et al., 2009). Thus, the location of the small-diameter loops shown in Figure 4A may coincide with a subepicardial vessel. By comparison, the looping architecture in the epicardial infarct border zone was markedly different from that in the normal heart with regard to both frequency and circumferential length of individual loops.
Previous studies, both experimental and computational, have provided evidence for the role of geometrical constraints in defining impulse conduction through a region of an axon (Swadlow et al., 1980; Ferrante et al., 2013). For example, morphological heterogeneities such as branch points and dilations may result in regions of low safety factor, i.e., regions where impulse propagation has a high likelihood of failing. Thus, the presence of multiple varicosities that are distributed in an ‘en passant’, bead-on-a-string manner along thin axonal branches as well as axonal branching are expected to significantly impact on axial conduction and, thus, to modify signal propagation within the network. Also, the formation of nested-loop axonal trajectories in the infarct border zone may promote high-frequency electrical reentry within the local network, giving rise to abnormal sympathetic signaling (Vladimirov et al., 2012).
The method presented here raises the intriguing possibility to directly interrogate the impact of sympathetic remodeling on functional aspects of underlying myocardium. For example, its combination with optical voltage- or calcium-imaging techniques (Bu et al., 2009; Rubart et al., 2003) and sympathetic activation in isolated perfused heart preparations with preserved postganglionic sympathetic innervation (Du et al., 1996) would enable direct assessment of the effects of altered axon density and/or connectivity on electrical or calcium-handling properties of the target tissue.
Overall, a 3-D reconstruction of the local sympathetic architecture using the approach presented here should facilitate theoretical analyses of the network’s electrical behavior as well as experimental assessment of the impact of network structure on target tissue function. Together, such analyses are likely to improve our understanding of neurocardiac coupling in normal and diseased states.
The depth to which useful images can be collected using two-photon fluorescence microscopy is limited due to light scattering, absorption and spherical aberration (Young et al., 2011), restricting image acquisition to superfical tissue layers. Future technical innovations (e.g., adaptive optics (Rueckel et al., 2006)) and tissue clearing (Shimozawa et al., 2013; Chung et al., 2013) may overcome some of these limitations.
The punctuated appearance of EGFP-labeled intracardiac axons, owing to intensely fluorescent varicosities alternating with thin, weakly fluorescent intervaricosal segments, poses limitations on the ability to image details of axonal connectivity in intact tissue. The resolution of diffraction-limited light microscopy may be insufficient even when used in conjunction with two-photon excitation to eliminate signals from out-of-focus structures. The advent of super-resolution techniques may provide a new avenue for imaging microstructures that have not been accessible with conventional light microscopy. Also, the overall fluorescence signal from and/or the EGFP labeling density in thin intervaricosal segments may have been insufficient to provide low noise images.
Imaging and analysis speeds limit the tissue volume for which a full circuit reconstruction can be obtained. Three nanoliters of heart volume resolved to the level of identifying every axon and bouton requires 3 hours to image and at least the same amount of time to analyze. Because the image volume contains axonal processes entering and leaving the volume, the local sympathetic circuit cannot be reconstructed completely. As a consequence, we were unable to determine the extent of myocardial territory innervated by indiviudal postganglionic neurons nor were we able to assess their spatial relationships to each other. Complete reconstruction of the cardiac sympathetic connectome will therefore require acquisition of larger image volumes as well as the abilty to identify the postganglionic sympathetic neurons from which individual intracardiac axons emanate. Use of a transgenic multicolor labeling strategy called Brainbow, which has been shown to result in marking of nearby neurons / axons with multiple distinct colors, may help characterize the spatial relationship between iso-neuronal and hetero-neuronal axons (Cai et al., 2013).
5. Conclusions
Overall, the present study demonstrates that the topological reconstruction of individual subarbors within finite volumes of the left ventricular epicardium enables derivation of sympathetic innervation micro-patterning in normal and diseased heart. Thus, the approach should be of general utility to unravel the mechanisms governing sympathetic remodeling during development and disease, as well as to gauge the impact of genetic factors on sympathetic axon spacing. In turn, unraveling the sympathetic mircoarchitetcture will aid in understanding network function. Finally, when combined with other cell lineage-restricted reporter systems, it will help delineate the topology between sympathetic nerves and different types of target cells in the intact heart.
Supplementary Material
Highlights.
Transgenic mice were generated to express EGFP under dopamine β-hydroxylase promoter
Intramyocardial EGFP distribution recapitulated the sympathetic microanatomy
In situ two-photon microscopy enabled 3-D imaging of local sympathetic networks
3-D tracking was used to generate the wiring topology of local sympathetic circuits
Useful approach to study the structural underpinnings of neurocardiac coupling
Acknowledgments
The study was supported by grants from NHLBI and Indiana CTSI to M. R. and a grant from Indiana CTSI to K. F. The hDβH promoter was kindly provided by Richard D. Palmiter, MD, University of Washington, Seattle, WA. This project was supported by a Project Development Team within the ICTSI NIH/NCRR Grant Number TR000006.
Non-standard Abbreviations
- EGFP
Enhanced Green Fluorescent Protein
- DβH
dopamine beta-hydroxylase
- TPLSM
two-photon laser scanning microscopy
- TH
tyrosine hydroxylase
Footnotes
Conflict of interest
The authors state that they have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bruce AF, Rothery S, Dupont E, Severs NJ. Gap junction remodelling in human heart failure is associated with increased interaction of connexin43 with ZO-1. Cardiovasc Res. 2008;77:757–65. doi: 10.1093/cvr/cvm083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu G, Adams H, Berbari EJ, Rubart M. Uniform action potential repolarization within the sarcolemma of in situ ventricular cardiomyocytes. Biophys J. 2009;96:2532–46. doi: 10.1016/j.bpj.2008.12.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Cohen KB, Luo T, Lichtman JW, Sanes JR. Improved tools for the Brainbow toolbox. Nature Methods. 2013;10:540–7. [PubMed] [Google Scholar]
- Cao JM, Fishbein MC, Han JB, Lai WW, Lai AC, Wu TJ, et al. Relationship between regional cardiac hyperinnervation and ventricular arrhythmia. Circulation. 2000;101:1960–9. doi: 10.1161/01.cir.101.16.1960. [DOI] [PubMed] [Google Scholar]
- Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–7. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke GL, Bhattacherjee A, Tague SE, Hasan W, Smith PG. β-adrenoceptor blockers increase cardiac sympathetic innervation by inhibiting autoreceptor suppression of axon growth. J Neurosci. 2010;30:12446–54. doi: 10.1523/JNEUROSCI.1667-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J. 2004;86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debanne D, Campanac E, Bialowas A, Carlier E, Alcaraz G. Axon physiology. Physiol Rev. 2011;9:555–602. doi: 10.1152/physrev.00048.2009. [DOI] [PubMed] [Google Scholar]
- Du XJ, Vincan E, Woodcock DM, Milano CA, Dart AM, Woodcock EA. Response to cardiac sympathetic activation in transgenic mice overexpressing beta 2-adrenergic receptor. American Journal of Physiology - Heart and Circulatory Physiology. 1996;271:H630–6. doi: 10.1152/ajpheart.1996.271.2.H630. [DOI] [PubMed] [Google Scholar]
- Evers JF, Schmitt S, Sibila M, Duch C. Progress in functional neuroanatomy: precise automatic geometric reconstruction of neuronal morphology from confocal image stacks. J Neurophysiol. 2005;93:2331–42. doi: 10.1152/jn.00761.2004. [DOI] [PubMed] [Google Scholar]
- Ferrante M, Migliore M, Ascoli GA. Functional impact of dendritic branch-point morphology. J Neurosci. 2013;33:2156–65. doi: 10.1523/JNEUROSCI.3495-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassankhani A, Steinhelper ME, Soonpaa MH, Katz EB, Taylor DA, Andrade-Rozental A, Factor SM, Steinberg JJ, Field LJ, Federoff HJ. Overexpression of NGF within the heart of transgenic mice causes hyperinnervation, cardiac enlargement, and hyperplasia of ectopic cells. Dev Biol. 1995;169:309–21. doi: 10.1006/dbio.1995.1146. [DOI] [PubMed] [Google Scholar]
- Herz J, Siffrin V, Hauser AE, Brandt AU, Leuenberger T, Radbruch H, et al. Expanding two-photon intravital microscopy to the infrared by means of optical parametric oscillator. Biophys J. 2010;98:715–23. doi: 10.1016/j.bpj.2009.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B, Costantini F, lacy L. Manipulating the Mouse Embryo - A Laboratory Manual. New York: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- Ieda M, Kanazawa H, Kimura K, Hattori F, Ieda Y, Taniguchi M, Lee JK, Matsumura K, Tomita Y, Miyoshi S, Shimoda K, Makino S, Sano M, Kodama I, Ogawa S, Fukuda K. Sema3a maintains normal heart rhythm through sympathetic innervation patterning. Nat Med. 2007;13:604–12. doi: 10.1038/nm1570. [DOI] [PubMed] [Google Scholar]
- Kanazawa H, Ieda M, Kimura K, Arai T, Kawaguchi-Manabe H, Matsuhashi T, et al. Heart failure causes cholinergic transdifferentiation of cardiac sympathetic nerves via gp130-signaling cytokines in rodents. J Clin Invest. 2010;120:408–21. doi: 10.1172/JCI39778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass M, Wittkin A, Terzopoulos D. Snakes: active contour models. Int J Comp Vis. 1988;1:321–31. [Google Scholar]
- Kimura K, Ieda M, Fukuda K. Development, Maturation, and Transdifferentiation of Cardiac Sympathetic Nerves. Circ Res. 2012;110:325–36. doi: 10.1161/CIRCRESAHA.111.257253. [DOI] [PubMed] [Google Scholar]
- Li W, Knowlton D, Van Winkle DM, Habecker BA, Li W, Knowlton D, et al. Am J Physiol Heart Circ Physiol. 2004;286:H2229–36. doi: 10.1152/ajpheart.00768.2003. [DOI] [PubMed] [Google Scholar]
- Long JB, Segal SS. Quantifying perivascular sympathetic innervation: regional differences in male C57BL/6 mice at 3 and 20 months. J Neurosci Methods. 2009;184:124–8. doi: 10.1016/j.jneumeth.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunkenheimer PP, Redmann K, Kling N, Jiang X, Rothaus K, Cryer CW, et al. Three-dimensional architecture of the left ventricular myocardium. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:565–78. doi: 10.1002/ar.a.20326. [DOI] [PubMed] [Google Scholar]
- Mercer EH, Hoyle GW, Kapur RP, Brinster RL, Palmiter RD. The dopamine [beta]-hydroxylase gene promoter directs expression of E.coli lacZ to sympathetic and other neurons in adult transgenic mice. Neuron. 1991;7:703–16. doi: 10.1016/0896-6273(91)90274-4. [DOI] [PubMed] [Google Scholar]
- Murry CE, Soonpaa MH, Reinecke H, Nakijima H, Nakijima HO, Rubart M, et al. Absence of cardiac transdifferentiation after direct injection of adult hematopoietic stem cells into myocardial infarcts. Nature. 2004;428:664–8. doi: 10.1038/nature02446. [DOI] [PubMed] [Google Scholar]
- Mühlfeld C, Papadakis T, Krasteva G, Nyengaard JR, Hahn U, Kummer W. An unbiased stereological method for efficiently quantifying the innervation of the heart and other organs based on total length estimations. J Appl Physiol. 2010;108:1402–09. doi: 10.1152/japplphysiol.01013.2009. [DOI] [PubMed] [Google Scholar]
- Nam J, Onitsuka I, Hatch J, Uchida Y, Ray S, Huang S, et al. Coronary veins determine the pattern of sympathetic innervation in the developing heart. Development. 2013;140:1475–85. doi: 10.1242/dev.087601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon y Cajal S. In: Histology of the nervous system of man and vertebrates. Swanson N, Swanson LW, translators. New York: Oxford University Press; 1995. [Google Scholar]
- Rubart M, Wang E, Dunn KW, Field LJ. Two-photon molecular excitation imaging of Ca2+ transients in Langendorff-perfused mouse hearts. Am J Physiol Cell Physiol. 2003;284:C1654–68. doi: 10.1152/ajpcell.00469.2002. [DOI] [PubMed] [Google Scholar]
- Rubart M, Pasumarthi KB, Nakajima H, Soonpaa MH, Nakajima HO, Field LJ. Physiological coupling of donor and host cardiomyocytes after cellular transplantation. Circ Res. 2003;92:1217–24. doi: 10.1161/01.RES.0000075089.39335.8C. [DOI] [PubMed] [Google Scholar]
- Rubart M. Two-photon microscopy of cells and tissue. Circ Res. 2004;95:1154–66. doi: 10.1161/01.RES.0000150593.30324.42. [DOI] [PubMed] [Google Scholar]
- Rueckel M, Mack-Bucher JA, Denk W. Adaptive wavefront correction in two-photon microscopy using coherence-gated wavefront sensing. Proc Natl Acad Sci U S A. 2006;103:17137–42. doi: 10.1073/pnas.0604791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh H, Delbridge LM, Blatter LA, Bers DM. Surface:volume relationship in cardiac myocytes studied with confocal microscopy and membrane capacitance measurements: species-dependence and developmental effects. Biophys J. 1996;70:1494–504. doi: 10.1016/S0006-3495(96)79711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S, Evers JF, Duch C, Scholz M, Obermayer K. New methods for the computer-assisted 3-D reconstruction of neurons from confocal image stacks. Neuroimage. 2004;23:1283–98. doi: 10.1016/j.neuroimage.2004.06.047. [DOI] [PubMed] [Google Scholar]
- Scriven DR, Lynch RM, Moore ED. Image acquisition for colocalization using optical microscopy. Am J Physiol Cell Physiol. 2008;294:C1119–22. doi: 10.1152/ajpcell.00133.2008. [DOI] [PubMed] [Google Scholar]
- Shcherbakova OG, Hurt CM, Xiang Y, Dell’Acqua ML, Zhang Q, Tsien RW, et al. Organization of beta-adrenoceptor signaling compartments by sympathetic innervation of cardiac myocytes. J Cell Biol. 2007;176:521–33. doi: 10.1083/jcb.200604167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimozawa T, Yamagata K, Kondo T, Hayashi S, Shitamukai A, Konno D, et al. Improving spinning disk confocal microscopy by preventing pinhole cross-talk for intravital imaging. Proc Natl Acad Sci U S A. 2013;110:3399–404. doi: 10.1073/pnas.1216696110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhelper ME, Cochrane KL, Field LJ. Hypotension in transgenic mice expressing atrial natriuretic factor fusion genes. Hypertension. 1990;16:301–7. doi: 10.1161/01.hyp.16.3.301. [DOI] [PubMed] [Google Scholar]
- Swadlow HA, Kocsis JD, Waxman SG. Modulation of impulse conduction along the axonal tree. Annu Rev Biophys Bioeng. 1980;9:143–79. doi: 10.1146/annurev.bb.09.060180.001043. [DOI] [PubMed] [Google Scholar]
- Vladimirov N, Tu Y, Traub RD. Shortest Loops are Pacemakers in Random Networks of Electrically Coupled Axons. Front Comput Neurosci. 2012;17:1–12. doi: 10.3389/fncom.2012.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young PA, Clendenon SG, Byars JM, Decca RS, Dunn KW. The effects of spherical aberration on multiphoton fluorescence excitation microscopy. J Microsc. 2011;242:157–65. doi: 10.1111/j.1365-2818.2010.03449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Chen LS, Miyauchi Y, Miyauchi M, Kar S, Kangavari S, et al. Mechanisms of cardiac nerve sprouting after myocardial infarction in dogs. Circ Res. 2004;95:76–83. doi: 10.1161/01.RES.0000133678.22968.e3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.