Abstract
The ubiquitin-proteasome system (UPS) is the primary selective degradation system in the nuclei and cytoplasm of eukaryotic cells, required for the turnover of myriad soluble proteins. The hundreds of factors that comprise the UPS include an enzymatic cascade that tags proteins for degradation via the covalent attachment of a poly-ubiquitin chain, and a large multimeric enzyme that degrades ubiquitinated proteins, the proteasome. Protein degradation by the UPS regulates many pathways and is a crucial component of the cellular proteostasis network. Dysfunction of the ubiquitination machinery or the proteolytic activity of the proteasome is associated with numerous human diseases. In this review we discuss the contributions of the proteasome to human pathology, describe mechanisms that regulate the proteolytic capacity of the proteasome, and discuss strategies to modulate proteasome function as a therapeutic approach to ameliorate diseases associated with altered UPS function.
Keywords: Proteasome, Ubiquitin, Protein degradation
1. Introduction
The cellular protein pool is in constant flux. Its composition is defined by the proteostasis network, comprised of ribosomes, chaperones and two proteolytic systems, the lysosome and UPS [1]. These adaptable systems sustain protein homeostasis under a large variety of different conditions such as during oxidative stress, cellular differentiation, varying nutrient supply, exposure to elevated or reduced temperature, and xenobiotic stress. The activities of the different components of the proteostasis network are functionally linked and compensatory strategies are in place to avoid proteostasis collapse if the activity of one or more of the network components declines. Failure or malfunction of the proteostasis network is often associated with human disease [1]. Modulating proteostasis has therefore emerged as a promising new avenue for the development of treatments for diverse diseases such as cancer, neurodegeneration, autoimmunity, cardiomyopathy, inherited diseases caused by partial loss of function mutations, such as cystic fibrosis, and inherited diseases associated with protein misfolding and toxic gain of function.
The UPS is the primary degradation system that mediates the degradation of short-lived regulatory proteins and the removal of damaged soluble proteins. The recognition signal for proteasomal degradation is a ubiquitin chain covalently attached to lysine residues in substrate proteins. Formation of an isopeptide bond between the ε-amino group of substrate lysines and the carboxyl group of the C-terminal glycine of ubiquitin is an ATP-dependent process and is achieved via an enzymatic cascade involving three distinct classes of enzymes: ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2) and ubiquitin ligases (E3) [2]. The same reaction links additional ubiquitin molecules to the primary ubiquitin via internal ubiquitin lysines, thus creating a ubiquitin chain. A chain of at least four ubiquitins linked via lysine 48 is the classical recognition motif for proteasomal degradation [3], though chains of other linkage types are now recognized as physiological targeting motifs, and for some substrates multiple monoubiquitin modifications are sufficient for targeting to the proteasome [4, 5]. The proteasome holocomplex or 26S proteasome consists of two entities: a central cylindrical structure with peptide hydrolysis activity (core particle (CP) or 20S proteasome) and a regulatory particle (RP, also known as the regulatory cap, 19S particle, or PA700), required for substrate recognition, removal of the ubiquitin chain and ATP-dependent unfolding.
Medical interest in modulating proteasome function for therapeutic purposes has significantly increased during the past decade since the first proteasome inhibitor, Velcade/Bortezomib, has been approved by the Food and Drug Association (FDA) in 2003 for the treatment of refractory multiple myeloma [6]. Studies that led to the FDA approval demonstrated that the proteasome can be transiently and safely inhibited in humans with anti-tumor activity especially against hematopoietic malignancies [6]. The efficacy of proteasome inhibition for the treatment of cancer is based on its role in regulating cell proliferation and on the exquisite reliance of cancer cells on proteasome function [7]. However, many diseases, especially neurodegenerative diseases are characterized by the accumulation of toxic misfolded proteins and eventual collapse of the proteostasis network. Thus, both proteasome inhibition and activation represent potential avenues for future drug development. During the last two decades many mechanisms that regulate the UPS have been unraveled. Mechanisms that specifically modulate the proteolytic activity of the proteasome are discussed in this review.
2. Major alternate forms of the proteasome
The central assembly of the proteasome is a cylindrical structure that houses the proteolytic active sites. The cylinder is formed by two different types of subunits, α and β, that are arranged in four stacked heptameric rings enclosing a central cavity. In eukaryotes, seven distinct α subunits are located in the two outer rings of the barrel, seven distinct β subunits form the two inner rings [8]. Three of the β subunits contain active sites with different peptide cleavage specificities. β1 has a caspase-like specificity, cleaving after acidic residues; β2 is trypsin-like, cleaving after basic residues; and β5 is chymotypsin-like in cleaving after large hydrophobic residues. Their active sites face the interior of the cylinder. Access to the central cavity is regulated by an adjustable gate that is formed by the N-terminal protrusions of the α subunits [9, 10]. The default status of the CP gate is closed and for proteasomal degradation and substrate access the N-termini need to be displaced from their axial position to reveal a continuous channel leading into the catalytic cavity. Modulation of the gate is a prerequisite for substrate entry into the proteolytic chamber and is mediated by proteasome activators. Several proteasome activators have been described, including the regulatory particle (RP/19S/PA700), activators of the PA28 protein family and Blm10/PA200 activators [11]. All activators share common binding sites on the two outer surfaces of the CP/20S (Figure 1). The binding pockets for activators are formed by interfaces of adjacent α subunits. The molecular mechanism of displacing the N-termini of the α subunit, which results in gate opening, appears to be different for the different activator families, and will be discussed below.
Figure 1. Modular structure of proteasome complexes.
The proteasome core particle can be capped with three different activator complexes: the hetero-oligomeric RP/19S/PA700 particle, the monomeric Blm10/PA200 proteins and the heptameric PA28/11S/REG rings (adapted from 11 and 9). Activators from all three families occupy the same binding site at the apical surface of the CP and open the CP gate.
It should be noted that in the absence of activators the free CP shows low but detectable activity towards small proteins with intrinsically unstructured regions in the absence of ubiquitination and ATP [12]. The significance of this activity is not understood but it may reflect the occurrence of spontaneous, short-lived opening of the gate at some frequency [13].
2.1. RP-CP: the proteasome holoenzyme
The dominant proteasome activator is the RP. It promotes ATP- and ubiquitin-dependent substrate turnover. The RP can bind to either one or two ends of the CP to form the proteasome holocomplex or 26S proteasome. The RP is composed of 19 integral subunits, which form two biochemically separable sub-complexes, lid and base [14]. The base subcomplex is situated proximal to the CP gate region. It contains six homologous ATPases (Rpt1-Rpt6), which form a hexameric ring. They belong to the AAA family of ATPases. The ring is not planar but has a spiral staircase-like topology, with Rpt3 at the highest and Rpt2 at the lowest position [15-17]. In addition to the ATPases, the base contains the two largest RP subunits, Rpn1 and Rpn2, and the two ubiquitin-receptors Rpn10 and Rpn13 [18]. The lid consists of nine non-ATPase subunits. One Rpn subunit, Rpn6, appears to contact both the base as well as the α ring and might be a crucial regulator of lid-base assembly [15, 19]. One of the non-ATPases, Rpn11, has deubiquitinating activity and is located in close proximity to the substrate entry pore formed by the ATPase ring [15]. Its deubiquitinating activity is required to promote the degradation of ubiquitinated proteasome substrates [16, 20-22].
Cryo-EM studies have defined two conformational states of the RP, ATP-h and ATP-γS. The ATP-h structure, the first to be defined [15, 17] is now seen as likely to represent a basal state of the enzyme. This structure is defined by the presence of ATP in the buffer and the absence of protein substrate. ATP can be hydrolyzed by these proteasomes, hence the designation ATP-h. However, the exact nucleotide state of ATP-h proteasomes is uncertain, since the attainable resolution at present does not allow for nucleotides to be visualized within the complex. Most likely some combination of ATP and ADP are present among the various ATPase sites of the complex.
ATP-γS proteasomes are defined by the presence of the nonhydrolyzable, or slowly hydrolyzed analog of ATP, ATP-γS. Major rearrangements of the RP are involved in the transition between the ATP-h and ATP-γS forms of the proteasome holoenzyme [17]. A common feature of these rearrangements is that they appear to poise the RP in a conformation better suited to drive substrate degradation. Because the proteasome may engage continuously in ATP hydrolysis, it is possible that it is never bound only to ATP, but rather to a mixture of ATP and ADP. This raises the possibility that the ATP-γS structure might not represent a physiological state of the enzyme. This, however, does not appear to be the case, since a similar form of the RP has been defined by trapping the RP in the presence of ATP and a ubiquitinated substrate that cannot be hydrolyzed due to mutational inactivation of Rpn11 [23]. Thus, the ATP-γS structure may represent the conformation of the proteasome as it is poised to degrade substrate. Among the signature features of the ATP-γS structure are (i) a movement of Rpn11 towards the substrate entry port of the RP, where it presumably releases ubiquitin from the substrate as the substrate is translocated into the RP channel, (ii) a widening of the translocation channel, and (iii) a repositioning of the RP channel so that it is more closely aligned to that of the CP [17, 23].
Gate opening, and thus proteasome activation by the RP, is promoted by the ATPase ring. Three of the ATPases, Rpt2, Rpt3, and Rpt5, dock into cognate binding pockets within the CP α ring [15, 16]. These subunits contain a characteristic motif at their C-termini, a HbYX-motif [24, 25], characterized by a hydrophobic residue, a conserved tyrosine at the penultimate position, and a variable C-terminal residue. Previous studies indicate that hepta-peptides containing this motif are able to activate latent isolated CP and thus are sufficient to induce gate opening [24, 26]. Based on these observations and on structural information obtained from a co-crystal structure with another HbYX-motif containing proteasome activator Blm10 (see below) [27] the most likely mechanism by which the HbYX-motif induces gate opening is through a displacement of conserved residues in the binding pockets of the α ring, which leads to long-range structural changes in the N-termini of the α subunits, which constitute the gate. This displacement opens a channel leading into the catalytic chamber. In several alternate forms of the proteasome, the open state of the CP gate is stabilized by interactions between the CP N-termini and elements within the axial pore of the activator complex. This has been shown for two complexes described in more detail below, PA28-CP [10] and Cdc48-CP [28]. These findings suggest that the open state of the CP N-termini may similarly be stabilized by interactions with the residues lining the axial pore of the Rpt ring in the proteasome holoenzyme.
Even though docking of the three HbYX motif-containing ATPases opens the proteasome gate, the resulting entry port does not allow for the passage of folded proteins. Prior to substrate entry, target proteins need to be unfolded, and this activity is provided by the proteasomal ATPases as the substrate is forced through the narrow translocation channel in an ATP-hydrolysis-dependent fashion [29, 30].
2.2. Blm10/PA200-CP
Blm10/PA200 proteasome activators are conserved from yeast to humans and represent monomeric proteins of ~250 kDa. They form hybrid complexes in which Blm10/PA200 binds to one end and the RP to the opposite end of the CP cylinder [31, 32]. Their cellular functions are emerging. Blm10 binds to the proteasome during the late phases of CP maturation and, like the RP, Blm10 contributes to the final maturation of CP complexes [33, 34]. One physiological target for Blm10-proteasome complexes has been identified to date, the transcription factor Sfp1, which regulates ribosomal protein genes [35]. Additional studies suggest a potential role for Blm10 in the maintenance of mitochondrial homeostasis [26, 27], a hypothesis that is supported by data obtained from mice that lack PA200, the ortholog of yeast Blm10. PA200−/− mice exhibit male fertility defects caused by impaired spermatogenesis [36], a process involving significant mitochondrial and metabolic changes. Further studies suggest a role for Blm10/PA200 activators in DNA or oxidative damage repair as well as in chromosome stability [31, 37, 38] most likely through ATP- and ubiquitin-independent degradation of acetylated histones in somatic cells [39]. Loss of PA200 in mice also leads to delayed core histone clearance in elongated spermatids [39]. Considering the impaired spermatogenesis of PA200 knock-out mice [36] these data suggest that PA200-mediated degradation of histones might be an essential mechanism for correct sperm formation.
The tertiary structure of Blm10/PA200 proteins is characterized by an array of HEAT repeats, which form an elongated solenoid [27]. The repeats make extensive contacts with the α ring surface. A co-crystal structure of reconstituted Blm10-CP complexes showed that Blm10 binding to the CP results in an at least partially open gate [27]. The conformations of the N-terminal residues of four of the α subunits are superimposable with the open gate conformation of PA26-CP complexes [40]. The remaining three N-termini could not be visualized, indicating they are also displaced from the closed gate conformation. Biochemical and crystallographic data also revealed that Blm10-CP association is mediated by the Blm10 C-terminus [26, 27]. As observed for the proteasomal ATPases, the C-terminus of Blm10 contains a HbYX-motif [26], and a heptameric peptide derived from the Blm10 C-terminus is sufficient to activate latent CP [26]. The position of the penultimate tyrosine within the CP binding pocket suggests a gate opening mechanism that is shared between the HbYX motif-containing proteasomal ATPases and Blm10.
Crucial for stabilizing the open gate conformation is the position of the conserved residue Pro17 within the α pocket, as first shown by work on the structure of the PA26-CP complex [40]. The penultimate tyrosine of Blm10 forms a hydrogen bond with the main chain oxygen of Gly19, which leads to a displacement of Pro17 and subsequent long-range alterations of the conformation of the α subunit N-termini. These conformational changes in the α subunit N-termini stabilize the open gate conformation. The structural changes within the proteasome gate are sufficient to allow passage of the small, unstructured in proteasome substrate tau-441 as demonstrated in vitro [26] and thus promote the degradation of certain proteins.
2.3. PA28-CP
The PA28/11S/REG proteasome activator family is expressed in higher eukaryotes and some unicellular eukaryotes, notably trypanosomes. In mammals, three different isoforms form two separate activators with different localization, induction and activation properties. PA28α and PA28β are 28-kDa proteins, which form hetero-heptameric rings in vertebrates [41, 42]. They bind to the CP in the cytoplasm or form hybrid complexes, where PA28 binds to one end of the CP and the 19S/PA700 complex to the opposing end [43-45]. Both subunits are inducible by interferon-γ, which suggested a potential role of PA28αβ in MHC Class I mediated antigen presentation [46]. Indeed, binding of PA28αβ affects the generation and presentation of a subset of viral antigens [47]. Although induced by cytokines, PA28αβ is present at a basal level in all tissues [48]. Thus, the cellular function of this class of proteasome activators has perhaps not been fully elucidated yet. A third PA28 isoform, PA28γ, forms homo-heptameric rings and is found in the nuclei of vertebrates as well as invertebrates [41, 42]. In mice, the loss of PA28γ results in reduced body size, and embryonic fibroblasts derived from these mice exhibit cell cycle defects [49]. Consistent with these findings, PA28γ promotes ATP- and ubiquitin-independent degradation of specific small regulatory proteins such as p21 and SRC-3 [50, 51].
The two PA28 complexes activate the CP via a common mechanism. As with the proteasomal ATPases, association with the CP is mediated via docking of the PA28 subunits’ C-termini into cognate binding pockets within the α subunits [40]. In contrast to the three proteasomal ATPases that dock into the CP surface, and Blm10, PA28 proteins lack the HbYX-motif. Thus, a direct structural change of the gate is not promoted by insertion of the C-termini. Instead, PA28 proteins utilize an internal loop structure, the activation loop, which contacts the 20S alpha subunits at Gly19 to stabilize Pro17 in a conformation, which promotes opening of the gate [40]. Three of the six proteasomal ATPases dock into the CP surface and the monomeric Blm10/PA200 activators insert into one pocket. Thus, the gate can be opened asymmetrically and without full occupancy of all seven binding pockets. PA28 activators, on the other hand, insert all of their seven C-termini.
2.4. Immunoproteasomes and Thymoproteasomes
In addition to proteasome activators, higher eukaryotes also express three alternate proteolytically active subunits, Lmp2, Lmp7 and MECL-1. Lmp2 and Lmp7 were initially identified because these genes were localized in a genomic region that is responsive to cytokines. Sequence analysis of Lmp2 and Lmp7 revealed homology to proteasome β subunits [52, 53]. Their cytokine-inducible expression suggested a potential role in the generation of antigenic peptides by the proteasome and they were accordingly called immunosubunits. Later a third subunit with homology to active site subunits was identified, MECL-1. Its expression is also sensitive to cytokines [54, 55]. All three immunosubunits are incorporated into the proteasome core to form immunoproteasomes or i-proteasomes. Lmp2 replaces β1, and is thus termed ß1i, whereas Lmp7 replaces β5, and MECL-1 β2. The two latter immunosubunits display essentially the same cleavage specificity as their housekeeping counterparts. Lmp2 on the other hand provides a chymotrypsin-like activity, in contrast to its constitutive counterpart, the caspase-like ß1. Thus, Lmp2 incorporation both attenuates the caspase-like activity of constitutive proteasomes and amplifies the chymotrypsin-like activity beyond that of ß5 or ß5i alone [56]. It was suggested that increased generation of peptides with hydrophobic residues should improve MHC Class I antigen presentation, since the preferred peptides for the MHC Class I molecules are peptides with a hydrophobic C-terminal residue. Despite this there were surprisingly modest effects on the immune response in mice deleted for either one, two, or all three immunosubunits as summarized in ref. [57]. Recent studies, however, with i-proteasome deficient mice and additional cell biological and biochemical data suggest that the function of the i-proteasome might go beyond antigen presentation. A role in the clearance of oxidatively damaged proteins was proposed [58, 59], especially during inflammation [59], in the regulation of tumor development, in lipid metabolism [60] and in NFκB signaling [58]. Whether immunoproteasomes have the capacity to alter the clearance of ubiquitinated proteins after interferon-Ɣ treatment, as has been proposed earlier [59] but is currently debated [61].
The newest member of alternate proteolytic active subunits of the proteasome has been identified during a homology search of the human and the mouse genome. The identified gene is homologous to β5 and is exclusively present in the thymus [62]. Incorporation of β5t results in a reduction of the chymotrypsin-like activity of the proteasome [62]. Current studies point to a specialized role of β5t-containing thymoproteasomes in the positive selection of CD8+ T cells [63].
3. Exchangeable and substoichiometric proteasome regulators
3.1. Ubiquitin receptors
Recognition of ubiquitinated target proteins by the proteasome is achieved by two intrinsic ubiquitin receptors: Rpn10 and Rpn13 [18, 64]. In addition, several ubiquitin binding proteins serve as shuttle factors, which deliver ubiquitinated substrates to the proteasome. The shuttle factors contain both a ubiquitin binding domain (UBD) and a ubiquitin-like domain (UBL). While the UBD promotes the binding of ubiquitinated substrate proteins, the UBL domain is required for interaction of the shuttle factors with the UBDs of intrinsic ubiquitin receptors or with Rpn1. The three known shuttle factors in yeast are Rad23 [65], Dsk2 [66], and Ddi1 [67].
3.2. Deubiquitinating enzymes and ubiquitin ligases
Two conserved deubiquitinating enzymes (DUBs) interact reversibly with the proteasome: Ubp6/Usp14 and UCH37/Uch2 [68, 69]. Usp14 antagonizes the degradation of ubiquitinated substrates by the proteasome [68, 70], and likely Uch37 as well [69, 71]. In contrast, the DUB activity of the intrinsic proteasome subunit Rpn11 promotes degradation. While inactivation of the deubiquitinating activity of Rpn11 prevented the degradation of proteasome substrates [20, 21], deletion of UBP6 results in enhanced proteasome activity [68]. These antagonistic effects of deubiquitination at the proteasome may reflect, at least in part, the nature of the cleavage site preferences of these DUBs. While Ubp6 and UCH37 are thought to remove ubiquitin preferentially from the distal end of the chain [68, 71], Rpn11 appears to cut at the base of the chain, thus removing the chain en bloc. Rpn11 is located within the proteasome RP close to the ATPase ring [15, 16], so that active translocation of the substrate by the ATPases should presumably present the chain to Rpn11, while at the same time carrying it away from the distally located Ubp6/Usp14. Once such translocation has initiated, the substrate is likely no longer dependent on the ubiquitin chain for proteasome binding, and therefore deubiquitination by Rpn11 does not suppress substrate degradation. Ubp6 on the other hand associates with Rpn1 at the outer surface of the proteasome [15, 72]. Thus, the effect of chain removal by Ubp6/Usp14 on proteasome-substrate affinity cannot be compensated by the interaction of substrate with the ATPases. Consequently, it is thought that the action of UBp6/Usp14 can result in premature release of the substrate from the proteasome.
The importance of DUB activity first became evident from studies on the cellular effects of loss of UBP6 in yeast. In the absence of Ubp6, cells acquire pleiotropic sensitivity to stress, accompanied by ubiquitin deficiency [73]. These data suggest that if ubiquitin chains are not trimmed from the distal end by DUBs, a high fraction of ubiquitin escapes the DUB activity of Rpn11 and is degraded along with the conjugated substrate, resulting in depletion of the cellular ubiquitin pool.
The inhibitory function of Ubp6 on the degradation of proteasomal substrates is antagonized by a HECT domain ligase, Hul5, which similarly associates with the proteasome [74]. Loss of HUL5 results in reduced degradation of ubiquitinated proteasome substrates and the data suggest that proteasome-associated Hul5 has an important role in extending existing ubiquitin chains, the defining characteristic of E4 ligases [74]. Longer ubiquitin chains show increased affinity for the proteasome [3], suggesting a simple mechanism by which Hul5 facilitates proteasome-mediated substrate degradation. This hypothesis is supported by a recent study, which demonstrated that Hul5's activity is crucial during heat stress, particularly for low-solubility substrates [75].
Several additional ubiquitin ligases have been found to interact with both yeast and mammalian proteasomes, such as Ubr1 and Ufd4 [76], RNF2 [77], SNEV [78], SCF and APC complexes [79, 80] and Parkin [81]. In most cases, proteasome association is thought to enhance substrate turnover by eliminating the need for a ubiquitinated protein to migrate to the proteasome [82]. If the interval between ubiquitination and proteasome binding is substantial, the ubiquitin chains could be released from the substrate by deubiquitinating enzymes. An exceptional case among proteasome-associated ligases is Parkin, which appears to modulate proteasome assembly [81].
3.3. Substoichiometric HbYX motif-containing proteasome modulators
As described above, the HbYX motif found in three of the six proteasomal ATPases, as well as in Blm10, mediates CP binding and gate opening. HbYX-motifs have been identified in the C-termini of other proteins as well. While the HbYX-motifs of the CP chaperones Pba1-Pba2 promote binding to inactive, immature CP subcomplexes and do not activate mature 20S [83], two other studies report on a proteasome activation function for proteins containing this motif: the Drosophila protein DmPI31, and Cdc48 isolated from the archaeon T. acidophilum. PI31 was previously described as a proteasome inhibitor [84, 85]. Recent studies, however, suggest alternate functions for PI31. Overexpression of PI31 in mammalian cells did not result in decreased proteasome activity and a role for PI31 specifically in immunoproteasome formation was proposed [86]. In Drosophila, overexpression of DmPI31 increased proteasome activity in vivo and rescued the adverse effects of proteasome inhibition [87]. In vitro studies with purified DmPI31 revealed that the protein activates 26S proteasomes but inhibits the CP. The ability of PI31 to stimulate proteasome activity is regulated by the F box protein Nutcracker, a component of the SCF E3 ligase complex in Drosophila. Nutcracker, however, does not promote turnover but rather stabilizes PI31 [87].
Cdc48/p97 is a conserved hexameric ring-shaped AAA ATPase complex whose function is intimately tied to the ubiquitin proteasome system. Its functional characteristics resemble those of the proteasomal ATPases: ubiquitin binding and protein unfoldase activity mediated by ATP-hydrolysis [88]. Cdc48 has been observed to remove ubiquitinated proteins from insoluble structures and from membrane-bound states [89]. Curiously, the Cdc48 C-terminus comprises a conserved HbYX-motif. A recent study revealed that archeal Cdc48 can dock onto the CP and can activate its peptidase activity [28]. The same activities have been shown for murine Cdc48, which can collaborate with murine CP [90]. Whereas Cdc48 has generally been envisaged as functioning upstream of the proteasome, delivering ubiquitinated proteins to the proteasome holoenzyme to be degraded, these studies [28, 90] raise the interesting possibility that Cdc48-CP can also function as an alternative form of the proteasome.
4. Transcriptional regulation of the proteasome
4.1. Rpn4
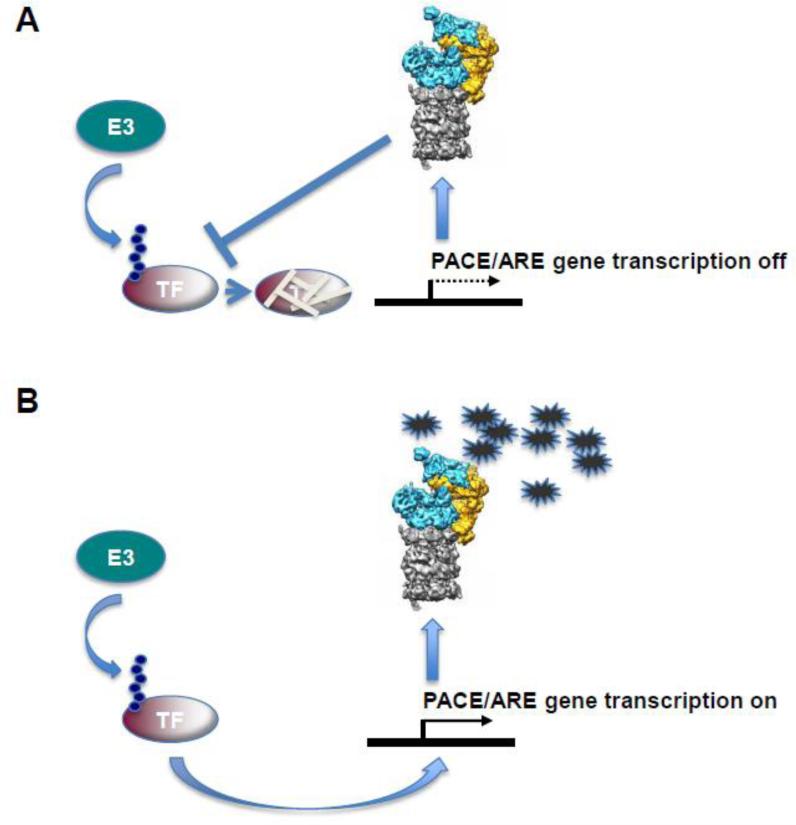
Proteasomes are abundant cellular constituents. However, under proteotoxic stress the basal level of proteasomes can become limiting for cell survival. To adapt proteasome levels to the cellular need for removal of damaged proteins, eukaryotic cells respond through transcriptional upregulation of proteasomal genes. This process involves coordinate upregulation of all proteasome genes, and is conserved across species [91, 92]. In S. cerevisiae the mechanisms that regulate proteasome expression are well understood. A single transcription factor, Rpn4, which binds to Proteasome Associated Control Elements (PACE) present in the promoter of all proteasome genes [93, 94], is required for the adaptive expression of proteasomal subunits (though not all Rpn4 target genes encode proteasome components) [95]. Rpn4 is a C2H2-type zinc finger-containing transcription factor. The transcriptional activity of Rpn4 is regulated by a negative feedback loop [96] (Figure 2). Under normal growth conditions Rpn4 has a very short half-life of ~2 min and is degraded by the proteasome via ubiquitin-dependent and ubiquitin-independent pathways [97]. Ubiquitin-dependent degradation of Rpn4 is mediated by the E3 ligase Ubr2 [98]. Additional regulation of Rpn4 abundance is achieved via transcriptional control of the RPN4 gene. Binding sites for Hsf1, the major regulator of proteins involved in alleviating heat stress, for Yap1, a transcription factor involved in upregulating detoxifying enzymes during oxidative stress, and for Pdr1 and Pdr3, which mediate pleiotropic drug resistance, are all present in the promoter of RPN4 [99]. Activation of RPN4 transcription by these stress-inducible transcription factors ensures that diverse proteotoxic stresses result in upregulation of proteasome levels.
Figure 2. Model for feedback regulation of proteasomal gene transcription.
A) Under normal growth conditions proteasomes are expressed at basal levels. Elevated expression of proteasomal genes is prevented via rapid ubiquitin-dependent degradation of a transcription factor that recognizes specific motifs in the promoter of proteasome genes: PACE and ARE elements in yeast and mammalian cells, respectively. B) Proteotoxic stress (indicated by accumulating damaged proteins symbolized as stars) saturates existing proteasome complexes, resulting in stabilization of the transcription factor and subsequent increased expression of proteasomal genes.
4.2. Nrf1/Nrf2/SKN-1
In mammals, short-term treatment of cells with proteasome inhibitors results in the upregulation of proteasomal genes. Two studies link the nuclear factor erythroid derived 2-related factors 1 (Nrf1) to the regulation of this response [100, 101], while other findings implicate Nrf2 in the stress-responsive regulation of proteasomal genes [102-105]. Both transcription factors belong to the Cap'n'Collar (CNC) transcription factor family, characterized by a unique basic leucine zipper domain. Nrf1 and Nrf2 regulate a large number of genes involved in antioxidant and xenobiotic defense [106]. A common promoter element in genes recognized by Nrf1 and Nrf2 is the antioxidant response element (ARE) [107]. They are therefore thought to have overlapping transcriptional activity, although differing strongly in their regulatory mechanisms, cellular localization, and in the phenotypes of the respective knockout mice. Nrf1 is an integral membrane protein of the endoplasmic reticulum (ER) [108], while Nrf2 is localized within the cytoplasm and at mitochondria under non-inducing conditions [109]. Nrf1 ablation in mice results in developmental defects and lethality caused by impaired liver erythropoiesis [110], while Nrf2−/− mice develop normally but are highly sensitive to oxidative stress. Nrf2−/− mice also develop neurodegenerative disorders, cancer, and autoimmune disease [111, 112]. Both Nrf proteins are regulated at the protein level via proteasome-mediated degradation. The ligases SCFβTrCP and Hrd1 have been found to be required for Nrf1 turnover [113] whereas a Cul3 family ligase that uses the KEAP protein for substrate recognition is required for regulated Nrf2 turnover [101, 114]. Although adaptive proteasome gene upregulation involves Nrf1 or Nrf2 activity, it is unclear yet whether these factors bind directly to proteasomal gene promoters. Putative AREs have been identified in proteasomal genes [100, 101], however, direct recruitment has not been shown, and a transcriptional analysis of liver-directed ablation of Nrf1 in mice did not alter the transcript levels of proteasomal genes [107].
Similarly to the response of mammalian cells to proteasome inhibition, reduced expression of proteasome genes in C. elegans activates SKN-1, the worm ortholog of Nrf1 and Nrf2 [115], and short-term proteasome inhibition leads to a compensatory upregulation of proteasomal genes via SKN-1 [116]. SKN-1 dependent upregulation of proteasomal genes is also observed upon treatment of C. elegans with H2O2 [117]. Furthermore, as described for Nrf2, SKN-1 levels are regulated by a cullin E3 ligase [118]. Interestingly, chromatin-IP (CHIP) experiments revealed that SKN-1 was bound to most proteasomal gene promoters during the L1 larval stage [119]. Recent studies suggest that the role of SKN-1 in regulating proteasome gene expression in response to proteotoxic stress is tissue specific and is coupled to the regulation of factors required for correct protein translation [116].
In summary, the data discussed provide compelling evidence that CNC transcription factors might be involved in the adaptive transcriptional regulation of proteasome genes. Furthermore, negative feedback regulation as described for Rpn4 in yeast appears to be conserved as well in higher eukaryotes (Figure 2).
4.3. FoxO/DAF-16
Recent evidence suggests that the role of SKN-1 in the regulation of proteasomal genes might depend on the reproductive state in worms. In germline-ablated (glp-1[e2141]) mutants, proteasomal gene transcription was independent of SKN-1. The mutants exhibited elevated proteasome levels, which were lost upon knocking down the FoxO transcription factor DAF-16 but not SKN-1 [120]. While previous studies demonstrated that FoxO transcription factors are regulated by proteasome-mediated degradation [121] the effect of DAF-16 on proteasome expression in germ line deleted worms represents the first evidence that DAF-16 might be involved in the regulation of proteasomal function. In contrast to Nrf1/2/SKN-1, which appear to be involved in the regulation of all proteasomal genes, however, only one proteasome subunit was found to be up-regulated in glp-1(e2141) mutants, RPN-6. Proteasome levels, however, remained unchanged. The authors argue that due to the specific topology of Rpn6 within the 26S proteasome, the positioning of which suggests a role in helping to bridge the RP and CP [15, 19], elevated RPN-6 levels might have a positive effect on the overall activity of the proteasome [120].
FoxO also induces increased proteolysis in atrophic muscles. While the major target for FoxO during muscle atrophy is the lysosomal arm of cellular proteolysis, it also activates certain E3 ligases, termed atrogenes and thus stimulates proteasomal degradation of muscle proteins [95, 122].
5. Post-translational modifications of the proteasome
Large-scale studies to define the phospho-proteome as well as other post-translational modifications both in yeast [123] and in higher eukaryotes [124] revealed that almost all proteasome subunits as well as the activators PA28 and Blm10 are phosphorylated. In addition, acetylation, myristoylation, ubiquitination, modification with O-linked N-acetyl-glucosamine (O-GlcNAc), S-glutathionylation and oxidation of proteasome subunits have been detected. Accumulating evidence suggests that these modifications might modulate proteasome activity. O-GlcNAc and oxidation results in proteasome inactivation [125-128]. Myristoylation of Rpt2 affects the localization of the proteasome [129]. Mono-ubiquitination of Rpn10 regulates substrate recruitment and interaction with proteasome shuttle factors [130, 131]. Reversible S-glutathionylation of cysteines in CP α subunits were found to activate isolated CP, an effect that was caused by gate opening [132]. Nuclear mammalian CP was also found to be poly-ADP ribosylated. This modification enhanced the activity of the CP towards fluorogenic peptides and oxidized histones [133] and ADP-ribosylation of PI31 regulates its interaction with the proteasome and affects proteasome assembly [134].
Several reports document both activating and inactivating functions for the phosphorylation of proteasome subunits, depending on the kinase. In vitro and in vivo studies propose a positive effect of protein kinase A (PKA) activity on proteasomal function [135-137]. PKA also influences proteasome assembly [138]. In neurons phosphorylation of Rpt6 by Ca2+/calmodulin-dependent protein kinase II α (CaMKIIα) results in increased proteasome activity and function [139]. Rpt6 phosphorylation has also been implicated in regulating proteasome assembly [140].
An inhibitory role for proteasome phosphorylation was proposed by a study that found the kinases c-Abl and Arg tyrosine kinase to interact with the proteasome and to phosphorylate α4, which led to a reduction in proteasome activity [141]. Similarly, phosphorylation of Rpn2 by p38 MAPK inhibited the activity of 26S proteasomes during osmotic stress [142]. Opposing effects of proteasome phosphorylation are also evident from studies focused on protein phosphatases. Phosphatase PP2A was reported to activate proteasome activity [143], whereas phosphatase UBLCP1, which contains a UBL domain, is recruited to the proteasome and inhibits proteasome assembly as well as its catalytic activity [144].
In summary, the post-translational modifications of the proteasome and their effects on its assembly, localization, and function are complex and provide a significantly increased layer of complexity to known proteasome regulatory mechanisms, which might fine tune proteasome function to specific cellular environments and cellular demands.
6. The proteasome as a therapeutic target
6.1. Cancer
Over the last decade, the potential applications of proteasome inhibition in disease have been explored extensively. Due to its broad impact on cellular functions, the proteasome was initially not considered to be a good target for the development of clinical drugs. Indeed, proteasome inhibition, if severe, disrupts the cell cycle and cellular protein homeostasis. Cells also activate cell death programs and apoptosis in response to prolonged proteasome inhibition. However, early studies demonstrated that certain types of cancers are exquisitely sensitive to proteasome inhibition [145, 146]. Subsequent work revealed that numerous cancer types exhibit aberrant function of the UPS [7, 147] and thus treatment with proteasome inhibitors might prove to be a successful strategy.
Initial studies with the peptide boronate proteasome inhibitor PS-341 (now called velcade or bortezomib) showed that the drug exhibited clinical activity especially towards hematologic malignancies [148, 149]. Bortezomib is a reversible inhibitor, which preferentially blocks the chymotryptic activity of the proteasome [150]. In 2003 bortezomib was approved by the US Food And Drug Administration (FDA) for the treatment of refractory multiple myeloma. It also exhibits efficacy in non-small cell lung cancer, mantle cell lymphomas and pancreatic cancer [151]. Bortezomib treatment, however, leads to the development of resistance. Adverse side effects include peripheral neuropathy, gastrointestinal problems, thrombocytopenia, asthenia, cardiac and pulmonary disorders and pain [151]. Thus, the development of new UPS-related drugs with reduced toxicity is desirable.
Several second-generation proteasome inhibitors are currently in clinical trials both for the treatment of hematologic malignancies and for solid tumors. Most of these newly developed inhibitors also inhibit the chymotrypsin-like activity of the proteasome, like bortezomib, but show increased chemical stability, different binding affinities for the proteasome as well as altered toxicities in the clinic [150]. Accelerated approval by the FDA was granted to the proteasome inhibitor carfilzomib in 2012 for the treatment of patients with multiple myeloma, which failed to respond to prior therapies. Carfilzomib has shown efficacy in bortezomib-resistant multiple myeloma patients [152] . In contrast to bortezomib, carfilzomib is an irreversible peptide epoxyketone inhibitor [152]. For a detailed description of current research on proteasome inhibitors see ref. [150]
6.2. Immune-related diseases
At the organismal level, proteasome inhibition results in decreased inflammatory and immune responses, with cell migration and cell adhesion being compromised [151]. The effects of proteasome inhibition on the immune system owe in part to the fundamental role of the proteasome in antigen presentation. A commonly observed toxicity during clinical administration of bortezomib is lymphopenia [149], thus bortezomib apparently has suppressive or toxic effects on the T-cell compartment. Subsequent studies showed that proteasome inhibition leads to T-cell apoptosis [153]. Activated T-cells are more susceptible to proteasome inhibition than resting T-cells [154], suggesting that proteasome inhibition might be a successful strategy to treat diseases involving activated T-cells, such as graft-versus-host disease (GVHD). Proteasome inhibition also results in depletion of alloreactive T-cells, whereas the immune defense against pathogens remains intact [154], suggesting that proteasome inhibition might be a viable strategy for the treatment of autoimmune diseases. These hypotheses are under investigation with several successful preclinical studies in mouse models for autoimmune encepahlomyelitis, GVHD, arthritis, colitis, systemic lupus erythematosus, myastenia gravis and allograft rejection [155].
Alternate forms of the three proteolytically active β subunits are incorporated into the proteasome CP in response to cytokines to generate the immunoproteasome (i-proteasomes). Recently the crystal structure of the i-proteasome has been solved [56], demonstrating that the active site architecture of the inducible subunits differs from that of the constitutive counterparts. This has allowed the synthesis of immunoproteasome specific inhibitors. Since i-proteasomes represent a minor fraction of the proteasome population and are found predominantly in cells involved in the immune response, it is expected that treating patients with inhibitors that specifically inhibit the i-β subunits, but not the constitutive proteasomes, should result in reduced toxicity [150]. In initial studies utilizing these novel proteasome inhibitors, they were found to inhibit the growth of multiple myeloma cell lines and exhibit anti-inflammatory and immunosuppressive effects [156, 157].
6.3. Neurodegenerative Diseases
A unifying characteristic of neurodegenerative diseases is the deposition of aggregated, misfolded proteins. This is true of Alzheimer's disease (AD), Parkinson's disease (PD), Huntington's disease (HD), Amyotrophic Lateral Sclerosis (ALS) and spongiform encephalopathies. Aggregation can occur in the cytoplasm, the nucleus or in the extracellular space. If unchecked, the accumulation of misfolded protein can disrupt cell and tissue function. Protein aggregation in general may be an indication of a compromised proteostasis network. Therefore, a wide body of research has assessed the role of the UPS in these diseases. This review will focus on the involvement of the proteasome in neurodegenerative diseases. The effect of the ubiquitination machinery has been discussed in detail previously [158].
Alzheimer's disease is the most prevalent neurodegenerative disease. It is estimated that ~30% of the human population over 85 years old show pathological signs of AD [159]. AD is characterized by progressive loss of cognitive function and dementia, resulting in memory loss, personality changes, psychosis, and language disturbancies. The progressive intellectual decline in AD patients is accompanied by an increase in the deposition of protein aggregates that eventually form intracellular neurofibrillar tangles (NFT) and extracellular senile/amyloid plaques [160]. The major component of NFTs is hyperphosphorylated tau, while senile/amyloid plaques contain mostly amyloid-β (Aβ). The latter is produced via endoproteolytic cleavage of the membrane-associated amyloid precursor protein (APP). Aβ aggregates in the extracellular space after incorrect cleavage of APP by inter-or extra-membrane protease family, the secretases [161]. While not directly participating in the generation of Aβ or the elimination of extracellular Aβ, the UPS may, according to previous reports, be involved in the degradation of APP via the endoplasmic reticulum-associated degradation (ERAD) arm of the UPS [162] and intracellular Aβ aggregates appear to inhibit the UPS [163].
The second protein associated with Alzheimer's disease, tau, is a structural protein, which associates with microtubules and has a role in axonal transport [164]. Tau is an intrinsically unstructured protein and in vitro degradation experiments with purified components showed that tau can be degraded by the CP without prior ubiquitination [26, 165]. ATP-dependent unfolding by the RP is also not required for in vitro proteasome-mediated degradation of tau. However the relevance of these observations to tau degradation within cells is not established. For example, in cells tau could be associated with other factors that serve to suppress this potential pathway of degradation. Proteasome activator Blm10 can also accelerate in vitro tau degradation [26]. Whether PA200, the human counterpart of Blm10, plays a similar role in tau degradation remains to be established.
Studies of tau turnover within cells point to roles for both autophagy and the UPS. There is little consensus on the relative importance of these pathways, possibly reflecting that, in the case of tau, the balance between autophagy and the proteasome will depend on cell type, tau overexpression, and similar experimental variables. Moreover, distinct pools of tau may be degraded differently. For example, hypoacetylated tau is a preferred substrate of the UPS [166, 167]. Also, when tau is not bound to microtubules it readily associates with Hsp70, where it may encounter the ubiquitin ligase CHIP, which can ubiquitinate tau and target it for the proteasome [168, 169]. Tau degradation can also be promoted when deubiquitination by the proteasome-associated deubiquitinating enzyme Usp14 is inhibited [166]. The dependence of tau degradation on CHIP and Usp14 indicates that in cells tau degradation by the proteasome is ubiquitin-dependent, although whether it is fully ubiquitin-dependent remains to be clarified. See reference [166] for a comprehensive discussion of the literature of tau degradation.
Co-immunoprecipitation experiments revealed that fibrillar tau co-precipitated with the proteasome, and proteasome activity was significantly reduced in AD patients, as compared to age-matched controls [170]. These data suggest that aggregated tau might inhibit proteasomal activity, which, due to the pleiotropic effects of proteasome function within cells, could contribute to the pathology of AD.
Parkinson's disease is a prevalent age-related neurodegenerative disorder affecting ~ 3% of the human population over 65 years. The typical symptoms of PD are rigidity, shaking and slowness of movement. The characteristic pathology found in PD patients involves Lewy Bodies in dopaminergic neurons in the substantia nigra of the brain. The major components of Lewy Bodies are α-synuclein, chaperones, ubiquitin, the E3 ligase parkin, and other proteins involved in the UPS. As tau, α-synuclein monomers are unfolded [171] and can be degraded by the 20S core particle without prior ubiquitination and in the absence of the RP [172]. Other studies, however, showed that depletion of the proteasome subunit Rpt2 results in accumulation of α-synuclein and the development of Lewy Body-like inclusions in mice, suggesting a role for the RP in α-synuclein degradation [173, 174]. This hypothesis is supported by a recent study of German PD patients, which found that variations in the gene PSMC4/Rpt3 correlated with the age of PD onset [175].
Huntington's disease is the best-studied example of a group of disorders, called polyglutamine (polyQ) diseases. Other diseases with the same pathological mechanism are spinocerebrellar ataxia and spinal and bulbular muscular atrophy [176]. HD is an autosomal dominant disease, which is characterized by motor dysfunction, cognitive decline and psychosis. The disease is caused by an expansion of a CAG triplet repeat region in the huntingtin (Htt) gene through out-of-register recombination between repeat elements, leading to an expansion of a poly-glutamine stretch in the N-terminal domain of Htt [177]. The Htt proteins of healthy individuals contain 6-35 glutamines, whereas a poly-glutamine stretch of >40 triggers the development of HD pathology [178]. Furthermore, disease progression correlates with the length of the polyQ expansion. At the structural level, expansion of glutamine stretches with more than 40 glutamines results in fibril formation and aggregation [179]. Aggregated mutant Htt is found in intracellular inclusion bodies in neurons. Although wild-type Htt has been associated with a variety of cellular functions such as transcriptional regulation and axonal transport, HD is most likely not caused by Htt loss of function, but rather by a toxic gain of function of the aggregated mutant form. This hypothesis is supported by the finding that expression of polyQ peptides in mice caused HD-like symptoms [180].
As in AD and PD, inclusions formed by mutant Htt contain both ubiquitinated proteins and proteasome subunits. Aggregated Htt itself was found to be ubiquitinated [181]. In contrast, soluble mutant Htt showed little cross reactivity with anti-ubiquitin antibodies [182]. Proteasome activity was reduced in brains derived from HD patients and in mice models, but the origin of proteasome dysfunction remains debated. Interestingly, while the chymotrypsin-like and the caspase-like activity were reduced, the trypsin-like activity was markedly enhanced in post-mortem HD brains and in a mouse knock-in model with Htt111Q [183]. The trypsin-like activity is required for the cleavage after positively charged amino acids but also after glutamine and asparagine. Apparently the upregulation of trypsin-like activity is unable to compensate for the overall loss of proteasome activity as ubiquitinated reporter proteins accumulate upon expression and aggregation of mutant Htt [184].
Several models for impaired proteasome function in HD have been proposed. Proteasomes were found sequestered in Htt inclusion bodies, which could explain the overall reduction in UPS function [185]. One in vitro study suggested that the proteasome might not be able to cleave within polyQ stretches and that the released polyQ stretch would clog the CP [186], while a second in vitro study was not in agreement [187]. A subsequent, carefully designed study found that Htt aggregates do not impair the general capacity of the proteasome to degrade ubiquitinated substrates [182]. The authors concluded that proteasome dysfunction might not originate from an inhibitory effect of the mutant Htt oligomers on the proteolytic capacity of the proteasome, but rather from a general proteostasis collapse. Whatever the cause of proteasome dysfunction, enhancement of proteasome activity may be beneficial in cells challenged by polyQ-Htt, since upregulation of PA28γ transcription improved cell survival in a cellular HD model [188].
Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disorder affecting motor neurons. Familial ALS has been linked to mutations in numerous genes [189]. Ubiquitinated inclusions are found within motor-neurons in both familial and sporadic forms of the disease, suggesting possible dysfunction of the ubiquitin-proteasome pathway in ALS, whether in a causative role or otherwise. Interestingly, a conditional knockout of proteasome subunit Rpt3 in motor neurons exhibited ALS-like pathology, particularly the accumulation of protein aggregates with signature components of ALS inclusions, such as the TDP-43 and FUS proteins [190]. These findings suggest that altered turnover of proteasome substrates may play a significant role in the pathogenesis of ALS. A parallel experiment in which autophagy was eliminated in motor neurons showed no such phenotypic effects [190], arguing for the specificity of the results and for a distinct importance of the proteasome in motor neurons.
There is currently no effective treatment available that could cure or substantially delay the progression of the neurodegenerative diseases described above. These diseases are however, generally thought to be characterized by decreased activity of the UPS. Therefore, drug development strategies resulting in an increase of the proteolytic capacity of the proteasome might be beneficial for disease progression. One specific approach to this involves small-molecule inhibitors of Usp14 activity [70]. Alternatively, one may envisage upregulation of proteasomal gene expression, upregulation of proteasomal activators such as PA28 or PA200 or the identification of small molecules that can activate the CP by inducing gate opening.
6.4. Cardiomyophathies
Similar to neurological toxicity and altered immune response, treatment of patients with bortezomib also led to an unexpected increase in cardiac dysfunction such as arrythmia or congestive heart failure [191]. These observations are supported by reports, which demonstrate that the UPS plays a major role in cardiac physiology and disease. Cardiac diseases, especially hypertrophic and dilated cardiomyopathies as well as ischemic heart diseases, are characterized by increased oxidative damage to proteins, elevated levels of ubiquitinated proteins and proteasome dysfunction [192]. A reduction of proteasome function is most clearly associated with myocardial ischemia/reperfusion (I/R) injury [126]. Short term and low level local treatment with proteasome inhibitors led to controversial results. Both beneficial and detrimental outcomes have been reported (reviewed in [192]).
In a recent study, a causal link between proteasome dysfunction and the progression of myocardial I/R injury has been established. The authors created a transgenic mouse line in which a catalytically inactive β5 subunit (β5T1A) was expressed in the heart at low levels. Under normal conditions there were no cardiac changes observed in the transgenic animals. However upon I/R injury increased cardiac damage was observed [193]. Interestingly, the same animal model was used to test the hypothesis that increased proteasome activity might ameliorate I/R injury. To that end the authors created a transgenic mouse line, which overexpressed the proteasome activator subunit PA28α in the heart and found that increased proteasome activity protected the animals against I/R injury [194]. In a mouse model for desmin-related cardiomyopathy (DRC) cardiac-restricted overexpression of PA28α also reduced cardiac hypertrophy and extended the lifespan of the animals [194]. These studies indicate a significant involvement of proteasome activity in cardiac function during I/R injury and in DRC.
6.5. Aging
A hallmark of aging is the progressive accumulation of damaged macromolecules and a progressive decline in the function of the cellular proteostasis network. A critical component of the proteostasis network in aging cells is the proteasome. Many reports describe a decline in proteasome function in aging cells, tissues and organisms (reviewed in [195]). Age-related proteasome dysfunction occurs at many levels, involving reduced expression of proteasome subunits [196], oxidative damage resulting in reduced proteolytic activity [126, 128], and dissociation of the holocomplex [128, 197, 198]. One view is that aggregated proteins might form nonproductive complexes with the proteasome and thus reduce its activity [199]. The importance of maintaining proteasome function in aging organisms is highlighted by a recent transgenic mouse model for proteasome dysfunction. In this model the housekeeping β5 subunit was replaced with β5t, a thymus-specific variant. As described above, proteasomes with incorporated β5t have reduced chymotrypsin-like activity. The transgenic mice exhibited signs of premature aging and had a significantly shorter lifespan [200].
Interestingly, several studies with exceptionally long-lived humans (centenarians) and animals reported increased proteasome activity. Proteasome expression and activity in fibroblasts derived from healthy centenarians were compared to fibroblasts from young and old control donors. Proteasome functionality in fibroblasts derived from centenarians resembled that of young donors [201]. Increased proteasome capacity was also observed in the exceptionally long-lived naked mole rat [202] a long-lived bat species [203] and in the giant clam, an exceptionally long-lived invertebrate [204].
The data above suggest that sustained proteasome activity might correlate with the lifespan of an organism. This hypothesis has found support through genetic approaches in several model organisms. Overexpressing of the proteasome chaperone Ump1 led to increased survival of yeast cells in stationary phase [205], a model for post-mitotic aging. Overexpression of β5 in primary human embryonic fibroblasts or of Rpn11 in Drosophila [206] extended lifespan. The latter reports suggest that increased expression of a single proteasome subunit can promote proteasome assembly, perhaps reflecting that these subunits are limiting for assembly in wild-type cells. Two recent studies utilized information on transcriptional regulation of the UPS to elevate the proteasome constitutively by activating or overexpressing transcription factors, which regulate the adaptive increase in proteasome expression in response to proteotoxic stress. In yeast deletion of UBR2, the E3 ligase, which regulates the levels of Rpn4, leads to constitutive upregulation of the UPS, enhanced proteasome capacity and a 70% increase in replicative lifespan [207]. These data suggest that enhanced proteasome might activity positively affect aging.
There are some common features of these studies that are worth noting. First, it does not as yet seem that any one method of enhancing proteasome function is unique in its ability to provide for enhanced stress resistance and lifespan. Secondly, enhanced proteasome activity has at least to date not been linked to detrimental effects on cell survival and regulation.
7. Perspectives
As the field of ubiquitination developed, the proteasome was seen as neither selective in its activity, nor rate limiting for degradation, nor subject to significant regulation. Prevailing views on these topics have changed radically, as proteasome levels and activity have over the past ten years been shown to be subject to fine control via a vast array of mechanisms, as described above. The existence of these mechanisms may be taken as a sign that the output of the proteasome is a central control point for protein degradation. The nature of these mechanisms also indicates that cellular regulation and survival can tolerate adjustments in proteasome activity, despite its importance in many aspects of cell function. These considerations have been highlighted especially by the success of proteasome inhibitors as anticancer agents.
Despite the clinical utility of proteasome inhibitors, one may imagine that enhancing proteasome activity could also have therapeutic benefits in the proper context. Tens of diseases, many of them major, are caused by toxic and often mutated proteins, the effects of which could be neutralized if their levels could be reduced through enhancing proteasome activity. Imbalanced protein homeostasis is common among neurodegenerative diseases and cardiomyopathies, and proteostasis collapse also is evident in aging cells. These diseases are frequently characterized by increased damage to the cellular protein pool, intracellular protein aggregation, and reduced proteasome activity. An attractive hypothesis for potential treatment of these diseases is to upregulate components of the proteostasis network. In the last few years increasing efforts have begun to test whether proteasome up-regulation might be a viable approach for alleviating these diseases and the published studies in different disease models and organisms are encouraging.
A number of distinct strategies may be taken to enhance proteasome activity. Among the most appealing is to inactivate endogenous proteins that serve to suppress proteasome activity. Since ubiquitin targets substrates to the proteasome, deubiquitinating enzymes are natural antagonists of the proteasome. In particular, deubiquitinating enzymes Uch37 and Usp14, being physically associated with the proteasome, may suppress proteasome activity through deubiquitinating a subset of ubiquitinated substrates, once the substrate is docked at the proteasome. Specific, small molecule mediated inhibition of proteasomal deubiquitinating enzyme Usp14 has been reported [70]. Proteasomes are also under negative control by the gate element of the CP, which is regulated by proteasome activators. Any compound or strategy that promotes the open state of this structure may stimulate the degradation of either canonical, ubiquitin-dependent proteasome substrates or noncanonical, ubiquitin-independent substrates. In general this approach may allow for substrates to bypass the proteasome holoenzyme, with differential activation of the free form of the CP, which is normally held under tight negative control. Direct activation of the CP would be predicted to favor the degradation of unfolded substrates of the proteasome. A final possible approach to stimulate proteasome activity is to increase its level via transcriptional up-regulation. It remains unclear however, whether transcriptional circuitry that is dedicated specifically to the proteasome exists. In summary, these various approaches to proteasome modulation will be distinguished in their relative feasibility and in the specificity of the effects that they achieve. Although our understanding of physiological proteasome regulation and of the potential therapeutic opportunities in up-regulating the proteasome with small molecules is still at an early stage, the next few years should provide exciting new information and a sharper vision of the implications of proteasome modulation for the treatment of human disease.
Highlights.
Different forms of the proteasome
How the proteasome is regulated
Role of the proteasome in disease
Therapeutic opportunities involving the proteasome
Acknowledgements
This work was supported by NIH grants RO1 GM084228 to M.S and R37 GM43601 and R01 GM65592 to D.F.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lindquist SL, Kelly JW. Chemical and biological approaches for adapting proteostasis to ameliorate protein misfolding and aggregation diseases: progress and prognosis. Cold Spring Harb Perspect Biol. 2011:3. doi: 10.1101/cshperspect.a004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. Embo J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dimova NV, Hathaway NA, Lee BH, Kirkpatrick DS, Berkowitz ML, Gygi SP, Finley D, King RW. APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1. Nat Cell Biol. 2012;14:168–176. doi: 10.1038/ncb2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shabek N, Herman-Bachinsky Y, Buchsbaum S, Lewinson O, Haj-Yahya M, Hejjaoui M, Lashuel HA, Sommer T, Brik A, Ciechanover A. The size of the proteasomal substrate determines whether its degradation will be mediated by mono- or polyubiquitylation. Mol Cell. 2012;48:87–97. doi: 10.1016/j.molcel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 6.Esseltine DL, Mulligan G. An historic perspective of proteasome inhibition. Semin Hematol. 2012;49:196–206. doi: 10.1053/j.seminhematol.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- 8.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 9.Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D. A gated channel into the proteasome core particle. Nat Struct Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 10.Whitby FG, Masters EI, Kramer L, Knowlton JR, Yao Y, Wang CC, Hill CP. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature. 2000;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- 11.Stadtmueller BM, Hill CP. Proteasome activators. Mol Cell. 2011;41:8–19. doi: 10.1016/j.molcel.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baugh JM, Viktorova EG, Pilipenko EV. Proteasomes can degrade a significant proportion of cellular proteins independent of ubiquitination. J Mol Biol. 2009;386:814–827. doi: 10.1016/j.jmb.2008.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osmulski PA, Hochstrasser M, Gaczynska M. A tetrahedral transition state at the active sites of the 20S proteasome is coupled to opening of the alpha-ring channel. Structure. 2009;17:1137–1147. doi: 10.1016/j.str.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 15.Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beck F, Unverdorben P, Bohn S, Schweitzer A, Pfeifer G, Sakata E, Nickell S, Plitzko JM, Villa E, Baumeister W, et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proc Natl Acad Sci U S A. 2012;109:14870–14875. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sledz P, Unverdorben P, Beck F, Pfeifer G, Schweitzer A, Forster F, Baumeister W. Structure of the 26S proteasome with ATP-gammaS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proc Natl Acad Sci U S A. 2013;110:7264–7269. doi: 10.1073/pnas.1305782110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pathare GR, Nagy I, Bohn S, Unverdorben P, Hubert A, Korner R, Nickell S, Lasker K, Sali A, Tamura T, et al. The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proc Natl Acad Sci U S A. 2012;109:149–154. doi: 10.1073/pnas.1117648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 21.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 22.Forster F, Lasker K, Beck F, Nickell S, Sali A, Baumeister W. An atomic model AAA-ATPase/20S core particle sub-complex of the 26S proteasome. Biochem Biophys Res Commun. 2009;388:228–233. doi: 10.1016/j.bbrc.2009.07.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matyskiela ME, Lander GC, Martin A. Conformational switching of the 26S proteasome enables substrate degradation. Nat Struct Mol Biol. 2013;20:781–788. doi: 10.1038/nsmb.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson D, Hakala K, DeMartino GN. Subcomplexes of PA700, the 19 S regulator of the 26 S proteasome, reveal relative roles of AAA subunits in 26 S proteasome assembly and activation and ATPase activity. J Biol Chem. 2009;284:24891–24903. doi: 10.1074/jbc.M109.023218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dange T, Smith D, Noy T, Rommel PC, Jurzitza L, Cordero RJ, Legendre A, Finley D, Goldberg AL, Schmidt M. Blm10 protein promotes proteasomal substrate turnover by an active gating mechanism. J Biol Chem. 2011;286:42830–42839. doi: 10.1074/jbc.M111.300178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sadre-Bazzaz K, Whitby FG, Robinson H, Formosa T, Hill CP. Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening. Mol Cell. 2010;37:728–735. doi: 10.1016/j.molcel.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barthelme D, Sauer RT. Identification of the Cdc48*20S proteasome as an ancient AAA+ proteolytic machine. Science. 2012;337:843–846. doi: 10.1126/science.1224352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braun BC, Glickman M, Kraft R, Dahlmann B, Kloetzel PM, Finley D, Schmidt M. The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat Cell Biol. 1999;1:221–226. doi: 10.1038/12043. [DOI] [PubMed] [Google Scholar]
- 30.Strickland E, Hakala K, Thomas PJ, DeMartino GN. Recognition of misfolding proteins by PA700, the regulatory subcomplex of the 26 S proteasome. J Biol Chem. 2000;275:5565–5572. doi: 10.1074/jbc.275.8.5565. [DOI] [PubMed] [Google Scholar]
- 31.Blickwedehl J, Agarwal M, Seong C, Pandita RK, Melendy T, Sung P, Pandita TK, Bangia N. Role for proteasome activator PA200 and postglutamyl proteasome activity in genomic stability. Proc Natl Acad Sci U S A. 2008;105:16165–16170. doi: 10.1073/pnas.0803145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt M, Haas W, Crosas B, Santamaria PG, Gygi SP, Walz T, Finley D. The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat Struct Mol Biol. 2005;12:294–303. doi: 10.1038/nsmb914. [DOI] [PubMed] [Google Scholar]
- 33.Marques AJ, Glanemann C, Ramos PC, Dohmen RJ. The C- terminal extension of the beta7 subunit and activator complexes stabilize nascent 20 S proteasomes and promote their maturation. J Biol Chem. 2007;282:34869–34876. doi: 10.1074/jbc.M705836200. [DOI] [PubMed] [Google Scholar]
- 34.Fehlker M, Wendler P, Lehmann A, Enenkel C. Blm3 is part of nascent proteasomes and is involved in a late stage of nuclear proteasome assembly. EMBO Rep. 2003;4:959–963. doi: 10.1038/sj.embor.embor938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez AD, Tar K, Krugel U, Dange T, Ros IG, Schmidt M. Proteasomal degradation of Sfp1 contributes to the repression of ribosome biogenesis during starvation and is mediated by the proteasome activator Blm10. Mol Biol Cell. 2011;22:528–540. doi: 10.1091/mbc.E10-04-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khor B, Bredemeyer AL, Huang CY, Turnbull IR, Evans R, Maggi LB, Jr., White JM, Walker LM, Carnes K, Hess RA, et al. Proteasome activator PA200 is required for normal spermatogenesis. Mol Cell Biol. 2006;26:2999–3007. doi: 10.1128/MCB.26.8.2999-3007.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doherty KM, Pride LD, Lukose J, Snydsman BE, Charles R, Pramanik A, Muller EG, Botstein D, Moore CW. Loss of a 20S proteasome activator in Saccharomyces cerevisiae downregulates genes important for genomic integrity, increases DNA damage, and selectively sensitizes cells to agents with diverse mechanisms of action. G3 (Bethesda) 2012;2:943–959. doi: 10.1534/g3.112.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blickwedehl J, McEvoy S, Wong I, Kousis P, Clements J, Elliott R, Cresswell P, Liang P, Bangia N. Proteasomes and proteasome activator 200 kDa (PA200) accumulate on chromatin in response to ionizing radiation. Radiat Res. 2007;167:663–674. doi: 10.1667/RR0690.1. [DOI] [PubMed] [Google Scholar]
- 39.Qian MX, Pang Y, Liu CH, Haratake K, Du BY, Ji DY, Wang GF, Zhu QQ, Song W, Yu Y, et al. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell. 2013;153:1012–1024. doi: 10.1016/j.cell.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol Cell. 2005;18:589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 41.Dubiel W, Pratt G, Ferrell K, Rechsteiner M. Purification of an 11 S regulator of the multicatalytic protease. J Biol Chem. 1992;267:22369–22377. [PubMed] [Google Scholar]
- 42.Ma CP, Slaughter CA, DeMartino GN. Identification, purification, and characterization of a protein activator (PA28) of the 20 S proteasome (macropain). J Biol Chem. 1992;267:10515–10523. [PubMed] [Google Scholar]
- 43.Tanahashi N, Murakami Y, Minami Y, Shimbara N, Hendil KB, Tanaka K. Hybrid proteasomes. Induction by interferon-gamma and contribution to ATP-dependent proteolysis. J Biol Chem. 2000;275:14336–14345. doi: 10.1074/jbc.275.19.14336. [DOI] [PubMed] [Google Scholar]
- 44.Kopp F, Dahlmann B, Kuehn L. Reconstitution of hybrid proteasomes from purified PA700-20 S complexes and PA28alphabeta activator: ultrastructure and peptidase activities. J Mol Biol. 2001;313:465–471. doi: 10.1006/jmbi.2001.5063. [DOI] [PubMed] [Google Scholar]
- 45.Cascio P, Call M, Petre BM, Walz T, Goldberg AL. Properties of the hybrid form of the 26S proteasome containing both 19S and PA28 complexes. Embo J. 2002;21:2636–2645. doi: 10.1093/emboj/21.11.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Realini C, Jensen CC, Zhang Z, Johnston SC, Knowlton JR, Hill CP, Rechsteiner M. Characterization of recombinant REGalpha, REGbeta, and REGgamma proteasome activators. J Biol Chem. 1997;272:25483–25492. doi: 10.1074/jbc.272.41.25483. [DOI] [PubMed] [Google Scholar]
- 47.Sijts A, Sun Y, Janek K, Kral S, Paschen A, Schadendorf D, Kloetzel PM. The role of the proteasome activator PA28 in MHC class I antigen processing. Mol Immunol. 2002;39:165–169. doi: 10.1016/s0161-5890(02)00099-8. [DOI] [PubMed] [Google Scholar]
- 48.Noda C, Tanahashi N, Shimbara N, Hendil KB, Tanaka K. Tissue distribution of constitutive proteasomes, immunoproteasomes, and PA28 in rats. Biochem Biophys Res Commun. 2000;277:348–354. doi: 10.1006/bbrc.2000.3676. [DOI] [PubMed] [Google Scholar]
- 49.Murata S, Kawahara H, Tohma S, Yamamoto K, Kasahara M, Nabeshima Y, Tanaka K, Chiba T. Growth retardation in mice lacking the proteasome activator PA28gamma. J Biol Chem. 1999;274:38211–38215. doi: 10.1074/jbc.274.53.38211. [DOI] [PubMed] [Google Scholar]
- 50.Li X, Amazit L, Long W, Lonard DM, Monaco JJ, O'Malley BW. Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGgamma-proteasome pathway. Mol Cell. 2007;26:831–842. doi: 10.1016/j.molcel.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 51.Chen X, Barton LF, Chi Y, Clurman BE, Roberts JM. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Mol Cell. 2007;26:843–852. doi: 10.1016/j.molcel.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown MG, Driscoll J, Monaco JJ. Structural and serological similarity of MHC-linked LMP and proteasome (multicatalytic proteinase) complexes. Nature. 1991;353:355–357. doi: 10.1038/353355a0. [DOI] [PubMed] [Google Scholar]
- 53.Ortiz-Navarrete V, Seelig A, Gernold M, Frentzel S, Kloetzel PM, Hammerling GJ. Subunit of the ‘20S’ proteasome (multicatalytic proteinase) encoded by the major histocompatibility complex. Nature. 1991;353:662–664. doi: 10.1038/353662a0. [DOI] [PubMed] [Google Scholar]
- 54.Groettrup M, Kraft R, Kostka S, Standera S, Stohwasser R, Kloetzel PM. A third interferon-gamma-induced subunit exchange in the 20S proteasome. Eur J Immunol. 1996;26:863–869. doi: 10.1002/eji.1830260421. [DOI] [PubMed] [Google Scholar]
- 55.Nandi D, Jiang H, Monaco JJ. Identification of MECL-1 (LMP-10) as the third IFN-gamma-inducible proteasome subunit. J Immunol. 1996;156:2361–2364. [PubMed] [Google Scholar]
- 56.Huber EM, Basler M, Schwab R, Heinemeyer W, Kirk CJ, Groettrup M, Groll M. Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell. 2012;148:727–738. doi: 10.1016/j.cell.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 57.Ferrington DA, Gregerson DS. Immunoproteasomes: structure, function, and antigen presentation. Prog Mol Biol Transl Sci. 2012;109:75–112. doi: 10.1016/B978-0-12-397863-9.00003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pickering AM, Davies KJ. Differential roles of proteasome and immunoproteasome regulators Pa28alphabeta, Pa28gamma and Pa200 in the degradation of oxidized proteins. Arch Biochem Biophys. 2012;523:181–190. doi: 10.1016/j.abb.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seifert U, Bialy LP, Ebstein F, Bech-Otschir D, Voigt A, Schroter F, Prozorovski T, Lange N, Steffen J, Rieger M, et al. Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell. 2010;142:613–624. doi: 10.1016/j.cell.2010.07.036. [DOI] [PubMed] [Google Scholar]
- 60.Kitamura A, Maekawa Y, Uehara H, Izumi K, Kawachi I, Nishizawa M, Toyoshima Y, Takahashi H, Standley DM, Tanaka K, et al. A mutation in the immunoproteasome subunit PSMB8 causes autoinflammation and lipodystrophy in humans. J Clin Invest. 2011;121:4150–4160. doi: 10.1172/JCI58414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nathan JA, Spinnenhirn V, Schmidtke G, Basler M, Groettrup M, Goldberg AL. Immuno- and constitutive proteasomes do not differ in their abilities to degrade ubiquitinated proteins. Cell. 2013;152:1184–1194. doi: 10.1016/j.cell.2013.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murata S, Sasaki K, Kishimoto T, Niwa S, Hayashi H, Takahama Y, Tanaka K. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science. 2007;316:1349–1353. doi: 10.1126/science.1141915. [DOI] [PubMed] [Google Scholar]
- 63.Nitta T, Murata S, Sasaki K, Fujii H, Ripen AM, Ishimaru N, Koyasu S, Tanaka K, Takahama Y. Thymoproteasome shapes immunocompetent repertoire of CD8+ T cells. Immunity. 2010;32:29–40. doi: 10.1016/j.immuni.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 64.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 65.Elsasser S, Chandler-Militello D, Muller B, Hanna J, Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem. 2004;279:26817–26822. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- 66.Matiuhin Y, Kirkpatrick DS, Ziv I, Kim W, Dakshinamurthy A, Kleifeld O, Gygi SP, Reis N, Glickman MH. Extraproteasomal Rpn10 restricts access of the polyubiquitin-binding protein Dsk2 to proteasome. Mol Cell. 2008;32:415–425. doi: 10.1016/j.molcel.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Voloshin O, Bakhrat A, Herrmann S, Raveh D. Transfer of Ho endonuclease and Ufo1 to the proteasome by the UbL-UbA shuttle protein, Ddi1, analysed by complex formation in vitro. PLoS One. 2012;7:e39210. doi: 10.1371/journal.pone.0039210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 69.Koulich E, Li X, DeMartino GN. Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Mol Biol Cell. 2008;19:1072–1082. doi: 10.1091/mbc.E07-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 72.Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Muller B, Feng MT, Tubing F, Dittmar GA, Finley D. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 73.Hanna J, Leggett DS, Finley D. Ubiquitin depletion as a key mediator of toxicity by translational inhibitors. Mol Cell Biol. 2003;23:9251–9261. doi: 10.1128/MCB.23.24.9251-9261.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- 75.Fang NN, Ng AH, Measday V, Mayor T. Hul5 HECT ubiquitin ligase plays a major role in the ubiquitylation and turnover of cytosolic misfolded proteins. Nat Cell Biol. 2011;13:1344–1352. doi: 10.1038/ncb2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xie Y, Varshavsky A. Physical association of ubiquitin ligases and the 26S proteasome. Proc Natl Acad Sci U S A. 2000;97:2497–2502. doi: 10.1073/pnas.060025497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee SJ, Choi D, Rhim H, Kang S. E3 ubiquitin ligase RNF2 interacts with the S6’ proteasomal ATPase subunit and increases the ATP hydrolysis activity of S6’. Biochem J. 2005;389:457–463. doi: 10.1042/BJ20041982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Loscher M, Fortschegger K, Ritter G, Wostry M, Voglauer R, Schmid JA, Watters S, Rivett AJ, Ajuh P, Lamond AI, et al. Interaction of U-box E3 ligase SNEV with PSMB4, the beta7 subunit of the 20 S proteasome. Biochem J. 2005;388:593–603. doi: 10.1042/BJ20041517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Seeger M, Hartmann-Petersen R, Wilkinson CR, Wallace M, Samejima I, Taylor MS, Gordon C. Interaction of the anaphase-promoting complex/cyclosome and proteasome protein complexes with multiubiquitin chain-binding proteins. J Biol Chem. 2003;278:16791–16796. doi: 10.1074/jbc.M208281200. [DOI] [PubMed] [Google Scholar]
- 80.Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, Deshaies RJ. Proteasomal proteomics: identification of nucleotide- sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Um JW, Im E, Lee HJ, Min B, Yoo L, Yoo J, Lubbert H, Stichel-Gunkel C, Cho HS, Yoon JB, et al. Parkin directly modulates 26S proteasome activity. J Neurosci. 2010;30:11805–11814. doi: 10.1523/JNEUROSCI.2862-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xie Y, Varshavsky A. UFD4 lacking the proteasome-binding region catalyses ubiquitination but is impaired in proteolysis. Nat Cell Biol. 2002;4:1003–1007. doi: 10.1038/ncb889. [DOI] [PubMed] [Google Scholar]
- 83.Kusmierczyk AR, Kunjappu MJ, Kim RY, Hochstrasser M. A conserved 20S proteasome assembly factor requires a C-terminal HbYX motif for proteasomal precursor binding. Nat Struct Mol Biol. 2011;18:622–629. doi: 10.1038/nsmb.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McCutchen-Maloney SL, Matsuda K, Shimbara N, Binns DD, Tanaka K, Slaughter CA, DeMartino GN. cDNA cloning, expression, and functional characterization of PI31, a proline-rich inhibitor of the proteasome. J Biol Chem. 2000;275:18557–18565. doi: 10.1074/jbc.M001697200. [DOI] [PubMed] [Google Scholar]
- 85.Zaiss DM, Standera S, Holzhutter H, Kloetzel P, Sijts AJ. The proteasome inhibitor PI31 competes with PA28 for binding to 20S proteasomes. FEBS Lett. 1999;457:333–338. doi: 10.1016/s0014-5793(99)01072-8. [DOI] [PubMed] [Google Scholar]
- 86.Zaiss DM, Standera S, Kloetzel PM, Sijts AJ. PI31 is a modulator of proteasome formation and antigen processing. Proc Natl Acad Sci U S A. 2002;99:14344–14349. doi: 10.1073/pnas.212257299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bader M, Benjamin S, Wapinski OL, Smith DM, Goldberg AL, Steller H. A conserved F box regulatory complex controls proteasome activity in Drosophila. Cell. 2011;145:371–382. doi: 10.1016/j.cell.2011.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yamanaka K, Sasagawa Y, Ogura T. Recent advances in p97/VCP/Cdc48 cellular functions. Biochim Biophys Acta. 2012;1823:130–137. doi: 10.1016/j.bbamcr.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 89.Buchberger A. Roles of cdc48 in regulated protein degradation in yeast. Subcell Biochem. 2013;66:195–222. doi: 10.1007/978-94-007-5940-4_8. [DOI] [PubMed] [Google Scholar]
- 90.Barthelme D, Sauer RT. Bipartite determinants mediate an evolutionarily conserved interaction between Cdc48 and the 20S peptidase. Proc Natl Acad Sci U S A. 2013;110:3327–3332. doi: 10.1073/pnas.1300408110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tan K, Shlomi T, Feizi H, Ideker T, Sharan R. Transcriptional regulation of protein complexes within and across species. Proc Natl Acad Sci U S A. 2007;104:1283–1288. doi: 10.1073/pnas.0606914104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Meiners S, Heyken D, Weller A, Ludwig A, Stangl K, Kloetzel PM, Kruger E. Inhibition of proteasome activity induces concerted expression of proteasome genes and de novo formation of Mammalian proteasomes. J Biol Chem. 2003;278:21517–21525. doi: 10.1074/jbc.M301032200. [DOI] [PubMed] [Google Scholar]
- 93.Mannhaupt G, Schnall R, Karpov V, Vetter I, Feldmann H. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 1999;450:27–34. doi: 10.1016/s0014-5793(99)00467-6. [DOI] [PubMed] [Google Scholar]
- 94.Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 95.Jelinsky SA, Estep P, Church GM, Samson LD. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol Cell Biol. 2000;20:8157–8167. doi: 10.1128/mcb.20.21.8157-8167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ju D, Wang L, Mao X, Xie Y. Homeostatic regulation of the proteasome via an Rpn4-dependent feedback circuit. Biochem Biophys Res Commun. 2004;321:51–57. doi: 10.1016/j.bbrc.2004.06.105. [DOI] [PubMed] [Google Scholar]
- 97.Ju D, Xie Y. Proteasomal degradation of RPN4 via two distinct mechanisms, ubiquitin-dependent and -independent. J Biol Chem. 2004;279:23851–23854. doi: 10.1074/jbc.C400111200. [DOI] [PubMed] [Google Scholar]
- 98.Wang L, Mao X, Ju D, Xie Y. Rpn4 is a physiological substrate of the Ubr2 ubiquitin ligase. J Biol Chem. 2004;279:55218–55223. doi: 10.1074/jbc.M410085200. [DOI] [PubMed] [Google Scholar]
- 99.Ma M, Liu ZL. Comparative transcriptome profiling analyses during the lag phase uncover YAP1, PDR1, PDR3, RPN4, and HSF1 as key regulatory genes in genomic adaptation to the lignocellulose derived inhibitor HMF for Saccharomyces cerevisiae. BMC Genomics. 2010;11:660. doi: 10.1186/1471-2164-11-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Radhakrishnan SK, Lee CS, Young P, Beskow A, Chan JY, Deshaies RJ. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol Cell. 2010;38:17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Steffen J, Seeger M, Koch A, Kruger E. Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol Cell. 2010;40:147–158. doi: 10.1016/j.molcel.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 102.Kraft DC, Deocaris CC, Wadhwa R, Rattan SI. Preincubation with the proteasome inhibitor MG-132 enhances proteasome activity via the Nrf2 transcription factor in aging human skin fibroblasts. Ann N Y Acad Sci. 2006;1067:420–424. doi: 10.1196/annals.1354.060. [DOI] [PubMed] [Google Scholar]
- 103.Pickering AM, Linder RA, Zhang H, Forman HJ, Davies KJ. Nrf2-dependent induction of proteasome and Pa28alphabeta regulator are required for adaptation to oxidative stress. J Biol Chem. 2012;287:10021–10031. doi: 10.1074/jbc.M111.277145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kapeta S, Chondrogianni N, Gonos ES. Nuclear erythroid factor 2-mediated proteasome activation delays senescence in human fibroblasts. J Biol Chem. 2010;285:8171–8184. doi: 10.1074/jbc.M109.031575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kwak MK, Wakabayashi N, Greenlaw JL, Yamamoto M, Kensler TW. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol Cell Biol. 2003;23:8786–8794. doi: 10.1128/MCB.23.23.8786-8794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sykiotis GP, Bohmann D. Stress-activated cap'n'collar transcription factors in aging and human disease. Sci Signal. 2010;3:re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ohtsuji M, Katsuoka F, Kobayashi A, Aburatani H, Hayes JD, Yamamoto M. Nrf1 and Nrf2 play distinct roles in activation of antioxidant response element-dependent genes. J Biol Chem. 2008;283:33554–33562. doi: 10.1074/jbc.M804597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang W, Chan JY. Nrf1 is targeted to the endoplasmic reticulum membrane by an N-terminal transmembrane domain. Inhibition of nuclear translocation and transacting function. J Biol Chem. 2006;281:19676–19687. doi: 10.1074/jbc.M602802200. [DOI] [PubMed] [Google Scholar]
- 109.Lo SC, Hannink M. PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Exp Cell Res. 2008;314:1789–1803. doi: 10.1016/j.yexcr.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chan JY, Kwong M, Lu R, Chang J, Wang B, Yen TS, Kan YW. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. Embo J. 1998;17:1779–1787. doi: 10.1093/emboj/17.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hubbs AF, Benkovic SA, Miller DB, O'Callaghan JP, Battelli L, Schwegler-Berry D, Ma Q. Vacuolar leukoencephalopathy with widespread astrogliosis in mice lacking transcription factor Nrf2. Am J Pathol. 2007;170:2068–2076. doi: 10.2353/ajpath.2007.060898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yoh K, Itoh K, Enomoto A, Hirayama A, Yamaguchi N, Kobayashi M, Morito N, Koyama A, Yamamoto M, Takahashi S. Nrf2-deficient female mice develop lupus-like autoimmune nephritis. Kidney Int. 2001;60:1343–1353. doi: 10.1046/j.1523-1755.2001.00939.x. [DOI] [PubMed] [Google Scholar]
- 113.Tsuchiya Y, Morita T, Kim M, Iemura S, Natsume T, Yamamoto M, Kobayashi A. Dual regulation of the transcriptional activity of Nrf1 by beta-TrCP- and Hrd1-dependent degradation mechanisms. Mol Cell Biol. 2011;31:4500–4512. doi: 10.1128/MCB.05663-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kahn NW, Rea SL, Moyle S, Kell A, Johnson TE. Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem J. 2008;409:205–213. doi: 10.1042/BJ20070521. [DOI] [PubMed] [Google Scholar]
- 116.Li X, Matilainen O, Jin C, Glover-Cutter KM, Holmberg CI, Blackwell TK. Specific SKN-1/Nrf stress responses to perturbations in translation elongation and proteasome activity. PLoS Genet. 2011;7:e1002119. doi: 10.1371/journal.pgen.1002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pickering AM, Staab TA, Tower J, Sieburth DS, Davies KJ. A conserved role for the 20S proteasome and Nrf2 transcription factor in oxidative- stress adaptation in mammals, C. elegans and D. melanogaster. J Exp Biol. 2012 doi: 10.1242/jeb.074757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Choe KP, Przybysz AJ, Strange K. The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans. Mol Cell Biol. 2009;29:2704–2715. doi: 10.1128/MCB.01811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Niu W, Lu ZJ, Zhong M, Sarov M, Murray JI, Brdlik CM, Janette J, Chen C, Alves P, Preston E, et al. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res. 2011;21:245–254. doi: 10.1101/gr.114587.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vilchez D, Morantte I, Liu Z, Douglas PM, Merkwirth C, Rodrigues AP, Manning G, Dillin A. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489:263–268. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- 121.Huang H, Tindall DJ. Regulation of FOXO protein stability via ubiquitination and proteasome degradation. Biochim Biophys Acta. 2011;1813:1961–1964. doi: 10.1016/j.bbamcr.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhao J, Brault JJ, Schild A, Goldberg AL. Coordinate activation of autophagy and the proteasome pathway by FoxO transcription factor. Autophagy. 2008;4:378–380. doi: 10.4161/auto.5633. [DOI] [PubMed] [Google Scholar]
- 123.Kikuchi J, Iwafune Y, Akiyama T, Okayama A, Nakamura H, Arakawa N, Kimura Y, Hirano H. Co- and post-translational modifications of the 26S proteasome in yeast. Proteomics. 2010;10:2769–2779. doi: 10.1002/pmic.200900283. [DOI] [PubMed] [Google Scholar]
- 124.Gomes AV, Zong C, Edmondson RD, Li X, Stefani E, Zhang J, Jones RC, Thyparambil S, Wang GW, Qiao X, et al. Mapping the murine cardiac 26S proteasome complexes. Circ Res. 2006;99:362–371. doi: 10.1161/01.RES.0000237386.98506.f7. [DOI] [PubMed] [Google Scholar]
- 125.Zhang F, Su K, Yang X, Bowe DB, Paterson AJ, Kudlow JE. O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell. 2003;115:715–725. doi: 10.1016/s0092-8674(03)00974-7. [DOI] [PubMed] [Google Scholar]
- 126.Bulteau AL, Lundberg KC, Humphries KM, Sadek HA, Szweda PA, Friguet B, Szweda LI. Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. J Biol Chem. 2001;276:30057–30063. doi: 10.1074/jbc.M100142200. [DOI] [PubMed] [Google Scholar]
- 127.Carrard G, Dieu M, Raes M, Toussaint O, Friguet B. Impact of ageing on proteasome structure and function in human lymphocytes. Int J Biochem Cell Biol. 2003;35:728–739. doi: 10.1016/s1357-2725(02)00356-4. [DOI] [PubMed] [Google Scholar]
- 128.Wang X, Yen J, Kaiser P, Huang L. Regulation of the 26S proteasome complex during oxidative stress. Sci Signal. 2010;3:ra88. doi: 10.1126/scisignal.2001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kimura A, Kato Y, Hirano H. N-myristoylation of the Rpt2 subunit regulates intracellular localization of the yeast 26S proteasome. Biochemistry. 2012;51:8856–8866. doi: 10.1021/bi3007862. [DOI] [PubMed] [Google Scholar]
- 130.Isasa M, Katz EJ, Kim W, Yugo V, Gonzalez S, Kirkpatrick DS, Thomson TM, Finley D, Gygi SP, Crosas B. Monoubiquitination of RPN10 regulates substrate recruitment to the proteasome. Mol Cell. 2010;38:733–745. doi: 10.1016/j.molcel.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lipinszki Z, Kovacs L, Deak P, Udvardy A. Ubiquitylation of Drosophila p54/Rpn10/S5a regulates its interaction with the UBA-UBL polyubiquitin receptors. Biochemistry. 2012;51:2461–2470. doi: 10.1021/bi3001006. [DOI] [PubMed] [Google Scholar]
- 132.Silva GM, Netto LE, Simoes V, Santos LF, Gozzo FC, Demasi MA, Oliveira CL, Bicev RN, Klitzke CF, Sogayar MC, et al. Redox control of 20S proteasome gating. Antioxid Redox Signal. 2012;16:1183–1194. doi: 10.1089/ars.2011.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ullrich O, Reinheckel T, Sitte N, Hass R, Grune T, Davies KJ. Poly-ADP ribose polymerase activates nuclear proteasome to degrade oxidatively damaged histones. Proc Natl Acad Sci U S A. 1999;96:6223–6228. doi: 10.1073/pnas.96.11.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cho-Park PF, Steller H. Proteasome Regulation by ADP Ribosylation. Cell. 2013;153:614–627. doi: 10.1016/j.cell.2013.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang F, Hu Y, Huang P, Toleman CA, Paterson AJ, Kudlow JE. Proteasome function is regulated by cyclic AMP-dependent protein kinase through phosphorylation of Rpt6. J Biol Chem. 2007;282:22460–22471. doi: 10.1074/jbc.M702439200. [DOI] [PubMed] [Google Scholar]
- 136.Zong C, Gomes AV, Drews O, Li X, Young GW, Berhane B, Qiao X, French SW, Bardag-Gorce F, Ping P. Regulation of murine cardiac 20S proteasomes: role of associating partners. Circ Res. 2006;99:372–380. doi: 10.1161/01.RES.0000237389.40000.02. [DOI] [PubMed] [Google Scholar]
- 137.Pereira ME, Wilk S. Phosphorylation of the multicatalytic proteinase complex from bovine pituitaries by a copurifying cAMP-dependent protein kinase. Arch Biochem Biophys. 1990;283:68–74. doi: 10.1016/0003-9861(90)90613-4. [DOI] [PubMed] [Google Scholar]
- 138.Asai M, Tsukamoto O, Minamino T, Asanuma H, Fujita M, Asano Y, Takahama H, Sasaki H, Higo S, Asakura M, et al. PKA rapidly enhances proteasome assembly and activity in in vivo canine hearts. J Mol Cell Cardiol. 2009;46:452–462. doi: 10.1016/j.yjmcc.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 139.Djakovic SN, Marquez-Lona EM, Jakawich SK, Wright R, Chu C, Sutton MA, Patrick GN. Phosphorylation of Rpt6 regulates synaptic strength in hippocampal neurons. J Neurosci. 2012;32:5126–5131. doi: 10.1523/JNEUROSCI.4427-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Satoh K, Sasajima H, Nyoumura KI, Yokosawa H, Sawada H. Assembly of the 26S proteasome is regulated by phosphorylation of the p45/Rpt6 ATPase subunit. Biochemistry. 2001;40:314–319. doi: 10.1021/bi001815n. [DOI] [PubMed] [Google Scholar]
- 141.Liu X, Huang W, Li C, Li P, Yuan J, Li X, Qiu XB, Ma Q, Cao C. Interaction between c-Abl and Arg tyrosine kinases and proteasome subunit PSMA7 regulates proteasome degradation. Mol Cell. 2006;22:317–327. doi: 10.1016/j.molcel.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 142.Lee SH, Park Y, Yoon SK, Yoon JB. Osmotic stress inhibits proteasome by p38 MAPK-dependent phosphorylation. J Biol Chem. 2010;285:41280–41289. doi: 10.1074/jbc.M110.182188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ramnanan CJ, Allan ME, Groom AG, Storey KB. Regulation of global protein translation and protein degradation in aerobic dormancy. Mol Cell Biochem. 2009;323:9–20. doi: 10.1007/s11010-008-9959-2. [DOI] [PubMed] [Google Scholar]
- 144.Guo X, Engel JL, Xiao J, Tagliabracci VS, Wang X, Huang L, Dixon JE. UBLCP1 is a 26S proteasome phosphatase that regulates nuclear proteasome activity. Proc Natl Acad Sci U S A. 2011;108:18649–18654. doi: 10.1073/pnas.1113170108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Delic J, Masdehors P, Omura S, Cosset JM, Dumont J, Binet JL, Magdelenat H. The proteasome inhibitor lactacystin induces apoptosis and sensitizes chemo- and radioresistant human chronic lymphocytic leukaemia lymphocytes to TNF-alpha-initiated apoptosis. Br J Cancer. 1998;77:1103–1107. doi: 10.1038/bjc.1998.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Orlowski RZ, Eswara JR, Lafond-Walker A, Grever MR, Orlowski M, Dang CV. Tumor growth inhibition induced in a murine model of human Burkitt's lymphoma by a proteasome inhibitor. Cancer Res. 1998;58:4342–4348. [PubMed] [Google Scholar]
- 147.Chen L, Madura K. Increased proteasome activity, ubiquitin conjugating enzymes, and eEF1A translation factor detected in breast cancer tissue. Cancer Res. 2005;65:5599–5606. doi: 10.1158/0008-5472.CAN-05-0201. [DOI] [PubMed] [Google Scholar]
- 148.Orlowski RZ, Stinchcombe TE, Mitchell BS, Shea TC, Baldwin AS, Stahl S, Adams J, Esseltine DL, Elliott PJ, Pien CS, et al. Phase I trial of the proteasome inhibitor PS-341 in patients with refractory hematologic malignancies. J Clin Oncol. 2002;20:4420–4427. doi: 10.1200/JCO.2002.01.133. [DOI] [PubMed] [Google Scholar]
- 149.Jagannath S, Barlogie B, Berenson J, Siegel D, Irwin D, Richardson PG, Niesvizky R, Alexanian R, Limentani SA, Alsina M, et al. A phase 2 study of two doses of bortezomib in relapsed or refractory myeloma. Br J Haematol. 2004;127:165–172. doi: 10.1111/j.1365-2141.2004.05188.x. [DOI] [PubMed] [Google Scholar]
- 150.Huber EM, Groll M. Inhibitors for the immuno- and constitutive proteasome: current and future trends in drug development. Angew Chem Int Ed Engl. 2012;51:8708–8720. doi: 10.1002/anie.201201616. [DOI] [PubMed] [Google Scholar]
- 151.Frankland-Searby S, Bhaumik SR. The 26S proteasome complex: an attractive target for cancer therapy. Biochim Biophys Acta. 2012;1825:64–76. doi: 10.1016/j.bbcan.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang Z, Yang J, Kirk C, Fang Y, Alsina M, Badros A, Papadopoulos K, Wong A, Woo T, Bomba D, et al. Clinical pharmacokinetics, metabolism, and drug-drug interaction of carfilzomib. Drug Metab Dispos. 2013;41:230–237. doi: 10.1124/dmd.112.047662. [DOI] [PubMed] [Google Scholar]
- 153.Nencioni A, Garuti A, Schwarzenberg K, Cirmena G, Dal Bello G, Rocco I, Barbieri E, Brossart P, Patrone F, Ballestrero A. Proteasome inhibitor-induced apoptosis in human monocyte-derived dendritic cells. Eur J Immunol. 2006;36:681–689. doi: 10.1002/eji.200535298. [DOI] [PubMed] [Google Scholar]
- 154.Blanco B, Sanchez-Abarca LI, Caballero-Velazquez T, Santamaria C, Inoges S, Perez-Simon JA. Depletion of alloreactive T-cells in vitro using the proteasome inhibitor bortezomib preserves the immune response against pathogens. Leuk Res. 2011;35:1412–1415. doi: 10.1016/j.leukres.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 155.Moran E, Carbone F, Augusti V, Patrone F, Ballestrero A, Nencioni A. Proteasome inhibitors as immunosuppressants: biological rationale and clinical experience. Semin Hematol. 2012;49:270–276. doi: 10.1053/j.seminhematol.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 156.Muchamuel T, Basler M, Aujay MA, Suzuki E, Kalim KW, Lauer C, Sylvain C, Ring ER, Shields J, Jiang J, et al. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat Med. 2009;15:781–787. doi: 10.1038/nm.1978. [DOI] [PubMed] [Google Scholar]
- 157.Basler M, Dajee M, Moll C, Groettrup M, Kirk CJ. Prevention of experimental colitis by a selective inhibitor of the immunoproteasome. J Immunol. 2010;185:634–641. doi: 10.4049/jimmunol.0903182. [DOI] [PubMed] [Google Scholar]
- 158.Huang Q, Figueiredo-Pereira ME. Ubiquitin/proteasome pathway impairment in neurodegeneration: therapeutic implications. Apoptosis : an international journal on programmed cell death. 2010;15:1292–1311. doi: 10.1007/s10495-010-0466-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hebert LE, Weuve J, Scherr PA, Evans DA. Alzheimer disease in the United States (2010-2050) estimated using the 2010 census. Neurology. 2013 doi: 10.1212/WNL.0b013e31828726f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Selkoe DJ. Alzheimer's disease. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Iwata N, Higuchi M, Saido TC. Metabolism of amyloid-beta peptide and Alzheimer's disease. Pharmacol Ther. 2005;108:129–148. doi: 10.1016/j.pharmthera.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 162.Kaneko M, Koike H, Saito R, Kitamura Y, Okuma Y, Nomura Y. Loss of HRD1-mediated protein degradation causes amyloid precursor protein accumulation and amyloid-beta generation. J Neurosci. 2010;30:3924–3932. doi: 10.1523/JNEUROSCI.2422-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tseng BP, Green KN, Chan JL, Blurton-Jones M, LaFerla FM. Abeta inhibits the proteasome and enhances amyloid and tau accumulation. Neurobiol Aging. 2008;29:1607–1618. doi: 10.1016/j.neurobiolaging.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Takei Y, Teng J, Harada A, Hirokawa N. Defects in axonal elongation and neuronal migration in mice with disrupted tau and map1b genes. The Journal of cell biology. 2000;150:989–1000. doi: 10.1083/jcb.150.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Grune T, Botzen D, Engels M, Voss P, Kaiser B, Jung T, Grimm S, Ermak G, Davies KJ. Tau protein degradation is catalyzed by the ATP/ubiquitin-independent 20S proteasome under normal cell conditions. Arch Biochem Biophys. 2010;500:181–188. doi: 10.1016/j.abb.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lee MJ, Lee JH, Rubinsztein DC. Tau degradation: The ubiquitin-proteasome system versus the autophagy-lysosome system. Prog Neurobiol. 2013 doi: 10.1016/j.pneurobio.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 167.Min SW, Cho SH, Zhou Y, Schroeder S, Haroutunian V, Seeley WW, Huang EJ, Shen Y, Masliah E, Mukherjee C, et al. Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron. 2010;67:953–966. doi: 10.1016/j.neuron.2010.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Petrucelli L, Dickson D, Kehoe K, Taylor J, Snyder H, Grover A, De Lucia M, McGowan E, Lewis J, Prihar G, et al. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum Mol Genet. 2004;13:703–714. doi: 10.1093/hmg/ddh083. [DOI] [PubMed] [Google Scholar]
- 169.Dickey CA, Koren J, Zhang YJ, Xu YF, Jinwal UK, Birnbaum MJ, Monks B, Sun M, Cheng JQ, Patterson C, et al. Akt and CHIP coregulate tau degradation through coordinated interactions. Proc Natl Acad Sci U S A. 2008;105:3622–3627. doi: 10.1073/pnas.0709180105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Keck S, Nitsch R, Grune T, Ullrich O. Proteasome inhibition by paired helical filament-tau in brains of patients with Alzheimer's disease. Journal of neurochemistry. 2003;85:115–122. doi: 10.1046/j.1471-4159.2003.01642.x. [DOI] [PubMed] [Google Scholar]
- 171.Bartels T, Choi JG, Selkoe DJ. alpha-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature. 2011;477:107–110. doi: 10.1038/nature10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tofaris GK, Layfield R, Spillantini MG. alpha-synuclein metabolism and aggregation is linked to ubiquitin-independent degradation by the proteasome. FEBS Lett. 2001;509:22–26. doi: 10.1016/s0014-5793(01)03115-5. [DOI] [PubMed] [Google Scholar]
- 173.Bedford L, Hay D, Devoy A, Paine S, Powe DG, Seth R, Gray T, Topham I, Fone K, Rezvani N, et al. Depletion of 26S proteasomes in mouse brain neurons causes neurodegeneration and Lewy-like inclusions resembling human pale bodies. J Neurosci. 2008;28:8189–8198. doi: 10.1523/JNEUROSCI.2218-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Paine SM, Anderson G, Bedford K, Lawler K, Mayer RJ, Lowe J, Bedford L. Pale body-like inclusion formation and neurodegeneration following depletion of 26S proteasomes in mouse brain neurones are independent of alpha-synuclein. PLoS One. 2013;8:e54711. doi: 10.1371/journal.pone.0054711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wahl C, Kautzmann S, Krebiehl G, Strauss K, Woitalla D, Muller T, Bauer P, Riess O, Kruger R. A comprehensive genetic study of the proteasomal subunit S6 ATPase in German Parkinson's disease patients. J Neural Transm. 2008;115:1141–1148. doi: 10.1007/s00702-008-0054-3. [DOI] [PubMed] [Google Scholar]
- 176.Schipper-Krom S, Juenemann K, Reits EA. The Ubiquitin- Proteasome System in Huntington's Disease: Are Proteasomes Impaired, Initiators of Disease, or Coming to the Rescue? Biochemistry research international. 2012;2012:837015. doi: 10.1155/2012/837015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 178.Snell RG, MacMillan JC, Cheadle JP, Fenton I, Lazarou LP, Davies P, MacDonald ME, Gusella JF, Harper PS, Shaw DJ. Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington's disease. Nature genetics. 1993;4:393–397. doi: 10.1038/ng0893-393. [DOI] [PubMed] [Google Scholar]
- 179.Sanchez I, Mahlke C, Yuan J. Pivotal role of oligomerization in expanded polyglutamine neurodegenerative disorders. Nature. 2003;421:373–379. doi: 10.1038/nature01301. [DOI] [PubMed] [Google Scholar]
- 180.Ordway JM, Tallaksen-Greene S, Gutekunst CA, Bernstein EM, Cearley JA, Wiener HW, Dure L.S.t., Lindsey R, Hersch SM, Jope RS, et al. Ectopically expressed CAG repeats cause intranuclear inclusions and a progressive late onset neurological phenotype in the mouse. Cell. 1997;91:753–763. doi: 10.1016/s0092-8674(00)80464-x. [DOI] [PubMed] [Google Scholar]
- 181.Kalchman MA, Graham RK, Xia G, Koide HB, Hodgson JG, Graham KC, Goldberg YP, Gietz RD, Pickart CM, Hayden MR. Huntingtin is ubiquitinated and interacts with a specific ubiquitin-conjugating enzyme. J Biol Chem. 1996;271:19385–19394. doi: 10.1074/jbc.271.32.19385. [DOI] [PubMed] [Google Scholar]
- 182.Hipp MS, Patel CN, Bersuker K, Riley BE, Kaiser SE, Shaler TA, Brandeis M, Kopito RR. Indirect inhibition of 26S proteasome activity in a cellular model of Huntington's disease. The Journal of cell biology. 2012;196:573–587. doi: 10.1083/jcb.201110093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hunter JM, Lesort M, Johnson GV. Ubiquitin-proteasome system alterations in a striatal cell model of Huntington's disease. Journal of neuroscience research. 2007;85:1774–1788. doi: 10.1002/jnr.21287. [DOI] [PubMed] [Google Scholar]
- 184.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 185.Holmberg CI, Staniszewski KE, Mensah KN, Matouschek A, Morimoto RI. Inefficient degradation of truncated polyglutamine proteins by the proteasome. Embo J. 2004;23:4307–4318. doi: 10.1038/sj.emboj.7600426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Venkatraman P, Wetzel R, Tanaka M, Nukina N, Goldberg AL. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol Cell. 2004;14:95–104. doi: 10.1016/s1097-2765(04)00151-0. [DOI] [PubMed] [Google Scholar]
- 187.Pratt G, Rechsteiner M. Proteasomes cleave at multiple sites within polyglutamine tracts: activation by PA28gamma(K188E). J Biol Chem. 2008;283:12919–12925. doi: 10.1074/jbc.M709347200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Seo H, Sonntag KC, Kim W, Cattaneo E, Isacson O. Proteasome activator enhances survival of Huntington's disease neuronal model cells. PLoS One. 2007;2:e238. doi: 10.1371/journal.pone.0000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Morris HR, Waite AJ, Williams NM, Neal JW, Blake DJ. Recent advances in the genetics of the ALS-FTLD complex. Curr Neurol Neurosci Rep. 2012;12:243–250. doi: 10.1007/s11910-012-0268-5. [DOI] [PubMed] [Google Scholar]
- 190.Tashiro Y, Urushitani M, Inoue H, Koike M, Uchiyama Y, Komatsu M, Tanaka K, Yamazaki M, Abe M, Misawa H, et al. Motor neuron-specific disruption of proteasomes, but not autophagy, replicates amyotrophic lateral sclerosis. J Biol Chem. 2012;287:42984–42994. doi: 10.1074/jbc.M112.417600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Schlossarek S, Carrier L. The ubiquitin-proteasome system in cardiomyopathies. Current opinion in cardiology. 2011;26:190–195. doi: 10.1097/HCO.0b013e32834598fe. [DOI] [PubMed] [Google Scholar]
- 192.Powell SR, Herrmann J, Lerman A, Patterson C, Wang X. The ubiquitin-proteasome system and cardiovascular disease. Prog Mol Biol Transl Sci. 2012;109:295–346. doi: 10.1016/B978-0-12-397863-9.00009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Tian Z, Zheng H, Li J, Li Y, Su H, Wang X. Genetically induced moderate inhibition of the proteasome in cardiomyocytes exacerbates myocardial ischemia-reperfusion injury in mice. Circ Res. 2012;111:532–542. doi: 10.1161/CIRCRESAHA.112.270983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Li J, Horak KM, Su H, Sanbe A, Robbins J, Wang X. Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia/reperfusion injury in mice. J Clin Invest. 2011;121:3689–3700. doi: 10.1172/JCI45709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Baraibar MA, Liu L, Ahmed EK, Friguet B. Protein oxidative damage at the crossroads of cellular senescence, aging, and age-related diseases. Oxid Med Cell Longev. 2012;2012:919832. doi: 10.1155/2012/919832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lee CK, Klopp RG, Weindruch R, Prolla TA. Gene expression profile of aging and its retardation by caloric restriction. Science. 1999;285:1390–1393. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- 197.Vernace VA, Arnaud L, Schmidt-Glenewinkel T, Figueiredo-Pereira ME. Aging perturbs 26S proteasome assembly in Drosophila melanogaster. Faseb J. 2007;21:2672–2682. doi: 10.1096/fj.06-6751com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Bajorek M, Finley D, Glickman MH. Proteasome disassembly and downregulation is correlated with viability during stationary phase. Curr Biol. 2003;13:1140–1144. doi: 10.1016/s0960-9822(03)00417-2. [DOI] [PubMed] [Google Scholar]
- 199.Grune T, Jung T, Merker K, Davies KJ. Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid, and ‘aggresomes’ during oxidative stress, aging, and disease. Int J Biochem Cell Biol. 2004;36:2519–2530. doi: 10.1016/j.biocel.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 200.Tomaru U, Takahashi S, Ishizu A, Miyatake Y, Gohda A, Suzuki S, Ono A, Ohara J, Baba T, Murata S, et al. Decreased proteasomal activity causes age-related phenotypes and promotes the development of metabolic abnormalities. Am J Pathol. 2012;180:963–972. doi: 10.1016/j.ajpath.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 201.Chondrogianni N, Petropoulos I, Franceschi C, Friguet B, Gonos ES. Fibroblast cultures from healthy centenarians have an active proteasome. Exp Gerontol. 2000;35:721–728. doi: 10.1016/s0531-5565(00)00137-6. [DOI] [PubMed] [Google Scholar]
- 202.Perez VI, Buffenstein R, Masamsetti V, Leonard S, Salmon AB, Mele J, Andziak B, Yang T, Edrey Y, Friguet B, et al. Protein stability and resistance to oxidative stress are determinants of longevity in the longest-living rodent, the naked mole-rat. Proc Natl Acad Sci U S A. 2009;106:3059–3064. doi: 10.1073/pnas.0809620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Salmon AB, Leonard S, Masamsetti V, Pierce A, Podlutsky AJ, Podlutskaya N, Richardson A, Austad SN, Chaudhuri AR. The long lifespan of two bat species is correlated with resistance to protein oxidation and enhanced protein homeostasis. Faseb J. 2009;23:2317–2326. doi: 10.1096/fj.08-122523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ungvari Z, Csiszar A, Sosnowska D, Philipp EE, Campbell CM, McQuary PR, Chow TT, Coelho M, Didier ES, Gelino S, et al. Testing predictions of the oxidative stress hypothesis of aging using a novel invertebrate model of longevity: the giant clam (Tridacna derasa). J Gerontol A Biol Sci Med Sci. 2013;68:359–367. doi: 10.1093/gerona/gls159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Chen Q, Thorpe J, Dohmen JR, Li F, Keller JN. Ump1 extends yeast lifespan and enhances viability during oxidative stress: central role for the proteasome? Free Radic Biol Med. 2006;40:120–126. doi: 10.1016/j.freeradbiomed.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 206.Tonoki A, Kuranaga E, Tomioka T, Hamazaki J, Murata S, Tanaka K, Miura M. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Molecular and cellular biology. 2009;29:1095–1106. doi: 10.1128/MCB.01227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kruegel U, Robison B, Dange T, Kahlert G, Delaney JR, Kotireddy S, Tsuchiya M, Tsuchiyama S, Murakami CJ, Schleit J, et al. Elevated proteasome capacity extends replicative lifespan in Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1002253. doi: 10.1371/journal.pgen.1002253. [DOI] [PMC free article] [PubMed] [Google Scholar]