Abstract
Lynch syndrome (LS) is a common cancer predisposition caused by an inactivating mutation in one of four DNA mismatch repair (MMR) genes. Frequently a variant of uncertain significance (VUS), rather than an obviously pathogenic mutation, is identified in one of these genes. The inability to define pathogenicity of such variants precludes targeted healthcare. Here, we have modified a cell-free assay to test VUS in the MMR gene PMS2 for functional activity We have analyzed nearly all VUS in PMS2 found thus far and describe loss of MMR activity for five, suggesting the applicability of the assay for diagnosis of LS.
Keywords: Lynch Syndrome, DNA Mismatch Repair, VUS, PMS2
LS predisposes to various cancers, most notably to colon and ovarian cancer [de la Chapelle, 2004]. LS is caused by heterozygous inactivating mutations in one of the MMR genes MSH2 (MIM# 609309), MSH6 (MIM# 600678), MLH1 (MIM# 120436) or PMS2 (MIM# 600259) [de la Chapelle, 2004]. Inadvertent loss of the second, wild type, allele in somatic cells results in MMR deficiency, which underlies the accumulation of spontaneous genomic mutations and the rapid development of cancer [de la Chapelle, 2004]. Confirmed LS patients enroll in lifelong preventive surveillance programs and may benefit from personalized chemoprevention and chemotherapy [Hewish et al., 2010; Burn et al., 2011]. Unfortunately, genetic diagnosis of LS patients is complicated by the fact that a significant fraction of all MMR gene alterations found are so-called Variants of Uncertain Significance (VUS) [de la Chapelle, 2004]. In the absence of data on the impact of the VUS on gene function, it is often difficult to interpret their pathogenicity. Moreover, the incidence of VUS is believed to increase steeply with the advent of personalized genomics [Rasmussen et al., 2012]. To enable personalized healthcare for carriers of pathogenic variants and to liberate unaffected relatives from the burden associated with the uncertain pathogenicity of the VUS, it is of great importance to develop procedures to evaluate their pathogenicity [Rasmussen et al., 2012].
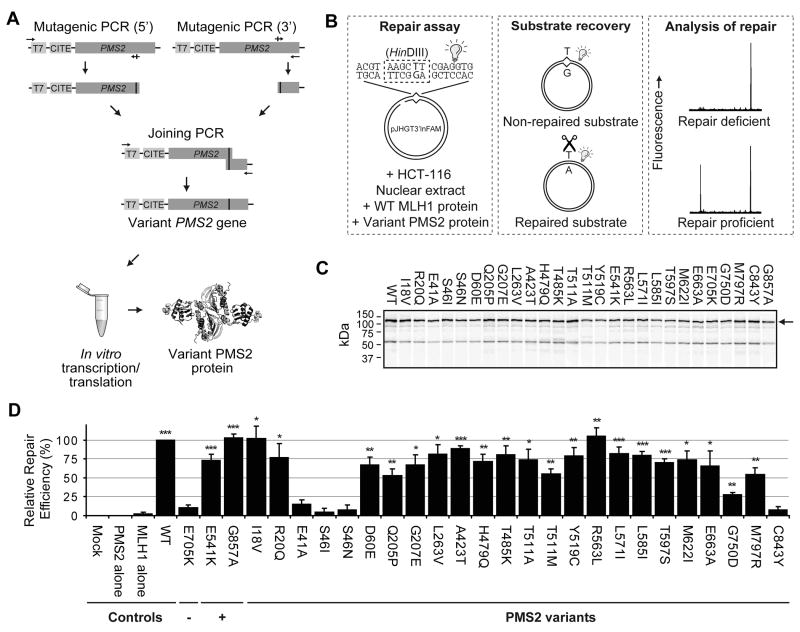
Of all MMR genes, VUS in PMS2 have the highest incidence, comprising ~49% of all alterations described in this gene (http://www.med.mun.ca/MMRvariants/statistics.aspx). We have recently described a cell-free assay to measure the functional activity of VUS in the MMR genes MSH2, MLH1 and MSH6 [Drost et al., 2010, 2012]. To facilitate the assessment of pathogenicity of VUS in PMS2 we have modified the cell-free assay to analyze their functional activity. In this assay the mutated cDNA is recreated by PCR, followed by in vitro transcription/translation of the variant PMS2 protein and of its wild type heterodimeric partner MLH1 (Figure 1A). To serve as template for the generation of the variant PMS2 alleles we used a wild type PMS2 fused to short S and thrombin tags, as this fusion protein displayed higher in vitro expression than the native PMS2 (Supp. Figure S1A, compare the first two lanes), while it did not affect its activity (Supp. Figure S1B). The variant MLH1/PMS2 heterodimer is added to an MLH1/PMS2-deficient cell extract and tested for its ability to restore a HinDIII restriction enzyme recognition site that is disrupted by an embedded G·T mismatch (Figure 1B). The inability to restore repair of the mismatch is indicative of the pathogenicity of a PMS2 VUS. All experimental procedures are described in the Supp. Materials & Methods. The assay appeared relatively insensitive to the amount of PMS2 included in the reaction, contributing to its robustness (Supp. Figure S1). The absolute repair efficiency of tagged wild type PMS2 under the conditions used in this assay is 43.5%±4.2 (mean±S.E.M.) and this is highly reproducible (Supp. Figure 1B). As the substrate concentration is in excess, absolute repair efficiencies are not a relevant measure for defects of a VUS. For this reason the in vitro MMR assay data is expressed as percentage of repair relative to wild type.
Figure 1. Mismatch repair activity of PMS2 VUS.
(A) Production of variant PMS2 alleles and proteins. All alleles, including template vector-derived T7 promoter and CITE sequences that are required for efficient transcription/translation in vitro, are generated by two sequential site-specific mutagenic PCR reactions. Variant PMS2 alleles are then used as a template in an in vitro transcription/translation reaction to produce variant PMS2 proteins.
(B) Flow scheme of the cell-free assay. Left: Fluorescently labelled (light bulb) substrate pJHGT3’lnFAM is incubated in HCT-116 nuclear extract and in vitro produced heterodimeric variant PMS2/wild type MLH1 protein. Middle: After incubation, the substrate is purified and digested. Right: Repair products are visualized by automated fragment analysis and quantified.
(C) Representative expression of 35S-Methionine-labeled variant PMS2 proteins, visualized after SDS-PAGE gel electrophoresis and autoradiography. Arrow: full-length PMS2 variants.
(D) Relative repair efficiencies for PMS2 VUS. (−): Repair-deficient control, (+): Repair-proficient controls. Results are shown as mean±S.E.M. of 3–4 independent experiments for all VUS and >6 experiments for controls. Mock: Mock expression. Asterisks: Significantly higher than repair-deficient control E705K. * p<0.05; ** p>0.01; *** p<0.001 (Student’s one-tailed t-test). For the “Mock” and “PMS2 only” reactions, no repair was detected in any of the experiments.
To test the applicability of the assay we have determined repair efficiencies of 27 VUS in PMS2, representing the large majority of all VUS registered in the Leiden Open Variation Database (LOVD; Table 1). Of these, variant E705K served as an MMR-deficient, pathogenic, control [Deschênes et al., 2007; van Oers et al., 2010]. Additionally, variants E541K and G857A were included as innocuous polymorphisms, as judged from their high allele frequencies (dbSNP rs2228006 and rs1802683, respectively). PMS2 variants registered in the LOVD as PMS2 pseudogene-derived were excluded from analysis. All alleles were recreated by PCR and protein was produced in vitro (Figure 1C).
Table 1.
Pathology data and references for PMS2 VUS tested in this work
| Mutation
|
In vitro MMR (This work) | Pathologyc,d
|
Referencesd | |||
|---|---|---|---|---|---|---|
| Proteina | DNAb | MSI | IHC
|
|||
| PMS2 | MLH1 | |||||
| I18V | c.52A>G | + | 1, 2 | |||
| R20Q | c.59G>A | + | H3 | 1, 2, 3, 4, 5 | ||
| E41A | c.122A>C | - | 5 | |||
| S46I | c.137G>T | - | H3, 6 | Neg3, 6 | Pos6 | 2, 3, 4, 6, 7, 8 |
| S46N | c.137G>A | - | 7, 8 | |||
| D60E | c.180C>G | + | LOVD | |||
| Q205P | c.614A>C | + | LOVD | |||
| G207E | c.620G>A | + | H9 | Neg9 | 9 | |
| L263V | c.787C>G | + | LOVD | |||
| A423T | c.1267G>A | + | 10 | |||
| H479Q | c.1437C>G | + | 11, 12 | |||
| T485K | c.1454C>A | + | H6 | Neg6 | Pos6 | 1, 2, 4, 6, 11 |
| T511A | c.1531A>G | + | H3 | 2, 3, 6, 11, 12 | ||
| T511M | c.1532C>T | + | LOVD | |||
| Y519C | c.1556A>G | + | LOVD | |||
| E541K | c.1621A>G | + | 1, 2, 5, 6 | |||
| R563L | c.1688G>T | + | 1, 2, 8 | |||
| L571I | c.1711C>A | + | 2 | |||
| L585I | c.1753C>A | + | 1 | |||
| T597S | c.1789A>T | + | L3 | 1, 3, 4, 5, 11 | ||
| M622I | c.1866G>A | + | 1, 2, 4, 6, 8, 11 | |||
| E663A | c.1988A>C | + | 8 | |||
| E705K | c.2113G>A | - | 2, 6, 8, 13, 14 | |||
| G750D | c.2249G>A | + | 8 | |||
| M797R | c.2390T>G | + | 2 | |||
| C843Y | c.2528G>A | - | 8 | |||
| G857A | c.2570G>C | + | 1, 2, 6, 12 | |||
Amino acid numbering is based on the PMS2 reference sequence NP_000526.1 with +1 corresponding to the translation initiation amino acid.
Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the translation initiation codon of the PMS2 GenBank reference sequence (NM_000535.5).
MSI=Microsatellite Instability. H=High, L=Low, IHC=Immunohistochemistry. Neg = Negative for staining, Pos = Positive for staining. In case cells are empty, the variant was not tested.
Variants have been selected from the LOVD (http://chromium.liacs.nl/LOVD2/colon_cancer/variants.php?select_db=PMS2&action=view_all&view=Prot_sub). Appropriate references are shown. References: 1 [Hendriks et al., 2006], 2 [Clendenning et al., 2006], 3 [Pastrello et al., 2011], 4 [Thompson et al., 2012], 5 [Borràs et al., 2013], 6 [Nakagawa, 2004], 7 [Jackson et al., 2008], 8 [Senter et al., 2008], 9 [Montazer Haghighi et al., 2009], 10 [Ganster et al., 2010], 11 [Wang et al., 1999], 12 [Basil et al., 1999], 13 [Deschênes et al., 2007], 14 [van Oers et al., 2010].
As the polymorphic PMS2 alleles enabled repair activities significantly higher than the known pathogenic control E705K, the assay has sufficient resolution to distinguish repair-proficient from repair-deficient VUS (Figure 1D). Variants with repair efficiencies not significantly higher than the pathogenic control (E41A, S46I, S46N and C843Y) were considered repair deficient and therefore presumably are pathogenic (Figure 1D). We conclude that this assay effectively identifies repair-deficient PMS2 variants. Variants such as Q205P, T511M and G750D that display repair efficiencies significantly higher than E705K but are compromised compared to wild type were not classified as repair deficient and we surmise that these variants might be pathogenic with reduced penetrance. Extensive calibration of the assay with clinical data is required to assess pathogenicity of alleles displaying such intermediate repair efficiencies.
For some of these VUS microsatellite instability (MSI), a hallmark of MMR deficient cancer, was previously investigated. Indeed, the MSI of a tumor carrying the S46I allele corresponds with its deficiency in MMR (Table 1). However, the MSI in tumors of carriers of the R20Q, G207E, T485K or T511A alleles is in apparent contrast with their normal MMR activity in vitro (Table 1). Possibly these patients carry another, yet unidentified MMR gene defect. Contrariwise, we cannot exclude that these alleles cause a defect that is only apparent in vivo. For this reason, all variants that are repair proficient in this assay cannot be classified as neutral, but may require additional analyses such as splicing assays, protein stability assays or nuclear localization assays [Rasmussen et al., 2012]. Ultimately, after the calibration of the assay, it may become a part of an integrated Bayesian analysis that determines pathogenicity of MMR gene VUS, as proposed by us and others [Goldgar et al., 2008; Rasmussen et al., 2012].
Supplementary Material
Acknowledgments
We thank Jacob G. Jansen for his comments on the manuscript. This work was supported by NIH grant 1R01CA164944-01A1.
Abbreviations used in this paper
- LS
Lynch syndrome
- MMR
DNA Mismatch Repair
- PMS2
Postmeiotic Segregation Increased 2
- VUS
Variant of Uncertain Significance
Footnotes
The authors declare no conflict of interest.
Supporting Information for this preprint is available from the Human Mutation editorial office upon request (humu@wiley.com)
References
- Basil JB, Swisher EM, Herzog TJ, Rader JS, Elbendary A, Mutch DG, Goodfellow PJ. Mutational analysis of the PMS2 gene in sporadic endometrial cancers with microsatellite instability. Gynecol Oncol. 1999;74:395–399. doi: 10.1006/gyno.1999.5486. [DOI] [PubMed] [Google Scholar]
- Borràs E, Pineda M, Cadiñanos J, Del Valle J, Brieger A, Hinrichsen I, Cabanillas R, Navarro M, Brunet J, Sanjuan X, Musulen E, van der Klift H, et al. Refining the role of pms2 in Lynch syndrome: germline mutational analysis improved by comprehensive assessment of variants. J Med Genet. 2013;50:552–63. doi: 10.1136/jmedgenet-2012-101511. [DOI] [PubMed] [Google Scholar]
- Burn J, Gerdes AM, Macrae F, Mecklin JP, Moeslein G, Olschwang S, Eccles D, Evans DG, Maher ER, Bertario L, Bisgaard ML, Dunlop MG, et al. Long-term effect of aspirin on cancer risk in carriers of hereditary colorectal cancer: an analysis from the CAPP2 randomised controlled trial. Lancet. 2011;378:2081–2087. doi: 10.1016/S0140-6736(11)61049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clendenning M, Hampel H, LaJeunesse J, Lindblom A, Lockman J, Nilbert M, Senter L, Sotamaa K, de la Chapelle A. Long-range PCR facilitates the identification of PMS2-specific mutations. Hum Mutat. 2006;27:490–495. doi: 10.1002/humu.20318. [DOI] [PubMed] [Google Scholar]
- de la Chapelle A. Genetic predisposition to colorectal cancer. Nat Rev Cancer. 2004;4:769–780. doi: 10.1038/nrc1453. [DOI] [PubMed] [Google Scholar]
- Deschênes SM, Tomer G, Nguyen M, Erdeniz N, Juba NC, Sepúlveda N, Pisani JE, Liskay RM. The E705K mutation in hPMS2 exerts recessive, not dominant, effects on mismatch repair. Cancer Lett. 2007;249:148–156. doi: 10.1016/j.canlet.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost M, Zonneveld JB, Van Dijk L, Morreau H, Tops CM, Vasen HFA, Wijnen J, De Wind N. A cell-free assay for the functional analysis of variants of the mismatch repair protein MLH1. Hum Mutat. 2010;31:247–253. doi: 10.1002/humu.21180. [DOI] [PubMed] [Google Scholar]
- Drost M, Zonneveld JB, van Hees S, Rasmussen LJ, Hofstra RM, De Wind N. A rapid and cell-free assay to test the activity of lynch syndrome-associated MSH2 and MSH6 missense variants. Hum Mutat. 2012;33:488–494. doi: 10.1002/humu.22000. [DOI] [PubMed] [Google Scholar]
- Ganster C, Wernstedt A, Kehrer-Sawatzki H, Messiaen L, Schmidt K, Rahner N, Heinimann K, Fonatsch C, Zschocke J, Wimmer K. Functional PMS2 hybrid alleles containing a pseudogene-specific missense variant trace back to a single ancient intrachromosomal recombination event. Hum Mutat. 2010;31:552–560. doi: 10.1002/humu.21223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldgar DE, Easton DF, Byrnes GB, Spurdle AB, Greenblatt MS Group IARCUGVW. Genetic evidence and integration of various data sources for classifying uncertain variants into a single model. Hum Mutat. 2008;29:1265–1272. doi: 10.1002/humu.20897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks YM, Jagmohan-Changur S, van der Klift HM, Morreau H, van Puijenbroek M, Tops C, van Os T, Wagner A, Ausems MG, Gomez E, Breuning MH, Bröcker-Vriends AH, et al. Heterozygous mutations in PMS2 cause hereditary nonpolyposis colorectal carcinoma (Lynch syndrome) Gastroenterology. 2006;130:312–322. doi: 10.1053/j.gastro.2005.10.052. [DOI] [PubMed] [Google Scholar]
- Hewish M, Lord CJ, Martin SA, Cunningham D, Ashworth A. Mismatch repair deficient colorectal cancer in the era of personalized treatment. Nat Rev Clin Oncol. 2010;7:197–208. doi: 10.1038/nrclinonc.2010.18. [DOI] [PubMed] [Google Scholar]
- Holmes J, Clark S, Modrich PL. Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc Natl Acad Sci USA. 1990;87:5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montazer Haghighi M, Radpour R, Aghajani K, Zali N, Molaei M, Zali M. Four novel germline mutations in the MLH1 and PMS2 mismatch repair genes in patients with hereditary nonpolyposis colorectal cancer. Int J Colorectal Dis. 2009;24:885–893. doi: 10.1007/s00384-009-0731-1. [DOI] [PubMed] [Google Scholar]
- Nakagawa H. Mismatch Repair Gene PMS2: Disease-Causing Germline Mutations Are Frequent in Patients Whose Tumors Stain Negative for PMS2 Protein, but Paralogous Genes Obscure Mutation Detection and Interpretation. Cancer Res. 2004;64:4721–4727. doi: 10.1158/0008-5472.CAN-03-2879. [DOI] [PubMed] [Google Scholar]
- Pastrello C, Pin E, Marroni F, Bedin C, Fornasarig M, Tibiletti M, Oliani C, De Leon M, Urso E, Puppa L, Agostini M, Viel A. Integrated analysis of unclassified variants in mismatch repair genes. Genet Med. 2011;13:115–124. doi: 10.1097/GIM.0b013e3182011489. [DOI] [PubMed] [Google Scholar]
- Rasmussen LJ, Heinen CD, Royer-Pokora B, Drost M, Tavtigian S, Hofstra RM, De Wind N. Pathological assessment of mismatch repair gene variants in Lynch syndrome: Past, present, and future. Hum Mutat. 2012;33:1617–1625. doi: 10.1002/humu.22168. [DOI] [PubMed] [Google Scholar]
- Senter L, Clendenning M, Sotamaa K, Hampel H, Green J, Potter JD, Lindblom A, Lagerstedt K, Thibodeau SN, Lindor N, Young J, Winship I, et al. The Clinical Phenotype of Lynch Syndrome Due to Germ-Line PMS2 Mutations. Gastroenterology. 2008;135:419–428. doi: 10.1053/j.gastro.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B, Greenblatt M, Vallee M, Herkert J, Tessereau C, Young E, Adzhubey I, Li B, Bell R, Feng B, Mooney S, Radivojac P, et al. Calibration of Multiple In Silico Tools for Predicting Pathogenicity of Mismatch Repair Gene Missense Substitutions. Hum Mutat. 2012;34:255–265. doi: 10.1002/humu.22214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oers JM, Roa S, Werling U, Liu Y, Genschel J, Hou H, Sellers RS, Modrich P, Scharff MD, Edelmann W. PMS2 endonuclease activity has distinct biological functions and is essential for genome maintenance. Proc Natl Acad Sci USA. 2010;107:13384–13389. doi: 10.1073/pnas.1008589107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Lasset C, Desseigne F, Saurin JC, Maugard C, Navarro C, Ruano E, Descos L, Trillet-Lenoir V, Bosset JF, Puisieux A. Prevalence of germline mutations of hMLH1, hMSH2, hPMS1, hPMS2, and hMSH6 genes in 75 French kindreds with nonpolyposis colorectal cancer. Hum Genet. 1999;105:79–85. doi: 10.1007/s004399900064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.