SUMMARY
The Unfolded Protein Response (UPR) is composed by homeostatic signaling pathways that are activated by excessive protein misfolding in the endoplasmic reticulum (ER). Ire1 signaling is an important mediator of the UPR, leading to the activation of the transcription factor Xbp1. Here, we show that Drosophila Ire1 mutant photoreceptors have defects in the delivery of Rhodopsin-1 to the rhabdomere and in the secretion of Spacemaker/Eyes shut into the inter-rhabdomeral space. However, these defects are not observed in Xbp1 mutant photoreceptors. Ire1 mutant retinas have higher mRNA levels for targets of regulated Ire1-dependent decay (RIDD), including for the fatty acid transport protein (fatp). Importantly, downregulation of fatp by RNA interference rescues the Rhodopsin-1 delivery defects observed in Ire1 mutant photoreceptors. Our results show that the role of Ire1 during photoreceptor differentiation is independent of Xbp1 function and demonstrate the physiological relevance of the RIDD mechanism in this specific paradigm.
INTRODUCTION
The endoplasmic reticulum (ER) is the cell organelle where secretory and membrane proteins are synthesized and folded. When the folding capacity of the ER is impaired, the presence of incorrectly folded (misfolded) proteins in the ER causes ER stress and activates the Unfolded Protein Response (UPR), which helps to restore homeostasis in the ER (Ron and Walter, 2007; Walter and Ron, 2011). In higher eukaryotes, the activation of the UPR is accomplished via three signaling pathways induced by ER-resident molecular ER stress sensors: Protein kinase (PKR)-like ER kinase (PERK), Activating transcription factor 6 (ATF6) and Inositol-requiring enzyme 1 (Ire1).
Being conserved in all eukaryotes, Ire1 contains an ER luminal domain, which is involved in the recognition of misfolded proteins (Credle et al., 2005; Gardner and Walter, 2011), and cytoplasmic endoribonuclease and kinase domains, which are involved in the activation of downstream pathways. Activated Ire1 mediates the non-conventional splicing of an intron from X-box binding protein 1 (Xbp1) mRNA (or HAC1 mRNA, the yeast Xbp1 ortholog), causing a frame-shift during translation, thereby introducing a new carboxyl domain in the Xbp1 protein (Cox and Walter, 1996; Mori et al., 1996; Yoshida et al., 2001; Calfon et al., 2002; Shen et al., 2001). Xbp1spliced is an effective transcription factor that regulates the expression of ER chaperones and other target genes (Acosta-Alvear et al., 2007).
In addition to mediating Xbp1 mRNA splicing, cell culture studies demonstrated that Ire1 promotes the degradation of mRNAs encoding ER-targeted proteins, a processed called RIDD (regulated Ire1-dependent decay), to reduce the load of ER client proteins during ER stress (Hollien and Weissman, 2006; Han et al., 2009; Hollien et al., 2009). The cytosolic domain of mammalian IRE1 binds Traf2 (tumour necrosis factor receptor-associated factor 2, Urano, 2000), an upstream activator of the c-Jun N-terminal Kinase (JNK) signaling pathway. This IRE1/Traf2 interaction is also independent of Xbp1 splicing and may lead to the activation of apoptosis after prolonged ER stress (Yoneda et al., 2001).
In the Drosophila photoreceptor cells, the rhabdomere is the light sensing organelle, a stack of photosensitive apical microvilli that is formed during the second half of pupal development (Cagan and Ready, 1989; Tepass and Harris, 2007). The rhabdomere is formed in the apical domain of each photoreceptor cell, which after a 90° rotation extends its apical domain along the proximal-distal axis of the retina. The growth of the rhabdomere requires the delivery of large amounts of membrane and proteins into this structure, imposing a considerable demand to the cellular mechanisms controlling protein folding and membrane production in the ER.
Among the proteins targeted to the developing rhabdomeres are the rhodopsins, the light sensitive proteins, and other proteins involved in the transduction of the light stimuli. Rhodopsin-1 (Rh1) is a seven transmembrane domain protein that starts to be expressed by 78% of pupal life (Kumar and Ready, 1995), and is delivered to the rhabdomeres of the outer photoreceptors (R1–R6), in a trafficking process that requires the activity of Rab11, MyosinV and dRip11 (Li et al., 2007; Satoh et al., 2005). The delivery of Rh1 to the rhabdomere is required for rhabdomere morphogenesis, since in Rh1 null mutants the rhabdomere does not form, causing degeneration of the photoreceptors (Kumar et al., 1997; Kumar and Ready, 1995).
In mammalians, the microRNA mir-708 is up-regulated by CHOP to control Rhodopsin expression levels and prevent an excessive Rhodopsin load into the ER (Behrman et al., 2011). In Drosophila, Ire1 signaling is activated in the photoreceptors upon expression of Rh1 folding mutants (Ryoo et al., 2007; Griciuc et al., 2010; Kang and Ryoo, 2009) or in ninaA mutations that cause the accumulation of misfolded Rh1 in the ER (Mendes et al., 2009). However, the role of Ire1 signaling during normal photoreceptor differentiation remains unknown. In this study, we show that Ire1 signaling is activated in the photoreceptors during pupal stages of Drosophila development. Ire1 mutant photoreceptors have defects in the delivery of Rh1 to the rhabdomere and the secretion of Spacemaker/Eyes shut (Spam/Eys) into the inter-rhabdomeral space. Surprisingly, Xbp1 null mutant photoreceptors have a milder phenotype with no defects in Rh1 delivery into the rhabdomere or spam/eys secretion. Targets of regulated Ire1-dependent decay (RIDD) are up-regulated in Ire1 mutant retinas, including the fatty acid transport protein (fatp), a known regulator of Rh1 protein levels (Dourlen et al., 2012). Finally, we show that the regulation of fatp levels by RIDD is critical for normal Rh1 delivery into the rhabdomere.
RESULTS
Ire1 signaling is activated in the photoreceptors during pupal stages
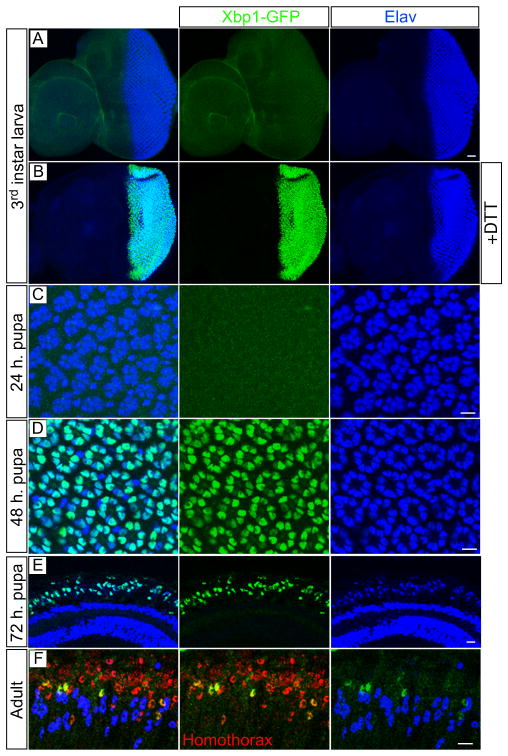
To detect Ire1 signaling activity in the photoreceptors during development, we used the Xbp1-EGFP reporter (Ryoo et al., 2007), in which EGFP becomes in frame with Xbp1 only upon Ire1-dependent mRNA splicing. We expressed Xbp1-EGFP in the developing eye under the control of the GMR-Gal4 driver, which is active during larval stages in the eye imaginal disc posterior to the morphogenetic furrow and in all cells of the eye during pupal and adult stages. During larval stages, endogenous Xbp1-EGFP was not observed in the photoreceptors (Figure 1A), but it could be induced upon culture of the imaginal discs in 2 mM dithiotheitol (DTT) for 4 hours (Figure 1B), a condition that interferes with oxidative protein folding in the ER and activates Ire1. During pupal stages, Xbp1-EGFP was not observed in the photoreceptors at 24 hours of pupal life (Figure 1C), but it became evident during the second half of pupal development (Figures 1D, 48 hours and 1E, 72 hours). In the adult, Xbp1-EGFP was observed overlapping with some Homothorax positive lattice cells (Wildonger et al., 2005), but not in the photoreceptors (Elav marker). These results show that Ire1 signaling is transiently activated in the photoreceptors during mid and late stages of pupal development.
Figure 1.
The Xbp1-EGFP Ire1 signaling reporter is activated in the photoreceptors during pupal stages. In all panels UAS-Xbp1-EGFP (green) is expressed under the control of GMR-Gal4 and Elav (blue) is a neuronal marker expressed in the photoreceptors. (A and B) Third instar larval stage eye/antenna imaginal disc (posterior is to the right). (A) Endogenous GFP expression is not observed in GMR-Gal4>UAS-Xbp1-EGFP. (B) Culture of GMR-Gal4>UAS-Xbp1-EGFP imaginal discs in 2 mM DTT for 4 hours activates Xbp1-EGFP. (C) Xbp1-EGFP is not observed at 24 h pupa. (D) Xbp1-EGFP is observed in the photoreceptors at 48 hours and (E) 72 hours of pupal development. (F) In the adult, Xbp1-EGFP is observed in some Homothorax (red) positive lattice cells, but not in the photoreceptors. (C and D) are tangential views of whole-mount pupal eyes and (E and F) are horizontal sections of the eye oriented with distal to the top. Scale bars, 10 μm.
Activation of Xbp1-EGFP in the photoreceptors requires Ire1
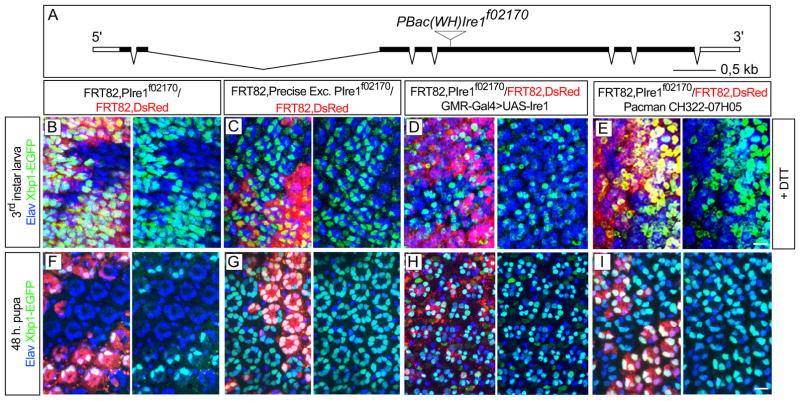
To test if the Xbp1-EGFP reporter expression observed in the photoreceptors corresponds to bona fide Ire1 activity, we did experiments where we tested Xbp1-EGFP expression in mosaic eyes containing Ire1 mutant clones. The homozygous lethal line PBac{WH}Ire1f02170 contains a piggyBac transposon insertion in the open reading frame of Ire1 (Figure 2A). We recombined PBac{WH}Ire1f02170 into FRT82B to make mosaic clones with the Flipase/FRT technique (Golic, 1991). Mosaic eyes were induced with eyeless-Flipase and Xbp1-EGFP was expressed under GMR-Gal4 control in order to be analyzed both in the 3rd larval instar (Figures 2B–2E) and pupal stages (Figures 2F–2I). Xbp1-EGFP was not observed in clones homozygous for PBac{WH}Ire1f02170 either in larval eye discs treated with 2 mM DTT (Figure 2B) or in the pupa (Figure 2F). Xbp1-EGFP expression was observed in clones of a precise excision of PBac{WH}Ire1f02170 that reverts the Ire1 genomic region to wild-type (Figures 2C and 2G). Expression of UAS-Ire1 under the control of GMR-Gal4 (Figures 2D and 2H) or a genomic construct covering the Ire1 region (Pacman CH322-07H05, Figures 2E and 2I) rescued Xbp1-EGFP expression in the background of PBac{WH}Ire1f02170 clones. Ire1 kinase (Ire1K576A) or RNase (Ire1H890A) mutants failed to rescue Xbp1-EGFP expression (Figure S1). These results demonstrate that the PBac{WH}Ire1f02170 insertion is a strong mutant allele of Ire1 and that Xbp1-EGFP activation in the photoreceptors requires Ire1 activity.
Figure 2.
Ire1 is required for Xbp1-EGFP activation in the Drosophila eye (A) Schematic of Ire1 genomic region with the PBac{WH}Ire1f02170 piggyBac transposon insertion in the open reading frame of Ire1. (B–E) Third instar stage larval eyes (posterior to the right) with eyeless-Flipase induced clones of PBac{WH}Ire1f02170 homozygous cells, labeled by the absence of UAS-DsRed, were treated with 2 mM DTT for 4 hours to activate Xbp1-EGFP (green). (B) Xbp1-EGFP is not observed in cells homozygous for PBac{WH}Ire1f02170. (C) Xbp1-EGFP is observed in clones of a precise excision of PBac{WH}Ire1f02170 that restores the genomic region. (D) Expression of UAS-Ire1 under the control of GMR-Gal4 or from (E) CH322-07H05 (Pacman) genomic construct covering Ire1 region rescues Xbp1-EGFP activation in PBac{WH}Ire1f02170 homozygous cells. (F–I) Pupal eyes (48 h.) with eyeless-Flipase induced clones of PBac{WH}Ire1f02170 homozygous cells, labeled by the absence of UAS-DsRed. (F) Endogenous Xbp1-EGFP in the photoreceptors is not observed in cells homozygous for PBac{WH}Ire1f02170. (G) Xbp1-EGFP is observed in clones of a precise excision of PBac{WH}Ire1f02170. (H) Expression of UAS-Ire1 under the control of GMR-Gal4 or from (I) CH322-07H05 (Pacman) genomic construct rescues Xbp1-EGFP activation in PBac{WH}Ire1f02170 homozygous cells. Elav is in blue. Scale bars, 10 μm.
Ire1 is not required for photoreceptor specification or for the establishment and maintenance of apical/basal polarity in the photoreceptors
The expression of Xbp1-EGFP in the photoreceptors during the second half of pupal life suggests that Ire1 signaling might be required for photoreceptor differentiation during these stages. However, it is also possible that the Xbp1-EGFP reporter is not sensitive enough to detect low levels of Ire1 activity that could have a role during earlier stages of photoreceptor development. To test this possibility, we analyzed the expression of photoreceptor specification markers in the 3rd larval instar stage in Ire1 mutant clones. Expression of Elav (pan-neural marker) and Runt (marker of R7 and R8, Kaminker et al., 2002) was normal in Ire1 homozygous mutant clones (Figure 3A). We also analyzed markers of apical/basal polarity, since rhabdomere morphogenesis in the pupa requires proper establishment and maintenance of apical/basal polarity in the photoreceptors (Pellikka et al., 2002; Izaddoost et al., 2002; Knust, 2007; Walther and Pichaud, 2010). In the 40h–50h pupa, Ire1 mutant clones exhibited normal localization of Armadillo (Figure 3B), DaPKC (Figure 3B), Crumbs (Figure 3C) and Cadherin (Figure S2) in the photoreceptors, indicating no defect in apical/basal polarity at these stages.
Figure 3.
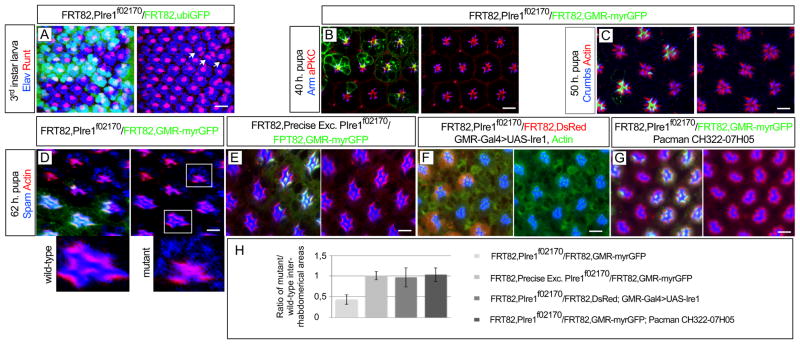
Ire1 is required for spam/eys secretion and formation of the inter-rhabdomeral space. (A) Third instar stage larval eyes (posterior to the right) with eyeless-Flipase induced clones of PBac{WH}Ire1f02170 homozygous cells, labeled by the absence of ubiGFP (green), show normal expression of the photoreceptor specification markers Elav (blue) and Runt (Red) (arrows). (B) 40 h pupal eye with PBac{WH}Ire1f02170 homozygous cells, labeled by the absence of myrGFP (green), show normal localization of arm (blue) and DaPKC (Red). (C) 50 h pupa with PBac{WH}Ire1f02170 homozygous cells, labeled by the absence of myrGFP (green), show normal localization of crumbs (blue). (D) By 62 h pupation, PBac{WH}Ire1f02170 homozygous ommatidia (absence of myrGFP, green) have low levels of Spam/Eys (blue) in the inter-rhabdomeral space (IRS) and present retention of Spam/Eys in the photoreceptors cell body. The rhabdomeres are stained with actin (red). The magnified mutant and control ommatidia are indicated with white squares in the figure. (E) Clones of a precise excision of PBac{WH}Ire1f02170 (absence of myrGFP, green) have normal localization of Spam/Eys (blue) in the IRS. (F) GMR-Gal4>UAS-Ire1 rescues Spam/Eys (blue) localization in PBac{WH}Ire1f02170 homozygous ommatidia (absence of DsRed). (G) CH322-07H05 genomic construct rescues Spam/Eys (blue) localization in PBac{WH}Ire1f02170 homozygous ommatidia (absence myrGFP, green). (H) Quantification of IRS size; average of the ratio of the IRS areas of adjacent homozygous mutant and wild-type ommatidia. Around 40 pairs of mutant/wild type ommatidia were quantified for each experimental condition indicated in the figure. Scale bars, 10 μm.
Ire1 is required for Spacemaker/Eyes shut secretion and formation of the inter-rhabdomeral space
The inter-rhabdomeral space (IRS) is an extracellular lumen between the rhabdomeres enclosed by the 4 cone cells that surround each ommatidial cluster. The formation of the IRS requires the secretion by the photoreceptors of the agrin/perclan-related protein Spacemaker/Eyes shut (Spam/Eys) (Husain et al., 2006; Zelhof et al., 2006). By 62 h pupation, Spam/Eys was observed in IRS of wild-type ommatidia. In contrast, Ire1 mutant ommatidia had low levels of Spam/Eys in the IRS, with Spam/Eys retained in the photoreceptor cell body (Figure 3D). As consequence of this defect in Spam/Eys secretion, Ire1 mutant ommatidia showed an IRS with reduced size, which we quantified by measuring the IRS area of adjacent wild-type and Ire1 mutant ommadia (Figure 3H). This defect in Spam/Eys secretion and IRS formation was not observed in clones of a precise excision of PBac{WH}Ire1f02170 (Figure 3E) and could be rescued by GMR-Gal4>UAS-Ire1 (Figure 3F) or the CH322-07H05 genomic construct covering the Ire1 region (Figure 3G).
Ire1 is required for Rh1 delivery into the rhabdomere
By 72h pupa, we observed that Ire1 mutant photoreceptors have defective delivery of Rh1 into the rhabdomeres, with retention of Rh1 in the photoreceptors cell body (Figure 4A). We observed this phenotype in later pupal stages and in the adult, with concomitant degeneration of the rhabdomeres (Figure S3). This defect in Rh1 delivery into the rhabdomere was not observed in clones of a precise excision of PBac{WH}Ire1f02170 (Figure 4B) and could be rescued by GMR-Gal4>UAS-Ire1 (Figure 4C) and CH322-07H05 genomic construct (Figure 4D). Ire1 kinase (Ire1K576A) or RNase (Ire1H890A) mutants failed to rescue Rh1 localization defect (Figure S1). We also observed that Ire1 mutant photoreceptors have reduced levels of ER markers, such as the ER chaperone BiP/GRP78 (Figures 4E, S4A and S4B) and the Rh1 specific chaperone ninaA (Figure 4F and S4A). The levels of the ER chaperone Calnexin (Cnx), however, were unchanged between wild-type and Ire1 mutant cells (Figure 4G).
Figure 4.
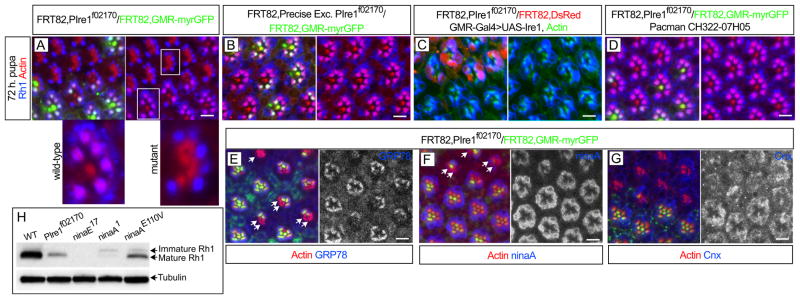
Ire1 is required for Rh1 delivery into the rhabdomere. (A) 72 h pupal eye with eyeless-Flipase induced clones of PBac{WH}Ire1f02170 homozygous cells, labeled by the absence of myrGFP (green), show defective delivery of Rh1 (blue) into the rhabdomeres (actin, red). The magnified mutant and control ommatidia are indicated with white squares. (B) Rh1 (blue) delivery into the rhabdomeres is normal in clones of a precise excision of PBac{WH}Ire1f02170 (labeled by the absence of myrGFP, green). (C) GMR-Gal4>UAS-Ire1 expression rescues Rh1 (blue) delivery into the rhabdomeres of PBac{WH}Ire1f02170 homozygous cells (labeled by the absence of DsRed). (D) The CH322-07H05 genomic construct rescues Rh1 (blue) delivery into the rhabdomeres of PBac{WH}Ire1f02170 homozygous cells (absence of myrGFP, green). (E–G) PBac{WH}Ire1f02170 homozygous cells (absence of myrGFP, green) have reduced levels (arrows) of (E) BiP/GRP78 (blue and monochrome) and (F) ninaA (blue and monochrome), but not (G) Cnx (blue and monochrome). (H) Western blot analysis of Rh1 mature (34 kDa) and immature (40 kDa) forms from adult head protein extracts of yw (WT, positive control), FRT82BPBac{WH}Ire1f02170/FRT82BCL,GMR-hid (whole eye is composed of homozygous Ire1 mutant cells as in (Stowers and Schwarz, 1999)), ninaE17 (Rh1 protein null) and 2 ninaA mutations (ninaA1=ninaAP228 and ninaAE110V). Tubulin is used as loading control. Scale bars, 10 μm.
The cytoplasmic retention of Rh1 and the lower levels of the ER chaperones BiP/GRP78 and ninaA, suggested that Rh1 could be accumulating as misfolded protein in Ire1 mutant photoreceptors. In ninaA mutations, Rh1 accumulates as an immature, glycosylated form, while the levels of mature Rh1 are reduced (Colley et al., 1991; Ondek et al., 1992; Baker et al., 1994). We performed Western Blot on protein extracts from Ire1 mutant eyes and although Rh1 mature form levels were reduced, no immature Rh1 form was observed (Figure 4H). This result indicates that, although ninaA levels are reduced in Ire1 mutant photoreceptors, residual ninaA activity could be sufficient to process Rh1 to the mature form.
Xbp1 is not required for Spacemaker/Eyes shut secretion or Rh1 delivery into the rhabdomere
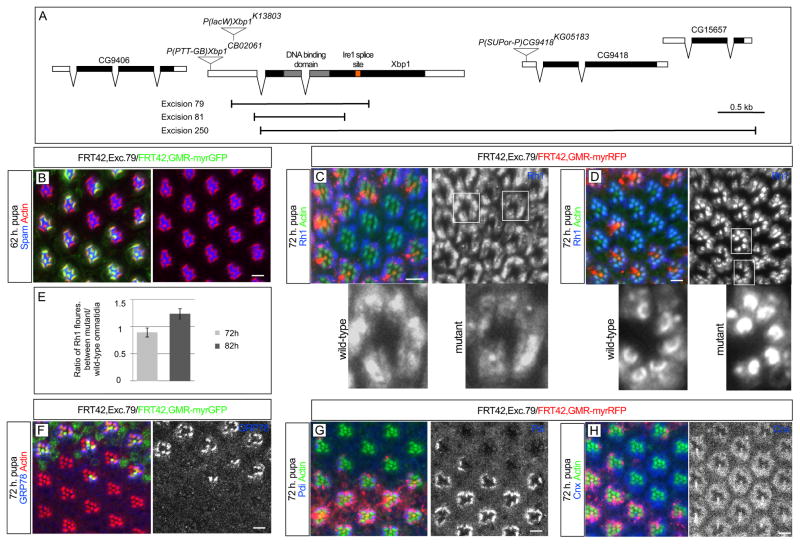
Next, we investigated if Xbp1 mutants also exhibit the same defects in Spam/Eys secretion and Rh1 delivery into the rhabdomere as observed in Ire1 mutant photoreceptors. We previously used a lethal P element insertion upstream of Xbp1 open reading frame, P{lacW}Xbp1K13803, which accelerates retinal degeneration induced by ninaE dominant mutations (Ryoo et al., 2007). Clones of P{lacW}Xbp1K13803 did not present the defects in Spam/Eys secretion and Rh1 delivery into the rhabdomere observed in the Ire1 mutation (data not shown). However, P{lacW}Xbp1K13803 still has residual expression of Xbp1 mRNA (Ryoo et al., 2007), that could be sufficient to rescue potential defects in the photoreceptors. For this reason, we decided to generate Xbp1 protein null deletion mutants with no residual function of the Xbp1 protein. Using Δ23 transposase, we induced imprecise excisions of 2 P elements (P[PTT-GB]Xbp1CB02061and P[SUPor-P]CG9418KG05183) neighboring Xbp1, which resulted in the identification of 3 different deletion mutations (Excisions 79, 81 and 250, Figure 5A). We recombined each deletion mutant into FRT42D chromosome to generate mosaic clones in the eye. We observed identical phenotypes with these 3 Xbp1 deletion mutants.
Figure 5.
Xbp1 is not required for Spacemaker/Eyes shut secretion or Rh1 delivery into the rhabdomere. (A) Schematic of Xbp1 genomic region with the localization of P[lacW]Xbp1K13803, P[PTT-GB]Xbp1CB02061and P[SUPor-P]CG9418KG05183. Excisions 81 and 79 delete the DNA binding domain of Xbp1 and were generated from jumps of P[PTT-GB]Xbp1CB02061. In excision 81, the sequence from 4bp to 622bp downstream from Xbp1 start codon (ATG) is deleted. In excision 79, the sequence between 260bp upstream and 748bp downstream from Xbp1 start codon (ATG) is deleted. In excision 250, the open reading frame of Xbp1 is deleted from 5bp after the start codon, together with CG9418 and CG15657, until 2R:17036107, in the inter-genic region between CG15657 and CG30389. Excision 250 was generated from a jump of P[SUPor-P]CG9418KG05183. (B) By 62 h pupation, eyeless-Flipase induced clones of cells homozygous for Excision 79, labeled by the absence of myrGFP (green), have normal levels of Spam/Eys (blue) in the IRS. The rhabdomeres are stained with actin (red). (C) At 72 h. pupa, Rh1 (blue) is delivered to the rhabdomere in photoreceptors homozygous for Excision 79, labeled by the absence of myrRFP (red), but Xbp1 mutant photoreceptors have lower levels of Rh1 in the rhabdomere in than wild type (monochrome magnifications with wild type and mutant ommatidia). (D) At 82 h. pupa, photoreceptors homozygous for Excision 79 present higher levels of Rh1 in the rhabdomere than wild-type. Excision 79 homozygous photoreceptors show even distribution of Rh1 in the rhabdomere, while wild-type show the typical crescent-shaped gradient of concentration with higher levels at the rhabdomere base (monochrome magnifications). (E) Quantification of Rh1 intensity. Average of the ratio of Rh1 fluorescence intensity divided by respective area between pairs of adjacent Excision 79 homozygous and wild-type ommatidia, at 72 and 82 hours of pupal development (FluoEx79/AreaEx79/FluoWT/AreaWT). Around 70 pairs of Excision 79 homozygous and wild-type ommatidia were quantified at each time point. At 72 h. pupa, Excision 79 homozygous photoreceptors have reduced levels of (F) BiP/GRP78 (blue and monochrome) and (G) ninaA (blue and monochrome) but not (H) Cnx (blue and monochrome). Scale bars, 10 μm.
In contrast to Ire1 mutants, Xbp1 mutant ommatidia presented normal levels of Spam/Eys secretion into the inter-rhabdomeral space with no retention of Spam/Eys in the cell body of the photoreceptors (Figure 5B). Likewise, Rh1 was delivered to the rhabdomere (Figures 5C and 5D), although at 72 h. pupa we observed lower levels of Rh1 in the rhabdomeres of Xbp1 mutant photoreceptors than in wild type (quantification in Figure 5E). In contrast, at 82 h. pupa, Xbp1 mutant rhabdomeres presented higher levels of Rh1 in the rhabdomere than wild-type (Figures 5D and 5E). Furthermore, at this stage, while wild-type showed the typical crescent-shaped gradient of concentration with higher levels at the rhabdomere base, Xbp1 mutant photoreceptors showed an even distribution of Rh1 in the rhabdomere (magnifications in Figure 5D). Our interpretation of these results is that in Xbp1 mutant ommatidia, Rh1 is delivered into the rhabdomere, although there is a developmental delay in this process. As in Ire1 mutant photoreceptors, Xbp1 mutants had reduced levels of the ER markers BiP/GRP78 (Figures 5F, S4A and S4B) and Protein disulphide isomerase (Pdi, Figure 5G). Cnx was unchanged between Xbp1 mutant and wild-type cells (Figure 5H).
The branches of the UPR are interconnected and loss of one pathway may lead to increased activation of parallel pathways during ER stress. Upregulation of ATF4 is induced by activation of the PERK pathway and we observed upregulation of ATF4 in Ire1 mutant clones but not in Xbp1 mutants (Figures S4C and S4D). Overall, these results demonstrate that Ire1 mutants have a more severe rhabdomere morphogenesis phenotype than Xbp1 mutants, indicating that Ire1 function in this context should be mediated by Xbp1-independent mechanisms.
Regulation of Fatp by RIDD is critical for rhabdomere morphogenesis
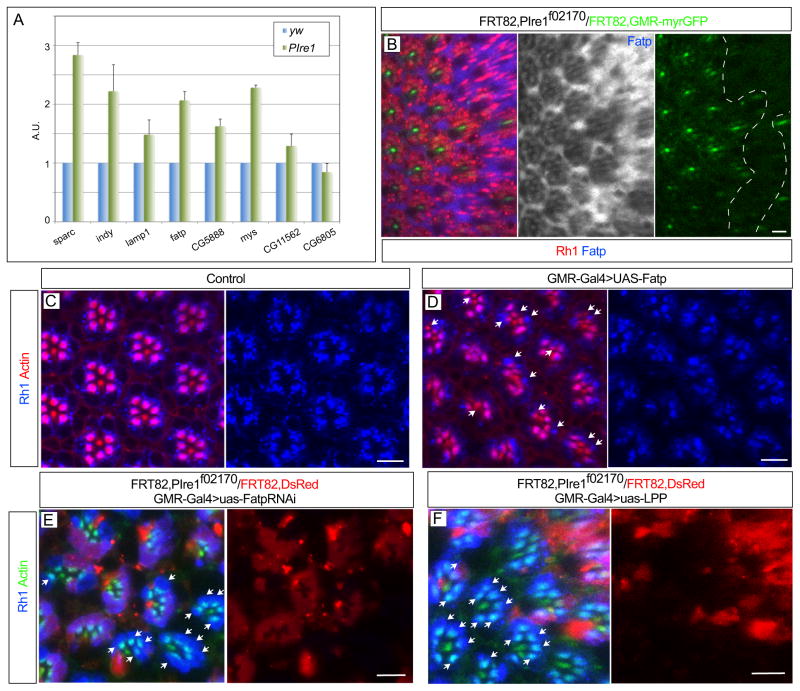
Studies using cell culture approaches showed that, during ER stress, Ire1 promotes the degradation of mRNAs encoding proteins that are translated in the ER (Hollien and Weissman, 2006; Han et al., 2009; Hollien et al., 2009). This Xbp1-independent function of Ire1 was denominated as Regulated Ire1-Dependent Decay (RIDD). To investigate if RIDD occurs in the Drosophila eye, we determined by quantitative RT-PCR the mRNA levels of specific RIDD targets in wild-type and Ire1 mutant eyes (Figure 6A). The mRNAs of genes such as Indy, Sparc and Fatp were 1,5 to 2,5 fold up-regulated in Ire1 mutant eyes, in comparison with controls. To confirm the quantitative RT-PCR results, we performed immuno-fluorescent staining in eyes with Ire1 mutant clones, using antibodies against Fatp and Sparc. We found that Fatp (Figure 6B) and Sparc (Figure S5A) protein levels were upregulated in Ire1 mutant clones. Furthermore, purified human Ire1 cleaved Drosophila Xbp1 and Fatp mRNAs in vitro (Figure S5B). We found two Ire1 cleavage sites in the mRNA of Fatp and mutagenesis of these sites abrogated cleavage of Fatp mRNA by human Ire1 (Figure S5B).
Figure 6.
Regulation of Fatp by RIDD is critical for rhabdomere morphogenesis (A) Quantitative RT-PCR comparing the mRNA levels (Y axis) of RIDD targets (X axis) in control (blue bars, yw) and Ire1 mutant eyes (green bars, eyFlp; FRT82BPBac{WH}Ire1f02170/FRT82B,CL,GMR-hid). The mRNA levels are presented in arbitrary units (fold change) relative to control (yw). (B) Clones of PBac{WH}Ire1f02170 homozygous ommatidia (absence of myrGFP, green, indicated by white dashed line) in the 72 h. pupa have higher levels of Fatp (blue and monochrome) than surrounding control tissue. (C) Adult control eyes (GMR-Gal4) have normal localization of Rh1 (blue) in the rhabdomere. (D) Adult GMR-Gal4>uas-Fatp eyes have defective delivery of Rh1 (blue) to the rhabdomere (arrows). (E) GMR-Gal4>uas-Fatp RNAi rescues the delivery of Rh1 (blue) to the rhabdomere (arrows) in clones of PBac{WH}Ire1f02170 homozygous ommatidia (compare with Figure 4A), as does (F) GMR-Gal4>uas-LPP. The rhabdomeres are stained with actin in red or green, as indicated. Scale bars, 10 μm.
Fatp is a regulator of Rh1 protein levels, since in Fatp loss-of-function mutants Rh1 levels are up-regulated, causing photoreceptor degeneration (Dourlen et al., 2012). Furthermore, fatty acids are precursors in the biosynthesis of phosphatidic acid and an increase in Fatp levels could lead to higher levels of phosphatidic acid, due to an increased import of fatty acids into the cell. High levels of phosphatidic acid cause downregulation of Rh1 protein levels and rhabdomere defects (Raghu et al., 2009), similar to the phenotypes observed in Ire1 mutant photoreceptors. Thus, we investigated if Fatp over-expression could lead to phenotypes similar to the ones observed in Ire1 mutant photoreceptors. In fact, photoreceptors where UAS-Fatp was expressed under the control of GMR-Gal4 showed defective delivery of Rh1 to the rhabdomere (Figure 6D). Furthermore, downregulation of Fatp levels by expression of Fatp RNAi in Ire1 mutant photoreceptors rescued the defective delivery of Rh1 to the rhabdomere (Figure 6E). Finally, we detected higher levels of phosphatidic acid in Ire1 mutant retinas than in controls (Figure S5C) and the defect of Rh1 delivery to the rhabdomere in Ire1 mutant photoreceptors was also rescued by overexpression of lipid phosphate phosphohydrolase (UAS-LPP (lazaro), Figure 6F), an enzyme that converts phosphatidic acid to diacylglycerol and is able to rescue the rhabdomere defects caused by excessive levels of phosphatidic acid (Raghu et al., 2009). We conclude that RIDD regulation of Fatp levels is an important part of Ire1 role during photoreceptor differentiation in the Drosophila eye.
DISCUSSION
Studies in mammalian systems revealed that the Ire1/Xbp1 signaling pathway is important during development for the differentiation of secretory cells. For example, Xbp1 “knock-out” mice have defects in the differentiation of antibody-secreting plasma cells (Reimold et al., 2001) and secretory cells of the exocrine glands of the pancreas (Lee et al., 2005). Presumably in these cases, activation of Ire1/Xbp1 signaling is required to increase the capacity of the ER to fold and process the high load of secreted proteins.
Our present results demonstrate that Ire1 signaling is required for photoreceptor differentiation and rhabdomere morphogenesis, a process that also imposes a high demand to the capacity of the ER to fold proteins such as Spam/Eys and Rh1. As shown, Ire1 mutant photoreceptors have defects in the secretion of Spam/Eys to the IRS and in the delivery of Rh1 to the rhabdomere. However, we see activation of the Xbp1-EGFP reporter starting at 48h of pupal development, well before when the Spam/Eys secretion and Rh1 delivery defects are observed. Presumably, the folding of other unidentified proteins during these earlier stages might also require Ire1 signaling. It is noteworthy though, that in mutant B lymphocytes modified to lack antibody production, Ire1 is still activated (and Xbp1 spliced) upon lymphocyte differentiation to plasma cells (Hu et al., 2009). Activation of Ire1/Xbp1 signaling in this context seems to be part of the process of plasma cell differentiation, independently of the accumulation of misfolded proteins in the ER lumen (van Anken et al., 2003; Hu et al., 2009).
Ire1 function is also required for the regulation of the membrane lipids. In mammalians, Ire1/Xbp1 signaling regulates the biosynthesis of phospholipids and other lipids (Sriburi et al., 2007; Lee et al., 2008). A study in yeast demonstrated that Ire1 is activated by “membrane aberrancy”, a condition of stress caused by the experimental depletion of inositol (Promlek et al., 2011). Activation of Ire1 in this case occurs by a mechanism that is distinct from the one involving the recognition of misfolded proteins by the luminal domain of Ire1 (Promlek et al., 2011). Furthermore, Ire1 can be activated by direct binding of flavonoids, such as quercetin, to a pocket present in the cytoplasmic domain of Ire1, in a mechanism that is also independent of the binding of misfolded proteins to Ire1 (Wiseman et al., 2010). Our present results do not clarify if Ire1 activation in the photoreceptors during pupal stages results from the accumulation of misfolded proteins in the ER lumen or an imbalance in the membrane lipids.
Our results demonstrate that Ire1 signaling is required for photoreceptor differentiation and rhabdomere morphogenesis in an Xbp1-independent manner. Studies using cell culture paradigms demonstrated that, in addition to mediating Xbp1 mRNA splicing, Ire1 also promotes RIDD (Regulated Ire1-Dependent Decay), the degradation of mRNAs encoding ER-targeted proteins (Hollien and Weissman, 2006); (Han et al., 2009) (Hollien et al., 2009), but the physiological significance of the RIDD mechanism is unknown. Our quantitative RT-PCR results show that RIDD targets are up-regulated in Ire1 mutant eyes, including fatty acid transport protein (fatp), a regulator of Rh1 protein levels (Dourlen et al., 2012). Our results show that regulation of fatp mRNA by RIDD is critical for rhabdomere morphogenesis, since the experimental downregulation of fatp mRNA by RNAi rescues the Rh1 rhabdomere delivery defect observed in Ire1 mutants.
Rh1 protein levels and Rh1 delivery to the rhabdomere is very sensitive to the levels of sphingolipids (Yonamine et al., 2011) and phosphatidic acid (Raghu et al., 2009). Increased Fatp levels may lead to an increase in the levels of fatty acids and subsequently, phosphatidic acid, which is known to downregulate Rh1 protein levels and cause rhabdomere morphogenesis defects (Raghu et al., 2009). High levels of phosphatidic acid disrupt the Arf1-dependent transport of membrane to the developing rhabdomere (Raghu et al., 2009). Our results show that phosphatidic acid levels are elevated in Ire1 mutant retinas and lowering phosphatidic acid levels by expression of LPP rescues the defects observed in Ire1 mutants, demonstrating that Ire1/Fatp dependent regulation of fatty and phosphatidic acids levels is important for rhabdomere morphogenesis in Drosophila. In addition, it is possible that the increase in phosphatidic acid levels in Ire1 mutant photoreceptors is also caused by the activation of PERK, since we observed upregulation of the PERK pathway mediator ATF4 in Ire1 mutant photoreceptors (Figure S4C) and in cell culture models it was shown that PERK is able to phosphorylate diacylglycerol and generate phosphatidic acid (Bobrovnikova-Marjon et al., 2012).
In conclusion, our results, using well-characterized genetic tools (Ire1 and Xbp1 null mutations) and a developmental paradigm (photoreceptor differentiation in the Drosophila pupa), demonstrate the physiological relevance of Xbp1-independent mechanisms downstream of Ire1 signaling.
EXPERIMENTAL PROCEDURES
Drosophila stocks and molecular biology
All Drosophila stocks were raised with standard cornmeal medium, at 25°C, in a 12 hour light/dark cycle. Clones of mutant eye tissue were generated by the Flp/FRT technique (Golic, 1991), with Flipase expression under the control of the eyeless promoter (Newsome et al., 2000). The open reading frame of Ire1 was amplified from larval cDNA with the forward primer 5’ggaagatctatgggcagtcttaagaagttacc and reverse primer 5’ cggggtacctcaatcctgcgttgaaggtgg and cloned into pUAST using BglII and Acc65I restriction sites. The injection of DNA into embryos to establish Drosophila transgenic lines was performed by BestGene. The CH322-07H05 (Pacman) genomic construct is a 22Kb fragment covering Ire1 and the neighbouring genes CG4662, CG11447, CG4572 and CG4686. The CH322-07H05 transgenic line was generated by using φC31 integrase-mediated transgenesis (BestGene Strain #9752, attP acceptor site in 22A3). The Xbp1 deletion mutants were created by crossing flies with the P elements P[PTT-GB]Xbp1CB02061or P{SUPor-P}CG9418KG05183 with Δ23 transposase flies and were characterized by PCR and sequencing. The precise excision of PBac{WH}Ire1f02170 was generated by crossing to PBac transposase and confirmed by sequencing. UAS-Fatp and UAS-Fatp RNAi were a gift from B. Mollereau and were described in (Dourlen et al., 2012). UAS-Lpp was a gift from P. Raghu and was described in (Raghu et al., 2009).
Quantitative RT-PCR
For quantitative reverse transcription-PCR (RT-PCR) analysis, total RNA was extracted from adult heads with Trizol and Direct-zol RNA mini-prep (Zymo Research). 0,5 μg of total RNA was retro-transcribed using RevertAid First Strand (Thermo/Fermentas). Each PCR reaction was performed on 1/40 of the cDNA obtained using SSoFast EvaGreen Supermix (Bio-Rad) according to the manufacturer’s instructions and using a Bio-Rad Cfx-96 detection system. All samples were analyzed in triplicates and from 3 independent RNA extractions. For each sample, the levels of mRNAs were normalized using rp49 as a loading control. PCR primers were: sparc (F: AGCTCCATCTGCATGTAGTGCTCG, R: ATCTGCCAGATGCACGACGAGAAG), indy (F:GCTTGCGGGTGAAGTACATCACAACC, R:TACCTTCAAGGGCATCTACGAGGC), lamp1 (F: CTTATTGGCTTGCCGAGAGTCGGT, R: TGGATGGTTGCGGATATGGTTGCG, fatp (F: CCTTGGAAGTCTGGTAGTACAGCG, R: AGAATGTTTCCACCAGCGAGGTGG), CG5888 (F: ACTCGGTTGGCACATTATCACCGC, R: GGAAGCAGTGATTTGCCCGGTAAC), mys (F: AGCAGGTGAGCTTCACAGCTCAGA, R: TCGATGTTAGATCCGTTGCCGAGC), CG11562 (F: TGAGACTGCACGATGGTGTCCTGA, R: CCATCGTGATTAGTCGTTCCACCC), CG6805 (F: ACCACTTGTCCGGATAACCAGCTG, R: GCGAAAGTCGAATGCTTACGGCAC), GRP78 (F: TGTCACCGATCTGGTTCTTCAGGC, R: GTCCCATGACCAAGGACAACCATC), ninaA (F: CGTGGCCAGTCTTCAC, R: CTCCACCGCCATCCTC), Pld (F: ACGGACTCAGCTTTCGGATCACGT, R: GTCTCAGTAGCTGTTAGACGGTGC), dgk (F: CTTCTGTTGCTGCTCGAACTGGAG, R: ATCACTAGCTGTCGAGGAAGCTGC), Lpp (lazaro) (F: CTCGTTGTCGTCCTCATTCTCTGC, R: TACGTGGAGCAGTTCCACTGCACT), cds (F:GAAGTTCCTGGTGACCTATCACCG, R: GTCTTCTTGGGGCTGAGCTTGATG) and rp49 (F: AGATCGTGAAGAAGCGCACCAAGC, R: GCACCAGGAACTTCTTGAATCCGG).
In vitro Ire1 mRNA cleavage assay
The in vitro Ire1 mRNA cleavage assay was performed as in (Cross et al., 2012). dXbp1 mRNA was produced in vitro with T7 RNA polymerase (Promega Ribomax Express RNA) from pOT2-dXbp1 after linearization with NotI. dFatp mRNA was produced from a PCR product obtained with F:TAATACGACTCACTATAGGGAGACCGG and R: CGGTGTGGTACAAAGGCAAGG. Fatp Mut1 was done by changing GTGGCCAATGTGctgcagGCTCAGGGCTAC into GTGGCCAATGTGctccacGCTCAGGGCTAC and Fatp Mut. 2 by changing TCCCTCCCTGctgcacAGCATCAC into TCCCTCCCTGctccaaAGCATCAC. The mRNAs (1μM) were incubated at 29°C with or without purified human Ire 1 (5 μM) and the Ire1 inhibitor 4μ8C (10 μM), kind gifts from Heather Harding. Cleavage products were resolved by electrophoresis on a 1% denaturing agarose gel stained with ethidium bromide.
Analysis of retinal phosphatidic acid
The analysis of retinal phosphatidic acid by LC-MS was performed following the protocol described in (Raghu et al., 2009). For each genotype 150 retinas were separated from the head after being frozen in liquid nitrogen and dehydrated in acetone. The retinas were homogenized using a glass piston in 1 mL methanol containing 500 ng of a 12:0/12:0 PA standard (Avanti Polar Lipids). The lipids were extracted with addition of 2 mL of chloroform and 1ml of 0.88% (w/v) KCl to split the phases. The lower organic phase was dried in a speedvac during 30 min and stored at −20°C until further analysis.
Phospholipids extracts were profiled using the LC-MS in order to identify and quantify the PA species present. The analysis was performed using a Surveyor HPLC system (Thermo Scientific) interfaced to an externally calibrated LTQ-Ion trap mass spectrometer (Thermo Scientific) with an electrospray ionization (ESI) source in negative mode. The capillary was operated at 5kV and source temperature was set to 300°C, the sheath gas and auxiliary gas setting was 20 (arbitrary units). For normal-phase chromatographic analysis, a 10 μL aliquot of phospholipid extract dissolved in 30 μL of chloroform was injected into a Luna, Phenomenex silica column (150 mm × 1.0 mm, 3um particle size). PA phospholipids species originating from fly extracts were separated using chloroform/methanol/water (90:9.5:0.5) containing 7.5 mM ethylamine changing to acetonitrile/chloroform/methanol/water (30:30:35:5) containing 10 mM ethylamine at flow rate of 50 μL/min. The column temperature was maintained at 25°C. The relative amount of each PA species was determined dividing the area of the corresponding m/z peak by that of the internal standard.
Immunohistochemistry and Electron microscopy
For pupal dissections, white pre-pupae (0h) were collected and maintained at 25°C until the required stage. For late pupal and adult eye dissections, pWIZ (a white gene RNAi line) was used to reduce the fluorescence background from the red eye pigment as in (Xie et al., 2007). Larval, pupal and adult eyes were dissected in 1xPBS, fixed in 1xPBS + 4% Formaldehyde for 40 minutes at room temperature and washed 3 times with 1xPBS + 0.3% Triton X-100. Primary antibodies were incubated in 1xPBS, 1% BSA, 0.1% Tween 20, 250 mM NaCl overnight at 4°C. Primary antibodies were as follows: rat anti-ELAV (7E8A10, DSHB), guinea pig anti-homothorax (Wildonger et al., 2005), guinea pig anti-Runt (Duffy et al., 1991), rat anti-cadherin (DCAD2, DSHB), mouse anti-armadillo (N2 7A1, DSHB), rabbit anti-aPKC (Santa Cruz, sc-216), mouse anti-Crumbs (Cq4, DSHB), mouse anti-spam (21A6, DSHB), mouse anti-Rh1 (4C5, DSHB), mouse anti-tubulin (12G10, DSHB), guinea pig anti-Bip/GRP78 (Hsc-3) (Ryoo et al., 2007), rabbit anti-Bip/GRP78 (StressMarq Biosciences, SPC-180), guinea pig anti-ATF4 (Kang et al., 2012), rabbit anti-calnexin (Rosenbaum et al., 2006), rabbit anti-ninaA (Baker et al., 1994), rabbit anti-Pdi (Abcam, ab2792), rabbit anti-fatp (Dourlen et al., 2012) and mouse anti-Sparc (Portela et al., 2010). Samples were washed 3 times with 1xPBS + 0.3% Triton X-100 and incubated with appropriate secondary antibodies (from Jackson Immuno-Research Laboratories) for 2 hours at room temperature. To stain actin in the rhabdomeres we used Phalloidin-TRITC or Phalloidin-FITC (Sigma). Samples were mounted in 80% glycerol in a bridge formed by two cover slips to prevent the samples from being crushed while analyzed on a Zeiss LSM 710 confocal microscope. Cryostat horizontal frozen sections of OCT embedded adult and pupal heads were performed as in (Mollereau et al., 2001). Measurement of the IRS area and quantification of the mean Rh1 fluorescence were performed using ImageJ. A ratio for each pair of adjacent wild-type and Ire1 mutant ommadia was obtained and expressed as a mean value.
For electron microscopy, adult eyes were fixed in 2,5% glutheraldehyde in 0,1 M cacodylate buffer (pH 7.4) and post-fixed with 1% osmium tetroxide. After dehydratation in ethanol and propylene oxide, the eyes were infiltrated with resin and mounted in silicone molds. Ultrathin sections (60nm) were cut and examined with electron transmission microscope (Jeol JEM).
Supplementary Material
HIGHLIGHTS.
Ire1 is activated in the photoreceptors during Drosophila pupal development.
Ire1 mutant photoreceptors have defects in rhabdomere morphogenesis.
The role of Ire1 in rhabdomere morphogenesis is independent of Xbp1 function.
Regulation of Fatp by RIDD is critical for rhabdomere morphogenesis.
Acknowledgments
We would like to thank Charles Zuker, Nansi Jo Colley, Tiffany Cook, Andrew Zelhof, Don Ready, Bertrand Mollereau, Eduardo Moreno, Padinjat Raghu, the Bloomington Stock Center and the Developmental Studies Hybridoma Bank (DSHB) for fly stocks and antibodies. We thank Heather Harding for purified human IRE1, the 4m8C inhibitor and advice on the mRNA cleavage protocol. We thank Padinjat Raghu, Trevor Pettitt and Michael Wakelam for advice on the LC-MS protocol. We thank Don Ready for comments on our manuscript. We thank CEDOC/FCM-UNL and IGC for access to the imaging and fly facilities. We thank Hermann Steller for all the support during the initial stages of this work. This work was supported by Fundação para a Ciência e a Tecnologia through grants PEst-OE/EQB/LA0004/2011, PTDC/BIA-BCM/105217/2008, PTDC/SAU-OBD/104399/2008 and PTDC/BEX-BCM/1217/2012, by a Marie Curie International Reintegration grant (PIRG03-GA-2008-230935), and by the National Institutes of Health grants R01EY020866 and R01DK047119. D.R. is a Principal Research Fellow of the Wellcome Trust. D.S.C was supported by a doctoral fellowship from the Gulbenkian Foundation.
References
- Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Baker EK, Colley NJ, Zuker CS. The cyclophilin homolog NinaA functions as a chaperone, forming a stable complex in vivo with its protein target rhodopsin. Embo J. 1994;13:4886–4895. doi: 10.1002/j.1460-2075.1994.tb06816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrman S, Acosta-Alvear D, Walter P. A CHOP-regulated microRNA controls rhodopsin expression. J Cell Biol. 2011;192:919–927. doi: 10.1083/jcb.201010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrovnikova-Marjon E, Pytel D, Riese MJ, Vaites LP, Singh N, Koretzky GA, Witze ES, Diehl JA. PERK utilizes intrinsic lipid kinase activity to generate phosphatidic acid, mediate Akt activation, and promote adipocyte differentiation. Mol Cell Biol. 2012;32:2268–2278. doi: 10.1128/MCB.00063-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagan RL, Ready DF. The emergence of order in the Drosophila pupal retina. Dev Biol. 1989;136:346–362. doi: 10.1016/0012-1606(89)90261-3. [DOI] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Colley NJ, Baker EK, Stamnes MA, Zuker CS. The cyclophilin homolog ninaA is required in the secretory pathway. Cell. 1991;67:255–263. doi: 10.1016/0092-8674(91)90177-z. [DOI] [PubMed] [Google Scholar]
- Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross BC, Bond PJ, Sadowski PG, Jha BK, Zak J, Goodman JM, Silverman RH, Neubert TA, Baxendale IR, Ron D, et al. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc Natl Acad Sci U S A. 2012;109:E869–878. doi: 10.1073/pnas.1115623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dourlen P, Bertin B, Chatelain G, Robin M, Napoletano F, Roux MJ, Mollereau B. Drosophila fatty acid transport protein regulates rhodopsin-1 metabolism and is required for photoreceptor neuron survival. PLoS Genet. 2012;8:e1002833. doi: 10.1371/journal.pgen.1002833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy JB, Kania MA, Gergen JP. Expression and function of the Drosophila gene runt in early stages of neural development. Development. 1991;113:1223–1230. doi: 10.1242/dev.113.4.1223. [DOI] [PubMed] [Google Scholar]
- Gardner BM, Walter P. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science. 2011;333:1891–1894. doi: 10.1126/science.1209126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic KG. Site-specific recombination between homologous chromosomes in Drosophila. Science. 1991;252:958–961. doi: 10.1126/science.2035025. [DOI] [PubMed] [Google Scholar]
- Griciuc A, Aron L, Roux MJ, Klein R, Giangrande A, Ueffing M. Inactivation of VCP/ter94 suppresses retinal pathology caused by misfolded rhodopsin in Drosophila. PLoS Genet. 2010:6. doi: 10.1371/journal.pgen.1001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, Backes BJ, Oakes SA, Papa FR. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- Hu CC, Dougan SK, McGehee AM, Love JC, Ploegh HL. XBP-1 regulates signal transduction, transcription factors and bone marrow colonization in B cells. Embo J. 2009;28:1624–1636. doi: 10.1038/emboj.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain N, Pellikka M, Hong H, Klimentova T, Choe KM, Clandinin TR, Tepass U. The agrin/perlecan-related protein eyes shut is essential for epithelial lumen formation in the Drosophila retina. Dev Cell. 2006;11:483–493. doi: 10.1016/j.devcel.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Izaddoost S, Nam SC, Bhat MA, Bellen HJ, Choi KW. Drosophila Crumbs is a positional cue in photoreceptor adherens junctions and rhabdomeres. Nature. 2002;416:178–183. doi: 10.1038/nature720. [DOI] [PubMed] [Google Scholar]
- Kaminker JS, Canon J, Salecker I, Banerjee U. Control of photoreceptor axon target choice by transcriptional repression of Runt. Nat Neurosci. 2002;5:746–750. doi: 10.1038/nn889. [DOI] [PubMed] [Google Scholar]
- Kang MJ, Chung J, Ryoo HD. CDK5 and MEKK1 mediate pro-apoptotic signalling following endoplasmic reticulum stress in an autosomal dominant retinitis pigmentosa model. Nat Cell Biol. 2012;14:409–415. doi: 10.1038/ncb2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MJ, Ryoo HD. Suppression of retinal degeneration in Drosophila by stimulation of ER-associated degradation. Proc Natl Acad Sci U S A. 2009;106:17043–17048. doi: 10.1073/pnas.0905566106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knust E. Photoreceptor morphogenesis and retinal degeneration: lessons from Drosophila. Curr Opin Neurobiol. 2007;17:541–547. doi: 10.1016/j.conb.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Kumar JP, Bowman J, O’Tousa JE, Ready DF. Rhodopsin replacement rescues photoreceptor structure during a critical developmental window. Dev Biol. 1997;188:43–47. doi: 10.1006/dbio.1997.8636. [DOI] [PubMed] [Google Scholar]
- Kumar JP, Ready DF. Rhodopsin plays an essential structural role in Drosophila photoreceptor development. Development. 1995;121:4359–4370. doi: 10.1242/dev.121.12.4359. [DOI] [PubMed] [Google Scholar]
- Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. Embo J. 2005;24:4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BX, Satoh AK, Ready DF. Myosin V, Rab11, and dRip11 direct apical secretion and cellular morphogenesis in developing Drosophila photoreceptors. J Cell Biol. 2007;177:659–669. doi: 10.1083/jcb.200610157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes CS, Levet C, Chatelain G, Dourlen P, Fouillet A, Dichtel-Danjoy ML, Gambis A, Ryoo HD, Steller H, Mollereau B. ER stress protects from retinal degeneration. Embo J. 2009;28:1296–1307. doi: 10.1038/emboj.2009.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollereau B, Dominguez M, Webel R, Colley NJ, Keung B, de Celis JF, Desplan C. Two-step process for photoreceptor formation in Drosophila. Nature. 2001;412:911–913. doi: 10.1038/35091076. [DOI] [PubMed] [Google Scholar]
- Mori K, Kawahara T, Yoshida H, Yanagi H, Yura T. Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells. 1996;1:803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- Ondek B, Hardy RW, Baker EK, Stamnes MA, Shieh BH, Zuker CS. Genetic dissection of cyclophilin function. Saturation mutagenesis of the Drosophila cyclophilin homolog ninaA. J Biol Chem. 1992;267:16460–16466. [PubMed] [Google Scholar]
- Pellikka M, Tanentzapf G, Pinto M, Smith C, McGlade CJ, Ready DF, Tepass U. Crumbs, the Drosophila homologue of human CRB1/RP12, is essential for photoreceptor morphogenesis. Nature. 2002;416:143–149. doi: 10.1038/nature721. [DOI] [PubMed] [Google Scholar]
- Portela M, Casas-Tinto S, Rhiner C, Lopez-Gay JM, Dominguez O, Soldini D, Moreno E. Drosophila SPARC is a self-protective signal expressed by loser cells during cell competition. Dev Cell. 2010;19:562–573. doi: 10.1016/j.devcel.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Promlek T, Ishiwata-Kimata Y, Shido M, Sakuramoto M, Kohno K, Kimata Y. Membrane aberrancy and unfolded proteins activate the endoplasmic reticulum stress sensor Ire1 in different ways. Mol Biol Cell. 2011;22:3520–3532. doi: 10.1091/mbc.E11-04-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghu P, Coessens E, Manifava M, Georgiev P, Pettitt T, Wood E, Garcia-Murillas I, Okkenhaug H, Trivedi D, Zhang Q, et al. Rhabdomere biogenesis in Drosophila photoreceptors is acutely sensitive to phosphatidic acid levels. J Cell Biol. 2009;185:129–145. doi: 10.1083/jcb.200807027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rosenbaum EE, Hardie RC, Colley NJ. Calnexin is essential for rhodopsin maturation, Ca2+ regulation, and photoreceptor cell survival. Neuron. 2006;49:229–241. doi: 10.1016/j.neuron.2005.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo HD, Domingos PM, Kang MJ, Steller H. Unfolded protein response in a Drosophila model for retinal degeneration. Embo J. 2007;26:242–252. doi: 10.1038/sj.emboj.7601477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh AK, O’Tousa JE, Ozaki K, Ready DF. Rab11 mediates post-Golgi trafficking of rhodopsin to the photosensitive apical membrane of Drosophila photoreceptors. Development. 2005;132:1487–1497. doi: 10.1242/dev.01704. [DOI] [PubMed] [Google Scholar]
- Shen X, Ellis RE, Lee K, Liu CY, Yang K, Solomon A, Yoshida H, Morimoto R, Kurnit DM, Mori K, et al. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- Sriburi R, Bommiasamy H, Buldak GL, Robbins GR, Frank M, Jackowski S, Brewer JW. Coordinate regulation of phospholipid biosynthesis and secretory pathway gene expression in XBP-1(S)-induced endoplasmic reticulum biogenesis. J Biol Chem. 2007;282:7024–7034. doi: 10.1074/jbc.M609490200. [DOI] [PubMed] [Google Scholar]
- Stowers RS, Schwarz TL. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics. 1999;152:1631–1639. doi: 10.1093/genetics/152.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U, Harris KP. Adherens junctions in Drosophila retinal morphogenesis. Trends Cell Biol. 2007;17:26–35. doi: 10.1016/j.tcb.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the ER to activation of JNK protein kinase by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- van Anken E, Romijn EP, Maggioni C, Mezghrani A, Sitia R, Braakman I, Heck AJ. Sequential waves of functionally related proteins are expressed when B cells prepare for antibody secretion. Immunity. 2003;18:243–253. doi: 10.1016/s1074-7613(03)00024-4. [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Walther RF, Pichaud F. Crumbs/DaPKC-dependent apical exclusion of Bazooka promotes photoreceptor polarity remodeling. Curr Biol. 2010;20:1065–1074. doi: 10.1016/j.cub.2010.04.049. [DOI] [PubMed] [Google Scholar]
- Wildonger J, Sosinsky A, Honig B, Mann RS. Lozenge directly activates argos and klumpfuss to regulate programmed cell death. Genes Dev. 2005;19:1034–1039. doi: 10.1101/gad.1298105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman RL, Zhang Y, Lee KP, Harding HP, Haynes CM, Price J, Sicheri F, Ron D. Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1. Mol Cell. 2010;38:291–304. doi: 10.1016/j.molcel.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie B, Charlton-Perkins M, McDonald E, Gebelein B, Cook T. Senseless functions as a molecular switch for color photoreceptor differentiation in Drosophila. Development. 2007;134:4243–4253. doi: 10.1242/dev.012781. [DOI] [PubMed] [Google Scholar]
- Yonamine I, Bamba T, Nirala NK, Jesmin N, Kosakowska-Cholody T, Nagashima K, Fukusaki E, Acharya JK, Acharya U. Sphingosine kinases and their metabolites modulate endolysosomal trafficking in photoreceptors. J Cell Biol. 2011;192:557–567. doi: 10.1083/jcb.201004098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda T, Imaizumi K, Oono K, Yui D, Gomi F, Katayama T, Tohyama M. Activation of caspase-12, an endoplastic reticulum (ER) resident caspase, through tumor necrosis factor receptor-associated factor 2-dependent mechanism in response to the ER stress. J Biol Chem. 2001;276:13935–13940. doi: 10.1074/jbc.M010677200. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Zelhof AC, Hardy RW, Becker A, Zuker CS. Transforming the architecture of compound eyes. Nature. 2006;443:696–699. doi: 10.1038/nature05128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.