Abstract
GH and GH receptors are expressed throughout life, and GH elicits a diverse range of responses, including growth and altered metabolism. It is therefore important to understand the full spectrum of GH signaling pathways and cellular responses. We applied mass spectrometry-based phosphoproteomics combined with stable isotope labeling with amino acids in cell culture to identify proteins rapidly phosphorylated in response to GH in 3T3-F442A preadipocytes. We identified 132 phosphosites in 95 proteins that exhibited rapid (5 or 15 min) GH-dependent statistically significant increases in phosphorylation by more than or equal to 50% and 96 phosphosites in 46 proteins that were down-regulated by GH by more than or equal to 30%. Several of the GH-stimulated phosphorylation sites were known (e.g. regulatory Thr/Tyr in Erks 1 and 2, Tyr in signal transducers and activators of transcription (Stat) 5a and 5b, Ser939 in tuberous sclerosis protein (TSC) 2 or tuberin). The remaining 126 GH-stimulated sites were not previously associated with GH. Kyoto Encyclopedia of Genes and Genomes pathway analysis of GH-stimulated sites indicated enrichment in proteins associated with the insulin and mammalian target of rapamycin (mTOR) pathways, regulation of the actin cytoskeleton, and focal adhesions. Akt/protein kinase A consensus sites (RXRXXS/T) were the most commonly phosphorylated consensus sites. Immunoblotting confirmed GH-stimulated phosphorylation of all seven novel GH-dependent sites tested [regulatory sites in proline-rich Akt substrate, 40 kDA (PRAS40), regulatory associated protein of mTOR, ATP-citrate lyase, Na+/H+ exchanger-1, N-myc downstream regulated gene 1, and Shc]). The immunoblot results suggest that many, if not most, of the GH-stimulated phosphosites identified in this large-scale quantitative phosphoproteomics analysis, including sites in multiple proteins in the Akt/ mTOR complex 1 pathway, are phosphorylated in response to GH. Their identification significantly broadens our thinking of GH-regulated cell functions.
GH, which binds to the ubiquitously expressed GH receptor, is an important regulator of body growth, development, and metabolism (1, 2). Patients with abnormally high levels of circulating GH throughout life (acromegaly) are extremely tall and may have enlarged liver, heart, kidneys, and/or other organs. Acromegalics often present with hypertension, cardiac, metabolic, and articular complications and have an increased risk of diabetes and malignant neoplasms (3). Patients with low levels of circulating GH display short stature, decreased lean body mass, and reduced bone density. GH-deficient adults report improved quality of life upon GH therapy (4). These findings suggest that GH affects multiple tissues and physiological functions.
GH elicits a diverse range of cellular responses, including secretion of IGF-I and promotion of cell proliferation and differentiation. GH signaling is initiated by GH binding to a GH receptor dimer, which leads to activation of the GH receptor-associated Janus kinase 2 (JAK2) (5, 6). The activated JAK2 phosphorylates tyrosines within itself and the GH receptor, forming binding sites for Src Homology (SH2) and phosphotyrosine binding domain-containing proteins, such as the signal transducers and activators of transcription (Stats) (reviewed in Refs. 7 and 8). In addition to signaling through Stats, GH activates the Ras/Raf/MAPK kinase (MEK)1/Erks 1/2 pathway (9, 10) and the phosphatidylinositol 3-kinase (PI3K)/Akt/mTORC1 pathway (11–14). GH has also been shown, often in a cell-specific manner, to activate members of the Src family of tyrosine kinases (15) and to stimulate the tyrosyl phosphorylation of a variety of other proteins, including transmembrane proteins (e.g. SIRP1α), phosphatases (e.g. SHP2), adapter proteins (e.g. SH2B1, CrkII), and proteins associated with the cytoskeleton (e.g. p130Cas/Bcar1) (16–18). Although investigation of these GH-regulated proteins has given us insight into some of the ways in which cells respond to GH, we hypothesized that these proteins most likely represent only a small subset of the proteins that undergo GH-dependent phosphorylation and thus contribute to cells' diverse responses to GH.
Recently, phosphoproteomic analysis has enabled large-scale, unbiased profiling of the sites of phosphorylation present in a population of cells and the ability to monitor dynamic changes in the phosphorylation at these sites after cellular stimulation (19–24). However, due to the low stoichiometry of phosphorylation and high complexity of samples isolated from whole cells, global analysis of protein phosphorylation in cells by liquid chromatography (LC)-mass spectrometry (MS) remains challenging. Enrichment of phosphopeptides before LC-MS using metal oxides (TiO2, ZrO2) (25, 26), immobilized metal affinity chromatography (27, 28), and phosphopeptide-specific antibodies (29) have been used to selectively enrich phosphopeptides from complex peptide mixtures and dramatically improve the detection of phosphopeptides. We employed a MS-based quantitative phosphoproteomic approach that combined immunoaffinity purification, ZrO2, and LC-tandem MS (MSMS) with SILAC (stable isotope labeling with amino acids in cell culture) (30) to identify and quantify rapid (5 and 15 min) GH-dependent changes in protein phosphorylation in the highly GH-sensitive 3T3-F442A preadipocyte cell line. SILAC-based protein quantification has been widely used to study dynamic changes in protein expression levels (31–34) because it enables samples to be combined and processed simultaneously, eliminating the differences within individual runs that are due to variation in sample handling during the extensive sample processing. This proteomic analysis enabled us to identify close to 1800 unique phosphorylation sites, of which 132 were found to be up-regulated by GH and 96 down-regulated by GH. The many new GH-regulated sites of phosphorylation identified by in this study suggest a broad array of proteins that are impacted by GH and are thus likely to contribute to the multiple and diverse physiological responses to GH. Western blotting confirmed GH stimulation of multiple phosphosites that lie downstream of Akt, underscoring the importance of the PI3K/Akt pathway in cellular responses to GH.
Materials and Methods
Antibodies
αPRAS40 (D23C7, catalog no. 2691), αpT247-PRAS40 (C77D7, catalog no. 2997), αpS184-PRAS40 (catalog no. 5936), αATP-citrate lyase (catalog no. 4332), αpS455-ATP-citrate lyase (ACLY) (catalog no. 4331), αS6 ribosomal protein (5G10, catalog no. 2217), αpS235/236-S6 ribosomal protein (catalog no. 4856), αShc (catalog no. 2432), αpY423-Shc (catalog no. 2431 labeled “phospho-Shc [Tyr317]” antibody), α N-myc downstream regulated gene (NDRG)1 (catalog no. 5196), αpS330-NDRG1 (catalog no. 5196), αAkt (catalog no. 9272), αpSer473-Akt (catalog no. 4058), αErk1/2 (catalog no. 9102) and αpErk1/2 (catalog no.9106, recognizes both Erk1 and Erk2 that are doubly phosphorylated on T203/Y205 in murine Erk1 and T183/Y185 in murine Erk2) were from Cell Signaling Technology (Danvers, MA). αNHE-1 (H-160, catalog no. Sc-28758) was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). αpS707-Na+/H+ exchanger-1 (NHE1) was from the Division of Signal Transduction Therapy (The University of Dundee, Dundee, Scotland) (35). α-Regulatory associated protein of mTOR (raptor) and αpS863-raptor were described previously (36).
Mass spectrometry
SILAC (reagents obtained from Invitrogen, Carlsbad, CA) was achieved by growing 3T3-F442A preadipocytes (cell stock from H. Green, Harvard Medical School, Boston, MA) in DMEM containing 5% dialyzed fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, 0.25 μg/ml amphotericin, and either [12C]Lys and [12C]Arg or [13C]Lys and [13C,15N]Arg for at least seven cell doublings. For the cells grown in [13C]Lys and [13C, 15N]Arg, heavy isotope incorporation was evaluated by running a pilot digestion of the heavy isotope [13C,15N]-labeled cell extract followed by LC-MSMS analysis. The heavy isotope-labeled amino acids were incorporated into more than 99% of the cellular protein. Cells were washed and incubated in DMEM (SILAC) containing 1% BSA, 100 U/ml penicillin, 100 μg/ml streptomycin, 0.25 μg/ml amphotericin, and the appropriate (either [12C], [13C], or [13C /15N]) Lys and Arg for 15 h before treatment. Cells were treated with vehicle or GH (500 ng/ml) for the indicated times. Cells were then washed with PBSV (10 mm sodium phosphate, 150 mm NaCl, 1 mm Na3VO4, pH 7.5), harvested, and centrifuged. The cell pellet was frozen and stored at −80 C until use. Lysis buffer [50 mm Tris-HCl (pH 8.2) and 8 m urea containing protease inhibitors (Roche, Indianapolis, IN; Complete Mini, EDTA-free tablet) and phosphatase inhibitors (1 mm sodium fluoride, 1 mm sodium vanadate, 10 mm sodium pyrophosphate, 25 mm β-glycerophosphate)] was added, and the cells were lysed on ice by sonication [Fisher sonic dismembrator (Thermo-Fisher Scientific, Pittsburgh, PA), model 100, three 30-sec bursts, power setting of 5 watts]. The protein content of the lysates was measured using the Bradford protein assay (37). Equal amounts of protein labeled with light isotope and heavy isotope were mixed, reduced, alkylated, and digested with TPCK-treated-trypsin (Worthington Biochemical Corp., Lakewood, NJ) at 37 C overnight.
Phosphotyrosine-containing peptides were isolated by immunoaffinity purification with immobilized αPY (PhosphoScan P-Tyr-100; Cell Signaling Technology, Danvers, MA) according to the vendor's protocol. Peptides that did not bind to the immobilized αPY were further fractionated by SCX HPLC, and the fractions were enriched for Ser-/Thr-phosphopeptides using ZrO2 (25). The peptides that bound to and eluted from the immobilized αPY, the SCX fractions enriched for Ser-/Thr-phosphopeptides using ZrO2, and the peptides which did not bind to the ZrO2 were analyzed by LC-MSMS on a nano LC system (Eksigent Technologies, Dublin, CA) interfaced with a linear trap quadrupole (LTQ)-Orbitrap XL mass spectrometer (Thermo-Fisher Scientific). The Thermo-Fisher Orbitrap was used for this study because its high mass accuracy and resolution reduces the false positives for peptide identification. Peptides were separated using a 75 μm × 15 cm column in-house packed with 3 μm C18 resin (Sepax HP-C18) over a 120-min gradient of 10–45% acetonitrile with 0.1% formic acid at a flow rate of 250 nl/min. Analytes were sprayed into the mass spectrometer via a chip-based nanoelectrospray source (Advion Triversa Nanomate) in positive ion mode. The LTQ-Orbitrap was operated in data-dependent mode by alternating single MS scans (300–1700 m/z) in the Orbitrap analyzer and sequential MSMS scans in the LTQ for the seven most intense ions from each MS survey scan. MS scans were acquired with the resolution (mass/peak width at half maximum) set at 60,000 at 400 m/z and an automatic gain control target of 1 × 106. MSMS scans were triggered on ions with signal intensity above 500. Recurring precursor ions were dynamically excluded for 30 sec. By applying charge-state monitoring, ions with 1+ or unassigned charge states were rejected to increase the fraction of ions producing useful fragmentation. The raw data files from LC-MSMS experiments were analyzed with MaxQuant software (version 1.0.13.13) (38) for peptide identification and quantification. Peak lists were generated from the raw data files using the Quant module in the MaxQuant program and searched by Mascot (39) against a concatenated target-decoy database of IPI-mouse version 3.63. Carbamylation (N terminus), oxidation (Met), and phosphorylation (Ser, Thr, Tyr) were searched as variable modifications. Carbamidomethylation (Cys) was specified as a fixed modification. Search results were maintained by applying 1% false discovery rate for peptide, protein, and site identification. Phosphopeptide pairs that have a normalized SILAC ratio with a significance B less than 0.05 (P value calculated by MaxQuant) were considered to have a statistically significant change in the level of phosphorylation. B less than 0.05 corresponded to a GH-dependent increase of approximately 50% or decrease of about 20% (exact percentage depended on the trial, Supplemental Tables 3–5 published on The Endocrine Society's Journals Online web site at http://mend.endojournals.org).
The datasets associated with this manuscript may be downloaded from the Tranche data repository (www.proteomecommons.org: under “browse data,” search for “Carter-Su C”).
Immunoblotting
3T3-F442A preadipocytes were grown in DMEM supplemented with 1 mm l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 0.25 μg/ml amphotericin, and 8% calf serum. Cells were incubated in serum-free medium overnight before GH treatment. In the inhibitor experiments, cells were pretreated with 100 nm wortmannin (Sigma, St. Louis, MO) and/or 100 nm CL-1040 (MEK inhibitor) (Selleck Chemicals, Houston, TX) for 30 min before GH treatment. After GH treatment, cells were washed with PBSV. Cells were solubilized with a buffer containing four parts lysis buffer [50 mm Tris (pH 7.5), 0.1% Triton X-100, 150 mm NaCl, 2 mm EGTA, supplemented with 1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride (PMSF), 10 μg/ml aprotinin, 10 μg/ml leupeptin (pH 7.5)] and one part SDS-PAGE sample buffer [50 mm Tris, 1% sodium dodecyl sulfate, 0.001% bromophenol blue, 10% glycerol, and 270 mm β-mercaptoethanol (pH 6.8)]. Samples were boiled and subjected to SDS-PAGE. Proteins in the gel were transferred to nitrocellulose, detected by immunoblotting with the indicated antibody, and visualized using an Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE). For experiments assessing raptor and phospho-raptor, cells were solubilized with lysis buffer, and cell lysates were clarified by centrifugation (10 min, 16,000 × g, 4 C) before SDS-PAGE.
Immunoprecipitation
3T3-F442A preadipocytes (15-cm culture dishes) were incubated in serum-free medium overnight before GH treatment. After GH treatment, cells were washed with PBSV, solubilized with 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate supplemented with 1 mm Na3VO4, 1 mm PMSF, 10 μg/ml aprotinin, 10 μg/ml leupeptin for 10 min on ice, and then centrifuged for 10 min at 16,000 × g at 4 C. Supernatants were incubated with αNHE1 and protein A-agarose beads overnight at 4 C. Beads were washed with lysis buffer, an 80:20 mixture of lysis buffer:SDS-PAGE sample buffer was added, and the samples were boiled for 5 min. Proteins were subjected to SDS-PAGE and immunoblotted as above. Band intensity was quantified using Odyssey software (LI-COR, version 2.0).
Results
Mass spectrometric analysis of GH signaling at 5 and 15 min
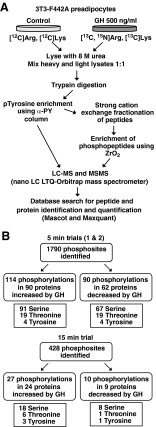
To expand our current view of GH-regulated proteins, we used a combined approach of SILAC, dual phosphopeptide enrichment using phosphoTyr antibodies, ZrO2, and LC-MSMS to quantify rapid GH-dependent changes in protein phosphorylation (Fig. 1A). 3T3-F442A preadipocytes, a highly GH-sensitive cell line with a relatively high GH receptor number and robust GH activation of JAK2, were used as a model system. 3T3-F442A preadipocytes have previously been used to identify and/or characterize multiple GH signaling proteins and pathways, including JAK2, Stats 1, 3, 5a, and 5b (5, 40–42), Shc/grb2/SOS/Ras/Raf/MEK/Erks1/2 (9, 43), and insulin receptor substrate 1/2/PI3K/Akt (10, 11, 44, 45). Light-labeled control cells grown in normal growth medium and heavy-labeled cells grown in medium containing [13C]Lys and [13C,15N]Arg were deprived of serum overnight, treated with (heavy-labeled) or without (light-labeled) GH for 5 or 15 min, and lysed. Equal (protein) amounts of cell lysates from GH-treated (+GH) and control (−GH) cell samples were mixed and digested with trypsin. Because of the low abundance of Tyr phosphorylation compared with Ser/Thr phosphorylation and its importance in post-GH receptor signaling pathways, Tyr-phosphorylated tryptic peptides were isolated by immunoaffinity purification using phosphoTyr-specific antibodies. Peptides present in the flow through from the immunoaffinity purification were fractionated using strong cationic exchange (SCX) HPLC and then enriched for Ser and Thr phosphopeptides using a ZrO2 column. The enriched phosphopeptides from the immunoaffinity purification/SCX/ZrO2-based purification were analyzed by LC-MSMS. The collected MSMS data were searched against a concatenated target-decoy database of IPI mouse database (version 3.63) using Mascot and processed with MaxQuant (version 1.0.13.13) (38) (www.maxquant.org) for identification and quantification of peptides and proteins. We set the threshold for a significant change at a significance B less than 0.05 (P value provided by MaxQuant) (38). A false discovery rate of 1% was used for peptide and protein identification. B less than 0.05 corresponded to a GH-dependent increase of approximately 50% or decrease of about 20% (Supplemental Tables 3–5).
Fig. 1.
A, Schematic of SILAC MS-based phosphoproteomics technique. B, Flowchart of results. The number of phosphosites in the various categories includes 1) unique localized phosphosites, and 2) unique unlocalized phosphosites that reside in peptides that do not contain a localized site (up to the total number of phosphosites in a peptide; see Supplemental Tables 1 and 2).
For the phosphosites for which phosphorylation decreased after GH treatment, the amount of the decrease required for significance was small (20% decrease). For borderline phosphosites, if the phosphosite was detected in a second trial, often the fold change fell below statistical relevance. To minimize the uncertainty associated with these peptides, for our analysis, we selected phosphosites that exhibited GH-dependent decreases of more than or equal to 30%.
In our first experiment, 3T3-F442A cells were treated without or with GH for 15 min. In two subsequent experiments, 3T3-F442A cells were treated without or with GH for 5 min because we wanted to increase the probability that very rapidly phosphorylated phosphosites were detected. In these three trials, we identified 428, 721, and 1406 phosphorylation sites, respectively (Fig. 1B). The increase in the number of phosphosites identified in each successive trial reflects, at least in part, an increase in the amount of cell lysate analyzed. In the first trial, 1.2 mg/condition were analyzed, 2.2 mg/condition were analyzed in the second trial, whereas 3.0 mg/condition were analyzed in the third trial. Localization probabilities and posttranslational modification scores were calculated by MaxQuant software. Phosphorylation sites with localization probabilities above 0.75 and a posttranslational modification score difference (between highest scoring site and second highest) larger than 5.0 (46) were considered to be localized phosphorylation sites (i.e. the specific phosphorylated amino acid(s) within the peptide could be identified with high confidence). In phosphopeptides with unlocalized phosphorylation sites, there is some ambiguity in the exact amino acid(s) that is (are) phosphorylated. Of the 1790 phosphorylation sites, we confidently localized 294, 500, and 869 phosphorylated residues on 240, 394, and 769 phosphopeptides in the 15-min and two 5-min trials, respectively. When the results of the three screens were combined, a total of 230 unique GH-dependent phosphosites (localized and unlocalized) on 141 different proteins were observed; 132 sites exhibited statistically significant GH-dependent increases (Table 1 and Supplemental Table 1) whereas the phosphorylation at 96 sites decreased by more than 30% (Supplemental Table 2). Ninety seven proteins had one GH-regulated phosphosite (68 increase, 29 decrease), 21 proteins had more than one site that showed increased phosphorylation, 17 had more than one site that showed decreased phosphorylation, and six had a combination of sites showing increased and decreased phosphorylation.
Table 1.
Phosphosites that exhibit increased phosphorylation after GH treatment
| Gene | Phosphosite2 | No. of Sites | Ratio (+GH/−GH) (5a/5b/15 min)3 | Function of phosphosite |
|---|---|---|---|---|
| 6330577E15Ri | [T70 = S71] | 1 | −/−/1.5 | Unknown |
| Acin1 | S710 | 1 | −/1.8/− | Unknown |
| Acly | S455 | 1 | 1.5/−/− | Increases catalytic activity (67) |
| Ahnak | S136 | 1 | 3.4/4.5/5.2 | Unknown |
| Ahnak | S217 | 1 | −/−/2.4 | Unknown |
| Ahnak | S4890 | 1 | 2.2/2.5/3.7 | Unknown |
| Ahnak | [S5557 >S5566 = T5567] | 1 | 2.1/−/− | Unknown |
| Ahnak | [T5569 >T5567 >T5571 >S5555 >S5566] | 2 | 2.5/−/− | Unknown |
| Ahnak2 | S652 | 1 | −/2.4/− | Unknown |
| Akap1 | S109 - isoforms 1,3,5,6 (S142 - isoforms 2,4) | 1 | 1.5/−/− | Unknown |
| Akap2 | S740 (S984) | 1 | −/1.9/1.6 | Unknown |
| Akap2 | [T738 >S740] [(T982 >S984)] | 1 | −/2.2/− | Unknown |
| Akt1s1 (PRAS40) | S184 (S255) | 1 | −/2.2/3.9 | Regulates PRAS40 interaction with raptor (87) |
| Akt1s1 (PRAS40) | T247 (T318) | 1 | 3.8/−/− | Suppresses PRAS40 inhibition of mTORC1 (60) |
| Arfgap2 | S145 | 1 | −/1.7/− | Unknown |
| Atp6v0a2 | S695 | 1 | 1.4/−/− | Unknown |
| Bat2 | S761 | 1 | −/1.5/− | Unknown |
| Bckdha | S334 (S338) | 1 | 1.6/−/− | Unknown |
| Bcl9l | [S116 >S118] | 1 | −/2.8/− | Unknown |
| Bcl9l | S118 | 1 | −/2.4/− | Unknown |
| Bclaf1 | S383 | 1 | 1.6/−/− | Unknown |
| Cad | S1859 | 1 | −/1.9/− | Unknown |
| Cast | S219 | 1 | −/1.5/− | Unknown |
| Cbx5 | S95 (S93) | 1 | −/2/− | Unknown |
| Cd2ap | S458 | 1 | −/1.5/− | Unknown |
| Cic | [S2280 >T2283 = S2284 >S2275 = T2277] | 2 | 1.4/−/− | Unknown |
| D930048N14Rik | T15 | 1 | −/4.1/− | Unknown |
| Dact3 | S409 | 1 | 2.2/−/− | Unknown |
| Dap | T56 | 1 | −/22/− | Unknown |
| Dcaf8 | S123, S124 | 2 | 1.5/−/− | Unknown |
| Dpysl3 | S101 | 1 | −/1.7/− | Unknown |
| Eif4b | [T420 >S422] | 1 | −/3.1/− | Increases interaction with Eif3 (88) and Eif4b activity (89) |
| Flnc | S2234 | 1 | 3.3/2.3/1.7 | Unknown |
| Fnbp1l | S295 | 1 | −/2.5/− | Unknown |
| Frs2 | T326 | 1 | 1.8/−/− | Unknown |
| Gm15128 | S384, S388 | 2 | −/1.5/− | Unknown |
| Gsta3 | T68, Y74 | 2 | −/−/1.8 | Unknown |
| Hmgn1 | [T70 = T71 >S84 = S87] | 2 | −/1.6/− | Unknown |
| Irf2bp2 | S421 | 1 | 2.2/−/− | Unknown |
| Irs2 | S556 | 1 | −/1.5/− | Unknown |
| Itpr3 | S934 | −/1.7/− | Unknown | |
| Kif21a | S855 | 1 | −/2.1/− | Unknown |
| Larp7 | [T251 >S253] | 1 | −/1.6/− | Unknown |
| Lima1 | S230 | 1 | −/1.5/− | Unknown |
| Map1b | S614 | 1 | −/1.6/− | Unknown |
| Map1b | S1793 | 1 | −/−/1.6 | Unknown |
| Mapk1 | [T179 >T183 = Y185 = T188] | 1 | −/8.7/− | pT183 and pY185 required for activation (90). Function of pT179 and pT188 unknown |
| Mapk1 | T183, Y185 | 2 | 11/−/− | |
| Mapk1 | T183 [Y185 = T188 >T179] | 2 | 9.5/−/− | |
| Mapk1 | Y185 [T179 = T183] | 2 | 5.4/−/− | |
| Mapk1 | Y185 | 1 | 2.4/2.4/− | |
| Mapk3 | T199 [T203 = Y205] | 2 | 6.7/−/− | pT203 and pY185 required for activation (90). Function of pT199 and pT208 unknown. |
| Mapk3 | Y205 | 1 | 3.5/2.9/− | |
| Mapk3 | T208 [T203 >Y205 >T199] | 2 | 4.8/−/− | |
| Mapk3 | [T208 >Y205] | 1 | −/3.0/− | |
| Matr3 | S188 | 1 | 1.7/1.8/− | Unknown |
| Matr3 | S195 | 1 | −/2/− | Unknown |
| Med19 | S226 >S234 = S235 = S238 = S239 = S240 = S241 = S242 | 7 | −/9.6/− | Unknown |
| Mia3 | S1915 | 1 | −/−/1.7 | Unknown |
| Myh9 | [T1938 = S1942] ([T1939 = S1943]) | 1 | 1.4/−/− | Unknown |
| Ncbp1 | S22 | 1 | −/2.2/− | Unknown |
| Ncoa5 | S29, S34 | 2 | −/1.6/− | Unknown |
| Ndrg1 | T328 | 1 | −/1.8/− | Primes NDRG1 for phosphorylation by GSK3 (65). Required for suppressive effect on NFκΒ signaling (66) |
| Ndrg1 | S330 | 1 | −/1.5/− | |
| Nr3c1 | T159 (T168) | 1 | −/1.8/− | Unknown |
| Oxr1 | S113 (S194) | 1 | 2.5/2.4/− | Unknown |
| Oxr1 | S114 (S195) | 1 | −/2.1/− | Unknown |
| Parva | T16, S19 | 2 | −/1.5/− | Unknown |
| Pdcd4 | S76 | 1 | −/3.5/− | Mediates Pdcd4 interaction with βTRCP (91) |
| Pdcd4 | S457 | 1 | −/3.8/− | Promotes in nuclear translocation of Pdcd4 (92) |
| Pgrmc1 | Y180 | 1 | 1.4/−/− | Unknown |
| Phf3 | [S658 >S660] | 1 | −/3.5/− | Unknown |
| Phldb2 | T468 | 1 | −/1.8/− | Unknown |
| Pi4k2a | [S460 >S462] | 1 | −/−/2.3 | Unknown |
| Pkn2 | [S581 = S582] | 1 | −/6.7/− | Unknown |
| Pkn2 | T957 | 1 | −/−/1.6 | Unknown |
| Plec1 | [T19 >S21] | 1 | −/−/1.6 | Unknown |
| Plec1 | [S4391 >S4392 >S4393] [(S4243 >S4244 >S4245)] | 1 | −/2.1/− | Unknown |
| Plec1 | S4393 (S4245) | 1 | 1.6/2.1/− | Unknown |
| Ppfibp1 | S957, S961 (S992, S996) | 2 | −/1.5/− | Unknown |
| Ppp1r12a | S507 | 1 | 6.9/8.6/− | Unknown |
| Ppp1r12a | S509 | 1 | 4.0/−/− | Unknown |
| Ppp1r12a | S861 | 1 | 1.5/−/− | Unknown |
| Ppp1r12a | [S870 >S877] | 1 | −/1.7/− | Unknown |
| Ptprn2 | S89 | 1 | −/104.9/− | Unknown |
| Rai14 | S914 | 1 | 1.5/−/− | Unknown |
| Ranbp3 | S58 | 1 | −/2.5/− | Important for nuclear export function (93) |
| Raver1 | S576 | 1 | −/1.9/− | Unknown |
| Rbm17 | S155 | 1 | 1.7/−/− | Unknown |
| Rbmx | S165 | 1 | −/1.5/− | Unknown |
| Rgs2 | S19 | 1 | −/12.2/− | Unknown |
| Rnf220 | T474 | 1 | −/−/5.6 | Unknown |
| Rptor | S863 | 1 | 1.5/2.0/1.9 | Required for hierarchical raptor phosphorylation. Plays a critical role in mTOR activity (57, 94) |
| Sdpr | S359 | 1 | 1.6/−/− | Unknown |
| Senp7 | S12 | 1 | −/2.7/− | Unknown |
| Sep9 | [S41 = T42] | 1 | 1.4/−/− | Unknown |
| Sgta | [S303 >T305] | 1 | −/2.0/− | Unknown |
| Shc1 | Y423 | 1 | −/3.7/− | Binds grb2 (61) |
| Sipa1l2 | S1461 | 1 | −/−/1.6 | Unknown |
| Slc9a1 (NHEI) | S707 | 1 | −/7.3/3.3 | Binds 14-3-3 and is required for Serum-induced activation of Nhe1 (95) |
| Srrm1 | [S461 >S463] | 1 | −/2.8/− | Unknown |
| Srrm2 | S2224 | 1 | −/2.9/− | Unknown |
| Stat5a | Y694 | 1 | −/+GH only/+GH only | Required for Stat5 activation (96) |
| Stat5b | Y699 | 1 | −/+GH only/+GH only | Required for Stat5 activation (96) |
| Stk10 | T950 | 1 | −/2/2.3 | Unknown |
| Stmn1 | S25 | 1 | −/7.6/− | Unknown |
| Tbc1d15 | S201 | 1 | −/−/1.6 | Unknown |
| Tnks1bp1 | S1022 | 1 | −/2.5/− | Unknown |
| Tnks1bp1 | S1375 | 1 | −/−/1.5 | Unknown |
| Tns3 | S769 | 1 | 2.0/−/− | Unknown |
| Top2b | S1387 (S1537) | 1 | 1.5/1.9/− | Unknown |
| Tpr | S2149 | 1 | −/6.3/− | Unknown |
| Trim28 | S471 | 1 | −/−/2.3 | Unknown |
| Trim28 | S473 | 1 | −/1.5/2.4 | Regulates Trim28 interaction with HP1β (97) |
| Trim47 | S591 (S592) | 1 | −/−/2.0 | Unknown |
| Trio | S2399, S2403 (S22575, 2579) | 2 | −/5.0/− | Unknown |
| Tsc2 | [S939 >T940 >S941] [(S937 >T938 >S939)] | 1 | −/1.9/− | Promotes interaction with 14-3-3 and relieves inhibition of mTOR signaling (98) |
| Twist1 | S68 | 1 | 1.6/−/− | Unknown |
| Twist2 | S55 | 1 | 1.6/−/− | Unknown |
| Usp10 | S223 | 1 | −/3.8/− | Unknown |
| Wdr48 | S586 | 1 | −/−/5.0 | Unknown |
| Zc3hc1 | S394 | 1 | −/−/1.8 | Unknown |
| Zc3hc1 | S406 | 1 | −/2.6/− | Unknown |
| Zyx | S336 | 1 | −/1.8/− | Unknown |
Numbering of residues is according to PhosphositePlus (http://www.phosphosite.org). Numbering in parentheses is according to MaxQuant. Numbering in brackets refers to unlocalized sites (localization probability <0.75 and >0.15).
5a and 5b refer to the first and second 5 min trials, respectively.
When the phosphorylation ratios (+GH/−GH) for the 327 phosphorylation sites present in both 5-min trials were compared, the ratios were very similar, with a correlation coefficient of 0.8766 (r2 = 76.84%) (Supplemental Tables 1–4). The consistency of the phosphorylation ratios for the sites present in both 5-min trials validates the SILAC results and underscores the fact that the relative peptide levels encoded in the isotopically labeled samples are relatively unaffected by variability introduced during sample preparation (SCX-fractionation, phosphopeptide enrichment, and multiple desalting steps) that precedes the analysis by MS.
In the first and second 5-min trials, 65 and 148 GH-regulated phosphorylation sites were observed, respectively, and 14 sites were present in both 5-min trials (Supplemental Tables 1 and 2). With the highly complex peptide mixtures obtained from cell lysates, a mass spectrometer obtains sequence from only a small fraction of the phosphopeptides present in the sample (47). For studies in which similar numbers of phosphosites are identified in the various individual trials, the fraction of the phosphosites identified in the replicate trials gives an indication of the extent to which the study has sampled the phosphoproteome of the cell type. Because only a small fraction of the phosphoproteome of the cell is normally sampled, the detection of a phosphorylated residue in even a single trial is noteworthy.
When compared with the 5-min screens, the 15-min screen identified a relatively small number of phosphosites, presumably because less lysate was analyzed in the 15-min screen (1.2 mg/condition vs. 2.2 and 3.0 mg/condition for the two 5-min screens). In the 15-min GH screen, more than 55% of the phosphosites identified, 238 of a total of 428 sites, were observed in at least one of the two 5-min GH screenings (Fig. 2 and Supplemental Tables 1–5). Of the 24 phosphosites that were observed in more than one of the three screens and fulfilled our criteria for a GH-dependent increase in at least one screen, 19 also fulfilled the criteria (with generally quite similar increases) in a second screen, three showed slightly reduced increases that fell just below our cut-off criteria in the second screen, and only two (<10%) showed an increase in one screen but not another (Table 1 and Supplemental Table 1). These findings suggest that the variability of this approach lies primarily in the ability to detect a particular phosphopeptide in any particular screen, rather than in the ability of GH to up-regulate phosphorylation of that amino acid. They also suggest that detection of a GH-dependent increase in a phosphosite observed in any one of the three screens has a high probability of being valid, even when the phosphopeptide was not detected in the other two screens.
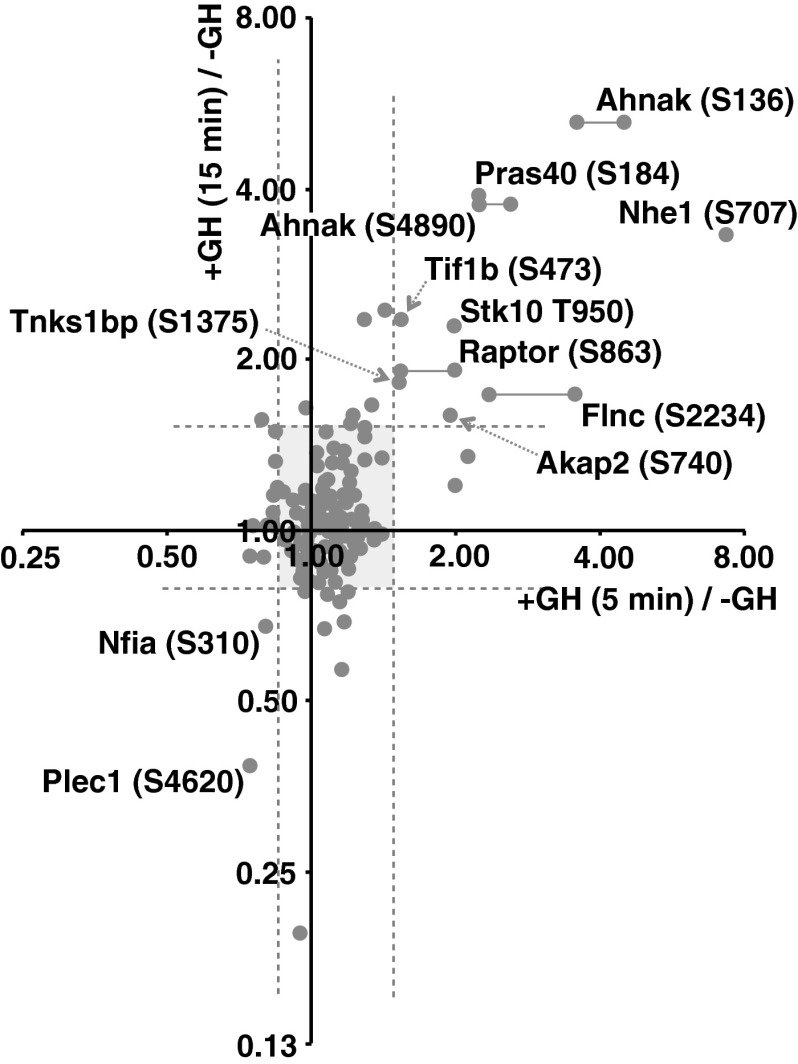
Fig. 2.
Scatter plot showing phosphorylation ratios of the 238 phosphosites observed in both the 15-min and 5-min GH screenings. The ratio (+GH/−GH) of phosphorylation of the phosphosites observed in either of the 5-min screens and the 15-min screen was graphed (log2 scale). The vertical and horizontal dotted lines indicate statistically significant (B < 0.05) changes in the GH-dependence for 5-min or 15-min samples, respectively. Phosphorylations that were either statistically significantly up- or down-regulated after both 5- and 15-min GH treatment are identified by protein name and phosphoamino acid site. Horizontal lines connecting dots link those sites that appeared in both 5-min GH screens. Most phosphorylations, shown in the shaded region, remained unchanged by GH treatment.
Some proteins that are phosphorylated in the basal state exhibit decreased phosphorylation in response to GH (Supplemental Table 2). GH presumably stimulates dephosphorylation of these target proteins by stimulating phosphatase activity or by inhibiting a constitutively active kinase. Increased phosphatase activity could be the result of a phosphatase being activated, e.g. by phosphorylation. Similarly, inhibition of a constitutively active kinase could occur as a consequence of a GH-dependent phosphorylation event. Alternatively, a protein could become more accessible to an already active phosphatase or less accessible to an already active kinase due to a GH-dependent change in subcellular localization or conformation. As one would predict for decreased phosphorylation, when a negatively regulated phosphopeptide was detected in both 5- and 15-min screens, the 15-min screen more often than not exhibited a more substantial decrease (Fig. 2). Of the 12 phosphosites that were observed in more than one of the three screens and fulfilled our criteria for a GH-dependent decrease in at least one screen, four were decreased after 15 min but not after 5 min, one fulfilled our criteria for a decrease in two separate screens, four showed a similar decrease in the second screen that fell just above our cut-off criteria, and three fulfilled the criteria in only one of the two 5-min screens. Thus, the data for the GH-dependent decreases in phosphorylation were not as reproducible as the data for phosphorylation increases. Because of the greater variability in and generally smaller magnitude of the GH-dependent decreases in phosphorylation and the unavailability of phosphospecific antibodies against the phosphosites with decreased phosphorylation, for subsequent analysis, we focused on the phosphosites that had GH-induced increases in phosphorylation.
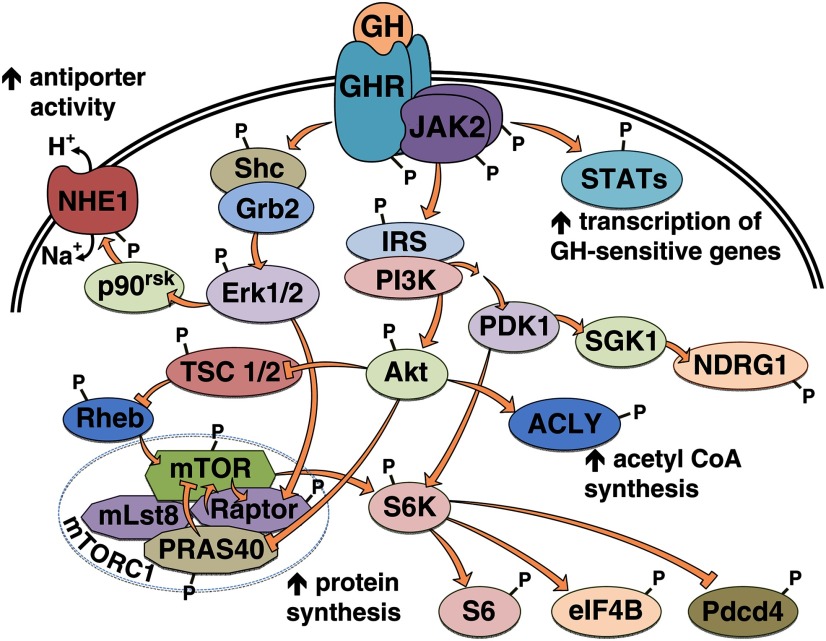
Of the 132 unique localized phosphosites showing GH-dependent increases in phosphorylation, only six were previously documented as GH responsive. These were the activating Tyr in the transcription factors Stat5a (Tyr694)1 and Stat5b (Tyr699) (only detected in GH-stimulated cells); the activating Thr and Tyr in the serine/threonine kinase Erk2 (Thr183/Tyr185) (+GH/−GH = 10.9); the activating Tyr in Erk1 (Tyr205) (+GH/−GH = 2.4); and the regulatory Ser939 in tuberous sclerosis protein 2 or tuberin (TSC2) (+GH/−GH = 1.9). TSC2 is a GTPase-activating protein, which inhibits Ras homolog enriched in brain (Rheb), and thereby inhibits mammalian target of rapamycin (mTOR) complex 1 (mTORC1) (Fig. 3; reviewed in Ref. 48). mTORC1 activity is required for rapid GH-stimulated protein synthesis (14). Phosphorylation of TSC2 Ser939 suppresses TSC2 function, resulting in increased mTORC1 signaling (49, 50). The remaining 126 phosphosites have not been identified previously as GH-regulated sites of phosphorylation. Of the newly identified GH-regulated sites, only 13 have been identified in the context of other stimuli and their function analyzed. The function and mechanism of phosphorylation is unknown for the remaining 113 sites.
Fig. 3.
Schematic of signaling pathways containing the newly identified GH-responsive phosphoproteins. The kinases in the AGC family (Akt, SGK1, and p90rsk) are colored light green. GHR, GH receptor; Rheb, Ras homolog enriched in brain.
Functional analysis of phosphoproteins identified after 5 and 15 min of GH treatment
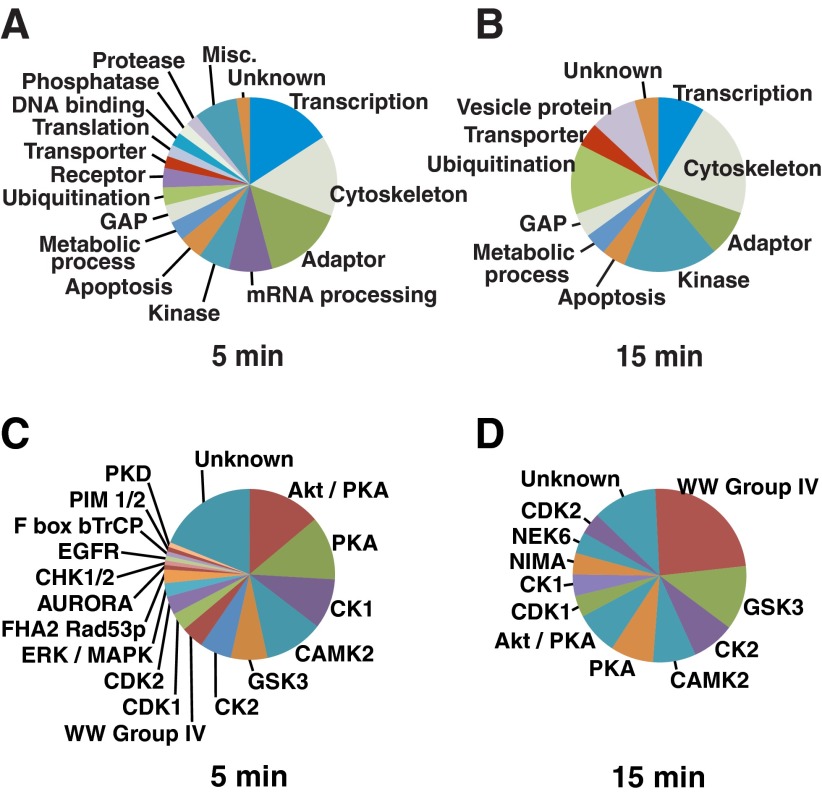
The phosphoproteins that exhibited increased phosphorylation in response to GH were assigned to functional groups based on manual annotation performed by the authors using information about protein function tabulated in PhosphositePlus (http://www.phosphosite.org) (Supplemental Table 1). During the early period (5–15 min) after GH treatment, the largest protein functional groups undergoing GH-dependent increases in phosphorylation were proteins that served as adaptors or were involved in regulating transcription, the cytoskeleton, or mRNA processing (Fig. 4, A and B). The broad range of functions for these 132 GH-responsive proteins, both metabolic and structural, suggests that further study of these proteins has the potential to provide a molecular basis for the many GH responses (51) that are only beginning to be understood.
Fig. 4.
Functions of the phosphoproteins and the kinase recognition motifs associated with the phosphosites that undergo increased GH-dependent phosphorylation. A and B, The phosphoproteins identified during the SILAC-based phosphoproteomic screens after 5-min (A) or 15-min (B) GH treatment were classified according to the information in Phosphosite Plus (http://www.phosphosite.org), which includes information from UniProtKB. The first function listed in the protein function column of Supplemental Table 1 was used to construct the pie charts. C and D, The GH-dependent phosphopeptides identified during the SILAC-based phosphoproteomics screens after 5-min (C) or 15-min (D) GH treatment were analyzed by MaxQuant software to predict the best-fit kinase motifs surrounding the phosphorylation sites (Best Motif column, Supplemental Tables 3–5). Akt/PKB; CDK,cell division protein kinase; PIM, proto-oncogene Ser/Thr-protein kinase; PKD, Ser/Thr-protein kinase D; NEK6, Ser/Thr-protein kinase Nek6; NIMA, never in mitosis A-related kinase 6; GAP, GTPase-activating protein; EGFR, epidermal growth factor receptor; CHK, csk homologous kinase; F box bTrCP, Fbox β-transducin repeat containing protein; FHA2 Rad53p, forkhead associated 2, radiation sensitive kinase 53.
To gain insight into pathways activated by GH, we used Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (http://www.genome.jp/kegg/pathway.html) to categorize the proteins that contain phosphosites that increased with GH treatment at 5 or 15 min (Table 2). Four pathways contained five or more proteins that exhibited GH-dependent phosphorylation. The four pathways identified by KEGG have obvious functional overlaps and not surprisingly, several proteins are present in two or more pathways. The identification of previously unknown, GH-regulated proteins in two of the categories (“regulation of the actin cytoskeleton,” and “focal adhesions”) highlights proteins that have the potential to broaden our understanding of the mechanistic basis underlying the ability of GH to promote changes in the actin cytoskeleton and cell motility (52–56). In the category “regulation of the actin cytoskeleton,” the phosphosites in protein phosphatase 1 regulatory subunit 12a (Ppp1r12a)/Mypt1/myosin-binding subunit of myosin phosphatase, NHE1, and myosin heavy chain 9 were not previously known to be GH regulated. In the category “focal adhesion,” the phosphosites in filamin, zyxin, parvin, and Ppp1r12a were not previously known to be GH regulated. The identification of proteins in the categories “insulin signaling pathway” and “mTOR signaling pathway” that exhibit increased levels of phosphorylation in GH-treated cells are consistent with the known role of GH in the regulation of metabolism and cell growth (1, 2). Erk1, Erk2, and TSC2 were known previously to be phosphorylated in response to GH on the sites we identified. The phosphosites in raptor and eukaryotic initiation factor 4B (eIF4B) were not previously known to be GH-regulated sites. Manual examination of the functions reported for proteins in our proteomic analysis revealed two additional proteins in the mTOR signaling pathway, each with two GH-up-regulated sites: PRAS40, which, like raptor, is part of the mTORC1 complex, and programmed cell death protein 4 (Pdcd4), which, like eIF4B, is a target of S6 kinase (Fig. 3). Thus, a total of 10 GH-dependent phosphosites in seven proteins lie within the mTOR signaling pathway. These newly identified GH-dependent phosphosites are all documented regulatory sites, i.e. sites whose phosphorylation in response to some stimulus has been reported to regulate the function of the protein (Table 1 and Supplemental Table 1), suggesting that GH-promoted phosphorylation at these phosphosites regulates function and is physiologically relevant. The 15 proteins in these four KEGG categories include only a small fraction of the 95 GH-regulated proteins identified in our SILAC/MS-based phosphoproteomic study that exhibit GH-dependent increases in phosphorylation. Future analysis of these latter proteins, most of whose GH-dependent phosphosites have no known function, stands to further increase our understanding of the physiological actions of GH.
Table 2.
Signaling pathways and cellular functions identified by KEGG analysis
| KEGG pathways | Identified proteins involved in pathway |
|---|---|
| Regulation of actin cytoskeleton | Erk1 (Thr199, Thr203, Tyr205, Thr208) |
| Erk2 (Thr179, Thr183, Tyr185) | |
| NHE1 (Ser707) | |
| Ppp1r12a (Ser507, Ser509, Ser870, Ser861) | |
| Myh9 (Thr1938) | |
| Focal adhesion | Filamin-C (Ser2234) |
| Zyxin (Ser336) | |
| Parvin (Thr16, Ser19) | |
| Ppp1r12a (Ser507, Ser509, Ser870, Ser861) | |
| Shc (Tyr 423) | |
| Erk1 (Thr199, Thr203, Tyr205, Thr208) | |
| Erk2 (Thr183, Tyr185, Thr179) | |
| Insulin-signaling pathway | IRS2 (Ser556) |
| Shc (Tyr 423) | |
| Raptor (Ser863) | |
| TSC2 (Ser939) | |
| Erk1 (Thr199, Thr203, Tyr205, Thr208) | |
| Erk2 (Thr179, Thr183, Tyr185) | |
| MTOR-signaling pathway | Erk1 (Thr199, Thr203, Tyr205, Thr208) |
| Erk2 (Thr179, Thr183, Tyr185) | |
| TSC2 (Ser939) | |
| Raptor (Ser863) | |
| eIF4B (Thr420) |
Proteins identified in the phosphoproteomic screens that contain phosphosites that increased with GH treatment at 5 or 15 min (Table 1 and Supplemental Table 1) were subjected to KEGG pathway analysis (http://www.genome.jp/kegg/pathway.html) to categorize the proteins by function and signaling pathway.
Motif analysis of GH-dependent phosphosites
To provide insight about the kinases and pathways activated in response to GH, the motif analysis provided by the MaxQuant software was used to predict which kinase(s) were likely to have phosphorylated each of the GH-dependent phosphorylation sites identified in our studies. After 5 min of GH treatment, 93 of the GH-stimulated phosphosites were in known consensus phosphorylation motifs. The majority of the 93 phosphosites were in consensus recognition sites for Akt/protein kinase A (PKA), PKA, casein kinase (CK)1, Ca2+/calmodulin-dependent protein kinase (CamK)2, glycogen synthase kinase-3 (GSK3), and CK2 (Fig. 4C). After 15 min of GH treatment, 24 of the GH-induced phosphorylation sites identified were in consensus phosphorylation motifs. For these sites, the most common consensus recognition motif was the WW Group IV (Fig. 4D). These findings suggest that the kinases Akt, PKA, CK1, CamK2, GSK3, and CK2, and kinases that recognize the WW Group IV motif (or kinases with similar consensus recognition sites) are important components in GH signaling pathways. Akt/PKA sites were the most prevalent; 16 phosphopeptides that contained Akt/PKA sites were detected in our screens, which raises the possibility that GH activation of Akt (or kinases with similar consensus recognition sites) plays a greater role in GH signaling than previously recognized. Because only a relatively small fraction of the sites fell within the consensus substrate sequences of known (e.g. ERK, Akt) GH-regulated kinases, this analysis also suggests that much work remains in determining the pathways by which the remaining sites are phosphorylated in response to GH.
Validation of mass spectrometric results
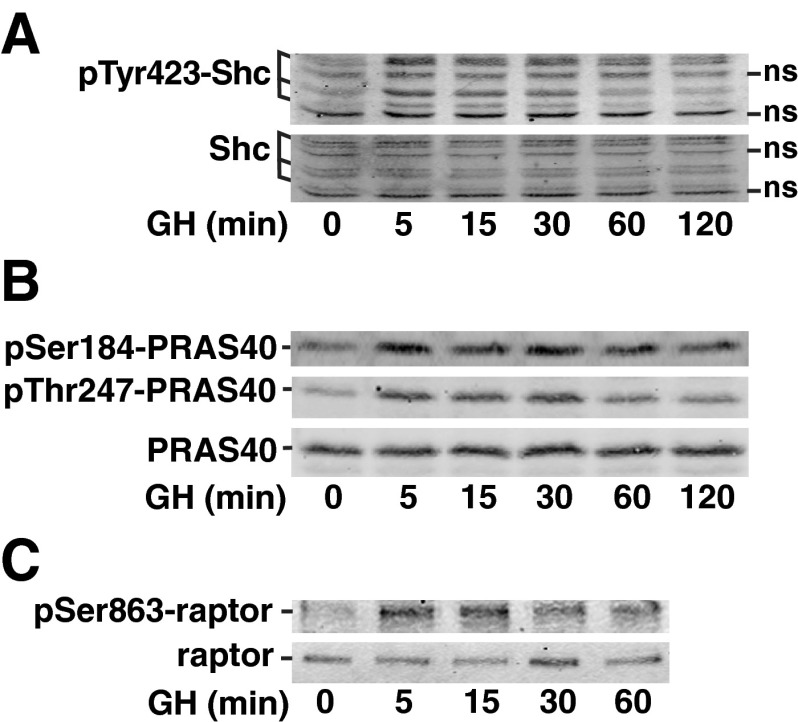
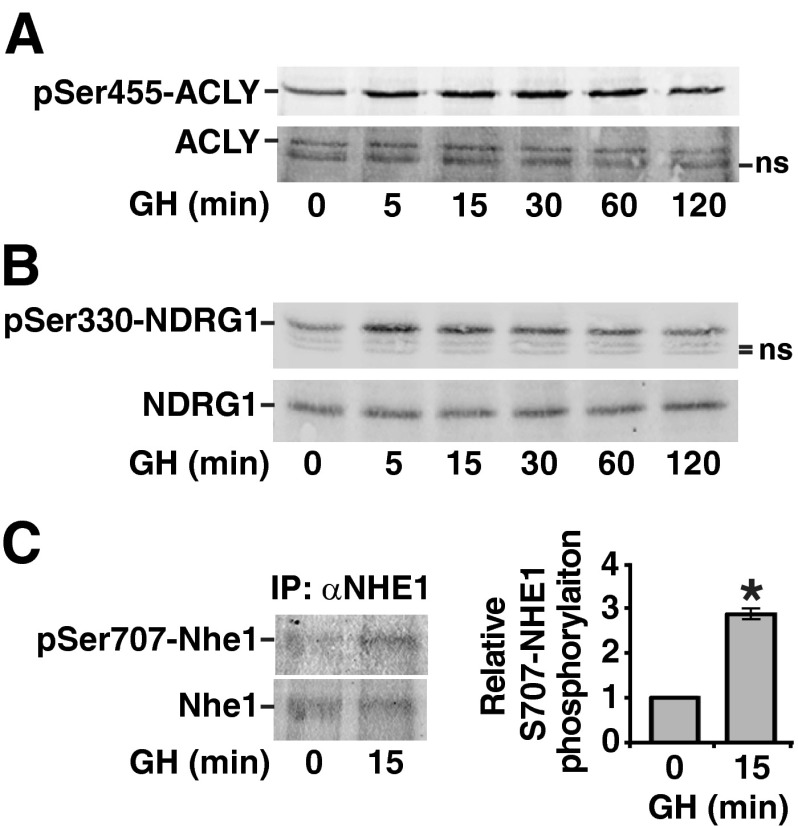
To gain insight into the reliability of the identification of the novel GH-dependent phosphosites, we performed immunoblotting with phosphospecific antibodies. Sites with available antibodies included three sites in proteins associated with the mTOR pathway (raptor Ser863, PRAS40 Ser184, and Thr247), one site identified in a protein in the insulin and focal adhesion categories (Shc Tyr423), and one site identified in a protein involved in regulation of actin cytoskeleton (NHEI Ser707). Of these, PRAS40 Thr247 and NHE1 Ser707 fell into the category of Akt/PKA substrate sites. We also tested for phosphorylation at two additional Akt/PKA substrate consensus sites, NDRG1 Ser330 and ACLY Ser455. All of these sites were previously identified as phosphosites with regulatory function in the context of other stimuli. Within the mTOR pathway, phosphorylation of raptor on Ser863 by mTOR within mTORC1 or by Erks 1/2 promotes phosphorylation of raptor on other sites and augments mTORC1 activity (36, 57, 58). Phosphorylation of PRAS40 Thr247 by Akt facilitates efficient phosphorylation of Ser184 in PRAS40 by mTORC1 (59), which relieves PRAS40-dependent inhibition of mTORC1 (60). Shc recruitment to GH receptor/JAK2 complexes and its subsequent phosphorylation by JAK2 and recruitment of grb2 is thought to initiate GH activation of a Shc/grb2/SOS/Ras/Raf/MEK/Erk pathway. Shc Tyr423 has been implicated as a grb2 binding site (61). NHE1 is a sodium/proton antiporter that catalyzes the exchange of Na+ and H+ down their concentration gradients. This exchange is important for control of intracellular pH, adhesion, migration, and proliferation. Phosphorylation of Ser707 of NHE1 has been reported to increase the antiporter activity of NHE1 (35). NDRG1 has been implicated in growth and differentiation and is necessary for p53-induced apoptosis (62–64). Phosphorylation of Ser330 (and Thr328) of NDRG1 primes NDRG1 for further phosphorylation by GSK3 (65) and is required for the suppressive effect of NDRG1 on the nuclear factor-κB signaling pathway (66). Finally, ACLY is the primary enzyme responsible for the synthesis of cytosolic acetyl-coenzyme A (CoA) in many tissues. Phosphorylation of ACLY Ser455 by Akt has been shown to activate ACLY (67).
To validate the results of our phosphoproteomic analysis, cell lysates from 3T3-F442A preadipocytes were treated with GH for varying amounts of time and then blotted with the appropriate phosphospecific antibodies. Immunoblotting with phosphospecific antibodies confirmed a rapid and robust GH-dependent phosphorylation of Shc Tyr423 (Fig. 5A), Ser184 and Thr247 in PRAS40 (Fig. 5B), and raptor Ser863 (Fig. 5C). A rapid, modest GH-dependent phosphorylation of Ser455 in ACLY (Fig. 6A) and Ser 330 in NDRG1 (Fig. 6B) was also observed. Similarly, when NHE1 was immunoprecipitated from 3T3-F442A preadipocytes that had been treated with GH for 0 or 15 min and immunoblotted with αpSer707-NHE1, a GH-dependent increase in phosphorylation of Ser707 was observed (Fig. 6C). In each case, blotting with antibody against the corresponding total protein confirmed that the change in signal was due to a change in the amount of protein phosphorylated and not in the amount of protein.
Fig. 5.
Shc, PRAS40, and raptor are phosphorylated in response to GH. 3T3–F442A preadipocytes were treated (A and C) with GH [500 ng/ml (23 nm)], or (B) with GH (100 ng/ml) for the times indicated. Whole-cell lysates were immunoblotted with αpY423-Shc, αShc, αpS184-PRAS40, αpT247-PRAS40, and αPRAS40 as indicated. Clarified cell lysates were immunoblotted with αpS863-raptor or α-raptor as indicated. The results are representative of three separate experiments. ns, Nonspecific.
Fig. 6.
ACLY, NDRG1 and NHE1 are phosphorylated in response to GH. 3T3–F442A preadipocytes were treated with GH (100 ng/ml) (panel A), or with GH (500 ng/ml) (panels B and C) for the times indicated. A and B, Whole-cell lysates were immunoblotted with αpS455-ACLY, αACLY, αpS330-NDRG, or αNDRG as indicated. C, NHE1 was immunoprecipitated from clarified cell lysates using αNHE1 and then immunoblotted with αpS707-NHE1 and αNHE1. αpS707-NHE1 band intensity was quantified and normalized to αNHE-1 band intensity. The average intensity of three experiments ± sem was graphed, *, P < 0.05. IP, Immunoprecipitation; ns, nonspecific.
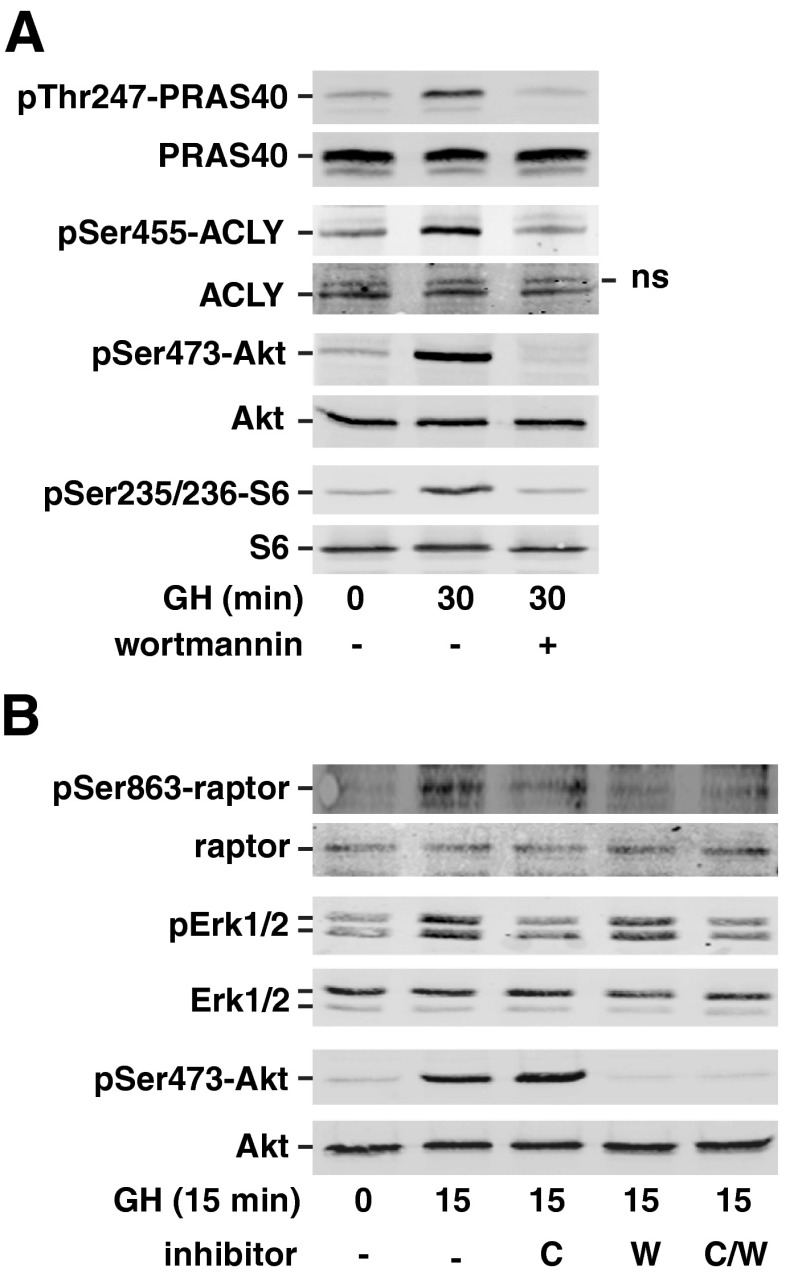
We also tested whether the GH-dependent phosphorylation detected at PRAS40 Thr247, ACLY Ser455, and raptor Ser863 is downstream of Akt as predicted by the motif analysis (Supplemental Tables 3–5). We treated 3T3-F442A preadipocytes for 30 min with wortmannin, a phosphatidylinositol 3-kinase (PI3K) inhibitor that blocks GH activation of Akt (68) and then with GH for 15 or 30 min. The efficacy of wortmannin at inhibiting Akt activity was determined by blotting with antibody to Akt pSer473 or with a phosphospecific antibody to the S6 kinase 1 substrate Ser235/Ser236 in ribosomal S6. We observed robust GH-dependent phosphorylation of PRAS40 Thr247, ACLY Ser455 (Fig. 7A), and raptor Ser863 (Fig. 7B). Phosphorylation at all of these sites was suppressed by wortmannin treatment, indicating that GH-induced phosphorylation of PRAS40 Thr247, ACLY Ser455, and raptor Ser863 lies downstream of PI3K/Akt. Phosphorylation of raptor Ser863 is catalyzed by several kinases (Fig. 3), including Erks1/2 (36, 57, 58). Because GH is known to stimulate Erk activation (9), we also investigated whether the MEK inhibitor CL-1040 inhibits GH-dependent phosphorylation of raptor Ser863. The efficacy of CL-1040 to inhibit MEK substrates Erk1 and Erk2 was assessed by blotting with antibody against the doubly phosphorylated activated form of Erk1/2. GH-dependent stimulation of raptor phosphorylation at Ser863 was suppressed when MEK/Erk activity was inhibited (Fig. 7B), suggesting that GH activation of both PI3K/Akt and Erk1/2 contributes to GH-induced phosphorylation of raptor Ser863. Taken together, these results provide strong support for the validity of the phosphoproteomics results and confirm a multifaceted role for the Akt pathway in GH signaling.
Fig. 7.
GH-induced phosphorylation of ACLY, PRAS40, and raptor is inhibited by wortmannin. A, 3T3–F442A preadipocytes were pretreated with 100 nm wortmannin or vehicle for 30 min and then treated with GH (500 ng/ml) for 30 min. Whole-cell lysates were immunoblotted with αpS455-ACLY, αACLY, αpT247-PRAS40, αPRAS40, αS473-Akt, αpS235/236-S6 kinase, or αS6 kinase as indicated (n = 3). B, 3T3–F442A preadipocytes were pretreated as indicated with vehicle, 100 nm wortmannin, and/or 500 nm CL-1040, for 30 min and then treated with GH (500 ng/ml) for 15 min. Clarified cell lysates were immunoblotted with αpS863-raptor, α-raptor, αpErk1/2, αErk1/2, αpS473-Akt, or αAkt (n = 3).
Discussion
The current study is the first to use SILAC and quantitative MS-based phosphoproteomics to study GH signaling. To our knowledge, it is also the first study to use SILAC and quantitative MS-based phosphoproteomics to investigate ligand-dependent signaling via any member of the cytokine receptor family. This approach allowed us to identify and quantify GH-dependent changes in more than 200 protein phosphorylation sites. Among the seven GH-stimulated sites identified by phosphoproteomics that were the focus of the validation portion of our analysis, the GH response determined by phosphoproteomics varied from robust to subtle. However, immunoblotting confirmed all seven sites tested, even those with the most subtle increases, including four that were picked up in only one of the three screens, supporting our contention that for the GH up-regulated phosphosites, identification in even a single screen makes that site likely to be a bona fide GH-regulated site. Clearly, complete coverage of the phosphoproteome was not obtained in any of our screens, as indicated by the absence of known GH-dependent phosphosites (e.g. in Akt, GH receptor, JAK2, SH2B1, Raf1, MEK1/2) (13, 45, 69–72) and the fact that many of the phosphosites were identified in only one of our three screens. We did not expect complete coverage for multiple reasons. First, a certain amount of variability originates in the multistep preparation that precedes the analysis by MS. Second, ZrO2 selects for a particular subpopulation of phosphopeptides in the phosphoproteome. Studies using yeast revealed that different modes of enriching for phosphopeptides sample quite different subpopulations of the phosphoproteome with little overlap among them. Use of four different enrichment methods significantly increased the coverage of the phosphopeptide population (Kweon and Andrews, manuscript in preparation), suggesting that future studies would benefit from combining the results from multiple, complementary enrichment schemes. Third, in the modern LC-mass spectrometer, for each successive peak fraction selected by the nano LC system for analysis, two steps are performed: first, the LTQ-Orbitrap mass spectrometer obtains a single MS survey scan in the Orbitrap analyzer, then for the seven most intense precursor ions from each MS survey scan, sequential MSMS scans are obtained in the LTQ to determine their peptide sequence. Due to the highly complex samples obtained from the cell lysates and the limited sequencing speed at the MSMS sequencing step, modern mass spectrometers are only capable of sequencing approximately 16% of the precursor ions present (47). Thus, the relative abundance of a phosphopeptide greatly influences the probability that the ion derived from a particular phosphopeptide will be selected for MSMS sequencing. It is not surprising that in analytical replicates, and certainly with biological replicates, the ions from less abundant phosphopeptides might not be consistently selected due to slight changes in chromatography at either the SCX, ZrO2, or LC-MS steps (73–75). As evidence of variation in selecting different precursor ions for sequencing, in our study, the inclusion of a technical replicate increased the number of identified peptides by about 20% on average. Furthermore, for low-abundance precursor ions, the quality of the MSMS spectra deteriorates, thus making it more difficult to obtain protein sequence. The fraction of identified peptides following the MSMS sequencing step has been determined to be approximately 60% for abundant precursor ions, but drops to about 10% for low-abundance ions (47). In our typical screen, of the approximately 10,000 spectra we obtained, about 30% were identified. Fourth, the low expression levels of regulatory proteins and low stoichiometry of their phosphorylation further increases the difficulty of detecting phosphorylation events (76). It is well known that compared with many growth factors, stimulation of cells with GH does not produce a very robust signaling response. In 3T3-F442A cells, the signaling response to GH is much less than for platelet-derived growth factor or epidermal growth factor (77). Therefore, identifying GH-dependent phosphosites is likely to be particularly challenging. Finally, many phosphorylation sites are not identifiable either because the tryptic fragments are too large or too small for identification or because their sequence-specific properties lead to low efficiency ionization or fragmentation. However, based on the validation that we did with our immunoblotting studies, even those phosphopeptides that were detected in only one screen have a high likelihood of being valid and certainly provide sufficient evidence to warrant further investigation of all the identified phosphosites.
Using immunoblot analysis, we confirmed the phosphoproteomics results for all seven of the GH-stimulated sites tested, which included nearly all of the phosphosites for which phosphospecific antibodies were available. The change in phosphorylation observed by immunoblotting was consistent with our MS data, even when the MS data revealed only a 50% increase in signal with GH, providing strong validation of our phosphoproteomics approach. We identified phosphorylation sites in proteins previously reported to be phosphorylated in response to GH (Erk1, Erk2, Stat5a, Stat5b, and Shc) but the vast majority of the identified phosphosites (125 of the 132 GH-up-regulated sites) were in proteins not previously known to be regulated by GH. Based on the high degree of reproducibility (>90%) for GH regulation when a phosphosite was detected in more than one screen, and the high degree of validation (100%) of sites by Western blotting (including sites identified in only one screen), SILAC-based phosphoproteomics analysis is a very powerful method to detect novel GH-regulated proteins. In a single experiment, we detected 1 order of magnitude more candidate GH-regulated proteins than we have detected using other techniques (yeast two-hybrid, GST pull-down, immobilized phosphopeptide pull-down, Tap-tagged and Flag-tagged coimmunoprecipitation, and cloning of ligand targets assays involving JAK2 and/or GH receptor) and observed a much higher rate of verification of candidate proteins (i.e. the proportion of false positives was substantially lower). This is probably at least in part due to: 1) our SILAC-based phosphoproteomics analysis method not requiring overexpression of any proteins or artificial treatment or manipulation of any protein, other than growing some of the cells in heavy-labeled amino acids, and 2) the fact that the GH-treated and untreated cell lysates were processed together in a combined sample.
Although GH signaling has been shown to phosphorylate and activate Akt (13, 45), the prominent representation of Akt/mTOR pathway components in our phosphoproteomics analysis (Table 3) was unexpected. The consensus substrate recognition sequence for Akt was one of the top motifs in our analysis of consensus phosphorylation motifs, and the Akt-regulated mTOR signaling pathway was one of the top four pathways in KEGG pathway analysis of our data. Given the shared roles of GH and mTORC1 in regulating protein synthesis as well as organ and body growth, it makes sense that GH activates Akt-regulated mTORC1 signaling. Before this work, only one GH-initiated pathway leading from Akt to the mTORC1 complex had been identified: phosphorylation of TSC2 Ser939 (14). Our phosphoproteomics analysis coupled with studies using wortmannin to inhibit the PI3K/Akt pathway revealed that GH stimulates the phosphorylation of numerous proteins in the Akt-mTORC1 pathway, including: 1) Akt-mediated TSC2 Ser939 phosphorylation, which suppresses TSC2 function and thus inhibits the activity of mTORC1 (49, 50); 2) Akt-mediated PRAS40 Thr247 phosphorylation, which is thought to contribute to relief of PRAS40-dependent inhibition of mTORC1 (59, 60); and 3) mTOR as well as Erk1/2-mediated raptor Ser863 phosphorylation, which has been shown to promote multisite raptor phosphorylation and augment mTORC1 activity (36, 57, 58).
Table 3.
GH-stimulated phosphosites identified by phosphoproteomics that are predicted4 to be phosphorylated by Akt/PKA
| Gene | Protein | Phosphosite5 | Ratio (+GH/−GH) (5a/5b/15 min)6 |
|---|---|---|---|
| Acly | ATP citrate lyase (ACLY) | Ser455 | 1.5/−/− |
| Akt1s1 | Proline-rich AKT1 substrate 1 (PRAS40) | Thr247 | 3.8/−/− |
| Bcl9l | B-cell CLL/lymphoma 9-like protein | Ser118 | −/2.4/− |
| Dact3 | Dapper homolog 3 | Ser409 | 2.2/−/− |
| Eif4b | Eukaryotic translation initiation factor 4B (eIF4B) | Thr420 | −/3.1/− |
| Flnc | Filamin-C | Ser2234 | 3.3/2.3/1.7 |
| Ndrg1 | N-myc downstream-regulated gene 1 protein (NDRG1) | Thr328 | −/1.8/− |
| Ndrg1 | N-myc downstream-regulated gene 1 protein (NDRG1) | Thr330 | −/1.5/− |
| Zc3hc1 | Nuclear-interacting partner of anaplastic lymphoma kinase (Nipa) | Ser406 | −/2.6/− |
| Pdcd4 | Programmed cell death protein 4 | Ser457 | −/3.8/− |
| Plec1 | Plectin-1 | Ser4391 (Ser4243) | −/2.1/− |
| Plec1 | Plectin-1 | Ser4393 (Ser4245) | 1.6/2.1/− |
| Ranbp3 | Ran-binding protein 3 | Ser58 | −/2.5/− |
| Slc9a1 | Sodium/hydrogen exchanger 1 (NHE1) | Ser707 | −/7.3/3.3 |
| Tns3 | Tensin-3 | Ser769 | 2.0/−/− |
GH-dependent phosphopeptides identified during the SILAC-based phosphoproteomics screens were analyzed by MaxQuant to predict phosphosites with best-fit motif consistent with phosphorylation by Akt.
Numbering is according to PhosphositePlus (http://www.phosphosite.org). Numbering in parentheses is according to MaxQuant.
5a and 5b refer to the first and second 5 min trials, respectively.
Additionally, among the sites in our phosphoproteomics analysis that we have verified by immunoblotting, several potential novel functions of GH have been revealed. Because phosphorylation of Ser455 in ACLY has been shown to increase ACLY-dependent synthesis of acetyl-CoA (67), our finding that GH stimulates phosphorylation of ACLY Ser455 suggests that GH may increase acetyl-CoA synthesis. Acetyl-CoA is carboxylated to malonyl-CoA in fatty acid synthesis. Thus, activation of ACLY would be consistent with the known rapid and transitory insulin-like stimulatory effects of GH on lipogenesis (78). Similarly, because phosphorylation of NHE1 at Ser707 has been shown to increase NHE1 activity (35), our finding that GH stimulates the phosphorylation of NHE1 Ser707 suggests that GH may activate the Na+/H+ exchange function of NHE1. Consistent with a multipronged regulation of proteins and pathways, GH may also increase NHE1 activity in a calmodulin-dependent manner. GH binds its receptor and activates the tyrosine kinase, JAK2. JAK2 activation has previously been reported to phosphorylate calmodulin, resulting in increased interaction of calmodulin with NHE1 and increased NHE1 activity (79). Although these proteins clearly merit and will receive more study, our SILAC and MS-based phosphoproteomic study reveals an additional 214 GH-dependent phosphosites (118 which undergo increased phosphorylation and 96 with decreased phosphorylation) in 133 proteins that have yet to be studied.
KEGG pathway analysis also identified the insulin-signaling pathway. The proteins in the KEGG category “insulin signaling pathway” include a number of signaling proteins shared by multiple growth factors that activate tyrosine kinases. In fact, the proteins associated with the insulin-signaling pathway fall within two known GH signaling pathways shared with insulin (and other growth factors): the insulin receptor substrate/PI3K/Akt/mTOR pathway and the Shc/Ras/MEK/Erk pathway. These two pathways undoubtedly contribute to the known rapid, insulin-like effects of GH that are observed in cells after a period of GH deprivation (80).
The KEGG pathways “regulation of actin cytoskeleton” and “focal adhesions” were also identified during the bioinformatics analysis. Previous studies have implicated GH in regulation of the actin cytoskeleton. GH has been shown to promote membrane ruffling of 3T3-F442A cells (54) and CHO cells expressing GH receptors (53). It has also been shown to increase the motility of 293T cells stably expressing GH receptor (56) and the directed migration of monocytes (52, 55) and RAW264.7 macrophages (Su, H.-W., and C. Carter-Su, manuscript in preparation). During cell migration, focal complexes assemble at the leading edge and then disassemble as a cell moves. Some focal complexes mature and recruit new proteins; these focal adhesions anchor cells to the matrix and are important for cell movement. Multiple focal adhesion proteins have been shown to undergo GH-dependent changes in tyrosyl phosphorylation (18, 81, 82). Of the seven proteins containing GH-enhanced phosphosites identified by KEGG analysis as “focal adhesion” proteins, four (filamin, zyxin, parvin, Ppp1r12a/regulatory subunit of myosin light chain phosphatase) had not been identified previously as GH-regulated proteins. Among the five GH-up-regulated phosphoproteins in the KEGG category “regulation of the actin cytoskeleton,” three (NHE1, myosin heavy chain 9, and Ppp1r12a) had not been identified previously as GH-regulated phosphoproteins.
Our motif analysis of the newly identified phosphosites that were up-regulated by GH determined that the dominant motifs phosphorylated were Akt/PKA, PKA, CK1, CamK2, GSK3, and CK2 consensus phosphorylation motifs. This analysis suggests that pathways that activate these kinases or kinases with similar substrate specificity may play an important role in GH signaling. Of these kinases, Akt is the only one known to be stimulated by GH (13). Our present findings and previous work of others (14) provide evidence that a number of the identified Akt phosphorylation consensus sites are phosphorylated by Akt in response to GH (e.g. TSC2 Ser939, PRAS40 Thr247, ACLY Ser455). However, because of overlap within kinase substrate consensus sequences and because the MaxQuant analysis of consensus phosphorylation motifs does not use a comprehensive list of kinases, some of the identified “consensus” phosphosites are likely to be phosphorylated primarily by a related kinase. For instance, GH signaling reportedly inhibits GSK3 (45). Thus, our finding that GH signaling increases phosphorylation of GSK3 consensus sites suggests that GSK3 does not represent the physiological kinase for these sites. Instead, another, perhaps related, kinase that responds positively to GH, seems more likely to be the responsible kinase. Akt is a member of the AGC family of kinases. Family members Akt and SGK1 are both reported to phosphorylate Ser and Thr that lie in RXRXXS/T motifs (83, 84). Rather than Akt/PKA, which was identified as the most likely kinase(s) by MaxQuant, the AGC kinase member SGK1 is thought to phosphorylate NDRG1 Thr328 and Ser330 (65). The AGC family member p90RSK (RXXS motif) is thought to be the kinase that phosphorylates NHE1 Ser707 (85).
At least one of the kinases identified in our motif analysis, CK2, is thought to be constitutively active (86). CK2 was recently reported to both associate with JAK2 and, in an in vitro assay, phosphorylate JAK2 (86). Additionally, CK2 inhibitors were found to depress GH-induced activation of STAT5, consistent with CK2 playing a role in GH signaling. With kinases like CK2 that are constitutively active, the regulation of ligand-dependent signaling likely requires a change in the conformation of protein substrates or altered subcellular localization of substrates or kinases to permit phosphorylation. Therefore, for potential substrates of CK2, it may be informative to determine whether any GH-dependent changes in subcellular localization of the substrates can be detected.
In summary, these studies are the first studies to use mass-spectrometry and SILAC-based phosphoproteomics to identify novel proteins involved in GH signaling. Using this approach, we have identified 132 GH-up-regulated phosphosites in 95 proteins and 96 GH-down-regulated phosphosites in 46 proteins. Of the seven sites subjected to further testing by immunoblotting, all were confirmed to be phosphorylated in response to GH, which suggests that the phosphoproteomic screen identified GH-dependent phosphorylation sites with high accuracy. Motif analysis suggested that Akt, PKA, CamK2, GSK3, CK1, and CK2 (or kinases with similar substrate specificity) are important kinases in GH signaling. Manual annotation (Supplemental Table 1) and KEGG pathway analysis revealed that the identified proteins included multiple adaptor proteins, focal adhesion proteins, proteins involved in the cytoskeleton, and proteins involved in transcription. Overall, these results provide multiple new candidates for study, some of which will extend our understanding of known GH-signaling pathways and functions, and others that have the potential to break new ground in our quest to understand the full range of GH responses.
Supplementary Material
Acknowledgments
We thank Drs. Jessica Schwartz, Ram Menon and Hsiao-Wen Su, and Michael Doche (University of Michigan, Ann Arbor, MI) for helpful discussions. We thank Joel Cline (University of Michigan, Ann Arbor, MI) for help with the cell culture and Barbara Hawkins (University of Michigan, Ann Arbor, MI) for help with the manuscript.
This work was supported by National Institutes of Health grants R01-DK34171 (to C.C.-S.), P41-RR18627 (to P.A.), and R01-DK-078135 (to D.F.) and a grant from the American Heart Association (to D.F.).
Because 3T3-F442A cells are of mouse origin, we have used the amino acid numbering for the mouse sequence used in PhosphositePlus (http://www.phosphosite.org) throughout the paper, unless specifically noted.
- ACLY
- ATP-citrate lyase
- CamK
- Ca2+/ calmodulin-dependent protein kinase
- CK
- casein kinase
- eIF4B
- eukaryotic initiation factor 4B
- GSK3
- glycogen synthase kinase-3
- LC
- liquid chromatography
- LTQ
- linear trap quadrupole
- mTOR
- mammalian target of rapamycin
- mTORC
- mTOR complex
- MS
- mass spectrometry
- MSMS
- tandem MS
- NDRG
- N-myc downstream regulated gene
- NHE1
- Na+/H+ exchanger-1
- PI3K
- phosphatidyl inositol 3-kinase
- PKA
- protein kinase A
- Ppp1r12a
- protein phosphatase 1 regulatory subunit 12a of myosin light chain phosphatase
- PRAS40
- proline-rich Akt substrate, 40 kDa
- raptor
- regulatory associated protein of mTOR
- SCX
- strong cationic exchange
- Ser
- S, serine
- SILAC
- stable isotope labeling with amino acids in cell culture
- Thr
- T, threonine
- Tyr
- Y, tyrosine
- Stat
- signal transducer and activator of transcription
- TSC2
- tuberous sclerosis protein 2 or tuberin.
References
- 1. Kaplan SL. 1999. Hormonal regulation of growth and metabolic effects of growth hormone. In: Kostyo JL, ed. Handbook of physiology. New York: Oxford University Press; 129–143 [Google Scholar]
- 2. Fryburg DA, Barrett EJ. 1999. The regulation of amino acid and protein metabolism by growth hormone. In: Kostyo JL, ed. Handbook of physiology. New York: Oxford University Press; 515–536 [Google Scholar]
- 3. Birzniece V, Nelson AE, Ho KK. 2011. Growth hormone and physical performance. Trends Endocrinol Metab 22:171–178 [DOI] [PubMed] [Google Scholar]
- 4. Kokshoorn NE, Biermasz NR, Roelfsema F, Smit JW, Pereira AM, Romijn JA. 2011. GH replacement therapy in elderly GH-deficient patients: a systematic review. Euro J Endocrinol 164:657–665 [DOI] [PubMed] [Google Scholar]
- 5. Argetsinger LS, Campbell GS, Yang X, Witthuhn BA, Silvennoinen O, Ihle JN, Carter-Su C. 1993. Identification of JAK2 as a growth hormone receptor-associated tyrosine kinase. Cell 74:237–244 [DOI] [PubMed] [Google Scholar]
- 6. Brown RJ, Adams JJ, Pelekanos RA, Wan Y, McKinstry WJ, Palethorpe K, Seeber RM, Monks TA, Eidne KA, Parker MW, Waters MJ. 2005. Model for growth hormone receptor activation based on subunit rotation within a receptor dimer. Nat Struct Mol Biol 12:814–821 [DOI] [PubMed] [Google Scholar]
- 7. Ceseña TI, Cui TX, Piwien-Pilipuk G, Kaplani J, Calinescu AA, Huo JS, Iñiguez-Lluhi JA, Kwok R, Schwartz J. 2007. Multiple mechanisms of growth hormone-regulated gene transcription. Mol Genet Metab 90:126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Herrington J, Smit LS, Schwartz J, Carter-Su C. 2000. The role of STAT proteins in growth hormone signaling. Oncogene 19:2585–2597 [DOI] [PubMed] [Google Scholar]
- 9. VanderKuur JA, Butch ER, Waters SB, Pessin JE, Guan KL, Carter-Su C. 1997. Signalling molecules involved in coupling growth hormone receptor to MAP kinase activation. Endocrinology 138:4301–4307 [DOI] [PubMed] [Google Scholar]
- 10. Wang X, Yang N, Deng L, Li X, Jiang J, Gan Y, Frank SJ. 2009. Interruption of growth hormone signaling via SHC and ERK in 3T3–F442A preadipocytes upon knockdown of insulin receptor substrate-1. Mol Endocrinol 23:486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Argetsinger LS, Hsu GW, Myers MG, Jr, Billestrup N, White MF, Carter-Su C. 1995. Growth hormone, interferon-γ, and leukemia inhibitory factor promoted tyrosyl phosphorylation of insulin receptor substrate-1. J Biol Chem 270:14685–14692 [DOI] [PubMed] [Google Scholar]
- 12. Ridderstråle M, Degerman E, Tornqvist H. 1995. Growth hormone stimulates the tyrosine phosphorylation of the insulin receptor substrate-1 and its association with phosphatidylinositol 3-kinase in primary adipocytes. J Biol Chem 270:3471–3474 [DOI] [PubMed] [Google Scholar]
- 13. Costoya JA, Finidori J, Moutoussamy S, Seãris R, Devesa J, Arce VM. 1999. Activation of growth hormone receptor delivers an antiapoptotic signal: evidence for a role of Akt in this pathway. Endocrinology 140:5937–5943 [DOI] [PubMed] [Google Scholar]
- 14. Hayashi AA, Proud CG. 2007. The rapid activation of protein synthesis by growth hormone requires signaling through mTOR. Am J Physiol Endocrinol Metab 292:E1647–E1655 [DOI] [PubMed] [Google Scholar]
- 15. Rowlinson SW, Yoshizato H, Barclay JL, Brooks AJ, Behncken SN, Kerr LM, Millard K, Palethorpe K, Nielsen K, Clyde-Smith J, Hancock JF, Waters MJ. 2008. An agonist-induced conformational change in the growth hormone receptor determines the choice of signalling pathway. Nat Cell Biol 10:740–747 [DOI] [PubMed] [Google Scholar]
- 16. Stofega MR, Wang H, Ullrich A, Carter-Su C. 1998. Growth hormone regulation of SIRP and SHP-2 tyrosyl phosphorylation and association. J Biol Chem 273:7112–7117 [DOI] [PubMed] [Google Scholar]
- 17. Rui L, Mathews LS, Hotta K, Gustafson TA, Carter-Su C. 1997. Identification of SH2-Bβ as a substrate of the tyrosine kinase JAK2 involved in growth hormone signaling. Mol Cell Biol 17:6633–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu T, Goh EL, LeRoith D, Lobie PE. 1998. Growth hormone stimulates the formation of a multiprotein signaling complex involving p130(Cas) and CrkII. Resultant activation of c-Jun N- terminal kinase/stress-activated protein kinase (JNK/SAPK). J Biol Chem 273:33864–33875 [DOI] [PubMed] [Google Scholar]
- 19. Bodenmiller B, Mueller LN, Pedrioli PG, Pflieger D, Jünger MA, Eng JK, Aebersold R, Tao WA. 2007. An integrated chemical, mass spectrometric and computational strategy for (quantitative) phosphoproteomics: application to Drosophila melanogaster Kc167 cells. Mol Biosyst 3:275–286 [DOI] [PubMed] [Google Scholar]
- 20. Wu CJ, Chen YW, Tai JH, Chen SH. 2011. Quantitative phosphoproteomics studies using stable isotope dimethyl labeling coupled with IMAC-HILIC-nanoLC-MS/MS for estrogen-induced transcriptional regulation. J Proteome Res 10:1088–1097 [DOI] [PubMed] [Google Scholar]
- 21. de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Fröhlich F, Walther TC, Mann M. 2008. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455:1251–1254 [DOI] [PubMed] [Google Scholar]
- 22. Dengjel J, Kratchmarova I, Blagoev B. 2009. Receptor tyrosine kinase signaling: a view from quantitative proteomics. Mol Biosyst 5:1112–1121 [DOI] [PubMed] [Google Scholar]
- 23. Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M. 2010. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal 3:ra3. [DOI] [PubMed] [Google Scholar]
- 24. Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villén J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, Blenis J. 2011. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332:1322–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kweon HK, Håkansson K. 2006. Selective zirconium dioxide-based enrichment of phosphorylated peptides for mass spectrometric analysis. Anal Chem 78:1743–1749 [DOI] [PubMed] [Google Scholar]
- 26. Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jørgensen TJ. 2005. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics 4:873–886 [DOI] [PubMed] [Google Scholar]
- 27. Posewitz MC, Tempst P. 1999. Immobilized gallium(III) affinity chromatography of phosphopeptides. Anal Chem 71:2883–2892 [DOI] [PubMed] [Google Scholar]
- 28. Yip TT, Hutchens TW. 2004. Immobilized metal-ion affinity chromatography. Methods Mol Biol 244:179–190 [DOI] [PubMed] [Google Scholar]
- 29. Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. 2005. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol 23:94–101 [DOI] [PubMed] [Google Scholar]
- 30. Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1:376–386 [DOI] [PubMed] [Google Scholar]
- 31. Tasaki S, Nagasaki M, Kozuka-Hata H, Semba K, Gotoh N, Hattori S, Inoue J, Yamamoto T, Miyano S, Sugano S, Oyama M. 2010. Phosphoproteomics-based modeling defines the regulatory mechanism underlying aberrant EGFR signaling. PLoS One 5:e13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bodenmiller B, Wanka S, Kraft C, Urban J, Campbell D, Pedrioli PG, Gerrits B, Picotti P, Lam H, Vitek O, Brusniak MY, Roschitzki B, Zhang C, Shokat KM, Schlapbach R, Colman-Lerner A, Nolan GP, Nesvizhskii AI, Peter M, Loewith R, von Mering C, Aebersold R. 2010. Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci Signal 3:rs4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Emmott E, Rodgers MA, Macdonald A, McCrory S, Ajuh P, Hiscox JA. 2010. Quantitative proteomics using stable isotope labeling with amino acids in cell culture reveals changes in the cytoplasmic, nuclear, and nucleolar proteomes in Vero cells infected with the coronavirus infectious bronchitis virus. Mol Cell Proteomics 9:1920–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Soufi B, Kumar C, Gnad F, Mann M, Mijakovic I, Macek B. 2010. Stable isotope labeling by amino acids in cell culture (SILAC) applied to quantitative proteomics of Bacillus subtilis. J Proteome Res 9:3638–3646 [DOI] [PubMed] [Google Scholar]
- 35. Chen S, Mackintosh C. 2009. Differential regulation of NHE1 phosphorylation and glucose uptake by inhibitors of the ERK pathway and p90RSK in 3T3–L1 adipocytes. Cell Signal 21:1984–1993 [DOI] [PubMed] [Google Scholar]
- 36. Foster KG, Acosta-Jaquez HA, Romeo Y, Ekim B, Soliman GA, Carriere A, Roux PP, Ballif BA, Fingar DC. 2010. Regulation of mTOR complex 1 (mTORC1) by raptor Ser863 and multisite phosphorylation. J Biol Chem 285:80–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bradford MM. 1976. A rapid and sensitive method for the quantitation of micrograms quantities of protein ultilizing the principle of protein dye-bing. Anal Biochem 72:248–254 [DOI] [PubMed] [Google Scholar]
- 38. Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372 [DOI] [PubMed] [Google Scholar]
- 39. Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551–3567 [DOI] [PubMed] [Google Scholar]
- 40. Meyer DJ, Campbell GS, Cochran BH, Argetsinger LS, Larner AC, Finbloom DS, Carter-Su C, Schwartz J. 1994. Growth hormone induces a DNA binding factor related to the interferon-stimulated 91kD transcription factor. J Biol Chem 269:4701–4704 [PubMed] [Google Scholar]
- 41. Smit LS, Meyer DJ, Billestrup N, Norstedt G, Schwartz J, Carter-Su C. 1996. The role of the growth hormone (GH) receptor and JAK1 and JAK2 kinases in the activation of Stats 1, 3, and 5 by GH. Mol Endocrinol 10:519–533 [DOI] [PubMed] [Google Scholar]
- 42. Smit LS, Vanderkuur JA, Stimage A, Han Y, Luo G, Yu-Lee LY, Schwartz J, Carter-Su C. 1997. Growth hormone-induced tyrosyl phosphorylation and DNA binding activity of Stat5A and Stat5B. Endocrinology 138:3426–3434 [DOI] [PubMed] [Google Scholar]
- 43. VanderKuur J, Allevato G, Billestrup N, Norstedt G, Carter-Su C. 1995. Growth hormone-promoted tyrosyl phosphorylation of Shc proteins and Shc association with Grb2. J Biol Chem 270:7587–7593 [DOI] [PubMed] [Google Scholar]
- 44. Argetsinger LS, Norstedt G, Billestrup N, White MF, Carter-Su C. 1996. Growth hormone, interferon-γ, and leukemia inhibitory factor utilize insulin receptor substrate-2 in intracellular signaling. J Biol Chem 271:29415–29421 [DOI] [PubMed] [Google Scholar]
- 45. Piwien-Pilipuk G, Van Mater D, Ross SE, MacDougald OA, Schwartz J. 2001. Growth hormone regulates phosphorylation and function of CCAAT/enhancer-binding protein β by modulating Akt and glycogen synthase kinase-3. J Biol Chem 276:19664–19671 [DOI] [PubMed] [Google Scholar]
- 46. Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. 2006. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127:635–648 [DOI] [PubMed] [Google Scholar]
- 47. Michalski A, Cox J, Mann M. 2011. More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J Proteome Res 10:1785–1793 [DOI] [PubMed] [Google Scholar]
- 48. Magnuson B, Ekim E, Fingar DC. 2012. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signaling networks. Biochem J 441:1–21 [DOI] [PubMed] [Google Scholar]
- 49. Inoki K, Li Y, Zhu T, Wu J, Guan KL. 2002. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol 4:648–657 [DOI] [PubMed] [Google Scholar]
- 50. Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. 2002. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell 10:151–162 [DOI] [PubMed] [Google Scholar]
- 51. Lanning NJ, Carter-Su C. 2006. Recent advances in growth hormone signaling. Rev Endocr Metab Disord 7:225–235 [DOI] [PubMed] [Google Scholar]
- 52. Wiedermann CJ, Reinisch N, Braunsteiner H. 1993. Stimulation of monocyte chemotaxis by human growth hormone and its deactivation by somatostatin. Blood 82:954–960 [PubMed] [Google Scholar]
- 53. Goh EL, Pircher TJ, Wood TJ, Norstedt G, Graichen R, Lobie PE. 1997. Growth hormone-induced reorganization of the actin cytoskeleton is not required for STAT5 (signal transducer and activator of transcription-5)-mediated transcription. Endocrinology 138:3207–3215 [DOI] [PubMed] [Google Scholar]
- 54. Herrington J, Diakonova M, Rui L, Gunter DR, Carter-Su C. 2000. SH2-B is required for growth hormone-induced actin reorganization. J Biol Chem 275:13126–13133 [DOI] [PubMed] [Google Scholar]
- 55. Kähler CM, Pischel AB, Haller T, Meierhofer C, Djanani A, Kaufmann G, Wiedermann CJ. 2001. Signal transduction pathways in directed migration of human monocytes induced by human growth hormone in vitro. Int Immunopharmacol 1:1351–1361 [DOI] [PubMed] [Google Scholar]
- 56. Diakonova M, Gunter DR, Herrington J, Carter-Su C. 2002. SH2-Bβ is a Rac-binding protein that regulates cell motility. J Biol Chem 277:10669–10677 [DOI] [PubMed] [Google Scholar]
- 57. Wang L, Lawrence JC, Jr, Sturgill TW, Harris TE. 2009. Mammalian target of rapamycin complex 1 (mTORC1) activity is associated with phosphorylation of raptor by mTOR. J Biol Chem 284:14693–14697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Carriere A, Romeo Y, Acosta-Jaquez HA, Moreau J, Bonneil E, Thibault P, Fingar DC, Roux PP. 2011. ERK1/2 phosphorylate Raptor to promote Ras-dependent activation of mTOR complex 1 (mTORC1). J Biol Chem 286:567–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nascimento EB, Snel M, Guigas B, van der Zon GC, Kriek J, Maassen JA, Jazet IM, Diamant M, Ouwens DM. 2010. Phosphorylation of PRAS40 on Thr246 by PKB/AKT facilitates efficient phosphorylation of Ser183 by mTORC1. Cell Signal 22:961–967 [DOI] [PubMed] [Google Scholar]
- 60. Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. 2007. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell 25:903–915 [DOI] [PubMed] [Google Scholar]
- 61. Salcini AE, McGlade J, Pelicci G, Nicoletti I, Pawson T, Pelicci PG. 1994. Formation of Shc-Grb2 complexes is necessary to induce neoplastic transformation by overexpression of Shc proteins. Oncogene 9:2827–2836 [PubMed] [Google Scholar]
- 62. Kurdistani SK, Arizti P, Reimer CL, Sugrue MM, Aaronson SA, Lee SW. 1998. Inhibition of tumor cell growth by RTP/rit42 and its responsiveness to p53 and DNA damage. Cancer Res 58:4439–4444 [PubMed] [Google Scholar]
- 63. Stein S, Thomas EK, Herzog B, Westfall MD, Rocheleau JV, Jackson RS, II, Wang M, Liang P. 2004. NDRG1 is necessary for p53-dependent apoptosis. J Biol Chem 279:48930–48940 [DOI] [PubMed] [Google Scholar]
- 64. van Belzen N, Dinjens WN, Diesveld MP, Groen NA, van der Made AC, Nozawa Y, Vlietstra R, Trapman J, Bosman FT. 1997. A novel gene which is up-regulated during colon epithelial cell differentiation and down-regulated in colorectal neoplasms. Lab Invest 77:85–92 [PubMed] [Google Scholar]
- 65. Murray JT, Campbell DG, Morrice N, Auld GC, Shpiro N, Marquez R, Peggie M, Bain J, Bloomberg GB, Grahammer F, Lang F, Wulff P, Kuhl D, Cohen P. 2004. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem J 384:477–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Murakami Y, Hosoi F, Izumi H, Maruyama Y, Ureshino H, Watari K, Kohno K, Kuwano M, Ono M. 2010. Identification of sites subjected to serine/threonine phosphorylation by SGK1 affecting N-myc downstream-regulated gene 1 (NDRG1)/Cap43-dependent suppression of angiogenic CXC chemokine expression in human pancreatic cancer cells. Biochem Biophys Res Commun 396:376–381 [DOI] [PubMed] [Google Scholar]
- 67. Potapova IA, El-Maghrabi MR, Doronin SV, Benjamin WB. 2000. Phosphorylation of recombinant human ATP:citrate lyase by cAMP-dependent protein kinase abolishes homotropic allosteric regulation of the enzyme by citrate and increases the enzyme activity. Allosteric activation of ATP:citrate lyase by phosphorylated sugars. Biochemistry 39:1169–1179 [DOI] [PubMed] [Google Scholar]
- 68. Arcaro A, Wymann MP. 1993. Wortmannin is a potent phosphatidylinositol 3-kinase inhibitor: the role of phosphatidylinositol 3,4,5-trisphosphate in neutrophil responses. Biochem J 296:297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mazurkiewicz-Munoz AM, Argetsinger LS, Kouadio JL, Stensballe A, Jensen ON, Cline JM, Carter-Su C. 2006. Phosphorylation of JAK2 at serine 523: a negative regulator of JAK2 that is stimulated by growth hormone and epidermal growth factor. Mol Cell Biol 26:4052–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. O'Brien KB, Argetsinger LS, Diakonova M, Carter-Su C. 2003. YXXL motifs in SH2-Bβ are phosphorylated by JAK2, JAK1, and platelet-derived growth factor receptor and are required for membrane ruffling. J Biol Chem 278:11970–11978 [DOI] [PubMed] [Google Scholar]
- 71. Argetsinger LS, Stuckey JA, Robertson SA, Koleva RI, Cline JM, Marto JA, Myers MG, Jr, Carter-Su C. 2010. Tyrosines 868, 966, and 972 in the kinase domain of JAK2 are autophosphorylated and required for maximal JAK2 kinase activity. Mol Endocrinol 24:1062–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Xu J, Liu Z, Clemens TL, Messina JL. 2006. Insulin reverses growth hormone-induced homologous desensitization. J Biol Chem 281:21594–21606 [DOI] [PubMed] [Google Scholar]
- 73. Ye J, Zhang X, Young C, Zhao X, Hao Q, Cheng L, Jensen ON. 2010. Optimized IMAC-IMAC protocol for phosphopeptide recovery from complex biological samples. J Proteome Res 9:3561–3573 [DOI] [PubMed] [Google Scholar]
- 74. Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, Rush J, Lauffenburger DA, White FM. 2005. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol Cell Proteomics 4:1240–1250 [DOI] [PubMed] [Google Scholar]
- 75. Cox J, Mann M. 2011. Quantitative, high-resolution proteomics for data-driven systems biology. Annu Rev Biochem 80:273–299 [DOI] [PubMed] [Google Scholar]
- 76. Villén J, Gygi SP. 2008. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc 3:1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Campbell GS, Christian LJ, Carter-Su C. 1993. Evidence for involvement of the growth hormone receptor-associated tyrosine kinase in actions of growth hormone. J Biol Chem 268:7427–7434 [PubMed] [Google Scholar]
- 78. Goodman HM. 1968. Growth hormone and the metabolism of carbohydrate and lipid in adipose tissue. Ann NY Acad Sci 148:419–440 [DOI] [PubMed] [Google Scholar]
- 79. Mukhin YV, Vlasova T, Jaffa AA, Collinsworth G, Bell JL, Tholanikunnel BG, Pettus T, Fitzgibbon W, Ploth DW, Raymond JR, Garnovskaya MN. 2001. Bradykinin B2 receptors activate Na+/H+ exchange in mIMCD-3 cells via Janus kinase 2 and Ca2+/calmodulin. J Biol Chem 276:17339–173346 [DOI] [PubMed] [Google Scholar]
- 80. Goodman HM. Effects of growth hormone on isolated adipose tissue. In: Muller EE, Pecile A, eds. Proc International Symposium on Growth Hormone Excerpta Medica Foundation, Amsterdam, The Netherlands, 1968, pp 153–171 [Google Scholar]
- 81. Zhu T, Goh EL, Lobie PE. 1998. Growth hormone stimulates the tyrosine phosphorylation and association of p125 focal adhesion kinase (FAK) with JAK2. Fak is not required for stat-mediated transcription. J Biol Chem 273:10682–10689 [DOI] [PubMed] [Google Scholar]
- 82. Ryu H, Lee JH, Kim KS, Jeong SM, Kim PH, Chung HT. 2000. Regulation of neutrophil adhesion by pituitary growth hormone accompanies tyrosine phosphorylation of Jak2, p125FAK, and paxillin. J Immunol 165:2116–2123 [DOI] [PubMed] [Google Scholar]
- 83. Alessi DR, Caudwell FB, Andjelkovic M, Hemmings BA, Cohen P. 1996. Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett 399:333–338 [DOI] [PubMed] [Google Scholar]
- 84. Kobayashi T, Cohen P. 1999. Activation of serum- and glucocorticoid-regulated protein kinase by agonists that activate phosphatidylinositide 3-kinase is mediated by 3-phosphoinositide-dependent protein kinase-1 (PDK1) and PDK2. Biochem J 339:319–328 [PMC free article] [PubMed] [Google Scholar]
- 85. Takahashi E, Abe J, Gallis B, Aebersold R, Spring DJ, Krebs EG, Berk BC. 1999. p90(RSK) is a serum-stimulated Na+/H+ exchanger isoform-1 kinase. Regulatory phosphorylation of serine 703 of Na+/H+ exchanger isoform-1. J Biol Chem 274:20206–20214 [DOI] [PubMed] [Google Scholar]
- 86. Zheng Y, Qin H, Frank SJ, Deng L, Litchfield DW, Tefferi A, Pardanani A, Lin FT, Li J, Sha B, Benveniste EN. 2011. A CK2-dependent mechanism for activation of the JAK-STAT signaling pathwayy. Blood 118:156–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Oshiro N, Takahashi R, Yoshino K, Tanimura K, Nakashima A, Eguchi S, Miyamoto T, Hara K, Takehana K, Avruch J, Kikkawa U, Yonezawa K. 2007. The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J Biol Chem 282:20329–20339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Shahbazian D, Roux PP, Mieulet V, Cohen MS, Raught B, Taunton J, Hershey JW, Blenis J, Pende M, Sonenberg N. 2006. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J 25:2781–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Raught B, Peiretti F, Gingras AC, Livingstone M, Shahbazian D, Mayeur GL, Polakiewicz RD, Sonenberg N, Hershey JW. 2004. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J 23:1761–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Robbins DJ, Zhen E, Owaki H, Vanderbilt CA, Ebert D, Geppert TD, Cobb MH. 1993. Regulation and properties of extracellular signal-regulated protein kinases 1 and 2 in vitro. J Biol Chem 268:5097–5106 [PubMed] [Google Scholar]
- 91. Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. 2006. S6K1- and βTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 314:467–471 [DOI] [PubMed] [Google Scholar]
- 92. Palamarchuk A, Efanov A, Maximov V, Aqeilan RI, Croce CM, Pekarsky Y. 2005. Akt phosphorylates and regulates Pdcd4 tumor suppressor protein. Cancer Res 65:11282–11286 [DOI] [PubMed] [Google Scholar]
- 93. von Knethen A, Tzieply N, Jennewein C, Brüne B. 2010. Casein-kinase-II-dependent phosphorylation of PPARγ provokes CRM1-mediated shuttling of PPARγ from the nucleus to the cytosol. J Cell Sci 123:192–201 [DOI] [PubMed] [Google Scholar]
- 94. Foster KG, Fingar DC. 2010. Mammalian target of rapamycin (mTOR): conducting the cellular signaling symphony. J Biol Chem 285:14071–14077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lehoux S, Abe J, Florian JA, Berk BC. 2001. 14–3-3 Binding to Na+/H+ exchanger isoform-1 is associated with serum-dependent activation of Na+/H+ exchange. J Biol Chem 276:15794–15800 [DOI] [PubMed] [Google Scholar]
- 96. Gouilleux F, Wakao H, Mundt M, Groner B. 1994. Prolactin induces phosphorylation of Tyr694 of Stat5 (MGF), a prerequisite for DNA binding and induction of transcription. EMBO J 13:4361–4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Chang CW, Chou HY, Lin YS, Huang KH, Chang CJ, Hsu TC, Lee SC. 2008. Phosphorylation at Ser473 regulates heterochromatin protein 1 binding and corepressor function of TIF1β/KAP1. BMC Mol Biol 9:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Cai SL, Tee AR, Short JD, Bergeron JM, Kim J, Shen J, Guo R, Johnson CL, Kiguchi K, Walker CL. 2006. Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J Cell Biol 173:279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.