Abstract
Many peptidases are thought to require non-active site interaction surfaces, or exosites, to recognize and cleave physiological substrates with high specificity and catalytic efficiency. However, the existence and function of protease exosites remain obscure owing to a lack of effective methods to identify and characterize exosite-interacting substrates. To address this need, we modified the cellular libraries of peptide substrates (CLiPS) methodology to enable the discovery of exosite-interacting peptide ligands. Invariant cleavage motifs recognized by the active sites of thrombin and caspase-7 were displayed on the outer surface of bacteria adjacent to a candidate exosite-interacting peptide. Exosite peptide libraries were then screened for ligands that accelerate cleavage of the active site recognition motif using two-color flow cytometry. Exosite CLiPS (eCLiPS) identified exosite-binding peptides for thrombin that were highly similar to a critical exosite interaction motif in the thrombin substrate, protease-activated receptor 1. Protease activity probes incorporating exosite-binding peptides were cleaved 10-fold faster than substrates without exosite ligands, increasing their sensitivity to thrombin activity in vitro. For comparison, screening with caspase-7 yielded peptides that modestly enhanced (twofold) substrate cleavage rates. The eCLiPS method provides a new tool to profile the ligand specificity of protease exosites and to develop improved substrates.
Keywords: CLiPS, exosite, protease, substrate, thrombin
Introduction
The ability of proteases to discriminate between many potential substrates and acquire high selectivity towards specific proteins is critical for regulating and localizing protein activity in diverse physiological processes. Misregulation of the activity of proteases has been implicated in many diseases including oncologic, inflammatory, neurodegenerative, and infectious diseases (Stamenkovic, 2000; Borgoño et al., 2004; Ala-aho and Kahari, 2005; Yasuda et al., 2005; Burrage et al., 2006; Lutgens et al., 2007; Tenore and Ferreira, 2009). Therefore, characterization of both substrate specificity and exosite interactions can provide valuable insights to understand biological pathways in health and disease, and to develop therapeutic inhibitors, protease-activated therapeutics, and diagnostic imaging probes.
Since protease function is largely defined by the physiological substrate repertoire, our limited understanding of protease substrate recognition mechanisms represents a major obstacle to understanding the biological functions of proteases. With few exceptions, the characterization of protease substrate recognition has been limited to defining the specificity determinants of the active site. However, many proteases utilize non-active site recognition surfaces to influence substrate selectivity and activity, including allosteric sites and exosites (Walker and Royston, 2002; Hardy et al., 2004). This view is supported by the typically large discrepancy in cleavage kinetics between active site recognition sequences, typically fewer than eight amino acids, and bona fide physiological substrates that are often cleaved orders of magnitude faster than these short peptide substrates (Cornille et al., 1997; Bock et al., 2007). Consequently, there remains a need for experimental strategies to identify protease exosite specificity determinants.
Methods to characterize protease substrate specificity typically involve the identification of short peptide substrates that interact with protease active sites. These methods include substrate phage display libraries, positional scanning peptide libraries, combinatorial fluorogenic substrate libraries, mixture-based oriented peptide libraries, and cellular libraries of peptide substrates (CLiPS) (Harris et al., 2000; Turk et al., 2001; Deperthes, 2002; Boulware and Daugherty, 2006; Schneider and Craik, 2009; Boulware et al., 2010). Using these methods, peptide substrates of five to ten amino acids have been identified for numerous proteases. However, typically these active site substrates are not highly selective and are cleaved by other proteases (Talanian et al., 1997; Turk et al., 2001). Thus far, the substrates identified using peptide library methods have not exploited non-active site surfaces, such as exosites, which could dramatically enhance substrate activity and selectivity.
Given the absence of techniques to profile protease exosites, a method was developed to identify exosite-interacting peptide ligands that enhance the cleavage kinetics of a known substrate. The cellular libraries of peptide substrates (CLiPS) method was modified to enable identification of peptide ligands that increase the cleavage rate of a fixed active site recognition motif. In this system, a defined cleavable substrate is expressed on the N-terminus of an E. coli outer membrane display scaffold (eCPX) (Rice and Daugherty, 2008) and a random peptide library is expressed as a fusion to the scaffold’s C-terminus. To demonstrate the utility of the exosite CLiPS (eCLiPS) approach, libraries were screened using human thrombin, which exhibits two distinct exosites removed from the active site, and caspase-7, which is not known to utilize a peptide-binding exosite. Thrombin is a serine protease that plays a central role in blood coagulation by modulating procoagulant and anticoagulant processes, including hydrolysis of fibrinogen into fibrin to promote clot formation. In addition to the active site, the specificity of thrombin toward physiological substrates is typically regulated by interactions with two surface epitopes termed exosite I and exosite II (Bode and Stubbs, 1993; Stubbs and Bode, 1993). Exosite I binds to fibrinogen and protease-activated receptor 1 (PAR1) (Verhamme et al., 2002; Huntington, 2005) and is targeted by the therapeutic inhibitors, hirudin and bivalirudin. Exosite II binds to the heparin-antithrombin complex as well as to glycoprotein Ibα, a protein in the platelet receptor complex and a cofactor for PAR1 cleavage (Huntington, 2005). Interactions with exosites are usually required for efficient cleavage of thrombin substrates by accelerating or stabilizing the formation of the initial thrombin-substrate complex (Huntington, 2005).
In contrast to thrombin, the manner by which most proteases increase their activity and specificity towards short peptide substrates is unknown. For example, the executioner proteases caspase-7 and caspase-3 of the cell apoptotic program were originally thought to have redundant function because they recognize identical substrate motifs (DEVD↑G); however, further investigation suggests that caspase-7 is more specific and the two proteases cleave distinct natural proteins (Walsh et al., 2008). Small molecule ligands that bind an allosteric site in a deep cavity at the dimer interface of caspase-7 can modulate the protease’s activity (Hardy et al., 2004), but peptide-binding motifs that enhance the activity of caspase-7 have not been reported. Although a candidate exosite cavity has been identified on caspase-7, ~30 Å from the two active sites, it has not been shown to interact with any physiological substrate (Agniswamy et al., 2007). In the present study, we applied eCLiPS to both thrombin and caspase-7 to investigate their exosite ligand preferences. Our results demonstrate that the eCLiPS method can be applied to determine the specificity of protease exosites and may be useful to develop more active and selective substrates for applications in basic research, therapeutics, and diagnostics.
Results
Development of an exosite library screening method using thrombin and PAR1
To validate the eCLiPS exosite library approach, thrombin was selected as a model target. Linear exosite-binding sequences within thrombin’s preferred physiological substrate PAR1 have been shown to accelerate the rate of substrate hydrolysis, kcat/KM, by 325-fold over the active site recognition motif alone. This effect was mediated by a 90-fold reduced KM value and an increase in kcat by 3.7-fold with the improved substrate binding contributing the most to the increased cleavage rate (Vu et al., 1991b). Given this substantial exosite mediated enhancement, only picomolar thrombin concentrations are required for platelet activation (Vu et al., 1991a). The active site recognition motif (LDPR↑SFL) and the exosite-binding region (LRNPNDKYEPFWEDEEK) of PAR1 were displayed on the outer surface of E. coli at the N-and C-terminus of the eCPX display scaffold, respectively (Rice and Daugherty, 2008). The extent of substrate conversion could be determined by labeling bacterial cells with fluorescent probes recognizing N- and C-terminal affinity tags. Specifically, N-terminal substrate cleavage was monitored with the red fluorescent probe streptavidin R-phycoerythrin (SA-PE) that binds to the intact N-terminus of the eCPX scaffold (Figure 1A). To ensure that the exosite ligand expressed on the C-terminus is accelerating substrate cleavage and not itself being cleaved, a distinct green fluorescent probe, AlajGFP-Mona SH3, was used to ensure that the C-terminal peptide remained intact. Using this labeling scheme, bacterial cells with cleaved substrates will exhibit primarily green fluorescence while clones with intact peptide substrates will exhibit both red and green fluorescence.
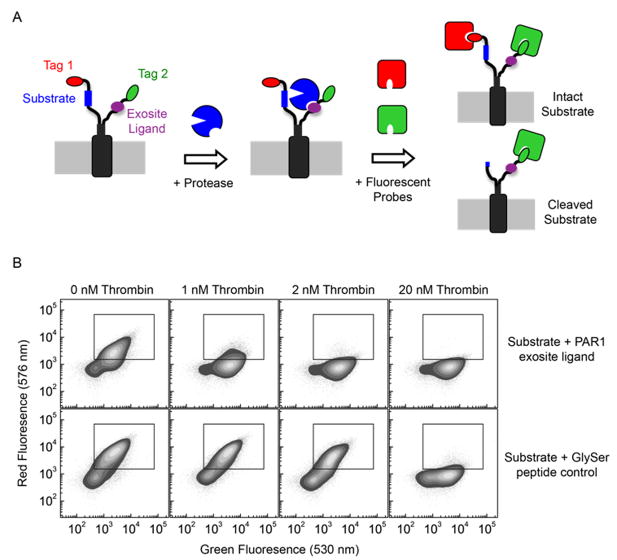
Figure 1. Method for the discovery of exosite ligands using eCLiPS.
A. Using a fixed substrate recombinantly fused to the N-terminus of the bacterial display scaffold, a library of candidate exosite binding peptides is constructed within the C-terminus and quantitatively screened for accelerated substrate cleavage using FACS based on a two-color labeling scheme. B. Flow cytometric analysis of cell populations displaying the PAR1 substrate and either the PAR1 exosite ligand or the control GlySer peptide, following incubation with three different thrombin concentrations.
Bacterial cells displaying the PAR1 substrate along with either the corresponding native exosite-binding sequence or a non-binding glycine-serine repeat peptide of equal length (GlySer control) were incubated with different concentrations of thrombin and the rate of substrate cleavage was measured using flow cytometry (Figure 1B). Following incubation with 1 nM thrombin for 60 min, cells displaying both the PAR1 substrate and exosite ligand exhibited a large decrease in red fluorescence, while significantly higher thrombin concentrations were necessary to achieve the same level of conversion with the GlySer control peptide. With 2 nM thrombin, the cells with the active site motif and PAR1 exosite ligand exhibited 90% substrate conversion. In contrast, cells displaying the active site recognition motif along with the GlySer peptide exhibited a slight decrease in red fluorescence with 2 nM thrombin, but required 20 nM thrombin for significant substrate cleavage, a 10-fold higher concentration. To test the time evolution of substrate cleavage, cells displaying the active site recognition motif with either the PAR1 exosite ligand or the GlySer control were incubated with 250 pM thrombin and the extent of substrate cleavage after 30, 45, and 60 min was measured by flow cytometry (Supplementary Figure 1). Cells displaying the GlySer control peptide did not exhibit significant substrate cleavage at any of the time points. In contrast, cells co-displaying the PAR1 substrate and exosite ligand exhibited 70% substrate conversion after 60 min, demonstrating that the exosite ligand again enhanced the rate of substrate cleavage. These results demonstrate that the PAR1 exosite ligand substantially enhanced the cleavage kinetics of the substrate when compared to a GlySer control exosite peptide.
Screening exosite peptide libraries with thrombin and caspase-7
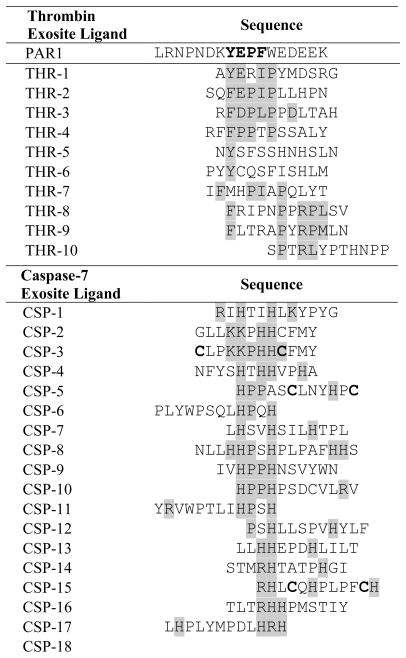
To identify exosite-interacting ligands for thrombin that enhance substrate hydrolysis rates of the PAR1-derived active site recognition sequence, an exosite ligand library displayed on the surface of E. coli was constructed. To co-display an active site binding sequence and a library of exosite recognition sequences, an invariant PAR1 substrate (LDPR↑SFL) was fused to the N-terminus of eCPX, along with a library of 5x107 random 12-mer peptides on the C-terminus of eCPX. The E. coli-displayed exosite peptide library was screened for exosite ligands that accelerate cleavage of the invariant active site sequence using FACS. A two-step screening process was applied to first enrich library members with cleaved substrates after incubation with thrombin (500 pM for 60 min) and labeling with the fluorescent probes SA-PE and AlajGFP-Mona SH3. In a second screening step without the thrombin incubation, cells exhibiting both green and red fluorescence were sorted to ensure that full-length proteins were enriched. To increase the stringency, sorting was repeated using 125 pM thrombin to isolate cells displaying ligands that enhance the hydrolysis rate of the thrombin substrate. The enriched library population was analyzed by DNA sequencing to determine the identities of exosite ligands that accelerated substrate cleavage. Several peptides in the enriched pool exhibited a consensus Y/F E/DP[F/I/L]P that is highly similar to the core YEPF exosite-binding motif in PAR1 (Gandhi et al., 2010) (Table 1). Individual identified peptides with the highest cleavage rates also included one or two anionic residues downstream of the core motif. Interestingly, however, none of the peptides identified by library screening included a string of four anionic residues similar to the PAR1 exosite ligand (EDEE).
Table 1.
Peptide sequences of potential exosite ligands from the bacterial display selections.
|
In an effort to identify caspase-7 exosite-interacting peptides, a library was similarly constructed wherein the active site recognition motif DEVD↑G was fixed on the N-terminus of eCPX, and a library of candidate exosite ligands was displayed at the C-terminus upstream of the affinity tag. Unlike the case of the thrombin-PAR1 interaction, a known exosite ligand was not available for comparison with the enriched clones. The caspase-7 library was sorted using 1 μM caspase-7 for the initial round followed by 100 nM caspase-7 in the later rounds, using alternating sorts for cleavage and substrate display. Individual isolated clones from the final rounds of screening encoded peptides enriched with basic amino acids including histidine, arginine, and lysine (Table 1), and several peptides exhibited a motif of the form HPxH. In addition, multiple exosite peptides had cysteines spaced by five or seven residues, and the two cysteines would likely form a disulfide bridge.
Measurement of substrate cleavage kinetics
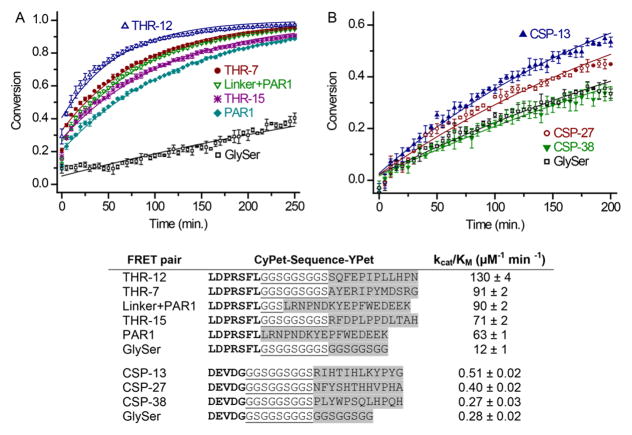
To determine whether peptides present in the enriched libraries accelerated substrate cleavage, the rate of cleavage was measured first for cell displayed substrates and then in the context of fluorescent protein FRET substrates. For thrombin, the rates of cleavage for three individual cell-displayed substrates, whose exosite sequences exhibited similarity with PAR1, were measured using flow cytometry. Similar to the PAR1 exosite ligand, each of the exosite ligand-displaying clones accelerated cleavage of the active site recognition motif by six- to ninefold over the GlySer control sequence with 250 pM thrombin (Figure 2). Thrombin at a concentration of 125 pM resulted in a cell-surface substrate conversion of 30–40%, when the exosite ligands were present. We next tested whether substrate-exosite ligand fusions, when reconstructed as contiguous linear FRET probes, would exhibit similar enhancement of hydrolysis rates. The soluble, protease activity probes were constructed using fluorescent proteins optimized for FRET, CyPet and YPet (Nguyen and Daugherty, 2005). The FRET probes were constructed by fusing the PAR1 thrombin substrate to the 12-mer exosite ligands with an intervening 9-mer GGS repeat linker. Two positive control FRET probes were constructed wherein the PAR1 active site motif was directly fused to the PAR1 exosite ligand, or separated by a three residue GGS linker from the PAR1 exosite ligand (Figure 3). As a non-functional exosite ligand control, the PAR1 active site motif was fused to the GlySer control peptide. The FRET probes were then used to measure second-order rate constants when cleaved by thrombin (Figure 3A). The exosite ligand THR-2 mediated the largest enhancement in hydrolysis rate (kcat/KM), which was roughly 10-fold greater than that of the substrate with the GlySer control peptide. The two other substrates analyzed were cleaved five- to eight-fold faster than the control. Interestingly, adding the GGS linker between the PAR1 substrate and exosite ligand improved the hydrolysis rate of the substrate by 40%. These results demonstrate that thrombin exosite-binding peptides identified using eCLiPS substantially increase the rate of substrate hydrolysis.
Figure 2. Measurement of the extent of conversion of bacterial clones with different exosite ligands.
The cells were incubated with 125 pM (white bars) and 250 pM thrombin (gray bars), and cellular red fluorescence was measured by flow cytometry in order to determine the extent of substrate conversion. The error bars are the standard deviation calculated from triplicate samples.
Figure 3. Determining the cleavage kinetics using FRET probes.
A. The FRET probes with the PAR1 substrate attached to the exosite ligands were incubated with 125 pM thrombin, and the conversion versus time was fit (solid lines) to calculate the second-order rate constants (kcat/KM). The error bars are the standard deviation from triplicate reactions. B. Similarly to the thrombin probes, the FRET probes with the caspase-7 substrate and exosite binders were incubated with 8 nM caspase-7. In the table, the sequences for the CyPet-YPet FRET pairs are listed with the substrate (bold), glycine-serine linker (underlined), and exosite or control ligand (highlighted in gray). The kcat/KM values are listed for each probe with the standard deviation of the value from triplicate reactions.
In contrast with the thrombin exosite ligands, the caspase-7 exosite ligands only modestly enhanced cleavage rates in the context of the FRET probes (Figure 3B). The best performing exosite ligand, CSP-1, conferred a two-fold increase in hydrolysis rate over the GlySer control peptide. Overall, the second-order rate constants for the caspase-7 FRET probes were approximately 100-fold lower than those observed for the thrombin substrates. Caspase-7 is a cysteine protease that exhibits higher activity in the presence of the reducing agent dithiothreitol (DTT). Because the streptavidin-binding peptide in the eCLiPS system contains a disulfide bond, DTT was not included during the screening procedure with caspase-7. Adding 10 mM DTT along with the FRET probe in caspase-7 reactions did increase the protease’s activity and promoted substrate cleavage at a lower caspase-7 concentration (0.5 nM versus 8 nM). Given the small enhancement mediated by caspase-7 selected exosite ligands, we then investigated whether adjusting the linker length between the substrate and the best exosite ligand, CSP-1, could enhance cleavage rates. The original glycine-serine linker (three GGS repeats) was shortened to a single GGS repeat and also lengthened to five or seven GGS repeats. Neither increasing nor decreasing the linker length significantly increased the cleavage kinetics for this substrate-exosite ligand pair with an approximately two- to three-fold difference between the FRET probes with the CSP-1 peptide versus the corresponding GlySer control. Thus, unlike thrombin, screening with caspase-7 did not yield peptides that substantially increase substrate cleavage kinetics. These results suggest that caspase-7 may not use an exosite to efficiently cleave extended linear peptides in a manner similar to that of thrombin.
Discussion
Here we have developed a bacterial surface display strategy to identify ligands for protease exosites and to develop substrates that exhibit higher catalytic turnover and are more selective for the target protease. Although peptide libraries are increasingly used to identify protease substrates, their application has been limited to characterizing the specificity of protease active sites (Harris et al., 2000; Turk et al., 2001; Deperthes, 2002; Boulware and Daugherty, 2006; Schneider and Craik, 2009). Cellular libraries of peptide substrates (CLiPS) and its predecessor, substrate phage display, are distinct from widely used positional scanning solution-phase libraries in that they enable screening or selection, respectively, for kinetically optimal substrates directly from very large libraries containing 108–109 members. These methodologies enable the identification of combinations of amino acids that can exhibit cooperativity in protease binding. CLiPS differs from the substrate phage method in that many copies of the substrate (~104) are displayed on the surface of E. coli, in contrast to the 4–5 copies displayed on phage when using the gpIII coat protein, such that screening is based upon average substrate conversion per cell. The use of cell surface display in conjunction with flow cytometry also enables real-time optimization and fine tuning of the library screening stringency. In the present study, the identification of exosite ligands required the use of cytometry to determine both effective protease concentrations and reaction times, along with comparison of the cleavage kinetics of the library population versus the control clones.
An important subtlety is that the eCLiPS system utilizes display on both the N- and C-termini of the display scaffold. Since the distance between the protease active site and a potential exosite is typically unknown, displaying the substrate and the candidate exosite ligand on different termini reduces spatial constraints to enable identification of ligands that bind exosites either close to, or far from, the active site. Instead, if both the substrate and potential exosite ligand were displayed on the N-terminus of eCPX, it would be difficult to differentiate between peptides that are acting as an additional substrate rather than binding to the protease to improve cleavage. By displaying the substrate on the N-terminus of the display scaffold and the potential exosite binder on the C-terminus, both termini can be labeled with different color fluorophores to enable sorting for bacterial clones with a cleaved substrate but intact C-terminal exosite binder. We also must consider the display efficiency of the library since some substrates and ligands display well when expressed on the same terminus while others work better using the biterminal approach. The optimal strategy can be determined empirically by testing display levels of the potential substrate and a glycine-serine control peptide both on the N-terminus, both on the C-terminus, and on separate termini. Although we demonstrated the utility of eCLiPS for enhancing thrombin and caspase-7 substrates, this method has the flexibility to be readily applied to identify and characterize exosite-interacting peptide substrates for other proteases as well.
While many experimental methods can identify preferred protease active site interaction motifs (e.g., P6-P5-P4-P3-P2-P1-↑-P1′-P2′), it has become increasingly clear that physiological substrate recognition frequently involves additional important protease-substrate interactions. There are now multiple examples of proteases that cleave physiological protein substrates orders of magnitude faster than the best known active site interacting peptide substrate. Urokinase-type plasminogen activator, for example, can cleave the physiological substrate plasminogen more than 100-fold faster than the best known peptide substrates selected using substrate phage display (e.g., PFGR↑SA) (Bergstrom et al., 2003). Although the precise mechanisms used by proteases to cleave physiological substrates so efficiently remain unclear, a subset of proteases can recognize an extended linear peptide substrate using multiple interacting exosites in a manner similar to thrombin. For example, the protease domain of botulinum neurotoxin A selectively and efficiently cleaves the substrate synaptosome-associated protein-25 kDa utilizing a remarkably extensive set of peptide-interacting exosites (Breidenbach and Brunger, 2004). Similarly, many other proteases may utilize one or more exosites to achieve high selectivity for physiological substrates and such mechanisms could dramatically restrict the repertoire of substrates that are effectively cleaved. The characterization of a larger number of protease exosites using eCLiPS could help identify the defining features of peptide-interacting exosites to enable their prediction from structural data. Further application of eCLiPS will provide an improved understanding of substrate recognition specificity to better elucidate the functions of proteases.
The substrate specificity of thrombin has been investigated previously using several methods including substrate phage libraries and positional scanning fluorogenic substrate libraries that target the active site (Backes et al., 2000; Ohkubo et al., 2001). In addition to the active site, thrombin exosites I and II are necessary for enhancing the specificity of thrombin for physiological substrates as shown for PAR1 (Figure 4A) (Vu et al., 1991b). Using eCLiPS, thrombin-binding peptides were identified that enhanced cleavage of the PAR1 thrombin substrate to a rate equivalent to, or greater than, the physiological thrombin exosite-binding peptide in the PAR1 protein. The commercially available peptide inhibitors of thrombin, bivalirudin, hirudin, and lepirudin (Warkentin, 2004), also utilize both the active site and exosite I to achieve high affinity and specificity. Using the exosite ligands identified here, we were able to construct highly sensitive thrombin-activity detection probes using fluorescent protein FRET probes (Boulware and Daugherty, 2006). The addition of exosite-binding peptides enhanced thrombin detection sensitivity by roughly ten-fold when compared to a conventional active site interacting substrate.
Figure 4. Location of the exosite on thrombin and allosteric site on caspase-7.

A. An extended exosite groove in human thrombin binds to the PAR1-derived peptide sequence (YEPFW highlighted in yellow). B. The structure of the caspase-7 dimer has a deep pocket (highlighted in yellow) located within 10 Å of the active site. The images shown were created using PDB:3LU9 (thrombin-PAR1) and PDB:1SHJ (caspase-7) with the Chimera software package.
Caspase-7 was selected for exosite profiling since it has been shown previously to possess an allosteric site, a deep pocket adjacent to the active site, that can bind to small molecules to modulate activity (Figure 4B) (Hardy et al., 2004). Although screening enriched peptides that accelerated catalysis, the effect was modest; substrates with exosite ligands identified by eCLiPS were cleaved two- to three-fold faster than the control substrate for the most effective linker length. One potential explanation for this small effect could be a non-optimal spacing between the active site motif and the exosite ligand. In our study, we attempted to optimize linker length C-terminal to the scissile bond, but did not explore the possibility of an exosite N-terminal to the scissile bond. Also, with the large diversity remaining in the caspase-7 final population, additional sorting with lower protease concentration may have improved the affinity of the exosite ligands or other peptides, like the constrained sequences, may have shown a larger improvement in cleavage kinetics.
One disadvantage of the biterminal display used here is that the length of linker between the substrate and exosite binder needs to be determined within the context of the soluble probe. With the thrombin ligands, the sequence of the native PAR1 substrate was used to estimate the needed linker length between the two entities. The native substrates of caspase-7, however, do not have known exosite-interacting regions so different linker lengths were tested; neither shorter nor longer linkers substantially improved the cleavage kinetic. While not explored here, further manipulation of the linker structure and length could potentially improve the cleavage rate. Alternatively, a linear peptide may not be able to mimic the complex protein structure utilized by caspase-7 to recognize native substrates. Nevertheless, the eCLiPS approach presented here can help to characterize the functions of protease exosites and identify peptide motifs that are critical to enhancing protease specificity for physiological protein substrates. Finally, protease-interacting peptides, such as those discovered here, could provide dramatically increased sensitivity for protease-activity detection in vitro and in vivo.
Materials and Methods
Reagents
Bacterial experiments were performed with E. coli strain MC1061 and grown in LB medium supplemented with 34 μg/ml chloramphenicol. Human α-thrombin was obtained from Haematologic Technologies, Inc. (Essex Junction, VT). Streptavidin-R-phycoerythrin (SAPE) was from Invitrogen (Carlsbad, CA). Oligonucleotides were from Operon Biotechnologies (Huntsville, AL) and restriction enzymes were from New England Biolabs (Ipswich, MA).
Caspase-7 mutant generation, expression and purification
Wild-type human caspase-7 was expressed from a two-plasmid expression system. A construct encoding residues 50–198 in the pRSET(AmpR) plasmid expresses the large subunit. A second construct encoding residues 199–303 plus one codon for Q plus a six-Histidine tag in the plasmid pBB75 (KanR) plasmid (Batchelor et al. 1998) expresses the small subunit. The recombinant large and small subunits were co-expressed in the BL21(DE3) strain of E. coli in 2xYT media grown for 18 h after induction with 1 mM Isopropyl β-D-1-thiogalactopyranoside at an OD600 of 0.6. Caspase-7 was purified using Ni-affinity liquid chromatography (HiTrap Chelating HP, GE Healthcare, Waukesha, WI). After binding protein to the affinity column, the protein was eluted with a step gradient from 2 mM imidazole to 250 mM imidazole. Protein was diluted to 50 mM NaCl and then purified using a Macro-Prep High Q ion exchange column (Bio-Scale Mini 5mL, Bio-Rad, Hercules, CA) with a linear gradient from 50 mM to 500 mM NaCl in 20mM Tris buffer pH 8.0, with 2mM DTT. Protein eluted in 120 mM NaCl and 20 mM Tris pH 8.0 was assessed for purity by SDS-PAGE to be 98%+ pure and stored in elution buffer at −80°C.
Clone and library construction
For the eCLiPS system, the pBAD33 plasmid consisted of a streptavidin-binding peptide (WVCHPMWEVMCLR), linker (GGSGQSGQGG), thrombin substrate from PAR1 (LDPRSFL) or a caspase substrate (DEVDG), and another linker (GSSGGSGGSGGSG) on the N-terminus of eCPX; on the C-terminus of eCPX, a linker (GGSGGSGGSGG) was followed by the exosite-interacting ligand, an additional linker (GQGGQGGSGGSGGS), and a Mona-SH3 domain binding peptide (HISQWKPKVPNREDKYKK, P2X). For a positive and negative control, either a thrombin-exosite I binding peptide from PAR1 (LRNPNDKYEPFWEDEEK) or a glycine-serine linker (GSGGSGGGSGGSGGSGG) was inserted in the exosite ligand position. For the thrombin and caspase-7 libraries, a 12-mer randomized region was constructed using NNS codons. Detailed cloning methods on the display constructs and library construction are included in the Supplementary Information.
Optimizing screening conditions for cell displayed thrombin substrates
To identify the screening conditions for the cell displayed substrates, the overnight bacterial cultures of the clones (thrombin substrate and glycine-serine or PAR1 control exosite ligands) were subcultured 1:50 (v:v) for 2 h at 37°C in LB (pH 5.5) and 0.5% glycerol, then induced with 0.2% (w:v) L-(+)-arabinose and 1 mM EDTA for 3.5 h at 37°C. Lower pH media was necessary to improve the display of substrates containing anionic residues. Cells were pelleted, washed with reaction buffer (50 mM Tris-Cl, pH 7.5 supplemented with 20 mM NaCl, 2mM CaCl2 and 10 μM ZnCl2), and pelleted again. The pelleted cells were then resuspended in 20 μl of reaction buffer and the volume was then divided in four 5 μl aliquots. The cells were incubated with thrombin concentrations of 20, 2, 1, and 0 nM in a total of volume of 10 μl. The reaction samples were incubated at 37°C with shaking for 1 h. The samples were then diluted 1:100 in phosphate buffered saline (PBS pH 7.4) to impede the reaction, pelleted, and then resuspended in PBS supplemented with 50 nM SA-PE and 250 nM AlajGFP-Mona SH3 to label the cells and were incubated at 4°C for 1 h. The labeled cells were then washed with PBS and analyzed on a FACSAria (BD Biosciences, San Jose, CA) with 488 nm excitation and fluorescence detection at 530 nm and 576 nm. To also optimize the thrombin incubation time, the cells were incubated with 250 pM thrombin at 37°C and washed after 30, 45, and 60 min. Then the cells were labeled and analyzed as described previously.
Screening the thrombin and caspase-7 exosite libraries
The libraries were grown and expression induced using the same conditions as the control constructs. For the library sorting against thrombin, approximately 108 cells were washed with reaction buffer and then pelleted. The pelleted cells were resuspended in 20 μl of reaction buffer and 10 μl were incubated either with 500 pM thrombin or no protease at 37°C with shaking for 1 h. The samples were washed with PBS and labeled with SA-PE and AlajGFP-Mona SH3 with the same protocol as the individual clones. Using a BD FACSAria, the cleaved cells (green(+) red(−)) were sorted. Following the initial round of sorting, eight additional sorts were performed. The rounds alternated between thrombin-free isolation of cells that properly displayed substrates (green(+) red(+)) and sorting for cells with cleaved substrates after incubating with 125 pM thrombin (green(+) red(−)).
Sorting of the caspase library for enhanced substrates was performed in a similar manner to the thrombin library. The caspase substrate with the control glycine-serine ligand did not display efficiently and was therefore not used for optimizing the sorting conditions. The reaction buffer used with caspase-7 was 25 mM HEPES pH 7.4, 20 mM NaCl, and 2 mM CaCl2. The library was sorted for a total of ten rounds, alternating between sorts for cleavage and sorts for display using 1 μM caspase-7 in the initial round and decreasing the concentration to 100 nM caspase-7 in the later rounds. During the sorting of both the thrombin and caspase-7 exosite libraries, approximately 5% of the library population was sorted for cleavage and 10% of the population showed high red and green fluorescence during the labeling sorts. Cells from the final round of sorting were plated to isolate single clones for DNA sequencing. The plasmids from approximately 100 clones from the final round of sorting of both the thrombin and caspase-7 exosite libraries were sequenced using the standard pBad reverse primer.
Cleavage kinetics of the cell displayed substrates
To measure the extent of conversion of cell displayed substrates, the overnight bacterial cultures of the clones from the final round of sorting were subcultured and induced as before in triplicate samples. Cells were washed and resuspended in reaction buffer (15 μl) and the volume was then divided in three (each 5 μl); the samples were incubated with 0.25, 0.125, and 0 nM thrombin for 1 h at 37°C. Cells were washed and labeled with 50 nM SA-PE for 1 h at 4°C. The labeled cells were again washed in PBS and their fluorescence was measured by flow cytometry. Conversion values were calculated using the following equation:
where FR− is the red fluorescence of the bacterial cell population without the protease, FR+ is the red fluorescence of the cells with protease, and FR0 is the fluorescence of unlabeled cells (Boulware and Daugherty, 2006).
Measurement of thrombin and caspase-7 activity using FRET probes
To determine the second-order rates constants of the substrates in soluble form, CyPet-YPet FRET pairs were constructed with the substrate followed by a linker (GGSGGSGGS) and the exosite ligand. The FRET pairs were expressed and purified as described previously (Jabaiah and Daugherty, 2011). The hydrolysis reaction for the thrombin substrates was performed in triplicate in 100 μl total volume with 80 nM FRET substrate and 125 pM of thrombin at 37°C for 250 min. With the caspase FRET substrates, the reactions included 120 nM FRET substrate and 8 nM caspase-7 and were run at 37°C for 200 min. To monitor the cleavage of the peptides, the fluorescence emissions of the FRET probes (485 nm and 527 nm for thrombin substrates, 475 nm and 532 nm for caspase substrates) were measured every 5 min upon excitation at 433 nm using a Tecan Safire™ fluorescence spectrophotometer (Tecan, Durham, NC). The second-order rate constants (kcat/KM) were then calculated by fitting the substrate conversion to the simplified Michaelis-Menten equation using non-linear regression:
where [E] is the protease concentration and t is time (Boulware and Daugherty, 2006).
Supplementary Material
Acknowledgments
This work was funded by grants 1R01GM097114-01A1 and 1R21CA159232-01. The authors also thank Amol Shivange for constructing the thrombin-PAR1 image.
References
- Agniswamy J, Fang B, Weber IT. Plasticity of S2–S4 specificity pockets of executioner caspase-7 revealed by structural and kinetic analysis. FEBS Journal. 2007;274:4752–4765. doi: 10.1111/j.1742-4658.2007.05994.x. [DOI] [PubMed] [Google Scholar]
- Ala-aho R, Kahari V. Collagenases in cancer. Biochimie. 2005;87:273–286. doi: 10.1016/j.biochi.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Backes BJ, Harris JL, Leonetti F, Craik CS, Ellman JA. Synthesis of positional-scanning libraries of fluorogenic peptide substrates to define the extended substrate specificity of plasmin and thrombin. Nat Biotechnol. 2000;18:187–193. doi: 10.1038/72642. [DOI] [PubMed] [Google Scholar]
- Bergstrom RC, Coombs GS, Ye S, Madison EL, Goldsmith EJ, Corey DR. Binding of Nonphysiological Protein and Peptide Substrates to Proteases: Differences between Urokinase-Type Plasminogen Activator and Trypsin and Contributions to the Evolution of Regulated Proteolysis†. Biochemistry. 2003;42:5395–5402. doi: 10.1021/bi027417x. [DOI] [PubMed] [Google Scholar]
- Bock PE, Panizzi P, Verhamme IMA. Exosites in the substrate specificity of blood coagulation reactions. J Thromb Haemostasis. 2007;5:81–94. doi: 10.1111/j.1538-7836.2007.02496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode W, Stubbs MT. Spatial structure of thrombin as a guide to its multiple sites of interaction. Semin Thromb Hemost. 1993;19:321–333. doi: 10.1055/s-2007-993283. [DOI] [PubMed] [Google Scholar]
- Borgoño CA, Michael IP, Diamandis EP. Human Tissue Kallikreins: Physiologic Roles and Applications in Cancer. Mol Cancer Res. 2004;2:257–280. [PubMed] [Google Scholar]
- Boulware KT, Daugherty PS. Protease specificity determination by using cellular libraries of peptide substrates (CLiPS) PNAS. 2006;103:7583–7588. doi: 10.1073/pnas.0511108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulware KT, Jabaiah A, Daugherty PS. Evolutionary optimization of peptide substrates for proteases that exhibit rapid hydrolysis kinetics. Biotechnol Bioeng. 2010;106:339–46. doi: 10.1002/bit.22693. [DOI] [PubMed] [Google Scholar]
- Breidenbach MA, Brunger AT. Substrate recognition strategy for botulinum neurotoxin serotype A. Nature. 2004;432:925–929. doi: 10.1038/nature03123. [DOI] [PubMed] [Google Scholar]
- Burrage PS, Mix KS, Brinckerhoff CE. Matrix metalloproteinases: role in arthritis. Front Biosci. 2006;11:529–543. doi: 10.2741/1817. [DOI] [PubMed] [Google Scholar]
- Cornille F, Martin L, Lenoir C, Cussac D, Roques BP, Fournie-Zaluski MC. Cooperative Exosite-Dependent Cleavage of Synaptobrevin by Tetanus Toxin Light Chain. J Biol Chem. 1997;272:3459–3464. doi: 10.1074/jbc.272.6.3459. [DOI] [PubMed] [Google Scholar]
- Deperthes D. Phage display substrate: a blind method for determining protease specificity. Biol Chem. 2002;383:1107–1112. doi: 10.1515/BC.2002.119. [DOI] [PubMed] [Google Scholar]
- Gandhi PS, Chen Z, Di Cera E. Crystal Structure of Thrombin Bound to the Uncleaved Extracellular Fragment of PAR1. J Biol Chem. 2010;285:15393–15398. doi: 10.1074/jbc.M110.115337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy JA, Lam J, Nguyen JT, O’Brien T, Wells JA. Discovery of an allosteric site in the caspases. PNAS. 2004;101:12461–12466. doi: 10.1073/pnas.0404781101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JL, Backes BJ, Leonetti F, Mahrus S, Ellman JA, Craik CS. Rapid and general profiling of protease specificity by using combinatorial fluorogenic substrate libraries. PNAS. 2000;97:7754–7759. doi: 10.1073/pnas.140132697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntington JA. Molecular recognition mechanisms of thrombin. J Thromb Haemostasis. 2005;3:1861–1872. doi: 10.1111/j.1538-7836.2005.01363.x. [DOI] [PubMed] [Google Scholar]
- Jabaiah A, Daugherty PS. Directed Evolution of Protease Beacons that Enable Sensitive Detection of Endogenous MT1-MMP Activity in Tumor Cell Lines. Chem Biol. 2011;18:392–401. doi: 10.1016/j.chembiol.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutgens SPM, Cleutjens KBJM, Daemen MJAP, Heeneman S. Cathepsin cysteine proteases in cardiovascular disease. FASEB J. 2007;21:3029–3041. doi: 10.1096/fj.06-7924com. [DOI] [PubMed] [Google Scholar]
- Nguyen AW, Daugherty PS. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat Biotechnol. 2005;23:355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- Ohkubo S, Miyadera K, Sugimoto Y, Matsuo K, Wierzba K, Yamada Y. Substrate phage as a tool to identify novel substrate sequences of proteases. Comb Chem High Throughput Screening. 2001;4:573–583. doi: 10.2174/1386207013330788. [DOI] [PubMed] [Google Scholar]
- Rice JJ, Daugherty PS. Directed evolution of a biterminal bacterial display scaffold enhances the display of diverse peptides. Protein Eng, Des Sel. 2008;21:435–442. doi: 10.1093/protein/gzn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider EL, Craik CS. Positional scanning synthetic combinatorial libraries for substrate profiling. Methods Mol Biol. 2009;539:59–78. doi: 10.1007/978-1-60327-003-8_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamenkovic I. Matrix metalloproteinases in tumor invasion and metastasis. Semin Cancer Biol. 2000;10:415–433. doi: 10.1006/scbi.2000.0379. [DOI] [PubMed] [Google Scholar]
- Stubbs MT, Bode W. A model for the specificity of fibrinogen cleavage by thrombin. Semin Thromb Hemost. 1993;19:344–351. doi: 10.1055/s-2007-993285. [DOI] [PubMed] [Google Scholar]
- Talanian RV, Quinlan C, Trautz S, Hackett MC, Mankovich JA, Banach D, Ghayur T, Brady KD, Wong WW. Substrate Specificities of Caspase Family Proteases. J Biol Chem. 1997;272:9677–9682. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- Tenore SB, Ferreira PRA. The place of protease inhibitors in antiretroviral treatment. Braz J Infect Dis. 2009;13:371–374. doi: 10.1590/S1413-86702009000500012. [DOI] [PubMed] [Google Scholar]
- Turk BE, Huang LL, Piro ET, Cantley LC. Determination of protease cleavage site motifs using mixture-based oriented peptide libraries. Nat Biotechnol. 2001;19:661–667. doi: 10.1038/90273. [DOI] [PubMed] [Google Scholar]
- Verhamme IM, Olson ST, Tollefsen DM, Bock PE. Binding of exosite ligands to human thrombin. J Biol Chem. 2002;277:6788–6798. doi: 10.1074/jbc.M110257200. [DOI] [PubMed] [Google Scholar]
- Vu TK, Hung DT, Wheaton VI, Coughlin SR. Molecular cloning of a functional thrombin receptor reveals a novel proteolytic mechanism of receptor activation. Cell. 1991a;64:1057–1068. doi: 10.1016/0092-8674(91)90261-v. [DOI] [PubMed] [Google Scholar]
- Vu TK, Wheaton VI, Hung DT, Charo I, Coughlin SR. Domains specifying thrombin-receptor interaction. Nature. 1991b;353:674–677. doi: 10.1038/353674a0. [DOI] [PubMed] [Google Scholar]
- Walker CP, Royston D. Thrombin generation and its inhibition: a review of the scientific basis and mechanism of action of anticoagulant therapies. Br J Anaesth. 2002;88:848–863. doi: 10.1093/bja/88.6.848. [DOI] [PubMed] [Google Scholar]
- Walsh JG, Cullen SP, Sheridan C, Lüthi AU, Gerner C, Martin SJ. Executioner caspase-3 and caspase-7 are functionally distinct proteases. PNAS. 2008;105:12815–12819. doi: 10.1073/pnas.0707715105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warkentin TE. Bivalent direct thrombin inhibitors: hirudin and bivalirudin. Best Pract Res, Clin Haematol. 2004;17:105–125. doi: 10.1016/j.beha.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Yasuda Y, Kaleta J, Brömme D. The role of cathepsins in osteoporosis and arthritis: Rationale for the design of new therapeutics. Adv Drug Delivery Rev. 2005;57:973–993. doi: 10.1016/j.addr.2004.12.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.