Abstract
Streptomycetes are mycelium-forming bacteria that produce two thirds of clinically relevant secondary metabolites. Secondary metabolite production is activated at specific developmental stages of Streptomyces life cycle. Despite this, Streptomyces differentiation in industrial bioreactors tends to be underestimated and the most important parameters managed are only indirectly related to differentiation: modifications to the culture media, optimization of productive strains by random or directed mutagenesis, analysis of biophysical parameters, etc. In this work the relationship between differentiation and antibiotic production in lab-scale bioreactors was defined. Streptomyces coelicolor was used as a model strain. Morphological differentiation was comparable to that occurring during pre-sporulation stages in solid cultures: an initial compartmentalized mycelium suffers a programmed cell death, and remaining viable segments then differentiate to a second multinucleated antibiotic-producing mycelium. Differentiation was demonstrated to be one of the keys to interpreting biophysical fermentation parameters and to rationalizing the optimization of secondary metabolite production in bioreactors.
Keywords: Streptomyces, Bioreactor, Differentiation, Antibiotics, Programmed cell death
1. Introduction
Streptomycetes are gram-positive, environmental soil bacteria that play important roles in the mineralization of organic matter. Streptomyces is extremely important in biotechnology, given that approximately two thirds of all clinical antibiotics and several other bioactive compounds are synthesized by members of this genus (Ruiz et al., 2010).
Streptomycetes are mycelial microorganisms with complex developmental cycles that include programmed cell death (PCD) and sporulation (reviewed in Claessen et al. (2006) and Yagüe et al. (2013)). In solid sporulating cultures, a compartmentalized mycelium (MI) initiates development. MI compartments are separated by septa formed by membranes which generally do not display thick cell walls (reviewed in Yagüe et al. (2013)). A fraction of MI cells undergo a highly ordered programmed cell death (PCD) (Yagüe et al., 2013), and remaining viable cells differentiate to a multinucleated mycelium that has only sporadic septa (MII). MII gradually begins to express the chaplin and rodlin proteins that assemble into the rodlet layer that, in turn, provides the surface hydrophobicity necessary to grow into the air (aerial mycelium) (reviewed in Claessen et al. (2006)). At the end of the cycle, hypha septation and sporulation take place. MI fulfills the vegetative role in Streptomyces and MII constitutes the reproductive stage that is destined to sporulate and also produces secondary metabolites (Yagüe et al., 2013). In previous works, it was reported that differentiation in non-sporulating liquid cultures (laboratory flasks) was similar to that occurring during the pre-sporulation stages in solid cultures (reviewed in Yagüe et al. (2013)): an initial compartmentalized mycelium (MI) undergoes PCD and the remaining viable segments of this mycelium differentiate to a multinucleated mycelium (MII), i.e. the antibiotic-producing mycelium (Yagüe et al., 2013).
Most processes for secondary metabolite production are performed in bioreactors. Nevertheless, Streptomyces differentiation under these conditions has barely been studied, mainly due to the fact that most Streptomyces strains do not sporulate under these conditions. Streptomyces fermentation analysis and optimization has mainly been empirical and focused on the analysis of biophysical parameters, such as mycelial grouping (pellets, clumps), media composition, oxygenation, pH, agitation, temperature and, of course, levels of secondary metabolite production. Several studies have tested the optimal composition of culture media (Wentzel et al., 2012), analyzing the kind of hyphae grouping that is best suited for secondary metabolite production (dispersed hyphae vs. clumps or pellets) (van Veluw et al., 2012; van Wezel et al., 2006), analyzing the effects of bioreactor hydrodynamics on the physiology of Streptomyces (reviewed in Olmos et al. (2013)), or optimizing productive strains by random or directed mutagenesis (van Wezel et al., 2006). However, the complex development of Streptomyces under these conditions has not been fully understood and, as a direct consequence, there is no general con-sensus as to how morphology and other biophysical parameters correlate with secondary metabolite production. Fermentation parameters need to be optimized empirically for each strain and compound. For example, pellet and clump formation has been described as essential for obtaining good production of retamycin or nikkomycin (Pamboukian and Facciotti, 2004), but in the case of virginiamycin, there is no relationship between morphology and secondary metabolite production (Yang et al., 1996); high dissolved oxygen tensions (DOT) have been reported as necessary for the production of vancomycin (Dunstan et al., 2000), but not for the production of erythromycin (Clark et al., 1995), just to name a few examples.
The main objective of this work is to extend understanding of Streptomyces differentiation to lab-scale bioreactors, defining the kind of differentiation present under these conditions, how differentiation, fermentation parameters and secondary metabolite production are correlated, and describing a general model applicable to improving secondary metabolite production in Streptomyces industrial fermentations. Streptomyces coelicolor is one of the best-characterized Streptomyces strains (Chater, 2001). It produces various secondary metabolites, including two well-characterized antibiotics: undecylprodigiosin and actinorhodin. In order to facilitate comparisons with differentiation and development in bioreactors and other developmental conditions (solid cultures and laboratory flasks), S. coelicolor was used in this work as a model strain.
2. Methods
2.1. Strains, media, and culture conditions
S. coelicolor M145 was the strain used in this work. Cultures were performed in R5A sucrose-free liquid media (Fernandez et al., 1998). This culture medium contains MOPS buffer in sufficient concentration (100 mM) to maintain pH stable during cultivations. Laboratory flasks of 500 ml were filled with 100 ml of culture medium and incubated at 200 rpm and 30 °C. Bioreactor cultivations were performed in a 2-L bioreactor (Bio-Flo 110, New Brunswick Scientific, NJ, USA) equipped with a pH meter (Mettler Toledo, Switzerland), a polarographic dissolved oxygen electrode (InPro 6830, Mettler Toledo, Switzerland), and rushton impellers. As described above, the effect of pitched blade impellers in fermentations was also tested (data not shown). An initial working volume of 1.3 L at 30 °C and aeration of 1 L/min were used. Dissolved oxygen tension was set to a minimum of 3.8 mg/L (50% saturation), using an agitation interval of between 300 and 800 rpm, and pH was set at 6.8 using a computer-controlled peristaltic pump via automatic addition of 2 M KOH and 1 M HCl.
Flasks and bioreactors were inoculated directly with freshly prepared spores at 1 × 107 (“dense cultures”) or 1 × 105 (“diluted cultures”) spores/ml. Where indicated, culture medium was supplemented with antifoam (Biospumex 153K, BASF) to a final concentration of 1%. The effect of the antifoam in preventing early massive fragmentation/lysis was not so evident at lower concentrations (data not shown). More than 5 biological replicates were performed for each culture, and monitored morphologically and biochemically. However, extensive quantitative measurements were performed in only two of these biological replicates, and the quantitative data presented in the figures of this work correspond to the average ± SD of these two independent fermentations (biological replicates).
2.2. S. coelicolor repeated batch cultivations
“Dense cultures” (107 spores/ml), growing in R5A sucrose-free medium amended with antifoam (1% of Biospumex 153K, BASF), and using the growth parameters indicated above were grown in the bioreactor for 66 h. After that time-point, the full bioreactor contents were extracted into a 2-L sterile bottle using a peristaltic pump connected to the inoculation port. Mycelial pellets were allowed to sediment in a static state for 5 min, after which supernatant was removed under sterile conditions. The volume of medium extracted was replaced by the same volume of fresh, sterile, R5A medium amended with antifoam, and the whole culture was reintroduced into the bioreactor using again a peristaltic pump connected to the inoculation port.
2.3. Determination of the oxygen uptake rate (OUR) and oxygen transfer rate (OTR)
OUR was obtained from the slope of the plot of dissolved oxygen concentration over time following a momentary interruption of the air supply to the bioreactor. OTR was estimated according to the slope of dissolved oxygen recovery after aeration (fixed value of 1 L/min) and agitation (interval of between 300 and 800 rpm) was restored to the values present at the time at which the air supply was interrupted. OUR and OTR values were only estimated at developmental time points at which DOT values were less than 7 mg/L (90% saturation).
2.4. Streptomyces sampling throughout the differentiation cycle
Samples of S. coelicolor obtained from liquid cultures were centrifuged (7740g, 10 min at 4 °C). Supernatants of the culture medium were used to estimate extracellular proteins. Cellular extracts were obtained as follows: the mycelium pellets were resuspended in a known volume of buffer A (Tris–HCl 20 mM, pH 8, EDTA 1 mM, β-mercaptoethanol 7 mM, and complete EDTA-free Protease Inhibitor Cocktail Tablets from Roche) and ruptured using Fast-Prep (MP™ Biomedicals) with ≤106 μm beads (Sigma, G8893500G) and three 20-s force 6.5 cycles, with 1 min on ice between each run. Finally, samples were centrifuged at 7740g in an Eppendorf microcentrifuge for 15 min at 4 °C; the resulting supernatant fraction was used as the cellular fraction.
2.5. Protein quantification
Determination of protein concentrations was carried out with the Bradford assay (Biorad) and a bovine serum albumin standard (Sigma). Protein measured in the supernatants of culture medium corresponded to extracellular protein; protein measured in the cellular extracts corresponded to intracellular protein; and the sum of both, corresponded to total protein.
2.6. Antibiotic quantification
Undecylprodigiosin and actinorhodin were quantified spectro-photometrically according to Tsao et al. (1985) and Bystrykh et al. (1996). In order to measure the total amount of actinorhodin (intracellular and extracellular), cells were ruptured in their culture medium by adding KOH 0.1 N. Cellular debris was discarded by centrifugation, and actinorhodin was quantified spectrophotometrically with a UV/visible spectrophotometer (Shimadzu, Model UV-1240), applying the linear Beer–Lambert relationship to estimate concentration (ε640 = 25,320). In the case of cultures with antifoam, acthinorhodin spectrophotometric measurements were performed at 4 °C, in order to prevent interference with turbidity due to the antifoam. Undecylprodigiosin was measured after vacuum drying the culture (including the mycelium and culture medium) followed by extraction with methanol, acidification with HCl (to 0.5 M), and spectrophotometric assay at 530 nm, again using the Beer–Lambert relationship to estimate concentration (ε530 = 100,500). In all cases, for the high concentration solutions, dilutions were performed to conduct the analysis in the linear Beer–Lambert region.
2.7. Viability staining
Culture samples were obtained and processed for microscopy at different incubation time points, as previously described (Manteca et al., 2008). Cells were stained with a cell-impermeant nucleic acid stain (propidium iodide, PI) in order to detect the dying cell population and with SYTO 9 green fluorescent nucleic acid stain (LIVE/DEAD Bac-Light Bacterial Viability Kit, Invitrogen, L-13152) to detect viable cells. The SYTO 9 green fluorescent stain labels all the cells, i.e. those with intact membranes, as well as those with damaged ones. In contrast, PI penetrates only bacteria with damaged membranes, decreasing SYTO 9 stain fluorescence when both dyes are present. Thus, in the presence of both stains, bacteria with intact cell membranes appear to fluoresce green, whereas bacteria with damaged membranes appear red. After leaving them for at least 10 min in the dark, the samples were examined under a Leica TCS-SP2-AOBS confocal laser-scanning microscope at a wavelength of either 488 nm or 568 nm excitation and 530 nm (green) or 630 nm (red) emission, respectively (optical sections about 0.2 lm). Images were mixed using Leica Confocal Software. In some cases, samples were also examined in differential interference contrast mode, available with the same equipment.
S. coelicolor unstained samples were used as controls to determine the minimum photomultiplier tube (PMT) gain necessary to detect autofluorescence in the confocal microscope. Green autofluorescence (Willemse and van Wezel, 2009) can interfere with green fluorescent fluorochromes as occurs, for instance, with GFP (Manteca et al., 2008; Willemse and van Wezel, 2009), and they can potentially interfere with SYTO9 green stain. However, in practice, there is no interference because the intensity of green autofluorescence is negligible when compared to the SYTO9 green fluorescence. Tenconi et al. (2013) have recently demonstrated the existence of red autofluorescence associated with undecylprodigiosin that displays an excitation-emission spectrum similar to PI. Under the experimental conditions used in this work, red autofluorescence was significantly less than PI fluorescence and the minimum PMT gain necessary to observe it was 860 V (using the 63 × objective), 60% more than the PMT gain used to observe PI fluorescence (535 V under the 63 × objective) (data not shown). Despite this, red autofluorescence was not negligible, and some of the red fluorescent background detected at later time points in the centers of mycelial pellets may be derived from undecylprodigiosin.
The antifoam used had green autofluorescence, however its intensity was very low in comparison with the SYTO9 green fluorescence. In addition, antifoam could be not confused with stained hyphae, because it was completely amorphous.
More than 30 images were analyzed for each developmental time point in a minimum of three independent cultures. The percentage of sporulation and MI compartmentalized hyphae were estimated by counting 200 hyphae, from different pictures, and different biological replicates, visualized independently in the same focal plane.
2.8. Nuclease activity gel analysis (zymograms)
Nucleases were separated in a 12% gel by SDS–PAGE containing 10 μg/ml of denatured calf thymus DNA (Sigma); 8.5 μg of protein were used per well. When necessary, protein samples were concentrated by filtration using Vivaspin 20 (10,000 molecular weight cutoff, Sartorius). After electrophoresis, the proteins were renatured by repeatedly washing the gel with the renaturation buffer (Tris–HCl 25 mM, pH 8.8, EDTA 1 mM, β-mercaptoethanol 7 mM) for 2 h at 4 °C. Nuclease activity was visualized by incubating the gels for 20 min at 37 °C in 20 mM Tris–HCl (pH 8.0), 7 mM 2-mercaptoethanol, 10 mM MgCl2, 5 mM CaCl2 and 10% DMSO buffer, as reported elsewhere (Nicieza et al., 1999), followed by staining with ethidium bromide and analysis under UV light. Micrococcal nuclease (16.7 kDa) and bovine pancreatic DNase I (31 kDa) (Amersham Pharmacia Biotech) were included as positive controls. The reproducibility of the data shown was corroborated by at least three independent cultures and nuclease analysis at various developmental time points.
3. Results and discussion
3.1. Differentiation of S. coelicolor M145 in 2 L-bioreactors
In order to facilitate comparison of Streptomyces differentiation in bioreactors with differentiation previously reported in laboratory flasks, a workflow similar to that used in flask cultures (Manteca et al., 2008) was utilized: R5A culture medium (without antifoam) were inoculated with either 107 spores/ml (“dense cultures”) (Fig. 1 and Supplementary Fig. S1) or 105 spores/ml (“diluted cultures”) (Supplementary Figs. S2 and S3). Despite the fact that some foam was formed in the bioreactor, it was not excessive, and cultures were initially performed without defoamers, in order to facilitate direct comparison with differentiation previously described in flask cultures (Manteca et al., 2008). Morphological differentiation was analyzed by means of confocal microscopy on cultures stained with the vital stains SYTO9 and propidium iodide (see Section 2 for details). At early time points, 100% of the hyphae presented the regular discontinuities and gaps (Supplementary Fig. S1E) previously described for MI hyphae (Manteca et al., 2008). MI differentiated into a second multinucleated mycelium (MII) that had only sporadic gaps (compare MI hyphae from Supplementary Fig. S1E with MII hyphae from Supplementary Fig. S1F). There was also a transition phase in which some segments of the MI started to grow in the form of hyphae with more widely spaced septa (MII) while other parts of the hyphae remained in the MI stage (data not shown). Cell death started in the centers of MI mycelial pellets (propidium iodide staining) (Supplementary Fig. S1A) and exhibited the hallmarks previously reported for Streptomyces PCD (reviewed in Yagüe et al. (2013)), including the activation of non-sequence specific nucleases involved in chromosomal DNA degradation (Supplementary Fig. S4A).
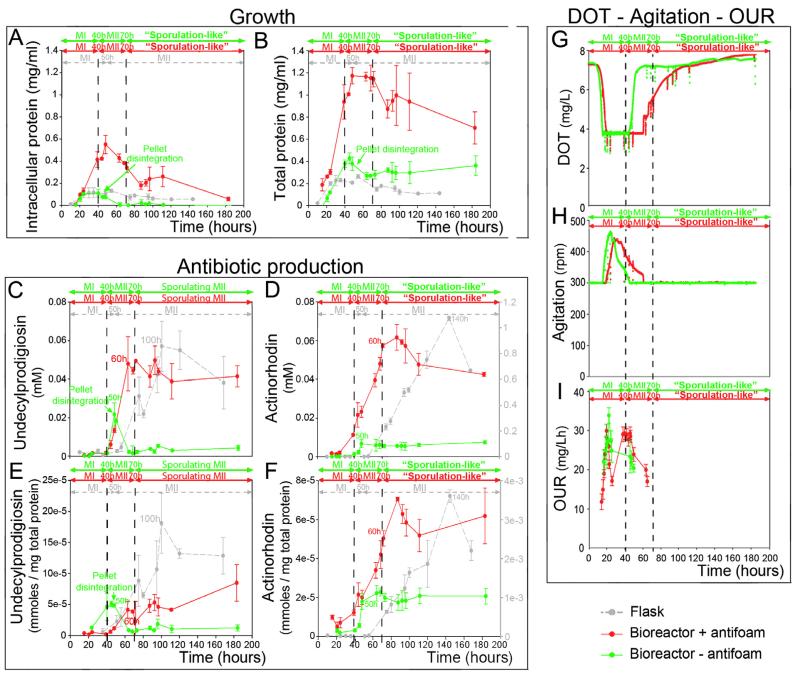
Fig. 1. Time-course of fermentation parameters.
The data correspond to “dense cultures” (107 spores/ml) of Streptomyces coelicolor M145. Green lines, 2-L bioreactors, R5A without antifoam; red lines, 2-L bioreactors, R5A with antifoam; gray lines, laboratory flasks, R5A without antifoam. (A) and (B) Growth curves (intracellular protein and total protein). (C)–(F) Antibiotic production (undecylprodigiosin and actinorhodin). Actinorhodin levels in the laboratory flasks in (D) and (F) have their own scales on the right. Time points at which maximum antibiotic productions was reached are indicated. (G)–(I) DOT, Agitation and OUR. Values are the average ± SD from two biological replicates.
One of the most important differences observed in the bioreactor with respect to laboratory flasks for the Streptomyces strain and culture conditions used in this work, was the existence of massive fragmentation and disintegration of mycelial pellets at around 50 h of fermentation. This massive disintegration was observed microscopically, in the irregular shape of the pellets (Supplementary Fig. S1B) or the fissures crossing them internally (Supplementary Fig. S1C), and macroscopically, in the form of the apparent clarification of the culture medium (notice that bioreactor probes are clearly visible in the bioreactor vessel) (Supplementary Fig. S1L and M). This pellet disintegration correlated with a sudden fall in intracellular protein levels (Fig. 1A), which decreased from 0.1 mg/ml prior to the massive pellet disintegration to 0.0002 mg/ml after that. The few pellets that did not lose their integrity continued to grow in diameter (up to 530–600 μm in the “dense cultures”) (Supplementary Fig. S1D). Most of the biomass in these pellets corresponded to dying cells (red staining) (Supplementary Fig. S1D) and, consequently, the number of remaining viable hyphae (green staining) in the bioreactor following massive pellet disintegration was extremely low. This kind of mycelial disintegration has been previously described as “massive lysis” in several Streptomyces fermentations, such as Streptomyces clavuligerus (reducing mycelium by more than 30%) (Roubos et al., 2001), Streptomyces spp. (Techapun et al., 2003), Streptomyces albulus (Shih and Shen, 2006), or S. coelicolor (Ozergin-Ulgen and Mavituna, 1993), to name just a few examples. This “massive lysis” differs from the “fragmentation of the mycelial clumps” described in some cases (van Wezel et al., 2006), which basically consists of the fragmentation of large clumps into small clumps, but without the early massive hyphal lysis reported in this work as well as others (Ozergin-Ulgen and Mavituna, 1993; Roubos et al., 2001; Shih and Shen, 2006; Techapun et al., 2003). The reason why this phenomenon occurred in some streptomycetes and not in others remains unknown. In S. coelicolor growing under the growth conditions used in this work, true lysis started in hyphae located at the centers of the pellets (beginning at times as early as 15 h) (Supplementary Fig. S1A), long before massive fragmentation took place (around 50 h) (Supplementary Fig. S1B and C). This massive pellet disintegration did not occur in laboratory flasks (Manteca et al., 2008), where it was observed neither macroscopically nor microscopically (data not shown), as reflected by the absence of the dramatic decrease in the intracellular protein described above in the bioreactor cultures (Fig. 1A). Hence, it seems that massive pellet disintegration depends on the hydrodynamics of the bioreactor combined with the tendency of S. coelicolor to form large pellets under the culture conditions used in this work.
Another important difference between bioreactor- and laboratory flask-cultured samples was the existence of a sporulation-like process, beginning at around 70 h, and affecting some 5% of hyphae that remained viable at these time points (see quantification criteria in Section 2). Two of the most important features of sporulation, division and separation of nucleoids (Supplementary Fig. S1G), and the physical strangulation of hypha forming chains of individual round segments (Supplementary Fig. S1H), were observed. Further work will be necessary to characterize if these round segments present the resistance properties characterizing Streptomyces spores formed in solid cultures (Lee and Rho, 1993).
Antibiotic production was accelerated in the bioreactor, peaking at 100–140 h in laboratory flasks vs. 50 h in the bioreactor (Fig. 1C and D). MII differentiation was slightly accelerated, from 50 h in laboratory flasks (Manteca et al., 2008) to around 40 h in the bioreactor (Supplementary Fig. S1 and data not shown). Antibiotic biosynthesis was halted after pellet disintegration, with maximum undecylprodigiosin production levels slightly lower in the bioreactor with respect to the laboratory flasks (0.02 mM vs. 0.06 mM observed in flasks) (Fig. 1C) and dramatically lower in the case of actinorhodin (0.007 mM vs. 1 mM observed in flasks) (Fig. 1D). These results match previous works reporting that undecylprodigiosin and actinorhodin are synthesized differentially in S. coelicolor fermentations (Sevcikova and Kormanec, 2004) and illustrates the important and well-known issue that, in addition to hyphal differentiation, there are specific regulatory mechanisms for different secondary metabolites.
Biophysical fermentation parameters, such as dissolved oxygen tension (DOT), agitation, and oxygen uptake rates (OUR), correlated well with differentiation (Fig. 1G–I): DOT fell from saturation to a fixed level (50% saturation), probably due to hyphal growth and respiration (Fig. 1G); there was a concomitant increase in agitation to maintain oxygen levels at the fixed level (Fig. 1H); once pellet disintegration started (see above), biological oxygen consumption and agitation decreased gradually, and dissolved oxygen levels increased suddenly to saturation. OUR values peaked at 20 h (MI stage) (Fig. 1I) and fell during the PCD of the MI; they also did not recover during the MII stage, probably due to early massive pellet disintegration/lysis described above. OUR values were consistently lower than oxygen transfer rates (OTR) (data not shown), indicating that oxygen did not limit growth at any time. Interestingly, Streptomyces coelicolor OUR values measured in this work were quite low, with peak values of 30 mg/L h (Fig. 1I). This might be a direct consequence of the unusual development of S. coelicolor, in which most of the biomass in the mycelial pellets is dying and therefore does not consume oxygen. This constitutes an elegant example of the importance of understanding Streptomyces differentiation, so as to be able to interpret classical biophysical fermentation parameters in the model strain S. coelicolor and conceivably in other industrial relevant streptomycetes, which is one of the most important conclusions of this work. Information concerning oxygen uptake kinetics of Streptomyces cultures is scarce despite their industrial importance. OUR values vary widely between strains, from 2.88 mg O2 g cell−1 h−1 in S. lividans (Magnolo et al., 1991) to 320 mg O2 g cell−1 h−1 in S. clavuligerus (Yegneswaran et al., 1991). The meaning of these differences is difficult to interpret due to the absence of any indication as to mycelium differentiation/PCD in most of these works. Ozergin-Ulgen and Mavituna (1998) described maximum OUR values for S. coelicolor of 192 mg/L h, 6.4-fold higher than the maximum OUR detected in this work for the same strain. This might be due to important differences in the bioreactor vessel used in Ozergin-Ulgen and Mavituna work (1998), which had large baffles instead of the smooth vessels used in this work. Baffles are in fact routinely used in laboratory flask cultures to prevent pellet formation in S. coelicolor (Kieser et al., 2000), and a dispersal growth could prevent cell death and increase OUR. An analysis of hyphae differentiation, development and PCD would be essential to address these differences in OURs between different Streptomyces strains and culture conditions.
Inoculation density is one of the well-known fermentation parameters that is usually conducive to modifications in growth and production. In order to test how this parameter would affect differentiation in the bioreactors, a 100-fold dilution, 105 spores/ml (“diluted cultures”), was used. The same kind of differentiation described above for “dense” culture, was also observed in “diluted” cultures (Supplementary Figs. S2 and S3): MI differentiated to MII (Supplementary Fig. S2) after a PCD that activated the non-sequence specific nucleases (Supplementary Fig. S4B); there was an early massive lysis of pellets (Supplementary Fig. S2), and sporulation-like processes were also observed (Supplementary Fig. S2) affecting approximately 5% of the hyphae. Massive pellet disintegration occurred at similar developmental time points in “dense” and “diluted” cultures (50 h, when the pellet diameter was 500 μm), but the biomass (number of pellets) was much lower in “diluted” compared to “dense” cultures: 0.016 mg/ml of intracellular protein in “diluted” (Supplementary Fig. S3A) vs. 0.07 mg/ml in “dense” cultures (Fig. 1A). Antibiotic production, DOT, agitation, and OUR also correlated well with differentiation (Supplementary Fig. S3G–I).
3.2. Differentiation of S. coelicolor M145 in 2-L bioreactors supplemented with antifoam
As commented above, the onset of antibiotic production in the bioreactor was more rapid than in laboratory flasks (Manteca et al., 2008), but the final levels of production were very low. In order to improve production in the bioreactor, growing conditions were modified to try to prevent the early massive lysis, so growth would more to resemble the development observed in flasks. The most obvious difference between bioreactors and laboratory flasks is the impellers used for agitation in the case of the bioreactor, so the first experimental approach to trying to prevent lysis was to reduce agitation to minimum levels (50 rpm); however, the same extension of pellet disintegration was observed (data not shown). Similar results were observed at different agitation rates (50, 100, 200 or 300 rpm) (data not shown). Second, Rushton impellers were replaced by a gentler impeller (pitched blade impellers), but similar results were again obtained (data not shown). Finally, the culture medium’s rheology was modified reducing surface tension by means of an antifoam agent (Biospumex 153K, BASF), at a concentration of 1%. Under these conditions, massive fragmentation of the pellets was avoided, as observed under the confocal microscope (compare Supplementary Fig. S1B and C with Supplementary Fig. S5B, noting the absence of fissures in the pellets) and macroscopically by the high turbidity of the cultures (compare Supplementary Fig. S5K and L with Supplementary Fig. S1L and M). The change was also reflected in the high levels of intracellular protein, which reached a maximum of 0.6 mg/ml (Fig. 1A) vs. the 0.1 mg/ml observed in cultures without antifoam (Fig. 1A). This effect of preventing early fragmentation/lysis was only observed at relatively high concentrations of antifoam (1%, see Section 2 for details). The reason why antifoam prevents pellet disintegration is as yet unknown; however, the antifoam tended to coat the mycelial pellets (as can observed by its own autofluorescence in Supplementary Fig. S5C) and the hydrophobic forces generated may have prevented this phenomenon. Antifoams are often used with S. coelicolor (Wentzel et al., 2012) as well as other Streptomyces fermentations to prevent foam formation, or even, in some cases to be used as carbon sources (Perlman and Wagman, 1952). They are usually added automatically in small amounts when foam is detected by a specific probe, and in some cases, they are added directly to the culture medium at concentrations up to 0.1% (Wentzel et al., 2012). However, to the best of our knowledge, this is the first time that early pellet fragmentation/lysis has been demonstrated to be prevented by adding antifoam to the culture media at relatively high concentrations, a fact that might be useful for preventing lysis in other industrial streptomycetes.
Under the conditions used in this work, antifoam led to a moderate increase in undecylprodigiosin production (0.05 mM vs. 0.02 mM in the case of cultures without antifoam) (Fig. 1C), but it was also slightly delayed when production was referenced to cellular protein (Fig. 1E). This level of production was comparable to the production observed in laboratory flasks (0.05 mM) (Fig. 1C). Actinorhodin production was also increased with respect to cultures without antifoam (0.06 mM in cultures with antifoam vs. 0.006 mM in cultures without antifoam), although it was far from the production levels obtained in laboratory flasks (1 mM) (Fig. 1D). Similar to the arrest in cellular metabolism associated with sporulation in solid sporulating cultures (Chater and Horinouchi, 2003), inhibition of actinorhodin production might be a consequence of a change in hyphae metabolism/differentiation, affecting most of the hyphae, and culminating in the sporulation-like processes which continued to affect some 5% of the hyphae in the case of “dense cultures” amended with antifoam (Supplementary Fig. S5F and G).
Biophysical fermentation parameters also correlated well with differentiation in cultures with antifoam (Fig. 1G–I): DOT fell from saturation (100%) to the set level (50% saturation) (Fig. 2G), leading to increased agitation (Fig. 1H); the absence of pellet disintegration might prolong the oxygen consumption phase, generating the two peaks of OUR separated by a stage of low oxygen consumption (Fig. 1I). The latter may reflect a transient arrest in metabolism (oxygen consumption) preceding MII differentiation similar to that previously reported in laboratory flasks (reviewed in Yagüe et al. (2013)). These two maxima in OUR were very unusual, and are another nice illustration of the necessity of understanding Streptomyces differentiation in order to interpret fermentation parameters. As in the case of fermentations without antifoam, oxygen levels did not limit growth; OUR levels were consistently lower than OTR values at any given point in time (data not shown).
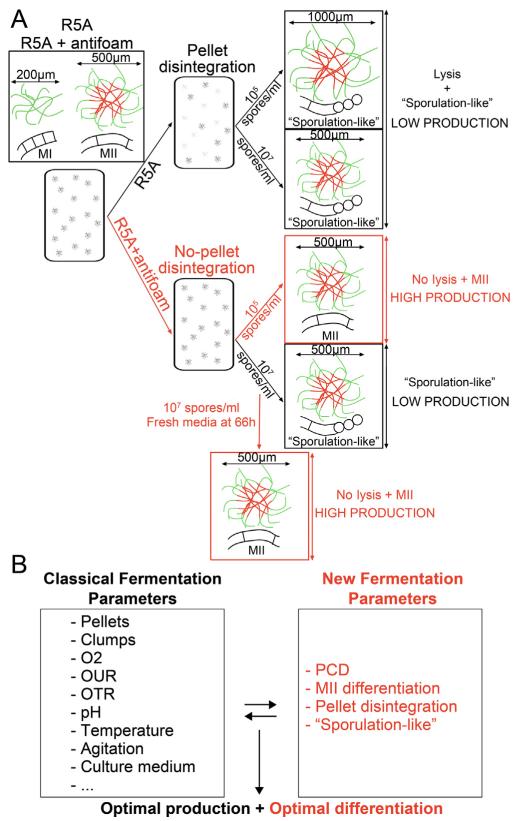
Fig. 2. Scheme illustrating S. coelicolor differentiation in bioreactors.
(A) S. coelicolor differentiation growing in R5A sucrose-free medium with and without antifoam, at two spore inoculations (105 and 107 spores/ml). Red corresponds to dying hyphae (PI staining) and green to viable hyphae (SYTO9 staining). The optimal fermentation workflow is highlighted in red. (B) Comparison between classic (black letters) and new (red letters) fermentation parameters established in this work. See text for details.
Antifoam also prevented pellet disintegration in the “diluted” cultures (Supplementary Figs S3 and S6). Interestingly, the sporulation-like processes observed in “dense” cultures amended with antifoam were not observed in the “diluted” cultures (Supplementary Fig. S6F). Sporulation in S. coelicolor liquid cultures is very unusual and to the best of our knowledge, has only been reported once before in laboratory flasks suffering nutritional downshifts (Daza et al., 1989). The differentiation signals activating sporulation in the bioreactors remain unknown. However, if it is considered that sporulation is triggered by environmental/biological stresses (Chater, 2001), the high growth rates achieved in the bioreactors together with pellet disintegration might approach the development occurring in stressed solid sporulating cultures. In the absence of pellet disintegration, putative differentiation diffusible signals (Chater et al., 2010; Horinouchi and Beppu, 1992) generated by stressed cells suffering PCD (Yagüe et al., 2013) would be hidden in the centers of the pellets.
The highest levels of antibiotic production were reached in the “diluted cultures” amended with antifoam. The maximum production levels were around 0.2 mM in the “diluted” cultures (Supplementary Fig. S3C and D) vs. 0.05 mM obtained in the “dense” cultures with antifoam (Fig. 1C and D) for both, undecylprodigiosin and actinorhodin. By contrast, growth in the “diluted” cultures, measured as total protein/ml, was half that in the “dense” cultures (compare Fig. 1B and Supplementary Fig. S3B). As a consequence, the sporulation-like processes observed in some 5% of the hyphae in the cultures without antifoam and in the “dense” cultures with antifoam correlate with a block in the production of secondary metabolites in the whole mycelium. This is another example of how growth/biomass production is not sufficient to guarantee secondary metabolite production in Streptomyces.
3.3. Optimization of MII differentiation and antibiotic production in S. coelicolor fermentations: repeated batch cultivations
As outlined in Fig. 2A, the highest levels of antibiotic production were reached by preventing pellet disintegration (using antifoams), blocking sporulation-like processes (using low inocula; i.e. slow growth rates), and prolonging the antibiotic-producing phase (MII). In spite of that, antibiotic production was faster in the “dense cultures” (maximum undecylprodigiosin levels at 60 h) (Fig. 1C) than in the “diluted cultures” (maximum undecylprodigiosin levels at 80 h) (Supplementary Fig. S3C). The possibility of combining the rate of antibiotic production reached in the “dense cultures” with the high yields obtained in the “diluted cultures” was tested. To do so, the possibility of repressing the sporulation-like processes observed in “dense cultures” was explored, by replacing culture medium with fresh medium once undecylprodigiosin reached maximum levels (66 h) (Fig. 1C) but prior to sporulation. Under these conditions, there was no significant increase in mycelial biomass (measured as either total or intracellular protein/ml; data not shown). The mycelium remained at the MII stage (Supplementary Fig. S7) and there was not sporulation (Supplementary Fig. S7). After an unsurprising delay of approximately 20 h, there was very rapid production of actinorhodin (0.004 mmol/L h, 4-fold faster than in the original culture) (Fig. 3A). In the case of undecylprodigiosin, there was also a delay of 20 h prior to production, but in this case, production was very weak (Fig. 3B), which once again demonstrates that in addition to differentiation there are specific regulators for different secondary metabolites. Moreover, the production of both antibiotics normalized by total protein was consistently higher in the repeated batch cultivation than in the original cultures (Fig. 3C and D). This kind of experimental workflow opens up the possibility of maintaining the mycelium in the productive stage (MII) indefinitely, replacing culture medium periodically after MII differentiation and once antibiotic production has already peaked.
Fig. 3. Time-course of antibiotic production in Streptomyces coelicolor repeated batch fermentations.
(A) Undecylprodigiosin (mM). (B) Actinorhodin (mM). (C) Undecylprodigiosin (mmol/mg total protein). (D) Actinorhodin (mmol/mg total protein). Rates of actinorhodin production (slopes) are indicated in the graphs. Antibiotic concentration values are the average ± SD from two biological replicates. See text for details.
3.4. Future perspectives
Different streptomycetes show different behaviors in liquid cultures: some species form large pellets, such as S. coelicolor, others growth more dispersed, as for instance S. clavuligerus (Roubos et al., 2001), while still others such as Streptomyces griseus sporulate (Kendrick and Ensign, 1983), etc. As a consequence, the effect of fermentation parameter modifications in different species cannot be easily predicted. The workflow proposed here for S. coelicolor (optimization of antibiotic-producing mycelium differentiation, prevention of sporulation) might be applied to rationalizing the biological effects of classical biophysical fermentation parameters, and to facilitating the optimization of secondary metabolite production in industrial streptomycetes. In addition, preventing early massive pellet fragmentation/lysis by adding antifoam directly to the culture medium at relatively high concentrations is novel, and may be useful for preventing lysis in other industrial streptomycetes.
4. Conclusions
The most important conclusions reached in this work, were: first, the existence of a progressive morphological differentiation in S. coelicolor growing in lab-scale bioreactors (PCD, MII differentiation, pellet disintegration, and a kind of sporulation) comparable to that occurring in solid cultures; second, it was demonstrated that this differentiation is one of the keys to interpreting typical fermentation parameters (growth, antibiotic production, dissolved oxygen tension, agitation and oxygen uptake rates) (outlined in Fig. 2B); third, a general consensus to improve secondary metabolite production in S. coelicolor was proposed: optimization of the differentiation of the antibiotic-producing mycelium (MII).
Supplementary Material
HIGHLIGHTS.
Differentiation in bioreactors is comparable to solid sporulating cultures.
Differentiation is linked to antibiotic production.
Differentiation is one of the keys to interpreting fermentation parameters.
General consensus: differentiation of the antibiotic-producing mycelium (MII).
Antifoams can prevent massive pellet fragmentation/lysis.
Acknowledgements
We wish to thank the European Research Council (ERC Starting Grant; Strp-differentiation 280304) for economic support; Dr. J.L. Gallego (Universidad de Oviedo, Área de Prospección e Investigación Minera) for providing the Bioreactor; Prof. L.A. Garcia and Dr. S. Alonso (Universidad de Oviedo, Dpto. Ingeniería Química) for helping with bioreactor management; A. Wentzel (Department of Biotechnology, SINTEF Materials and Chemistry, Norway) for discussions about antifoams, Beatriz Gutierrez Magan (Universidad de Oviedo, Dpto. Biología Funcional, Área de Microbiología) for laboratory assistance, and Proof-Reading-Service.com for proofreading the text.
Abbreviations
- PCD
programmed cell death
- MI
first compartmentalized mycelium
- MII
second multinucleated mycelium
- PI
propidium iodide
- DOT
dissolved oxygen tension
- OTR
oxygen transfer rates
- OUR
oxygen uptake rate
- PMT
photomultiplier tube
References
- Bystrykh LV, Fernandez-Moreno MA, Herrema JK, Malpartida F, Hopwood DA, Dijkhuizen L. Production of actinorhodin-related “blue pigments” by Streptomyces coelicolor A3(2) J. Bacteriol. 1996;178:2238–2244. doi: 10.1128/jb.178.8.2238-2244.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater KF. Regulation of sporulation in Streptomyces coelicolor A3(2), a checkpoint multiplex? Curr. Opin. Microbiol. 2001;4:667–673. doi: 10.1016/s1369-5274(01)00267-3. [DOI] [PubMed] [Google Scholar]
- Chater KF, Horinouchi S. Signalling early developmental events in two highly diverged Streptomyces species. Mol. Microbiol. 2003;48:9–15. doi: 10.1046/j.1365-2958.2003.03476.x. [DOI] [PubMed] [Google Scholar]
- Chater KF, Biró S, Lee KJ, Palmer T, Schrempf H. The complex extracellular biology of Streptomyces. FEMS Microbiol. Rev. 2010;34:171–198. doi: 10.1111/j.1574-6976.2009.00206.x. [DOI] [PubMed] [Google Scholar]
- Claessen D, de Jong W, Dijkhuizen L, Wösten HA. Regulation of Streptomyces development, reach for the sky! Trends Microbiol. 2006;14:313–319. doi: 10.1016/j.tim.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Clark GJ, Langley D, Bushell ME. Oxygen limitation can induce microbial secondary metabolite formation, investigations with miniature electrodes in shaker and bioreactor culture. Microbiology. 1995;141:663–669. [Google Scholar]
- Daza A, Martín JF, Dominguez A, Gil JA. Sporulation of several species of Streptomyces in submerged cultures after nutritional downshift. J. Gen. Microbiol. 1989;135:2483–2491. doi: 10.1099/00221287-135-9-2483. [DOI] [PubMed] [Google Scholar]
- Dunstan GH, Avignone-Rossa C, Langley D, Bushell ME. The vancomycin biosynthetic pathway is induced in oxygen-limited Amycolatopsis orientalis (ATCC 19795) cultures that do not produce antibiotic. Enzyme Microb. Technol. 2000;27:502–510. doi: 10.1016/s0141-0229(00)00238-6. [DOI] [PubMed] [Google Scholar]
- Fernandez E, Weissbach U, Sanchez-Reillo C, Braña AF, Mendez C, Rohr J, Salas JA. Identification of two genes from Streptomyces argillaceus encoding glycosyltransferases involved in transfer of a disaccharide during biosynthesis of the antitumor drug mithramycin. J. Bacteriol. 1998;180:4929–4937. doi: 10.1128/jb.180.18.4929-4937.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S, Beppu T. Autoregulatory factors and communication in actinomycetes. Annu. Rev. Microbiol. 1992;46:377–398. doi: 10.1146/annurev.mi.46.100192.002113. [DOI] [PubMed] [Google Scholar]
- Kendrick KE, Ensign JC. Sporulation of Streptomyces griseus in submerged culture. J. Bacteriol. 1983;155:357–366. doi: 10.1128/jb.155.1.357-366.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. John Innes Foundation; Norwich: 2000. [Google Scholar]
- Lee KJ, Rho YT. Characteristics of spores formed by surface and submerged cultures of Streptomyces albidoflavus SMF301. J. Gen. Microbiol. 1993;139:3131–3137. [Google Scholar]
- Magnolo SK, Leenutaphong DL, DeModena JA, Curtin JE, Bailey JE, Galazzo JL, Hughes DE. Actinorhodin production by Streptomyces coelicolor and growth of Streptomyces lividans are improved by the expression of a bacterial haemoglobin. Biotechnology. 1991;9:473–476. doi: 10.1038/nbt0591-473. [DOI] [PubMed] [Google Scholar]
- Manteca A, Alvarez R, Salazar N, Yagüe P, Sánchez J. Mycelium differentiation and antibiotic production in submerged cultures of Streptomyces coelicolor. Appl. Environ. Microbiol. 2008;74:3877–3886. doi: 10.1128/AEM.02715-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicieza RG, Huergo J, Connolly BA, Sánchez J. Purification, characterization, and role of nucleases and serine proteases in Streptomyces differentiation: analogies with the biochemical processes described in late steps of eukaryotic apoptosis. J. Biol. Chem. 1999;274:20366–20375. doi: 10.1074/jbc.274.29.20366. [DOI] [PubMed] [Google Scholar]
- Olmos E, Mehmood N, Haj Husein L, Goergen JL, Fick M, Delaunay S. Effects of bioreactor hydrodynamics on the physiology of Streptomyces. Bioprocess Biosyst. Eng. 2013;36:259–272. doi: 10.1007/s00449-012-0794-1. [DOI] [PubMed] [Google Scholar]
- Ozergin-Ulgen K, Mavituna F. Actinorhodin production by Streptomyces coelicolor A3(2): kinetic parameters related to growth, substrate uptake and production. Appl. Microbiol. Biotechnol. 1993;40:457–462. [Google Scholar]
- Ozergin-Ulgen K, Mavituna F. Oxygen transfer and uptake in Streptomyces coelicolor A3(2) culture in a batch bioreactor. J. Chem. Technol. Biotechnol. 1998;73:243–250. [Google Scholar]
- Pamboukian CRD, Facciotti MCR. Production of the antitumoral retamycin during continuous fermentations of Streptomyces olindensis. Process Biochem. 2004;39:2249–2255. doi: 10.1385/abab:112:2:111. [DOI] [PubMed] [Google Scholar]
- Perlman D, Wagman GH. Studies on the utilization of lipids by Streptomyces griseus. J. Bacteriol. 1952;63:253–262. doi: 10.1128/jb.63.2.253-262.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roubos JA, Krabben P, Luiten RG, Verbruggen HB, Heijnen JJ. A quantitative approach to characterizing cell lysis caused by mechanical agitation of Streptomyces clavuligerus. Biotechnol. Prog. 2001;17:336–347. doi: 10.1021/bp0001617. [DOI] [PubMed] [Google Scholar]
- Ruiz B, Chávez A, Forero A, García-Huante Y, Romero A, Sánchez M, Rocha D, Sánchez B, Rodríguez-Sanoja R, Sánchez S, Langley E. Production of microbial secondary metabolites, regulation by the carbon source. Crit. Rev. Microbiol. 2010;36:146–167. doi: 10.3109/10408410903489576. [DOI] [PubMed] [Google Scholar]
- Sevcikova B, Kormanec J. Differential production of two antibiotics of Streptomyces coelicolor A3(2), actinorhodin and undecylprodigiosin, upon salt stress conditions. Arch. Microbiol. 2004;181:384–389. doi: 10.1007/s00203-004-0669-1. [DOI] [PubMed] [Google Scholar]
- Shih IL, Shen MH. Optimization of cell growth and poly(ε-lysine) production in batch and fed-batch cultures by Streptomyces albulus IFO 14147. Process Biochem. 2006;41:1644–1649. [Google Scholar]
- Techapun C, Poosaran N, Watanabe M, Sasaki K. Optimization of aeration and agitation rates to improve cellulase-free xylanase production by thermotolerant Streptomyces sp Ab106 and repeated fed-batch cultivation using agricultural waste. J. Biosci. Bioeng. 2003;95:298–301. doi: 10.1016/s1389-1723(03)80033-6. [DOI] [PubMed] [Google Scholar]
- Tenconi E, Guichard P, Motte P, Matagne A, Rigali S. Use of red autofluorescence for monitoring prodiginine biosynthesis. J. Microbiol. Methods. 2013;93:138–143. doi: 10.1016/j.mimet.2013.02.012. [DOI] [PubMed] [Google Scholar]
- Tsao SW, Rudd BA, He XG, Chang CJ, Floss HG. Identification of a red pigment from Streptomyces coelicolor A3(2) as a mixture of prodigiosin derivatives. J. Antibiot. 1985;38:128–131. doi: 10.7164/antibiotics.38.128. [DOI] [PubMed] [Google Scholar]
- van Veluw GJ, Petrus ML, Gubbens J, de Graaf R, de Jong IP, van Wezel GP, Wösten HA, Claessen D. Analysis of two distinct mycelial populations in liquid-grown Streptomyces cultures using a flow cytometry-based proteomics approach. Appl. Microbiol. Biotechnol. 2012;96:1301–1312. doi: 10.1007/s00253-012-4490-5. [DOI] [PubMed] [Google Scholar]
- van Wezel GP, Krabben P, Traag BA, Keijser BJ, Kerste R, Vijgenboom E, Heijnen JJ, Kraal B. Unlocking Streptomyces spp. for use as sustainable industrial production platforms by morphological engineering. Appl. Environ. Microbiol. 2006;72:5283–5288. doi: 10.1128/AEM.00808-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzel A, Bruheim P, Øverby A, Jakobsen ØM, Sletta H, Omara WA, Hodgson DA, Ellingsen TE. Optimized submerged batch fermentation strategy for systems scale studies of metabolic switching in Streptomyces coelicolor A3(2) BMC Syst. Biol. 2012;6:59. doi: 10.1186/1752-0509-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemse J, van Wezel GP. Imaging of Streptomyces coelicolor A3(2) with reduced autofluorescence reveals a novel stage of FtsZ localization. PLoS ONE. 2009;4:e4242. doi: 10.1371/journal.pone.0004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagüe P, López-García MT, Rioseras B, Sánchez J, Manteca A. Pre-sporulation stages of Streptomyces differentiation, state-of-the-art and future perspectives. FEMS Microbiol. Lett. 2013;342:79–88. doi: 10.1111/1574-6968.12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YK, Morikawa M, Shimizu H, Shioya S, Suga KI, Nihira T, Yamada Y. Image analysis of mycelial morphology in virginiamycin production by batch culture of Streptomyces virginiae. J. Ferm. Bioeng. 1996;81:7–12. [Google Scholar]
- Yegneswaran PK, Gray MR, Thompson BG. Experimental simulation of dissolved oxygen fluctuations in large fermentors: effect on Streptomyces clavuligerus. Biotechnol. Bioeng. 1991;38:1203–1209. doi: 10.1002/bit.260381012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.