Abstract
In the extensive network of interdependent biochemical processes required for cell growth and division, there is mounting evidence that ribosomal DNA transcription by RNA polymerase I (pol I) not only drives cell growth via its direct role in production of the ribosomal RNA (rRNA) component of the protein-synthesis machinery, but that it is also crucial in determining the fate of the cell. Considerable progress has been made in recent years towards understanding both the function of components of the pol I transcription machinery and how cells accomplish the tight control of pol I transcription, balancing the supply of rRNA with demand under different growth conditions.
Introduction
A remarkable 50% of nascent RNA synthesis in a cell is accounted for by the transcription of ribosomal RNA (rRNA) genes [1–7], which direct and support the production of several millions of ribosomes [8]. Eukaryotic cells have evolved a ribosomal DNA (rDNA) transcription machinery that incorporates RNA polymerase I (pol I), an enzyme dedicated to this pursuit. rDNA transcription is confined to the nucleolus, which is the site of ribosome biogenesis. There are hundreds of copies of rRNA genes in mammalian and yeast cells; they are arranged in clusters as tandem head-to-tail repeats, and constitute the nucleolar organizing regions (NORs; Figure 1). The primary rRNA transcript synthesized by mammalian pol I is processed into the mature 18S, 5.8S and 28S rRNAs which, together with the 5S rRNA transcribed by RNA polymerase III (pol III), constitute the major catalytic and architectural components of the ribosome [9]. Crucially, there is a fine balance between the growth status of the cell and the accumulation of rRNAs, which is largely controlled at the level of rDNA transcription. Signalling pathways that affect cell growth in response to nutrients and growth factors and during the cell cycle have a direct influence on rRNA synthesis, with the downstream effectors of such pathways converging at the pol I transcription cycle. Here, we review recent progress in this area of research, primarily focusing on mammalian cells, and also touching on the potential impact of altered rRNA synthesis on the fate of the cell.
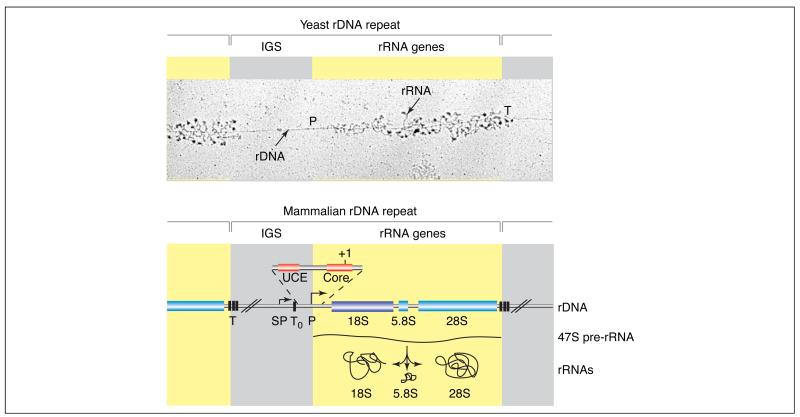
Figure 1. Organization of the rRNA genes.
The repetitive nature of the rDNA is illustrated in an electron microscopic image of a yeast nuclear chromatin spread (‘Miller spread’). Progressively longer rRNAs (stained for associated proteins) emanate from the many pol I complexes as they transcribe the rDNA, beginning at the promoter (P) and finishing at the terminator (T). Beneath, a representative mammalian rDNA repeat is outlined (not drawn to scale). Each human rDNA repeat unit (GenBank accession number: U13369) of ~43 kb contains an intergenic spacer (IGS) of ~30 kb (grey), which contains the transcription regulatory elements, and ~13 kb of sequences encoding the precursor rRNA (yellow). The rRNA genes are present in a single transcription unit, transcribed by pol I to yield a 47S precursor rRNA that is, in part, co-transcriptionally processed and modified by methylation and pseudo-uridinylation to produce the mature 18S, 5.8S and 28S rRNAs. Pol I initiates transcription at the human rDNA promoter (P), which contains an essential core element from −45 to +18 relative to the start site (+1), and an upstream control element (UCE) from −156 to −107 [7]. A spacer-promoter (SP) upstream of the gene promoter directs pol-I-dependent transcription of short-lived transcripts of unknown function. Several transcription-termination elements are located at the 3′ end of the transcribed region of the rRNA genes (T) and immediately upstream of the rRNA gene transcription start site (T0), between the spacer and gene promoters [3].
The pol I transcription cycle
Pre-initiation complex formation
Transcription commences with the recruitment and assembly of pol I and other transcription factors into a pre-initiation complex (PIC) at the rRNA gene promoter. The mammalian rRNA gene promoter contains a core element, which is essential for accurate transcription initiation, and an upstream control element (UCE), which has a modulatory role; the spacing between these sequences is crucial, as is their relative orientation (Figure 1). In addition to these elements, there are distal enhancer-like sequences, which might function by increasing the probability of stable PIC formation on the rRNA gene promoter [3].
‘Basal’ levels of transcription in vitro can be achieved in the presence of a PIC comprising only pol I and human selectivity factor 1 [SL1; also known as mouse transcription initiation factor (TIF)-IB] at the rDNA promoter, in which SL1 is a complex of the TATA-box-binding protein (TBP) and at least three TBP-associated factors including TAFI110, TAFI63 and TAFI48 [10,11] (Figure 2a). Recombinant mammalian SL1 that comprises only these three TAFs and TBP does not support efficient promoter-specific pol I transcription in an in vitro transcription system [12,13]. For the assembly of the PIC, SL1 recruits pol I to the promoter via interaction of its TAFI63 and TAFI110 subunits with the pol-I-associated factor RRN3 [14,15] (or mouse TIF-IA [16]); RRN3, in turn, is tethered to the pol I core-subunit A43 (Figure 2b,d) and pol-I-associated factor of 67 kDa (PAF67) [17,18]. Remarkably, this network of interactions is conserved from mammals to yeast. In yeast, Rrn3p bridges pol I and the promoter-bound core factor, which is the functional equivalent of SL1 [14,19,20]. Yet, intriguingly, although Rrn3p is essential in yeast, a TIF-IA (RRN3) mouse knockout survives until day 9.5, but the embryos are small because they contain fewer cells (Ingrid Grummt, personal communication). SL1, through the TAFIs, displays promoter- and polymerase-selectivity and, we consider, is fundamental in nucleating PIC formation specifically at the rDNA promoter and in mediating the recruitment of pol I, which itself has no sequence-specific DNA-binding activity. Activated transcription requires, in addition to pol I and SL1, the upstream binding factor UBF [21] (Figure 2c). SL1 interacts with the activator UBF via the highly acidic C terminus of UBF, and this might involve subunits TAFI48 and TBP of SL1 [5,7,10] (Figure 2d).
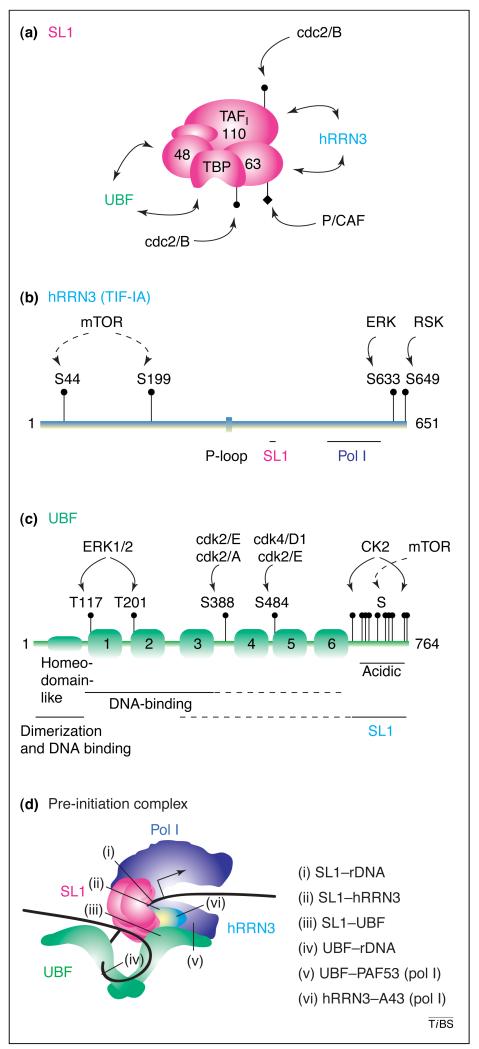
Figure 2. The molecular architecture of the mammalian pol I transcription factors, modification sites and interactions in the transcription pre-initiation complex.
(a) Selectivity factor SL1 is an ~300-kDa complex of the TATA-binding protein (TBP) and at least three pol-I-specific TBP-associated factors (TAFIs) of 110, 63 and 48 kDa (GenBank accession numbers: NM_003194, NM_005679, NM_005680 and NM_005681). The mouse complex is TIF-IB and the homologous TAFIs are 95, 68 and 48 kDa, respectively (GenBank accession numbers: Y09974, Y09973 and Y09972). The SL1 subunits that interact with UBF and hRRN3 and post-translational modifications of TIF-IB [cdc2-cyclin B-mediated phosphorylation and p300/CBP-associated factor (P/CAF) acetylation] are indicated. (b) Human RRN3, a 651-amino acid polypeptide (GenBank accession number: NM_018427), and mouse homologue TIF-IA have sequence and functional homology to Saccharomyces cerevisiae Rrn3p. Conserved regions important for the interactions with SL1 (411–415) and Pol I (512–609) [18] and an ATP/GTP-binding site motif (P-loop; 333–340), of as yet unknown function, are indicated. TIF-IA activity can be regulated by the mTOR kinase and ERK-MAPK signalling pathways (ERK and RSK). (c) Human upstream binding factor UBF (also referred to as UBF1; GenBank accession number: NM_014233) is a 764-amino acid polypeptide of 97 kDa, which is highly conserved in vertebrates and is an activator of pol I transcription. The N-terminal domain is involved in dimerization (essential for the transcription activation function of UBF) and contains a homeo-domain-like motif (possibly involved in DNA binding). In addition, the protein has six high-mobility group (HMG) boxes (labelled 1–6). HMG boxes 1, 2 and 3 are involved in DNA binding through the minor groove. The C terminus, which contains 77% acidic amino acids and is rich in serine residues, can interact with SL1 and, in addition, might influence the binding of HMG box 1 to DNA in a UBF dimer. Human UBF2 is a 94-kDa protein of unknown function encoded by the same gene as UBF1, and is identical to UBF1 except for its lack of 37 amino acids from HMG box 2 due to alternative splicing [80]. Post-translational modifications of UBF include acetylation and phosphorylation via PI3K, cyclin-dependent kinases, the ERK–MAPK pathway and the mTOR pathway [signalling via p70 ribosomal S6 kinase to serine residues in the C terminus, also shown to be consensus phosphorylation sites for Casein Kinase 2 (CK2)]. (d) In the RNA polymerase I (pol I) pre-initiation complex (PIC), there are a multitude of protein–protein and protein–ribosomal promoter–DNA interactions (i) –(vi). For simplicity, only a single dimer of UBF contacting the rDNA is shown, but there might be a dimer at both the UCE and core sequences. UBF binds pol I, in part, through Pol-I-subunit PAF53, a homologue of yeast pol-I-subunit A49. Also involved in PIC formation, but of unknown identity, are TIF-IC, a mouse Pol-I-associated factor [41], and p70 [81]. The polymerase core subunits and pol-I-associated factors, other than hRRN3, are not detailed.
UBF binds as a dimer to the UCE and core regions of the promoter, and displays cooperativity with SL1 in binding DNA. UBF binds DNA via its high-mobility-group (HMG) boxes. HMG boxes share significant homology to ~80 amino acid domains in the nuclear non-histone HMG proteins HMG1 and HMG2, which are important structural elements of chromatin and chromosomes [21,22]. A role for UBF as an architectural protein is suggested by its ability to induce formation of an ‘enhancesome’, in which a dimer of UBF organizes ~140 base pairs of enhancer DNA into a single 360° turn as a result of six in-phase bends generated by three of the six HMG boxes in each UBF monomer [23]. Formation of such structures at the UCE and at the core would juxtapose these precisely spaced promoter elements, thereby, presumably, supporting interaction between SL1 and UBF. Interaction of UBF with individual protein components of the pol I machinery at sites other than the promoter throughout the rDNA repeat [24] might alter the chromatin structure and enable access of such factors to the promoter and/or increase the local concentration of such factors, and thereby indirectly enhance the efficiency of PIC formation in vivo.
Facilitated binding of components of the PIC has also been proposed to explain the intriguing positive effect that the binding of transcription termination factor I (TTF-I) to the terminator sequence upstream of the rRNA gene promoter has on transcription (Figure 1). TTF-I interacts with histone acetyltransferase p300/calcium response element-binding protein (CREB)-binding protein (CBP)-associated factor (P/CAF), which can acetylate the TAFI68 subunit of TIF-IB (SL1) and thereby enhance the binding of this SL1 subunit to the rDNA promoter [25]. In crude extracts, TTF-I induces chromatin remodelling, perhaps increasing the accessibility of transcription factors to the promoter [26]. Paradoxically, TTF-I is involved in the establishment of rDNA silencing and, perhaps, its epigenetic inheritance via recruitment of NoRC (nucleolar remodelling complex), which induces nucleosome sliding, and via association with a co-repressor complex, including histone deacetylase 1 (HDAC1) and a DNA methyltransferase [27,28].
Pol I is the most complex component of the PIC. Yeast Pol I comprises 14 subunits, and there are mammalian homologues for all but RNA polymerase A (yeast pol I) 14-kDa subunit (RPA14) [29,30] (K. Panov and J. Zomerdijk, unpublished). A structural model of the native yeast enzyme, based on cryo-electron microscopy, resembles the Saccharomyces cerevisiae pol II (Δ4 and 7) crystal structure, but differs in the pol-I-specific subunit locations [31]. There is more complexity in human pol I, which, notably, is found as part of several distinct complexes of >1 MDa, each with discrete functions [15]. Pol Iα, which comprises the majority (>90%) of the total cellular pol I, is unable to initiate accurate transcription at a rDNA promoter even in the presence of SL1, yet is catalytically active and can initiate transcription at random from DNA ends. These properties are consistent with a role for pol Iα in elongation of transcription, although, equally, pol Iα could be a partially assembled initiation-competent complex. Pol Iβ is the distinct subpopulation of transcriptionally active and initiation-competent pol I enzymes. Pol Iβ comprises <10% of cellular polymerases and is distinguishable from pol Iα, both in its ability to direct accurate initiation of transcription at the rDNA promoter and by the presence of RRN3 [15].
There has been much debate over whether assembly of the polymerase and other components of the PIC occurs stepwise (subunit by subunit), at the rDNA promoter or, alternatively, whether there is pre-assembly into holoenzyme complexes or subcomplexes that dock at the promoter. The dynamics of the recruitment of components of the pol I transcription machinery to the rDNA gene locus at the fibrillar centre (FC) of the nucleolus suggests that pol I subunits are brought to the FCs individually rather than as part of a common complex [32]. This could indicate a stepwise assembly of pol I during formation of a functional PIC in vivo, but only if the majority of the overexpressed components detected in the FCs become incorporated into productive PICs at the rDNA promoter, and this is difficult to ascertain. Moreover, the isolation of pol I holoenzyme complexes from extracts of mammalian cells, for instance, contests the stepwise model for PIC formation, although the functional status of these complexes in the transcription cycle has yet to be established. The holoenzymes contain the core enzyme plus various combinations of the following factors: the pol I transcription factors SL1 and UBF, enzymes such as kinases and histone acetyltransferases, and components of the DNA replication, recombination and repair machineries [15,33,34].
Initiation and promoter escape
Productive PIC formation leads to promoter opening, whereupon the first ribonucleotide becomes incorporated and transcription initiates. The RNA polymerase then stutters over the synthesis of the first few nucleotides until the inhibitory interactions between the enzyme and the transcription factors at the promoter are surmounted, and pol I escapes the promoter to engage in elongation of the transcript (Figure 3). Interestingly, promoter escape – or clearance – is the rate-limiting step in a reconstituted pol I transcription system [35], and we believe that activation of transcription by UBF occurs at this step in vitro (K. Panov, J.K. Friedrich, J. Russell and J. Zomerdijk, unpublished). Conversion of the Pol Iβ initiation-competent polymerase into the elongating form of the polymerase involves the loss of RRN3, which is inactivated following its release from the polymerase [36,37].
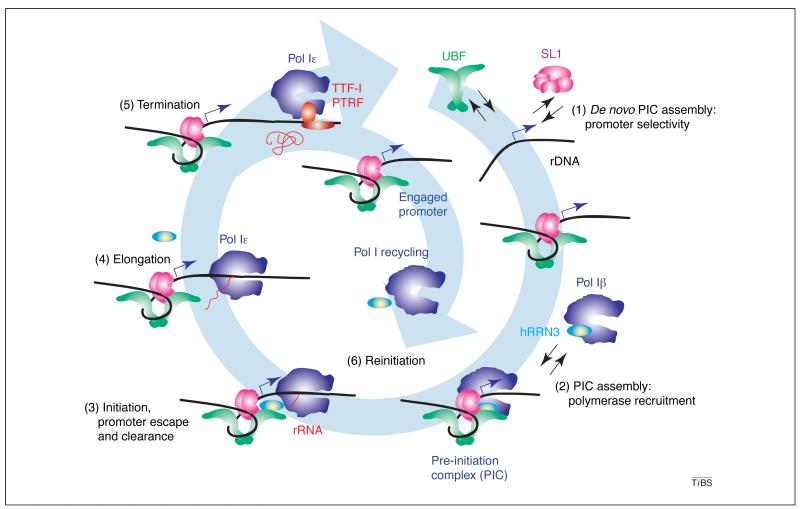
Figure 3. The RNA polymerase I (pol I) transcription cycle: pre-initiation complex formation (PIC), transcription initiation, promoter escape and clearance, elongation, termination and reinitiation.
(1) De novo PIC formation involves the selective binding of selectivity factor 1 (SL1) to the rDNA promoter, the incorporation of activator upstream binding protein (UBF) and (2) the recruitment of Pol Iβ by SL1. (3) Pol I initiates transcription upon promoter opening and, following promoter escape (3), pol I is converted into a processive enzyme (pol Iε), which elongates the nascent rRNA (4). (5) Transcription by pol I terminates at the 3′ end of the gene at specific sequences bound by termination factor TTF-I and transcript-release factor PTRF, with the concomitant release of pol I and the nascent rRNA. (6) SL1 and UBF remain promoter-bound following promoter clearance by pol I, and form a reinitiation scaffold onto which a pol Iβ complex, perhaps generated from recycled pol I and hRRN3, is recruited, and the resultant productive PIC can initiate another cycle of transcription.
Elongation of transcription
As pol I escapes and clears the promoter, UBF and SL1 remain promoter-bound in vitro, poised to recruit the next pol I complex and reinitiate transcription from the same promoter, and so support multiple rounds of transcription [35] (Figure 3). ‘Miller spreads’ indicate an extraordinarily high density of loading of pol I on rRNA genes in rapidly growing cells (Figure 1). Whereas most pol-II transcribed genes are associated with only one polymerase [38]; estimates suggest a pol I complex every ~100 base pairs in yeast [39]. Following promoter escape of the polymerase, elongation proceeds in vitro on a naked DNA template largely unimpeded.
The situation is likely to be different in the context of chromatin. Nucleosomes seem to be distributed throughout the rRNA genes, although the exact nature of actively transcribed rDNA chromatin is unknown. Psoralen cross-linking and nucleolar-dominance analyses have established a link between chromatin structure and the transcriptional activity of an rRNA gene, with a correlation between lack of regularly spaced nucleosomes (an ‘open’ configuration) and actively transcribed genes [40]. Pol I transcribes through nucleosomes, either disrupting or bypassing the structures, perhaps assisted in this process by chromatin-remodelling activities. In addition, UBF – which is distributed throughout the rDNA [24] – might influence elongation by pol I positively via its putative anti-repressor function, or negatively as recently proposed [6]. An elusive mouse pol-I-associated factor, TIF-IC, can stimulate the overall rate of transcription elongation and suppress pausing of pol I [41].
Efficient elongation of nascent rRNA might require cleavage of the 3′ end of transcripts at stalled polymerases to enable backtracking and resumption of elongation. TFIIS executes this role for pol II, and can also stimulate elongation by pol I [42], although a distinct RNase activity has been identified that might otherwise fulfill this function for pol I transcription [43].
Topological changes in the rDNA during transcription include bending and supercoiling; these changes are induced by the interactions of UBF and other PIC components with the promoter region, and produced by pol I on the template as transcription proceeds [44]. The ability of UBF to contort DNA into structures that resemble the core nucleosome both in mass and DNA content raises the possibility that UBF impedes the dissipation of torsional strain. The torsional strain accrued, with positive supercoils ahead of the polymerase and negative supercoils behind, might be resolved with the aid of DNA topoisomerases, which transiently break one or both of the DNA strands and enable swivelling of the strands. Indeed, the activity of topoisomerase (Topo) I or II is required for the efficient transcription of chromosomal rDNA in yeast [44,45].
Elongation is likely to be interrupted at sites of DNA damage in the rDNA template. Transcription-coupled repair occurs in pol-I-transcribed genes [46] and this might explain the presence of DNA-repair proteins TFIIH, Cockayne syndrome B (CSB) and xeroderma pigmentosum (XP) group G (XPG) in a complex with pol I that participates in rDNA transcription in the nucleolus of mammalian cells. Mutations in CSB, and the XPB and XPD helicase subunits of TFIIH (all of which confer Cockayne’s syndrome), disrupt the interaction of TFIIH with pol I in this complex [47]. TFIIH is also found associated with subpopulations of TIF-IB (SL1), in addition to pol I, and is required for productive – but not abortive – rDNA transcription, which implies a post-initiation role in transcription [48]. Curiously, this does not require its ATPase, helicase and/or kinase activities because ATP hydrolysis is not essential for pol I transcription. Werner’s syndrome helicase WRN (and yeast DNA helicases SGS1 and SRS2) also accelerates rDNA transcription when part of a pol-I-associated complex in mammalian cells [49], and, given its ability to resolve aberrant DNA structures arising from DNA metabolic processes (including repair) might affect the elongation phase of the transcription cycle. Whether such proteins operate solely in a DNA-repair capacity or perform other functions in pol I transcription is not yet fully resolved.
Termination and reinitiation of transcription
Transcription termination elements are located at the 3′ end of the transcribed region of the rRNA gene and upstream of the rDNA transcription start site, between the spacer and gene promoters (Figure 1). Mammalian TTF-I binds and bends the termination site at the 3′ end of the transcribed region, forces pol I to pause, and cooperates with pol I and transcript-release factor PTRF in conjunction with a T-rich DNA sequence, to induce transcription termination and dissociate the elongating pol I and transcript from the template DNA [50] (Figure 3). The high density of pol I loading on the rDNA (Figure 1) indicates that, under certain conditions, termination of transcription might be rate-limiting. Following termination of transcription, the components of the released polymerase are likely to be recycled to produce initiation competent pol I; indeed, the elongating form of pol I can be converted back into the initiation-competent form in vitro [35]. Therefore, in facilitating release of the polymerase, TTF-I and PTRF indirectly stimulate the reinitiation of transcription [51] (Figure 3).
Control of rDNA transcription
The rate of cell growth and proliferation is directly proportional to the rate of protein synthesis, which is intricately linked to ribosome biogenesis and controlled at the level of rDNA transcription by pol I [1,6,52]. Intracellular signals must coordinate the synthesis of rRNA with that of other ribosome building blocks and components of the protein translation machinery. Control of pol I transcription could involve either adjustments to the number of genes actively engaged in transcription because only a fraction (~50%) of the rRNA genes is actively transcribed in an interphase cell, or to the rate of transcription from each active gene, and there is evidence for both (Box 1). In mammalian cells, but not in yeast, rDNA transcription is a cell-cycle regulated process; transcription is absent during mitosis and gradually increases during G1, peaking in the S and G2 phases (Box 2). rRNA synthesis is delicately balanced to support the required levels of protein synthesis in response to extracellular signals that influence cell growth and proliferation, in addition to cellular stress signals.
Box 1. Mechanisms in the control of polymerase I transcription.
Control of polymerase I (pol I) transcription could involve either of two mechanisms (that are not mutually exclusive): adjustments to the number of genes actively engaged in transcription (approximately half of the multiple copies of the rRNA genes are actively transcribed in an interphase cell) or to the rate of transcription from each active gene. There is evidence for both, and the extent to which each is employed is likely to depend on the physiological or differentiation status of the cell and its external environment.
The number of active genes
In mammalian cells, the number of active rRNA genes varies between different cell types with the same genotype [82], which suggests that adjustments to the number of active copies are made during development and cellular differentiation. In most cases, differentiation is accompanied by a decrease in rRNA synthesis [3]. During spermatogenesis, for instance, in the development from diploid to haploid spermatocytes, this reduction is the result of a decrease in the number of active rRNA genes. By contrast, phytohemagglutinin stimulation of lymphocytes, which leads to active cell division, causes an increase in rRNA synthesis with an increase in the number of active rRNA genes, implying that formerly inactive rRNA genes are recruited for transcription [82].
Stimulated rDNA transcription correlates with an increased level of promoter occupancy by selectivity factor 1 (SL1) in the rapid response to insulin-like growth factor-1 (IGF-1) in human embryonic kidney cells 293, as determined by chromatin immunoprecipitation analysis [53]. Because SL1 directs pre-initiation complex (PIC) formation and seems to be stably associated with already active rRNA gene promoters [35], poised for reinitiation events, it could be argued that this increase in SL1 bound to rDNA promoters reflects the activation of previously silent genes rather than an increased frequency of initiation from each active gene.
There is also evidence that budding yeast can regulate rRNA synthesis in response to variations in environmental conditions by altering the number of rRNA genes that are active [83]. Approximately 50% of the rRNA genes in each yeast cell are transcribed and maintained in an ‘open’ psoralen-accessible conformation during log phase, whereas, during the stationary phase, the percentage of ‘open’ rRNA genes is reduced owing to changes in chromatin. Rpd3–Sin3 histone deacetylase (HDAC) complex is required to inactivate (‘close’) the individual rRNA genes via its association with rDNA chromatin and deacetylation of histone H4 [59,84].
The rate of transcription from active genes
Alterations in the extent of rDNA transcription can also occur via changes to the rate of transcription from each active rRNA gene. In the downregulation of rDNA transcription in mammalian cells, during nutrient starvation or growth to stationary phase, the number of active gene promoters in the psoralen-accessible ‘open’ configuration remains constant [40].
That cells can regulate rRNA levels by adjusting the rate of transcription from active rRNA genes has been demonstrated most convincingly in the Saccharomyces cerevisiae system. Yeast cells engineered to contain only ~40 copies of the rRNA gene produce equivalent levels of rRNAs and grow at a rate similar to cells with the usual complement of ~140 copies, of which ~75 are active. To achieve this, the frequency of transcription initiation events on each gene was increased, as determined by direct measurement, in ‘Miller’ spreads, of the number of active genes per nucleolus and the number of polymerases per gene [39]. Furthermore, rRNA synthesis was reduced in yeast cells entering stationary phase even when they harboured an Rpd3 deletion mutation that maintained the same number of rDNA genes active, in an ‘open’ psoralen-accessible configuration. This correlated with a reduction in the number of polymerases transcribing each ‘open’ rRNA gene, again suggesting that the cell is able to adjust rRNA synthesis by regulating the rate of transcription at individual active genes [84].
Box 2. Mechanisms in the cell-cycle regulation of RNA polymerase I transcription.
In mammalian cells, but not in yeast, rDNA transcription is cell-cycle regulated; with no transcription in mitosis, a gradual increase in transcription in G1 and peak transcription in S and G2. Only a proportion of the rRNA genes are active during interphase and transcription from these genes is downregulated during mitosis. Epigenetic silencing of rRNA genes, which is influenced by rDNA chromatin structure and modification, ensures re-establishment of the same proportion of active genes as the cell exits mitosis [28]. Mitotic silencing of RNA polymerase I (pol I) transcription and reactivation during the transition from mitosis to the G1 phase of the cell cycle are controlled at multiple levels. Selectivity factor 1 (SL1) and upstream binding factor (UBF) remain associated with the rDNA throughout mitosis and for most of mitosis pol I also remains associated with the rDNA chromatin [85,86]. However, time-lapse analysis of single live cells revealed a window during metaphase in which pol I is no longer associated with the rDNA chromatin [85]. The phosphorylation status of SL1 fluctuates during the cell cycle. Phosphorylation of the SL1 subunit TAFI110 by cdc2-cyclin B during metaphase correlates with the inactivation of SL1, an inability of SL1 to interact with UBF and mitotic repression of rDNA transcription [87]. Therefore, productive pre-initiation complex (PIC) formation, initiation and/or promoter escape and clearance might be downregulated in mitosis. UBF inactivation during mitosis also correlates with its phosphorylation status [88] and stronger binding to the rDNA [89]. Phosphorylation of termination factor TTF-I by cdc2-cyclin B correlates with a decreased chromatin-binding affinity of TTF-I in mitotic chromosomes compared with that at interphase [89]. Given the role of TTF-I in chromatin remodelling [26] and the architectural role for UBF, the differential binding is likely to influence rDNA chromatin structure at mitosis, which might contribute to mitotic repression. In mitosis, however, psoralen cross-linking analyses have shown that the ‘open’ configuration seems to be maintained in the metaphase chromosomes in the absence of rDNA transcription [40]. Activation of rDNA transcription, upon serum stimulation of serum starved NIH3T3 cells, correlates with phosphorylation of UBF by the G1-specific complexes cyclin-dependent kinase (cdk)4-cyclin D1 and cdk2-cyclin E at S484 and, later in G1, by cdk2-cyclin E and cdk2-cyclin A at S388, required for the interaction of UBF with pol I [1].
Other proteins with known effects on the cell cycle that regulate rRNA synthesis include: p53, which prevents the interaction between SL1 and UBF and represses Pol I transcription [1]; Rb, which disrupts the interaction between UBF and SL1 and downregulates pol I transcription [65] (P. Cabart and J. Zomerdijk, unpublished); and TAF1 (TAFII250), a subunit of TFIID involved in the transcription of cell cycle and growth regulatory genes [90], which binds UBF and stimulates transcription.
The phosphatidyl inositol-3 kinase (PI3K) mammalian target of rapamycin (mTOR) and mitogen-activated protein kinase (MAPK) signalling cascades, which are pivotal to the control of cell growth and proliferation, have been implicated in the regulation of pol I transcription in the response to mitogens, growth factors and nutrient availability [1,6]. However, it seems that the contribution of each of these pathways can vary according to cell type and/or cellular environment. For example, for nutrient or insulin-like growth factor 1 (IGF-1)-stimulated pol I transcription in human embryonic kidney cells 293, PI3K is essential, the mTOR pathway has a modulatory role and the MAPK cascade has a minor role [53], whereas, for epidermal growth factor (EGF)-stimulated pol I transcription in human neuroepithelioma cells, the extracellular signal-regulated kinase (ERK)–MAPK pathway predominates [54]. There is an integrated response to growth factors and nutrients, for example, there is no stimulation of pol I transcription in response to growth factors when nutrients are limiting, and this implies cross-talk between the pathways [53].
rDNA transcription could be regulated at any stage during the transcription cycle (Figure 3) – at PIC assembly, initiation, promoter escape, elongation, termination or reinitiation – or via chromatin remodelling. The phosphorylation status of pol I transcription factors can influence the activity and interactions of these proteins in the transcription cycle. Various complementary modifications affecting pol I factors, such as (de-)acetylation, further increase the complexity and network of regulation. Aside from control by post-translational modification of components of the transcription machinery, pol I transcription can be regulated at the level of abundance of the individual transcription components, such as SL1 subunits and UBF. Several other factors outside the pol I transcription machinery have been implicated in modulating pre-rRNA synthesis, directly or indirectly (see Supplementary Table).
Control of pol I and pol-I-associated factors
Little is known of the regulation of the pol I enzyme itself. An established point of control of pre-rRNA synthesis is the association of hRRN3 (TIF-IA) with pol I, to generate Pol Iβ [14–16,19]. In stationary phase, nutrient-limited or cycloheximide-treated mammalian cells, cellular rRNA synthesis is downregulated and the interaction between RRN3 (TIF-IA) and pol I is impaired [17,18]. There is evidence that, in mammalian cells, the phosphorylation state of RRN3 regulates rDNA transcription by determining the steady-state concentration of the RRN3–pol I complex pol Iβ [17], and that RRN3 is inactivated during the process of rDNA transcription, possibly as a result of the reversal of such phosphorylations [36]. The ERK–MAPK and mTOR signalling cascades have been implicated in the control of TIF-IA (RRN3) activity and its interaction with pol I. ERK and p90 ribosomal S6 kinase (RSK) phosphorylation of TIF-IA (Figure 2b) in growth factor-stimulated mouse NIH3T3 cells correlates with increased TIF-IA activity, rRNA gene transcription and cell proliferation [55]. Inactivation of mTOR by rapamycin treatment of NIH3T3 cells has been reported to downregulate TIF-IA activity (Figure 2b) by preclusion of interactions of TIF-IA with pol I and SL1 in addition to translocation of TIF-IA to the cytoplasm, which implicates the mTOR pathway in PIC formation [56]. However, Hannan et al. [57] have reported that treatment of the same cell type with rapamycin does not affect RRN3 (TIF-IA) activity. The physiological status of the cell, therefore, probably dictates the requirement for mTOR in the control of RRN3 activity and PIC formation.
The TOR signalling pathway has also been implicated in regulation of the Rrn3p–pol I interaction and the Rrn3-dependent recruitment of pol I to the promoter in yeast [58]. Inactivation of TOR by rapamycin is accompanied by the release of pol I from the nucleolus and inhibition of rDNA transcription [59]. In yeast, however, phosphorylation of pol I – and not Rrn3p – is required to form a stable pol I–Rrn3p complex for efficient in vitro transcription initiation, and the association of pol I with Rrn3p correlates with a change in the in vivo phosphorylation state of pol I [60].
Control of SL1
Although there are no reports of SL1 phosphorylation in response to growth-stimulatory factors, some SL1 subunits are phosphoproteins. SL1 activity might be controlled at the level of acetylation because P/CAF acetylation of mouse SL1 (TIF-IB) subunit TAFI68 enhances its binding to the rDNA promoter and stimulates pol I transcription [25] (Figure 2a). Furthermore, expression of TBP can be upregulated via the Ras-MAPK pathway, leading to stimulation of transcription from pol I and pol III promoters, implying a coordinated regulation of rRNA and tRNA synthesis [61,62].
Control of UBF
There are reports of the regulated phosphorylation of UBF affecting its interaction with DNA or other PIC components. UBF can be phosphorylated by casein kinase 2 (CK2) at serine residues within the C-terminal acidic domain in vitro and this contributes to, but is not sufficient for, transcriptional activation (Figure 2c); C-terminal phosphorylation of UBF increases its ability to interact with SL1 [5]. Direct phosphorylation by PI3K of UBF, predominantly in the C terminus, occurs upon IGF-1 treatment of serum-starved mouse cells following the nucleolar translocation of insulin-receptor substrate 1 (IRS-1) and the interaction of IRS-1 with UBF and PI3K, and this also correlates with increased rRNA synthesis [63]. Upon serum induction of rDNA transcription in NIH3T3 cells, C-terminal phosphorylation of UBF by the mTOR signalling pathways (Figure 2c), through p70 ribosomal S6 kinase 1, promotes interaction between UBF and SL1 [57].
UBF is also a target of the ERK–MAPK pathway upon EGF stimulation of pol I transcription in human neuroepithelioma cells, and ERK1/2 phosphorylation influences the interaction of UBF with DNA [54] (Figure 2c). DNA-bound UBF is not a substrate for this kinase in vitro [54], therefore, UBF binding to DNA has to be dynamic to enable phosphorylation by ERK. How the alteration in DNA binding contributes to activated transcription is unknown, but one model proposes that cyclic-phosphorylation events promote passage of the elongating pol I through an altered UBF–DNA complex, perhaps immediately downstream of the transcription start site [54]. This would be consistent with a role for UBF in promoter escape or clearance of pol I.
Competitive acetylation and deacetylation of UBF regulates its activity without affecting its ability to bind DNA [64]. The CBP acetyltransferase upregulates UBF and its binding to UBF precludes the binding of tumour-suppressor Rb (and associated histone deacetylases), which has a negative effect on UBF binding to SL1 [65]. Moreover, UBF can be acetylated by the Tip60 histone acetyltransferase (HAT) subunit in vitro [66].
UBF seems limiting in some cells [67], thus pol I transcription could be regulated at the level of UBF expression. Indeed, UBF overexpression occurs before increased rRNA synthesis in mitogen-stimulated lymphocytes and UBF expression decreases upon induced differentiation of promyelocytic leukaemia HL-60 cells [68]. UBF expression is upregulated by serum induction via the mTOR pathway [57] and by the Myc proto-oncogene [69], and UBF is expressed at high levels in cardiac hypertrophy and liver cancer cells, which support high levels of rRNA synthesis [70,71].
rRNA synthesis, cell growth, cell-cycle progression and cell fate
Certainly, there is a crucial role for rRNA synthesis in normal cell growth and in the adjustment of cell growth in response to growth factors and nutrient availability. Furthermore, rRNA levels influence cellular differentiation, cell fate and the development of an organism [72].
Cell growth (increase in size and mass) and cell proliferation (increase in cell number) are intricately linked in most cells. Cell growth is essential for cell-cycle progression and sustained cell proliferation, and the attainment of a particular cell mass is a prerequisite for cell division [73]. As (pre)rRNA levels reflect the capacity of the cell to grow and achieve this cell mass, it is conceivable that (pre)rRNA levels are decisive for cell-cycle progression.
Predictably, rRNA levels also reflect cell size. Cell growth and cell-cycle progression can be uncoupled and, for instance, cell growth without cell division leads to hypertrophy (the production of large cells with a single nucleus). This correlates with increased levels of rRNA, ribosome accumulation and protein synthesis, which must rise proportionally to support the increased cell mass. In cardiac hypertrophy, increased rRNA levels could be a consequence of the increased expression of UBF [70]. Downregulation of rRNA synthesis (reflected in a reduced nucleolar size) and protein synthesis, and a reduced cell-size phenotype occur upon disruption of the mTOR pathway in metazoan cells. This implies a role for mTOR in coupling cell growth and cell-cycle progression [74,75], perhaps by its regulation of rRNA levels.
In transformed cells, cell growth and cell-cycle progression are linked, but cell growth is not coupled to the requirement for growth factors. The levels of rDNA transcription are increased in transformed cells and, indeed, the corresponding increase in nucleolar size is often used as a prognostic marker for the rapidity of cancer-cell proliferation [76]. The increased rRNA levels support the rise in protein synthesis required to accommodate the escalated cell growth and proliferation in transformed cells. rDNA transcription might be upregulated in transformed cells by overexpression or increased activity of components of the pol I transcription machinery or proteins that directly affect rDNA transcription. UBF, for instance, is overexpressed in 70% of hepatocellular carcinomas [71]. Constitutively active growth-factor-independent signalling via Ras–MAPK, mTOR or PI3K pathways in cancer cells can activate pol I transcription and drive ribosome biogenesis. Tumour-suppressor proteins such as p53 and Rb, and the proto-oncogene Myc protein, all affect rRNA synthesis, so rRNA levels are also likely to be influenced by mutations in these proteins, leading to cellular transformation. Recently the links between increased ribosome biogenesis, protein synthesis and cancerhave been explored [52]. However, the question remains as to whether the deregulation of rRNA synthesis itself could trigger the runaway growth that is the signature of transformed cells, or whether increased rRNA synthesis plays a secondary, but necessary, part in supporting the increased cell growth.
Another crucial link between the fate of the cell and the extent of ribosome biogenesis has been derived from observation that reagents that perturb the structure or function of the nucleolus, the centre of rDNA transcription and ribosome production, can cause p53 accumulation [77,78], which can lead to cell-cycle arrest or apoptosis [79]. Because these reagents produce a negative effect on rRNA transcription or (pre)rRNA, we suggest that the level of (pre)rRNA could influence p53 accumulation and the decision to trigger cell-cycle arrest, or even apoptosis.
In addition, we propose that rRNA gene transcription is a direct sensor for DNA damage as the repetitive nature of the rRNA genes, in combination with the dense loading of the pol I complexes on the active templates, provides a high probability of a polymerase encountering, and stalling at, a site of DNA damage. The accretion of signals associated with such stalled polymerases and/or reduced (pre)rRNA levels, could activate p53 and the DNA-damage response pathways.
Concluding remarks and future perspectives
Significant advances have been made towards an understanding of the pol I transcription machinery (works) in the transcription cycle. Recent studies have shown that regulation of rDNA transcription is manifested via multiple pathways and mechanisms operating in parallel with distinct kinetics. The exact combination is likely to be dependent upon cell type and physiological status. Future research could indicate whether rDNA transcription and rRNA levels couple the capacity of a cell for growth to its progression through the cell cycle and/or directly influence the decision of a cell to arrest or exit from the cell cycle in response to DNA damage, stress or malfunction. Whether or not overproduction of rRNA transcripts can drive cell proliferation and cause cancer, upregulation of rRNA synthesis, is characteristic of the transformed cell phenotype. Thus, drugs that would target components of the pol I transcription machinery in cancer cells should have a dramatic effect on the growth and proliferation of these cells. Certainly, deregulation of rRNA synthesis can have an enormous impact on the fate of individual cells and the ability of a cell to sustain life.
Supplementary Material
Acknowledgements
We thank Ann Beyer for providing the Miller spread image in Figure 1. We also thank Julian Blow, Taciana Kasciukovic, Angus Lamond, Kostya Panov, Shalini Patak, Neil Perkins and reviewers for insightful comments, and Ingrid Grummt for allowing us to quote unpublished data. Work in our laboratory is supported by the Wellcome Trust Senior Research Fellowship of J.C.B.M.Z. We apologize to colleagues whose work could not be included in this review owing to space constraints.
References
- 1.Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003;17:1691–1702. doi: 10.1101/gad.1098503R. [DOI] [PubMed] [Google Scholar]
- 2.Moss T, Stefanovsky VY. At the center of eukaryotic life. Cell. 2002;109:545–548. doi: 10.1016/s0092-8674(02)00761-4. [DOI] [PubMed] [Google Scholar]
- 3.Reeder RH. Regulation of RNA polymerase I transcription in yeast and vertebrates. Prog. Nucleic Acid Res. Mol. Biol. 1999;62:293–327. doi: 10.1016/s0079-6603(08)60511-5. [DOI] [PubMed] [Google Scholar]
- 4.Hannan KM, et al. Transcription by RNA polymerase I. Front. Biosci. 1998;3:d376–d398. doi: 10.2741/a282. [DOI] [PubMed] [Google Scholar]
- 5.Comai L. Mechanism of RNA polymerase I transcription. Adv. Protein Chem. 2004;67:123–155. doi: 10.1016/S0065-3233(04)67005-7. [DOI] [PubMed] [Google Scholar]
- 6.Moss T. At the crossroads of growth control; making ribosomal RNA. Curr. Opin. Genet. Dev. 2004;14:210–217. doi: 10.1016/j.gde.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Zomerdijk JCBM, Tjian R. Initiation of transcription on human rRNA genes. In: Paule MR, editor. Transcription of Eukaryotic Ribosomal RNA Genes by RNA Polymerase I. Springer-Verlag; 1998. pp. 121–134. [Google Scholar]
- 8.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 9.Moore PB, Steitz TA. The involvement of RNA in ribosome function. Nature. 2002;418:229–235. doi: 10.1038/418229a. [DOI] [PubMed] [Google Scholar]
- 10.Zomerdijk JCBM, Tjian R. Structure and assembly of human selectivity factor SL1. In: Paule MR, editor. Transcription of Eukaryotic Ribosomal RNA Genes by RNA Polymerase I. Springer-Verlag; 1998. pp. 67–73. [Google Scholar]
- 11.Grummt I. Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog. Nucleic Acid Res. Mol. Biol. 1999;62:109–154. doi: 10.1016/s0079-6603(08)60506-1. [DOI] [PubMed] [Google Scholar]
- 12.Zomerdijk JCBM, et al. Assembly of transcriptionally active RNA polymerase I initiation factor SL1 from recombinant subunits. Science. 1994;266:2015–2018. doi: 10.1126/science.7801130. [DOI] [PubMed] [Google Scholar]
- 13.Heix J, et al. Cloning of murine RNA polymerase I-specific TAF factors: conserved interactions between the subunits of the species-specific transcription initiation factor TIF-IB/SL1. Proc. Natl. Acad. Sci. U. S. A. 1997;94:1733–1738. doi: 10.1073/pnas.94.5.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moorefield B, et al. RNA polymerase I transcription factor Rrn3 is functionally conserved between yeast and human. Proc. Natl. Acad. Sci. U. S. A. 2000;97:4724–4729. doi: 10.1073/pnas.080063997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller G, et al. hRRN3 is essential in the SL1-mediated recruitment of RNA polymerase I to rRNA gene promoters. EMBO J. 2001;20:1373–1382. doi: 10.1093/emboj/20.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bodem J, et al. TIF-IA, the factor mediating growth-dependent control of ribosomal RNA synthesis, is the mammalian homolog of yeast Rrn3p. EMBO Rep. 2000;1:171–175. doi: 10.1093/embo-reports/kvd032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cavanaugh AH, et al. Rrn3 phosphorylation is a regulatory checkpoint for ribosome biogenesis. J. Biol. Chem. 2002;277:27423–27432. doi: 10.1074/jbc.M201232200. [DOI] [PubMed] [Google Scholar]
- 18.Yuan X, et al. Multiple interactions between RNA polymerase I, TIF-IA and TAFI subunits regulate preinitiation complex assembly at the ribosomal gene promoter. EMBO Rep. 2002;3:1082–1087. doi: 10.1093/embo-reports/kvf212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peyroche G, et al. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J. 2000;19:5473–5482. doi: 10.1093/emboj/19.20.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nomura M. Ribosomal RNA genes, RNA polymerases, nucleolar structures, and synthesis of rRNA in the yeast Saccharomyces cerevisiae. Cold Spring Harb. Symp. Quant. Biol. 2001;66:555–565. doi: 10.1101/sqb.2001.66.555. [DOI] [PubMed] [Google Scholar]
- 21.Reeder RH, et al. UBF, an architectural element for RNA polymerase I promoters. In: Eckstein F, Lilley DMJ, editors. Nucleic Acids and Molecular Biology. Vol. 9. Springer-Verlag; 1995. pp. 251–263. [Google Scholar]
- 22.Thomas JO, Travers AA. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 2001;26:167–174. doi: 10.1016/s0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- 23.Bazett Jones DP, et al. Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science. 1994;264:1134–1137. doi: 10.1126/science.8178172. [DOI] [PubMed] [Google Scholar]
- 24.O’Sullivan AC, et al. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol. Cell. Biol. 2002;22:657–668. doi: 10.1128/MCB.22.2.657-668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muth V, et al. Acetylation of TAF(I)68, a subunit of TIF-IB/SL1, activates RNA polymerase I transcription. EMBO J. 2001;20:1353–1362. doi: 10.1093/emboj/20.6.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langst G, et al. TTF-I determines the chromatin architecture of the active rDNA promoter. EMBO J. 1998;17:3135–3145. doi: 10.1093/emboj/17.11.3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strohner R, et al. Recruitment of the nucleolar remodeling complex NoRC establishes ribosomal DNA silencing in chromatin. Mol. Cell. Biol. 2004;24:1791–1798. doi: 10.1128/MCB.24.4.1791-1798.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grummt I, Pikaard CS. Epigenetic silencing of RNA polymerase I transcription. Nat. Rev. Mol. Cell Biol. 2003;4:641–649. doi: 10.1038/nrm1171. [DOI] [PubMed] [Google Scholar]
- 29.Shematorova EK, Shpakovskii GV. Structure and function of eukaryotic nuclear DNA-dependent RNA polymerase I. Mol Biol (Mosk) 2002;36:3–26. [PubMed] [Google Scholar]
- 30.Yamamoto K, et al. Multiple protein-protein interactions by RNA polymerase I-associated factor PAF49 and role of PAF49 in rRNA transcription. Mol. Cell. Biol. 2004;24:6338–6349. doi: 10.1128/MCB.24.14.6338-6349.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bischler N, et al. Localization of the yeast RNA polymerase I-specific subunits. EMBO J. 2002;21:4136–4144. doi: 10.1093/emboj/cdf392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dundr M, et al. A kinetic framework for a mammalian RNA polymerase in vivo. Science. 2002;298:1623–1626. doi: 10.1126/science.1076164. [DOI] [PubMed] [Google Scholar]
- 33.Hannan RD, et al. Identification of a mammalian RNA polymerase I holoenzyme containing components of the DNA repair/replication system. Nucleic Acids Res. 1999;27:3720–3727. doi: 10.1093/nar/27.18.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seither P, et al. Mammalian RNA polymerase I exists as a holoenzyme with associated basal transcription factors. J. Mol. Biol. 1998;275:43–53. doi: 10.1006/jmbi.1997.1434. [DOI] [PubMed] [Google Scholar]
- 35.Panov KI, et al. A step subsequent to preinitiation complex assembly at the ribosomal RNA gene promoter is rate limiting for human RNA polymerase I-dependent transcription. Mol. Cell. Biol. 2001;21:2641–2649. doi: 10.1128/MCB.21.8.2641-2649.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirschler-Laszkiewicz I, et al. Rrn3 becomes inactivated in the process of ribosomal DNA transcription. J. Biol. Chem. 2003;278:18953–18959. doi: 10.1074/jbc.M301093200. [DOI] [PubMed] [Google Scholar]
- 37.Milkereit P, Tschochner H. A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription. EMBO J. 1998;17:3692–3703. doi: 10.1093/emboj/17.13.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson DA, et al. Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol. Biol. Cell. 1998;9:1523–1536. doi: 10.1091/mbc.9.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.French SL, et al. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol. 2003;23:1558–1568. doi: 10.1128/MCB.23.5.1558-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conconi A, et al. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989;57:753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- 41.Schnapp G, et al. TIF-IC, a factor involved in both transcription initiation and elongation of RNA polymerase I. EMBO J. 1994;13:4028–4035. doi: 10.1002/j.1460-2075.1994.tb06719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schnapp G, et al. TFIIS binds to mouse RNA polymerase I and stimulates transcript elongation and hydrolytic cleavage of nascent rRNA. Mol. Gen. Genet. 1996;252:412–419. doi: 10.1007/BF02173006. [DOI] [PubMed] [Google Scholar]
- 43.Tschochner H. A novel RNA polymerase I-dependent RNase activity that shortens nascent transcripts from the 3′ end. Proc. Natl. Acad. Sci. U. S. A. 1996;93:12914–12919. doi: 10.1073/pnas.93.23.12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brill SJ, et al. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature. 1987;326:414–416. doi: 10.1038/326414a0. [DOI] [PubMed] [Google Scholar]
- 45.Schultz MC, et al. Topoisomerases and yeast rRNA transcription: negative supercoiling stimulates initiation and topoisomerase activity is required for elongation. Genes Dev. 1992;6:1332–1341. doi: 10.1101/gad.6.7.1332. [DOI] [PubMed] [Google Scholar]
- 46.Conconi A, et al. Transcription-coupled repair in RNA polymerase I-transcribed genes of yeast. Proc. Natl. Acad. Sci. U. S. A. 2002;99:649–654. doi: 10.1073/pnas.022373099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bradsher J, et al. CSB is a component of RNA pol I transcription. Mol. Cell. 2002;10:819–829. doi: 10.1016/s1097-2765(02)00678-0. [DOI] [PubMed] [Google Scholar]
- 48.Iben S, et al. TFIIH plays an essential role in RNA polymerase I transcription. Cell. 2002;109:297–306. doi: 10.1016/s0092-8674(02)00729-8. [DOI] [PubMed] [Google Scholar]
- 49.Shiratori M, et al. WRN helicase accelerates the transcription of ribosomal RNA as a component of an RNA polymerase I-associated complex. Oncogene. 2002;21:2447–2454. doi: 10.1038/sj.onc.1205334. [DOI] [PubMed] [Google Scholar]
- 50.Jansa P, Grummt I. Mechanism of transcription termination: PTRF interacts with the largest subunit of RNA polymerase I and dissociates paused transcription complexes from yeast and mouse. Mol. Gen. Genet. 1999;262:508–514. doi: 10.1007/s004380051112. [DOI] [PubMed] [Google Scholar]
- 51.Jansa P, et al. The transcript release factor PTRF augments ribosomal gene transcription by facilitating reinitiation of RNA polymerase I. Nucleic Acids Res. 2001;29:423–429. doi: 10.1093/nar/29.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat. Rev. Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 53.James MJ, Zomerdijk JC. Phosphatidylinositol 3-kinase and mTOR signaling pathways regulate RNA polymerase I transcription in response to IGF-1 and nutrients. J. Biol. Chem. 2004;279:8911–8918. doi: 10.1074/jbc.M307735200. [DOI] [PubMed] [Google Scholar]
- 54.Stefanovsky VY, et al. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol. Cell. 2001;8:1063–1073. doi: 10.1016/s1097-2765(01)00384-7. [DOI] [PubMed] [Google Scholar]
- 55.Zhao J, et al. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol. Cell. 2003;11:405–413. doi: 10.1016/s1097-2765(03)00036-4. [DOI] [PubMed] [Google Scholar]
- 56.Mayer C, et al. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004;18:423–434. doi: 10.1101/gad.285504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hannan KM, et al. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol. Cell. Biol. 2003;23:8862–8877. doi: 10.1128/MCB.23.23.8862-8877.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Claypool JA, et al. Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes. Mol. Biol. Cell. 2004;15:946–956. doi: 10.1091/mbc.E03-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsang CK, et al. Chromatin-mediated regulation of nucleolar structure and RNA pol I localization by TOR. EMBO J. 2003;22:6045–6056. doi: 10.1093/emboj/cdg578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fath S, et al. Differential roles of phosphorylation in the formation of transcriptional active RNA polymerase I. Proc. Natl. Acad. Sci. U. S. A. 2001;98:14334–14339. doi: 10.1073/pnas.231181398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhong S, et al. Epidermal growth factor enhances cellular TATA binding protein levels and induces RNA polymerase I- and III-dependent gene activity. Mol. Cell. Biol. 2004;24:5119–5129. doi: 10.1128/MCB.24.12.5119-5129.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.White RJ. RNA polymerase III transcription and cancer. Oncogene. 2004;23:3208–3216. doi: 10.1038/sj.onc.1207547. [DOI] [PubMed] [Google Scholar]
- 63.Drakas R, et al. Control of cell size through phosphorylation of upstream binding factor 1 by nuclear phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9272–9276. doi: 10.1073/pnas.0403328101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pelletier G, et al. Competitive recruitment of CBP and Rb-HDAC regulates UBF acetylation and ribosomal transcription. Mol. Cell. 2000;6:1059–1066. doi: 10.1016/s1097-2765(00)00104-0. [DOI] [PubMed] [Google Scholar]
- 65.Hannan KM, et al. Rb and p130 regulate RNA polymerase I transcription: Rb disrupts the interaction between UBF and SL1. Oncogene. 2000;19:4988–4999. doi: 10.1038/sj.onc.1203875. [DOI] [PubMed] [Google Scholar]
- 66.Halkidou K, et al. Putative involvement of the histone acetyltransferase Tip60 in ribosomal gene transcription. Nucleic Acids Res. 2004;32:1654–1665. doi: 10.1093/nar/gkh296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hannan R, et al. Cellular regulation of ribosomal DNA transcription: both rat and Xenopus UBF1 stimulate rDNA transcription in 3T3 fibroblasts. Nucleic Acids Res. 1999;27:1205–1213. doi: 10.1093/nar/27.4.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cabart P, Kalousek I. Early gene expression of both RNA polymerase I transcription factors UBF1 and UBF2 precedes ribosomal RNA synthesis during lymphocyte mitogenic stimulation. Cell Mol Biol. 1998;44:343–350. [PubMed] [Google Scholar]
- 69.Poortinga G, et al. MAD1 and c-MYC regulate UBF and rDNA transcription during granulocyte differentiation. EMBO J. 2004;23:3325–3335. doi: 10.1038/sj.emboj.7600335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brandenburger Y, et al. Increased expression of UBF is a critical determinant for rRNA synthesis and hypertrophic growth of cardiac myocytes. FASEB J. 2001;15:2051–2053. doi: 10.1096/fj.01-0853fje. [DOI] [PubMed] [Google Scholar]
- 71.Huang R, et al. Upstream binding factor up-regulated in hepatocellular carcinoma is related to the survival and cisplatin-sensitivity of cancer cells. FASEB J. 2002;16:293–301. doi: 10.1096/fj.01-0687com. [DOI] [PubMed] [Google Scholar]
- 72.Valdez BC, et al. The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10709–10714. doi: 10.1073/pnas.0402492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saucedo LJ, Edgar BA. Why size matters: altering cell size. Curr. Opin. Genet. Dev. 2002;12:565–571. doi: 10.1016/s0959-437x(02)00341-6. [DOI] [PubMed] [Google Scholar]
- 74.Fingar DC, et al. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16:1472–1487. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang H, et al. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 2000;14:2712–2724. doi: 10.1101/gad.835000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Derenzini M, et al. Nucleolar function and size in cancer cells. Am. J. Pathol. 1998;152:1291–1297. [PMC free article] [PubMed] [Google Scholar]
- 77.Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Olson MOJ. Sensing cellular stress: another new function for the nucleolus? Sci. STKE. 2004;2004:pe10. doi: 10.1126/stke.2242004pe10. [DOI] [PubMed] [Google Scholar]
- 79.Vogelstein B, et al. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 80.McStay B, et al. The Xenopus RNA polymerase I transcription factor, UBF, has a role in transcriptional enhancement distinct from that at the promoter. EMBO J. 1997;16:396–405. doi: 10.1093/emboj/16.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yamamoto K, et al. Identification of a novel 70 kDa protein that binds to the core promoter element and is essential for ribosomal DNA transcription. Nucleic Acids Res. 2000;28:1199–1205. doi: 10.1093/nar/28.5.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Haaf T, et al. Quantitative determination of rDNA transcription units in vertebrate cells. Exp. Cell Res. 1991;193:78–86. doi: 10.1016/0014-4827(91)90540-b. [DOI] [PubMed] [Google Scholar]
- 83.Dammann R, et al. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:2331–2338. doi: 10.1093/nar/21.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sandmeier JJ, et al. RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J. 2002;21:4959–4968. doi: 10.1093/emboj/cdf498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Leung AKL, et al. Quantative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J. Cell Biol. 2004;166:787–800. doi: 10.1083/jcb.200405013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jordan P, et al. In vivo evidence that TATA-binding protein/SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J. Cell Biol. 1996;133:225–234. doi: 10.1083/jcb.133.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heix J, et al. Mitotic silencing of human rRNA synthesis: inactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. EMBO J. 1998;17:7373–7381. doi: 10.1093/emboj/17.24.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Klein J, Grummt I. Cell cycle-dependent regulation of RNA polymerase I transcription: the nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc. Natl. Acad. Sci. U. S. A. 1999;96:6096–6101. doi: 10.1073/pnas.96.11.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sirri V, et al. The mitotically phosphorylated form of the transcription termination factor TTF-1 is associated with the repressed rDNA transcription machinery. J. Cell Sci. 1999;112:3259–3268. doi: 10.1242/jcs.112.19.3259. [DOI] [PubMed] [Google Scholar]
- 90.Lin CY, et al. The cell cycle regulatory factor TAF1 stimulates ribosomal DNA transcription by binding to the activator UBF. Curr. Biol. 2002;12:2142–2146. doi: 10.1016/s0960-9822(02)01389-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.