Abstract
Reporters secreted into the conditioned medium in culture or into blood in vivo have shown to be useful tools for simple and non-invasive monitoring of biological processes in real-time. Here, we characterize the naturally secreted Vargula luciferase as a secreted blood reporter and show that this reporter can be multiplexed with the secreted Gaussia luciferase and alkaline phosphatase for simultaneous monitoring of three different cellular processes in the same biological system. We applied this system to monitor the response of three different subsets of glioma cells to a clinically-relevant chemotherapeutic agent in the same well in culture or animal in vivo. This system could be applied to any field to detect multiple biological phenomena in the same biological system and is amenable for high-throughput screening to find drugs that affect multiple cellular populations/phenomena simultaneously.
Introduction
Reporters secreted in blood are valuable tools for sensitive and fast detection, quantification and non-invasive ex vivo monitoring of biological processes in in vivo models.1 Currently, the three most commonly used blood reporters are the secreted embryonic alkaline phosphatase (SEAP),2-5 soluble peptides derived from human carcioembryonic antigen (hCEA) and human chorionic gonadotropin (βhCG),6-9 and Gaussia luciferase (Gluc).10-14 The level of these secreted reporters can be measured over time in blood, serum and/or urine to generate multiple data sets without the need to sacrifice the animal, since only a small amount of fluid is required. In contrast to other tools for monitoring of cellular processes, secreted blood reporters are suitable to follow biological parameters in the course of time, providing new insights in the factors contributing to disease development and progression.1 During the last two decades, secreted blood reporters have proven their value in a wide variety of medical fields including the study of embryo development, viral dissemination, the fate of (stem) cells, gene transfer, tumorigenesis and response to therapy.14-16 The contribution of secreted blood reporters to understanding of these complicated processes would increase even further, if instead of one, multiple parameters could be measured simultaneously and over time.
The discovery of new secreted reporters with different substrate specificities, emission spectra and/or detection assays will allow the development of multiplex assays that are capable in simultaneously monitoring several processes, given that their separate reactions remain distinguishable. Here, we characterized the naturally secreted luciferase from the marine ostracod Vargula (Cypridina) hilgendorfi (Vluc)17,18 as a blood reporter and multiplexed it with Gluc and SEAP to develop a triple blood reporter system to monitor three distinct biological processes. As a proof of concept, we successfully monitored the response of three different subsets of glioma cells to the chemotherapeutic agent temozolomide19 in the same animal. This multiplex system can be extended and applied to many different fields for simultaneous monitoring of multiple parameters in the same biological system.
Experimental Section
Lentivirus vectors
Lentivirus vectors expressing CMV-SEAP, CMV-Gluc and CMV-Fluc were previously described.14 Codon-optimized Vluc cDNA for mammalian gene expression was amplified by polymerase chain reaction (PCR) from pCMV-Vluc (Targeting Systems) and cloned in a similar vector backbone as CMV-SEAP creating CMV-Vluc. Lentivirus vectors were packaged as previously described.14
Cell culture and reagents
U87 human glioma cells were obtained from ATCC. U87R1 and U87R2 cells were generated by long-term exposure (twice per week over 7 weeks) of U87 parental cells (in duplicates) with a clinically-relevant concentration of temozolomide (TMZ; 100 μM). Resistant cells in culture were regularly challenged with 100 μM TMZ to maintain the resistant phenotype. All cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Sigma, St. Louis, MO, USA), 100 U penicillin, and 0.1 mg streptomycin (Sigma) per milliliter, at 37 °C and 5% CO2 in a humidified atmosphere. Temozolomide was obtained from Sigma.
Reporter substrates
Coelenterazine was obtained from NanoLight™ Technology (Pinetop, AZ) and resuspended at 5 mg/ml in acidified methanol. Vargulin substrate was obtained from NanoLight™ Technology and was resuspended at 5 mg/ml in acidified methanol. D-luciferin was purchased from Gold Biotechnology® (St. Louis, MO) and resuspended at 25 mg/ml in PBS. SEAP was detected using the Great EscAPe SEAP kit (Clontech) as per manufacturer's instructions.
In vitro experiments
Seven thousands of either U87-Gluc, U87R1-Vluc, or U87R2-SEAP/Fluc cells (for singleplex assay) or a combination of all three cells (2500 of each line for multiplex) were plated in a 96-well plate in a total volume of 100 μl. Twenty-four hours later, cells were treated with either temozolomide (100 μM) or DMSO vehicle. At different time points, 10 μl aliquots (in triplicates) of cell-free conditioned medium was collected and assayed for either Gluc, Vluc or SEAP using either coelenterazine (5 μg/ml in PBS), vargulin (0.25 μg/ml in PBS) or the Great EscAPe SEAP respectively. We refreshed the media of cells 6 hours before each measurement to avoid accumulation of the reporter.
Animal studies and blood collection
All animal studies were approved by the Massachusetts General Hospital Review Board. U87 human glioma cells, U87R1 and U87R2 were transduced with each lentivirus vector for stable expression by adding the vector directly to the cells using 10 transducing units of each vector per cell leading to >90% transduction efficiency. To generate subcutaneous tumors, different amount of these cells (in 50 μl) were mixed with equal volume of Matrigel and injected in the flanks of athymic nude mice. For brain tumor model, a mixture of these cells (25,000 per cell line, in 2 μl Opti-MEM) was intracranially injected in the left midstriatum of nude mice using the following coordinates from bregma in mm: anterior-posterior +0.5, medio-lateral +2.0, dorso-ventral −2.5. These injections were performed using a Micro 4 Microsyringe Pump Controller (World Precision Instruments, Sarasota, FL) attached to a Hamilton syringe with a 33-gauge needle (Hamilton, Rena, NV) at a rate of 0.2 μl/min. One week later, mice were randomized in 2 groups (n=6/group) and treated with either 10 mg/kg temozolomide or DMSO vehicle. Blood samples were collected from these mice as well as from mice without tumors (negative control) by making a small incision in the tail and directly adding it to an eppendorf tube containing EDTA as an anti-coagulant (10 mM final concentration).
Ex vivo multiplex blood reporter assays
For Gluc assay, activity was measured by injecting 100 μl 100 μM coelenterazine (nanolight, pinetop, AZ) to 5 μl of blood and acquiring photon counts over 10 seconds using a luminometer (Dynex). The SEAP chemiluminescence activity was measured in 5 μl serum using the Great EscAPe SEAP kit (Clontech) as per manufacturer's instructions. For Vluc assay, activity was measured in 5 μl blood or serum after injecting 100 μl 0.25 μg/ml of vargulin (diluted in PBS) and acquiring photon counts for 10 sec using a luminometer.
Triple in vivo imaging
For Fluc imaging, mice were injected i.p. with 200 mg/kg body weight of D-luciferin solution and imaging was performed 10 min later. For Gluc imaging, mice were injected i.v. (via retro-orbital route) with 5 mg/kg body weight of coelenterazine solution diluted in PBS and imaging was performed immediately. For Vluc in vivo imaging, mice were injected i.v. with 4 mg/kg body weight (diluted in PBS) and imaging was performed immediately. For sequential imaging of all three reporters, we imaged Gluc first followed by Vluc 4 hours later and Fluc on the following day to allow enough time for the signal to reach background levels between different imaging sessions. Imaging was performed using an IVIS® Spectrum optical imaging system fitted with an XGI-8 Gas Anesthesia System (Caliper Life Sciences, Hopkinton, MA). Bioluminescent images were acquired using the auto-exposure function. Data analysis for signal intensities and image comparisons were performed using Living Image® software (Caliper Life Sciences). To calculate radiance for each animal, regions of interest (ROIs) were carefully drawn around each signal in the brain which is expressed as radiance (photons/sec/cm2/steradian).
Results and Discussion
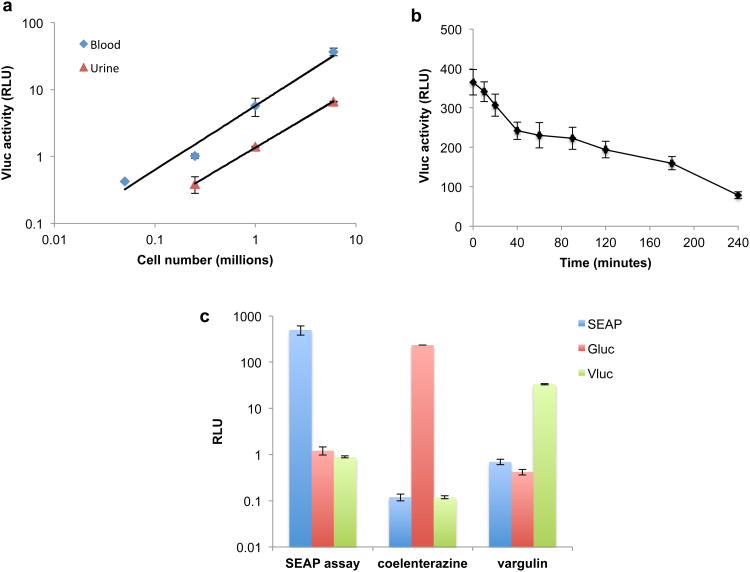
We first characterized a codon-optimized Vluc variant for mammalian gene expression as a blood reporter. Different amounts of U87 human glioma cells stably expressing Vluc were implanted subcutaneously in nude mice. Three days post-implantation, 5 μl blood samples (in triplicates) were withdrawn, mixed with EDTA (as an anti-coagulant), and assayed for Vluc activity using a luminometer after addition of the vargulin substrate. Several optimization steps for the detection of Vluc in the blood were first performed using different concentrations of the vargulin substrate in blood and serum. We found that 100 μl of 0.25 μg/ml vargulin (diluted in PBS) gave the best signal-to-background (S/B) ratio (supplementary Fig. 1a). Since hemoglobin in whole blood is known to interfere with bioluminescence, we compared the activity of Vluc in blood versus serum and observed that serum gave higher S/B ratio at a concentration of 0.25 μg/ml vargulin (supplementary Fig. 1a). EDTA anticoagulant did not have any effect on the Vluc activity (Supplementary Fig. 1b). Using these optimized conditions, we observed that Vluc activity in blood is linear with respect to cell number in a range covering at least 3 orders of magnitudes (Fig. 1a). We then checked the possibility of detecting Vluc in urine, similar to Gluc.13,14. We found that Vluc activity in urine was also linear with respect to cell number, albeit less sensitive, showing that Vluc is cleared by the kidneys (Fig. 1a). We next evaluated the half-life of Vluc in circulation by intravenously injecting Vluc-containing conditioned medium of cells in nude mice and assaying blood for Vluc activity at different time points. We found that the Vluc half-life in circulation to be around 3 hours (Fig. 1b), similar to previously reported SEAP4. Gluc on the other hand has a very short half-life of 20 minutes14. This relatively fast clearance of these reporters allow them to be used as markers for dynamic events.
Figure 1.
Triple secreted reporter system. (a) Linearity of Vluc blood reporter with respect to cell number. Different U87 glioma cells expressing Vluc were implanted subcutaneously in nude mice (n=4/group). One week later, 5 μl blood or urine (in triplicates) was assayed for Vluc activity using 100 μl of the vargulin substrate. (b) half-life of Vluc in circulation. Conditioned medium from U87 cells expressing Vluc were filtered, and 150 μl was i.v. injected in nude mice (n=4). At different time points, 5 μl blood (in triplicates) were assayed for Vluc activity. (c) Specificity of each secreted reporter for its substrate. Mice were injected subcutaneously with U87 cells expressing either Gluc, Vluc or SEAP (n=4/group). Three days later, 5 μl blood or serum (in triplicates) was assayed for Gluc, Vluc, and SEAP activity. A significant signal is obtained in blood/serum only with the proper reporter/substrate combination.
Since it is of uttermost importance that the chemical reaction of the individual reporters used for multiplex application can be distinguished, we determined the specificity of each reporter to its substrate in the blood. U87 glioma cells stably expressing Gluc, Vluc or SEAP (all under the control of CMV promoter) were implanted subcutaneously in different nude mice. Three days later, 5 μl blood was assayed for each reporter activity using coelenterazine, vargulin or SEAP substrate. Significant signal from blood was obtained only when the proper reporter/substrate combination was used, showing no substrate overlap or cross reaction among the different reporters, and indicating that Gluc, Vluc and SEAP can be used together for multiplex applications (Fig. 1c).
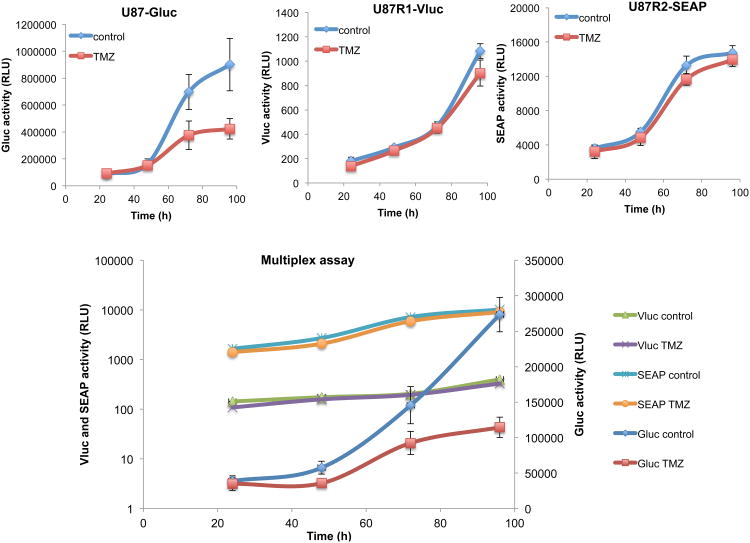
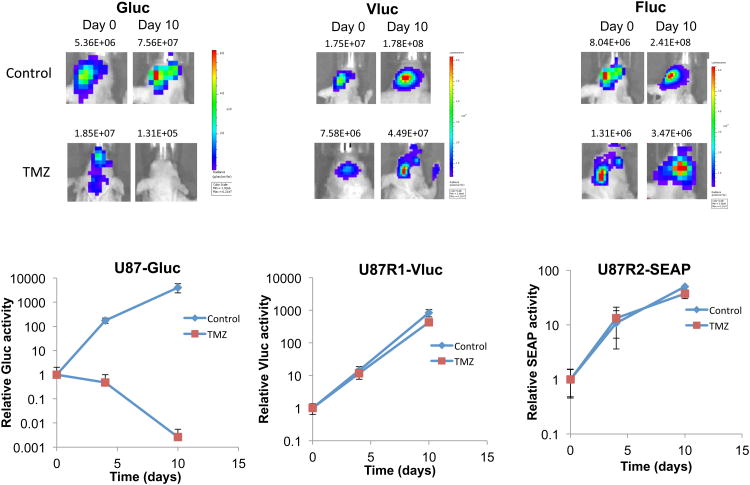
Finally, we applied the triple reporter system for non-invasive monitoring of three different subsets of U87 glioma cells in response to temozolomide (TMZ), the chemotherapeutic agent of choice for the treatment of grade IV glioma19. We used U87 parent cells as well as two TMZ-resistant subclones of these cells, U87R1 and U87R2 (Supplementary Fig. 2). We first confirmed that each of the three reporters (Gluc, Vluc and SEAP) could be used to monitor cell viability. U87 parent cells (sensitive to TMZ) were transduced with a lentivirus vector to express either Gluc, Vluc or SEAP. These cells were plated in a 96-well plate and treated with either DMSO or 100 μM TMZ. At different time points, aliquots of conditioned medium were assayed for each reporter activity. A substantial differences between TMZ-treated and control samples was observed with all three reporters confirming that Gluc, Vluc and SEAP can be used as markers for cell viability (Supplementary Fig. 3). We then applied the triple reporter for multiplex applications. U87 parent cells were engineered by a lentivirus vector to express Gluc (U87-Gluc), while U87R1 were engineered to express Vluc (U87R1-Vluc) and U87R2 to express both SEAP and Fluc (U87R2-SEAP/Fluc). Fluc here is used for in vivo localization of U87R2 cells with bioluminescence imaging (since SEAP reporter cannot be imaged in vivo) while Gluc and Vluc are used to localize U87 and U87R1 cells respectively. First, the triple secreted reporter system was confirmed in culture by plating 7,000 cells of either U87-Gluc, U87R1-Vluc, U87R2-SEAP/Fluc (singleplex) or combination of all three cells (2,500 of each line; multiplex) in a 96-well plate. The next day, cells were treated with TMZ (100 μM) or DMSO control. Aliquots of the conditioned medium were then assayed for each luciferase activity every day over 4 days. Using the singleplex assay, we observed that U87-Gluc cells responded very well to TMZ (>50% cell death) while this drug had no significant effect on U87R1-Vluc and U87-R2-SEAP/Fluc as expected (Fig. 2). The multiplex assay showed the exact same phenomena proving that these three reporters can be used together for simultaneous monitoring of three distinct biological processes over time. We then mixed these three cell lines equally and intracranially implanted 75,000 cells (25,000 of each line) in the brain of nude mice. One week later (time zero), a sample of blood was withdrawn and a group of mice (n=6) was injected with 10 mg/kg TMZ, while the other group (n=6) was injected with DMSO vehicle (control). Blood was collected at different time points and 5 μl of blood (for Gluc) or serum (for Vluc and SEAP) in triplicates was assayed for each reporter activity using coelenterazine, vargulin or SEAP assay respectively. As expected, a continuous decrease in Gluc level in the blood in response of parental U87 cells to TMZ was observed over time in the treated group, while an increase in Gluc signal was observed in the control group (Fig. 3). On the other hand, both Vluc and SEAP levels increased in serum over time in both treated and control groups showing that U87R1 and U87R2 cells are resistant to TMZ (Fig. 3). Before (day zero) and 10 days post-treatment, mice were imaged for Gluc, Vluc and Fluc after injection of coelenterazine, vargulin and D-luciferin confirming the blood assays data (Fig. 3). Gluc imaging was performed first followed by Vluc imaging (4 hours later) and then Fluc imaging 24 hours later. This sequential imaging ensured that the signal of the first reporter reached background level before imaging the second reporter (Supplementary Fig. 4). All together, these results show that Gluc, Vluc and SEAP can be multiplexed together as blood reporters for non-invasive monitoring of biological processes, in real time.
Figure 2.
Triple secreted reporter system in culture. U87-Gluc, U87R1-Vluc, U87R2-SEAP/Fluc cells (singleplex) or a combination of all three cell lines (multiplex) were plated in a 96-well plate and treated with either TMZ (100 μM) or DMSO control. At different time points, aliquots of conditioned medium were assayed for Gluc, Vluc or SEAP reporters. Only U87 parental cells responded to TMZ (as observed by Gluc assay) but not U87R1 or U87R2 cells using both the singleplex and multiplex assay. Data presented as average RLU ± SD (n=5). Typical background signal from non-transduced cells for Gluc 0.15±0.012; Vluc 57.61±2.98; SEAP 7.91±0.83.
Figure 3.
Multiplex blood reporter system. A mixture of U87-Gluc, U87-Vluc and U87-SEAP/Fluc cells (25,000 of each line) were intracranially injected in the brain of nude mice. One week later, mice were randomized in two groups which received either 10mg/kg TMZ or DMSO vehicle (n=6/group). Before and at different time points post-treatment, 5 μl blood was assayed for Gluc activity. Likewise, 5 μl of serum was assayed for Vluc and SEAP activity. Before and at day 10 post-TMZ treatment, signal was localized to tumors by in vivo triple bioluminescence imaging of Gluc, Vluc and Fluc after injection of coelenterazine, vargulin and D-luciferin substrates respectively. Gluc imaging was performed first, followed by Vluc imaging 4 hours later and then Fluc imaging, 24 hours later. Representative mouse with brain tumor total flux signal from each group is shown. Data presented as normalized reporter activity in which the signal obtained before treatment (one week post-cells implantation) is set at 1. Typical background signal from naïve mice with no tumor for Gluc 0.096±0.010; Vluc 4.273±0.542; SEAP 1.012±0.117
Luciferase-mediated bioluminescence imaging is widely used as a reporting tool for monitoring various biological processes in vitro and in vivo. In vivo bioluminescence imaging has several disadvantages including photon absorption by tissues as a function of depth and the inefficiency of luciferase substrates in crossing the blood-brain/tumor barrier. Further, it is time-consuming and involves some inconvenience due to frequent anesthesia, need for transport of animals, and repeated systemic substrate injections, thereby limiting the ability to monitor relatively large cohorts in small time intervals. In addition, certain biological processes cannot be efficiently measured using in vivo bioluminescence imaging due to dispersion of the bioluminescence signal throughout the animal or simply because the signal is below the detection limit of the CCD camera. These include for instance monitoring of circulating cells such as stem cells or immune cells, and the measurement of systemic responses such as the immune or stress responses. The multiplex reporter system described here provides a simple, non-invasive, and sensitive method for dynamic analysis of cell viability both in vitro and ex vivo complementing in vivo bioluminescence imaging which has the unique ability to localize the signal, and thereby greatly facilitating non-invasive monitoring of biological processes.
Conclusion
In summary, we have developed a multiplexed blood reporter system for simultaneous monitoring of multiple biological parameters in the same experimental animal in real time. This system could be applied in many different fields facilitating the understanding of disease development and expedites findings of novel therapeutics and translation into the clinic.
Supplementary Material
Acknowledgments
This work was supported by grants from NIH/NINDS P30NS045776, 1R01NS064983, and NIH/NCI 1R01CA166077 (BAT). M Sarah Bovenberg was supported by a Fulbright scholarship, the Huygens Scholarship Program, the VSB Foundation and the Saal van Zwanenberg Foundation. M Hannah Degeling was supported by a Fulbright scholarship, the Saal van Zwanenberg Foundation, the VSB Foundation, the Dr. Hendrik Muller Vaderlandsch fonds, the Dutch Cancer Society (KWF Kankerbestrijding), the Hersenstichting/Dutch Brain Foundation as well as the Jo Keur Foundation (Leiden University Medical Center, The Netherlands).
Footnotes
References
- 1.Tannous BA, Teng J. Biotechnology advances. 2011;29:997. doi: 10.1016/j.biotechadv.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blacklock J, You YZ, Zhou QH, Mao G, Oupicky D. Biomaterials. 2009;30:939. doi: 10.1016/j.biomaterials.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cutrera J, Dibra D, Xia X, Hasan A, Reed S, Li S. Mol Ther. 2011 doi: 10.1038/mt.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiramatsu N, Kasai A, Hayakawa K, Yao J, Kitamura M. Nucleic Acids Res. 2006;34:e93. doi: 10.1093/nar/gkl515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hughes TS, Langer SJ, Johnson KW, Chavez RA, Watkins LR, Milligan ED, Leinwand LA. Mol Ther. 2009;17:88. doi: 10.1038/mt.2008.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galanis E, Hartmann LC, Cliby WA, Long HJ, Peethambaram PP, Barrette BA, Kaur JS, Haluska PJ, Jr, Aderca I, Zollman PJ, Sloan JA, Keeney G, Atherton PJ, Podratz KC, Dowdy SC, Stanhope DR, Wilson TO, Federspiel MJ, Peng KW, Russell SJ. Cancer Res. 2010;70:875. doi: 10.1158/0008-5472.CAN-09-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iankov ID, Hillestad ML, Dietz AB, Russell SJ, Galanis E. Mol Ther. 2009;17:1395. doi: 10.1038/mt.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu C, Sarkaria JN, Petell CA, Paraskevakou G, Zollman PJ, Schroeder M, Carlson B, Decker PA, Wu W, James CD, Russell SJ, Galanis E. Clin Cancer Res. 2007;13:7155. doi: 10.1158/1078-0432.CCR-07-1306. [DOI] [PubMed] [Google Scholar]
- 9.Peng KW, Facteau S, Wegman T, O'Kane D, Russell SJ. Nat Med. 2002;8:527. doi: 10.1038/nm0502-527. [DOI] [PubMed] [Google Scholar]
- 10.Griesenbach U, Vicente CC, Roberts MJ, Meng C, Soussi S, Xenariou S, Tennant P, Baker A, Baker E, Gordon C, Vrettou C, McCormick D, Coles R, Green AM, Lawton AE, Sumner-Jones SG, Cheng SH, Scheule RK, Hyde SC, Gill DR, Collie DD, McLachlan G, Alton EW. Biomaterials. 2011;32:2614. doi: 10.1016/j.biomaterials.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Nakanishi H, Higuchi Y, Kawakami S, Yamashita F, Hashida M. Molecular therapy : the journal of the American Society of Gene Therapy. 2010;18:707. doi: 10.1038/mt.2009.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niers JM, Kerami M, Pike L, Lewandrowski G, Tannous BA. Mol Ther. 2011 doi: 10.1038/mt.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tannous BA. Nat Protoc. 2009;4:582. doi: 10.1038/nprot.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wurdinger T, Badr C, Pike L, de Kleine R, Weissleder R, Breakefield XO, Tannous BA. Nat Methods. 2008;5:171. doi: 10.1038/nmeth.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maelandsmo GM, Ross PJ, Pavliv M, Meulenbroek RA, Evelegh C, Muruve DA, Graham FL, Parks RJ. J Gene Med. 2005;7:307. doi: 10.1002/jgm.666. [DOI] [PubMed] [Google Scholar]
- 16.Msaouel P, Dispenzieri A, Galanis E. Curr Opin Mol Ther. 2009;11:43. [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson EM, Nagata S, Tsuji FI. Proc Natl Acad Sci U S A. 1989;86:6567. doi: 10.1073/pnas.86.17.6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson EM, Nagata S, Tsuji FI. Gene. 1990;96:257. [PubMed] [Google Scholar]
- 19.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO. The New England journal of medicine. 2005;352:987. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.