Abstract
More than 130 million people world-wide chronically infected with hepatitis C virus (HCV) are at risk of developing severe liver disease. Antiviral treatments are only partially effective and a vaccine is not available. Development of more efficient therapies has been hampered by the lack of a small animal model. Building on the observation that CD81 and occludin (OCLN) comprise the minimal set of human factors required to render mouse cells permissive to HCV entry1 we previously showed that transient expression of these two human genes is sufficient to allow viral uptake into fully immunocompetent inbred mice2. Here, we demonstrate that transgenic mice stably expressing human CD81 and OCLN also support HCV entry but innate and adaptive immune responses restrict HCV infection in vivo. Blunting antiviral immunity in genetically humanized mice infected with HCV results in measurable viremia over several weeks. In mice lacking the essential cellular co-factor cyclophilin A (CypA), HCV RNA replication is markedly diminished, providing genetic evidence that this process is faithfully recapitulated. Using a cell-based fluorescent reporter activated by the NS3-4A protease we visualize HCV infection in single hepatocytes in vivo. Persistently infected mice produce de novo infectious particles, which can be inhibited with directly acting antiviral drug treatment, thereby providing for the first time evidence for the completion of the entire HCV life-cycle in inbred mice. This genetically humanized mouse model opens new opportunities to genetically dissect HCV infection in vivo and provides an important preclinical platform for testing and prioritizing drug and vaccine candidates.
The narrow species tropism of HCV is incompletely understood. Mouse cells do not support viral entry, and inefficiently replicate HCV RNA, but do support virion assembly and release. HCV utilizes numerous cellular factors to enter hepatocytes in a coordinated multistep process, including glycosaminoglycans3, low-density lipoprotein receptor4, scavenger receptor class B type I (SCARB1)5, the tetraspanin CD816, the tight junction proteins, claudin-1 (CLDN1) 7 and occludin (OCLN)1,8, the receptor tyrosine kinases epidermal growth factor receptor9 and ephrin receptor A29, and the cholesterol uptake receptor Niemann Pick C1 like 110. Of these, CD81 and OCLN comprise the minimal human factors needed for HCV uptake into rodent cells1.
We recently demonstrated that adenoviral delivery of HCV entry factors renders mice susceptible to HCV infection2. This transient approach is high-throughput and allows the possibility of rapidly evaluating mutant versions of HCV entry factors. However, adenoviral gene delivery strongly induces interferon-stimulated genes (ISGs) creating an environment that may antagonize HCV replication. In order to limit variability and to prevent vector-mediated immune activation, we generated transgenic mice stably expressing human CD81, SCARB1, CLDN1 and/or OCLN under the control of a liver-specific albumin promoter. Transgenic expression of the human orthologues of the HCV entry factors resulted in similar mRNA levels of the human and endogenous mouse genes in the murine liver (Suppl. Fig. 1) and expression of all four proteins (Suppl. Fig. 2) with the expected subcellular distribution in the liver (Suppl. Fig. 3). Next, we aimed to test the susceptibility of entry factor transgenic (EFT) mice to HCV infection. To identify founder lines supporting viral entry we took advantage of a previously generated, highly sensitive detection system which is based on the activation of a loxP-flanked STOP-luciferase reporter in the genome of Rosa26-Fluc mice by Cre recombinase encoded in recombinant HCV genomes2. We crossed EFT mice to a Rosa26-Fluc background and challenged these animals with a bicistronic HCV genome expressing Cre (HCV-Cre). Consistent with previous data2, the bioluminescent reporter was activated in mice expressing human CD81 and OCLN (Fig. 1a and suppl. Fig. 4a). The addition of human SCARB1 and CLDN1 (Suppl. Fig. 4b) did not increase the entry signal, demonstrating that their murine orthologues are functional for HCV entry in vivo. For subsequent experiments, founder lines Alb-hCD81/hOCLN#941 (2hEF) and Alb-hCD81/hSCARB1/hCLDN1/hOCLN#100 (4hEF), which supported the most efficient viral uptake (Suppl. Fig. 4a), were used. To estimate the number of HCV-infected liver cells, we used an indicator mouse strain in which Cre leads to activation of a nuclear-localized green fluorescent protein/β-galactosidase (GNZ) reporter (Rosa26-GNZ)11. Similar to our previous observations2, HCV-CRE infection resulted in reporter activation in approximately 1–1.5% of murine hepatocytes in 2hEF or 4hEF mice (Fig. 1b and suppl. Fig. 5). To provide additional evidence that viral uptake into EFT mice is mediated by the specific interaction of HCV glycoproteins with host entry factors, we administered antibodies directed against the HCV envelope glycoprotein complex E1E2 or the host entry factor CD81. Delivery of anti-human CD81 or anti-E1E2 (AR4A12) antibodies resulted in a dose-dependent inhibition of HCV-Cre infection (Fig. 1c), whereas isotype control immunoglobulins had no effect. These data further affirm that HCV is taken up in a viral glycoprotein-specific fashion in vivo and underscore the utility of this model for evaluation of passive immunization strategies.
Figure 1. Transgenic expression of human CD81 and OCLN renders mice permissive to HCV entry.

(a) Longitudinal bioluminescence imaging of Rosa26-Fluc mice expressing either hCD81, hOCLN, hSCARB1 and hCLDN1, hCD81 and hOCLN or all four HCV entry factors (4hEF). (b) Quantification of viral uptake into murine hepatocytes expressing hCD81 and OCLN or all four human entry factors determined by flow cytometry 72 h post infection with BiCre-Jc1. (c) Blocking of HCV infection in vivo by either blocking antibodies against CD81 (JS-81) or neutralizing antibodies against HCV E1E2 (AR4A). Rosa26-Fluc 4hEF mice were injected with the indicated amounts of antibodies 24 hours and 4 hours prior to infection with BiCre-Jc1. (d, e) Longitudinal quantification of HCV RNA by RT-qPCR in (d) serum and (e) liver of either wt or mice expressing all four HCV entry factors. (f, g) expression of the interferon-stimulated genes (f) ifi27, ifi44, mx1, pkr, (g) viperin, 2’oas and ip10 in the liver following infection with BiCre-Jc1 in wt or 4hEF mice. (h) Serum levels of IFNγ in wt or 4hEF mice infected with BiCre-Jc1. (i) Analysis of liver-infiltrating IFNγ-secreting NKp46-positive NK cells in BiCre-Jc1-infected wild-type or 4hEF mice. Data shown are mean ± SD of 4 independent experiments. For panels (e-g) four mice were used per time-point. *P < 0.05.
Direct measurement of HCV genome levels by quantitative reverse transcription (qRT)-PCR demonstrated a slight increase in HCV RNA in the serum (at 4h) and liver (at 3h and 24h) of inoculated mice expressing the human entry factors; at 72h, however, the signal was reduced to background (Fig. 1d, e). HCV infection resulted in the upregulation of several ISGs (Fig. 1f, g and suppl. Fig. 6), infiltration of immune cells, especially NK cells into the liver (Fig. 1i) and elevated proinflammatory cytokine levels in the serum (Fig. 1h and suppl. Fig. 7), which could antagonize HCV replication. This hypothesis is further supported by the previous observation that HCV replicons, selectable HCV RNA genomes, replicate more efficiently in murine cells with impaired antiviral signaling 13,14.
To identify a murine environment more conducive to HCV replication we crossed 4hEF R26-Fluc mice to strains carrying targeted disruptions in PKR, MAVS, IRF-1, IRF-3, IRF-7, IRF-9, STAT1 or the IFNαβ receptor (Fig. 2). These strains are viable and known to be hyper-susceptible to RNA viruses, due to impaired innate immune responses. The luminescent reporter signal was slightly elevated during the early phase following infection with HCV-Cre in most EFT strains impaired in antiviral signaling as compared to EFT mice on a wild-type background (Fig. 2a–d). 20–40 days after infection there was a marked increase in the luciferase signal, particularly in IRF-1 (8-fold), IRF-7 (16-fold) (Fig. 2c), IFNαβR (20-fold), and STAT1 (40-fold) deficient mice (Fig. 2d) compared to non-transgenic littermate controls. The increased reporter signal, which presumably reflects a transient burst in viral replication and spread, eventually returned to background levels possibly marking clearance of HCV infected cells by the murine immune system. The elevated luminescent signal correlated with increases in serum HCV RNA levels at peak time points (Fig. 2e).
Figure 2. Blunting of antiviral immune responses in mice expressing HCV entry factors augments HCV RNA replication.
(a–d) Bioluminescence kinetic of BiCre-Jc1 infected Rosa26-Fluc mice expressing all four HCV entry factors and entry-factor negative controls (a) with fully intact innate immune system or impaired in (b) PKR, MAVS, (c) IRF1, IRF3, IRF7, IRF9, (d) IFNαβR or STAT1. (e) HCV RNA levels in the serum of 4hEF mice or 4hEF mice deficient in STAT1, IRF1, IRF3, IRF7, MAVS or IRF9 6 weeks post infection with BiCre-Jc1.
To validate that the elevated signal is indeed due to increased HCV RNA replication in mice with blunted antiviral immunity, we crossed cyclophilin A (CypA) deficient mice (PPIA−/−) to the EFT Rosa26-Fluc STAT1−/− background. CypA is a critical host factor for HCV RNA replication in human cells15. In PPIA−/− 4hEF Rosa26-Fluc STAT1−/− mice HCV RNA (Fig. 3c) and the luminescent reporter signal at peak times (day 31) was more than 60% lower than that of PPIA+/+ and PPIA+/− mice (Fig. 3d) likely as a direct consequence of the murine PPIA deficiency. These data provide the first direct genetic evidence that CypA is a bona fide HCV replication factor in vivo. Furthermore, treatment of 4hEF STAT1−/− mice with an NS5A inhibitor for 3 weeks following infection suppressed HCV RNA loads to below the limit of detection (Fig. 3e) providing additional evidence that HCV RNA does indeed replicate in these mice.
Figure 3. Visualization and genetic and pharmacological interference with HCV infection.

(a, b) Quantification of murine hepatocytes actively replicating HCV in wild-type, 4hEF mice and 4hEF STAT1−/− mice as measured by the HCV NS3-4a dependent cleavage of the TagBFPnlsMAVS transgenic reporter construct by ImageStream X analysis. (c, d) Longitudinal HCV RNA levels in 4hEF STAT1−/− mice, (c) 4hEF STAT1−/− mice lacking PPIA or (d) 4hEF STAT1−/− mice treated with an HCV NS5A inhibitor (BMS-790052) for 20 days. Data shown are mean ± SD of n=10–18 mice from 2 independent experiments. **P < 0.01.
Unambiguous detection of HCV antigens in situ is notoriously difficult. Therefore, we constructed a transgenic mouse line expressing a modified version of a previously described cell based fluorescent reporter system 16 to directly visualize infection in the liver of infected mice (Suppl. Fig. 8). This highly sensitive reporter, whose activity directly correlates with the level of HCV RNA replication, is based on cleavage of a TagBFP/MAVS fusion protein by the HCV NS3-4A serine protease and translocation of the mitochondrially-anchored fluorescent protein to the nucleus. The chimeric TagBFPnlsMAVS protein was widely expressed in the liver as assessed by histology and flow-cytometry (Suppl. Fig. 8a, b). To accurately quantify HCV protease-mediated nuclear relocalization we subjected single hepatocyte suspensions to ImageStreamX analysis. Adenoviral delivered overexpression of NS3-4A activated the reporter in a majority of cells (Suppl. Fig. 9), demonstrating the functionality of the reporter in vivo. In EFT+ but not EFT− (Fig 3a) TagBFPnlsMAVS transgenic mice HCV infection causes overlap of TagBFP fluorescence with the nuclear counterstain in approximately 0.2% of the cells (Fig. 3b). This low but HCV-specific reporter activation is likely due to the limited amounts of NS3-4A being translated from the incoming viral RNA, which cannot accumulate as a consequence of the abortive presumably single cycle infection on a fully immunocompetent background. In contrast crossing the EFT-TagBFPnlsMAVS strain to a STAT1-null background increased the frequency of cells displaying HCV infection two-fold over STAT1-positive controls (to 0.4%). Importantly, while the reporter was activated only transiently in STAT1 sufficient mice, the signal persisted at least until day 10 in mice on a STAT1−/− background (Fig. 3b). Taken together, these results suggest that HCV can replicate in immunocompromised mice expressing human CD81 and OCLN and highlight the value of the model for studying HCV replication in vivo.
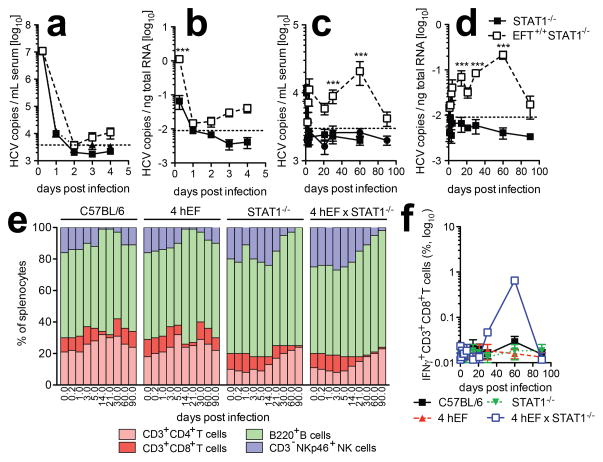
We further characterized viral kinetics in mice infected with a monocistronic HCVcc. HCV RNA rose approximately 10-fold over the limit of quantitation in serum (Fig. 4a, c) and liver (Fig. 4b, d) of EFT STAT1−/− mice as compared to non-transgenic STAT1−/− controls. Animals remained persistently infected for a majority of the observation period, with HCV RNA becoming nearly undetectable after 90 days (Fig. 4c, d). Sequence analysis of HCV RNA detected at late time points in mice infected with HCV-Cre revealed mutations in some viral genomes, but none were shared among the 5 mice that were analyzed. (Suppl. Fig. 12). Whether any of these are adaptive mutations that increase viral fitness in vivo will be the subject of future studies. HCV infection caused splenomegaly in some innate immune deficient mice expressing HCV entry factors, but not in non-EFT mice (Suppl. Fig. 10a) indicative of HCV specific immune activation. Indeed we observed increased frequencies of NK cells and subsequently B cells in the spleens of 4hEF STAT−/− mice in the early and late phases of infection, respectively (Fig. 4e and Suppl. Fig. 10b). Consistent with results in humans and chimpanzees primarily NK and IFN-γ-producing CD8+ T cells (Fig. 4f) infiltrated the livers of 4hEF STAT−/− mice with a skewing towards an effector memory phenotype near the end of the time course (Suppl. Fig. 10c). These data suggest that HCV infection elicits cellular immune responses, albeit potentially confounded by the STAT1 deficiency, which may contribute to eventual viral clearance.
Figure 4. HCV infection in 4hEF STAT1−/− mice leads to immune activation.
(a–d) HCV RNA copies in (a, c) serum and (b, d) liver of 4hEF STAT1−/− mice and STAT1−/− mice (a, b) early or (c, d) late during infection with Con1/Jc1. (e) Relative frequencies of the indicated lymphocyte subsets in spleen of wild-type, 4hEF, STAT1−/− or STAT1−/− 4hEF mice isolated at the indicated time-points post infection with Con1/Jc1. (f) Analysis of liver-infiltrating IFNγ-producing CD3+CD8+ T cells of wild-type, 4hEF, STAT1−/− and 4hEF STAT1−/− mice following infection with Con1/Jc1. ***P < 0.001.
To determine whether primary hepatocytes in EFT mice on immunodeficient backgrounds were capable of producing infectious virions, sera collected at day 40 following infection were used to inoculate naïve Huh-7.5 cells. Infectious virus was detected in sera of 4hEF mice deficient for STAT1, IRF-1 and IRF-7 (Fig. 5a), consistent with the increased levels of luminescent reporter activity (Fig 2c, d) and HCV RNA load (Fig. 2e) in these strains. In HCV infected 4hEF STAT1−/− mice titers reached approximately 100 TCID50/ml (Fig. 5b). Sera from HCV infected non-EFT littermates, DAA treated 4hEF STAT1−/− or 4hEF STAT1−/− PPIA−/− mice collected at the same timepoints, did not yield NS5A-positive cells (data not shown), indicating de novo virus production rather than carry over of the inoculum. To demonstrate that the lack of HCV NS5A positive cells in the infectivity assay cannot simply be attributed to residual quantities of the DAA in circulation we spiked the sera from the BMS-790052-treated animals with tissue culture produced BiCreJc1 virus (Fig. 5b) and found no inhibitory effect. Furthermore, in Huh-7.5 cells expressing and TagRFPnlsMAVS reporter16 infection with sera from infected mice resulted in relocalization of the reporter signal whereas inoculation with sera from non-infected, DAA treated or PPIA−/− mice did not (Suppl. Fig. 11). These data are in accordance with previous in vitro observations demonstrating that mouse hepatoma cells support late stages of the HCV life-cycle17.
Figure 5. Evidence for production of infectious particles.

(a) STAT1−/−, IRF1−/−, IRF3−/−, IRF7−/−, IRF9−/− and MAVS−/− mice expressing all four human HCV entry factors were infected with BiCre-Jc1. Sera were collected 6 weeks following infection and were used to infect naïve Huh-7.5 cells. NS5A staining was performed 72 h post infection and the frequency of HCV antigen bearing cells quantified by flow cytometry. (b) HCV infectious particles released into the serum of 4hEF STAT1−/− mice, 4hEF STAT1−/− mice lacking PPIA or 4hEF STAT1−/− mice treated with BMS-790052 for 20 days as determined by limiting dilution assay. * = not detectable.
This study represents an important step forward in developing an animal model for HCV infection and immunity. To our knowledge this is the first time that the entire HCV life-cycle has been recapitulated in inbred mice with inheritable susceptibility to HCV. Previously developed xenotransplant mouse models can also be infected with HCV. However, these models are hampered by intra- and interexperimental variability, donor-to-donor variability, low throughput, and high costs. The inbred mouse model presented here can overcome many of these challenges, is amendable to genetic manipulations and can be employed for preclinical assessment of the efficacy of antiviral drug candidates and entry inhibitors. In order to study unperturbed HCV-specific immune responses and HCV-associated pathogenesis it will be necessary to establish persistent HCV infection in fully immunocompetent mice. By harnessing the remarkable genetic plasticity of HCV it may be possible to select for viral variants that replicate more robustly in sufficiently immunocompromised rodent strains. High titer sera could subsequently be passaged through progressively more immunocompetent hosts to produce this outcome. Future studies will address the capacity of different, genetically diverse HCV genotypes to establish chronicity in genetically humanized mice.
Methods summary
For more detailed methods see Supplementary Methods.
Mice
Construction of all mice expressing HCV entry factors under the control of a liver specific albumin promoter or CAGGS-TagBFPnlsMAVS and the source of all other mouse mutant strains is described in the supplementary methods. Mice were bred and maintained at the Comparative Bioscience Center of The Rockefeller University according to guidelines established by the Institutional Animal Committee.
Hepatitis C virus
Plasmids encoding chimeric HCV genomes, including Jc1, Con1-Jc1 and bicistronic HCV genomes expressing CRE, were linearized with XbaI and in vitro transcribed. RNA was electroporated into Huh-7.5 cells and infectious virus was collected from supernatants 48–72 h after transfection18.
RT-PCR quantification of HCV RNA
Total RNA was isolated from mouse brain, liver and sera using the RNAeasy kit (Qiagen, Valencia, CA). HCV genome copy number was quantified by one step RT-PCR.
SUPPLEMENTARY METHODS (Online only)
Animals and cell lines
Gt(ROSA)26Sortm1(Luc)Kaelin19 (Rosa26-Fluc), B6;129-Gt(ROSA)26Sortm1Joe/J20 (Rosa26-GNZ), and C57BL/6 (wild-type) mice were obtained from The Jackson Laboratory (Bar Harbor, ME). John Colgan (University of Iowa, Iowa City, IA, USA) kindly made the Ppiatm1Lubn (PPIA−/−) 21 available. Rosa26-Fluc mice contain the firefly luciferase (luc) gene inserted into the Gt(ROSA)26Sor locus. Expression of the luciferase gene is blocked by a loxP-flanked STOP fragment placed between the luc sequence and the Gt(ROSA)26Sor promoter. Cre recombinase mediated excision of the transcriptional stop cassette results in luciferase expression in Cre-expressing tissues. Rosa26-GNZ knock-in mice have widespread expression of a nuclear-localized green fluorescent protein/beta-galactosidase fusion protein (GFP-NLS-GNZ) once an upstream loxP-flanked STOP sequence is removed. When Cre recombinase is introduced into cells the resulting GNZ fusion protein expression allows for enhanced (single cell level) visualization. Mice were bred and maintained at the Comparative Bioscience Center of the Rockefeller University according to guidelines established by the Institutional Animal Committee. Huh-7.522, Huh-7.5.123 and Huh-7.5 stably expressing the TagRFPnlsMAVS reporter24 were maintained in DMEM with 10% fetal bovine serum (FBS) and 1% nonessential amino acids (NEAA).
Mutant mice with targeted disruptions in genes involved in antiviral defenses
Irf1tm1Mak (IRF1−/−)25 were obtained from the Jackson Laboratory (Bar Harbor, Maine, USA), Ifnar1tm1Agt (IFNαβ R−/−) 26 from B&K Universal Ltd (Hull, UK) and Stat1tm1Dlv (STAT1−/−) 27 from Taconic (Hudson, NY, USA). Bcl2l12/Irf3tm1Ttg (IRF3−/−) 28, Irf7tm1Ttg (IRF7−/−) 29 and Irf9tm1Ttg (IRF9−/−) 30 were kindly provided by Tadasugo Taniguchi. Takashi Satoh and Shizuo Akira kindly provided Dhx58tm1(A30K)Aki (LGP2A30K/A30K) 31 and Taro Kawai and Shizuo Akira kindly provided Mavstm1Aki (MAVS−/−) 32 mice. Adolfo Garcia-Sastre (Mount Sinai School of Medicine, New York, NY, USA) kindly made the Eif2ak2tm1Cwe (PKR−/−) 33 and Anton Berns (Netherlands Cancer Institute, Amsterdam, Netherlands) the (Rag2−/− IL2RNULL) 34 mice available.
Generation of HCV entry factor transgenic mice
cDNAs encoding the human CD81, SCARB1, CLDN1 or OCLN were cloned into a vector between a chimeric intron and the 3′ flanking regions of human growth hormone (GH1), in which the mouse albumin enhancer/promoter drives gene expression 35. Vector-free human CD81, SCARB1, CLDN1 and/or OCLN expression fragments were prepared by NotI and KpnI digestion and microinjected alone or in combination into fertilized C57Bl/6 mouse eggs. Transgenic offspring were mated with C57BL/6 wild-type animals to select for founder lines stably inheriting the transgene(s). In some mice that were co-injected with multiple expression constructs, transgenes did not segregate in the F1 generation, indicating separate insertions in close proximity or insertion as concatemers. The founder lines were designated as follows: C57BL/6-Tg(Alb-hCD81)976Plo/RU (Alb-hCD81), C57BL/6-Tg(Alb-hSCARB1)Plo/RU (Alb-hSCARB1), C57BL/6-Tg(Alb-hOCLN)70Plo/RU (Alb-hOCLN), C57BL/6-Tg(Alb-hCD81/hOCLN)941Plo/RU (Alb-hCD81/hOCLN), C57BL/6-Tg(Alb-hSCARB1/hCLDN1)935Plo/RU (Alb-hSCARB1/hCLDN1), C57BL/6-Tg(Alb-hCD81/hSCARB1/hCLDN1/hOCLN)100Plo/RU (Alb-EFT4x).
Generation of TagBFPnlsMAVS reporter mice (C57BL/6-Tg(TagBFPnlsMAVS)4065)
The TagRFPnlsMAVS(wt) cassette24 was inserted into pCR2.1-TOPO (Invitrogen) and modified to contain TagBFP in place of TagRFP. The resulting TagBFPnlsMAVS cassettes was inserted into the pCAGGS vector (Addgene) to yield pCAGGS-TagBFPnlsMAVS(wt). The pCAGGS backbone drives transgene expression from a ubiquitously active chimeric CMV/β actin promoter36 and has been used to successfully create transgenic mice (for example 37). The pCAGGS-TagBFPnlsMAVS(wt) internal transgenesis cassette was isolated by linearization with SalI/PstI enzymes and injected into C57BL/6 pronuclei. Founder animals were identified by PCR and bred with congenic C57BL/6J animals.
HCV generation and infections
Construction of BiCre-Jc138, Jc139, Con1-Jc140 and Jc1 5AB Ypet41 was described elsewhere. Huh-7.5.123 or Huh-7.542 cells were electroporated with in vitro transcribed full-length HCV RNA. 72h post-electroporation, the medium was replaced with DMEM containing 1.5% FBS and supernatants were harvested every 6h starting from 72h. Pooled supernatants were clarified by centrifugation at 1,500 xg, filtered through a 0.45 μm bottle top filter (Millipore, Billerica, MA) and concentrated using a stirred cell (Millipore). Viral titers (TCID50) were determined using Huh-7.5 cells as previously described43. All infections of mice with the indicated genotypes were performed intravenously.
Antibodies and inhibitors
Blocking antibodies against CD81 (JS81) and IgG1 control antibodies were obtained from BD Biosciences (Franklin Lakes, NJ). Antibodies against NS5A43 and E2 (clone AR4A)44 and the human IgG1 isotype control (b1245, kindly provided by Dennis Burton, The Scripps Research Institute) have been described previously. Antibodies for the detection of human CD81 were purchased from BD Biosciences, OCLN from BD Biosciences (for histology) and from Invitrogen (Carlsbad, CA, for Western blotting) CLDN1 from from Invitrogen (for Western blotting) and Abcam (Cambridge, MA for histology) and SCARB1 from Genetex (Irvine, CA, for histology) and from BD Biosciences (for Western blotting). The HCV NS5A inhibitor BMS-79005246 was obtained from Selleck Chemicals (Houston, TX).
Adenovirus constructs
Adenoviral constructs encoding HCV JFH1 NS3-4A were created using the AdEasy™ Adenoviral Vector System (Agilent Technologies, Santa Clara, CA) according to the manufacturer’s instructions. Briefly, JFH1 NS3-4A cDNA was PCR-amplified from pCR3.1-NS3/4A and inserted into the pShuttle-CMV™ using KpnI/NotI sites. Recombinant pShuttle-CMV plasmids were linearized with PmeI and ligated to pAdEasy™ by homologous recombination followed by electroporation into BJ5183 cells (Agilent). Recombinant pShuttle-pAdEasy constructs were identified by PacI restriction analysis. All plasmid constructs were verified by DNA sequencing.
Histologic detection of HCV entry factors
Liver and spleen of mice injected with adenoviruses encoding human entry factors were harvested 24h post-injection and fixed using 4% paraformaldehyde. Tissue sections (8 μm) were deparaffinized and subjected to antigen retrieval by boiling for 30 min in citrate buffer (10mM sodium citrate, 0.05% Tween 20, pH 6.0). Entry factors were stained with human-specific primary antibodies for 16h at 4°C followed by secondary antibody staining using Alexa 488 or Alexa 633-conjugated antibodies for 2h at RT. For in situ detection of eGFP fluorescence, mouse tissue was immediately frozen in O.C.T. (Optimal Cutting Temperature) compound at −80°C. Tissue sections (~5–6 μm) were cut on poly-L-lysine coated slides. Secondary antibodies goat-anti-mouse or goat-anti-rabbit Alexa 488- or rhodamine-conjugates (Invitrogen; 1:1000) were used for immunofluorescence. Nuclei were detected using DAPI in VectaShield Mounting medium (Vector Laboratories, Burlingame, CA). Images were captured on an Axioplan 2 imaging fluorescence microscope (Zeiss, Thornwood, NY) using Metavue Software (Molecular Devices, Sunnyvale, CA). Images were processed using ImageJ software (NIH, Bethesda, MD).
Isolation of murine hepatocytes
Mice were anaesthetized by intraperitoneal injection of a mixture of 100mg/kg ketamine and 10mg/kg xylazine. Livers were perfused through the inferior vena cava for 5 minutes each with chelating buffer (0.5mM EGTA, 0.05M HEPES pH7.3 in Ca/Mg-free HBSS) at a flow rate of 2mL/min followed by collagenase solution (4.8mM CaCl2, 100 U/mL Collagenase type IV, 0.05M HEPES pH 7.3 in Ca/Mg-free HBSS). The resulting cell suspension was passed through a 100 μm cell strainer, washed twice in HBSS and was fixed in 4% paraformaldehyde. Purity of isolated hepatocytes was over 90% in all preparations as confirmed by intracellular staining for murine albumin.
Immune activation
Lymphocytes were isolated from liver and spleen by digestion with 0.1% collagenase (Sigma) for 30 min at 37°C. Lymphocytes were then isolated from the cell suspensions as well as from peripheral blood by density gradient centrifugation. Cells were stained with directly fluorochrome-conjugated antibodies against CD3, CD4, CD8, B220 (eBioscience) and NKp46 (BD Biosciences). Following cell surface staining, cells were fixed and permeabilized using BD Cytofix/Cytoperm (BD Biosciences) and stained with antibodies against IFNγ and TNFα. Samples were measured using a BD LSR2 flow cytometer (BD Biosciences) and data were analyzed using Flowjo (Treestar Software).
Western blotting
Perfused murine liver tissue was homogenized in lysis buffer containing 1% Triton X100, 50 mM Tris-HCl pH 8, 150 mM NaCl, and Mini EDTA-free Protease Inhibitor Cocktail (Roche, Basel, Switzerland) for 30 min on ice. 30 μg of protein lysate was separated on 4–12% Bis/Tris NuPage polyacrylamide gels (Invitrogen). Proteins were transferred to nitrocellulose membranes and entry factors were detected using antibodies against human SCARB1 (1:500) and CLDN1 (1:200. β-actin (1:10000) was probed as a loading control. Following secondary antibody staining with HRP-conjugated anti-mouse IgG Fc (JIR, 1:10000), Western blots were visualized using SuperSignal West Pico (Thermo Scientific).
RT-PCR quantification of HCV entry factors and interferon-stimulated genes (ISGs)
To quantify expression of human and murine genes (entry factors and ISGs), the livers of FVB/NJ mice were harvested at the indicated time points. Total liver RNA was isolated using RNeasy isolation kit (Qiagen, Valencia, CA) and cDNA was synthesized from 0.5 μg RNA using a SuperScript® VILO™ cDNA Synthesis Kit (Invitrogen, Carlsbad, CA) according to manufacturers instructions. Quantitative PCR was performed with a light cycler LC480 (Roche Applied Science, Indianapolis, IN) using an Applied Biosystems SYBR Green PCR Master Mix (Warrington, UK) and the following primer pairs:
| Human Gene | Forward Primer | Reverse Primer |
|---|---|---|
| CD81 | TGTTCTTGAGCACTGAGGTGGTC | TGGTGGATGATGACGCCAAC |
| SCARB1 | CGGATTTGGCAGATGACAGG | GGGGGAGACTCTTCACACATTCTAC |
| CLDN1 | CACCTCATCGTCTTCCAAGCAC | CCTGGGAGTGATAGCAATCTTTG |
| OCLN | CGGCAATGAAACAAAAGGCAG | GGCTATGGTTATGGCTATGGCTAC |
| Mouse Gene | Forward Primer | Reverse Primer |
|---|---|---|
| cd81 | GGCTGTTCCTCAGTATGGTGGTAG | CCAAGGCTGTGGTGAAGACTTTC |
| scarb1 | CAAAAAGCATTTCTCCTGGCTG | AATCTGTCAAGGGCATCGGG |
| cldn1 | TTATGCCCCCAATGACAGCC | ATGAGGTGCCTGGAAGATGATG |
| ocln | ACTAAGGAAGCGATGAAGCAGAAG | GCTCTTTGGAGGAAGCCTAAACTAC |
| gapdh | ACGGCCGCATCTTCTTGTGCA | ACGGCCAAATCCGTTCACACC |
| viperin | TGCTGGCTGAGAATAGCATTAGG | GCTGAGTGCTGTTCCCATCT |
| ifi27 | GCTTGTTGGGAACCCTGTTTG | GGATGGCATTTGTTGATGTGGAG |
| ifi44 | AACTGACTGCTCGCAATAATGT | GTAACACAGCAATGCCTCTTGT |
| mx1 | GACCATAGGGGTCTTGACCAA | AGACTTGCTCTTTCTGAAAAGCC |
| pkr | ATGCACGGAGTAGCCATTACG | TGACAATCCACCTTGTTTTCGT |
| 2’oas | ATGGAGCACGGACTCAGGA | TCACACACGACATTGACGGC |
| ifn-β | CAGCTCCAAGAAAGGACGAAC | GGCAGTGTAACTCTTCTGCAT |
| ip10 | CCAAGTGCTGCCGTCATTTTC | GGCTCGCAGGGATGATTTCAA |
RT-PCR quantification of HCV RNA
Total RNA was isolated from the indicated mouse tissues using the RNAeasy kit (Qiagen, Valencia, CA). HCV genome copy number was quantified by one step RT-PCR using Multicode-RTx HCV RNA Kit Eragen (Madison, WI) and a light cycler LC480 (Roche Applied Science, Indianapolis, IN), according to manufacturers’ instructions.
ImageStream
Hepatocytes were isolated as described above. Hepatocytes were fixed in 4% paraformaldehyde, permeabilized using 0.1% Triton X-100 and stained with antibodies against mouse albumin (Cedarlane) and DRAQ5 (eBioscience) prior to Imagestream acquisition using an Imagestream X with Multimag (Amnis).
Bioluminescence Imaging
Unless otherwise specified, mice were injected with 1011 adenoviral particles 24h prior to intravenous injection with 2×107 TCID50 HCV-Cre. At 72h post infection, mice were anaesthetized using ketamin/xylazine and injected intraperitoneally with 1.5 mg Luciferin (Caliper Lifesciences, Hopkinton, MA). Bioluminescence was measured using an IVIS Lumina II platform (Caliper Lifesciences).
HCV genome sequencing
Total RNA isolated from mouse sera was used to synthesize cDNA covering the HCV genome using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen Life Technologies, Grand Island, NY). Briefly, cDNA fragments were synthesized covering the BiCre HCV genome, using the following primer sets: 10526/17376, 16768/12470, 5928/17377, and 6993/15692, or 10526/17375, 15356/17376, 15695/6986, 3949/5996, 17119/12470, 5928/6815, 6993/16864, 16863/17388, depending on the mouse (see table).
Primers used for cDNA synthesis
| Primer | Sequence | Orientation | Position |
|---|---|---|---|
| RU-O-10526 | 5′-ACCTGCCCCTAATAGGGGCGAC-3′ | Sense | 5′ UTR |
| RU-O-17376 | 5′-TGCTGGCGTTGAAGTCAGCTC-3′ | Antisense | E2 |
| RU-O-16768 | 5′-CGCACCCATACTGTTGGGGG-3′ | Sense | E2 |
| RU-O-12470 | 5′-AAGCCTCATACAGGACCTCC-3′ | Antisense | NS4A |
| RU-O-5928 | 5′-GATGCTACCTCCATTCTCG-3′ | Sense | NS5B |
| RU-O-17377 | 5′-GCATTGTGGGCGCAACTATCC-3′ | Antisense | NS5B |
| RU-O-6993 | 5′-CCGCCCTCACCAGTCCGTTGT-3′ | Sense | NS4B |
| RU-O-15692 | 5′-TATTACCGCCTTTGAGTGAGCTGA-3′ | Antisense | 3′ UTR |
| RU-O-17375 | 5′-CCGAACCACGGGGACGTGGTT-3′ | Antisense | EMCV |
| RU-O-15356 | 5′-GCAAGGTCTGTTGAATGTCG-3′ | Sense | EMCV |
| RU-O-15695 | 5′-CGAATGTGGCCGTGCAGCGGC-3′ | Sense | E1 |
| RU-O-6986 | 5′-AGTCTTTGGAGCCGTGCAAGT-3′ | Antisense | NS3 |
| RU-O-3949 | 5′-GCATCCTGATACCACTTACCTC-3′ | Sense | E2 |
| RU-O-5996 | 5′-CAGGTAGAGGAAGACAGGGCA-3′ | Antisense | NS3 |
| RU-O-17119 | 5′-GCTCCCATCACTGCTTATGCC-3′ | Sense | NS3 |
| RU-O-6815 | 5′-CAACACCTATGACGTGGACATG-3′ | Antisense | NS5A |
| RU-O-16864 | 5′-GTAACTCGCTGTTGCGATACC-3′ | Antisense | NS5B |
| RU-O-16863 | 5′-CAGGTAGAGCTTCAACCTCCC-3′ | Sense | NS5A |
| RU-O-17388 | 5′-ATTGCCGGAGGCGCGCCTACT-3′ | Antisense | 3′ UTR |
Resulting cDNA fragments were then amplified by PCR using Taq polymerase and a combination of the PCR primers listed below. m44 and m55 primer pairs: 3949/5996, 14317/17435, 7086/17376, 15355/16490, 16253/17377, 6993/6815, 3949/6986, 17119/5996. m46 and m64 primer pairs: 3949/5996, 16768/6986, 15354/16490, 6993/16864, 10526/17374, 15356/17376, 17119/12470, 14317/17435, 5928/6815, 7086/17376. m54 primer pairs: 15355/17375, 7086/17376, 3949/6986, 3949/10215, 17119/5996, 5928/12470, 6993/6815, 17391/16864, 14317/17388.
Primers used for sub-fragment PCR amplification
| Primer | Sequence | Orientation | Position |
|---|---|---|---|
| RU-O-3949 | 5′-GCATCCTGATACCACTTACCTC-3′ | Sense | E2 |
| RU-O-5996 | 5′-CAGGTAGAGGAAGACAGGGCA-3′ | Antisense | NS3 |
| RU-O-14317 | 5′-CGGGTGGAGTATCTCTTGAA-3′ | Sense | NS5B |
| RU-O-17435 | 5′-CTGTGTGAAATTGTTATCCGC-3′ | Antisense | 3′ UTR |
| RU-O-7086 | 5′-TGGGCAGGATGGCTCCTGTC-3′ | Sense | Core |
| RU-O-17376 | 5′-TGCTGGCGTTGAAGTCAGCTC-3′ | Antisense | E2 |
| RU-O-15355 | 5′-TGGCAGAACGAAAACGCTG-3′ | Sense | nlsCre |
| RU-O-16490 | 5′-CGCTGCCGAAGTGAAGAACA-3′ | Antisense | E1 |
| RU-O-17377 | 5′-GCATTGTGGGCGCAACTATCC-3′ | Antisense | NS5B |
| RU-O-6993 | 5′-CCGCCCTCACCAGTCCGTTGT-3′ | Sense | NS4B |
| RU-O-6815 | 5′-CAACACCTATGACGTGGACATG-3′ | Antisense | NS5A |
| RU-O-16253 | 5′-CATAGGTTTGCACCCACA-3′ | Sense | NS5A |
| RU-O-6986 | 5′-AGTCTTTGGAGCCGTGCAAGT-3′ | Antisense | NS3 |
| RU-O-17119 | 5′-GCTCCCATCACTGCTTATGCC-3′ | Sense | NS3 |
| RU-O-16768 | 5′-CGCACCCATACTGTTGGGGG-3′ | Sense | E2 |
| RU-O-15354 | 5′-CAAGAAGAAGAGGAAGGTGTC-3′ | Sense | nlsCre |
| RU-O-10526 | 5′-ACCTGCCCCTAATAGGGGCGAC-3′ | Sense | 5′ UTR |
| RU-O-16864 | 5′-GTAACTCGCTGTTGCGATACC-3′ | Antisense | NS5B |
| RU-O-17374 | 5′-CGGTCCCAGCCACGTGGAAGGC -3′ | Antisense | Core |
| RU-O-15356 | 5′-GCAAGGTCTGTTGAATGTCG-3′ | Sense | EMCV |
| RU-O-12470 | 5′-AAGCCTCATACAGGACCTCC-3′ | Antisense | NS4A |
| RU-O-5928 | 5′-GATGCTACCTCCATTCTCG-3′ | Sense | NS5B |
| RU-O-17375 | 5′-CCGAACCACGGGGACGTGGTT-3′ | Antisense | EMCV |
| RU-O-10215 | 5′-CATCTATGACCACCTCACACC-3′ | Antisense | NS2 |
| RU-O-17391 | 5′-CATAGGTTTGCACCCACACCA-3′ | Sense | NS5A |
| RU-O-17388 | 5′-ATTGCCGGAGGCGCGCCTACT-3′ | Antisense | 3′ UTR |
The resulting PCR amplicons were cloned into the pCR2.1 vector using the TOPO TA Cloning Kit (Invitrogen Life Technologies, Grand Island, NY). Resulting clones were screened for proper amplicon insertion size by EcoRI digestion, and sequenced using M13F/R primers, as well as additional internal primers for full genomic coverage (MacrogenUSA, New York, NY).
Statistical analysis
Statistical analyses were performed using Graphpad Prism Software. Statistics were calculated using Kruskal–Wallis one-way analysis of variance. P values below 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank J. Sable, E. Castillo, B. Flatley, S. Shirley, A. Webson and Erick Giang for laboratory support. Alison North and the Rockefeller University (RU) Bioimaging Core Facility, Svetlana Mazel and the RU Flowcytometry Core Facility, Chingwen Yang and the Gene Targeting Center and Ravi Tolwani and the staff of the Comparative Bioscience Center provided outstanding technical support. This study was supported in part by award number RC1DK087193 (to C.M.R. and A.P.) from the National Institute of Diabetes and Digestive and Kidney Diseases, R01AI072613, R01AI099284, (to C.M.R.), R01AI079031 (to M.L.) from the National Institute for Allergy and Infectious Disease, R01CA057973 (to C.M.R) from the National Cancer Institute, The Starr Foundation, the Greenberg Medical Research Institute, the Richard Salomon Family Foundation, the Ronald A. Shellow, M.D. Memorial Fund, the MGM Mirage Voice Foundation, Gregory F. Lloyd Memorial contributions, and anonymous donors. M.D. was supported by postdoctoral fellowship from the German Research Foundation (Deutsche Forschungsgesellschaft). M.T.C. is a recipient of The Rockefeller University Women & Science Fellowship. A.P. is a recipient of the Astella Young Investigator Award from the Infectious Disease Society of America and a Liver Scholar Award from the American Liver Foundation. The funding sources were not involved in the study design, collection, analysis and interpretation of data or in the writing of the report.
Footnotes
Author contributions M.D. and J.A.H. planned and performed experiments and contributed to writing the manuscript. B.M.D., R.N.L., W.C.B, T.F., A.V. and M.T.C. performed the experimental work; T.K., T.S., S.A. and M.L. provided reagents. C.M.R. provided laboratory infrastructure, space, reagents, advice and edited the manuscript. A.P. planned and performed experiments and wrote the manuscript.
Competing financial interests. The following conflicts of interest are managed under University policy: C.M.R. has equity in Apath, LLC, which holds commercial licenses for the Huh-7.5 cell line, HCV cell culture system, the use of OCLN to construct HCV animal models and the fluorescent cell-based reporter system to detect HCV infection.
References
- 1.Ploss A, et al. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–886. doi: 10.1038/nature07684. nature07684 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dorner M, et al. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474:208–211. doi: 10.1038/nature10168. nature10168 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barth H, et al. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J Biol Chem. 2003;278:41003–41012. doi: 10.1074/jbc.M302267200. [DOI] [PubMed] [Google Scholar]
- 4.Agnello V, Abel G, Elfahal M, Knight GB, Zhang QX. Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci USA. 1999;96:12766–12771. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scarselli E, et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO Journal. 2002;21:5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pileri P, et al. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 7.Evans MJ, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. doi: 10.1038/nature05654. nature05654 [pii] [DOI] [PubMed] [Google Scholar]
- 8.Liu S, et al. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J Virol. 2009;83:2011–2014. doi: 10.1128/JVI.01888-08. JVI.01888-08 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lupberger J, et al. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med. 2011;17:589–595. doi: 10.1038/nm.2341. nm.2341 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sainz B, Jr, et al. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med. 2012 doi: 10.1038/nm.2581. nm.2581 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Awatramani R, Soriano P, Mai JJ, Dymecki S. An Flp indicator mouse expressing alkaline phosphatase from the ROSA26 locus. Nat Genet. 2001;29:257–259. doi: 10.1038/ng1101-257. ng1101-257 [pii] [DOI] [PubMed] [Google Scholar]
- 12.Giang E, et al. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6205–6210. doi: 10.1073/pnas.1114927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin LT, et al. Replication of subgenomic hepatitis C virus replicons in mouse fibroblasts is facilitated by deletion of interferon regulatory factor 3 and expression of liver-specific microRNA 122. J Virol. 2010;84:9170–9180. doi: 10.1128/JVI.00559-10. JVI.00559-10 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang KS, et al. Replication of hepatitis C virus (HCV) RNA in mouse embryonic fibroblasts: protein kinase R (PKR)-dependent and PKR-independent mechanisms for controlling HCV RNA replication and mediating interferon activities. J Virol. 2006;80:7364–7374. doi: 10.1128/JVI.00586-06. 80/15/7364 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang F, et al. Cyclophilin A is an essential cofactor for hepatitis C virus infection and the principal mediator of cyclosporine resistance in vitro. J Virol. 2008;82:5269–5278. doi: 10.1128/JVI.02614-07. JVI.02614-07 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones CT, et al. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol. 2010;28:167–171. doi: 10.1038/nbt.1604. nbt.1604 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long G, et al. Mouse hepatic cells support assembly of infectious hepatitis C virus particles. Gastroenterology. 2011 doi: 10.1053/j.gastro.2011.06.010. S0016-5085(11)00767-0 [pii] [DOI] [PubMed] [Google Scholar]
- 18.Lindenbach BD, et al. Complete Replication of Hepatitis C Virus in Cell Culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 19.Safran M, et al. Mouse reporter strain for noninvasive bioluminescent imaging of cells that have undergone Cre-mediated recombination. Mol Imaging. 2003;2:297–302. doi: 10.1162/15353500200303154. [DOI] [PubMed] [Google Scholar]
- 20.Stoller JZ, et al. Cre reporter mouse expressing a nuclear localized fusion of GFP and beta-galactosidase reveals new derivatives of Pax3-expressing precursors. Genesis. 2008;46:200–204. doi: 10.1002/dvg.20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colgan J, et al. Cyclophilin A regulates TCR signal strength in CD4+ T cells via a proline-directed conformational switch in Itk. Immunity. 2004;21:189–201. doi: 10.1016/j.immuni.2004.07.005. S107476130400192X [pii] [DOI] [PubMed] [Google Scholar]
- 22.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong J, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones CT, et al. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol. 2010;28:167–171. doi: 10.1038/nbt.1604. nbt.1604 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuyama T, et al. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell. 1993;75:83–97. 0092-8674(93)90681-F [pii] [PubMed] [Google Scholar]
- 26.Muller U, et al. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 27.Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. S0092-8674(00)81289-1 [pii] [DOI] [PubMed] [Google Scholar]
- 28.Sato M, et al. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. S1074-7613(00)00053-4 [pii] [DOI] [PubMed] [Google Scholar]
- 29.Honda K, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. nature03464 [pii] [DOI] [PubMed] [Google Scholar]
- 30.Kimura T, et al. Essential and non-redundant roles of p48 (ISGF3 gamma) and IRF-1 in both type I and type II interferon responses, as revealed by gene targeting studies. Genes Cells. 1996;1:115–124. doi: 10.1046/j.1365-2443.1996.08008.x. [DOI] [PubMed] [Google Scholar]
- 31.Satoh T, et al. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci U S A. 2010;107:1512–1517. doi: 10.1073/pnas.0912986107. 0912986107 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar H, et al. Essential role of IPS-1 in innate immune responses against RNA viruses. J Exp Med. 2006;203:1795–1803. doi: 10.1084/jem.20060792. jem.20060792 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang YL, et al. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gimeno R, et al. Monitoring the effect of gene silencing by RNA interference in human CD34+ cells injected into newborn RAG2−/− gammac−/− mice: functional inactivation of p53 in developing T cells. Blood. 2004;104:3886–3893. doi: 10.1182/blood-2004-02-0656. 2004-02-0656 [pii] [DOI] [PubMed] [Google Scholar]
- 35.Suemizu H, et al. Establishment of a humanized model of liver using NOD/Shi-scid IL2Rgnull mice. Biochem Biophys Res Commun. 2008;377:248–252. doi: 10.1016/j.bbrc.2008.09.124. S0006-291X(08)01892-5 [pii] [DOI] [PubMed] [Google Scholar]
- 36.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 37.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. S0014-5793(97)00313-X [pii] [DOI] [PubMed] [Google Scholar]
- 38.Dorner M, et al. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474:208–211. doi: 10.1038/nature10168. nature10168 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pietschmann T, et al. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc Natl Acad Sci U S A. 2006;103:7408–7413. doi: 10.1073/pnas.0504877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gottwein JM, et al. Development and characterization of hepatitis C virus genotype 1–7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology. 2009;49:364–377. doi: 10.1002/hep.22673. [DOI] [PubMed] [Google Scholar]
- 41.Horwitz JA, et al. Expression of heterologous proteins flanked by NS3-4A cleavage sites within the hepatitis C virus polyprotein. Virology. 2013;439:23–33. doi: 10.1016/j.virol.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for hepatitis C virus genomic and subgenomic RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindenbach BD, et al. Complete Replication of Hepatitis C Virus in Cell Culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 44.Giang E, et al. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6205–6210. doi: 10.1073/pnas.1114927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burton DR, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 46.Gao M, et al. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature. 2010;465:96–100. doi: 10.1038/nature08960. nature08960 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.