Abstract
Vibrio parahaemolyticus, autochthonous to estuarine, marine, and coastal environments throughout the world, is the causative agent of food-borne gastroenteritis. More than 80 serotypes have been described worldwide, based on antigenic properties of the somatic (O) and capsular (K) antigens. Serovar O3:K6 emerged in India in 1996 and subsequently was isolated worldwide, leading to the conclusion that the first V. parahaemolyticus pandemic had taken place. Most strains of V. parahaemolyticus isolated from the environment or seafood, in contrast to clinical strains, do not produce a thermostable direct hemolysin (TDH) and/or a TDH-related hemolysin (TRH). Type 3 secretion systems (T3SSs), needle-like apparatuses able to deliver bacterial effectors into host cytoplasm, were identified as triggering cytotoxicity and enterotoxicity. Type 6 secretion systems (T6SS) predicted to be involved in intracellular trafficking and vesicular transport appear to play a role in V. parahaemolyticus virulence. Recent advances in V. parahaemolyticus genomics identified several pathogenicity islands (VpaIs) located on either chromosome in both epidemic and pandemic strains and comprising additional colonization factors, such as restriction-modification complexes, chemotaxis proteins, classical bacterial surface virulence factors, and putative colicins. Furthermore, studies indicate strains lacking toxins and genomic regions associated with pathogenicity may also be pathogenic, suggesting other important virulence factors remain to be identified. The unique repertoire of virulence factors identified to date, their occurrence and distribution in both epidemic and pandemic strains worldwide are described, with the aim of highlighting the complexity of V. parahaemolyticus pathogenicity as well as its dynamic genome.
Keywords: V. parahaemolyticus, O3:K6, tdh, trh, PAI, virulence markers, pandemic strains, epidemic strains
Introduction
Some pathogenic bacteria occur naturally in coastal waters worldwide, of which a few can cause disease outbreaks, depending on specific environmental conditions. A good example is Vibrio parahaemolyticus, a Gram-negative halophilic bacterium autochthonous to estuarine, marine, and coastal environments. Occupying a variety of niches, V. parahaemolyticus can exist in a free-swimming state, with its motility conferred by a single polar flagellum, or sessile, attached to inert and animate surfaces, such as suspended particulate matter, zooplankton, fish, and shellfish (McCarter, 1999). Depending on environmental conditions, V. parahaemolyticus can produce a capsule with a number of different somatic (O) and capsular (K) antigens and these are employed as a primary basis of strain classification (Nair et al., 2007). Distribution of V. parahaemolyticus in the marine environment is related to water temperature and it is rarely isolated from seawater until the temperature rises to 15°C and higher (Kaneko and Colwell, 1973). Contamination of raw shellfish by V. parahaemolyticus is also known to be related to water temperature and it is more likely to occur during spring and summer months.
Since its discovery in 1950 (Fujino et al., 1953), V. parahaemolyticus has been recognized as a leading cause of seafood-derived food poisoning throughout the world. Virulent strains transmitted via consumption of raw or undercooked seafood are a common cause of acute gastroenteritis (Newton et al., 2012). Although the gastroenteritis may be self-limited, the infection can cause septicemia that is life-threatening to those with a preexisting medical conditions (Su and Liu, 2007). In Japan, V. parahaemolyticus is responsible for 20–30% of all food poisoning cases (Alam et al., 2002) and is the common cause of seafood-borne illness in many Asian countries (Koralage et al., 2012; Yu et al., 2013). V. parahaemolyticus has also become the leading agent of human gastroenteritis associated with seafood consumption in the United States (Newton et al., 2012).
The global occurrence of V. parahaemolyticus emphasizes the importance of understanding its many virulence factors and their effect on the human host. The objective of this study, therefore, was to review V. parahaemolyticus virulence associated factors that have been identified and described to date, and employ this information to provide a better understanding of V. parahaemolyticus pathogenesis.
V. parahaemolyticus distinctive virulence factors, tdh and trh
Although V. parahaemolyticus is a natural inhabitant of the estuarine and marine environment, only some strains have proven to be pathogenic (Oliver and Kaper, 2007). Almost all environmental isolates are negative in experimental animal diarrhogenic tests, whereas isolates from patients are positive (Shinoda, 2011). To date, the most distinctive factors of virulent strains are the thermostable direct hemolysin (TDH) (Nishibuchi et al., 1992) and TDH-related hemolysin (TRH) (Honda et al., 1988). Almost all V. parahaemolyticus strains isolated from clinical samples possess beta-hemolytic activity attributed to these two genes. Such isolates are able to lyse human erythrocytes when plated on high-salt Wagatsuma agar, a process designated as the Kanagawa phenomenon (KP) (Miyamoto et al., 1969; Joseph et al., 1982).
TDH is an amyloid toxin with two potential activities, one of which is disruption of lipid microdomains, abrogating cytotoxicity in the cell (Matsuda et al., 2010). The fairly large pore size allows both water and ions to flow through the membrane (Honda et al., 1992). These alterations in ion flux in the intestine ultimately prove to be the mechanism of the diarrhea observed during infection. However, even in the absence of these thermostable hemolysins V. parahaemolyticus remains pathogenic, indicating other virulence traits exist (Nishibuchi et al., 1992; Xu et al., 1994). Several tdh alleles sharing high nucleotide homology (>97%) have been identified on both chromosomes and plasmids (Nishibuchi and Kaper, 1995). The intensity of the hemolysis has also been associated with different transcriptional control and tdh gene copy in strains of V. parahaemolyticus (Nishibuchi and Kaper, 1990). The tdh gene is absent in most environmental strains of V. parahaemolyticus, but present in some strains of V. mimicus, V. cholerae non-O1/non-O139 and V. hollisae (Nishibuchi and Kaper, 1995; Shinoda, 2011).
KP-negative clinical strains of V. parahaemolyticus not possessing the tdh gene have been shown to produce a second hemolysin, TRH, which, unlike TDH, is heat labile and immunologically similar to TDH (Honda et al., 1988). EB101, the type strain of V. parahaemolyticus (Fujino et al., 1953) was reported to be KP-negative and later identified as tdh−/trh+ by molecular biological analysis (Shinoda, 2011). Both hemolysins (trh and tdh) share approximately 70% homology, with trh having higher nucleotide variability among alleles (Kishishita et al., 1992). TRH, believed to act similarly to TDH by activating Cl− channels, resulting in altered ion flux (Takahashi et al., 2000), was recently identified in Aeromonas veronii isolates (Raghunath et al., 2010).
Although correlation between pathogenicity of V. parahaemolyticus and presence of tdh and trh is well accepted, it should also be noted that not all clinical strains possess these genes. Several studies have reported about 10% of clinical strains do not contain tdh and trh, with the result that the overall mechanism of V. parahaemolyticus pathogenesis remains unclear (Miyamoto et al., 1969; Shirai et al., 1990; Okuda et al., 1997a). Association of a predominant serotype with human disease, namely V. parahaemolyticus O3:K6, and achievement of the first complete genome sequencing have shed some light on the virulence and epidemiological characteristics of V. parahaemolyticus.
V. parahaemolyticus O3:K6 as landmark of epidemiology
At the beginning of 1996 in Kolkata, India, during ongoing surveillance, an increase in patients with V. parahaemolyticus gastroenteritis was observed. Analysis of the strains revealed a new unique serotype, O3:K6, which accounted for 50–80% of infections during the following months. All O3:K6 isolates were tdh+/trh− and shared nearly identical genotype profiles (Okuda et al., 1997b). Within a few months thereafter, strains of the same serogroup were isolated in the neighboring countries of Vietnam, Indonesia, Bangladesh, Laos, Japan, Korea, and Thailand (Nair et al., 2007). Specific methods to identify the new clone, based on variation in nucleotide sequence of the toxRS region, were employed (Matsumoto et al., 2000) and the results showed existence of strains almost indistinguishable from the “new” O3:K6 clone, even though the isolates belonged to different serovars (Chowdhury et al., 2000a). To date, 21 serotypes have been identified, collectively referred to as serovariants of O3:K6, the most common being O4:K68, O1:K25, O1:K41, and O1:KUT (Nair et al., 2007). As anticipated by Chowdhury and colleagues, these serotypes differ from O3:K6 in altered O:K antigens, but are of clonal origin (Chowdhury et al., 2000a). By the end of 2006, V. parahaemolyticus O3:K6 and its serovariants were being isolated in Europe, Mozambique, the United States, Mexico, and South American countries (Nair et al., 2007), marking what has been claimed to be the beginning of the first pandemic of V. parahaemolyticus, bringing this pathogen to the forefront of the global public health agenda.
It is not obvious what environmental factors were associated with the global reach of these pandemic strains. Several efforts were made to determine factor(s) giving V. parahaemolyticus O3:K6 the ability to cause an increase in hospitalizations and to become the dominant serotype. Increased production of TDH was rapidly disproven since no substantial difference was detected between pandemic and non-pandemic strains (Okuda et al., 1997b). The V. parahaemolyticus O3:K6 and earlier isolates did not differ in survival under the same environmental conditions, such as extreme temperature, low pH, and/or elevated salinity (Wong et al., 2000). Yeung and colleagues suggested enhanced adherence and cytotoxicity may contribute to the pathogenic potential of V. parahaemolyticus O3:K6 isolates (Yeung et al., 2002), but the real advantage of O3:K6 over other strains remains unclear. Strains belonging to pandemic O3:K6 or related serogroups have been isolated from environmental samples in several countries, including Bangladesh (Islam et al., 2004), Japan (Hara-Kudo et al., 2003), India (Deepanjali et al., 2005), and Italy (Caburlotto et al., 2010a) suggesting they may prove endemic, if favorable niches occur in the environment.
Pandemic vs. non-pandemic strains and virulence characteristics
What has clearly emerged from analysis of V. parahaemolyticus O3:K6 strains and comparison with other serotypes is that the genetic organization of V. parahaemolyticus is more complex than expected. Results of molecular analysis, including ribotyping, AP-PCR, and PFGE (Okuda et al., 1997b; Chowdhury et al., 2000a,b; Matsumoto et al., 2000) showed O3:K6 isolates are genetically similar to each other and distinct from the O3:K6 strains isolated before 1995 and non-O3:K6 strains. Post-1995 V. parahaemolyticus O3:K6 isolates carry tdh but not trh and are defined by pandemic group-specific PCR (PGS-PCR) (Okura et al., 2003, 2004) and by a unique/new ToxR sequence within the toxRS operon, showing a slightly different nucleotide sequence compared to non-pandemic strains and likely involved in regulation of conserved virulence-associated genes in species of the genus Vibrio (Matsumoto et al., 2000).
These initial findings were the basis of the definition of pre- and post-pandemic V. parahaemolyticus strains of other serogroups beside O3:K6 (or serovariants) isolated before and after 1995. Differences observed among and between O3:K6 strains led to the definition of non-pandemic O3:K6 (or serovariants) as strains isolated between 1980–1990 in several Asian countries, including India, Taiwan, Japan, Thailand, and Bangladesh (Okuda et al., 1997b; Matsumoto et al., 2000; Osawa et al., 2002; Wong et al., 2005).
Nasu and colleagues reported that a wide collection of O3:K6 isolates had in common the filamentous phage f237 and suggested a specific association between the phage and widespread O3:K6 serotype (Nasu et al., 2000). Furthermore, they suggested that ORF8, located in the phage and encoding a putative adherence protein specific to post-1995 V. parahaemolyticus O3:K6 strains, might have played a significant role in increasing the virulence of O3:K6 isolates by their being more adhesive to host intestinal cells. However, many post-1995 V. parahaemolyticus strains lack phage f237 (Chowdhury et al., 2000b; Bhuiyan et al., 2002). The possible role of phages in pandemic V. parahaemolyticus infection was also suggested by identification of VfO4K68 in V. parahaemolyticus O4:K68 strains (Chan et al., 2002).
Okura and colleagues discovered that pandemic strains share the gene sequence VP2905, while non-pandemic group strains do not (Okura et al., 2005). This gene is located in a 16-kb region inserted in the open reading frame of the histone-like DNA-binding protein HU-α, causing a frameshift in the amino acid sequence (Williams et al., 2004). In enteric bacteria, this class of proteins has been associated with modification in drug resistance (Nishino and Yamaguchi, 2004). In Streptococcus it is related to tissue inflammation (Stinson et al., 1998). The effect on V. parahaemolyticus pathogenicity is unknown at this time.
Although these genetic traits appeared to be suitable markers for identification of pandemic strains, inconsistencies have been noted whereby pandemic O3:K6 strains with atypical profiles (lack of tdh or f237) have been isolated in Taiwan, Bangladesh, Japan, and Thailand (Bhuiyan et al., 2002; Osawa et al., 2002; Chowdhury et al., 2004; Jones et al., 2012) highlighting the complexity of pandemic V. parahaemolyticus, leaving genotyping an open question.
The first sequenced V. parahaemolyticus genome and discovery of T3SS
Pandemic V. parahaemolyticus O3:K6 strain RIMD 2210633 was isolated in Japan at the Kansai International Airport quarantine station in 1996 from a patient with travelers' diarrhea (Nasu et al., 2000). Makino and colleagues described its genome as comprising two circular chromosomes, with 4832 genes and showing rearrangements within and between the two chromosomes and the V. cholerae genome (Makino et al., 2003). A new type III secretion system, T3SS1, located on chromosome 1 was identified. Type 3 secretion systems (T3SSs) are a needle-like bacterial machine used to inject bacterial effectors directly into the membrane and cytoplasm of eukaryotic cells without encountering the extracellular environment (Cornelis, 2006). The T3SS1 gene cluster is composed of 42 genes, of which 30 are similar to those of other T3SS, e.g., Pseudomonas aeruginosa and Yersinia sp. (Makino et al., 2003; Park et al., 2004a). Three main effectors from T3SS1 were described: (i) VopQ induces rapid induction of autophagy in target cells preventing phagocytosis of the infecting bacteria (Sreelatha et al., 2013); (ii) VPA0450 destabilizes the cell by detachment of the plasma membrane from the actin cytoskeleton (Broberg et al., 2010); and (iii) VopS promotes collapse of the actin cytoskeleton, leading to cell rounding and shrinkage (Yarbrough et al., 2009). The role of a fourth effector located within the T3SS1 gene locus, VopR, has not been determined (Broberg et al., 2011). Collectively, T3SS1 effectors help V. parahaemolyticus to evade the host immune response, inducing autophagy followed by cell rounding and cell lysis (Higa et al., 2013). T3SS1 activities appear to be highly regulated by a dual system similar to the ExsACDE regulatory cascade of P. aeruginosa and negative regulation by H-NS (Kodama et al., 2010). Sharing a certain degree of homology with Yersinia spp. and other Vibrio species systems, the T3SS1 high sequence homology suggests these genes were ancestrally acquired and have been evolutionarily conserved (Ham and Orth, 2012). T3SS1 is well conserved and widespread in both clinical and environmental strains of V. parahaemolyticus (Park et al., 2004a; Meador et al., 2007; Noriea et al., 2010), being an essential characteristic to this species.
In addition to T3SS1, Makino et al. (2003) identified a pathogenicity island (Vp-PAI, currently referred to as VPaI-7) on chromosome 2 encoding several virulence-related genes, such as homologs of the Escherichia coli cytotoxic necrotizing factor (CNF) and Pseudomonas exoenzyme T (Makino et al., 2003). Associated with VPaI-7, typically flanked by two tdh genes, is a second type III secretion system, T3SS2-α, not similar to any T3SS of other bacteria, suggesting it is intrinsic to the species. Recent findings demonstrated that, along with its six effectors, T3SS2-α allows V. parahaemolyticus to invade, survive, and replicate in non-phagocytic host cells (Zhang et al., 2012). VopC has homologies to CNF1 described in pathogenic E. coli, and activates small GTPases Rac and CDC42 to induce changes in the actin cytoskeleton and facilitate V. parahaemolyticus entry into nonphagocytic host cells (Zhang et al., 2012). VopT is an ADP-ribosyltransferase able to modify the small GTPase Ras (Kodama et al., 2007) and is likely to have many effects during host infection such as cytotoxicity. VopA/P blocks activation of the MAPK signaling pathway which prevents induction of cytokines (Trosky et al., 2004). VopL polymerization activity is responsible for the strong actin filament nucleation observed in the host cell (Namgoong et al., 2011). VopV encodes an F-actin-binding protein involved in V. parahaemolyticus enterotoxicity (Hiyoshi et al., 2011). Finally, recently identified VopZ inhibits kinase TAK1 activation and is essential for V. parahaemolyticus induced diarrhea and tissue disruption (Zhou et al., 2013).
The co-presence of tdh genes and T3SS2-α on Vp-PAI was investigated and tdh was found to be co-regulated with T3SS2-α genes (Park et al., 2004a,b; Gotoh et al., 2010), suggesting a specific role of T3SS2-α and tdh in V. parahaemolyticus pathogenicity. Interestingly, T3SS2 was found only in Kanagawa-phenomenon-positive isolates and associated with enterotoxicity, whereas T3SS1 is ubiquitous in V. parahaemolyticus strains and correlated with cytotoxic activity (Park et al., 2004a,b; Meador et al., 2007; Izutsu et al., 2008).
Following the observation that trh could be located on a pathogenicity island (Iida et al., 1998), Okada and colleagues analyzed the surrounding region of the trh gene in strain TH3996 and discovered a novel T3SS2-α homolog inserted in pathogenicity island Vp-PAITH3996 (a.k.a. tdhPAI), T3SS2-β (Okada et al., 2009; Chen et al., 2011). They concluded that the new T3SS-related gene cluster does not occur in KP-positive strains, indicating a distinct lineage of T3SS2-related genes in tdh− and trh+ V. parahaemolyticus strains.
It was believed T3SS2-α and tdh were distinctive features of the TDH-producing (Kanagawa-phenomenon-positive) strains, exclusive to clinical isolates and associated with pandemic V. parahaemolyticus strains (Baker-Austin et al., 2010; Broberg et al., 2011). As in tdh+ strains, trh is linked with T3SS2-β, identified only in trh+ V. parahaemolyticus strains (Ottaviani et al., 2012). Recent findings question a direct correlation between T3SSs and tdh/trh genes. T3SS2-α and T3SS2-β, respectively, were found in tdh+/trh−andtdh−/trh+ strains, as expected, but also in V. parahaemolyticus environmental and clinical isolates, for which presence of tdh and trh was variable (Jones et al., 2012; Paranjpye et al., 2012).
Understanding the relationship between tdh, trh, and type III secretion systems in environmental isolates is of vital importance for validating hemolysin genes as virulence markers, as well as understanding the pathogenic potential of environmental strains.
Comparative genomics: pathogenicity islands and the dynamic genome of V. parahaemolyticus
Unlike V. cholerae, for which the genomes of well over 100 strains have been sequenced over the past 13 years, only a few V. parahaemolyticus strains have been completely sequenced and/or annotated to date (Table 1) (Boyd et al., 2008; Chen et al., 2011; Gonzalez-Escalona et al., 2011; Jensen et al., 2013; Jun et al., 2013; Liu and Chen, 2013). Nevertheless, since the first strain, RIMD 2210633, was sequenced (Makino et al., 2003), several comparative studies have identified new genomic islands (GIs). Acquisition of new genetic material by horizontal transfer plays a pivotal role in shaping the V. parahaemolyticus genome.
Table 1.
V. parahaemolyticus strains that have been sequenced.
| Strain | Isolation (date and location) | Serotype | tdh/trh | T3SSs/T6SS | Origin | Sequencing status | References |
|---|---|---|---|---|---|---|---|
| PRE-PANDEMIC | |||||||
| BB22OP | 1980s, Bangladesh | O4:K8 | +/− | +/+ | Env | Complete | Jensen et al., 2013 |
| AQ3810 | 1983, Singapore | O3:K6 | +/− | +/− | Clin | Draft | Boyd et al., 2008 |
| AQ4037 | 1985, Maldive | O3:K6 | −/+ | +/+ | Clin | Draft | Chen et al., 2011 |
| PANDEMIC | |||||||
| RIMD 2210633 | 1996, Japan | O3:K6 | +/− | +/+ | Clin | Complete | Makino et al., 2003 |
| Peru466 | 1996, Peru | O3:K6 | +/− | +/+ | Clin | Draft | Chen et al., 2011 |
| K5030 | 2005, India | O3:K6 | +/− | +/+ | Clin | Draft | Chen et al., 2011 |
| NON-PANDEMIC | |||||||
| 10329 | 1998, USA | O4:K12 | +/+ | ND/ND | Clin | Draft | Gonzalez-Escalona et al., 2011 |
| AN5034 | 1998, Bangladesh | O4:K68 | +/− | +/+ | Clin | Draft | Chen et al., 2011 |
| SNUVpS-1 | 2009, Korea | ND | −/− | ND/ND | Env | Draft | Jun et al., 2013 |
| v110 | 2010, Hong Kong | ND | −/− | +*/ND | Env | Draft | Liu and Chen, 2013 |
Env, environmental; Clin, clinical; ND, not determined.
Only T3SS1.
GIs are chromosomal regions usually acquired by horizontal gene transfer. They carry genes that can confer fitness advantage to their bacterial host. GIs encoding virulence determinants or colonization factors promote pathogenicity or survival of the bacterium in the host and are referred to as pathogenicity islands (Dobrindt et al., 2004).
Today it is known that V. parahaemolyticus contains nine GIs, VPaI-1 to VPaI-9, located on both chromosomes and variably distributed among different isolates (Hurley et al., 2006; Boyd et al., 2008). The nine GIs possess a phage-like integrase gene and are flanked by direct repeat sequences, with the exception of VPaI-7 that lacks an integrase but contains several transposase genes (Hurley et al., 2006). GIs range in size between 10 and 81 kb and their G+C content is lower than the overall genome G+C content (~45%) suggesting that these regions were acquired by horizontal gene transfer (Hurley et al., 2006; Boyd et al., 2008). The possible origin of the VPaIs is still not clearly understood. Blast analysis revealed the presence of homolog ORFs to some of the proteins encoded in VPaI-1, VPaI-2, VPaI-3, and VPaI-5 in different strains of V. cholerae, V. harveyi, and Shewanella sp. (Boyd et al., 2008). VPaIs have no close homologs in other Vibrionaceae but share the same insertion sites into tRNA-Met, tRNA-Ser or tmRNA loci as key pathogenicity islands of V. cholerae O1 (VPI-1, VPI-2, and VSP-2) and V. vulnificus (VVI-1, VVI-3, and VVI-7) (Hurley et al., 2006).
Analysis of VPaI-1 revealed a 22.79 kb pathogenicity island containing 24 open reading frames encoding proteins involved in DNA replication, transcription regulation, signal transduction, and general metabolism, as well as a type I restriction-modification complex and a DNA methyltransferase gene (Wang et al., 2006). By analogy with the mannose-fucose-resistant hemagglutinin (MFRHA) of V. cholerae O1 (Franzon et al., 1993), it is assumed that the DNA methyltransferase VP0394 comprises an additional colonization factor. Interestingly, the presence of this gene is strongly associated with tdh+ pandemic isolates, suggesting that VP0394 may confer unique virulence or fitness traits to these strains (Hurley et al., 2006; Wang et al., 2006). Comparison of VPaI-1 with other bacterial genomes revealed that an 8 kb region containing genes VP0389, VP0390, VP0391, and VP0392 is syntenic with chromosomal regions found in V. vulnificus CMCP6 (VVI-2) and Shewanella sp. MR-7 (Nishioka et al., 2008). The possible role of VP0390 and VP0392 in the cold adaptation of post-1995 V. parahaemolyticus strains has been hypothesized but further work needs to be done.
VPaI-2 to VPaI-6 range in size between 10 and 32 kb and encode putative virulence genes potentially involved in V. parahaemolyticus pathogenicity: outer membrane proteins and resolvases (VPaI-2); methyl accepting chemotaxis proteins (VPaI-3); putative pore forming cytotoxin integrase and M proteins likely involved in classical bacterial surface virulence factors (VPaI-4); phage-like protein that may encode a phage (VPaI-5); and putative colicin proteins (VPaI-6) (Hurley et al., 2006).
Involvement of VPaI-7 (formerly known as Vp-PAI, 8 kb) in cytotoxicity and enterotoxicity of V. parahaemolyticus has been described and correlated with presence of tdh and T3SS2-α (Makino et al., 2003; Park et al., 2004a,b; Sugiyama et al., 2008). Microarray analysis demonstrated association of VpaI-7 with pandemic strains (Izutsu et al., 2008). Comparison of V. parahaemolyticus O3:K6 and O4:K68 pandemic strains showed their genomes are similar in content. Furthermore, comparison between KP-positive and KP-negative strains highlighted that only the former contain VPaI-7, strongly suggesting that not only tdh but the entire region is required for pathogenicity of KP-positive clinical V. parahaemolyticus strains (Izutsu et al., 2008). Absence of VPaI-7 in KP-negative strains is compensated by Vp-PAITH3996 typical of tdh−/trh+ strains, encoding T3SS2-β (Okada et al., 2009). Recent studies suggest that, similar to clinical strains, marine isolates of V. parahaemolyticus carrying the T3SS2 effectors VopT and VopB2 and other genes included in VPaI-7 are capable of adhering to human cells and also causing cytoskeletal disruption and loss of membrane integrity in infected cells (Caburlotto et al., 2010b). These findings support the consideration of environmental V. parahaemolyticus strains as a risk to human health.
GIs VPaI-8 and VPaI-9 have recently been described and appear to be a feature of tdh+ pre-pandemic strains, namely AQ3810 (Boyd et al., 2008). VPaI-8 is 17 kb long and contains several ORFs encoding hypothetical proteins, two integrases, and homologs of KAP proteins. The latter are implicated in nearly all biochemical and mechanical processes in the cell, including replication and repair, intracellular trafficking, membrane transport, and activation of various metabolites (Aravind et al., 2004). VPaI-9 is a 22 kb region and, among other functions, encodes an excisionase, an helicase and a type I restriction modification system (Boyd et al., 2008). The role of these two GIs in V. parahaemolyticus pathogenicity has not been determined.
During the past few years GIs have been used as genetic markers to identify pandemic strains, with VPaI-1, VPaI-4, VPaI-5, and VPaI-6 found to be unique to post-1995 pandemic V. parahaemolyticus (Hurley et al., 2006), as well as VPaI-3 (Izutsu et al., 2008). VPaI-2 was described, not only in isolates post-1995 but also in isolates before 1995, prior to acquisition by the pandemic strains (Boyd et al., 2008).
Unsurprisingly, the scenario is more complex, as established by several exceptions. Not all pandemic strains contain VPaI-1 and VPaI-5 and tdh+ non-pandemic strains can be associated with VPaI-7 (Chao et al., 2009). Pandemic strains devoid of VPaI-4 have also been described, whereas VPaI-6 is common in non-pathogenic, pathogenic, and pandemic strains, indicating non-uniqueness of VPaI-6 to the pandemic clone (Chao et al., 2010). The presence of genomic regions characteristic of the pandemic clone in other non-pandemic strains provides evidence of genetic transfer, undoubtedly a major force in shaping virulence of V. parahaemolyticus.
T6SS, the ultimate weapon to compete for new niches
Comparison between pandemic and non-pandemic strains of V. parahaemolyticus led to identification of type VI secretion systems, T6SS1 and T6SS2, located on chromosome 1 and 2 of V. parahaemolyticus RIMD 2210633, respectively (Boyd et al., 2008; Izutsu et al., 2008). Homologs of Type 6 secretion system (T6SS) are present in V. alginolyticus, V. harveyi, and V. cholerae and are predicted to be involved in intracellular trafficking and vesicular transport (Boyd et al., 2008).
The role of T6SS2 is under analysis, but preliminary data suggest that it is not involved in cytotoxicity, as is the case for other bacterial T6SSs (Pukatzki et al., 2007), but is functional in adhesion to host cells (Yu et al., 2012). Functionality of T6SS1 has not yet been demonstrated. Since T6SS2 and T3SS2 co-exist, it was proposed that the two systems might cooperate during infection. T6SS2 plays its role in adhesion, the first step of infection, and T3SS2 exports effectors by inducing enterocytotoxicity (Park et al., 2004a; Yu et al., 2012).
A role for T6SSs in environmental fitness of V. parahaemolyticus has also been proposed. Salomon and colleagues elegantly established that T6SS1 is most active under warm marine-like conditions, while T6SS2 is active under low salt conditions and that surface sensing and quorum sensing differentially regulate both systems (Salomon et al., 2013). Moreover, they confirmed that V. parahaemolyticus employs T6SS1 to compete against other bacterial species, as well as against strains of its own species. This represents an advantage during summer months, when coastal waters are warm and increase in marine bacterial populations requires V. parahaemolyticus to compete for a niche (Salomon et al., 2013).
V. parahaemolyticus T6SS was used as a virulence marker to differentiate strains. Chao and colleagues reported that most pandemic strains isolated in China had the complete set of T6SS genes, whereas the majority of non-pathogenic strains had a partial set of T6SS genes (Chao et al., 2010). These results are consistent with the hypothesis that the entire set of T6SS genes is associated with pandemic strains (Boyd et al., 2008).
Conclusion and future perspectives
It is evident that the genome of V. parahaemolyticus is highly versatile, and the presence of pandemic genomic regions in non-pandemic strains provides evidence for horizontal gene transfer, shaping virulence, and evolution of V. parahaemolyticus.
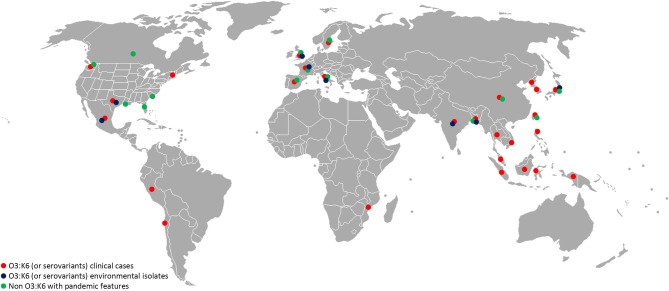
Virulence genes and pathogenicity islands are present in environmental strains and acquisition of these genes very likely occurs in the aquatic environment. Non-pathogenic Vibrio strains isolated in the Venetian Lagoon were found to contain remnants of V. parahaemolyticus VPaI-7, along with V. cholerae neuraminidase nanH and a modified version of pathogenicity island VPI-2 (Gennari et al., 2012). Also, environmental strains of non-O3:K6 V. parahaemolyticus carrying genetic markers associated with pandemic strains (tdh, orf8, toxRS/new) were recently isolated in Europe, the United States, Mexico, and Bangladesh (Alam et al., 2009; Caburlotto et al., 2010a; Jones et al., 2012; Velazquez-Roman et al., 2012) (Figure 1). These findings support the view that estuarine and marine bacteria comprise a significant reservoir of virulence and fitness genes. Such genes may provide selective advantage in the aquatic environment, enabling the bacteria to persist in the environment, with greater chance of encountering a susceptible host or exchange genetic material with other members of marine microbial communities in the same ecological niche. Exchange of genetic material occurs among autochthonous estuarine and marine bacteria and also human pathogens released via anthropogenic activities to the aquatic environment. Coastal water contamination with V. parahaemolyticus O3:K6 has been reported worldwide (Myers et al., 2003; Islam et al., 2004; Quilici et al., 2005; Ottaviani et al., 2010a; Powell et al., 2013). Clearly pathogenic strains of V. parahaemolyticus can be considered ubiquitous in the marine environment.
Figure 1.
Worldwide distribution of V. parahaemolyticus featuring virulence markers. Red, O3:K6 (or serovariants) clinical isolates; blue, O3:K6 (or serovariants) environmental isolates (water, seafood); green, non O3:K6 strains showing pandemic features (tdh, trh, PAI, T3SS, and/or T6SS). References to these data are a small portion of the published studies and should guide as an example. (Myers et al., 2003; Islam et al., 2004; Quilici et al., 2005; Nair et al., 2007; Ellingsen et al., 2008; Ottaviani et al., 2008, 2010b; Chao et al., 2009, 2010; Baker-Austin et al., 2010; Jones et al., 2012; Velazquez-Roman et al., 2012; Powell et al., 2013).
The rapid emergence of V. parahaemolyticus non-O3:K6 serogroups carrying pandemic markers (Figure 1) indicates predisposition of V. parahaemolyticus to genetic change. Jones and colleagues recently reported isolating clinical isolates negative for tdh, trh, and T3SS2, indicating the tdh and/or tdh and T3SS2 genes are not necessarily predictive of pathogenic potential (Jones et al., 2012).
In summary, review of the literature shows a higher level of complexity in virulence potential for V. parahaemolyticus than previously known and draws attention to the value of reliable virulence markers. How can a strain be identified as pandemic or merely pathogenic? The search for pandemic strains might well prove to be very complicated if horizontal gene transfer is as extensive in V. parahaemolyticus as it is in V. cholerae. Strains representing pandemic, non-pandemic but pathogenic, and non-pathogenic strains of V. parahaemolyticus must be rigorously analyzed to assess the role of mobile genetic elements in V. parahaemolyticus virulence. This is especially important as horizontal transfer of mobile genetic elements may lead to emergence of new pandemic clones with expanded ecological persistence, infectivity, and dispersion.
Author contributions
The project was conceived and designed by Daniela Ceccarelli, Nur A. Hasan, Anwar Huq, and Rita R. Colwell. The paper was written by Daniela Ceccarelli. All authors discussed, read, contributed to and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Work was supported in part by National Institutes of Health Grant No. 2RO1A1039129-11A2, and National Science Foundation Grant No. 0813066.
References
- Alam M., Chowdhury W. B., Bhuiyan N. A., Islam A., Hasan N. A., Nair G. B., et al. (2009). Serogroup, virulence, and genetic traits of Vibrio parahaemolyticus in the estuarine ecosystem of Bangladesh. Appl. Environ. Microbiol. 75, 6268–6274 10.1128/AEM.00266-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M., Tomochika K., Miyoshi S., Shinoda S. (2002). Environmental investigation of potentially pathogenic Vibrio parahaemolyticus in the Seto-Inland Sea, Japan. FEMS Microbiol. Lett. 208, 83–87 10.1111/j.1574-6968.2002.tb11064.x [DOI] [PubMed] [Google Scholar]
- Aravind L., Iyer L., Leipe D., Koonin E. (2004). A novel family of P-loop NTPases with an unusual phyletic distribution and transmembrane segments inserted within the NTPase domain. Genome Biol. 5, R30 10.1186/gb-2004-5-5-r30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker-Austin C., Stockley L., Rangdale R., Martinez-Urtaza J. (2010). Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: a European perspective. Environ. Microbiol. Rep. 2, 7–18 10.1111/j.1758-2229.2009.00096.x [DOI] [PubMed] [Google Scholar]
- Bhuiyan N. A., Ansaruzzaman M., Kamruzzaman M., Alam K., Chowdhury N. R., Nishibuchi M., et al. (2002). Prevalence of the pandemic genotype of Vibrio parahaemolyticus in Dhaka, Bangladesh, and significance of its distribution across different serotypes. J. Clin. Microbiol. 40, 284–286 10.1128/JCM.40.1.284-286.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd E., Cohen A., Naughton L., Ussery D., Binnewies T., Stine O., et al. (2008). Molecular analysis of the emergence of pandemic Vibrio parahaemolyticus. BMC Microbiol. 8:110 10.1186/1471-2180-8-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broberg C. A., Calder T. J., Orth K. (2011). Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect. 13, 992–1001 10.1016/j.micinf.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broberg C. A., Zhang L., Gonzalez H., Laskowski-Arce M. A., Orth K. (2010). A Vibrio effector protein is an inositol phosphatase and disrupts host cell membrane integrity. Science 329, 1660–1662 10.1126/science.1192850 [DOI] [PubMed] [Google Scholar]
- Caburlotto G., Gennari M., Ghidini V., Tafi M., Lleo M. M. (2010a). Serological and molecular characterization of Vibrio parahaemolyticus marine strains carrying pandemic genetic markers. ISME J. 4, 1071–1074 10.1038/ismej.2010.34 [DOI] [PubMed] [Google Scholar]
- Caburlotto G., Lleò M. M., Hilton T., Huq A., Colwell R. R., Kaper J. B. (2010b). Effect on human cells of environmental Vibrio parahaemolyticus strains carrying type III secretion system 2. Infect. Immun. 78, 3280–3287 10.1128/IAI.00050-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan B., Miyamoto H., Taniguchi H., Yoshida S. (2002). Isolation and genetic characterization of a novel filamentous bacteriophage, a deleted form of phage f237, from a pandemic Vibrio parahaemolyticus O4:K68 strain. Microbiol. Immunol. 46, 565–569 10.1111/j.1348-0421.2002.tb02734.x [DOI] [PubMed] [Google Scholar]
- Chao G., Jiao X., Zhou X., Wang F., Yang Z., Huang J., et al. (2010). Distribution of genes encoding four pathogenicity islands (VPaIs), T6SS, biofilm, and type I pilus in food and clinical strains of Vibrio parahaemolyticus in China. Foodborne Pathog. Dis. 7, 649–658 10.1089/fpd.2009.0441 [DOI] [PubMed] [Google Scholar]
- Chao G., Jiao X., Zhou X., Yang Z., Huang J., Pan Z., et al. (2009). Serodiversity, pandemic O3:K6 clone, molecular typing, and antibiotic susceptibility of foodborne and clinical Vibrio parahaemolyticus isolates in Jiangsu, China. Foodborne Pathog. Dis. 6, 1021–1028 10.1089/fpd.2009.0295 [DOI] [PubMed] [Google Scholar]
- Chen Y., Stine O. C., Badger J. H., Gil A. I., Nair G. B., Nishibuchi M., et al. (2011). Comparative genomic analysis of Vibrio parahaemolyticus: serotype conversion and virulence. BMC Genomics 12:294 10.1186/1471-2164-12-294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A., Ishibashi M., Thiem V., Tuyet D., Tung T., Chien B., et al. (2004). Emergence and serovar transition of Vibrio parahaemolyticus pandemic strains isolated during a diarrhea outbreak in Vietnam between 1997 and 1999. Microbiol. Immunol. 48, 319–327 10.1111/j.1348-0421.2004.tb03513.x [DOI] [PubMed] [Google Scholar]
- Chowdhury N., Chakraborty S., Eampokalap B., Chaicumpa W., Chongsa-Nguan M., Moolasart P., et al. (2000a). Clonal dissemination of Vibrio parahaemolyticus displaying similar DNA fingerprint but belonging to two different serovars (O3:K6 and O4:K68) in Thailand and India. Epidemiol. Infect. 125, 17–25 10.1017/S0950268899004070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury N. R., Chakraborty S., Ramamurthy T., Nishibuchi M., Yamasaki S., Takeda Y., et al. (2000b). Molecular evidence of clonal Vibrio parahaemolyticus pandemic strains. Emerg. Infect. Dis. 6, 631–636 10.3201/eid0606.000612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G. R. (2006). The type III secretion injectisome. Nat. Rev. Microbiol. 4, 811–825 10.1038/nrmicro1526 [DOI] [PubMed] [Google Scholar]
- Deepanjali A., Kumar H. S., Karunasagar I., Karunasagar I. (2005). Seasonal variation in abundance of total and pathogenic Vibrio parahaemolyticus bacteria in oysters along the southwest coast of India. Appl. Environ. Microbiol. 71, 3575–3580 10.1128/AEM.71.7.3575-3580.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrindt U., Hochhut B., Hentschel U., Hacker J. (2004). Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2, 414–424 10.1038/nrmicro884 [DOI] [PubMed] [Google Scholar]
- Ellingsen A. B., Jørgensen H., Wagley S., Monshaugen M., Rørvik L. M. (2008). Genetic diversity among Norwegian Vibrio parahaemolyticus. J. Appl. Microbiol. 105, 2195–2202 10.1111/j.1365-2672.2008.03964.x [DOI] [PubMed] [Google Scholar]
- Franzon V. L., Barker A., Manning P. A. (1993). Nucleotide sequence encoding the mannose-fucose-resistant hemagglutinin of Vibrio cholerae O1 and construction of a mutant. Infect. Immun. 61, 3032–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino T., Okuno Y., Nakada D., Aoyama A., Fukai K., Mukai T., et al. (1953). On the bacteriological examination of shirasu-food poisoning. Med. J. Osaka Univ. 4, 299–304 [Google Scholar]
- Gennari M., Ghidini V., Caburlotto G., Lleo M. M. (2012). Virulence genes and pathogenicity islands in environmental Vibrio strains nonpathogenic to humans. FEMS Microbiol. Ecol. 82, 563–573 10.1111/j.1574-6941.2012.01427.x [DOI] [PubMed] [Google Scholar]
- Gonzalez-Escalona N., Strain E. A., De Jesus A. J., Jones J. L., Depaola A. (2011). Genome sequence of the clinical O4:K12 serotype Vibrio parahaemolyticus strain 10329. J. Bacteriol. 193, 3405–3406 10.1128/JB.05044-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh K., Kodama T., Hiyoshi H., Izutsu K., Park K.-S., Dryselius R., et al. Iida T. (2010). Bile acid-induced virulence gene expression of Vibrio parahaemolyticus reveals a novel therapeutic potential for bile acid sequestrants. PLoS ONE 5:e13365 10.1371/journal.pone.0013365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham H., Orth K. (2012). The role of type III secretion system 2 in Vibrio parahaemolyticus pathogenicity. J. Microbiol. 50, 719–725 10.1007/s12275-012-2550-2 [DOI] [PubMed] [Google Scholar]
- Hara-Kudo Y., Sugiyama K., Nishibuchi M., Chowdhury A., Yatsuyanagi J., Ohtomo Y., et al. (2003). Prevalence of pandemic thermostable direct hemolysin-producing Vibrio parahaemolyticus O3:K6 in seafood and the coastal environment in Japan. Appl. Environ. Microbiol. 69, 3883–3891 10.1128/AEM.69.7.3883-3891.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa N., Toma C., Koizumi Y., Nakasone N., Nohara T., Masumoto J., et al. (2013). Vibrio parahaemolyticus effector proteins suppress inflammasome activation by interfering with host autophagy signaling. PLoS Pathog. 9:e1003142 10.1371/journal.ppat.1003142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyoshi H., Kodama T., Saito K., Gotoh K., Matsuda S., Akeda Y., et al. (2011). VopV, an F-actin-binding type III secretion effector, is required for Vibrio parahaemolyticus-induced enterotoxicity. Cell Host Microbe 10, 401–409 10.1016/j.chom.2011.08.014 [DOI] [PubMed] [Google Scholar]
- Honda T., Ni Y., Miwatani T., Adachi T., Kim J. (1992). The thermostable direct hemolysin of Vibrio parahaemolyticus is a pore-forming toxin. Can. J. Microbiol. 38, 1175–1180 10.1139/m92-192 [DOI] [PubMed] [Google Scholar]
- Honda T., Ni Y. X., Miwatani T. (1988). Purification and characterization of a hemolysin produced by a clinical isolate of Kanagawa phenomenon-negative Vibrio parahaemolyticus and related to the thermostable direct hemolysin. Infect. Immun. 56, 961–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley C., Quirke A., Reen F., Boyd E. (2006). Four genomic islands that mark post-1995 pandemic Vibrio parahaemolyticus isolates. BMC Genomics 7:104 10.1186/1471-2164-7-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida T., Park K.-S., Suthienkul O., Kozawa J., Yamaichi Y., Yamamoto K., et al. (1998). Close proximity of the tdh, trh and ure genes on the chromosome of Vibrio parahaemolyticus. Microbiology 144, 2517–2523 10.1099/00221287-144-9-2517 [DOI] [PubMed] [Google Scholar]
- Islam M. S., Tasmin R., Khan S. I., Bakht H. B. M., Mahmood Z. H., Rahman M. Z., et al. (2004). Pandemic strains of O3:K6 Vibrio parahaemolyticus in the aquatic environment of Bangladesh. Can. J. Microbiol. 50, 827–834 10.1139/w04-072 [DOI] [PubMed] [Google Scholar]
- Izutsu K., Kurokawa K., Tashiro K., Kuhara S., Hayashi T., Honda T., et al. (2008). Comparative genomic analysis using microarray demonstrates a strong correlation between the presence of the 80-kilobase pathogenicity island and pathogenicity in Kanagawa phenomenon-positive Vibrio parahaemolyticus strains. Infect. Immun. 76, 1016–1023 10.1128/IAI.01535-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R. V., Depasquale S. M., Harbolick E. A., Hong T., Kernell A. L., Kruchko D. H., et al. (2013). Complete genome sequence of prepandemic Vibrio parahaemolyticus BB22OP. Genome Announc. 1, 00002–00012 10.1128/genomeA.00002-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. L., Ludeke C. H. M., Bowers J. C., Garrett N., Fischer M., Parsons M. B., et al. (2012). Biochemical, serological, and virulence characterization of clinical and oyster Vibrio parahaemolyticus Isolates. J. Clin. Microbiol. 50, 2343–2352 10.1128/JCM.00196-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph S., Colwell R., Kaper J. (1982). Vibrio parahaemolyticus and related halophilic Vibrios. Crit. Rev. Microbiol. 10, 77–124 10.3109/10408418209113506 [DOI] [PubMed] [Google Scholar]
- Jun J. W., Kim J. H., Choresca C. H., Shin S. P., Han J. E., Park S. C. (2013). Draft genome sequence of Vibrio parahaemolyticus SNUVpS-1 isolated from Korean seafood. Genome Announc. 1, 00132–00112 10.1128/genomeA.00132-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T., Colwell R. R. (1973). Ecology of Vibrio parahaemolyticus in Chesapeake Bay. J. Bacteriol. 113, 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishishita M., Matsuoka N., Kumagai K., Yamasaki S., Takeda Y., Nishibuchi M. (1992). Sequence variation in the thermostable direct hemolysin-related hemolysin (trh) gene of Vibrio parahaemolyticus. Appl. Environ. Microbiol. 58, 2449–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama T., Rokuda M., Park K.-S., Cantarelli V. V., Matsuda S., Iida T., et al. (2007). Identification and characterization of VopT, a novel ADP-ribosyltransferase effector protein secreted via the Vibrio parahaemolyticus type III secretion system 2. Cell. Microbiol. 9, 2598–2609 10.1111/j.1462-5822.2007.00980.x [DOI] [PubMed] [Google Scholar]
- Kodama T., Yamazaki C., Park K.-S., Akeda Y., Iida T., Honda T. (2010). Transcription of Vibrio parahaemolyticus T3SS1 genes is regulated by a dual regulation system consisting of the ExsACDE regulatory cascade and H-NS. FEMS Microbiol. Lett. 311, 10–17 10.1111/j.1574-6968.2010.02066.x [DOI] [PubMed] [Google Scholar]
- Koralage M., Alter T., Pichpol D., Strauch E., Zessin K., Huehn S. (2012). Prevalence and molecular characteristics of Vibrio spp. isolated from preharvest shrimp of the North Western Province of Sri Lanka. J. Food Prot. 75, 1846–1850 10.4315/0362-028X.JFP-12-115 [DOI] [PubMed] [Google Scholar]
- Liu M., Chen S. (2013). Draft genome sequence of Vibrio parahaemolyticus V110, isolated from shrimp in Hong Kong. Genome Announc. 1, 00300–00313 10.1128/genomeA.00300-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino K., Oshima K., Kurokawa K., Yokoyama K., Uda T., Tagomori K., et al. (2003). Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361, 743–749 10.1016/S0140-6736(03)12659-1 [DOI] [PubMed] [Google Scholar]
- Matsuda S., Kodama T., Okada N., Okayama K., Honda T., Iida T. (2010). Association of Vibrio parahaemolyticus thermostable direct hemolysin with lipid rafts is essential for cytotoxicity but not hemolytic activity. Infect. Immun. 78, 603–610 10.1128/IAI.00946-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto C., Okuda J., Ishibashi M., Iwanaga M., Garg P., Rammamurthy T., et al. (2000). Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J. Clin. Microbiol. 38, 578–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter L. (1999). The multiple identities of Vibrio parahaemolyticus. J. Mol. Microbiol. Biotechnol. 1, 51–57 [PubMed] [Google Scholar]
- Meador C. E., Parsons M. M., Bopp C. A., Gerner-Smidt P., Painter J. A., Vora G. J. (2007). Virulence gene- and pandemic group-specific marker profiling of clinical Vibrio parahaemolyticus isolates. J. Clin. Microbiol. 45, 1133–1139 10.1128/JCM.00042-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y., Kato T., Obara Y., Akiyama S., Takizawa K., Yamai S. (1969). In vitro hemolytic characteristic of Vibrio parahaemolyticus: its close correlation with human pathogenicity. J. Bacteriol. 100, 1147–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers M. L., Panicker G., Bej A. K. (2003). PCR detection of a newly emerged pandemic Vibrio parahaemolyticus O3:K6 pathogen in pure cultures and seeded waters from the Gulf of Mexico. Appl. Environ. Microbiol. 69, 2194–2200 10.1128/AEM.69.4.2194-2200.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair G. B., Ramamurthy T., Bhattacharya S. K., Dutta B., Takeda Y., Sack D. A. (2007). Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 20, 39–48 10.1128/CMR.00025-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namgoong S., Boczkowska M., Glista M. J., Winkelman J. D., Rebowski G., Kovar D. R., et al. (2011). Mechanism of actin filament nucleation by Vibrio VopL and implications for tandem W domain nucleation. Nat. Struct. Mol. Biol. 18, 1060–1067 10.1038/nsmb.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasu H., Iida T., Sugahara T., Yamaichi Y., Park K.-S., Yokoyama K., et al. (2000). A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3:K6 strains. J. Clin. Microbiol. 38, 2156–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton A., Kendall M., Vugia D. J., Henao O. L., Mahon B. E. (2012). Increasing rates of vibriosis in the United States, 1996–2010: review of surveillance data from 2 systems. Clin. Infect. Dis. 54, S391–S395 10.1093/cid/cis243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishibuchi M., Fasano A., Russell R. G., Kaper J. B. (1992). Enterotoxigenicity of Vibrio parahaemolyticus with and without genes encoding thermostable direct hemolysin. Infect. Immun. 60, 3539–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishibuchi M., Kaper J. B. (1990). Duplication and variation of the thermostable direct haemolysin (tdh) gene in Vibrio parahaemolyticus. Mol. Microbiol. 4, 87–99 10.1111/j.1365-2958.1990.tb02017.x [DOI] [PubMed] [Google Scholar]
- Nishibuchi M., Kaper J. B. (1995). Thermostable direct hemolysin gene of Vibrio parahaemolyticus: a virulence gene acquired by a marine bacterium. Infect. Immun. 63, 2093–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino K., Yamaguchi A. (2004). Role of histone-like protein H-NS in multidrug resistance of Escherichia coli. J. Bacteriol. 186, 1423–1429 10.1128/JB.186.5.1423-1429.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka T., Kamruzzaman M., Nishibuchi M., Satta Y. (2008). On the origin and function of an insertion element VPaI-1 specific to post-1995 pandemic Vibrio parahaemolyticus strains. Genes Genet. Syst. 83, 101–110 10.1266/ggs.83.101 [DOI] [PubMed] [Google Scholar]
- Noriea N. F., 3rd., Johnson C. N., Griffitt K. J., Grimes D. J. (2010). Distribution of type III secretion systems in Vibrio parahaemolyticus from the northern Gulf of Mexico. J. Appl. Microbiol. 109, 953–962 10.1111/j.1365-2672.2010.04722.x [DOI] [PubMed] [Google Scholar]
- Okada N., Iida T., Park K.-S., Goto N., Yasunaga T., Hiyoshi H., et al. (2009). Identification and characterization of a novel type III secretion system in trh-positive Vibrio parahaemolyticus strain TH3996 reveal genetic lineage and diversity of pathogenic machinery beyond the species level. Infect. Immun. 77, 904–913 10.1128/IAI.01184-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda J., Ishibashi M., Abbott S., Janda J., Nishibuchi M. (1997a). Analysis of the thermostable direct hemolysin (tdh) gene and the tdh-related hemolysin (trh) genes in urease-positive strains of Vibrio parahaemolyticus isolated on the West Coast of the United States. J. Clin. Microbiol. 35, 1965–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda J., Ishibashi M., Hayakawa E., Nishino T., Takeda Y., Mukhopadhyay A. K., et al. (1997b). Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J. Clin. Microbiol. 35, 3150–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okura M., Osawa R., Arakawa E., Terajima J., Watanabe H. (2005). Identification of Vibrio parahaemolyticus pandemic group-specific DNA sequence by genomic subtraction. J. Clin. Microbiol. 43, 3533–3536 10.1128/JCM.43.7.3533-3536.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okura M., Osawa R., Iguchi A., Arakawa E., Terajima J., Watanabe H. (2003). Genotypic analyses of Vibrio parahaemolyticus and development of a pandemic group-specific multiplex PCR assay. J. Clin. Microbiol. 41, 4676–4682 10.1128/JCM.41.10.4676-4682.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okura M., Osawa R., Iguchi A., Takagi M., Arakawa E., Terajima J., et al. (2004). PCR-based identification of pandemic group Vibrio parahaemolyticus with a novel group-specific primer pair. Microbiol. Immunol. 48, 787–790 10.1111/j.1348-0421.2004.tb03596.x [DOI] [PubMed] [Google Scholar]
- Oliver J. D., Kaper J. (2007). Vibrio species, in Food Microbiology: Fundamentals and Frontiers, 3rd Edn., eds Doyle M. P., Beuchat L. R., Montville T. J. (Washington, DC: ASM; ), 343–379 [Google Scholar]
- Osawa R., Iguchi A., Arakawa E., Watanabe H. (2002). Genotyping of pandemic Vibrio parahaemolyticus O3:K6 still open to question. J. Clin. Microbiol. 40, 2708–2709 10.1128/JCM.40.7.2708-2709.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani D., Leoni F., Rocchegiani E., Canonico C., Potenziani S., Santarelli S., et al. (2010a). Prevalence, serotyping and molecular characterization of Vibrio parahaemolyticus in mussels from Italian growing areas, Adriatic Sea. Environ. Microbiol. Rep. 2, 192–197 10.1111/j.1758-2229.2010.00134.x [DOI] [PubMed] [Google Scholar]
- Ottaviani D., Leoni F., Rocchegiani E., Canonico C., Potenziani S., Santarelli S., et al. (2010b). Vibrio parahaemolyticus-associated gastroenteritis in Italy: persistent occurrence of O3:K6 pandemic clone and emergence of O1:KUT serotype. Diagn. Microbiol. Infect. Dis. 66, 452–455 10.1016/j.diagmicrobio.2009.11.015 [DOI] [PubMed] [Google Scholar]
- Ottaviani D., Leoni F., Rocchegiani E., Santarelli S., Canonico C., Masini L., et al. (2008). First clinical report of pandemic Vibrio parahaemolyticus O3:K6 infection in Italy. J. Clin. Microbiol. 46, 2144–2145 10.1128/JCM.00683-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani D., Leoni F., Serra R., Serracca L., Decastelli L., Rocchegiani E., et al. (2012). Nontoxigenic Vibrio parahaemolyticus strains causing acute gastroenteritis. J. Clin. Microbiol. 50, 4141–4143 10.1128/JCM.01993-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjpye R., Hamel O. S., Stojanovski A., Liermann M. (2012). Genetic diversity of clinical and environmental Vibrio parahaemolyticus strains from the Pacific Northwest. Appl. Environ. Microbiol. 78, 8631–8638 10.1128/AEM.01531-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K.-S., Ono T., Rokuda M., Jang M.-H., Okada K., Iida T., et al. (2004a). Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect. Immun. 72, 6659–6665 10.1128/IAI.72.11.6659-6665.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K., Ono T., Rokuda M., Jang M., Iida T., Honda T. (2004b). Cytotoxicity and enterotoxicity of the thermostable direct hemolysin-deletion mutants of Vibrio parahaemolyticus. Microbiol. Immunol. 48, 313–318 10.1111/j.1348-0421.2004.tb03512.x [DOI] [PubMed] [Google Scholar]
- Powell A., Baker-Austin C., Wagley S., Bayley A., Hartnell R. (2013). Isolation of pandemic Vibrio parahaemolyticus from UK water and shellfish produce. Microb. Ecol. 65, 924–927 10.1007/s00248-013-0201-8 [DOI] [PubMed] [Google Scholar]
- Pukatzki S., Ma A. T., Revel A. T., Sturtevant D., Mekalanos J. J. (2007). Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U.S.A. 104, 15508–15513 10.1073/pnas.0706532104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilici M., Robert-Pillot A., Picart J., Fournier J. (2005). Pandemic Vibrio parahaemolyticus O3:K6 spread, France. Emerg. Infect. Dis. 11, 1148–1149 10.3201/eid1107.041008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunath P., Maiti B., Shekar M., Karunasagar I., Karunasagar I. (2010). Clinical isolates of Aeromonas veronii biovar veronii harbor a nonfunctional gene similar to the thermostable direct hemolysin-related hemolysin (trh) gene of Vibrio parahaemolyticus. FEMS Microbiol. Lett. 307, 151–157 10.1111/j.1574-6968.2010.01974.x [DOI] [PubMed] [Google Scholar]
- Salomon D., Gonzalez H., Updegraff B. L., Orth K. (2013). Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS ONE 8:e61086 10.1371/journal.pone.0061086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda S. (2011). Sixty years from the discovery of Vibrio parahaemolyticus and some recollections. Biocontrol Sci. 16, 129–137 10.4265/bio.16.129 [DOI] [PubMed] [Google Scholar]
- Shirai H., Ito H., Hirayama T., Nakamoto Y., Nakabayashi N., Kumagai K., et al. (1990). Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect. Immun. 58, 3568–3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreelatha A., Bennett T. L., Zheng H., Jiang Q.-X., Orth K., Starai V. J. (2013). Vibrio effector protein, VopQ, forms a lysosomal gated channel that disrupts host ion homeostasis and autophagic flux. Proc. Natl. Acad. Sci. U.S.A. 110, 11559–11564 10.1073/pnas.1307032110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinson M. W., McLaughlin R., Choi S. H., Juarez Z. E., Barnard J. (1998). Streptococcal histone-like protein: primary structure of hlpA and protein binding to lipoteichoic acid and epithelial cells. Infect. Immun. 66, 259–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y. C., Liu C. (2007). Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol. 24, 549–558 10.1016/j.fm.2007.01.005 [DOI] [PubMed] [Google Scholar]
- Sugiyama T., Iida T., Izutsu K., Park K.-S., Honda T. (2008). Precise region and the character of the pathogenicity island in clinical Vibrio parahaemolyticus strains. J. Bacteriol. 190, 1835–1837 10.1128/JB.01293-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A., Kenjyo N., Imura K., Myonsun Y., Honda T. (2000). Cl- secretion in colonic epithelial cells induced by the Vibrio parahaemolyticus hemolytic toxin related to thermostable direct hemolysin. Infect. Immun. 68, 5435–5438 10.1128/IAI.68.9.5435-5438.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trosky J. E., Mukherjee S., Burdette D. L., Roberts M., McCarter L., Siegel R. M., et al. (2004). Inhibition of MAPK signaling pathways by VopA from Vibrio parahaemolyticus. J. Biol. Chem. 279, 51953–51957 10.1074/jbc.M407001200 [DOI] [PubMed] [Google Scholar]
- Velazquez-Roman J., León-Sicairos N., Flores-Villaseñor H., Villafaña-Rauda S., Canizalez-Roman A. (2012). Association of pandemic Vibrio parahaemolyticus O3:K6 present in the coastal environment of Northwest Mexico with cases of recurrent diarrhea between 2004 and 2010. Appl. Environ. Microbiol. 78, 1794–1803 10.1128/AEM.06953-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.-Z., Wong M. M. L., O'Toole D., Mak M. M. H., Wu R. S. S., Kong R. Y. C. (2006). Identification of a DNA methyltransferase gene carried on a pathogenicity island-like element (VPAI) in Vibrio parahaemolyticus and its prevalence among clinical and environmental isolates. Appl. Environ. Microbiol. 72, 4455–4460 10.1128/AEM.02095-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T. L., Musser S. M., Nordstrom J. L., Depaola A., Monday S. R. (2004). Identification of a protein biomarker unique to the pandemic O3:K6 clone of Vibrio parahaemolyticus. J. Clin. Microbiol. 42, 1657–1665 10.1128/JCM.42.4.1657-1665.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H. C., Chen C. H., Chung Y. J., Liu S. H., Wang T. K., Lee C. L., et al. (2005). Characterization of new O3:K6 strains and phylogenetically related strains of Vibrio parahaemolyticus isolated in Taiwan and other countries. J. Appl. Microbiol. 98, 572–580 10.1111/j.1365-2672.2004.02478.x [DOI] [PubMed] [Google Scholar]
- Wong H.-C., Liu S.-H., Wang T.-K., Lee C.-L., Chiou C.-S., Liu D.-P., et al. (2000). Characteristics of Vibrio parahaemolyticus O3:K6 from Asia. Appl. Environ. Microbiol. 66, 3981–3986 10.1128/AEM.66.9.3981-3986.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Yamamoto K., Honda T., X M. (1994). Construction and characterization of an isogenic mutant of Vibrio parahaemolyticus having a deletion in the thermostable direct hemolysin-related hemolysin gene (trh). J. Bacteriol. 176, 4757–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarbrough M. L., Li Y., Kinch L. N., Grishin N. V., Ball H. L., Orth K. (2009). AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science 323, 269–272 10.1126/science.1166382 [DOI] [PubMed] [Google Scholar]
- Yeung P. S. M., Hayes M. C., Depaola A., Kaysner C. A., Kornstein L., Boor K. J. (2002). Comparative phenotypic, molecular, and virulence characterization of Vibrio parahaemolyticus O3:K6 isolates. Appl. Environ. Microbiol. 68, 2901–2909 10.1128/AEM.68.6.2901-2909.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W. T., Jong K. J., Lin Y. R., Tsai S. E., Tey Y. H., Wong H. C. (2013). Prevalence of Vibrio parahaemolyticus in oyster and clam culturing environments in Taiwan. Int. J. Food Microbiol. 160, 185–192 10.1016/j.ijfoodmicro.2012.11.002 [DOI] [PubMed] [Google Scholar]
- Yu Y., Yang H., Li J., Zhang P., Wu B., Zhu B., et al. (2012). Putative type VI secretion systems of Vibrio parahaemolyticus contribute to adhesion to cultured cell monolayers. Arch. Microbiol. 194, 827–835 10.1007/s00203-012-0816-z [DOI] [PubMed] [Google Scholar]
- Zhang L., Krachler A. M., Broberg C. A., Li Y., Mirzaei H., Gilpin C. J., et al. (2012). Type III effector VopC mediates invasion for Vibrio species. Cell Reports 1, 453–460 10.1016/j.celrep.2012.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Gewurz B. E., Ritchie J. M., Takasaki K., Greenfeld H., Kieff E., et al. (2013). A Vibrio parahaemolyticus T3SS effector mediates pathogenesis by independently enabling intestinal colonization and inhibiting TAK1 activation. Cell Rep. 3, 1690–1702 10.1016/j.celrep.2013.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]