Abstract
The restart of a stalled replication fork is a major challenge for DNA replication. Depending upon the nature of the damage, different repair processes might be triggered; one is template switching that is bypass of a leading strand lesion via fork regression. Using magnetic tweezers to study the T4 bacteriophage enzymes, we have reproduced in vitro the complete process of template switching. We show that the UvsW DNA helicase in cooperation with the T4 holoenzyme can overcome leading strand lesion damage by a pseudo stochastic process periodically forming and migrating a four ways Holliday junction. The initiation of the repair process requires partial replisome disassembly via the departure of the replicative helicase. The results support the role of fork regression pathways in DNA repair.
DNA damage causes the replication fork to stall or to collapse and is responsible for illegitimate recombination and cellular dysfunction (1, 2). While a damage in the lagging strand is less likely to block fork progression; a damage in the leading strand is a particular challenge due to the continuous nature of the leading strand synthesis. The fork regression pathway is one means to overcome leading strand lesions as generally described in terms of four steps (fig. S1): 1) replisome disassembly, 2) stalled fork regression to form a Holliday junction (HJ) structure (also called a chicken-foot structure), 3) either (II) lesion excision repair or (I) template switching (3,4), that is polymerase extension of the 3′ end of the leading strand, now annealed to an intact template, downstream of the lesion (referred here as a lesion bypass), 4) restoration of the fork and reloading of the replisome (2). However, the details of such a pathway and the biological role of regressed forks in vivo are poorly understood (2, 5).
The T4 bacteriophage lacks translesion polymerases and unlike E. coli cannot reinitiate synthesis by re-priming the leading strand (6, 7). Hence the T4 phage may exclusively use fork regression to bypass leading strand lesions via an active helicase driven pathway involving the UvsW protein (5). UvsW is a functional analog of E. coli RecG helicase and has properties similar to the RecQ helicase family that includes human enzymes whose defects contribute to various disease states (8–10). With its dual unwinding and annealing activities (11), UvsW is capable of regressing forks in vitro (5, 6). In the T4 system, when the replisome encounters a leading strand lesion, replication in the lagging strand continues at least one Okazaki fragment beyond the lesion site (6). This uncoupled synthesis results in a DNA structure that can be effectively regressed by UvsW helicase to generate the HJ structure required for template switching (fig. S1(I)). However, the reconstruction of the full template switching pathway is missing..
We have investigated lesion bypass via template switching using magnetic tweezers to manipulate a DNA hairpin and follow the activity of the UvsW helicase alone and in collaboration with different components of the replisome. Experiments were carried out by tethering the DNA hairpin between a glass surface and a magnetic bead (Fig. 1A). A pair of magnets was used to apply tension to the ends of the hairpin while the extension of the DNA molecule Ze(t) was obtained from tracking the position of the bead (12, 29). Three different hairpins provided either adapted sequences or modified bases (fig. S2 and (29)): 7 Kbp Long (Lh), 1.2 Kbp Medium (Mh) and 100 bp Short (Sh) hairpins. By applying a force of ~ 15 pN we trapped Mh molecules in an intermediate configuration where the initial stem was denatured but the GC rich region before the apex remained intact (fig. S2D). This partially denatured hairpin was an ideal substrate to simultaneously test UvsW annealing and unwinding activities (Fig. 1B). The changes in Ze(t) were converted to number of unwound or annealed base pairs using the ssDNA elasticity (fig. S3)
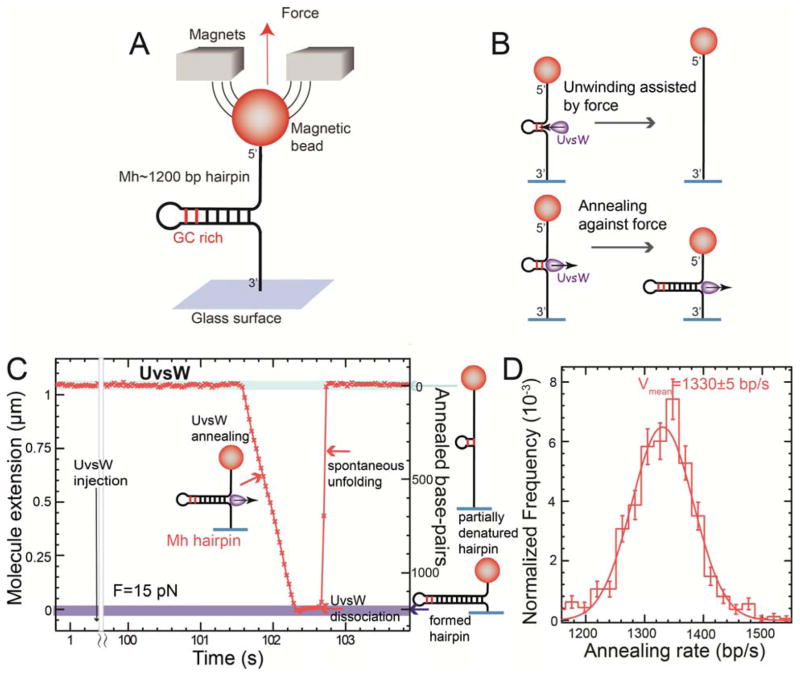
Fig. 1.
Single-molecule characterization of UvsW annealing activity (A) Schematic representation of the experimental set-up. A DNA hairpin substrate was tethered between the glass surface and a magnetic bead held in a MT. The GC rich region in Mh is shown in red. (B) Schematics of the assay allowing for unwinding and annealing detection. (C) Experimental trace showing Ze(t) obtained in the MT assays with the partial denatured hairpin at F = 15 pN with ATP and UvsW. Annealing bursts were detected as decreases in Ze. (D) Distribution of annealing rates dZe/dt (N==766 events) with a Gaussian fit. Error bars are inversely proportional to the square root of the number of points for each bin.
In the presence of UvsW and ATP, we observed events corresponding to the zipping of the hairpin, followed by the spontaneous recovery of the initial molecular extension (Fig. 1C). After the annealing burst UvsW remained bound maintaining the formed hairpin for a fraction of a second before dissociating. The rate of annealing and the enzyme’s processivity measured at 15 pN were very large, ~1300 bp/s and ~9 Kbp respectively (Fig. 1D and fig. S4). UvsW annealing activity was also tested with an ensemble FRET-based assay (fig. S5). We found that the UvsW annealing activity is an active process that requires ATP hydrolysis, with KM and kcat of 57 μM and 1280 bp/s, respectively (fig. S6). Unwinding by UvsW was only observed when annealing was prevented (closed hairpin at low forces, fig. S7) or in resolving branched structures requiring combined annealing and unwinding activities (fig. S8). These results showed that UvsW possessed both unwinding and annealing activities, but favored annealing.
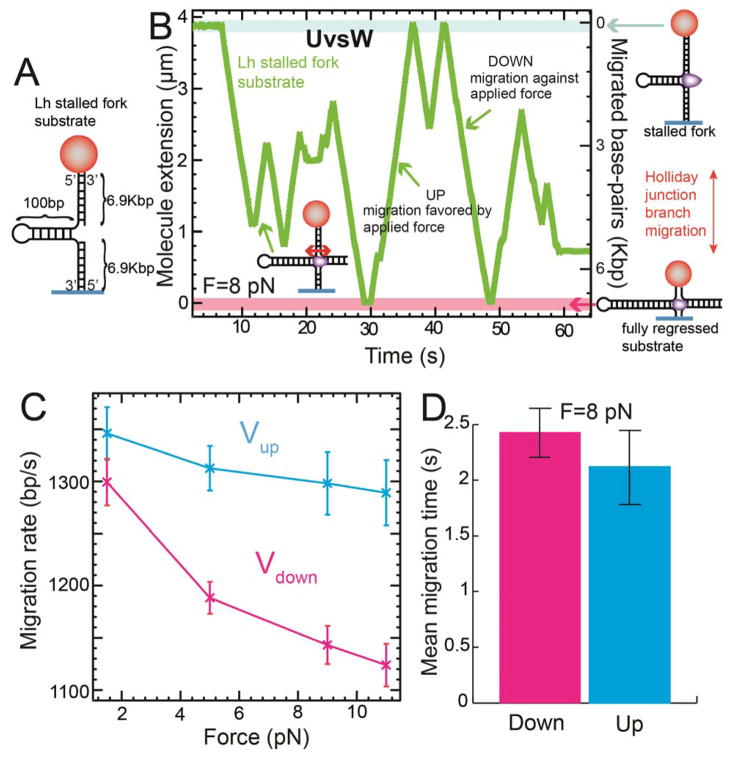
Ensemble kinetic studies employing a synthetic branched HJ (13) confirmed that UvsW resolved branched structures efficiently (fig. S9). To investigate in real-time the generation and migration of HJ by UvsW with magnetic tweezers, we constructed a DNA substrate mimicking a stalled fork (Fig. 2A) in situ by using either Mh or Lh (fig. S2 & S10A). The fork regression was then initiated by introducing UvsW and ATP. HJ migration was followed by monitoring changes in Ze(t) (Fig. 2B). The transient decreases and increases in Ze(t) corresponded to the migration of the formed HJ in the downward and upward directions. By using dsDNA elasticity (fig. S3) we converted Ze(t) into migrated base-pairs and measured a rate of HJ migration close to the annealing rate,1000 to 1300 bp/s, with a small force dependent asymmetry between the branch migration rates against or favored by the applied force (Fig. 2C and fig. S10). Frequent instantaneous random switches in the migration direction, mediated by a single enzyme complex (fig. S11 and S12), were observed with a characteristic time of ~2 s (Fig. 2D). The ability of UvsW to switch migration direction is essential during the remodeling of stalled forks since it provides the means to revert the HJ back to a normal fork.
Fig. 2.
Single-molecule characterization of UvsW HJ migration activity (A) Schematics of the Lh stalled fork substrate. (B) Experimental HJ migration trace showing Ze(t) with the Lh stalled fork substrate, ATP and UvsW at F = 8 pN. (C) Mean HJ migration rate measured in the downward (against the applied force) and upward (favored by the applied force) migration direction as a function of the applied force (N=54 to 248). Error bars are SEM. (D) Mean migration time in the downward and upward directions (N=69). Error bars are SEM.
The template switching pathway (fig. S1(I)) requires the sequential action of several proteins. We investigated whether the minimal system consisting of UvsW and T4 holoenzyme (composed of gp43 polymerase, gp45 sliding clamp and gp44/62 clamp loader (14)) was sufficient to reproduce such a pathway in vitro. Ensemble experiments on a pGEM vector had shown a lesion in the leading strand regressed a fork by the minimal protein system (6). We prepared a stalled fork substrate presenting a lesion on the leading strand (Fig. 3A) using a 100 bp hairpin with a built-in primer substrate (fig. S13) which contains three locked nucleic acid (LNA) nucleotides in the template leading strand (40 bp before the apex) presenting an efficient roadblock for the holoenzyme (15) that arrested 98 % of the molecules (Fig. 3C).
Fig. 3.
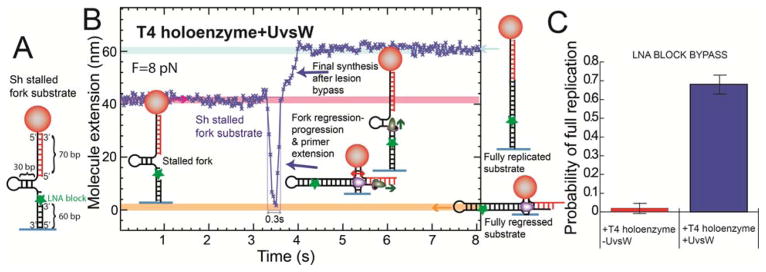
Reconstruction of the template switching pathway (A) Sh stalled fork substrate with a LNA block at the leading strand. (B) Experimental trace at F = 8 pN showing Ze(t) with ATP, dNTPs, UvsW and T4 holoenzyme starting with the stalled fork and ending with the fully replicated substrate. (C) Frequency of events showing the full synthesis of the hairpin in presence (N=58) and absence (N=95) of UvsW. Error bars are SEM.
Addition of UvsW, T4 holoenzyme, ATP and nucleotides first produced a transient decrease associated with fork regression, and next an increase associated with the recovery of the replication fork (Fig. 3B). When the HJ structure is formed, the holoenzyme has the ability to extend the leading strand using the exogenous oligonucleotide as a substrate. We did not detect directly this initial elongation of the primer, since it was not associated with a change in Ze(t), but its further extension by the holoenzyme after fork progression by UvsW had restored the fork with the LNA lesion bypassed (Fig. 3B). Template switching and final hairpin synthesis was further confirmed by checking the disappearance of spontaneous fluctuations of the hairpin caused by its conversion to a full DNA duplex (fig. S14). The frequency of full replicated substrates was increased by more than a factor 30 in presence of UvsW (Fig. 3C), confirming the need for fork regression, and thus demonstrating template switching process in vitro. The fact that the time required for lesion bypass (~0.3 s) was much smaller than the typical holoenzyme loading time (~20 s) (fig. S15) suggests that the holoenzyme probably remained bound to execute both HJ primer extension and final strand displacement synthesis. The random switching of UvsW between fork regression and progression appears as a simple means to coordinate its action with the holoenzyme without a strict synchronization mechanism.
How does UvsW coordinate its action with the different replisome components so as not to interfere with the normal replication process? Firstly, we tested the effect of T4 gp32 single-stranded DNA binding protein (16) on UvsW activity on a partially denatured hairpin. Normal annealing activity was detected in those assays (fig. S16) demonstrating the ability of UvsW to catalyze protein displacement. Next, we performed competition experiments between UvsW and holoenzyme and/or replicative helicase. We found that UvsW annealed a partially replicated hairpin, even when the T4 holoenzyme was replicating the leading strand supporting the hypothesis that UvsW annealing promotes the shift of the leading strand holoenzyme from the fork to the formed HJ (fig. S17A). In contrast, no UvsW activity was detected when the replicative helicase (17) was unwinding the dsDNA (moving along the lagging strand) isolated or coupled to the T4 holoenzyme during leading strand synthesis (figs. S17B–C), demonstrating that gp41 helicase dissociation was required for initiation of the UvsW catalyzed reversal. Thus UvsW activity was inhibited during coupled replication (fig. S17D). Blockage of the leading strand holoenzyme at the lesion site leads to the helicase uncoupling from the holoenzyme and to helicase dissociation (fig. S18 and (6)), then allowing for UvsW loading.
Following the activities of UvsW together with the replisome we have reconstructed the steps for the complete template switching pathway in vitro. The overall results provide the basis for a stepwise model for a lesion bypass mechanism in T4 (Fig. S19): (i) DNA lesion induced partial uncoupling of the replisome that allows for UvsW loading after the departure of the helicase; (ii) fork regression by UvsW shifting the leading strand holoenzyme to the formed HJ; (iii) leading strand primer extension by the holoenzyme while UvsW is migrating the HJ; (iv) UvsW randomly switching direction; (v) fork progression by UvsW to recover the replication fork where the lesion is now bypassed. Several helicases, such as E. coli RecG or human BLM and WRN helicases, present the ability to remodel stalled forks (18, 19) and can potentially play the role of UvsW in fork regression pathways in E. coli and human cells respectively.
Supplementary Material
Acknowledgments
We would like to thank Bénédicte Michel and David Bensimon for useful comments on the manuscript and M.M. Spiering for DNA substrates. This work was supported by a Human Frontier Science Program grant (to V.C. and S.J.B.), US National Institutes of Health grant GM013306 (to S.J.B.), ERC grant Magreps 267 862 (to V.C.) and Juan de la Cierva Program MICINN-JDC (to M.M). M. M. conducted single molecule assays and performed the analysis. S.P conducted ensemble assays, analyzed data and prepared proteins. V.C built the magnetic tweezers. M.M. S.P. V.C and S.J.B wrote the paper. Materials and methods are available as Supplementary Online Material.
References
- 1.Cox MM, et al. Nature. 2000;404:37. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 2.Atkinson J, McGlynn P. Nucl Acids Res. 2009;37:3475. doi: 10.1093/nar/gkp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higgins NP, Kato K, Strauss B. J Mol Biol. 1976;101:417. doi: 10.1016/0022-2836(76)90156-x. [DOI] [PubMed] [Google Scholar]
- 4.Fujiwara Y, Tatsumi M. Mutat Res Fundam Mol Mech Mugag. 1976;37:91. doi: 10.1016/0027-5107(76)90058-0. [DOI] [PubMed] [Google Scholar]
- 5.Long DT, Kreuzer KN. EMBO Rep. 2009;10:394. doi: 10.1038/embor.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson SW, Benkovic SJ. J Mol Biol. 2010;401:743. doi: 10.1016/j.jmb.2010.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeeles JTP, Marians KJ. Science. 2011;334:235. doi: 10.1126/science.1209111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carles-Kinch K, Georg JW, Kreuzer KN. EMBO J. 1997;16:4142. doi: 10.1093/emboj/16.13.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. Nucl Acids Res. 1989;17:4713. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu WK, Hickson ID. Nat Rev Cancer. 2009;9:644. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 11.Nelson SW, Benkovic SJ. J Biol Chem. 2006;282:407. doi: 10.1074/jbc.M608153200. [DOI] [PubMed] [Google Scholar]
- 12.Gosse C, Croquette V. Biophys J. 2002;82:3314. doi: 10.1016/S0006-3495(02)75672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Webb MR, Plank JL, Long DT, Hsieh T, Kreuzer KN. J Biol Chem. 2007;282:34401. doi: 10.1074/jbc.M705913200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hacker KJ, Alberts BM. J Biol Chem. 1994;269:24221. [PubMed] [Google Scholar]
- 15.Manosas M, et al. Nucl Acids Res. 2012;40:6174. doi: 10.1093/nar/gks253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chase JW, Williams KR. Annu Rev Biochem. 1986;55:103. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- 17.Venkatesan M, Silver LL, Nossal NG. J Biol Chem. 1982;257:12426. [PubMed] [Google Scholar]
- 18.McGlynn P, Lloyd RG. Proc Natl Acad Sci USA. 2001;98:8227. doi: 10.1073/pnas.111008698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Machwe A, Xiao L, Groden J, Orren DK. Biochemistry. 2006;45:13939. doi: 10.1021/bi0615487. [DOI] [PubMed] [Google Scholar]
- 20.Nelson SW, Perumal SK, Benkovic SJ. Biochemistry. 2009;48:1036. doi: 10.1021/bi801792q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frey MW, Nossal NG, Capson TL, Benkovic SJ. Proc Natl Acad Sci USA. 1993;90:2579. doi: 10.1073/pnas.90.7.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nossal NG. J Biol Chem. 1979;254:6026. [PubMed] [Google Scholar]
- 23.Rush J, et al. J Biol Chem. 1989;264:10943. [PubMed] [Google Scholar]
- 24.Valentine AM, Ishmael FT, Shier VK, Benkovic SJ. Biochemistry. 2001;40:15074. doi: 10.1021/bi0108554. [DOI] [PubMed] [Google Scholar]
- 25.Nelson SW, Kumar R, Benkovic SJ. J Biol Chem. 2008;283:22838. doi: 10.1074/jbc.M802762200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manosas M, Spiering MM, Zhuang Z, Benkovic SJ, Croquette V. Nat Chem Biol. 2009;5:904. doi: 10.1038/nchembio.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manosas M, et al. Meth Enz. 2010;475:297. doi: 10.1016/S0076-6879(10)75013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manosas M, Spiering MM, Ding F, Croquette V, Benkovic SJ. Nucl Acids Res. 2012;40:6187. doi: 10.1093/nar/gks254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.