Abstract
We explore the influence of genetic variation on subjective well-being by employing a twin design and genetic association study. In a nationally-representative twin sample, we first show that about 33% of the variation in life satisfaction is explained by genetic variation. Although previous studies have shown that baseline happiness is significantly heritable, little research has considered molecular genetic associations with subjective well-being. We study the relationship between a functional polymorphism on the serotonin transporter gene (5-HTTLPR) and life satisfaction. We initially find that individuals with the longer, transcriptionally more efficient variant of this genotype report greater life satisfaction (n=2,545, p=0.012). However, our replication attempts on independent samples produce mixed results indicating that more work needs to be done to better understand the relationship between this genotype and subjective well-being. This work has implications for how economists think about the determinants of utility, and the extent to which exogenous shocks might affect individual well-being.
Keywords: life satisfaction, twin study, genetic association, serotonin transporter gene, 5-HTTLPR, rs2020933
Happiness research has become one of the liveliest subjects in economics in recent years1. Its main goal is to explain the determinants of individual life satisfaction or subjective well-being (often loosely called happiness). Economists have mainly dealt with economic influences, in particular, income and its distribution, labor market regulation, unemployment and inflation. For example, Di Tella et al. (2001) used happiness surveys to determine the welfare costs of inflation and unemployment, showing that unemployment depresses reported well-being more than does inflation. In fact, their longitudinal study of life satisfaction self-reports enabled these authors to estimate that people would trade off a 1 percentage-point increase in the unemployment rate for a 1.7 percentage-point increase in the inflation rate. Systematic influences on life satisfaction have also been found for socio-demographic factors (age, gender, race, marital status, children, and social networks) as well as for political and cultural factors (such as democracy, decentralization, and religiosity). While variables like socio-economic status, income, marriage, education, and religiosity are significantly associated with individual happiness, none typically accounts for more than 3% of the variation (Layard, 2005;Frey, 2008). Moreover, changes in these variables appear to yield only short term changes to happiness. For example, the “Easterlin Paradox” (Easterlin, 1974; Clark et al., 2008) suggests that increases in real income either have no lasting effect on happiness, or only a quite small one (Stevenson and Wolfers, 2008). A reason appears to be that happiness levels tend to revert toward what psychologists describe as a “set point” or “baseline” of happiness that is partly shaped by personality and genetic predispositions (Kahneman et al., 1999; Diener and Lucas, 1999). This in turn has implications for how economists think about the determinants of utility, and the extent to which exogenous shocks might affect an individual's well-being.
Although previous studies have shown that baseline happiness is significantly heritable (Lykken and Tellegen, 1996), little research has considered molecular genetic associations with subjective well-being. Here, we first corroborate the earlier work showing that happiness is significantly influenced by genetic variation in a nationally-representative twin sample and subsequently we present mixed evidence for a candidate gene association with life satisfaction. In our discovery sample we initially found that individuals with a transcriptionally more efficient version of the serotonin transporter gene (SLC6A4, more commonly referred to as 5-HTT or SERT for the protein it encodes) are significantly more likely to report higher levels of life satisfaction. However, our replication efforts on independent samples produce mixed results. This combination of economics and genetics is of rising salience (Benjamin et al., 2007; Beauchamp et al., 2011; Benjamin et al., 2012).
Before we detail our genetic association approach and results, we explore the general influence that genes may have on happiness through a twin study design. A growing number of studies use twin research techniques to gauge the relative importance of genetic and environmental influences on economic behaviors (e.g. Cesarini et al. (2009), Fowler et al. (2009). We estimate the heritability of subjective well-being at 33%, indicating that about one-third of the variance in individual life satisfaction can be attributed to genetic influences.
Although twin studies are an important step in establishing the influence of genes in subjective well-being, they do not identify the specific genes involved. The increasing availability of genotypic information now allows us to test hypotheses about targeted genes and their effects. One place to start the search for such genes is among those that have already been shown to account for variation in emotional states. Among these, 5-HTT is a prime candidate. The 5-HTT gene encodes a transporter in the cell wall that absorbs serotonin into the presynaptic neuron in parts of the brain that influence mental states (Hariri et al., 2002; Bertolino et al., 2005; Heinz et al., 2005; Canli and Lesch, 2007). 5-HTT has been studied for more than twenty years and much is known about the way different versions of this gene influence transcription, metabolism, and signal transfers between neurons, all of which may influence personality. In particular, less transcriptionally efficient variants of this gene have been shown to moderate the influence of life stress on depression (Caspi et al., 2003); and the more transcriptionally efficient alleles have been linked to optimism (Fox et al., 2009). As a result, economists have specifically identified 5-HTT as a candidate gene for further study (Benjamin et al., 2007).
Using data from two independent sources, the National Longitudinal Study of Adolescent Health (Add Health) and the Framingham Heart Study (FHS), we analyze the relationship between variants of 5-HTT and life satisfaction. We find some evidence of significant association in both data sets, suggesting that the 5-HTT gene may play a role in explaining life satisfaction yet more work needs to be done in order to verify and better understand the relationship between this genotype and subjective well-being. We do not claim that 5-HTT determines happiness, nor do we exclude the likely possibility that several other genes may play a role in accounting for the influence of genes on happiness.
1 The Add Health data
This research is based on genetic and survey data collected as part of the National Longitudinal Study of Adolescent Health (Add Health). The study was initially designed to explore the health-related behavior of adolescents in grades 7 through 12, but it has been employed widely across disciplines and has made recent contributions in economics (Echenique et al., 2006; Echenique and Fryer, 2007; Alcott et al., 2007; Norton and Han, 2009). In the first wave of the Add Health study (1994–1995) 80 high schools were selected from a sampling frame of 26,666 based on their size, school type, census region, level of urbanization, and percent of the population that was white. Participating high schools were asked to identify junior high or middle schools that served as feeder schools to their school. This resulted in the participation of 145 middle, junior high, and high schools. From those schools, 90,118 students completed a 45-minute questionnaire and each school was asked to complete at least one School Administrator questionnaire. This process generated descriptive information about each student, the educational setting, and the environment of the school. From these respondents, a core random sample of 12,105 adolescents in grades 7–12 were drawn, along with several over-samples, totaling more than 27,000 adolescents. These students and their parents were administered in-home surveys in the first wave.
Wave II (1996) was comprised of another set of in-home interviews of more than 14,738 students from the Wave I sample and a follow-up telephone survey of the school administrators. Wave III (2001–02) consisted of an in-home interview of 15,170 Wave I participants. Finally, Wave IV (2008) consisted of an in-home interview of 15,701 Wave I participants. The result of this sampling design is that Add Health is a nationally representative study. Women make up 49% of the study's participants, Hispanics 12.2%, Blacks 16.0%, Asians 3.3%, and Native Americans 2.2%. Participants in Add Health also represent all regions of the United States.
In Wave I of the Add Health study, researchers created a sample of sibling pairs including all adolescents that were identified as twin pairs, half-siblings, or unrelated siblings raised together. Twin pairs were sampled with certainty. The sibling-pairs sample is similar in demographic composition to the full Add Health sample (Jacobson and Rowe, 1998). The number of identical (monozygotic) and non-identical (dizygotic) twins who participated in Wave III was 1,098 (434 MZ and 664 DZ), with 872 twins (434 MZ and 438 DZ) in same sex pairs. The Add Health data has been widely used for twin studies (Harris et al., 2006; Fowler et al., 2008).
Allelic information for a number of genetic markers were collected for 2,574 individuals as part of Wave III. The candidate genotypes were chosen for inclusion in the study because they are known to affect brain development, neurotransmitter synthesis and reception, and hormone regulation. Details of the DNA collection and genotyping process are available at the Add Health website (Add Health Biomarker Team, 2007). These candidate genes include markers that identify alleles (variants) of the serotonin transporter gene or 5-HTT. The promotor region of 5-HTT contains a variable number tandem repeat (VNTR) sequence that influences transcriptional activity—the “long” 528 base-pair allele is associated with a higher basal activity than the “short” 484 base-pair allele. This functional polymorphism on the 5-HTT gene is commonly referred to as 5-HTTLPR (5-HTT-linked polymorphic region). Allele frequency for the short allele is 43% and for the long allele is 57%.
In 2012, Add Health released a second batch of genotypical data that extended to almost all Wave III participants and included the 5-HTTLPR genotype. The distributional frequencies of this new sample (used here for replication purposes) closely align with the earlier discovery sample.
In Wave III, subjects were asked “How satisfied are you with your life as a whole?” Answer categories ranged from very dissatisfied, dissatisfied, neither satisfied nor dissatisfied, satisfied, to very satisfied. Alternative answers were “refused” or “don't know” and these were discarded for the purpose of this study (less than 1% of interviewees gave such a response). This question and answer formulation is standard in the economics of happiness literature (Di Tella et al., 2001, 2003; Kahneman and Krueger, 2006; Frey, 2008). The distribution of answers to the life satisfaction question is shown in Appendix. In line with the happiness literature, a large majority of respondents report being satisfied or very satisfied (Frey and Stutzer, 2002a). That most people, in fact, report a positive level of subjective well-being is the object of a paper by Diener and Diener (1996), where the authors find this distribution to be representative in a wide cross-national analysis.
2 Twin design
2.1 Methods
Twin studies compare the traits, behaviors, and other outcomes (called “phenotypes”) of twins who share 100% of their genetic material (identical or monozygotic twins) to those who share 50% of their genetic material (fraternal or dizygotic twins) in order to estimate the relative importance of genetic and environmental influences (Taubman, 1976; Ashenfelter and Krueger, 1994). Following Benjamin et al. (2012), if we assume that the residual factors co-vary equally for monozygotic (MZ) and dizygotic (DZ) twins (informally called the “equal environments” assumption), and there are no gene-environment interactions, then the variance in happiness can be decomposed into additive genetic effects (A), common or shared environmental influences (C), and unshared or unique environmental influences (E). The ACE model does not allow us to observe environmental and genetic influences directly, but it does allow us to estimate these effects by observing the covariance across MZ and DZ twins.
Although the assumptions underlying the ACE model are strong, the method produces results that have been validated in numerous other studies. For example, studies of twins reared apart generate similar heritability estimates to those generated by studies of twins raised together (Bouchard, 1998). More recently, Visscher et al. (2006) utilize the small variance in percentage of shared genes among DZ twins to estimate heritability without using any MZ twins, and they are able to replicate findings from studies of MZ and DZ twins reared together. Moreover, personality and cognitive differences between MZ and DZ twins persist even among twins whose zygosity has been miscategorized by their parents, indicating that being mistakenly treated as an identical twin by ones parents is not sufficient to generate a difference in concordance (Scarr and Carter-Saltzman, 1979; Kendler et al., 1993; Xian et al., 2000).
The ACE model can be formally expressed as:
where y is the measure of the phenotype, j denotes the family, i denotes the individual twin in the family, µ is the mean of this phenotype across all observations, is the additive genetic component, is the shared environment component, and is the unshared environment component. Notice that these assumptions imply:
If we further assume that the unshared environment is uncorrelated between twins (COV (E1j, E2j = 0), that genes are perfectly correlated between MZ twins and the covariance between DZ twins who share half their genes on average is half that of identical twins then we have two additional equations
,
The covariance equations reflect the fact that DZ twins share on average 50% of their genes whereas MZ twins share all of their genes. Based on these equations, we can estimate the ACE model via a random effects regression model with the 2 × 2 variance-covariance matrix specified as:
where R is the genetic relatedness of the twin pair equaling 1 for MZ twins and for DZ twins. We use the variances of the random effects to generate estimates of heritability, common environment, and unshared environment.2
To generate the ACE estimates, we use the structural equation modeling program OpenMx developed by Neale et al. (2010). In addition to estimating ACE models, we estimate all of the possible submodels to compare model fit. These include an AE model, which assumes only genes and unshared environment influence the phenotype (C=0), a CE model which assumes only common and unshared environment influence the phenotype (A=0), and an E model (A=0 and C=0). If a submodel fits better than the general ACE model, this suggests the parameters left out of the submodel are not significantly contributing to model fit. To compare the submodels, we use the Akaike Information Criterion (AIC) in maximum likelihood estimation, where smaller values indicate better fit.
2.2 Twin results
When assessing the role of genetic influences, the first step is to compare the correlation in phenotype among MZ twin pairs to that of DZ twin pairs. For life satisfaction, the correlation coefficient for MZ twins is 0.334 and for DZ twins is 0.132. The difference in correlations is significant (p = 0.013, one sided). These correlations show that identical twins are significantly more similar in their level of happiness than fraternal twins, which suggests that genetic factors might play a role in this trait.
In Table 1 we report results from several variance decomposition models described above. Note that the ACE model yields a heritability estimate of 33% (SE = 0.044) and the estimate for unshared environment is 67% (SE = 0.029) while a null estimate is returned for common environment. In other words, about a third of the variance in happiness in our sample can be attributed to variance in genetic factors. We also examine the submodels and find that the models with lowest AIC all include A, suggesting that the finding that happiness is heritable is robust to different model specifications.3.
Table 1. Summary of ACE twin model results.
| Life satisfaction | Fit statistics | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| a2 | c2 | e2 | ep | −2ll | df | AIC | diff −2ll | diff df | p | |
| ACE | 0.331 | 0.000 | 0.669 | 4 | 1878.9 | 795 | 288.9 | - | - | - |
| AE | 0.331 | - | 0.669 | 3 | 1878.9 | 796 | 286.9 | 0 | 1 | 1 |
| CE | - | 0.257 | 0.743 | 3 | 1882.9 | 796 | 290.9 | 4 | 1 | 0.05 |
| E | - | - | 1 | 2 | 1907.2 | 797 | 313.2 | 28.3 | 2 | 0 |
Note: The models consist of additive genetic factors (A), shared or common environmental factors (C), and unshared environ- mental factors (E). The model includes 217 MZ and 219 DZ same-sex twin pairs.
Compared to previous studies of happiness, our heritability estimate of 33% is on the lower end of reported estimates. In fact, the seminal paper by Lykken and Tellegen (1996) estimated heritability at about 50%, and subsequent estimates ranged from 38% (Stubbe et al., 2005) to 36–50% (Bartels and Boomsma, 2009) to 42–56% (Nes et al., 2006). However, the Add Health study includes other questions that suggest the heritability of happiness rises as people age. The standard life satisfaction question used in this paper is only asked of Add Health subjects in Wave III (2001–02), but in other interview waves the following question is asked of participants: “How often was the following true during the past seven days? You felt happy. ” Answers range from “never or rarely” to “most of the time or all of the time.” Figure 3 shows the MZ and DZ twin pair correlations of the time series that combines the “life satisfaction” and “You felt happy” questions. The basic heritability estimates that result from comparing MZ and DZ correlations range from 22% in Wave I (1994) to 54% in Wave IV (2008). This longitudinal analysis is consistent with a growing body of longitudinal twin research that shows that the heritability of a number of traits (e.g. intelligence) increases with age (Plomin et al., 2008). It also shows that the finding that happiness is heritable is robust to a variety of measures and time periods over the life course. These findings are generally taken to mean that genes and environment can play differing roles in explaining experience at different points in the life course.
3 Genetic association
Twin studies are important because they allow us to gauge the relative influence of our genetic makeup on subjective well-being. However, twin studies do not give insight into which specific genes may be involved in explaining the heritability of traits. Because Add Health collected a number of specific genetic markers, it presents us with a unique opportunity to move beyond a twin design study. Below we introduce some basic concepts in genetics, our genetic association research design, and present discovery and replication results for our candidate gene study.
3.1 Basic concepts in genetics
Human DNA is composed of an estimated 21,000 genes that form the blueprint for molecules that regulate the development and function of the human body. Genes are distinct regions of human DNA that are placed in the 23 pairs of chains, or chromosomes, that make up all human DNA. Almost all human cells contain the same DNA they inherited at the moment of conception.
Individuals inherit one half of their DNA from each parent, with one copy of each gene coming from the mother and one copy from the father. Some genes come in different versions, known as “alleles”—for example, sickle cell disease results from a particular allele coding for abnormal rather than normal hemoglobin. Each parent has two separate copies of an allele at each “locus”, or location, on the chromosome, but each sperm or egg cell contains only one of these alleles. Thus a child has a 50% chance of receiving a particular allele from a particular parent. For example, suppose that at a given locus there are two possible alleles, A and B. If both parents are “heterozygous” at that locus, meaning they each have an A and a B allele (AB or BA—order is irrelevant), then a given offspring has a 25% chance of being “homozygous” for A (AA), a 25% chance of being homozygous for B (BB) and a 50% chance of being heterozygous (AB or BA). If an individual is heterozygous at a locus, a “dominant” allele may impose itself on the “recessive” allele and the expression of the latter allele will not be observed.
Genes transcribe proteins that begin a cascade of interactions that regulate bodily structure and function. Many of the observable traits and behaviors of interest, referred to as “phenotypes” are far downstream from the original “genotypes” present in the DNA. While in some cases one allele can single-handedly lead to a disease (such as Sickle Cell Anemia, Huntingtons disease, or cystic fibrosis), the vast majority of phenotypes are “polygenic”, meaning they are influenced by multiple genes (Mackay, 2001; Plomin et al., 2008), and are shaped by a multitude of environmental forces. As a result, association models between genotypes and phenotypes are an important first step, but they are not the end of the story. It is also important to investigate the extent to which genetic associations are moderated by environmental factors and other genes.
3.2 5-HTTLPR, serotonin, and happiness
One strategy in behavioral genetics is to start with a “candidate” gene that is thought to influence behaviors or processes in the body that are related to the phenotype of interest. For subjective well-being, this means focusing on genes that affect brain development, neurotransmitter synthesis and reception, hormone regulation, and transcriptional factors (Damberg et al., 2001; Benjamin et al., 2007).
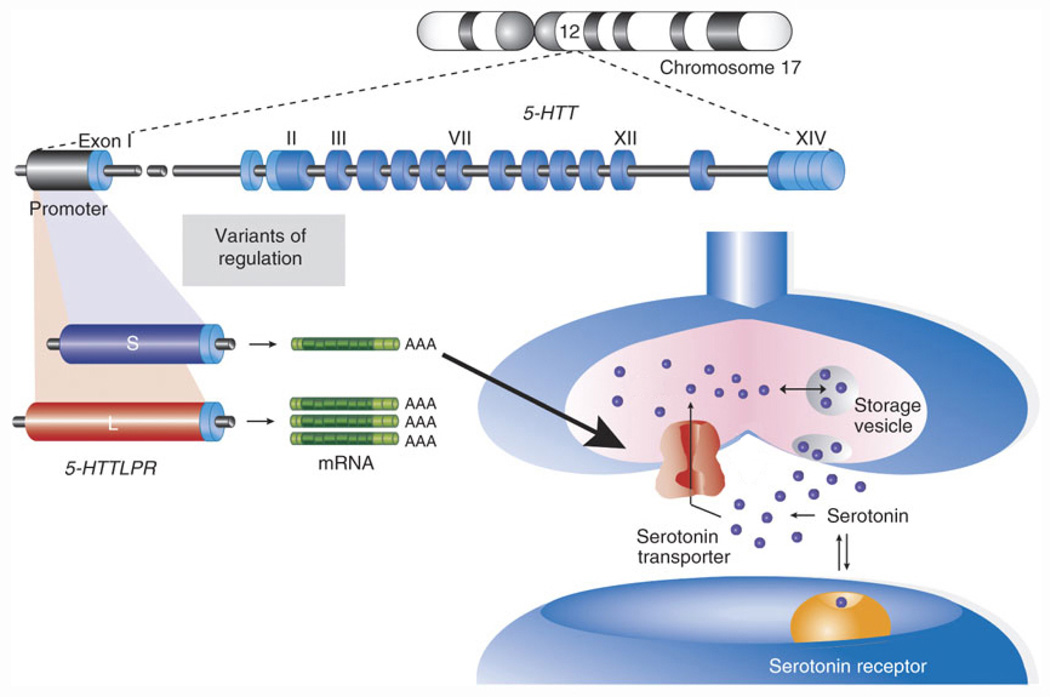
We choose a candidate gene that has already received a great deal of attention by biologists and social scientists for its association with mental states. The 5- HTT gene is critical to the metabolism of serotonin in the brain. As shown in Figure 1, serotonin is a chemical that is released by a neuron and sensed by a receptor on the receiving neuron, passing an electric potential across a gap called a nerve synapse (the nerve that emits the serotonin is on the “pre-synaptic” side of the gap). Signals are carried throughout the body by the sequential release of a neurotransmitter by one neuron after another across these synapses. The 5-HTT gene codes for the serotonin transporters that are placed in the cell wall and reabsorb the neurotransmitter serotonin from the synaptic cleft. Most serotonin is recycled after use and the serotonin transporter allows serotonergic neurons to restock. The serotonin transporter gene has been studied extensively and much is known about the way different versions of this gene influence serotonergic neurotransmission which, in turn, is found to influence personality and mental health (Hariri et al., 2002; Hariri and Holmes, 2006; Canli and Lesch, 2007).
Figure 1.
Representation of the long/short functional variant on the 5-HTT gene and the release, reception, and recycling of serotonin in neurons. Adapted from Canli & Lesch (2007), with permission from the Nature Publishing Group.
The 5-HTT gene contains a 44 base-pair variable-number tandem repeat (VNTR) polymorphism4 in the promoter region5(5-HTTLPR) that is believed to be responsible for variation in transcriptional efficiency. The “long” (528 bp) and “short” (484 bp) polymorphism produce the same protein, but the long allele is associated with an approximately three times higher basal activity than the shorter allele. Consequently, the long variant produces significantly more 5-HTT mRNA6 and protein (Heils et al., 1996; Lesch et al., 1996; Little et al., 1998; Glatz et al., 2003; Canli and Lesch, 2007). The long polymorphism thus results in increased gene expression and more serotonin transporters in the cell membrane. In turn, more serotonin is reintroduced into the pre-synaptic cell. This process is also shown in Figure 1.
Functional variation in the serotonin transporter gene is increasingly understood to exert influence on parts of the brain regulated by serotonergic neurotransmission. In particular, research shows increased amygdala activation to negative emotional stimuli among carriers of short alleles (Hariri et al., 2002; Heinz et al., 2005; Munafò et al., 2008; Pezawas et al., 2005; Canli et al., 2005). A morphometrical study of this genetic association reports reduced gray matter volume in short-allele carriers in limbic regions critical for processing of negative emotion, particularly perigenual cingulate and amygdala (Pezawas et al., 2005). These authors conclude that 5-HTTLPR induced variation in anatomy and function of an amygdala-cingulate feedback circuit critical for emotion regulation indicates one mechanism for a genetic susceptibility for depression (Pezawas et al., 2005). Another morphometrical study corroborates the finding that short-allele carriers show decreased volume in the affective division of the anterior cingulate and decreased gray matter density in its pregenual region (Canli et al., 2005). The same study also finds that the 5-HTTLPR polymorphism is associated with activation changes to positive stimuli, suggesting a general role in emotional regulation, rather than negative valence specifically (Canli et al., 2005).
Myriad behavioral studies also suggest that serotonin and 5-HTT play an important role in emotional regulation (Heils et al., 1996; Hariri et al., 2002; Hariri and Holmes, 2006). Specifically, variance in 5-HTTLPR was found to be be associated with variation in mental health outcomes (Lesch et al., 1996) and subsequent studies report that about 10% of the variance in anxiety-related traits depends on variation in serotonin transporters (Sen et al., 2004; Munafò et al., 2005). A recent study by Fox et al. (2009) also suggests that 5-HTTLPR may influence optimism. The authors obtained DNA from about 100 participants and compared reaction times to pictures with positive, negative, and neutral emotional valence (replicating a common experiment in psychopathology research). The results show that individuals with the transcriptionally more efficient 5-HTTLPR alleles display a significant bias towards processing positive information and selectively avoiding negative information. This emotionally self-protective pattern does not obtain in individuals carrying one or both short alleles. It is important to note, however, that positive and negative emotions are not different sides to the same coin. A score of zero on a depression or anxiety scale may be indicative of the absence of such mental health issues but it is not indicative of the presence of happiness (McGreal and Stephen, 1993).
Not all studies show a direct relationship between a gene variant and a phenotype. Instead, developmental or concurrent environments may moderate an association between genes and phenotypes. A study by Caspi et al. (2003) suggests a gene-environment interaction for the influence of life stress on depression. The authors find that individuals with short 5-HTTLPR alleles gene are more vulnerable to stress-induced depression. Among those individuals that had experienced a relatively large number of stressful life events, about 33% of the carriers of the less efficient short allele were cases of diagnosed depression as compared to only 17% of the individuals that carried both long alleles. Thus, in the Caspi et al. (2003) study, the gene itself is not associated with depression. Rather, it is the combination of both gene and environment that yields a significant association. In this study we do not report on a gene-environment interaction, but the direct association between the number of long 5-HTTLPR alleles and life satisfaction.
It is important to highlight, however, that later studies often fail to replicate candidate gene discoveries. For example, a meta-analysis by Risch et al. (2009) that incorporated 14 studies yielded no evidence for a direct effect between 5-HTTLPR and depression nor an indirect effect moderated by stressful life events.
3.3 Association methods
Genetic association studies test whether an allele or genotype occurs more frequently within a group exhibiting a particular phenotype than those without the phenotype. However, a significant association can mean one of three things: (1) The allele itself inuences subjective well-being; (2) the allele is in “linkage disequilibrium” with an allele at another locus that influences subjective well-being; or (3) the observed association is a false positive signal due to population stratification.7
Population stratification occurs because groups may have different allele frequencies due to their genetic ancestry. Subjective well-being in these groups may be the product of their environments, alleles other than the one of interest, or some unobserved reason. For example, two groups may not have mixed in the past for cultural reasons. Through the process of local adaptation or genetic drift, these groups may develop different frequencies of a particular allele. At the same time, the two groups may also develop divergent behaviors that are not influenced by the allele but solely by the environment in which they live. Once these two groups mix in a larger population, simply comparing the frequency of the allele to the observed behavior would lead to a spurious association.
There are two main research designs employed in association studies, case-control designs and family-based designs. Case-control designs compare the frequency of alleles or genotypes among subjects that exhibit a phenotype of interest to subjects who do not. As a result, case-control designs are vulnerable to population stratification if either group is especially prone to selection effects. A typical way to control for this problem is to include controls for the race or ethnicity of the subject or to limit the analysis to a specific racial or ethnic group. Family-based designs eliminate the problem of population stratification by using family members, such as parents or siblings, as controls. Tests using family data compare whether offspring exhibiting the trait receive a risk allele from their parents more often than would be expected by chance. This design is very powerful in minimizing type I error but also suffers from much lower power in detecting a true association. Xu and Shete (2006) show, based on extensive simulation work, that a case-control association study using mixed-effects regression analysis outperforms family-based designs in detecting an association while at the same time effectively limiting type I error.
To test for genetic association we employ a mixed-effects OLS regression model:8
where i and j index subject and family respectively. For the 5-HTT gene, G = 2 if the subject's genotype is LL, G = 1 for genotypes LS or SL, and G = 0 if the subject's genotype is SS (where L represents having a copy of a 528 base-pair “long” allele, and S represents having a copy of a 484 base-pair “short” allele). Z is a matrix of variables to control for the underlying population structure of the Add Health sample as well as potentially mediating factors such as age, gender, education, religiosity, marriage, job, welfare, or medication that may all influence subjective well-being. Finally, the variable U is a family random effect that controls for potential genetic and environmental correlation among family members, and ε is an individual-specific error.
To control for the effects of the underlying population structure, we include indicator variables for whether a subject self-reported as Black, Hispanic, or Asian (base category is White). Following the policy of the United States Census, Add Health allows respondents to mark more than one race. Since this complicates the ability to control for stratification, we exclude these individuals (N = 117), but a supplementary analysis including them yields substantively equal results. Population stratification is a pertinent challenge in our sample. The Hardy-Weinberg equilibrium (HWE) test indicates a significant deviation from the expected frequencies (χ2 = 7.66, p = 0.005). On the other hand, when considering the separate HWE test statistics by ethnicity there are no longer significant deviations from the expected HWE frequencies (see Appendix). As such, introducing ethnicity controls and running a whites-only case control test (Xu and Shete, 2006) may adequately control for population stratification.
3.4 Association results: Add Health discovery sample
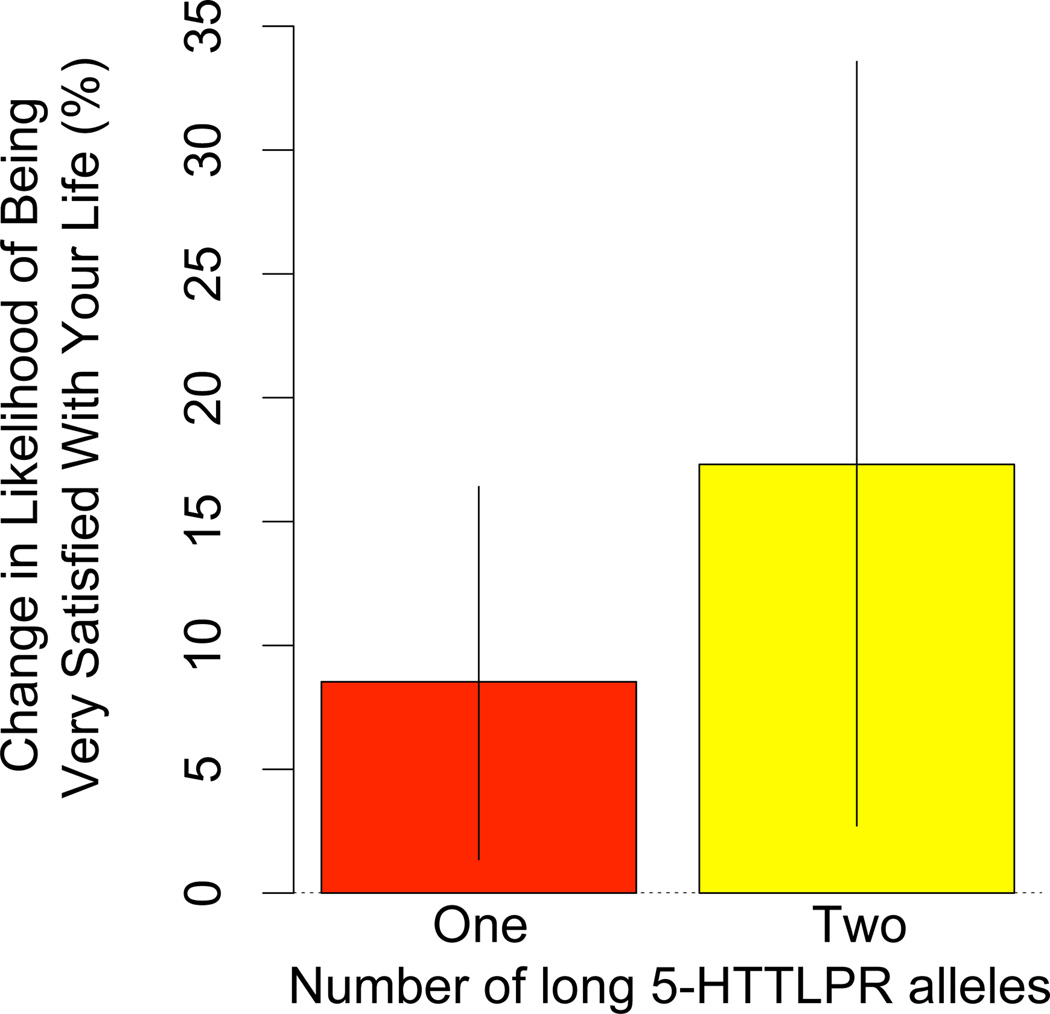
Table 2 shows the results of several specifications of the model to test the hypothesis that the 5-HTTLPR long allele is associated with subjective well-being in the original Add Health discovery sample.9 Each of these specifications includes variables for age, gender, and race to control for population stratification. Model 1 shows that the long allele is significantly associated with increased life satisfaction (p = 0.012). In Figure 2, we summarize the results for 5-HTTLPR by simulating first differences from the coefficient covariance matrix of Model 1. Holding all else constant and changing the 5-HTTLPR variant for all subjects from zero to one long allele would increase the reporting of being very satisfied with one's life in this population by about 8.5%. Similarly, changing the 5-HTTLPR variant from zero to two long alleles would increase the reporting of being very satisfied by about 17.3%.
Table 2.
OLS models of association between 5-HTTLPR and life satisfaction (discovery sample). Variable definitions are in the Appendix. Standard errors (SE) and P-values are also presented.
| Model 1 | Model 2 | Model 3 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Coeff. | SE | P-value | Coeff | SE | P-value | Coeff. | SE | P-value | |
| 5-HTTLPR long | 0.059 | 0.023 | 0.012 | 0.065 | 0.023 | 0.005 | 0.070 | 0.029 | 0.017 |
| Black | −0.111 | 0.048 | 0.021 | −0.114 | 0.049 | 0.020 | |||
| Hispanic | 0.198 | 0.117 | 0.092 | 0.216 | 0.118 | 0.067 | |||
| Asian | −0.196 | 0.073 | 0.007 | −0.221 | 0.071 | 0.002 | |||
| Age | 0.004 | 0.009 | 0.705 | −0.011 | 0.009 | 0.262 | −0.031 | 0.012 | 0.008 |
| Male | 0.014 | 0.033 | 0.682 | 0.028 | 0.033 | 0.406 | 0.039 | 0.041 | 0.341 |
| Job | 0.093 | 0.041 | 0.024 | 0.104 | 0.057 | 0.071 | |||
| College | 0.115 | 0.033 | 0.001 | 0.238 | 0.042 | 0.000 | |||
| Married | 0.232 | 0.041 | 0.000 | 0.318 | 0.050 | 0.000 | |||
| Divorced | −0.313 | 0.153 | 0.041 | −0.310 | 0.155 | 0.047 | |||
| Religiosity | 0.103 | 0.017 | 0.000 | 0.082 | 0.023 | 0.000 | |||
| Welfare | −0.236 | 0.098 | 0.017 | −0.121 | 0.153 | 0.432 | |||
| Medication | −0.045 | 0.032 | 0.162 | −0.095 | 0.041 | 0.021 | |||
| Intercept | 4.078 | 0.208 | 0.000 | 4.096 | 0.210 | 0.000 | 4.514 | 0.262 | 0.000 |
| N | 2,545 | 2,528 | 1,446 | ||||||
| R2 | 0.01 | 0.06 | 0.08 | ||||||
Figure 2.
Increasing the number of “long,” more efficient 5-HTTLPR alleles is associated with higher life satisfaction in the Add Health discovery sample. First differences based on simulations of Model 1 parameters are presented along with 95% confidence intervals. All other variables are held at their means.
Model 2 includes a number of socio-economic factors that are known to influence subjective well-being. In particular, having a job, education, marriage, divorce, religiosity, welfare assistance, and being on medication. This model also suggests that there is a statistically significant association (p = 0.005) between the 5-HTTLPR long variant and the reporting of life satisfaction. Notice also that the coefficient actually increases a bit, suggesting that the association cannot be explained by a mediation effect this genotype may have on any other variables included in the model.10
Following Xu and Shete (2006), as a robustness test for population stratification, we also include Model 3 that is a case-control association model for those subjects that uniquely identified themselves as being white. The coefficient on 5-HTTLPR and its p-value (p = 0.017) suggest that population stratification between self-reported racial categories is not driving the association between 5-HTTLPR and life satisfaction in the Add Health discovery sample.
3.5 Replication studies
Specific genotypes usually only account for a very small amount of the variance in complex social behaviors, which means the tests often have low power. As a result, it is very important to replicate results in independent samples (Beauchamp et al., 2011; Benjamin et al., 2012). Such efforts to replicate a significant genetic association result, as well as increasing the sample sizes, are key in addressing the possibility that the original association would be a spurious result or false positive. Below we first report on our replication effort in the Framingham Heart Study. More recently, the release of new genotypical data for the Add Health data allowed for another replication effort which we also detail below. Although the association between functional variation on the 5-HTT gene and life satisfaction found in the Add Health discovery sample replicates in the Framingham sample it does not replicate in the new Add Health sample.
3.5.1 Replication study 1: Framingham Heart Study
Here we utilize the Framingham Heart Study (FHS), a population-based, longitudinal, observational cohort study that was initiated in 1948 to prospectively investigate risk factors for cardiovascular disease. Since then, the FHS has come to be composed of four separate but related cohort populations: (1) the Original Cohort enrolled in 1948 (N=5,209); (2) the Offspring Cohort (the children of the Original Cohort and spouses of the children) enrolled in 1971 (N=5,124); (3) the Omni Cohort enrolled in 1994 (N=508); and (4) the Generation 3 Cohort (the grandchildren of the Original Cohort) enrolled beginning in 2002 (N=4,095). Published reports provide details about sample composition and study design for all these cohorts (Cupples and D'Agnostino, 1988; Kannel et al., 1979).
The Framingham Heart Study makes available genetic markers for its participants. Out of the 14,428 members of the three main cohorts, a total of 9,237 individuals have been genotyped (4,986 women and 4,251 men) for single nucleotide polymorphisms (SNPs). These are specific locations on human DNA where a single pair of nucleotides varies for some part of the human population. FHS makes available a data set of expected genotypes for all 2,543,887 SNPs in the European ancestry HapMap sample that was computed from the 550,000 observed SNPs from an Affymetrix array using the program MACH (for information on how this data set was constructed, see De Bakker (2008). Although this data does not contain the same VNTR polymorphism marker for 5-HTT that we analyze in Add Health, it does contain a nearby marker called “rs2020933”, and the “A” allele of this marker is known to be associated with higher transcriptional efficiency of serotonin transporters (Martin et al., 2007; Wendland et al., 2006; Lipsky et al., 2009; Fahad et al., 2010). It is also known to be in positive linkage disequilibrium with the long allele of 5-HTTLPR (Huezo-Diaz et al., 2009). The FHS also asked 3,460 participants in the offspring cohort a variant of the life satisfaction question: “Indicate where you think you belong between these two extremes … satisfied with job or home life OR ambitious, want change.” Respondents were given a 7 point scale to choose from, and we reverse coded the scale so that higher values indicated greater satisfaction with life (mean=4.7, SD=1.7). Although this question is not exactly like the one asked in Add Health, if there is a real association between 5-HTT and happiness, we expect it to show up in spite of variations in the way the question is asked.
We merged the gene and life satisfaction data and conducted an association test using a linear regression with a general estimating equations (GEE) approach to account for within-family correlation of errors. As shown in Model 1 in Table 3, this association is significant (p = 0.05) and in the expected direction. In Model 2 we include additional controls for age and gender. We also include the first ten principal components of a singular value decomposition of the subject-genotype matrix in the regression (see Appendix), which has been shown to effectively control for population stratification (Price et al., 2006). Once again, the association is significant (p = 0.05).
Table 3.
GEE models of association between rs2020933 and life satisfaction (Framingham Heart Study). Standard errors (SE) and P-values are also presented.
| Model 1 | Model 2 | ||||||
|---|---|---|---|---|---|---|---|
| Coeff. | SE | P-value | Coeff. | SE | P-value | ||
| rs2020933 “A” alleles | 0.22 | 0.11 | 0.05 | 0.21 | 0.11 | 0.05 | |
| Age | 0.04 | 0.00 | 0.00 | ||||
| Male | −0.00 | 0.06 | 0.99 | ||||
| Principal Component 1 | −0.88 | 1.57 | 0.58 | ||||
| Principal Component 2 | 0.04 | 6.43 | 0.99 | ||||
| Principal Component 3 | −3.32 | 2.21 | 0.13 | ||||
| Principal Component 4 | −1.08 | 2.33 | 0.64 | ||||
| Principal Component 5 | −3.30 | 2.64 | 0.21 | ||||
| Principal Component 6 | 1.13 | 2.45 | 0.65 | ||||
| Principal Component 7 | 2.21 | 1.97 | 0.26 | ||||
| Principal Component 8 | −2.10 | 2.21 | 0.34 | ||||
| Principal Component 9 | −0.52 | 2.06 | 0.80 | ||||
| Principal Component 10 | −1.82 | 2.26 | 0.42 | ||||
| Intercept | 4.68 | 0.04 | 0.00 | 2.90 | 0.16 | 0.00 | |
| N | 2,843 | 2,831 | |||||
| R2 | 0.01 | 0.05 | |||||
3.5.2 Replication study 2: new Add Health data
The staggered release of genotypical data by Add Health provides another opportunity to test the association between the 5-HTTLPR genotype and life satisfaction in an independent replication sample as well as the larger combined sample. Table 4 (replication sample) and Table 5 (full sample) present results for association models that are identical to those run in the discovery sample and shown in Table 2. While all model specifications return coefficients on 5-HTTLPR that indicate a positive correlation between the long alleles of this genotype and life satisfaction, no model specification obtains statistical significance. We thus fail to replicate the original significant association result in the newly available Add Health data.
Table 4.
Replication sample OLS models of association between 5-HTTLPR and life satisfaction. Variable definitions are in the Appendix. Standard errors (SE) and P-values are also presented.
| Model 1 | Model 2 | Model 3 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Coeff. | SE | P-value | Coeff. | SE | P-value | Coeff | SE | P-value | |
| 5-HTTLPR long | 0.003 | 0.012 | 0.823 | 0.000 | 0.012 | 0.985 | 0.003 | 0.015 | 0.827 |
| Black | −0.127 | 0.022 | 0.000 | −0.117 | 0.023 | 0.000 | |||
| Hispanic | −0.067 | 0.033 | 0.044 | −0.067 | 0.033 | 0.042 | |||
| Asian | −0.081 | 0.034 | 0.017 | −0.093 | 0.034 | 0.007 | |||
| Age | 0.002 | 0.005 | 0.614 | −0.010 | 0.005 | 0.030 | −0.007 | 0.006 | 0.249 |
| Male | 0.010 | 0.016 | 0.542 | 0.024 | 0.017 | 0.154 | −0.009 | 0.023 | 0.680 |
| Job | 0.092 | 0.021 | 0.000 | 0.115 | 0.029 | 0.000 | |||
| College | 0.200 | 0.017 | 0.000 | 0.238 | 0.042 | 0.000 | |||
| Married | 0.237 | 0.022 | 0.000 | 0.278 | 0.028 | 0.000 | |||
| Divorced | −0.080 | 0.061 | 0.187 | −0.056 | 0.071 | 0.429 | |||
| Religiosity | 0.075 | 0.009 | 0.000 | 0.068 | 0.012 | 0.000 | |||
| Welfare | −0.112 | 0.043 | 0.009 | −0.214 | 0.062 | 0.001 | |||
| Medication | −0.041 | 0.018 | 0.019 | −0.046 | 0.025 | 0.063 | |||
| Intercept | 4.119 | 0.102 | 0.000 | 4.107 | 0.105 | 0.000 | 4.029 | 0.139 | 0.000 |
| N | 10,163 | 10,030 | 5,335 | ||||||
| R2 | 0.004 | 0.042 | 0.055 | ||||||
Table 5.
Full sample OLS models of association between 5-HTTLPR and life satisfaction. Variable definitions are in the Appendix. Standard errors (SE) and P-values are also presented.
| Model 1 | Model 2 | Model 3 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Coeff | SE | P-value | Coeff. | SE | P-value | Coeff. | SE | P-value | |
| 5-HTTLPR long | 0.012 | 0.011 | 0.249 | 0.012 | 0.011 | 0.267 | 0.015 | 0.014 | 0.287 |
| Black | −0.127 | 0.020 | 0.000 | −0.120 | 0.021 | 0.000 | |||
| Hispanic | −0.077 | 0.030 | 0.010 | −0.070 | 0.030 | 0.018 | |||
| Asian | −0.102 | 0.032 | 0.001 | −0.114 | 0.032 | 0.000 | |||
| Age | 0.002 | 0.004 | 0.599 | −0.011 | 0.004 | 0.013 | −0.012 | 0.006 | 0.029 |
| Male | 0.011 | 0.015 | 0.440 | 0.024 | 0.015 | 0.114 | −0.000 | 0.020 | 0.984 |
| Job | 0.091 | 0.019 | 0.000 | 0.117 | 0.026 | 0.000 | |||
| College | 0.181 | 0.015 | 0.000 | 0.229 | 0.020 | 0.000 | |||
| Married | 0.233 | 0.019 | 0.000 | 0.283 | 0.025 | 0.000 | |||
| Divorced | −0.124 | 0.057 | 0.031 | −0.122 | 0.069 | 0.076 | |||
| Religiosity | 0.085 | 0.008 | 0.000 | 0.075 | 0.011 | 0.000 | |||
| Welfare | −0.136 | 0.040 | 0.001 | −0.198 | 0.058 | 0.001 | |||
| Medication | −0.045 | 0.032 | 0.162 | −0.060 | 0.022 | 0.006 | |||
| Intercept | 4.122 | 0.092 | 0.000 | 4.113 | 0.095 | 0.000 | 4.134 | 0.124 | 0.000 |
| N | 12,391 | 12,232 | 6,639 | ||||||
| R2 | 0.004 | 0.044 | 0.058 | ||||||
4 Discussion
Our main objective here has been to provide empirical evidence that genes matter for subjective well-being and to encourage economists to consider the importance of biological differences. The results we present address one possible source of the “baseline” or “set point” for happiness that prior work has identified (Kahneman et al., 1999; Graham, 2008). The existence of a baseline does not mean that the socio-economic influences on happiness so far identified by researchers are unimportant. Rather, our results complement these studies and suggest a new direction for research.
As indicated by the R2 values in Table 2—where the 5-HTTLPR genotype explains less than one percent of the variation in life satisfaction—genotype effect sizes tend to be very small. Still, contrary to a single nucleotide polymorphism (SNP), a functional polymorphism such as the 5-HTTLPR variable number tandem repeat (VNTR) covers a broader fragment of the genome and is understood to have a potentially larger phenotypical influence (Redon et al., 2006; Can et al., 2011). Following on this we also note that because the twin analysis suggests that all genes together account for about a third of the total variance that it is therefore highly likely that many other genes, in conjunction with environmental factors, help to explain how baseline happiness varies from one person to another.
Another use of work such as this is to address the problem of omitted variable bias (OVB). A missing variable might be linked to multiple parameters and thus bias the estimate of the causal effect of X on Y. To the extent that genetic attributes are a source of OVB, and to the extent that they can be added to models of economic outcomes and behaviors, accounting for such variables will improve causal estimates of other attributes.
While the Add Health study presents us with a valuable opportunity to explore a genetic basis of subjective well-being, we want to emphasize a limitation of the data. The Add Health sample is restricted to individuals who are 18–26 years old during Wave III, so our results apply only to the subjective well-being of young adults and not to people in different age categories. However, the strong similarity in the distribution of answers in the Add Health data as compared to other life satisfaction surveys used in the happiness literature suggests that the age limits are not likely to gravely distort our results (Di Tella et al., 2001, 2003; Kahneman and Krueger, 2006; Frey, 2008). Our analyses in the Framingham Heart Study, which has a much wider age range, further suggests a degree of generalizability.
A second important limitation is that we use a case-control method that is vulnerable to population stratification. Because of limited mobility, local adaptation, and genetic drift, it is possible that people from different cultures have a different incidence of certain genotypes, which could lead to a spurious association between genotype and cultural attributes. We limit this potential threat to the validity of our results by including controls for race and limiting the analysis to a specific racial or ethnic group in Add Health. Our related association analysis in the Framingham Heart Study—that controls for the first ten principal components of a singular value decomposition of the subject-genotype matrix—has been shown to effectively deal with the problem of population stratification (Price et al., 2006).
The estimates of the influence of socio-demographic, economic, and cultural covariates on life satisfaction in Table 2 corroborate the generally identified systematic effects of these variables in the literature (for a survey, see Dolan et al. (2008)). In particular, gender does not systematically affect happiness. Higher age has a negative, though not statistically significant effect (this is not surprising considering that our sample refers to young adults). African Americans and Asian Americans are systematically less happy than are Whites, while Latinos are somewhat happier, but not in a statistically significant way. Better educated and married individuals report having significantly higher life satisfaction, while divorced people are more unhappy. Having a job strongly raises life satisfaction. This reflects the psychic benefits of being occupied and integrated into society. At the same time it suggests that having an income raises life satisfaction. In contrast, persons on welfare are much less happy than those employed which reflects the psychic costs of unemployment. Religious individuals are significantly more happy than those without religious beliefs. Persons with less good health, as measured by the need to be on medication, are also less happy. As is the case with most research on happiness, these estimates identify correlations, not causality, given the difficulty in disentangling endogeneity. Once again, consistency with previous studies suggests that results using the Add Health data may generalize to other populations and a wider demography in terms of age.
The life satisfaction question and answer formulation used in Add Health is standard in the economics and psychology literatures (Diener and Diener, 1996; Di Tella et al., 2001; Kahneman and Krueger, 2006; Frey, 2008). This question has been cross-validated with alternative measures that gauge subjective well-being (Kahneman and Krueger, 2006; Bartels and Boomsma, 2009) and Oswald and Wu (2010) provide objective confirmation of life satisfaction as a measure of subjective well-being. Still, the life satisfaction question has been criticized for inducing a focussing illusion by drawing attention to people's relative standing rather than moment-to-moment hedonic experience (Kahneman et al., 2006).
5 Conclusion
Our results corroborate prior research in suggesting that genetic factors significantly influence individual subjective well-being. Using twin study techniques we estimate that genetic variation explains about 33% of the variance in individual happiness. Moreover, using molecular genetic methods we studied the relationship between a functional polymorphism on the serotonin transporter gene (5-HTTLPR) and life satisfaction. Generally, our results provide mixed evidence for a positive association between the “long, ” more efficient 5-HTTLPR alleles and self-reported life satisfaction in a discovery sample and two replication samples. By moving beyond a twin study and focusing on specific genes, our analysis is among the first to consider pathways through which genes may influence happiness levels.
Given prior research linking the “short,” less transcriptionally efficient, alleles of the 5-HTT gene to mood disorders, and the “long,” more efficient alleles to optimism bias, we hypothesized that carriers of the “long” alleles would be more likely to report being satisfied with their lives. We find some support for this intuition in both the Add Health and Framingham Heart Study data but more work needs to be done to better understand the relationship between this genotype and subjective well-being.
We have stressed that genetic factors complement, rather than substitute for, the existing studies showing the influence of socio-demographic, economic and cultural variables on life satisfaction. Future work could attempt to identify other genes or gene-environment interactions that are implicated in subjective well-being. Finding out which genes they are and what physical function they have will improve our understanding of the biological processes that underlie economic outcomes like well-being and may also shed light on their evolutionary origin (Fitzpatrick et al., 2005). While the 5-HTT gene may be a good candidate gene for further study, it is important to re-emphasize that there is no single “happiness gene.” Instead, there is likely to be a set of genes whose expression, in combination with environmental factors, influences subjective well-being.
More broadly, these results suggest that integrating the unique biology of each individual, in addition to studying experience and environment, may usefully complement existing models and increase their explanatory power (Caplin and Dean, 2008). We also believe that genetic association studies such as ours may be a new catalyst for two important lines of research. First, economics places a high premium on causal inference. Provided that robust genetic associations are available and that exclusion restrictions are met, genotypes could function as instrumental variables to disentangle the reverse causality in important relationships that have been plagued by endogeneity. First attempts at using genes as instruments have been tried on the link between health and educational attainment (Fletcher and Lehrer, 2009; Norton and Han, 2009; von Hinke Kessler Scholder et al., 2010; Beauchamp et al., 2011; O'Malley et al., 2010). Second, integrating genetic variation and neuroscientific research may further advance our understanding of the biological underpinnings of individual behavior. For example, the work by Urry et al. (2004) presents neural correlates of subjective well-being. Some of the neurological variation they observe may result from differences in genotypes and could thus inform and stimulate new studies. Since genes are upstream from neurological processes, a better understanding of genetic variation may bring us closer to understanding the objective sources of subjective well-being.
Supplementary Material
Acknowledgment
This research uses data from Add Health, a program project directed by Kathleen Mullan Harris and designed by J. Richard Udry, Peter S. Bearman, and Kathleen Mullan Harris at the University of North Carolina at Chapel Hill, and funded by grant P01-HD31921 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, with cooperative funding from 23 other federal agencies and foundations. Special acknowledgment is due Ronald R. Rindfuss and Barbara Entwisle for assistance in the original design. Information on how to obtain the Add Health data files is available on the Add Health website (http://www.cpc.unc.edu/addhealth). No direct support was received from grant P01-HD31921 for this analysis.
Appendix
Variable Definitions
5-HTTLPR long is a variable for having 0, 1, or 2 of the 528 base-pair alleles of the 5-HTT gene (as opposed to the 484 base-pair version). The race/ethnicity indicator variables are based on the questions “Are you of Hispanic or Latino origin?” and “What is your race? [white/black or African American/American Indian or Native American/Asian or Pacific Islander]”. Age is self-reported age and Male is an indicator taking the value of 1 if the respondent is a male and 0 for a female. Job is the response to the question “Do you currently have a job?“ College is an indicator variable taking the value 1 if the respondent completed at least one year of college and 0 for no college. It is based on the question “What is the highest grade or year of regular school you completed?” Married and Divorced are dummies derived from the population subset that have married and answered “Are you still married?” Religiosity relies on “To what extent are you a religious person?” and takes a value between 0 and 3 for very religious. Welfare is a dummy for “Are you receiving welfare?” Medication is a dummy for “In the past 12 months, have you taken any prescription medication—that is, a medicine that must be prescribed by a doctor or nurse?” DRD4 is the number of r7 alleles (0, 1, or 2) as opposed to r4 alleles. DRD2 is the number of a2 alleles (0, 1, or 2) as opposed to a1 alleles. DAT1 is the number of r9 alleles (0, 1, or 2) as opposed to r10 alleles. MAOA is the number of “High” alleles (0, 1, or 2) as opposed to “Low” alleles. rs2304297 is the number of G alleles (0, 1, or 2) for this SNP on CHRNA6 (as opposed to C alleles). rs892413 is the number of C alleles (0, 1, or 2) for this SNP on CHRNA6 (as opposed to A alleles). rs4950 is the number of G alleles (0, 1, or 2) for this SNP on CHRNB3 (as opposed to A alleles). rs13280604 is the number of G alleles (0, 1, or 2) for this SNP on CHRNB3 (as opposed to A alleles). rs2020933 is the number of A alleles (0, 1, or 2) for this SNP on 5-HTT (as opposed to T alleles).
Principal Component 1–10 is the individual loading for each individual on the 10 principal components associated with the 10 largest eigenvalues of a singular value decomposition of the subject-genotype matrix. These 10 values contain information about population structure, so including them in an association test helps to control for population stratification (Price et al., 2006). Because principal component analysis assumes independent observations, we did not use our entire (family-based) FHS sample to construct the principal components. Instead we used a subsample of 2,507 unrelated individuals to calculate the principal components of the genotypic data and then projected the other individuals in the sample onto those principal components, thus obtaining the loadings of each individual on each of the top 10 principal components.
Footnotes
This paper expands on De Neve (2011)—a research note that reports on the genetic association result in the Add Health discovery sample. The authors thank the anonymous reviewers and Dan Benjamin, Chris Chabris, Chris Dawes, Pete Hatemi, David Laibson, Andrew Oswald, Richard Layard, Jaime Settle, Albert Vernon Smith, and Piero Stanig. De Neve benefited from the generous hospitality of the Institute for Empirical Research in Economics (IEW) at the University of Zurich and CREMA. Research was supported by National Institute on Aging grant P-01 AG-031093 and National Science Foundation grant SES-0719404.
Books are e.g. Kahneman et al. (1999), Graham and Pettinato (2002), Frey and Stutzer (2002a), Van Praag and Ferrer-I-Carbonell (2004), Layard (2005), or Frey (2008); articles are e.g. Easterlin (1974), Clark and Oswald (1996), Frey and Stutzer (2002b), Di Tella et al. (2003), Luttmer (2005), Di Tella and MacCulloch (2006), Rayo and Becker (2007), Dolan et al. (2008), Fowler and Christakis (2008), Urry et al. (2004) or Clark et al. (2008).
They are defined as respectively.
When we split our twin sample by sex we find that there are significant differences between men and women. As in Table 1, Table 3 in the Appendix shows that the AE models fit happiness best according to the AIC values. However, the heritability estimate for males is 39%, whereas for females it is 26%.
A VNTR polymorphism is a repeated segment of DNA that varies among individuals in a population.
A promoter region is the regulatory region of DNA that tells transcription enzymes where to begin. These promoter regions typically lie upstream from the genes they control.
Messenger ribonucleic acid (mRNA) is a type of RNA that carries information from DNA to ribosomes. In turn, these ribosomes “read” messenger RNAs and translate their information into proteins.
Given our data, we cannot differentiate between 1 and 2. In order to do so, we would need additional genetic information about loci in close proximity to the locus of interest. Thus, a significant association means that either a particular allele, or one likely near it on the same gene, significantly influences subjective well-being.
The choice between OLS and ordered probit regression analysis rests on whether the categories of the life satisfaction are considered cardinal or ordinal. Economists typically consider these happiness scores as ordinal and have mainly opted for the ordered type of analysis. Psychologists and sociologists interpret happiness categories as cardinal and therefore use OLS. Ferrer-i-Carbonell and Frijters (2004) survey and test both empirical literatures to conclude that assuming cardinality or ordinality of happiness surveys makes little difference in studies where the dependent variable is measured at a single point in time. We opted for OLS, but other analyses using ordered probit reveal no meaningful differences in coefficients or significance.
This genetic association result in the Add Health discovery sample is also reported in De Neve (2011).
We also report the results of association tests with 5-HTTLPR for each of these socio-economic factors in the appendix. An association with medication is nearly significant (p = 0.08) but loses its significance (p = 0.17) when controlling for age, gender, and race. Hence, medication cannot be considered a mediating variable (Baron and Kenny, 1986).
References
- Add Health Biomarker Team. Biomarkers in wave iii of the add health study. 2007. http://www.cpc.unc.edu/projects/addhealth/files/biomark.pdf. [Google Scholar]
- Alcott H, Karlan D, Mobius M, Rosenblat T, Szeidl A. Community size and network closure. American Economic Review. 2007;97(2):80–85. [Google Scholar]
- Ashenfelter O, Krueger AB. Estimates of the economic return to schooling from a new sample of twins. American Economic Review. 1994;84(5):1157–1173. [Google Scholar]
- Baron R, Kenny D. The moderator-mediator variable distinction in social psychological research: Conceptual, strategic, and statistical considerations. Journal of Personality and Social Psychology. 1986;51(6):1173–1182. doi: 10.1037//0022-3514.51.6.1173. [DOI] [PubMed] [Google Scholar]
- Bartels M, Boomsma DI. Born to be happy? the etiology of subjective well-being. Behavior Genetics. 2009;39(6):605–615. doi: 10.1007/s10519-009-9294-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp J, Cesarini D, Johannesson M, van der Loos M, Koellinger P, Groenen P, Fowler J, Rosenquist J, Thurik A, Christakis N. Molecular genetics and economics. Journal of Economic Perspectives. 2011;25(4):57–82. doi: 10.1257/jep.25.4.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin DJ, Cesarini D, Chabris CF, Glaeser EL, Laibson DI, Gunason V, Harris TB, Launer LJ, Purcell S, Smith AV, Johannesson M, Magnusson PK, Beauchamp JP, Christakis NA, Atwood CS, Hebert B, Freese J, Hauser RM, Hauser TS, Grankvist A, Hultman CM, Lichtenstein P. The promises and pitfalls of genoeconomics. Annual Review of Economics. 2012 Sep;4 doi: 10.1146/annurev-economics-080511-110939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin DJ, Chabris CF, Glaeser EL, Gudnason V, Harris TB, Laibson DI, Launer L, Purcell S. Genoeconomics. In: Weinstein M, Vaupel JW, Wachter KW, editors. Biosocial Surveys. Washington, DC: The National Academies Press; 2007. [Google Scholar]
- Bertolino A, Arciero G, Rubino V, Latorre V, Candia MD, Mazzola V. Variation of human amygdala response during threatening stimuli as a function of 5httlpr genotype and personality style. Biological Psychiatry. 2005;57(12):1517–1525. doi: 10.1016/j.biopsych.2005.02.031. [DOI] [PubMed] [Google Scholar]
- Bouchard T. Genetic and environmental inuences on adult intelligence and special mental abilities. Human Biology. 1998;70(2):257–279. [PubMed] [Google Scholar]
- Can A, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nature Reviews Genetics. 2011;12(5):363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canli T, Lesch K. Long story short: the serotonin transporter in emotion regulation and social cognition. Nature Neuroscience. 2007;10(9):1103–1109. doi: 10.1038/nn1964. [DOI] [PubMed] [Google Scholar]
- Canli T, Omura K, Haas BW, Fallgatter A, Constable RT. Beyond affect: a role for genetic variation of the serotonin transporter in neural activation during a cognitive attention task. Proceedings of the National Academy of Sciences USA. 2005;102:12224–12229. doi: 10.1073/pnas.0503880102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplin A, Dean M. Dopamine, reward prediction error, and economics. Quarterly Journal of Economics. 2008;123(2):663–701. [Google Scholar]
- Caspi A, Sugden K, Moffitt T, Taylor A, Craig I, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, Poulton R. Inuence of life stress on depression: Moderation by a polymorphism in the 5-htt gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Cesarini D, Dawes CT, Johannesson M, Lichtenstein P, Wallace B. Genetic variation in preferences for giving and risk-taking. Quarterly Journal of Economics. 2009;124(2):809–842. [Google Scholar]
- Clark AE, Frijters P, Shields MA. Relative income, happiness, and utility: An explanation for the easterlin paradox and other puzzles. Journal of Economic Literature. 2008;46(1):95–144. [Google Scholar]
- Clark AE, Oswald AJ. Satisfaction and comparison income. Journal of Public Economics. 1996;61(3):359–381. [Google Scholar]
- Cupples L, D'Agnostino R. Survival following initial cardiovascular events: : 30 year follow-up. In: Kannel WB, Wolf PA, Garrison RJ, editors. The Framingham Study: An epidemiological investigation of cardiovascular disease. Bethesda, MD: National Heart, Lung and Blood Institute; 1988. pp. 88–2969. [Google Scholar]
- Damberg M, Garpenstrand H, Hallman J, Oreland L. Genetic mechanisms of behavior: don't forget about the transcription factors. Molecular Psychiatry. 2001;6(5):503–510. doi: 10.1038/sj.mp.4000935. [DOI] [PubMed] [Google Scholar]
- De Bakker P. Imputation in the framingham heart study. Harvard Medical School; Mimeo: 2008. [Google Scholar]
- De Neve J-E. Functional polymorphism (5-httlpr) in the serotonin transporter gene is associated with subjective well-being: Evidence from a us nationally representative sample. Journal of Human Genetics. 2011;56(6):456–459. doi: 10.1038/jhg.2011.39. [DOI] [PubMed] [Google Scholar]
- Di Tella R, MacCulloch R. Some uses of happiness data in economics. Journal of Economic Perspectives. 2006;20:25–46. [Google Scholar]
- Di Tella R, MacCulloch R, Oswald AJ. Preferences over ination and unemployment: Evidence from surveys of happiness. American Economic Review. 2001;91(1):335–341. [Google Scholar]
- Di Tella R, MacCulloch R, Oswald AJ. The macroeconomics of happiness. Review of Economics and Statistics. 2003;85(4):809–827. [Google Scholar]
- Diener E, Diener C. Most people are happy. Psychological Science. 1996;7(3):181–185. [Google Scholar]
- Diener E, Lucas R. Personality and subjective well-being. In: Kahneman D, Diener E, Schwarz N, editors. Well-being: The foundations of hedonic psychology. New York, NY: Sage; 1999. [Google Scholar]
- Dolan P, Peasgood T, White M. Do we really know what makes us happy? a review of the economic literature on the factors associated with subjective well-being. Journal of Economic Psychology. 2008;29:94–122. [Google Scholar]
- Easterlin R. Does economic growth improve the human lot? some empirical evidence. In: David P, Reder M, editors. Nations and Households in Economic Growth: Essays in Honour of Moses Abramowitz. Academic Press; 1974. [Google Scholar]
- Echenique F, Fryer RG. A measure of segregation based on social interactions. Quarterly Journal of Economics. 2007;122(2):441–485. [Google Scholar]
- Echenique F, Fryer RG, Kaufman A. Is school segregation good or bad? American Economic Review. 2006;96(2):265–269. [Google Scholar]
- Fahad AR, Vasiliou SA, Haddley K, Paredes UM, Roberts JC, Miyajima F, Klenova E, Bubb VJ, Quinn J. Combinatorial interaction between two human serotonin transporter gene variable number tandem repeats and their regulation by ctcf. Journal of Neurochemistry. 2010;112(1):296–306. doi: 10.1111/j.1471-4159.2009.06453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-i-Carbonell A, Frijters P. How important is methodology for the estimates of the determinants of happiness. The Economic Journal. 2004 Jul;114:641–659. [Google Scholar]
- Fitzpatrick M, Ben-Shahar Y, Smid H, Vet L, Robinson G, Sokolowski M. Candidate genes for behavioural ecology. Trends in Ecology and Evolution. 2005;20(2):96–104. doi: 10.1016/j.tree.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Fletcher J, Lehrer S. Using genetic lotteries within families to examine the causal impact of poor health on academic achievement; Prepared for the Annual Meeting of the American Economic Association; 2009. [Google Scholar]
- Fowler JH, Baker LA, Dawes CT. Genetic variation in political participation. American Political Science Review. 2008;101(2):233–248. [Google Scholar]
- Fowler JH, Christakis NA. Dynamic spread of happiness in a large social network: Longitudinal analysis over 20 years in the framingham heart study. British Medical Journal. 2008;337(2338):1–9. doi: 10.1136/bmj.a2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler JH, Dawes CT, Christakis NA. Model of genetic variation in human social networks. Proceedings of the National Academy of Sciences USA. 2009;106(6):1720–1724. doi: 10.1073/pnas.0806746106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox E, Ridgewell A, Ashwin C. Looking on the bright side: biased attention and the human serotonin transporter gene. Proceedings of the Royal Society B. 2009;276:1747–1751. doi: 10.1098/rspb.2008.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey BS. Happiness: A Revolution in Economics. MIT Press; 2008. [Google Scholar]
- Frey BS, Stutzer A. Happiness and Economics: How the Economy and Institutions Affect Well-Being. Princeton University Press; 2002a. [Google Scholar]
- Frey BS, Stutzer A. What can economists learn from happiness research? Journal of Economic Literature. 2002b;40(2):402–435. [Google Scholar]
- Glatz K, Mössner R, Heils A, Lesch K. Glucocorticoid-regulated human serotonin transporter (5-htt) expression is modulated by the 5-htt genepromotor-linked polymorphic region. J. Neurochem. 2003;85(5):1072–1078. doi: 10.1046/j.1471-4159.2003.01944.x. [DOI] [PubMed] [Google Scholar]
- Graham C. Economics of happiness. In: Durlauf SN, Blume LE, editors. The New Palgrave Dictionary of Economics. Palgrave Macmillan; 2008. [Google Scholar]
- Graham C, Pettinato S. Happiness and Hardship: Opportunity and Insecurity in New Market Economics. Brookings Institution Press; 2002. [Google Scholar]
- Hariri A, Holmes A. Genetics of emotional regulation: the role of the serotonin transporter in neural function. Trends Cognitive Science. 2006;10:182–191. doi: 10.1016/j.tics.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Hariri A, Mattay V, Tessitore A, Kolachana B, Fera F, Goldman D. Serotonin transporter genetic variation and the response of the human amygdala. Science. 2002;297(5580):400–403. doi: 10.1126/science.1071829. [DOI] [PubMed] [Google Scholar]
- Harris KM, Halpern CT, Smolen A, Haberstick BC. The national longitudinal study of adolescent health (add health) twin data. Twin Research and Human Genetics. 2006;9(6):988–997. doi: 10.1375/183242706779462787. [DOI] [PubMed] [Google Scholar]
- Heils A, Teufel A, Petri S, Stober G, Riederer P, Bengel D, Lesch K. Allelic variation of human serotonin transporter gene expression. J. Neurochem. 1996;66(6):2621–2624. doi: 10.1046/j.1471-4159.1996.66062621.x. [DOI] [PubMed] [Google Scholar]
- Heinz A, Braus D, Smolka M, Wrase J, Puls I, Hermann D, et al. Amygdala-prefrontal coupling depends on a genetic variation of the serotonin transporter. Nat Neurosci. 2005;8(1):20–21. doi: 10.1038/nn1366. [DOI] [PubMed] [Google Scholar]
- Huezo-Diaz P, Rietschel M, Henigsberg N, Marusic A, Mors O, Maier W, Hauser J, Souery D, Placentino A, Zobel A, Larsen ER, Czerski PM, Gupta B, Hoda F, Perroud N, Farmer A, Craig I, Aitchison KJ, McGuffin P. Moderation of antidepressant response by the serotonin transporter gene. British Journal of Psychiatry. 2009;195:30–38. doi: 10.1192/bjp.bp.108.062521. [DOI] [PubMed] [Google Scholar]
- Jacobson K, Rowe D. Genetic and shared environmental inuences on adolescent bmi: Interactions with race and sex. Behavior Genetics. 1998;28(4):265–278. doi: 10.1023/a:1021619329904. [DOI] [PubMed] [Google Scholar]
- Kahneman D, Diener E, Schwarz N. Well-being: The Foundations of Hedonic Psychology. New York: Russel Sage; 1999. [Google Scholar]
- Kahneman D, Krueger AB. Developments in the measurement of subjective well-being. Journal of Economic Perspectives. 2006;20:3–24. [Google Scholar]
- Kahneman D, Krueger AB, Schkade D, Schwarz N, Stone AA. Would you be happier if you were richer? a focusing illusion. Science. 2006;312(5782):1908–1910. doi: 10.1126/science.1129688. [DOI] [PubMed] [Google Scholar]
- Kannel W, Feinleib M, McNamara P, Garrison R, Castelli W. An investigation of coronary heart disease in families. American Journal of Epidemiology. 1979;110:281–290. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Kessler RC, Heath AC, Eaves LJ. A test of the equal-environment assumption in twin studies of psychiatric illness. Behavior Genetics. 1993;23:21–27. doi: 10.1007/BF01067551. [DOI] [PubMed] [Google Scholar]
- Layard R. Happiness: Lessons from a New Science. Penguin; 2005. [Google Scholar]
- Lesch K, Bengel D, Heils A, Sabol S, Greenberg B, Petri S, et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274:1527–1531. doi: 10.1126/science.274.5292.1527. [DOI] [PubMed] [Google Scholar]
- Lipsky RH, Hu X-Z, Goldman D. Additional functional variation at the slc6a4 gene. American Journal of Medical Genetics B Neuropsychiatric Genetics. 2009;150B(1):153. doi: 10.1002/ajmg.b.30766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little K, McLaughlin D, Zhang L, Livermore C, Dalack G, McFinton P. Cocaine, ethanol, and genotype effects on human midbrain serotonin transporter binding sites and mrna levels. American Journal of Psychiatry. 1998;155:207–213. doi: 10.1176/ajp.155.2.207. [DOI] [PubMed] [Google Scholar]
- Luttmer EF. Neighbors as negatives: Relative earnings and well-being. Quarterly Journal of Economics. 2005;120(3):963–1002. [Google Scholar]
- Lykken D, Tellegen A. Happiness is a stochastic phenomenon. Psychological Science. 1996;7(3):186–189. [Google Scholar]
- Mackay T. The genetic architecture of quantitative traits. Annual Review of Genetics. 2001;35:303–339. doi: 10.1146/annurev.genet.35.102401.090633. [DOI] [PubMed] [Google Scholar]
- Martin J, Cleak J, Willis-Owen S, Flint J, Shifman S. Mapping regulatory variants for the serotonin transporter gene based on allelic expression imbalance. Molecular Psychiatry. 2007;12(5):421–422. doi: 10.1038/sj.mp.4001952. [DOI] [PubMed] [Google Scholar]
- McGreal R, Stephen J. The depression-happiness scale. Psychological Reports. 1993;3:1279–1282. doi: 10.2466/pr0.1993.73.3f.1279. [DOI] [PubMed] [Google Scholar]
- Munafò MR, Brown SM, Hariri AR. Serotonin transporter (5httlpr) genotype and amygdala activation: a meta-analysis. Biological Psychiatry. 2008;63:852–857. doi: 10.1016/j.biopsych.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafò MR, Clark T, Flint J. Does measurement instrument moderate the association between the serotonin transporter gene and anxietyrelated personality traits? a meta-analysis. Molecular Psychiatry. 2005;10:415–419. doi: 10.1038/sj.mp.4001627. [DOI] [PubMed] [Google Scholar]
- Neale M, Boker S, Xie G, Maes H. OpenMx advanced structural equation modeling. 2010. ( http://openmx.psyc.virginia.edu/) [Google Scholar]
- Nes RB, Roysamb E, Tambs K, Harris JR, Reichborn-Kjennerud T. Subjective well-being: genetic and environmental contributions to stability and change. Psychological medicine. 2006;36(7):1033–1042. doi: 10.1017/S0033291706007409. [DOI] [PubMed] [Google Scholar]
- Norton E, Han E. How smoking, drugs, and obesity affect education, using genes as instruments; Prepared for the Annual Meeting of the American Economic Association; 2009. [Google Scholar]
- O'Malley A, Rosenquist J, Zaslavsky A, Christakis N. Estimation of peer effects in longitudinal models using genetic alleles as instrumental variables. Mimeo: Harvard University; 2010. [Google Scholar]
- Oswald AJ, Wu S. Objective confirmation of subjective measures of human well-being: Evidence from the u.s.a. Science. 2010;327:576–579. doi: 10.1126/science.1180606. [DOI] [PubMed] [Google Scholar]
- Pezawas L, Meyer-Lindenberg A, Drabant EM, Verchinski BA, Munoz KE, Kolachana BS, Egan MF, Mattay VS, Hariri AR, Weinberger DR. 5-httlpr polymorphism impacts human cingulate-amygdala interactions: a genetic susceptibility mechanism for depression. Nature Neuroscience. 2005;8:828–834. doi: 10.1038/nn1463. [DOI] [PubMed] [Google Scholar]
- Plomin R, DeFries JC, McClearn GE, McGuffin P. Behavioral genetics. 5th ed edition New York, NY: Worth Publishers; 2008. [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Rayo L, Becker GS. Habits, peers, and happiness: An evolutionary perspective. American Economic Review. 2007;97(2):487–491. [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, et al. Global variation in copy number in the human genome. Nature. 2006;444(7118):444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Herrell R, Lehner T, Liang K, Eaves L, Hoh J, Griem A, Kovacs M, Ott J, Merikangas K. Interaction between the serotonin transporter gene (5-httlpr), stressful life events, and risk of depression: a meta-analysis. Journal of the American Medical Association. 2009;301:2462–2471. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarr S, Carter-Saltzman L. Twin method: Defense of a critical assumption. Behavior Genetics. 1979;9:527–542. doi: 10.1007/BF01067349. [DOI] [PubMed] [Google Scholar]
- Sen S, Burmeister ML, Ghosh D. Meta-analysis of the association between a serotonin transporter promoter polymorphism (5-httlpr) and anxiety related personality traits. AmJMed. Genet. B. 2004;127:85–89. doi: 10.1002/ajmg.b.20158. [DOI] [PubMed] [Google Scholar]
- Stevenson B, Wolfers J. Economic growth and subjective well-being: Reassessing the easterlin paradox. Brookings Papers on Economic Activity. 2008;2:1–87. [Google Scholar]
- Stubbe J, Posthuma D, Boomsma D, De Geus E. Heritability of life satisfaction in adults: a twin-family study. Psychological Medicine. 2005;35(11):1581–1588. doi: 10.1017/S0033291705005374. [DOI] [PubMed] [Google Scholar]
- Taubman P. The determinants of earnings: Genetics, family, and other environments: A study of white male twins. American Economic Review. 1976;66:858–870. [Google Scholar]
- Urry HL, Nitschke JB, Dolski I, Jackson DC, Dalton KM, Mueller CJ, Rosenkranz MA, Ryff CD, Singer BH, Davidson RJ. Making a life worth living: Neural correlates of well-being. Psychological Science. 2004;15(6):367–372. doi: 10.1111/j.0956-7976.2004.00686.x. [DOI] [PubMed] [Google Scholar]
- Van Praag B, Ferrer-I-Carbonell A. Happiness Quantified: A Satisfaction Calculus Approach. Oxford University Press; 2004. [Google Scholar]
- Visscher P, Medland S, Ferreira M, Morley K, Zhu G, Cornes B, Montgomery G, Martin N. Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between full siblings. PLoS Genetics. 2006;2:e41. doi: 10.1371/journal.pgen.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hinke Kessler Scholder S, Smith GD, Lawlor DA, Propper C, Windmeijer F. Genetic markers as instrumental variables: An application to child fat mass and academic achievement. University of Bristol, Centre for Market and Public Organisation Working Paper 10/229. 2010. [Google Scholar]
- Wendland J, Martin B, Kruse M, Lesch K, Murphy D. Simultaneous genotyping of four functional loci of human slc6a4, with a reappraisal of 5-httlpr and rs25531. Molecular Psychiatry. 2006;11(3):224–226. doi: 10.1038/sj.mp.4001789. [DOI] [PubMed] [Google Scholar]
- Xian H, Scherrer JF, Eisen SA, True WR, Heath AC, Goldberg J, Lyons MJ, Tsuang MT. Self-reported zygosity and the equalenvironments assumption for psychiatric disorders in the vietnam era twin registry. Behavior Genetics. 2000;30:303–310. doi: 10.1023/a:1026549417364. [DOI] [PubMed] [Google Scholar]
- Xu H, Shete S. Mixed-effects logistic approach for association following linkage scan for complex disorders. Annals of Human Genetics. 2006;71(2):230–237. doi: 10.1111/j.1469-1809.2006.00321.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.