Abstract
Nucleic acid (NA) extraction and purification has become a common technique in both research and clinical laboratories. Current methods require repetitive wash steps with a pipette that is laborious and time consuming making it inefficient for clinical settings. We present here a simple technique that relies on spontaneous biphasic plug flow inside a capillary to achieve sample preparation. By filling the sample with oil, aqueous contaminants were displaced from the captured NA without pipetting wash buffers or use of external force and equipment. mRNA from mammalian cell culture was purified and PCR amplification showed similar threshold cycle values as those obtained from a commercially available kit. HIV viral like particles were spiked into serum and a 5-fold increase in viral RNA extraction yield was achieved compared to the conventional wash method. In addition, viral RNA was successfully purified from human whole blood, and a limit of detection of approximately 14 copies of RNA extracted per sample. The results demonstrate the utility of the current technique for nucleic acid purification for clinical purposes, and the overall approach provides a potential method to implement nucleic acid testing in low resource settings.
INTRODUCTION
Nucleic acid (NA) testing provides a rapid and sensitive way of monitoring and diagnosing a wide range of diseases. Currently approved by the US Food and Drug Administration (FDA) are NA tests that are used to diagnose tuberculosis, chlamydia, gonorrhea, and human immunodeficiency virus (HIV). For viral diseases, NA tests based on polymerase chain reaction (PCR) offer faster disease confirmation prior to seroconversion as well as important quantitative information (e.g., viral load) not available from antibody-based diagnostics. While these PCR based tests provide sensitive quantitative measurements, the reliability of PCR results is heavily dependent on the quality of the nucleic acid template and the presence of inhibitors.1 As such, effective sample preparation procedures are critical to ensure reliable amplification and accurate test results.
Sample preparation begins with disruption of the cell or virus membrane by chemical or mechanical methods. The majority of commercially available kits employ solid phase extraction (SPE), which utilize functionalized surfaces or silica coated particles (i.e. paramagnetic particles, PMPs) to capture NA. This is followed by extensive and repetitive washing or centrifugation steps to ensure removal of contaminants and inhibitors.2 These procedures are time consuming, laborious and costly, making NA extraction especially difficult to perform in rural clinics and low resource settings. As such, sample preparation and PCR are usually performed in a centralized laboratory with specialized instruments and skilled technicians. As a result, implementation of NA testing remains difficult and the impact of these tests is limited in these regions.
To address these issues, a number of research groups have focused on developing microfluidic based diagnostic devices with sample preparation technologies that minimize complex equipment and infrastructure requirements. In particular, paper-based microfluidic devices have received substantial interest due to their inherent low cost, ease of fabrication and simplicity of operation.3,4,5 In contrast to lateral flow assays, these devices were prepared by patterning paper with different hydrophobic materials to create confined geometry and channels to facilitate multi-analyte detection.4 Other groups have combined papers with different properties together to facilitate sample filtration and detection.5 While a few paper based systems have demonstrated applications for NA testing,6 the method is predominately used for protein and small molecule detection that are present in blood or serum which do not require complicated sample preparation method to extract.
Other microfluidic NA tests have focused on using immiscible phases to facilitate sample preparation.7,8 Specifically, Shen et al. performed qPCR to quantify HIV/HCV viral load using the slipchip system and Bordelon et al. performed RNA extraction using a moving magnet that pulls PMPs across air and liquid interfaces. In addition, Sur et al. performed NA purification in a single pass through a liquid wax filtration device. Our group demonstrated the use of “pinned” immiscible fluids to purify nucleic acids.7 By moving PMPs with captured nucleic acids across oil/aqueous interfaces, a purified sample is achieved in a single step without repetitive washing.
Here, we describe a sample preparation method that utilizes the ability to generate spontaneous biphasic plug flow in capillaries for NA purification. First introduced by Marangoni and later explained by Bico et al., immiscible fluids connected inside a capillary generate a spontaneous motion without external force.9,10 Pompano and colleagues took advantage of this phenomenon to control initiation and rate of fluid flow in microfluidic channels on SlipChip.19 To purify NA, we rely on an imbalance of surface tensions between two immiscible fluids developed inside a capillary tube to spontaneously displace the aqueous phase contaminants from the NA bound solid phase. To operate, lysed sample containing PMPs functionalized to bind NA are introduced to the glass capillary via capillary filling. A magnet then holds the PMPs stationary as an immiscible oil is introduced to spontaneously “displace” the cell debris and contaminants by leveraging an asymmetry between the fluids. Finally, an elution buffer is introduced to release the NA’s bound to the PMPs. This method improves assay efficiency by reducing the number of wash steps and eliminates the need for complicated external equipment, potentially making NA testing more accessible in resource limited regions.
EXPERIMENTAL SECTION
Capillary Operation
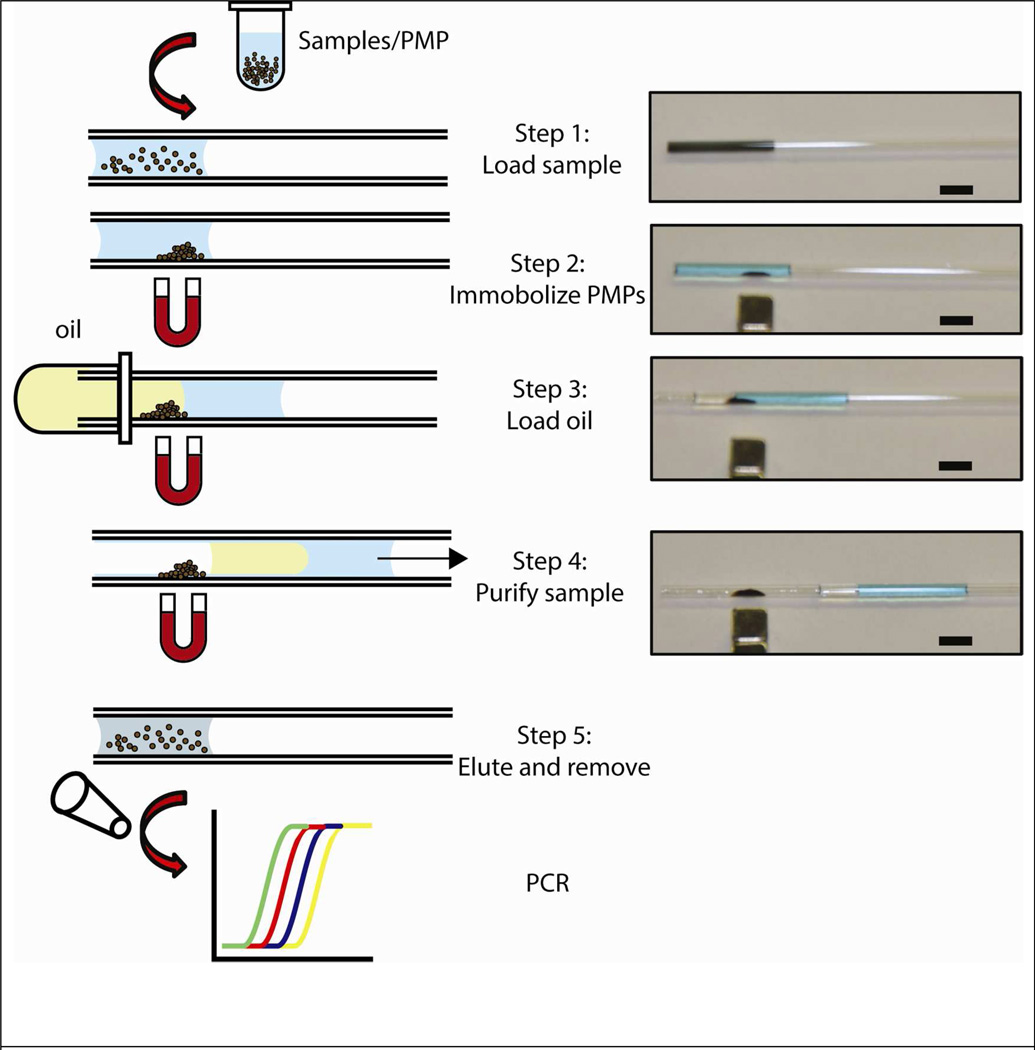
First, a 10 µL of lysed sample (cell lysate, viral particles in serum or whole blood) with NA capturing PMPs was introduced into a glass capillary tube via capillary action (Fig 1, Step 1). A magnet was held parallel to the capillary to immobilize the beads to the capillary wall surface (Fig 1, Step 2). Next, 10 µL of FC-40 oil containing 1 % PFO was drawn into the capillary connecting the two fluids (Fig 1, Step 3). Once the plug of oil was filled, the juxtaposition of immiscible phases (oil and aqueous lysate) created an asymmetry in the overall surface tension-generated forces. This imbalance generated an overall force that spontaneously moved the fluid plugs towards the end of the capillary where they were removed via a wicking pad (Fig 1, Step 4). Finally, the magnet was removed and 10 µL of elution buffer (EB) was pulled into the tube to re-suspend the PMPs (Fig 1, Step 5). After 5 minutes, the eluted NA was collected from the capillary using a single pipetting step for downstream detection via RT-qPCR.
Figure 1.
Capillary based nucleic acid purification device. Sample with lysis buffer and PMP was pulled into a tube via capillary forces (Step 1). PMPs with captured nucleic acids were immobilized on the tube surface with a magnet (Step 2). Oil passively pulled into the tube by capillary action (Step 3) creates a surface tension asymmetry between the two phases displacing the entire fluid from one end of the capillary to the other (Step 4). This results in exclusion of PMPs from the uncaptured components of the lysate and purification of the captured nucleic acids (Step 4). Captured nucleic acids were then eluted from PMPs for downstream analysis (Step 5). Pictures on left correspond to each of the steps shown in the schematic. In these images, the aqueous phase is colored with blue dye for visualization. Scale bar is 5 mm.
Residual Sample Characterization
2% (v/v) red fluorescent protein (RFP) in PBS with varying concentration of Triton-X 100 was used to characterize the amount of residual sample on untreated or bovine serum albumin (BSA) treated capillary walls. BSA treated capillaries were prepared by immersing the glass capillary tubes (0.81 mm ID, Fisher Scientific, Pittsburgh PA) in 3% (w/v) BSA in PBS overnight and drying under N2 to remove any remaining solution prior to experiments. To characterize sample deposition on capillary walls, 10 µL of RFP solution was first drawn into the capillary followed by the addition of 10 µL FC-40 with 1 % 1H,1H,2H,2H-perfluoro-1-octanol (PFO, Sigma Aldrich, St. Louis, MO) to facilitate liquid displacement. Fluorescent images of the capillary before and after the solution traveled across the capillary were captured using a fluorescence microscope (Olympus IX70, Center Valley, PA). NIH Image J software was used to determine the amount of residual RFP on the wall surface.
Cellular mRNA Purification and Amplification
Breast cancer epithelial cells (MCF-7) were cultured in Dulbecco's Modified Eagle Medium (DMEM; Mediatech, Herndon, VA) containing 10 % fetal bovine serum (FBS) and 1 % Pen-Strep at 37°C, and maintained under 5 % CO2 in polystyrene flasks until confluent. Cells were released using a 0.25% trypsin/EDTA solution and collected via centrifugation. 106 cells were pelleted and lysed in 1× RIPA buffer (Millipore, Billerica, MA) for 1 minute with constant mixing followed by serial dilution with RIPA buffer. For each purification, 10 µL of the lysate was mixed with 15 µg mRNA Dynabeads Oligo(dT) PMPs (Life Technologies, Carlsbad, CA) for 5 minutes at room temperature. The standard wash protocol from Dynabeads® mRNA Direct™ Kit (Life Technologies) was compared to the capillary purification method. Briefly, after lysis and mixing, the supernatant was removed and the PMPs were sequentially washed with the buffers provided by the manufacturer. Nuclease free (NF) water was used to elute the nucleic acids from PMPs and collected for downstream analysis. For capillary based purification, the device was operated as previously described using fluorinated oil FC-40 containing 1 % PFO instead of the buffers.
Isolated mRNA was reverse transcribed using the High Capacity RNA to cDNA Reverse Transcription kit (ABI, Foster City, CA) at 37°C for 60 minutes followed by 95°C for 5 minutes. 20 µl qPCR reactions were setup by mixing, 4 µL of cDNA template with SYBR® Green PCR Master Mix (Bio-Rad, Hercules, CA) and NF water along with forward (5’-GACAATGGCAGCATCTACAAC-3’) and reverse (5’-GCAGACAGACACTGGCAAC-3’) primers for human ribosomal protein P0 to achieve a final primer concentration of 5 µM. The reaction was amplified at 95°C for 15 seconds (denaturing) followed by 60°C for 1 minute (annealing) for 45 cycles using a MyiQ thermal cycler (Bio-Rad). Data were analyzed using software provided by the manufacturer to determine threshold cycle values (Ct) that correspond to gene expression levels.
HIV Viral Like Particles (VLPs) Production
HIV viral like particles expressing p24 capsid protein were produced by transfecting HEK293T cells with Gag-Pol-Vif and Rev plasmids (generous gift from Dr. Nate Sherer). 4×106 cells were plated onto a 100 mm cell culture dish one day prior to transfection. Cells were cultured in DMEM supplemented with 4.5 g/L D-Glucose, L-Glutamine (Life Technologies), and 10% FBS and maintained under humidified air with 5% CO2 (v/v) at 37 °C. Transfection medium was prepared by mixing 30 µl of FuGENE®6 Transfection Reagent (Promega, Fitchburg, WI) with 7.5 µg of p24 Gag-Pol-Vif expression plasmid and 2.5 µg of pRev expression plasmid; the mixture was added to the cells. Cells were incubated for 4 days with medium being changed every 24 hr. On days 3 and 4, culture medium was collected and filtered through a 0.2 µm filter (Nalgene, Rochester, NY). The filtered medium was centrifuged for 2 hours at 21,000 × g at 4°C through a 20% sucrose gradient (Sigma Aldrich). The supernatant was discarded and the pellet containing the HIV VLPs was resuspended in 1× PBS containing 10 % FBS. VLPs were stored at −80°C until use in an isolation experiment.
VLP Spike-in Experiment
HIV VLPs were spiked into either fetal bovine serum or human heparinized blood (Valley Biomedical, Winchester, VA) to simulate clinical samples. Nucleic acids from VLPs were purified using the MagAttract Viral M48 RNA kit (Qiagen, Germantown, MD). Lysis, elution buffers, and PMPs were used from the MagAttract kit. Briefly, heparinized blood that was tested negative for HIV was mixed with MLF lysis buffer at a 1:2 ratio. 10 µL of VLP stock solution at a concentration of 140000 copies/µL was spiked into the mixture. From there 4, 1:10 dilutions were performed using the initial 1:2 heparinized blood and MLF lysis buffer solution to keep the buffer to sample ratio consistent. At the lowest concentration, a total of 14 copies of viral RNA were extracted in each procedure. For each reaction, 10 µL of each dilution was added to 1 µL MagAttract Suspension F PMPs. Each sample was incubated for 5 minutes at room temperature. Purification was executed as described above using FC 40 oil containing 1 % PFO instead of wash buffers.
Isolated RNA from VLPs was reverse transcribed as previously described. For qPCR amplification, 4 µL cDNA template was mixed with 10 µL of TaqMan Gene Expression Master Mix (ABI), 5 µL NF water and 1 µL each of the p24 primers and 0.5 ul of probe (forward 5’-GGCCAGGAATTTTCTTCAGA-3’, reverse 5’-TTGTCTCTTCCCCAAACCTGA-3', probe 5’-ACCAGAGCCAACAGCCCCACCAGA-3’). The reaction was amplified for 40 cycles (denaturing at 95°C for 15 sec followed by annealing at 60°C for 1 minute) using the Roche LightCycler 480.
To quantify the amount of extracted viral RNA, a standard curve was generated by amplifying known copy numbers of p24 Gag-Pol-Vif and pREV expression plasmid to determine the corresponding threshold cycle values.
RNA Quantification
RNA samples were quantified using the QuantiFluor RNA Dye System (Promega). Following the manufacturer’s protocol, a standard curve was first prepared in order to determine the RNA samples. Briefly, 1.2 Kb RNA Standards (provided by the manufacturer) were serially diluted (0 ng/mL to 250 ng/mL) in TE buffer and mixed with the QuantiFluor RNA Dye at a 1:1 ratio. Mixtures were incubated in the dark for 5 minutes, and 2 µL was added to a Nanodrop (Thermo Fisher Scientific, Waltham, MA). Samples were illuminated at 450 nm and intensity captured at 540 nm. Extracted RNA were diluted and mixed with the Quantifluor Dye at a 1:1 ratio allowing for the concentration to be calculated from the standard curve.
RESULTS AND DISCUSSION
The extraction and purification of NA is a ubiquitous technique performed in both research and clinical laboratories. Current methods utilizing PMPs and other SPE methods require multiple wash steps with specialized buffers to sufficiently remove any contaminants or inhibitors from the sample. To overcome these obstacles, we have developed a simple method that utilizes spontaneous biphasic plug flow inside a capillary to effectively separate aqueous contaminants from captured nucleic acids on paramagnetic particles (PMPs) without external application of force or equipment. In contrast to conventional methods which involve repeated pipetting of fluids through the capture beads, with our method, the entire purification was completed passively by capitalizing on capillary action and surface tension.
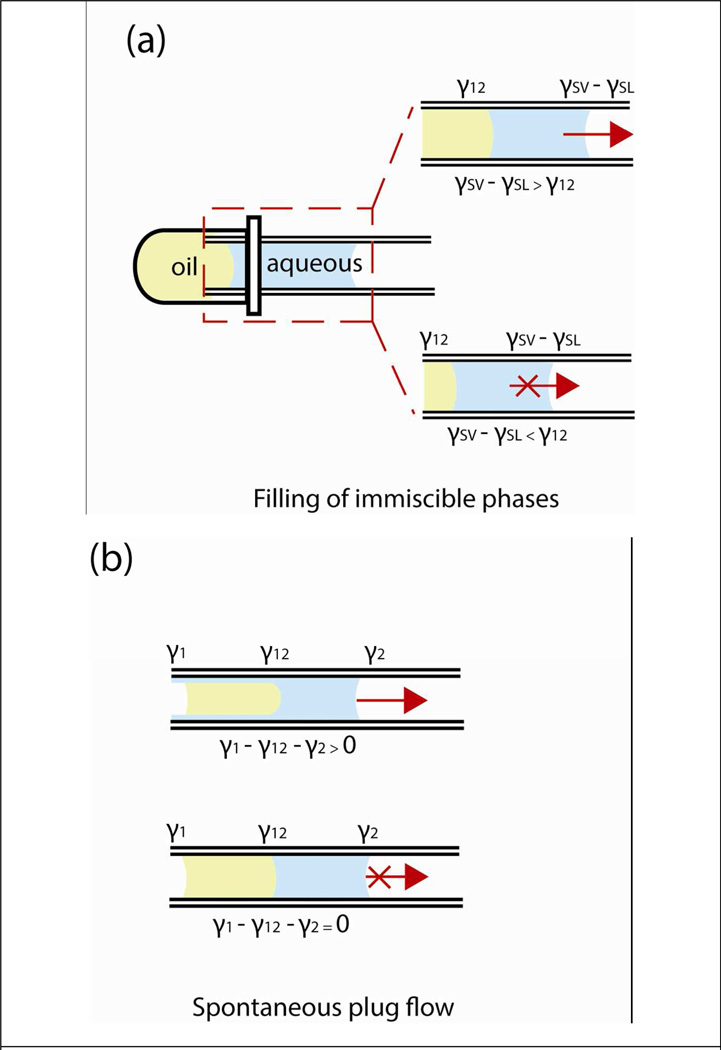
To operate the system, there are two distinct processes that need to occur. First, both phases must fill the tube by capillary action from a reservoir of liquid (Fig 2a). Second, an imbalance in the overall forces generated by surface tension must exist between the immiscible fluids (Fig 2b). To fill the oil behind the aqueous phase, the pressure drop induced by oil-aqueous interface must be smaller than the pressure drop generated by the aqueous-air interface. Using the Young-Laplace equation, the following equation was derived which characterized this phenomenon,
| (1) |
where γSV is the surface tension between glass and air, γSL is the surface tension between glass and aqueous phase liquid, and γ12 is the interfacial tension between the immiscible phases. Only when the interfacial tension γ12 is lower than the differences of glass-air and glass-liquid surface tension (γSV − γSL) will both fluids fill the capillary (Fig 2a, also see Supplementary Section Table S1 for empirical data and derivation of the equation).
Figure 2.
Characterization of the driving force behind fluid displacement in capillary. To fill the capillary with both oil and aqueous phase, the interfacial tension between the two fluids (γ12) has to be less than the energy required to wet the capillary (γSV − γSL), otherwise it will not be energetically favorable for both fluids to be pulled in the capillary (a). Once filling has occurred, the force to displace the fluids within the capillary is the result of an imbalance in the surface tension (b). When surface tensions are balanced, the fluids remain stationary. γ1 and γ2 are the surface tension of aqueous and oil phase, γSV and γSL are the surface tension of glass in air and glass in liquid, respectively.
To induce fluid displacement of the combined liquids (aqueous, oil), asymmetry of the forces generated by surface tension between the advancing and receding interfaces must occur (Fig 2b). This spontaneous plug-flow phenomenon creates a forward displacement of fluids was first observed by Marangoni and later explained by Bico9,10 which is described as:
| (2) |
where γ1 is the surface tension of the aqueous phase, γ2 is the surface tension of oil, and γ12 is the interfacial tension between the fluids. The generation of fluid motion in our system does not require any external forces (i.e. pressure) and is solely dependent on the force imbalance created by surface tension. When the forces generated by surface tension are balanced, the fluids remain stationary (Fig 2b, second condition pictured).
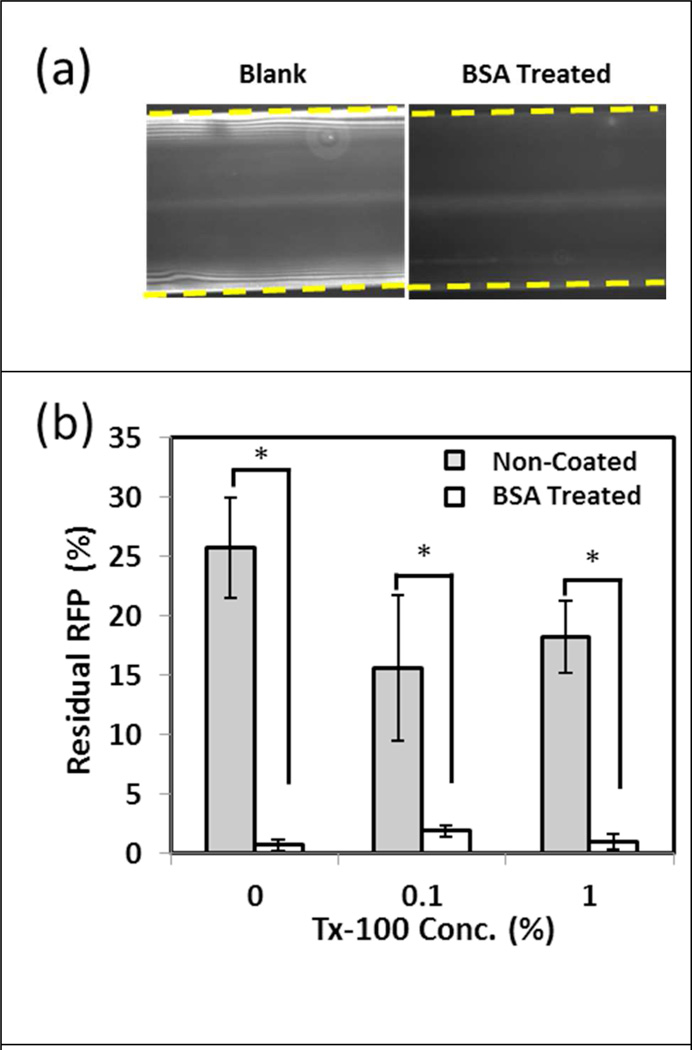
We first examined the ability of the current technique to displace aqueous contaminants. Previous studies have shown that the aqueous phase acts as a lubricant, leaving behind a thin film on the capillary walls.10 RFP was added to the aqueous phase to visualize and determine the amount of residual sample in the capillary after purification. In untreated glass capillaries, a layer of residual RFP solution was observed after the displacement of the liquid (Fig. 3a). After repeated washes with PBS, residual RFP remained unchanged indicating that the protein had adsorbed onto the wall surface. To reduce the effects of non-specific adsorption, capillaries were pre-treated with BSA; thus resulting in decreased RFP intensity (Fig 3a). In untreated capillaries, approximately 25% of the RFP remained within the capillary due to protein adsorption on the wall surface. BSA treated capillaries minimized this effect with less than 3% residual RFP after purification; indicating that protein absorption was further eliminated (Fig 3b). Additionally, increasing the surfactant concentration (Triton-X 100) in the aqueous phase did not affect the overall quantity of residual RFP within the capillary, a potentially useful feature for applications requiring higher aqueous surfactant concentrations especially during lysis step.
Figure 3.
Characterization of residual sample on capillary walls during purification. Red fluorescent protein (RFP) spiked into PBS was used as a model system to quantify the amount of residual sample after the purification step. In both untreated and BSA treated capillaries, RFP was observed on the surface after liquid had traveled across the capillary, with less residual RFP observed in the BSA capillary (a). Dashed lines demonstrate the outer edges of the capillary. Untreated capillaries showed higher residual RFP signal compared to those that were pre-treated with BSA (b). Two tailed t-tests were performed to compare BSA treated and non-treated samples (* indicates p < 0.05).
Non-specific adsorption of molecules onto glass surfaces have been extensively studied and attributed to electrostatic interactions. In the context of our current system, minimizing the adsorption onto capillary walls with BSA treatment allowed the quantification of residual fluid after purification. In addition, this method could also serve to reduce potential sample loss of nucleic acids adsorbed on glass surfaces, a commonly used method to capture NA. As such, all experiments from this point were performed using BSA treated glass capillary tubes.
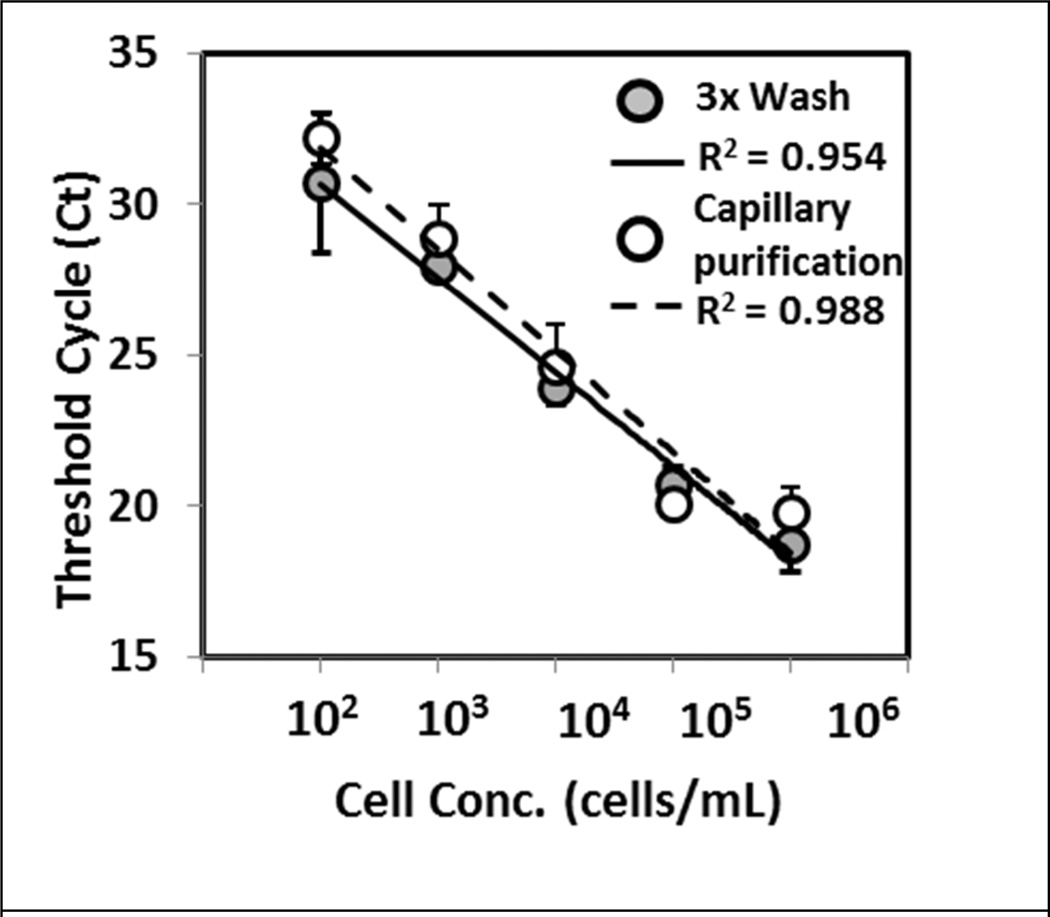
The utility of the current technique to extract NA was examined using different biological samples. First, mRNA from a breast cancer epithelial cell lysate (MCF-7) was extracted and purified. Expression levels of ribosomal protein P0 were amplified and compared to those obtained using the Invitrogen mRNA Direct kit method. The amount of mRNA recovered, as determined by threshold cycle, from both methods showed no statistical difference across the different cell concentrations using the two-tailed Student’s t-test (Fig 4). Furthermore, the efficiencies of PCR were calculated to be 99 % and 110 % for capillary purification and the commercial kit, both within the acceptable range for PCR assays.11 In addition, the current method successfully purified mRNA from samples containing a single cell (10 µL input sample at 100 cell/mL), and the threshold cycle agreed with those obtained using the commercial kit (data not shown). These results show the ability of the current technique to effectively purify mRNA across a range of cell numbers.
Figure 4.
Nucleic acid extraction and purification from cell lysate. RNA from MCF-7 cell lysate was purified using capillary and conventional washing methods. Amplification of ribosomal protein P0 demonstrated similar threshold cycle values between the two purification methods. A linear response was observed across the different cell concentration with both techniques as shown by the R2 values. Data and error bars represent the mean and the standard deviation of the triplicate experiment.
NA purification kits typically require multiple washing and mixing steps to separate and dilute aqueous contaminants. Depending on the SPE provided from the manufacturer, different equilibration conditions from wash buffers as well as vigorous washing can remove PMPs or bound samples, reducing the overall yield. We compared the amount of extracted viral RNA using the current capillary purification method with from the Qiagen MagAttract Viral RNA kit. The beads provided from this kit are silica coated PMPs and bind to RNA through electrostatic interaction in the presence of chaotropic salts. To demonstrate relevance for clinical testing, HIV viral-like particles (VLP), each containing two copies of viral RNA, were spiked into fetal bovine serum. HIV viral-like particles were prepared by transfecting HEK293T cells and the particles contain only the specific genetic material that was included during production without the envelope proteins required for viral infection. The sample was lysed and split in half to be purified using our capillary method and the conventional kit-based method. Higher concentrations of viral RNA were consistently extracted with the capillary purification method compared to the protocol from the kit (Table 1). The multiple buffers in the purification kit require the user to pipette repeatedly creating high shear, thus potentially resulting in sample loss during the wash and transfer processes. In comparison, the capillary method completes purification in a single step without repeated sample mixing thus improving the overall yield of extracted RNA by 5 fold. While this potential sample loss could be due to multiple transfer and pipette steps it could also be a dilutive effect from the wash buffers in comparison to the capillary based exclusion method.
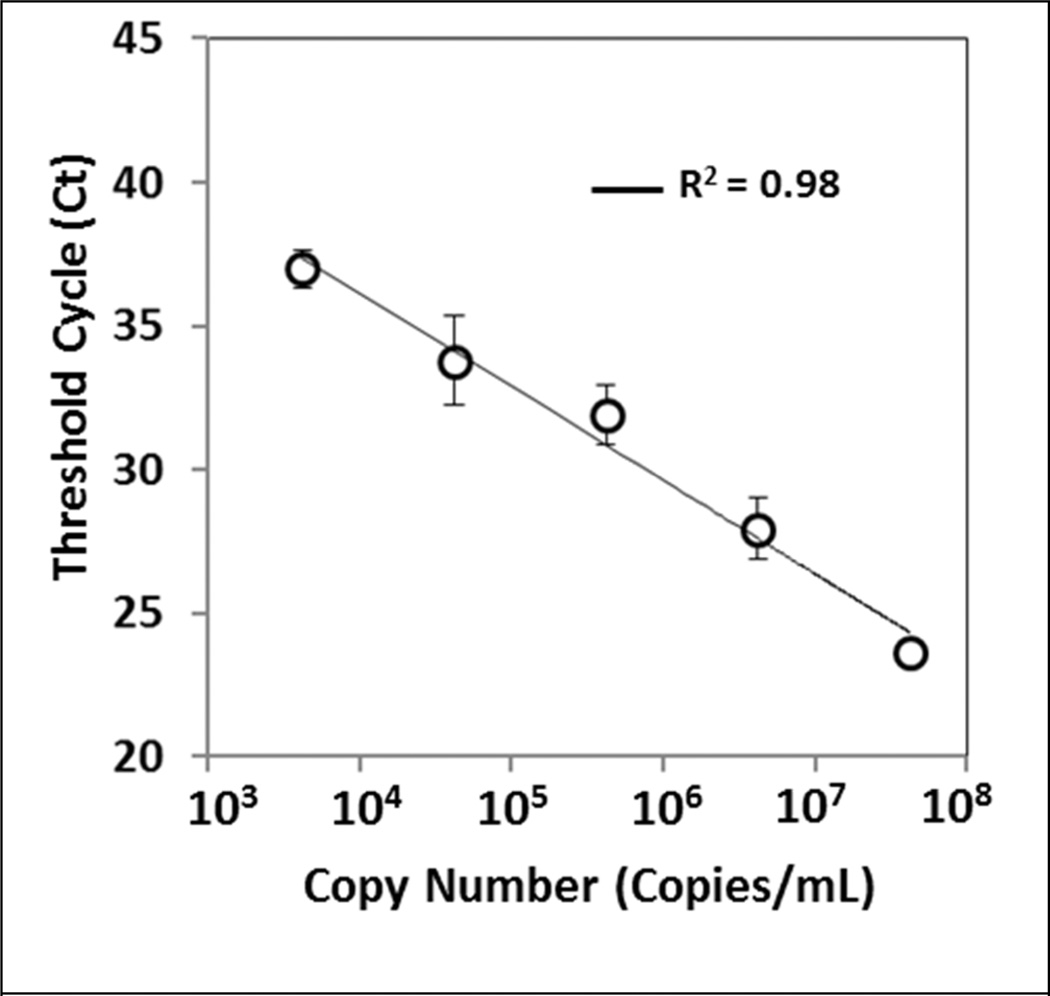
Next, we examined the ability to purify NA from complex human samples. Commercially available viral load tests require blood samples to be centrifuged to first separate the plasma from whole blood prior to analysis. To determine the impact of sample background on the purified sample, we tested the current method to extract viral RNA from human blood. VLPs were spiked into commercially available HIV negative heparinized whole blood. Viral RNA was extracted as previously described, and RT-qPCR performed looking at p24 capsid protein expression to quantify the amount of captured viral RNA. The resulting threshold cycle showed a linear response across the ranges of viral copy numbers and a limit of detection was determined to be approximately 4200 copies/mL (based on the total blood volume) (Fig 5). At this current copy number, a total of 14 copies of viral RNA are in each sample and the data demonstrate consistent extraction and purification. Even in the presence of heparin, which is a known inhibitor of PCR reaction,1 the sample was successfully processed, indicating that the capillary method is a reliable method to extract nucleic acids from complex samples.
Figure 5.
Purification of viral RNA from human blood. HIV viral like particles were spiked into heparinized human whole blood. RNA was extracted and purified using capillary method and RT-qPCR was performed to amplify the p24 capsid protein from the samples. A linear response was observed across the range of viral copy number (shown by R2 value), and a limit of detection of 4200 copies/mL was obtained using the current method which corresponds to 14 copies per extraction. Data represent the mean and the standard deviation of three to six experiments.
CONCLUSION
We have developed a nucleic acid sample preparation method based on surface tension imbalance generated by immiscible phases inside capillary tubes. By filling the capillary with oil and aqueous sample, asymmetry of surface forces creates a spontaneous plug flow which separates contaminants and inhibitors in the aqueous phase from the NA bound PMPs. This method allows the user to achieve sample purification through the simple procedure of filling the capillary with immiscible fluids, thereby greatly reducing the number of pipetting and dilutive steps associated with conventional extraction methods. The overall approach provides a potential solution to address the issues of complicated extraction methods commonly associated with NA testing that limit its use outside of the lab.
Supplementary Material
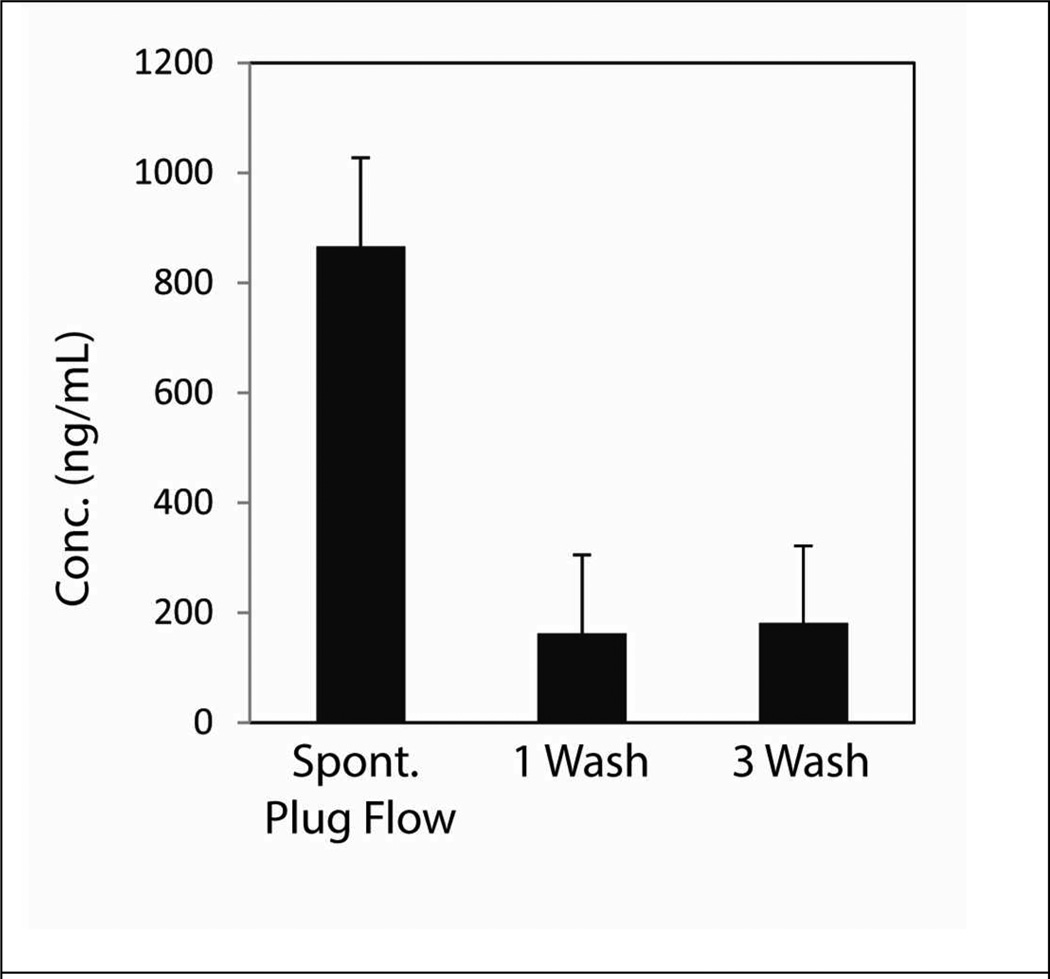
Figure 6.
Extraction of viral RNA from fetal bovine serum. HIV viral like particles were spiked into fetal bovine serum. Sample was divided into two aliquots and extracted using MagAttract PMPs. Samples were purified using either the capillary or 1 or 3 conventional wash steps. Extracted RNA was quantified using the fluorescent Nanodrop method. Higher concentrations of RNA were consistently extracted using the capillary method compared to the manufacturer’s method. No statistical differences was seen between 1 or 3 wash steps using a Student’s t-test (p>0.29).
ACKNOWLEDGMENT
This work was supported by a grant from the Bill & Melinda Gates Foundation through the Grand Challenges in Global Health initiative, NIH Grant #5R33CA137673 and NSF GRFP DGE-0718123. ABT was supported by the UW-Madison Molecular and Environmental Toxicology Center, NIH grant T32ES007015. David J. Beebe holds equity in Bellbrook Labs, LLC and Ratio, Inc. We would like to thank Dr. Nathan Sheer for all his help and guidance on the production of HIV viral-like particles.
Footnotes
Supporting Information.
Additional information is available as noted in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Schrader C, Schielke A, Ellerbroek L, Johne R. Journal of Applied Microbiology. 2012;113:1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- 2.Price CW, Leslie DC, Landers JP. Lab on a Chip. 2009;9:2484–2494. doi: 10.1039/b907652m. [DOI] [PubMed] [Google Scholar]
- 3.Martinez AW, Phillips ST, Whitesides GM, Carrilho E. Analytical Chemistry. 2010;82:3–10. doi: 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]; Klasner SA, Price AK, Hoeman KW, Wilson RS, Bell KJ, Culbertson CT. Analytical and Bioanalytical Chemistry. 2010;397:1821–1829. doi: 10.1007/s00216-010-3718-4. [DOI] [PubMed] [Google Scholar]; Fu E, Lutz B, Kauffman P, Yager P. Lab on a Chip. 2010;10:918–920. doi: 10.1039/b919614e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinez AW, Phillips ST, Butte MJ, Whitesides GM. Angewandte Chemie-International Edition. 2007;46:1318–1320. doi: 10.1002/anie.200603817. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lu Y, Shi W, Qin J, Lin B. Analytical Chemistry. 2010;82:329–335. doi: 10.1021/ac9020193. [DOI] [PubMed] [Google Scholar]
- 5.Lafleur L, Stevens D, McKenzie K, Ramachandran S, Spicar-Mihalic P, Singhal M, Arjyal A, Osborn J, Kauffman P, Yager P, Lutz B. Lab on a Chip. 2012;12:1119–1127. doi: 10.1039/c2lc20751f. [DOI] [PubMed] [Google Scholar]
- 6.Jangam SR, Yamada DH, McFall SM, Kelso DM. Journal of Clinical Microbiology. 2009;47:2363–2368. doi: 10.1128/JCM.r00092-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; Govindarajan AV, Ramachandran S, Vigil GD, Yager P, Boehringer KF. Lab on a Chip. 2012;12:174–181. doi: 10.1039/c1lc20622b. [DOI] [PubMed] [Google Scholar]
- 7.Berry SM, Alarid ET, Beebe DJ. Lab on a Chip. 2011;11:1747–1753. doi: 10.1039/c1lc00004g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sur K, McFall SM, Yeh ET, Jangam SR, Hayden MA, Stroupe SD, Kelso DM. Journal of Molecular Diagnostics. 2010;12:620–628. doi: 10.2353/jmoldx.2010.090190. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shen F, Sun B, Kreutz JE, Davydova EK, Du W, Reddy PL, Joseph LJ, Ismagilov RF. Journal of the American Chemical Society. 2011;133:17705–17712. doi: 10.1021/ja2060116. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bordelon H, Adams NM, Klemm AS, Russ PK, Williams JV, Talbot HK, Wright DW, Haselton FR. Acs Applied Materials & Interfaces. 2011;3:2161–2168. doi: 10.1021/am2004009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marangoni C. Ann. Phy. Chem. 1871;143:337. [Google Scholar]; Bico J, Quere D. Europhysics Letters. 2000;51:546–550. [Google Scholar]
- 10.Bico J, Quere D. Journal of Fluid Mechanics. 2002;467:101–127. [Google Scholar]
- 11.PCR Primer: A Laboratory Manual. 2 ed. CSHL Press; 2003. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.