Abstract
Pancreatic cancer is the fourth leading cause of cancer death in the United States because most patients are diagnosed too late in the course of the disease to be treated effectively. Thus, there is a pressing need to more clearly understand how gene expression is regulated in cancer cells and to identify new biomarkers and therapeutic targets. Translational regulation is thought to occur primarily through non-SMAD directed signaling pathways. We tested the hypothesis that SMAD4-dependent signaling does play a role in the regulation of mRNA entry into polysomes and that novel candidate genes in pancreatic cancer could be identified using polysome RNA from the human pancreatic cancer cell line BxPC3 with or without a functional SMAD4 gene. We found that (i) differentially expressed whole cell and cytoplasm RNA levels are both poor predictors of polysome RNA levels; (ii) for a majority of RNAs, differential RNA levels are regulated independently in the nucleus, cytoplasm, and polysomes; (iii) for most of the remaining polysome RNA, levels are regulated via a “tagging” of the RNAs in the nucleus for rapid entry into the polysomes; (iv) a SMAD4-dependent pathway appears to indeed play a role in regulating mRNA entry into polysomes; and (v) a gene list derived from differentially expressed polysome RNA in BxPC3 cells generated new candidate genes and cell pathways potentially related to pancreatic cancer.
Keywords: polysomes, differential gene expression, pancreatic cancer, BxPC3, SMAD4
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of cancer death in the United States because most patients are diagnosed and treated too late in the course of the disease resulting in a 5-yr survival rate of less than 5% [1,2]. The reasons for the grim statistics include lack of symptoms, a strong resistance to conventional treatments, and the tendency for the cancer to metastasize to the lymphatic system, lung, liver, and other vital organs. Although diagnostic approaches such as imaging and endoscopy continue to improve, progress is still slow in the development of effective diagnoses and treatments [3]. Consequently, there is an urgent need to identify accurate diagnostic biomarkers and new therapeutic targets and to more clearly understand the underlying biology of pancreatic cancer.
At the molecular level, the etiology and progression of PDAC is complex and multi-faceted, in which oncogenes are activated while tumor suppressor genes are lost. PDAC typically harbors several mutated genes, including K-RAS, p53, p16, and SMAD4 [4]. SMAD4 is an integral member of the transforming growth factor-beta (TGF-β) signaling pathway and is mutated or deleted in 50% of PDACs [5,6]. The members of the SMAD family are divided into three functional groups. R-SMADs (SMAD2 and 3) are receptor regulated transcription factors, co-SMAD (SMAD4) translocates SMAD2 and 3 to the nucleus, and I-SMADs (SMAD6 and 7) interact with SMADs2/3 and with the activated TGF-β type I receptor to inhibit signaling. Following phosphorylation of SMAD2 or 3 at the carboxy terminus by the TGF-β ligand/TGF-β type II/type I receptor complex, the R-SMADs partner with SMAD4, translocate to the nucleus, and bind as a heterodimer to SMAD-response elements on various gene promoters to regulate transcription. Thus, without a functional SMAD4, TGF-β signaling is disrupted. In addition to the deletion or mutation of SMAD4 in PDAC, TGF-β, and the SMAD6 and 7 inhibitors are often expressed at higher levels [4,7-9]. Thus, the inhibition of cell cycle progression normally carried out by TGF-β signaling is rendered ineffectual in many PDACs and other cancers.
TGF-β signaling may also play a role in the translational regulation of gene expression. TGF-β signaling is known to induce epithelial to mesenchymal transition (EMT) during development and cancer progression [10-13]. In a study of EMT using mouse NMuMG cells, TGF-β signaling induced the phosphorylation of an mRNP protein (hnRNPE1) on two mRNAs encoding two key EMT proteins (Dab2 and ILEI) and subsequent release from translational inhibition [14]. The study also showed that TGF-β caused an increase in polysome formation. Another study of the role of TGF-β signaling in EMT showed that TGF-β induced a global increase in translation via activation of the mTOR pathway [15]. Both EMT studies concluded that the translational regulation by TGF-β signaling was via non-SMAD pathways, that is, PIK3/AKT pathway, known to be activated by TGF-β.
The prevailing thought is that transcriptional and translational regulation is via SMAD-dependent and SMAD-independent pathways, respectively [14,15]. Evidence from our laboratory suggested that there are multiple regulation points in RNA expression influenced by SMAD4-dependent signaling, in which we showed that differential RNA levels among whole cell, nucleus, and cytoplasm were extraordinarily different in PDAC cells with or without the SMAD4 gene [16]. To build on that, we wanted to test the hypothesis that SMAD4 also had a role in regulating mRNA entry into polysomes. By characterizing polysome RNA in two pancreatic cancer cell lines in which the sole difference was the absence or presence of a functional SMAD4, we could determine whether a SMAD4-dependent signaling carried out a role in the regulation of polysome RNA levels. We also wanted to identify novel candidate disease-causing genes, biomarkers, pathways, and therapeutic targets from the polysome RNA profiles that would not ordinarily be identified by conventional approaches.
The premise that differential polysome RNA levels may be a rich source of new candidate genes and pathways was that (i) except for relatively few examples, cell structure and functions are carried out by proteins, thus, the proteome provides a more accurate view of a cell’s disease and functional states; (ii) because polysome RNA are those RNAs in the process of translation into proteins, polysome RNA is representative of a cell’s proteome, and (iii) therefore, polysome RNA profiles are an accurate reflection of the proteome and may uncover new key genes involved in the underlying disease biology. The premise is supported by a study of irradiated U87 cells that compared polysome RNA and whole cell RNA levels to protein levels [17]. Using Western immunoblotting, 14 of 16 (87.5%) proteins showed analogous changes to polysome RNA levels while whole cell RNA levels showed little correlation.
An excellent model system in which to study the roles of SMAD-dependent TGF-β signaling in pancreatic cancer is the BxPC3 pancreatic cancer cell line (SMAD4–/–, K-ras+/+, p53 mutant), which was isolated from a PDAC patient [18]. In order to help define the role of SMAD4 in pancreatic cancer, BxPC3 cells were engineered to express SMAD4 (SMAD4+), while BxPC3 cells harboring the corresponding sham vector (SMAD4–/–) were engineered to serve as a control [19]. Using the BxPC3 cell lines, we present results showing that differentially expressed cytoplasmic RNA and whole cell RNA levels are poor predictors of polysome RNA levels; that polysome RNA levels are regulated via multiple mechanisms; that in addition to non-SMAD TGF-β signaling pathways, mRNA entry into polysomes is also regulated via SMAD-dependent signaling; and that polysome RNA profiling may be a powerful means to identify new genes and pathways involved in disease states and underlying biology.
MATERIALS AND METHODS
Cell Cultures
BxPC3 cells were grown in RPMI-1640, supplemented with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 5% fungizone [19]. Cells were maintained in monolayer cultures at 37°C in humidified air with 5% CO2, and cell numbers were determined by counting the cells with a hemocytometer.
Nuclear and Cytoplasmic Fractionation
The nuclear and cytoplasmic fractions were isolated as described [16,19]. BxPC3 cells were incubated for 30 min at 4°C in hypotonic buffer (20 mM HEPES, pH 7.6, 20% glycerol, 10 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.1% Triton X-100, 25 mM NaF, 25 mM β-glycerophosphate, 1 mM phenylmethanesulfonyl fluoride (PMSF), 1 mM sodium orthovanadate, 1 mM dithiothreitol, 1 μg/mL leupeptin, and 1 μg/mL aprotinin). Following centrifugation at 1700 rpm for 5 min, supernatants were used for the cytoplasmic fraction. The pelleted nuclei were washed with 1 × PBS (137 mM NaCl, 2.7 mM KCl, 10 mM sodium phosphate dibasic, 2 mM potassium phosphate monobasic, pH of 7.4) and incubated for 10 min at 4°C in nuclear extraction buffer (hypotonic buffer containing 500 mM NaCl) and cleared by centrifugation.
Polysome Isolation
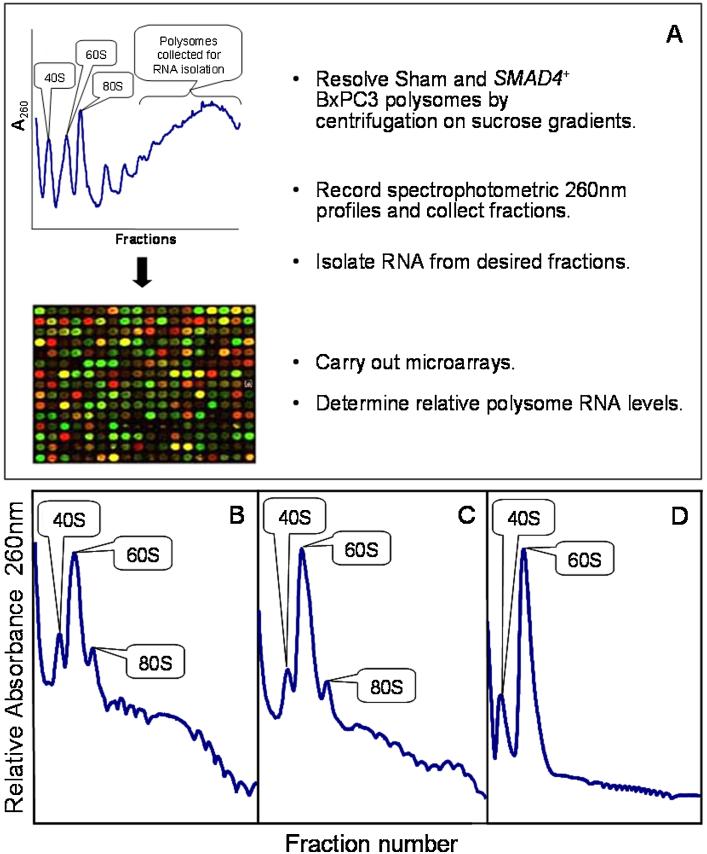
The BxPC3 cells were harvested at 4°C, washed with 1× PBS containing 100 μg/mL cycloheximide, and lysed in 1 mL of polysome buffer (10 mM Tris–HCl, pH 8, 140 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40, 10 mM dithiothreitol, 100 μg/ mL cycloheximide, 500 μg/mL heparin, 1 mM PMSF, and 500 U/mL RNasin [Promega, Madison, WI]). The lysates were incubated for 10 min on ice, centrifuged at 10 000g at 4°C for 10 min, and 400–500 μL of the resulting supernatant layered onto a 10–50% sucrose gradient (10% and 50% sucrose in 10 mM Tris, pH 7.5, 1.5 mM MgCl2, 140 mM NaCl, 1 mM DTT, 100 μg/mL cycloheximide, 0.5 μg/mL heparin [Sigma, St. Louis, MO]). The gradients were centrifuged at 164 000g at 4°C for 3.5 h in a Superspeed 630 rotor using a Sorvall Discovery 90SE centrifuge (Thermo Scientific, Waltham, MA). Fractions (0.5 mL) were collected using a Gilson gradient fraction collector system (Middleton, WI) with continuous monitoring at A254 with a Bio-Rad UV monitor (Hercules, CA) and Windaq software (Akron, OH). The fractions corresponding to polysome RNA (Figure 1) were pooled and prepared for RNA purification.
Figure 1.
Experimental design (A) and representative polysome profiles of SMAD4–/– (Sham) BxPC3 cells (B), SMAD4+ BxPC3 cells (C), and polysomes from SMAD4–/– BxPC3 cells treated with 100 μM EDTA for 10 min (D). [Color figure can be seen in the online version of this article, available at http://wileyonlinelibrary.com/journal/mc]
RNA Purification
Cells and cell fractions were homogenized in TRIzol Reagent (Invitrogen Corp., Carlsbad, CA). The RNA was further purified to remove heparin using a LiCl precipitation method [20]. RNA purity, quantity, and quality were determined using a NanoDrop spectrophotometer ND-1000 (Thermo Scientific) and Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA) [21].
Microarrays
The RNA gene expression microarray experiments were carried out by the Dartmouth Genomics & Microarray Laboratory (DGML). The HumanHT-12 v4 Expression BeadChip array (Illumina, San Diego, CA) with approximately 47 000 annotated RefSeq derived probes were used for the RNA profiling. Fluorescent images were obtained with an Illumina 500GX scanner in the DGML and processed with the BeadScan software (Illumina). Following the accompanying protocol, reverse transcription was carried out using an oligo(dT) primer bearing a T7 promoter and the high yield ArrayScriptTM reverse transcriptase to make cDNA. The cDNA was made double-stranded with DNA polymerase, and the dsDNA was purified to use as a template for in vitro transcription with T7 RNA polymerase and the included biotin–NTP mix. The labeled cRNA was purified and 1.5 μg used for hybridization to the beadarrays for 16 h at 55°C. Following hybridization, the beadarrays were washed and stained with streptavidin–Cy3 (GE Healthcare, Piscataway, NJ). Fluorescent images were obtained with an Illumina 500GX scanner in the DGML and processed with the BeadScan software (Illumina).
Quantitative Polymerase Chain Reaction (QPCR)
QPCR analysis using SYBR green and designed primers (Table S1) was carried out as described [22] to verify the microarray results (Table S2). Approximately 2 μg of total RNA (the same RNA used for the microarrays) from the nucleus, cytoplasm, or polysomes was used as template for cDNA synthesis. The QPCR reactions were preformed on a DNA Engine Opticon Monitor System using software version 3.1 (Bio-Rad) set at 35–40 cycles. Agarose gel electrophoresis showed that each PCR produced a single band of the predicted size. Assays to determine DNA contamination were carried out by omitting reverse transcriptase from the reactions. Except for the OAS1 gene, which encodes at least three isoforms, the QPCR results were in agreement with the microarray results (Table S2).
Microarray Data Analysis
Four biological replicates per experimental condition were carried out. The data analysis was carried out by performing Quantile normalization [23] without background correction to pre-process the image files from the Illumina software and an intensity-based, modified t-test [24] to characterize the significance level of each feature using limma R packages from Bioconductor [25]. The data were also analyzed using Pathway Studio (Ariadne, Rockville, MD) to generate gene lists with designated P values and false discovery rates (FDR) [26,27]. Statistically significant, differentially expressed genes were annotated with functional assignments to help determine category enrichment using the biological process (BP-FAT) branch of the Gene Ontology (GO) database [28] via the DAVID program of the National Institute of Allergy and Infectious Diseases [29,30]. Venn diagrams of the statistical results were constructed using the GeneVenn software package [31]. Lists of all the differentially expressed genes from whole cells, nucleus, cytoplasm, and polysomes are shown in Tables S3-S8. The network associations among the proteins encoded by the differentially expressed genes of the polysomes were drawn using Pathway Studio [32].
RESULTS
Cytoplasm RNA Levels and Whole Cell RNA Levels Are Both Poor Predictors of Polysome RNA Levels
The SMAD4 gene is deleted in the human pancreatic cancer cell line BxPC3 [18]. To help define the role of SMAD4 in pancreatic cancer, BxPC3 cells were engineered to express SMAD4 (SMAD4), while BxPC3 cells harboring the corresponding sham vector (SMAD4–/–) were engineered to serve as a control [19]. We followed the scheme shown in Figure 1A to determine differentially expressed RNA levels in polysomes between sham and SMAD4+ BxPC3 cells. Cell lysates from cultures grown to 80% confluence were placed on 10–50% sucrose gradients, and the polysomes were resolved by ultracentrifugation. Polysomes from four sham and four SMAD4+ BxPC3 biological replicates were examined. Fractions of RNA representing the heavier two-thirds of the polysomes (those polysomes under the bracket in Figure 1A) were pooled, purified, and prepared for microarray analysis. Representative polysome profiles are shown in Figure 1B and C. Intact polysomes were isolated because treatment with 100 μM EDTA, which chelates Mg++ necessary for polysome formation [33], disassociated the polysomes to form exclusively the 40S and 60S ribosomal subunits (Figure 1D).
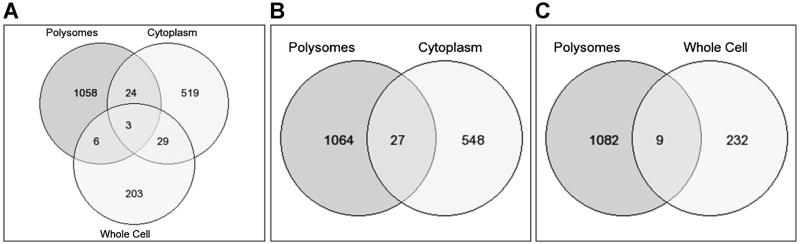
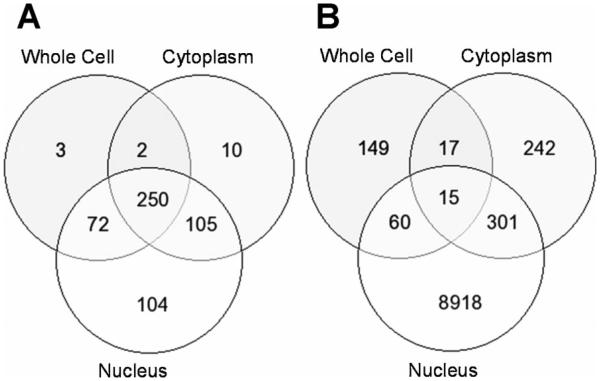
In order to accurately measure steady-state RNA levels at each stage of gene expression, all passages of the BxPC3 cells were cultured continuously in RMPI-1640, supplemented with 10% FBS, without addition of TGF-β. The omission of TGF-β in the culture medium ensured that RNA levels at each point of gene expression were at steady-state levels allowing valid comparisons. Addition of TGF-β would have perturbed cellular systems such that steady-state RNA levels at the whole cell, nuclear, cytoplasmic, and polysome levels would have peaked at different and difficult to determine times. The profiling of differentially expressed polysome RNA between sham and SMAD4+ BxPC3 cells allowed us to ask several questions. Because whole cell RNA levels poorly correlate with protein levels [34-37], we reasoned in turn that if polysome RNA is representative of the proteome then there should also be poor correlation between whole cell RNA and polysome RNA levels. We had already shown that there was little association between cytoplasmic RNA levels and whole cell RNA levels in BxPC3 cells [16]. The first question asked was whether cytoplasm RNA levels are representative of polysome RNA levels. Of the 1091 differentially expressed polysome RNAs (P-value 0.05), only 27 (4.7% of differentially expressed genes from cytoplasm) and nine (3.7% of differentially expressed genes from whole cells) differentially expressed genes were shared between the polysomes and cytoplasm and between the polysomes and whole cell, respectively (Table 1, Figure 2). Thus, although there is little correlation between whole cell and cytoplasm RNA levels [16], there was also little correlation between polysome RNA and cytoplasm RNA levels, and the differential RNA levels of the cytoplasm were no better in predicting polysome RNA levels than were whole cell RNA levels.
Table 1.
Differential RNA Levels From the Whole Cell, Nucleus, and Cytoplasm Compartments Are All Poor Predictors of Differential Polysome RNA Levels
| Relative RNA level measurement | Cell compartment |
|||
|---|---|---|---|---|
| Whole cell | Nucleus | Cytoplasm | Polysomes | |
| Number of differentially expressed genes (P-value <0.05) |
241 | 9294 | 575 | 1091 |
| Number of differentially expressed genes shared with polysomes (percentage of differentially expressed genes shared with polysomes) |
9 (3.7) | 593 (6.4) | 27 (4.7) | NA |
| Number of differentially expressed genes shared with polysomes with same fold change direction (percentage of differentially expressed genes with same fold change of polysomes) |
2 (0.8) | 124 (1.4) | 8 (1.4) | NA |
| Number of differentially expressed genes shared with polysomes with opposite fold change direction (Percentage of differentially expressed genes with opposite fold change of polysomes) |
7 (2.9) | 469 (5.0) | 19 (3.3) | NA |
NA, not applicable.
Figure 2.
Venn diagram depicting the numbers of differentially expressed genes shared among the cytoplasm, polysomes, and whole cells (A), between polysomes and cytoplasm (B), and between polysomes and whole cells (C) from SMAD4–/– versus SMAD4+ BxPC3 cells.
RNA Levels Are Differentially Regulated at Multiple Cellular Junctures
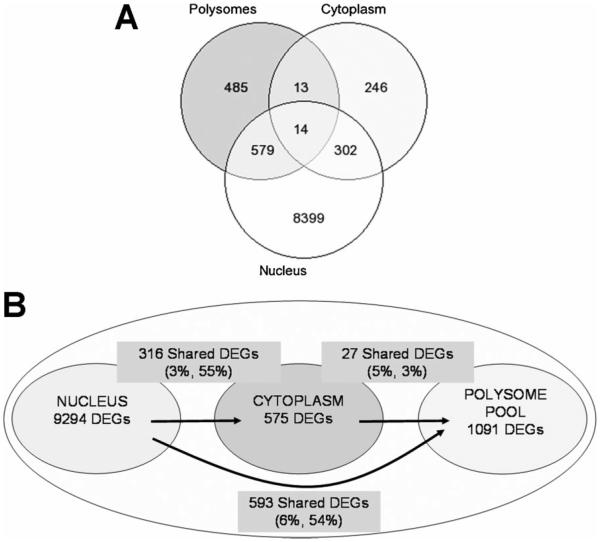
The second question asked was whether polysome RNA levels are differentially regulated. Differential gene expression profiles from the nucleus, cytoplasm, and polysomes of sham and SMAD4+ BxPC3 cells were compared. We found that the number of differentially expressed genes in the nucleus (9294) eclipsed those of the cytoplasm and polysomes (575 and 1091, respectively), and that the number and percentage of shared differentially expressed genes between nucleus and cytoplasm were much greater than that of the cytoplasm to polysomes (Figure 3). Of the 575 differentially expressed genes of the cytoplasm, 316 of the genes were also differentially expressed in the nucleus (55%), whereas, only 27 genes were shared with the differentially expressed genes of the polysomes (4.7%).
Figure 3.
The numbers of differentially expressed genes (DEGs) between, among, and exclusively of the nucleus, cytoplasm, and polysomes from SMAD4–/– versus SMAD4+ BxPC3 cells shown in a Venn diagram (A) and in the different cell compartments (B). The percentages in parentheses are the percent of shared DEGs for the two corresponding RNA populations.
Contrary to our expectation that a sizable proportion of the differential polysome RNA levels would have originated from differential expression in the nucleus and cytoplasm, only 14 common genes were differentially regulated from nucleus to cytoplasm to polysomes. Lists of all the unique and shared genes between and among the nucleus, cytoplasm, and polysome are displayed in Table S9. The surprise was the relatively large number and high percentage of shared genes between the nucleus and polysomes (but not cytoplasm). Over half of the differentially expressed genes in the polysomes (54%, 593 of 1091) were also differentially expressed in the nucleus. From these results, we concluded that (i) there is extensive and independent gene regulation in the nucleus, cytoplasm, and polysomes and (ii) a large proportion of differentially expressed RNAs in polysomes are “tagged” in the nucleus to directly enter the polysomes once transported to the cytoplasm.
Biological Processes and Pathways
The third question asked was whether there was any commonality in the cell pathways encoded by the differentially expressed RNAs of the nucleus and cytoplasm to that of the polysomes. Pathway analyses of the significantly changed genes from the different cell compartments were performed using the biological process (BP-FAT) branch of the GO database [28] to identify enriched cellular pathways. Tables S10 (P-value 0.02) and S11 (P-value 0.05) lists the complete, shared, and unique GO BP-FAT based biological processes for the nucleus, cytoplasm, and polysomes.
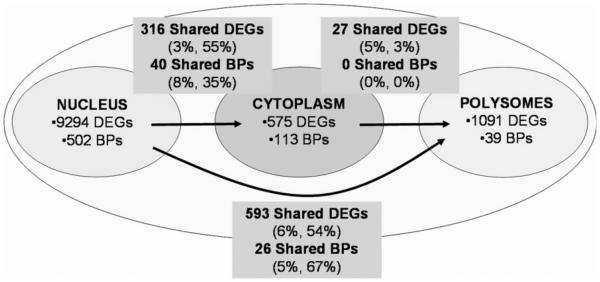
As with the genes, in which there were relatively few shared differentially expressed genes among polysomes, nucleus, and cytoplasm; similarly, there were relatively few shared biological processes (Figure 4). For example, there were 593 differentially expressed genes shared between the nucleus and polysomes, which represented 6% and 53% of the total number of differentially expressed genes in the two cellular compartments, respectively. Likewise, there were 26 GO biological processes shared between the nucleus and polysome compartments representing 5% and 67% of the respective cell compartments. Thus, as with the differentially expressed genes in the nucleus, cytoplasm, and polysomes, the encoded proteins were in large part unshared between cell compartments. In regard to the polysomes, the analysis revealed that (i) RNA processing pathways dominated in total polysomes, (ii) translation dominated the pathways of those unique to polysomes, and (iii) protein and RNA processing were the dominant pathways in those shared between nucleus and polysomes (Tables S10 and S11).
Figure 4.
The numbers of differentially expressed genes (DEGs) and corresponding pathways derived from Gene Ontology Biological Processes-FAT (BPs) in the different cell compartments between and exclusively of the nucleus, cytoplasm, and polysomes in SMAD4–/– versus SMAD4+ BxPC3 cells. The percentages in parentheses are the percent of shared DEGs and shared BPs, respectively, for the two corresponding RNA populations.
New SMAD4-Dependent BxPC3 Genes
Almost all published studies using high-throughput microarray approaches rely on RNA from whole cells in the belief that whole cell RNA is a valid measure of steady-state mRNA levels [16]. In a study to determine whether whole cell RNA is indeed representative of steady-state mRNA levels, we showed that nuclear RNA greatly distorts mRNA profiles and that precise mRNA profiling is possible only by the preparation of unadulterated cytoplasmic RNA [16]. A comparison of relative RNA levels for the 25 genes with the greatest fold change in differential expression between polysomes and the conventionally used whole cells (Table 2) illustrates how disparate the results can be for different RNA sources from the same cells. Two points can be made. One, there are no shared genes between polysomes and whole cells, and two, the differential changes are much more pronounced in the polysomes. Thus, depending on the RNA source, very different results are obtained. For example, NEDD8, which encodes a ubiquitin-like protein involved in many cancers [38], is one of the genes that would have been identified as a differentially expressed gene using polysome RNA but would have been missed using whole cell RNA. The marked increased of NEDD8 expression in sham cells to that of SMAD4-expressing cells suggests that SMAD4 suppresses NEDD8 levels and that SMAD4 may interfere with tumor progression, in part, by attenuating NEDD8-mediated activation of cullin-RING ubiquitin E3 ligases which are known to promote cancer cell survival [39].
Table 2.
Top 25 Differentially Expressed RNAs in Polysomes and Whole Cell of Sham Versus SMAD4+ BxPC3 Pancreatic Cancer Cells (P-Value <0.05)
| Polysomes |
Whole cell |
||||
|---|---|---|---|---|---|
| Gene symbol | Sham/SMAD4+ fold change |
Gene bank IDs | Gene symbol | Sham/SMAD4+ fold change |
Gene bank IDs |
| HMGN1a | 7.70 | NM_004965 | COBL | 1.81 | NM_015198 |
| RPS27 | 7.03 | NM_001030 | TLN2 | 1.79 | NM_015059 |
| NQO1b | 6.12 | NM_000903 | CDH5 | 1.65 | NM_001795 |
| SFRS6a | 5.92 | NM_006275 | GFPT2 | 1.64 | NM_005110 |
| EEF1B2 | 5.72 | NM_001037663 | RTN1 | 1.55 | NM_206857 |
| C19orf2a | 5.69 | NM_134447 | CSPG2 | 1.55 | NM_004385 |
| NEDD8 | 5.66 | NM_006156 | TBL1X | 1.51 | NM_005647 |
| SNRPD2 | 5.61 | NM_004597 | LRIG1 | 1.47 | NM_015541 |
| ANP32Ba | 5.53 | NM_006401 | RSAD2 | 1.45 | NM_080657 |
| SNRPG | 5.35 | NM_003096 | IGFL1d | 1.45 | NM_198541 |
| ATP5I | 5.27 | NM_007100 | VIM | 1.43 | NM_003380 |
| POLE3a | 5.24 | NM_017443 | MCTP1 | 1.42 | NM_001002796 |
| TOMM7 | 5.21 | NM_019059 | GPR37 | 1.41 | NM_005302 |
| SS18L2 | 5.07 | NM_016305 | SGPP2 | 1.40 | NM_152386 |
| ATF4a | 4.84 | NM_001675 | PCDHA13 | 1.37 | NM_031865 |
| TBCA | 4.72 | NM_004607 | FLJ20701 | 1.36 | NM_017933 |
| C10orf116c | 4.69 | NM_006829 | COL8A1 | 1.34 | NM_001850 |
| GLRX5a | 4.67 | NM_016417 | BCL11B | 1.34 | NM_022898 |
| EIF3S6a | 4.65 | NM_001568 | OAS2d | 1.34 | NM_002535 |
| POLR1D | 4.61 | NM_015972 | BDKRB1d | 1.33 | NM_000710 |
| C16orf61 | 4.52 | NM_020188 | SPINK5 | 1.32 | NM_006846 |
| HNRPDa | 4.49 | NM_031369 | TMEM92 | 1.31 | NM_153229 |
| PRR13a | 4.48 | NM_018457 | FLJ33860 | 1.30 | NM_173644 |
| C2orf26a | 4.43 | NM_023016 | TFF2 | 1.30 | NM_005423 |
| SHFM1 | 4.31 | NM_006304 | SUSD2 | 1.29 | NM_019601 |
Also differentially expressed in the nucleus.
Also differentially expressed in the nucleus and cytoplasm.
Also differentially expressed in the cytoplasm.
Also differentially expressed in polysomes.
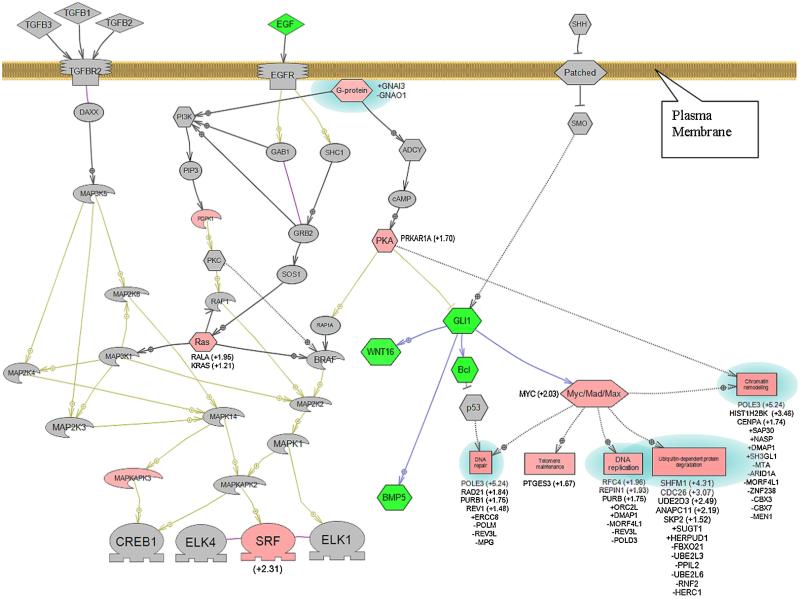
Network analysis of the proteins encoded by the differentially expressed genes of the polysomes showed known key players involved in PDAC (Figure 5). For example, the relative RNA levels of the MYC and K-RAS genes were greater in the Sham versus SMAD4–/– BxPC3 cells; while the RNA levels of the WNT16, GLI1, and BCL genes were lower [40]. Two novel genes revealed by the network analysis that were highly differentially expressed and maybe worthy of further investigation were SRF (+2.31-fold) and POLE3 (+5.24-fold). The SRF (serum response factor) gene is a transcription factor found in all tissues involved in the regulation of hundreds of genes [41]. SRF is usually associated with cardiac and skeletal muscle development and angiogenesis; however, SRF inactivation in the pancreas leads to severe pancreatitis [42], which is often a precursor to PDAC. The POLE3 (polymerase-DNA directed, epsilon 3, p17 subunit) gene is regulated by MYC [43] and is part of a complex with ACF1 involved in the repair of DNA double-strand breaks [44], and its dysregulation may contribute to the genomic instability found in BxPC3 cells and PDAC.
Figure 5.
Network map of key signaling pathways associated with PDAC and encoded by differentially expressed mRNA from polysomes of SMAD4−/− versus SMAD4+ BxPC3 cells. Pink shapes represent mRNAs expressed at relatively higher levels, those in green at relatively lower levels, and those in gray are not differentially expressed (P-value ≤0.05). Proteins and biological processes with blue halos represent a set of genes in a biological process that include genes expressed at both relatively higher and lower levels. Values in parentheses represent relative changes in RNA levels of greater than 1.2-fold. Fold changes less than 1.2-fold are represented with a + or − sign preceding the gene symbol. Hexagon, signaling protein; Rectangle, biological process; Diamond, ligand; Oval, small molecule; Boomerang, kinase;  , receptor;
, receptor;  , transcription factor. [Color figure can be seen in the online version of this article, available at http://wileyonlinelibrary.com/journal/mc]
, transcription factor. [Color figure can be seen in the online version of this article, available at http://wileyonlinelibrary.com/journal/mc]
A portion of the studies reported here was a replication of earlier work. In the earlier study, we attempted to determine whether whole cell RNA was an accurate representation of cytoplasm steady-state mRNA levels [16]. Using the Agilent microarray platform (Agilent Technologies), we showed that whole cell RNA did not accurately measure steady-state mRNA levels because the nuclear RNA contribution strongly biased the results, and that the cytoplasm RNA fraction produced a more precise mRNA steady-state profile. For the current study, the Illumina bead array system (Illumina) was used with the same nucleus, cytoplasm, and whole cell RNA preparations (Figure 6A and B). Although the numbers and proportions of differentially expressed genes for the two microarray platforms are markedly different for some of the cell fractions, for example, the nucleus, it was reassuring that the same conclusions could be drawn from both studies. That is, the RNA profiles for the whole cell and cytoplasm fractions are very different, and the nuclear RNA has a large impact on the whole cell RNA profile.
Figure 6.
Venn diagrams depicting the numbers of differentially expressed genes between, among, and exclusively of the cytoplasm, polysomes, and whole cells from SMAD4–/– versus SMAD4+ BxPC3 cells using the Agilent microarray platform (A) and the Illumina bead array platform (B).
DISCUSSION
A major intent of our work was to identify novel genes and pathways that may be involved in pancreatic cancer. Based on the premise that the proteome is a better measure of a cell’s disease state than the transcriptome, we sought to determine the differential expression of a subset of cytoplasmic RNAs that are in the active process of being translated, that is, the polysomes. Thus, polysome profiling using microarrays was carried out that revealed new genes and pathways that may play a role in pancreatic cancer. Table 2 lists the genes with polysome and whole cell RNA levels of the greatest fold change. Most of the genes differentially expressed in polysomes would not have been detected with conventionally used whole cell RNA, including the NEDD8 and POLE3 genes described above.
We found as with whole cells, there were few shared differentially expressed genes between cytoplasm and polysomes. This result was somewhat of a surprise. As described above, there is little association between whole cell and cytoplasm RNA levels [16], and it has been long known that whole cell RNA levels and protein levels are poorly associated [34-37]. We reasoned that these results may be indicative of a positive association between cytoplasm RNA and polysome RNA levels, especially given the fact that the polysomes reside in the cytoplasm. Instead, we found that the correlation between cytoplasm and polysomes was one of the weaker associations. These results are similar to results we obtained using vehicle control versus dioxin-treated mouse Hepa1c1c7 cells [45].
TGF-β signaling is fully functional in SMAD4-expressing BxPC3 cells [19], and the only known difference between the two BxPC3 cell types used in our studies is the presence of a functional SMAD4 gene. Therefore, on the findings reported here, we concluded that SMAD4-dependent activity has a large impact on all levels of differential gene expression, including polysomes. This conclusion is supported by a study showing that gene expression in pancreatic cancer cells (as well as another cell type) have SMAD4-dependent and SMAD4-independent sets of genes regulated by TGF-β signaling [46]. Although the study used a different pancreatic cancer cell line (Colo-357) and were exposed to TGF-β, three of the SMAD4-dependent, differentially expressed genes were identified in our work and expressed in polysomes (FOXQ1) and whole cells (TGM1 and PCDH7). It should be kept in mind that BMP-induced signaling of the TGF-β pathway may also contribute to the multi-layered regulation of SMAD4-dependent gene expression [47].
Our studies also showed that for many genes, RNA expression at the level of the nucleus, cytoplasm, and polysomes appear to be regulated independently (Figures 3 and 4), again, similar to results we obtained with the mouse Hepa1c1c7 - cells [45]. The findings suggest that the independent control of gene expression at multiple cell junctions may be widespread. The categorized genes in Figures 3 and 4 were sorted further and are shown in Table 3, which lists the nine possible combinations of the relative nuclear and cytoplasmic RNA levels and corresponding genes associated with an increase or decrease in polysome RNA levels. Two major classes of differentially regulated polysome RNAs were identified. Those polysome RNAs in which there was an association between an increase in nuclear RNA levels and an increase (62.1%) or decrease (34.7%) in differentially expressed polysome RNA levels; and those RNAs regulated independently at the polysome level (44.5% of total differentially expresses polysome RNAs). RNAs regulated similarly among nucleus, cytoplasm, and polysomes (0.2%), and RNAs regulated similarly between cytoplasm and polysomes (1.5%) were rare. There was a modest association between a decrease in nuclear RNA levels and a decrease in differentially expressed polysome RNA levels (9.2%).
Table 3.
Number of Genes With Similar and/or Dissimilar Differential RNA Expression Levels Among Cell Compartments in SMAD4–/– Versus SMAD4+ BxPC3 Cells (P-Value ≤0.05)
| Cell compartments | ↑ Polysome RNA levels |
Percent | ↓ Polysome RNA levels |
Percent | Total | Percent |
|---|---|---|---|---|---|---|
| Nuc↑, Cyto↑ | 1 | 0.2 | 9 | 1.6 | 10 | 0.9 |
| Nuc↑, Cyto↓ | 2 | 0.4 | 0 | 0 | 2 | 0.2 |
| Nuc↑, Cyto↔ | 319 | 62.1 | 200 | 34.7 | 519 | 47.6 |
| Nuc↓, Cyto↑ | 0 | 0 | 0 | 0 | 0 | 0 |
| Nuc↓, Cyto↓ | 2 | 0.4 | 0 | 0 | 2 | 0.2 |
| Nuc↓, Cyto↔ | 7 | 1.4 | 53 | 9.2 | 60 | 5.5 |
| Nuc↔, Cyto↑ | 4 | 0.8 | 3 | 0.5 | 7 | 0.6 |
| Nuc↔, Cyto↓ | 3 | 0.6 | 3 | 0.5 | 6 | 0.5 |
| Nuc↔, Cyto↔ | 176 | 34.2 | 309 | 53.6 | 485 | 44.5 |
| Total | 514 | 100 | 577 | 100 | 1091 | 100 |
↑, relative increase in RNA levels in SMAD4–/– versus SMAD4+ BxPC3 cells; ↓, relative decrease in RNA levels in SMAD4–/– versus SMAD4+ BxPC3 cells; ↔, no difference in RNA levels between SMAD4–/– versus SMAD4+ BxPC3 cells.
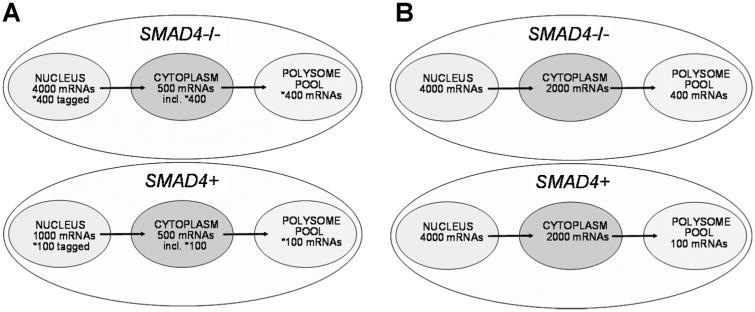
The two major polysome RNA classes described above are illustrated by the models presented in Figure 7, in which a single representative RNA species in a given polysome RNA class is depicted. Model 1 (Figure 7A) represents those RNAs with the largest number of polysome RNAs. These polysome RNAs showed an association between an increase in nuclear RNA levels and an increase or decrease in polysome RNA levels. The results suggest that these polysome RNAs are somehow “tagged” in the nucleus for polysome entry or for the blockage into polysomes. The apparent nuclear mRNA tagging could be based on the status of the 3′ terminus poly (A) tract [48]; miRNA targeting [49]; and/or via specific sequence motifs, such as internal ribosome entry sites that are utilized in stress responses [50], possibilities we are currently investigating. Model 2 (Figure 7B) represents those RNAs regulated independently at the polysome level. A large proportion of polysome RNAs (44.5%) were differentially expressed only in the polysomes but not nucleus and cytoplasm.
Figure 7.
Models depicting how differentially expressed polysome RNA populations may be distributed in the cell. A single RNA species is depicted in each model to be representative of a given polysome RNA population. Model 1 (A) represents the common RNAs differentially regulated between nucleus and polysomes but not cytoplasm (579/1091 = 53.1%). Model 2 (B) represents those RNAs regulated independently at the polysome level and are differentially expressed only in the polysomes but not nucleus and cytoplasm (485/1091 = 44.5%).
How are polysome RNA levels regulated? It is well known that cell location plays a large role in the regulation of gene expression [51]. In a study of liver regeneration in rats, at least 85 miRNAs were associated with three different polysome types (free, cytoskeleton-bound, and membrane-bound), and differential protein expression of c-Myc and p53 among the different polysome populations appeared to be due, at least in part, to regulation by miRNAs [49]. Other studies using genome-wide approaches to examine mRNA and protein levels in parallel showed that miRNA can repress the expression of many proteins by directly repressing translation without affecting mRNA levels, although the same studies found that for the majority of mRNAs, the major effect of miRNAs on mRNA expression is by promoting mRNA turnover [52-54].
The loss of SMAD4 may directly impact other points of gene expression as well as polysome RNA levels. TGF-β and BMP signaling was shown to induce DROSHA in the processing of pri-miR-21 and pri-miR-199a in human smooth muscle cells [55]. Later studies showed that the post-transcriptional processing for as many as 20 pri-miRNAs are regulated by the direct interaction of R-SMADs to a consensus sequence called R-SBE in the stem region of pri-miRs [56]. The authors speculated that R-SMAD/SMAD4 complexes are preferentially recruited to DNA to regulate transcription and that SMAD4-independent R-SMADs are preferentially recruited to pri-miRNAs and other RNAs to regulate DROSHA processing and other events. R-SMADs can bind to both DNA and RNA, as a result, R-SBE-containing pri-miRNAs may interfere with SMAD-mediated transcription by sequestering SMADs. In conclusion, it is easy to envision that the loss of SMAD4 in PDAC and BxPC3 cells could drastically alter global gene expression at many levels and that regulation of gene expression relies heavily on several SMAD4-dependent mechanisms.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Yolanda Sanchez and Emily K. Schaeffer of the Dartmouth Medical School and Norris Cotton Cancer Center for their critical reading of the manuscript, the Dartmouth Genomics & Microarray Laboratory for the microarray studies, the Bioinformatics Shared Resource for the microarray data analyses, and the reviewers for their thoughtful comments. This work was supported by NIH/NIEHS grant R21ES013827 to CRT.
Abbreviations
- PDAC
pancreatic ductal adenocarcinoma
- TGF-β
transforming growth factor-beta
- EMT
epithelial to mesenchymal transition
- DGML
Dartmouth Genomics & Microarray Laboratory
- QPCR
quantitative polymerase chain reaction
- GO
Gene Ontology
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- 1.DiMagno EP, Reber HA, Tempero MA. AGA technical review on the epidemiology, diagnosis, and treatment of pancreatic ductal adenocarcinoma. American Gastroenterological Association. Gastroenterology. 1999;117:1464–1484. doi: 10.1016/s0016-5085(99)70298-2. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 3.Tomlinson CR, Korc M. Squeezing data from pancreatic juice. Cancer Biol Ther. 2006;5:1390–1391. doi: 10.4161/cbt.5.10.3486. [DOI] [PubMed] [Google Scholar]
- 4.Hansel DE, Kern SE, Hruban RH. Molecular pathogenesis of pancreatic cancer. Annu Rev Genomics Hum Genet. 2003;4:237–256. doi: 10.1146/annurev.genom.4.070802.110341. [DOI] [PubMed] [Google Scholar]
- 5.Cowgill SM, Muscarella P. The genetics of pancreatic cancer. Am J Surg. 2003;186:279–286. doi: 10.1016/s0002-9610(03)00226-5. [DOI] [PubMed] [Google Scholar]
- 6.Hahn SA, Schutte M, Hoque AT, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 7.Korc M. Biology of pancreatic cancer. In: Rustgi AK, Crawford J, editors. Gastrointestinal cancers. W.B. Saunders Co.; Oxford, UK: 2003. [Google Scholar]
- 8.Kleeff J, Maruyama H, Friess H, Buchler MW, Falb D, Korc M. Smad6 suppresses TGF-beta-induced growth inhibition in COLO-357 pancreatic cancer cells and is overexpressed in pancreatic cancer. Biochem Biophys Res Commun. 1999;255:268–273. doi: 10.1006/bbrc.1999.0171. [DOI] [PubMed] [Google Scholar]
- 9.Kleeff J, Ishiwata T, Maruyama H, et al. The TGF-beta signaling inhibitor Smad7 enhances tumorigenicity in pancreatic cancer. Oncogene. 1999;18:5363–5372. doi: 10.1038/sj.onc.1202909. [DOI] [PubMed] [Google Scholar]
- 10.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 11.Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 12.Massague J. TGFbeta in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bierie B, Moses HL. TGF-beta in cancer. Cytokine Growth Factor Rev. 2006;17:29–40. doi: 10.1016/j.cytogfr.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL, Howe PH. TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat Cell Biol. 2010;12:286–293. doi: 10.1038/ncb2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamouille S, Derynck R. Cell size and invasion in TGF-beta-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J Cell Biol. 2007;178:437–451. doi: 10.1083/jcb.200611146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trask HW, Cowper-Sal·lari R, Sartor MA, et al. Microarray analysis of cytoplasmic versus whole cell RNA reveals a considerable number of missed and false positive mRNAs. RNA. 2009;15:1917–1928. doi: 10.1261/rna.1677409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu X, de la Pena L, Barker C, Camphausen K, Tofilon PJ. Radiation-induced changes in gene expression involve recruitment of existing messenger RNAs to and away from polysomes. Cancer Res. 2006;66:1052–1061. doi: 10.1158/0008-5472.CAN-05-3459. [DOI] [PubMed] [Google Scholar]
- 18.Farivar RS, Gardner-Thorpe J, Ito H, et al. The efficacy of tyrosine kinase inhibitors on human pancreatic cancer cell lines. J Surg Res. 2003;115:219–225. doi: 10.1016/s0022-4804(03)00246-4. [DOI] [PubMed] [Google Scholar]
- 19.Yasutome M, Gunn J, Korc M. Restoration of Smad4 in BxPC3 pancreatic cancer cells attenuates proliferation without altering angiogenesis. Clin Exp Metastasis. 2005;22:461–473. doi: 10.1007/s10585-005-2891-x. [DOI] [PubMed] [Google Scholar]
- 20.del Prete MJ, Vernal R, Dolznig H, Mullner EW, Garcia-Sanz JA. Isolation of polysome-bound mRNA from solid tissues amenable for RT-PCR and profiling experiments. RNA. 2007;13:414–421. doi: 10.1261/rna.79407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Zhu W, Levy DE. Nuclear and cytoplasmic mRNA quantification by SYBR green based real-time RT-PCR. Methods. 2006;39:356–362. doi: 10.1016/j.ymeth.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 22.Schwanekamp JA, Sartor MA, Karyala S, Halbleib D, Medvedovic M, Tomlinson CR. Genome-wide analyses show that nuclear and cytoplasmic RNA levels are differentially affected by dioxin. Biochim Biophys Acta. 2006;1759:388–402. doi: 10.1016/j.bbaexp.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 23.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 24.Sartor MA, Tomlinson CR, Wesselkamper SC, Sivaganesan S, Leikauf GD, Medvedovic M. Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinformatics. 2006;7:538. doi: 10.1186/1471-2105-7-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smyth GK. Limma: Linear models for microarray data. In: Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S, editors. Bioinformatics and computational biology solutions using R and bioconductor. Springer-Verlag; New York: 2005. pp. 397–420. [Google Scholar]
- 26.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Royal Statist Soc. 1995;B57:289–399. [Google Scholar]
- 27.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 28.Ashburner M, Ball CA, Blake JA, et al. Gene Ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Pirooznia M, Nagarajan V, Deng Y. GeneVenn—A web application for comparing gene lists using Venn diagrams. Bioinformation. 2007;1:420–422. doi: 10.6026/97320630001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikitin A, Egorov S, Daraselia N, Mazo I. Pathway studio-the analysis and navigation of molecular networks. Bioinformatics. 2003;19:2155–2157. doi: 10.1093/bioinformatics/btg290. [DOI] [PubMed] [Google Scholar]
- 33.Pestka S, Hintikka H. Studies on the formation of ribonucleic acid-ribosome complexes. XVI. Effect of ribosomal translocation inhibitors on polyribosomes. J Biol Chem. 1971;246:7723–7730. [PubMed] [Google Scholar]
- 34.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen G, Gharib TG, Huang CC, et al. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics. 2002;1:304–313. doi: 10.1074/mcp.m200008-mcp200. [DOI] [PubMed] [Google Scholar]
- 36.Tew KD, Monks A, Barone L, et al. Glutathione-associated enzymes in the human cell lines of the National Cancer Institute Drug Screening Program. Mol Pharmacol. 1996;50:149–159. [PubMed] [Google Scholar]
- 37.Haynes PA, Gygi SP, Figeys D, Aebersold R. Proteome analysis: Biological assay or data archive? Electrophoresis. 1998;19:1862–1871. doi: 10.1002/elps.1150191104. [DOI] [PubMed] [Google Scholar]
- 38.Soucy TA, Smith PG, Milhollen MA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 39.Soucy TA, Smith PG, Rolfe M. Targeting NEDD8-activated cullin-RING ligases for the treatment of cancer. Clin Cancer Res. 2009;15:3912–3916. doi: 10.1158/1078-0432.CCR-09-0343. [DOI] [PubMed] [Google Scholar]
- 40.Morris JP, Wang SC, Hebrok M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat Rev Cancer. 2010;10:683–695. doi: 10.1038/nrc2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miano JM. Role of serum response factor in the pathogenesis of disease. Lab Invest. 2010;90:1274–1284. doi: 10.1038/labinvest.2010.104. [DOI] [PubMed] [Google Scholar]
- 42.Miralles F, Hebrard S, Lamotte L, et al. Conditional inactivation of the murine serum response factor in the pancreas leads to severe pancreatitis. Lab Invest. 2006;86:1020–1036. doi: 10.1038/labinvest.3700457. [DOI] [PubMed] [Google Scholar]
- 43.Bolognese F, Forni C, Caretti G, Frontini M, Minuzzo M, Mantovani R. The Pole3 bidirectional unit is regulated by MYC and E2Fs. Gene. 2006;366:109–116. doi: 10.1016/j.gene.2005.07.046. [DOI] [PubMed] [Google Scholar]
- 44.Lan L, Ui A, Nakajima S, et al. The ACF1 complex is required for DNA double-strand break repair in human cells. Mol Cell. 2010;40:976–987. doi: 10.1016/j.molcel.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 45.Thornley J, Trask H, Ridley C, et al. Differential regulation of polysome mRNA levels in mouse Hepa-1C1C7 cells exposed to dioxin. Toxicol In Vitro. 2011. in press. [DOI] [PMC free article] [PubMed]
- 46.Levy L, Hill CS. Smad4 dependency defines two classes of transforming growth factor {beta} (TGF-{beta}) target genes and distinguishes TGF-{beta}-induced epithelialmesenchymal transition from its antiproliferative and migratory responses. Mol Cell Biol. 2005;25:8108–8125. doi: 10.1128/MCB.25.18.8108-8125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki Y, Ohga N, Morishita Y, Hida K, Miyazono K, Watabe T. BMP-9 induces proliferation of multiple types of endothelial cells in vitro and in vivo. J Cell Sci. 2010;123:1684–1692. doi: 10.1242/jcs.061556. [DOI] [PubMed] [Google Scholar]
- 48.Munroe D, Jacobson A. mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol Cell Biol. 1990;10:3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kren BT, Wong PY-P, Shiota A, Zhang X, Zeng Y, Steer CJ. Polysome trafficking of transcripts and microRNAs in regenerating liver after partial hepatectomy. Am J Physiol Gastrointest Liver Physiol. 2009;297:G1181–G1192. doi: 10.1152/ajpgi.90636.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fitzgerald KD, Semler BL. Bridging IRES elements in mRNAs to the eukaryotic translation apparatus. Biochim Biophys Acta. 2009;1789:518–528. doi: 10.1016/j.bbagrm.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corral-Debrinski M. mRNA specific subcellular localization represents a crucial step for fine-tuning of gene expression in mammalian cells. Biochim Biophys Acta. 2007;1773:473–475. doi: 10.1016/j.bbamcr.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 52.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 53.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hendrickson DG, Hogan DJ, McCullough HL, et al. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by drosha. Mol Cell. 2010;39:373–384. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.