Abstract
In utero electroporation (IUE) has become a method of choice for rapid gain and loss of function studies in embryonic cerebral cortex. In this review we highlight some of the proven and recent advances in IUE technology that make it applicable to an increasingly wide array of experiments requiring spatial and temporal control of gene expression. Recently, cell-type–specific promoters and tamoxifen-gated cre-recombinase have been shown to work effectively with IUE. Experiments can now be designed and carried out to test whether and which cell-type–specific mechanisms operate within defined periods of neuronal migration and maturation. We have recently adapted this conditional expression approach to implement conditional rescue experiments. In conditional rescue, expression of an RNA interference (RNAi) target is restored by tamoxifen-induced cre-mediated recombination. An initial disruption in migration, and resultant malformation, caused by DCX RNAi was reversed by delayed re-expression of Dcx. In the future, combinations of spatially directed, cell-type–specific, and tamoxifen-gated transgene expression can be used to address the complex mechanisms likely to operate during development of cerebral cortex.
Keywords: migration, neocortex, transgenesis, stem cell
Introduction
In order to investigate cell and region specific properties of the cerebral cortex it will be necessary to conduct experiments that combine both loss and gain of function in defined cell populations at defined times in development. For example, different neuronal types between and within cortical lamina are likely to migrate to their appropriate positions and form cell-specific connections by distinct and shared mechanisms. The only way to study these mechanisms selectively will be to alter expression in defined cell populations at select developmental time points. Mouse genetic approaches have advanced to the point where such controlled gene expression is possible, however, it can require the production and maintenance of multiple lines of mice at an increasing expense to laboratories. Over the past several years in utero electroporation (IUE) has emerged as an efficient means to transfect and to manipulate cerebral cortical precursor cells and neurons.
In 2001, 3 papers demonstrated successful transfection by electroporation of cells in developing neocortex in mouse and rat embryos. (Fukuchi-Shimogori and Grove 2001; Saito and Nakatsuji 2001; Tabata and Nakajima 2001) The advantages of this approach became apparent in studies of mechanisms of migration (Takahashi et al. 2002; Kawauchi et al. 2003) and regional patterning (Fukuchi-Shimogori and Grove 2001). By combining IUE with short-hairpin RNA (shRNA) plasmids to create RNA interference (RNAi) (Bai et al. 2003) both gain and loss of function studies became possible. In addition, IUE is applicable to species for which genetic manipulation is not easily implemented. In the past few years, new approaches have been applied to IUE that make it suitable for increasingly sophisticated experiments requiring cellular and temporal control of transgene expression. In this review we discuss 5 features of IUE that make it a suitable method for targeting select populations of cells- as defined by position, promoter, or progenitor type- to manipulate molecular pathways that function in different periods of development. Finally, we introduce a conditional rescue approach that can be used to address questions regarding the reversibility of developmental disruptions and disorders that may occur during formation of cortex.
Spatially Directed IUE
The first published report using IUE in developing cortex (Fukuchi-Shimogori and Grove 2001) made use of spatially directed electroporation. By visually directing the position of the positive electrode, a defined region of the cortical surface was transfected, and a secondary patch of FGF8 expression was localized to parietal cortex. This in turn shifted the position of somatosensory cortex caudally and in some cases resulted in a duplication of the barrel fields (Fukuchi-Shimogori and Grove 2001). Regionalized transfection in this study required a challenging surgery involving intrauterine insertion of 2 electrodes in young (E11–12) embryos. Since then, several investigators have shown that it is possible to direct transfection to different regions of the lateral ventricles by orienting the position of extra-uterine paddle electrodes. For example, directed electroporation of the hippocampus is possible by positioning the positive paddle electrode contralateral to the ventricle injected with plasmid, and thereby drive electroporation into medial regions of the embryonic telencephalon that give rise to hippocampus (Nakahira and Yuasa 2005; Navarro-Quiroga et al. 2007). Similarly, electroporation can be directed to ventral progenitors that give rise to interneurons in the mouse cortex (Borrell et al. 2005). Dorsal or lateral pallium that gives rise to neocortical pyramidal neurons, or to piriform cortex and amygdala (Remedios et al. 2007; Bai et al. 2008) can be transfected by placing the positive electrode at dorsal–lateral or lateral–ventral positions respectively (Fig. 1). In addition, although still not as spatially precise as insertion of electrodes into the embryonic cortex, we have found that reducing the size of the positive paddle electrode to 2–3 mm in diameter can result in localized patches of electroporation. In the future, more spatially restricted expression may be possible by combining electrode position with plasmid vectors containing promoters for genes that show regionalized expression.
Figure 1.
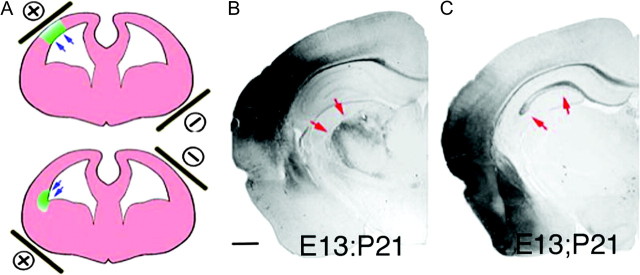
Spatial control of transfection. (A) Schematic showing how positioning the paddle electrodes during IUE can be used to direct the location of transfection. (B) Example of a dorsal lateral transfection delivered at E13 and assessed at P21. Cells are primarily neocortical pyramidal neurons that project axons into the thalamus (red arrows). (C) Example of a lateral ventral transfection at E13 that resulted in transfection of primarily entorhinal cortex, piriform cortex and amygdala. Note that in contrast to B axons in C (red arrows) are found primarily in the hippocampus and not in the thalamus.
Cell-Type–Specific Expression
Recent studies show the IUE can be used to preferentially express transgenes in different subsets of progenitor cells at the ventricular zone (VZ) surface. One simple way for such progenitor-specific transfection takes advantage of both the temporal order of cell generation in cortex and the fact that plasmids transfected by IUE appear to be lost in cells that undergo cell division. Thus, IUE can be used similarly to mitotic birth dating. In a systematic comparison of neuronal birth-dating by IUE versus BrdU, Langevin et al. (2007), demonstrated that IUE can be used to specifically label neurons occupying different cortical lamina depending on the time of electroporation (Langevin et al. 2007). Hatanaka et al. (2004), effectively used the approach of differing the time of electroporation to determine that Cdk5 functions more critically in the migration of late generated upper layer neurons than in early generated deep layer neurons (Hatanaka et al. 2004). We have found similarly that changing the time of electroporation can not only be used to transfect pyramidal neurons in different layers, but also can be used to transfect astrocyte progenitors and astrocytes. As shown in Figure 2, if IUE is administered at E18 in the rat, then astrocytes are primarily transfected (Fig. 2).
Figure 2.
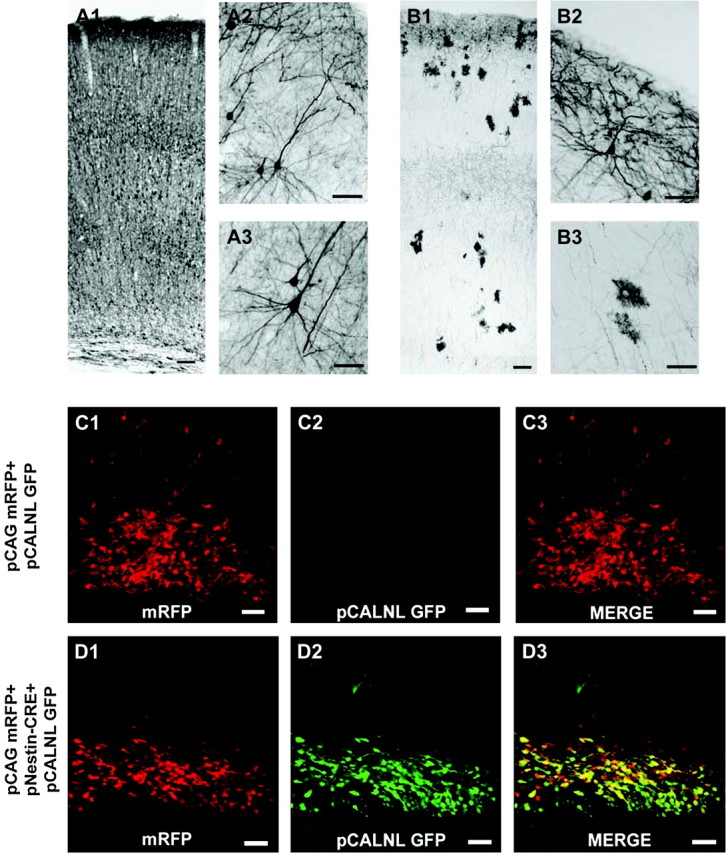
Temporal and cre-recombinase induced control of gene expression by IUE. (A) (A1–A3) Coronal section of rat neocortex transfected with GFP at embryonic day (E) 16 and fixed at postnatal day (P) 21. (A1) GFP labeled neurons are present across deep and upper cortical layers. (A2) Higher magnification micrograph of layer 2/3 cortical pyramidal neurons labeled with GFP. (A3) Micrograph of GFP+ deep layer pyramidal neuron. (B) Coronal section of rat neocortex transfected at E18 with GFP and fixed at P21. A distinct band of labeled upper layer 2 neurons are present as are a preponderance of astrocytes spanning multiple laminae. Higher magnification image of GFP+ superficial layer 2 cortical pyramidal neuron located at the pial surface (B2), and of GFP labeled parenchymal astrocytes (B3). (C) An E19 harvested rat embryo following electroporation at E17 with pCAG mRFP + pCALNL GFP shows only mRFP expression. The pCANLl-GFP plasmid contains a loxp-neostop-loxp sequence upstream from GFP sequence (see Matsuda and Cepko 2007). (D) E19 harvested rat embryo following E17 electroporation with pCAG mRFP + pNestin-CRE (nestin enhancer element driving CRE recombinase expression) + pCALNL GFP shows robust GFP and mRFP expression within ventricular zone progenitors. Scale bars: A1 and B1 = 100 μm; A2, A3, B2, and B3 = 50 μm; C1–D3 = 40 μm.
Figure 3.
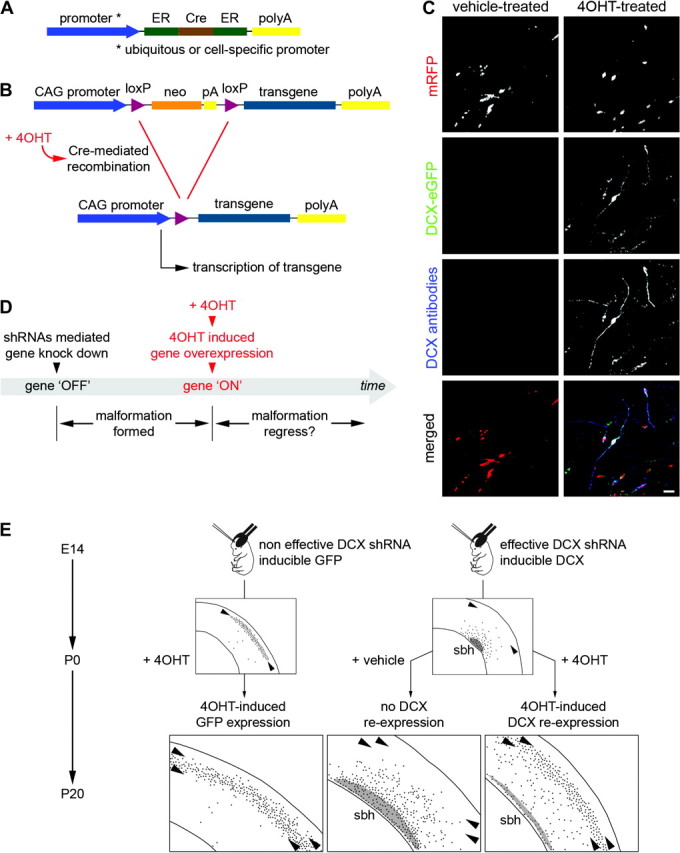
Conditional RNAi rescue strategy to investigate reversibility of neuronal migration disorders. (A, B) Schematic showing the strategy for conditional RNAi rescue. (A) 4-OHT-activatable Cre recombinase composed of 2 ER binding domains is expressed under the control of the strong ubiquitous CAG promoter or a cell-specific promoter. (B) Cre-dependent inducible expression vectors contain sequence for the neomycin resistance gene (neo-stop and polyA), flanked by 2 loxP sites. Cre-mediated recombination would remove the neo cassette and allow expression of the transgene. (C) An example of conditional DCX-eGFP expression. Neurons were transfected with 4 plasmids encoding mRFP, 4-OHT–activatable Cre recombinase, inducible DCX-eGFP and shRNAs against DCX. In the 4-OHT-treated condition (left panels), DCX-eGFP is expressed and DCX is detected with antibodies in transfected neurons. In the vehicle-treated condition (right panels), no signal is detected in the green channel or with DCX antibodies. Scale bar, 50 μm. (D) Proposed experimental paradigm to investigate the possibility of reactivating migration using a conditional RNAi rescue strategy. (E) An example of conditional RNAi rescue strategy applied to the model of subcortical band heterotopia (sbh) induced by in utero RNAi against DCX. Two groups of animals were electroporated at E14 with either ineffective (malformation-free animals) or effective shRNA vectors against Dcx (animals with sbh), together with vectors coding mRFP. Malformation-free animals were co-electroporated with an inducible eGFP expression vector and animals with sbh with an inducible DCX-eGFP expression vector, together with vectors coding 4-OHT–activatable Cre recombinase. At birth, transfected neurons were normally located in upper cortical layers in malformation-free animals and were ectopically accumulated at deep positions, forming a band heterotopia, in sbh animals. At birth, animals were injected with 4-OHT or its vehicle solution to activate Cre and induce Cre-mediated recombination. At P15, in sbh animals in which Dcx re-expression was induced with 4-OHT at P0, previously mispositioned transfected neurons were relocated to upper cortical layers similar to malformation-free animals that received induced GFP expression. Size of sbh (gray areas) was also greatly reduced as compared with sbh animals that received no induction.
Cell-type–specific promoters have also been used to drive expression selectively in IUE experiments. Promoters for the transcription factors, ER81 and NGN2, have been used to drive expression of fluorescent proteins selectively in neocortical neurons (Langevin et al. 2007). Similarly, Wang et al recapitulated doublecortin expression in immature, migratory neurons of cortex by using a 3.7-kb genomic fragment upstream of the doublecortin start codon as the promoter to drive expression of green fluorescent protein (GFP) or DsRED. In addition, the authors used electroporation of different constructs to quickly identify the essential regulatory sequences in the DCX promoter. The authors showed that a 2-kb sequence within the 3.7-kb region was critical for reporter expression in DCX expressing cells (Wang et al. 2007). IUE can therefore be used not only to direct cell-specific expression but also as a relatively rapid screen to identify promoter regions.
As a clear demonstration of using promoters to label subsets of neocortical progenitors, Gal et al. (2006), used different promoters to drive GFP and DsRED2 in populations of ventricular zone progenitors (Gal et al. 2006). Promoters that are putatively active specifically in neuronal progenitors (Tα1) and radial glia (BLBP and GLAST) were used to differentially drive expression of fluorescent proteins in cells at the VZ surface with distinct morphologies. The plasmid pTα1-EGFP labeled rounded cells at the ventricular surface that lacked long radial processes, whereas pBLBP-EGFP and pGLAST-EGFP labeled cells that had typical radial glia-like morphologies (Gal et al. 2006). With coelectroporation experiments, the authors further showed a lack of overlap between cells transfected with the different promoter constructs. Although it remains to be determined whether these VZ promoters are active in progenitors that give rise to different cell types, the results suggest differences in promoter activities can be used to label subsets of cells at the VZ surface. Overall, the use of time of transfection and different promoters will be extremely useful in defining potential mechanistic heterogeneity amongst different neocortical progenitors. For example, such promoters can be used to drive differential expression of shRNAs for RNAi or dominant negative proteins to determine whether there are differences amongst progenitor types in the mechanisms that regulate migration or proliferation.
Cross Rescue and Combinatorial RNAi
In addition to cell-specific expression, IUE has been used to identify functional interactions between genes. As up to at least 4 plasmids cotransfect at rates of over 80%, it is possible to combine multiple RNAis and expression of multiple transgenes in the same population of cells. By combining RNAis directed against different targets it has been possible to test for synergistic, antagonistic, or saturating effects of knocking down combinations of gene targets. Similarly, cross rescue experiments (i.e., rescuing an RNAi phenotype with expression of a different transgene) can be used to test whether another gene is downstream from a particular RNAi target. The results of combinatorial RNAi and cross rescue allowed Young-Pearse et al. (2007) to show that App functions in neuronal migration, at least in part, though interaction with Dab1. RNAi of App and Dab1 synergistically impaired migration, and overexpression of Dab1 partially rescued the impairment in migration caused by RNAi targeted against App (Young-Pearse et al. 2007). Similarly, Tsai et al. (2007) combined RNAi against Lis1 with expression of dynein mutants to show that that Lis1 and dynein play dual roles in radial migration (Tsai et al. 2007). Rescue of RNAi phenotypes can also be used to rapidly define the domains of a protein that are necessary and sufficient to neuronal migration. For example, Wang et al. (2006) transfected shRNA expressing plasmids targeting Dyx1c1 along with different truncations of Dyx1c1 coding sequence (Wang et al. 2006). The approach was used to narrow the functionally necessary and sufficient region of Dyx1c1 in neuronal migration to 120 amino acids of the C-terminus (Wang et al. 2006).
Conditional Expression and Conditional RNAi
Conditional expression systems have been developed and applied to IUE experiments to induce cell and time specific expression of transgenes (Matsuda and Cepko 2007). In this approach, cre-recombinase expressed from a transfected plasmid can be used to gate the expression of shRNAs for RNAi or any other transgene in a cell and time specific manner. This 2 plasmid system uses one plasmid to drive the expression of Cre-recombinase and another plasmid that contains loxp sequence flanking neo-stop-polyA sequences that must be removed by recombination before the product of a second transgene can be expressed (Matsuda and Cepko 2007). If cre-recombinase expression is under the control of a cell-type–specific promoter, then expression of the second transgene is gated on in only those cells that can activate the cell-specific promoter. As shown in Figure 2, this approach can be used to effectively drive expression of fluorescent proteins in cortical VZ progenitors. As shown in Figure 2, the system shows virtually no leak as only when the conditional reporter plasmid (pCALNL-GFP) is cotransfected with the Cre expression plasmid (pNestin-Cre) is the eGFP reporter expressed. This approach can also be used to gate RNAi in subsets of progenitors or to fate map progenitors that have promoters transiently active. Because the recombined construct has a ubiquitous promoter, even after the progenitor promoters are no longer active as cells mature the cells will continue to express reporter protein from the ubiquitous promoter. In addition, unlike virus or genetically based fate mapping, because IUE plasmids are episomal and lost after successive cell division, the fate mapping with this approach could be restricted to newly postmitotic cells and not to all subsequent members of the lineage. This could be an advantage if the goal is to track the fates of cells that undergo a final division at nearly the same point in time.
In order to design experiments that define the role of particular genes at arbitrarily selected times in development it becomes necessary to be able to conditionally activate the expression of transgenes by addition of some pharmacological agent. One such system, extensively used in transgenic animals, is based on a fusion protein of Cre-recombinase with a tamoxifen-binding site. Matsuda and Cepko (2007) developed and proved an elegant conditional expression system appropriate for IUE in retina and cortex that is based on a modified version of tamoxifen receptor-Cre fusions. In their system they show that it is important to use a version of Cre fused to a tamoxifen receptor located at both the N and C terminus in order to prevent leakiness from active Cre in the absence of tamoxifen. The resulting protein ERt2-Cre-ERt2 will only be transported to the nucleus by receptor mediated transport when it is bound to tamoxifen. Once in the nucleus it can induce directed recombination at loxp sites. Matsuda and Cepko showed that in the absence of tamoxifen there was no recombination as evidenced by expression from a reporter plasmid. They further showed that the inducible system could be used to gate the expression of shRNAs for RNAi experiments. In a demonstration of the power and versatility of the approach they showed that by expressing ERt2-Cre-ERt2 by promoters active specifically in rod photoreceptor progenitors that it was possible to create cell-type–specific, temporally inducible expression of transgenes. Another important and remarkable feature of the system developed is that inducible expression is achieved by a single injection of tamoxifen, and this allows for temporally precise expression of an shRNA or other transgene. With the addition of this set of tools to IUE, it now becomes theoretically possible to control the expression of shRNAs or other transgenes specifically in virtually any cortical neuronal cell type at nearly any time in development.
Conditional RNAi Rescue
Conditional expression systems make it possible to test new developmental possibilities and properties. As an example, we have recently combined conditional expression with RNAi to ask whether there is a critical period during cortical neuronal migration, when stalled migration, can be reactivated. More specifically, we applied this approach to our previously developed model of subcortical band heterotopia created by in utero RNAi against Dcx (Bai et al. 2003). We conditionally re-expressed Dcx using the ERt2-Cre-ERt2 system reviewed above, to investigate whether delayed reintroduction of Dcx, following RNAi of Dcx could re-initiate migration to normal positions even after a cortical malformation had formed in the perinatal cerebral cortex (Fig. 3). Our results revealed that migration can indeed be successfully restarted even after a protracted delay, causing previously mis-positioned neurons to settle into appropriate upper cortical layers (Manent et al. 2009). In addition, significant re-migration was achieved at ages up to P5 indicating that there is an extended period where developmental plasticity can be manipulated to ameliorate cortical malformations. In addition, neurons could be shifted towards appropriate migration targets by the re-expression of Dcx even as late as P10. Of potential clinical significance, this study also revealed that a functional impairment, increased seizure sensitivity, was restored to control levels following the reactivation of migration. The conditional rescue approach serves as a general example for a class of experiments that can be performed to define the points of developmental reversibility and vulnerability in developing cortex. We anticipate that future experiments will be used to define classes of disruptions that show varying degrees of reversibility, and to identify signaling pathways that when activated may ameliorate the functional deficits associated with developmental malformations of cortex.
Future Prospects
One potential limitation of IUE, similar to DNA viruses such as herpes virus, is that the transgenes introduced by IUE appear to remain episomal and are lost from cells following successive cell divisions. We and others have found that in postmitotic cells the episomal transgenes remain active for months after transfection (Ramos et al. 2006). Nevertheless it could be desirable to deliver transgenes by IUE that would integrate into the genome. One possible strategy for this may be piggyBac transposase which has been shown to work efficiently in mammalian cells to integrate transgenes (Ding et al. 2005; Shinohara et al. 2007). Another area of potential improvement in IUE will be to increase the number of cells that can be transfected. Currently, many thousands of cells can be transfected but there are still many cells at the VZ surface that remain untransfected. Possibilities for increasing transfection rates could include combining electroporation with transfection reagents, ultrasound pulses, or brief laser pulses (Wells 2004). In addition, increasingly IUE is being used in combination with mouse knock out models. One particularly powerful combination of IUE and conditional mouse knockouts is electroporating conditionally active cre plasmids, pCAGGS-ERt2-Cre-ERt2, into mice harboring conditional knock out alleles. Such a mosaic analysis will be extremely useful in distinguishing cell autonomous from noncell autonomous mechanisms of migration and differentiation.
Acknowledgments
Conflict of Interest: None declared.
References
- Bai J, Ramos RL, Ackman JB, Thomas AM, Lee RV, LoTurco JJ. RNAi reveals doublecortin is required for radial migration in rat neocortex. Nat Neurosci. 2003;6:1277–1283. doi: 10.1038/nn1153. [DOI] [PubMed] [Google Scholar]
- Bai J, Ramos RL, Paramasivam M, Siddiqi F, Ackman JB, LoTurco JJ. The role of DCX and LIS1 in migration through the lateral cortical stream of developing forebrain. Dev Neurosci. 2008;30:144–156. doi: 10.1159/000109859. [DOI] [PubMed] [Google Scholar]
- Borrell V, Yoshimura Y, Callaway EM. Targeted gene delivery to telencephalic inhibitory neurons by directional in utero electroporation. J Neurosci Methods. 2005;143:151–158. doi: 10.1016/j.jneumeth.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Fukuchi-Shimogori T, Grove EA. Neocortex patterning by the secreted signaling molecule FGF8. Science. 2001;294:1071–1074. doi: 10.1126/science.1064252. [DOI] [PubMed] [Google Scholar]
- Gal JS, Morozov YM, Ayoub AE, Chatterjee M, Rakic P, Haydar TF. Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. J Neurosci. 2006;26:1045–1056. doi: 10.1523/JNEUROSCI.4499-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka Y, Hisanaga S, Heizmann CW, Murakami F. Distinct migratory behavior of early- and late-born neurons derived from the cortical ventricular zone. J Comp Neurol. 2004;479:1–14. doi: 10.1002/cne.20256. [DOI] [PubMed] [Google Scholar]
- Kawauchi T, Chihama K, Nabeshima Y, Hoshino M. The in vivo roles of STEF/Tiam1, Rac1 and JNK in cortical neuronal migration. EMBO J. 2003;22:4190–4201. doi: 10.1093/emboj/cdg413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin LM, Mattar P, Scardigli R, Roussigne M, Logan C, Blader P, Schuurmans C. Validating in utero electroporation for the rapid analysis of gene regulatory elements in the murine telencephalon. Dev Dyn. 2007;236:1273–1286. doi: 10.1002/dvdy.21126. [DOI] [PubMed] [Google Scholar]
- Manent JB, Wang Y, Chang Y, Paramasivam M, Loturco JJ. Dcx reexpression reduces subcortical band heterotopia and seizure threshold in an animal model of neuronal migration disorder. Nat Med. 2009;15:84–90. doi: 10.1038/nm.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci USA. 2007;104:1027–1032. doi: 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira E, Yuasa S. Neuronal generation, migration, and differentiation in the mouse hippocampal primoridium as revealed by enhanced green fluorescent protein gene transfer by means of in utero electroporation. J Comp Neurol. 2005;483:329–340. doi: 10.1002/cne.20441. [DOI] [PubMed] [Google Scholar]
- Navarro-Quiroga I, Chittajallu R, Gallo V, Haydar TF. Long-term, selective gene expression in developing and adult hippocampal pyramidal neurons using focal in utero electroporation. J Neurosci. 2007;27:5007–5011. doi: 10.1523/JNEUROSCI.0867-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos RL, Bai J, LoTurco JJ. Heterotopia formation in rat but not mouse neocortex after RNA interference knockdown of DCX. Cereb Cortex. 2006;16:1323–1331. doi: 10.1093/cercor/bhj074. [DOI] [PubMed] [Google Scholar]
- Remedios R, Huilgol D, Saha B, Hari P, Bhatnagar L, Kowalczyk T, Hevner RF, Suda Y, Aizawa S, Ohshima T, et al. A stream of cells migrating from the caudal telencephalon reveals a link between the amygdala and neocortex. Nat Neurosci. 2007;10:1141–1150. doi: 10.1038/nn1955. [DOI] [PubMed] [Google Scholar]
- Saito T, Nakatsuji N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev Biol. 2001;240:237–246. doi: 10.1006/dbio.2001.0439. [DOI] [PubMed] [Google Scholar]
- Shinohara ET, Kaminski JM, Segal DJ, Pelczar P, Kolhe R, Ryan T, Coates CJ, Fraser MJ, Handler AM, Yanagimachi R, et al. Active integration: new strategies for transgenesis. Transgenic Res. 2007;16:333–339. doi: 10.1007/s11248-007-9077-z. [DOI] [PubMed] [Google Scholar]
- Tabata H, Nakajima K. Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex. Neuroscience. 2001;103:865–872. doi: 10.1016/s0306-4522(01)00016-1. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Sato K, Nomura T, Osumi N. Manipulating gene expressions by electroporation in the developing brain of mammalian embryos. Differentiation. 2002;70:155–162. doi: 10.1046/j.1432-0436.2002.700405.x. [DOI] [PubMed] [Google Scholar]
- Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci. 2007;10:970–979. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- Wang X, Qiu R, Tsark W, Lu Q. Rapid promoter analysis in developing mouse brain and genetic labeling of young neurons by doublecortin-DsRed-express. J Neurosci Res. 2007;85:3567–3573. doi: 10.1002/jnr.21440. [DOI] [PubMed] [Google Scholar]
- Wang Y, Paramasivam M, Thomas A, Bai J, Kaminen-Ahola N, Kere J, Voskuil J, Rosen GD, Galaburda AM, Loturco JJ. DYX1C1 functions in neuronal migration in developing neocortex. Neuroscience. 2006;143:515–522. doi: 10.1016/j.neuroscience.2006.08.022. [DOI] [PubMed] [Google Scholar]
- Wells DJ. Gene therapy progress and prospects: electroporation and other physical methods. Gene Ther. 2004;11:1363–1369. doi: 10.1038/sj.gt.3302337. [DOI] [PubMed] [Google Scholar]
- Young-Pearse TL, Bai J, Chang R, Zheng JB, LoTurco JJ, Selkoe DJ. A critical function for beta-amyloid precursor protein in neuronal migration revealed by in utero RNA interference. J Neurosci. 2007;27:14459–14469. doi: 10.1523/JNEUROSCI.4701-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]


