Abstract
The development of a transgenic mouse model carrying the human poliovirus receptor has made it possible to investigate the molecular mechanisms of the viral dissemination process in a whole organism. Studies on this have provided an insight into the mechanisms for viral permeation through the blood-brain barrier and retrograde axonal transport of the virus. In addition, strain-specific neurovirulence levels are shown to depend mainly on the replicating capacity of the virus in the central nervous system rather than the efficiency of the 2 dissemination pathways indicated above. Studies of poliovirus-induced cytopathic effects on neural cells revealed that neural cells possess anti-poliovirus characteristics that may offer a new avenue for investigating the molecular mechanisms of poliovirus neurovirulence.
Keywords: poliovirus, transgenic mouse, poliovirus receptor, dissemination, neurovirulence
Introduction
Poliovirus, a human enterovirus that belongs to the family Picornaviridae, is the causative agent of poliomyelitis. Humans are the only natural hosts of poliovirus. The virus, however, can be transferred to monkeys when it is directly inoculated into the central nervous system (CNS). The species specificity of this virus is governed by a specific cell surface molecule that serves as the poliovirus receptor (PVR).1),2) Indeed, transgenic mice carrying the human poliovirus receptor (hPVR/CD155) gene show susceptibility to poliovirus infection, although mice in general are not susceptible to poliovirus.3),4)
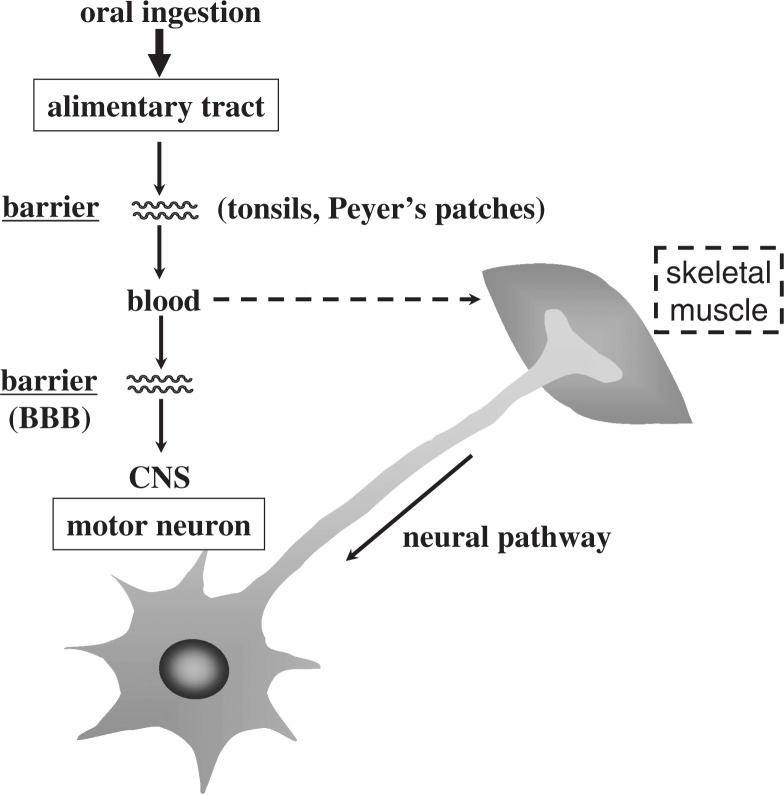
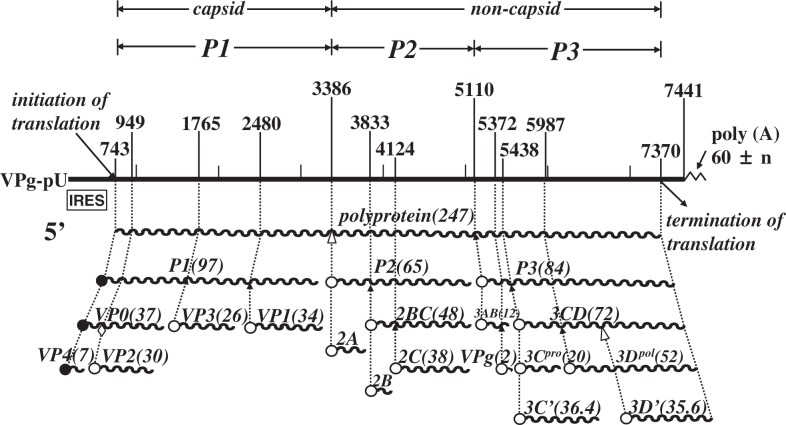
In humans, poliovirus infection usually begins with oral ingestion of the virus. After oral ingestion, the virus multiplies in the alimentary mucosa, and possibly in the tonsils and Peyer’s patches. The virus then moves into the blood stream (viremia) through the putative barrier(s) that virulent poliovirus strains cross more efficiently than attenuated strains. The circulating virus invades the CNS and replicates in neurons, particularly motor neurons. There are 2 possible dissemination routes through which the circulating polio-virus can enter the CNS. One is virus permeation through the blood-brain barrier (BBB), and the other is virus transmission via peripheral nerves (Fig. 1). Paralytic poliomyelitis occurs as a result of neuronal destruction by lytic replication of the poliovirus, although paralysis develops in less than 1% of those infected.5),6) A poliovirion consists of a single-stranded RNA genome of positive polarity and a non-enveloped capsid that comprises 60 copies of each of 4 capsid proteins: VP1, VP2, VP3, and VP4. The three-dimensional structure has been elucidated,7) and the study revealed depressions called “canyons” on the virion surface, which have been shown to be attachment sites for PVR.8)–10) The genome of the poliovirus functions as mRNA in the cytoplasm of infected cells. The virus-specific translation process begins with the entry of ribosomes into the internal ribosome entry site (IRES) within the 5′ noncoding sequence of the RNA,11) and all viral proteins are translated as a large precursor protein from a single open reading frame that is cotranslationally processed into functional viral proteins (Fig. 2).
Fig. 1.
Dissemination pathways of poliovirus in humans.
Fig. 2.
Genome organization of poliovirus type 1. Genomic RNA and its gene organization are shown. VPg is a small protein attached to the 5′ end of the genome; poly(A) is 3′ terminal. Nucleotide numbers are shown over the genome RNA. P1 represents the protein region of viral capsid; P2 and P3 non-capsid protein. Gene products are indicated by wavy lines, and figures in parentheses are molecular mass of the corresponding viral gene products. IRES stands for internal ribosome entry site.
Poliovirus is classified into 3 stable serotypes (types 1, 2, and 3), and each serotype can cause poliomyelitis. To control poliomyelitis, attenuated poliovirus strains of all 3 serotypes (Sabin 1, Sabin 2, and Sabin 3) have been developed and used effectively as oral poliovaccines (OPV).12) After oral administration, these Sabin strains can replicate to a sufficiently high level in the human alimentary tract to elicit neutralizing antibodies, although the virus has little capacity to move into the blood stream and replicate in the CNS.
The result of viral dissemination studies using polio-sensitive transgenic mice is reviewed here. In addition, the anti-poliovirus response indicated by poliovirus-infected neural cells is described as a new aspect of poliovirus neurovirulence.
A mouse model for poliomyelitis
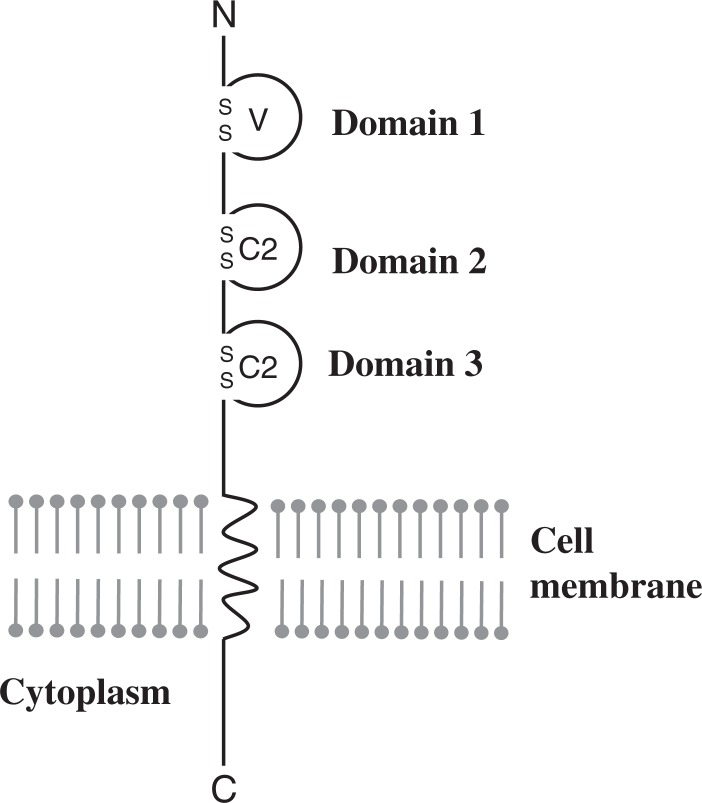
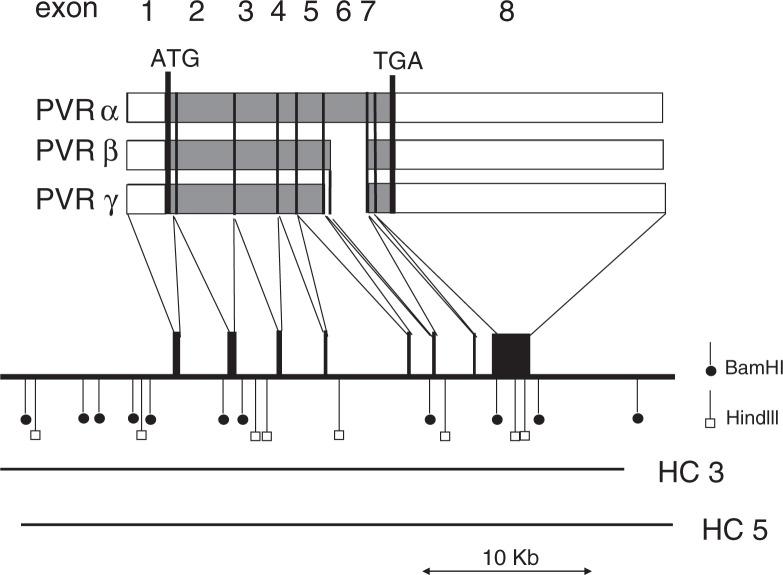
The hPVR (CD155) is a member of the immunoglobulin (Ig) superfamily, with 3 linked extracellular Ig-like domains followed by a membrane-spanning domain and a cytoplasmic domain1),2) (Fig. 3). Molecular genetic analysis revealed that the amino-terminal Ig-like domain is essential for poliovirus binding and infection.13)–16) Currently, 4 mRNA isoforms are known to be generated by alternative splicing2) and translated into 4 different forms of hPVRs, namely, hPVRα, hPVRβ, hPVRγ, and hPVRδ (Fig. 4). Of these, hPVRβ and hPVRδ lack the transmembrane domain and therefore soluble forms. Two other hPVRs, namely, hPVRα and hPVRβ are functional poliovirus receptors. They differ only in a region of the amino-acid sequence of their carboxyl-termini of the cytoplasmic domain (Fig. 5). Although hPVR is reported to bind CD226, a cell surface molecule of NK cells, and activate NK cells,17),18) it is not known whether activation of NK cells is the only physiological function of the hPVRs and why 4 different forms of hPVRs are generated.
Fig. 3.
Schematic structure of human poliovirus receptor (hPVR). Three linked extracellular Ig-like domains (V, C2, and C2 types) are followed by a membrane-spanning domain and a cytoplasmic domain.
Fig. 4.
Restriction map of the human PVR gene and multiple splicing of its transcript. Structures of mRNAs for hPVRα, hPVRβ, and hPVRγ are shown by open (untranslated sequences) and shaded (translated sequences) boxes. Numbers of exons are indicated at the top of the figure. Receptor regions are connected by thin lines to exons contained on a gene map shown underneath. Restriction cleavage sites of BamHI and HindIII are indicated by vertical bars with closed circles and open boxes, respectively. Sequences of the human PVR gene cloned into cosmids HC3 and HC5 are indicated by bars. A scale for length of nucleotides of the PVR gene is shown by bar with arrowheads at both ends at the bottom of the figure. (quoted from ref. 2).
Fig. 5.

Amino acid sequences of cytoplasmic domains of hPVRα and hPVRδ. KXXR; Tctex-1 binding motif.
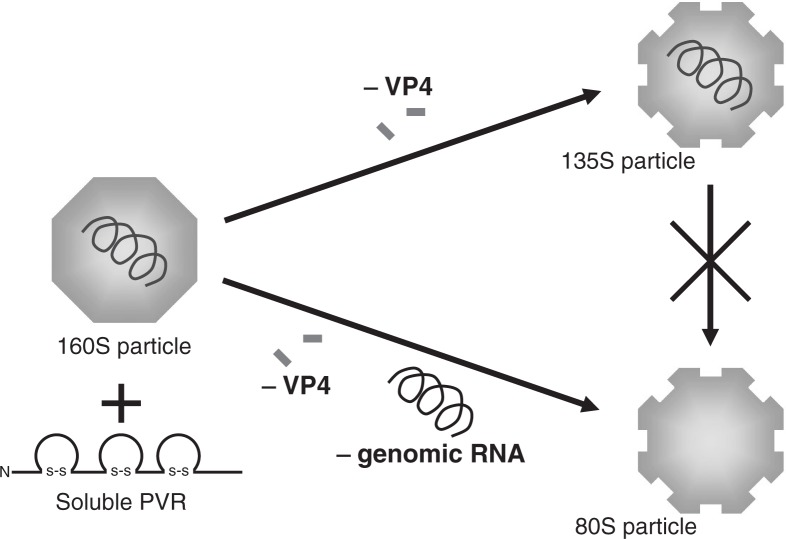
Poliovirus alters its conformation when hPVR binds the virus, and hPVR is thought to be the only molecule that is involved in the uncoating process of poliovirus. Indeed, infectious poliovirus (160S) converts to 135S and 80S particles when the virus (160S) is incubated with the extracellular domain of hPVR in vitro.19),20) A 135S particle has lost the internal protein VP4, and an 80S particle, the RNA genome as well as VP4 (Fig. 6). It is indicated that poliovirus uncoating proceeds independently of an acidic pH environment in cultured cells.19)–21)
Fig. 6.
Conformational alteration of 160S (intact) poliovirus particle induced by the addition of soluble hPVR. Infectious poliovirus (160S) converts to 135S and 80S particles when the virus (160S) is incubated with the extracellular domain of hPVR in vitro.
A transgenic (Tg) mouse carrying the hPVR gene (PVR-Tg mouse) showed susceptibility to poliovirus by intravenous (IV), intramuscular (IM), intracerebral (IC), or intraspinal (ISp) inoculation.3),4),22)–26) Transgenic mice infected via any of these 4 inoculation routes show signs of paralysis that resemble those of poliomyelitis in humans and monkeys. The results of histocytochemical examinations of the CNS of paralyzed mice are also similar to those of humans and monkeys.4),24) Furthermore, mouse neurovirulence levels of individual poliovirus strains using the PVR-Tg21 mouse model with an ISp or IC inoculation route are parallel with monkey neurovirulence.23),24) Based on these results, PVR-Tg21 mice, in addition to monkeys, are recognized by the World Health Organization (WHO) as an alternative poliomyelitis animal model. The susceptibility of PVR-Tg21 mice to orally administered poliovirus is, if any, very low (similar to that of monkeys), probably because of a strong response by interferon signaling in the gut of mice. This notion was recently proved by using PVR-Tg mice that are deficient in the type I interferon receptor.27) In addition, poliovirus was shown to be inactivated by the low pH of the gastric contents in mice.27)
The development of a mouse model for poliomyelitis makes it considerably simpler to investigate the efficiency of poliovirus dissemination in an entire organism. For example, the efficiencies of viral BBB permeation25) and retrograde axonal transport26) can be estimated after viral IV inoculation and viral IM inoculation, respectively. The results of these experiments performed using transgenic mice are described below.
Blood-brain barrier permeation
Physiological pharmacokinetic analysis was employed to investigate the fate of poliovirus inoculated into the tail vein of mice.25) The inoculated virus is distributed to various tissues, such as the spleen, liver, kidney, small intestine, heart, lung, muscle, and CNS tissues. The amount of poliovirus delivered to the CNS tissues, including the cerebellum and cerebrum, are the smallest. However, this amount is significantly greater than the theoretical amount estimated within the vascular volume of the brain. In contrast, the amount of poliovirus distributed to other tissues almost equals the theoretical amount predicted based on vascular volume. These data suggest that poliovirus passes through the BBB.
Unexpectedly, the distribution profiles of the virus 5 h after the inoculation to transgenic and non-transgenic mice are similar (Fig. 7). This suggests that specific distribution of poliovirus to the CNS tissues is not due to the expression of hPVR. This is supported by results of experiments in which the pre-administration of anti-hPVR monoclonal antibody (mAb) p286 did not affect the distribution profile of poliovirus in transgenic mice (Fig. 7). MAb p286 as well as mAb p40316) has an ability to block binding and infection of poliovirus to susceptible cells. These results indicate that hPVR expression in transgenic mice does not play a significant role in the tissue distribution profile of poliovirus until 5 h after IV inoculation.
Fig. 7.
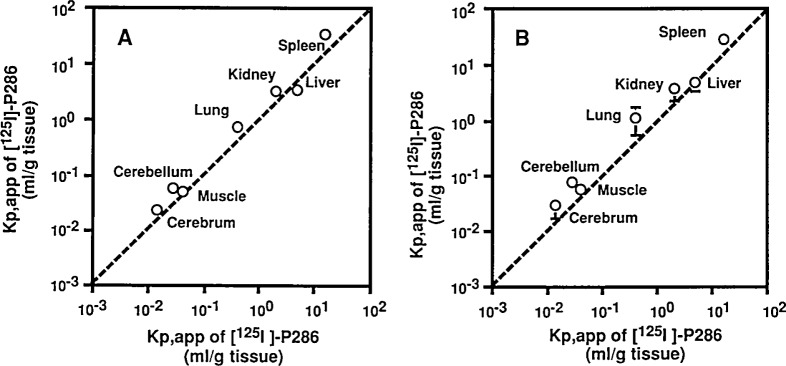
Tissue distribution of poliovirus in mice after intravenous inoculation. (A) Kp,app values of [35S]methionine-labeled MSM (a recombinant poliovirus Mahoney strain having the Sabin 1 derived capsid proteins) in various tissues were calculated in transgenic mice (vertical axis) and non-transgenic mice (horizontal axis) 5 h after the IV inoculation. (B) Transgenic mice were IV-injected with mAb p286 5 h before the IV inoculation of [35S]methionine-labeled MSM.Kp,app values of MSM in various tissues were calculated (vertical axis) and transgenic mice pretreated with the mAb (horizontal axis) 5 h after the inoculation. (quoted from ref. 25).
The linear accumulation of poliovirus in the CNS tissues was observed at least 7.5 h after inoculation.25) The accumulation rates in the brains of transgenic and non-transgenic mice were compared for the virulent and attenuated strains of poliovirus type 1 and control materials (Table 1). The difference in accumulation rates between the virulent Mahoney and attenuated Sabin 1 strains were not clearly significant. In addition, the accumulation rate observed in transgenic mice was similar to that in non-transgenic mice, supporting the notion that the hPVR does not play an important role in delivering circulating poliovirus to the tissues.
Table 1.
| Control materials | Tg mouse | Non-Tg mouse | Rat |
|---|---|---|---|
| Virulent Mahoney | 164 | 123 | — |
| Attenuated Sabin 1 | 223 | 246 | — |
| Albumin | — | 1c | — |
| Cationized RSA | — | — | 123 |
Abbreviation: Tg, transgenic.
RSA, rat serum albumin.
Modified from Ref 25.
0.001 μl min−1 g−1 tissue is regarded as 1.
The rates of accumulation of the virus in the brain is more than 100 times higher than that of albumin, which is not thought to permeate the BBB via a specific transport system, and is similar to that of cationized rat serum albumin, which is known to efficiently permeate the BBB (Table 1). These data suggest that polioviruses permeate the BBB at a fairly high degree of efficiency, independent of the hPVR or virus strains. Thus, some host cell molecules other than hPVR must be involved in the BBB permeation of poliovirus.
To elucidate mechanisms for the BBB permeation of poliovirus in vitro, an experimental system is required. We are currently investigating the BBB permeation mechanism of poliovirus by using mouse vascular endothelial cells cultured in a transwell.
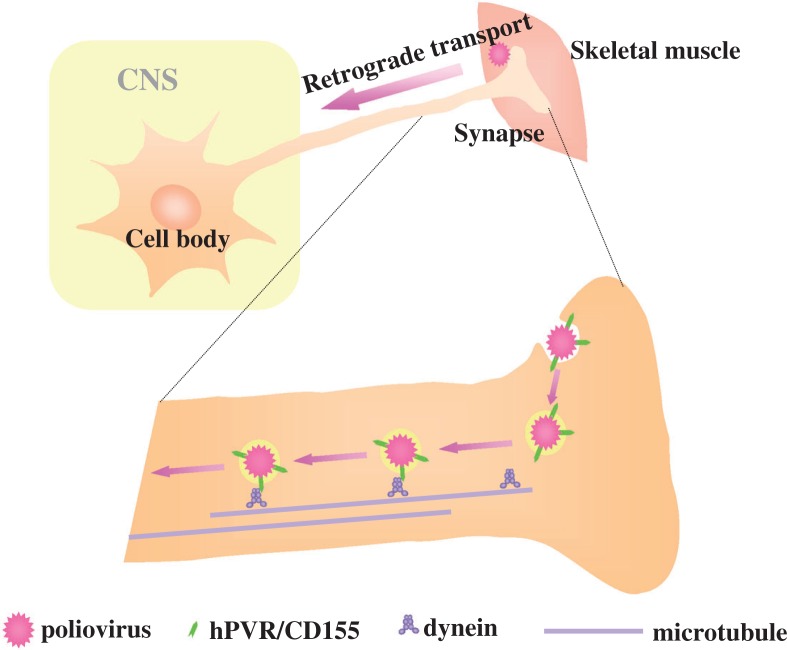
Retrograde axonal transport
A pathway of the neural dissemination caused by poliovirus has been reported for humans, monkeys, and transgenic mice. A high frequency of initial paralysis was observed in the inoculated limb in the Cutter vaccine incident, in which children received incompletely inactivated polio vaccine prepared from virulent poliovirus strains.28) In addition, experimental evidence has indicated that poliovirus can spread to the CNS through the sciatic nerve of monkeys and transgenic mice.29) Using transgenic mice, mouse neurovirulence induced by this dissemination route was shown to require hPVR. Furthermore, provocation poliomyelitis, a phenomenon resulting from physical trauma during poliovirus infection, can be experimentally reproduced in transgenic mice,30) suggesting that skeletal muscle injury stimulates retrograde axonal transport of poliovirus and thereby facilitates viral invasion of the CNS, resulting in spinal cord damage. These findings afford renewed interest in studying the neural pathway of poliovirus.
Experiments involving transection of the sciatic nerve after IM-inoculation of poliovirus into the calves of PVR-Tg mice revealed that some of the inoculated viruses move along an axon via retrograde transport at a rate of more than 12 cm per day26) (Fig. 8). This velocity is classified into fast retrograde axonal transport.31) This suggests that poliovirus is packed in endosomes during transportation through the axon because many substances that are carried retrogradely by the fast transport system are usually packed in endosomes. Indeed, an electron microscopic study detected such endosomes containing poliovirus at synapses in the vicinity of inoculation points.32) Thus, it is possible that poliovirus is enclosed by endosomes that result from hPVR-mediated endocytosis of the virus at synapses, and then conveyed through the axon retrogradely.
Fig. 8.
Poliovirus antigens in axons. The sections of the sciatic nerves were prepared from a poliovirus-infected Tg mouse. The sections were immunostained with rabbit anti-poliovirus hyperimmune serum. (Left panel) Myelin sheaths can be seen. (Center panel) Bright fluorescence is visible. (Right panel) Merging picture of the left and center panels. Poliovirus antigens existing in axons surrounded by myelin sheath. Bar =50 μm. (modified from ref. 26).
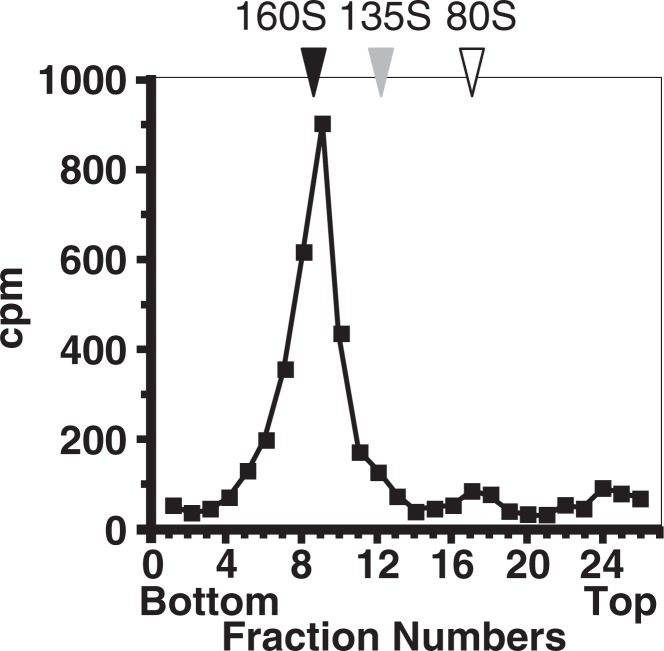
The involvement of hPVR in poliovirus pathogenesis via the neural pathway suggested that poliovirus-related materials in the sciatic nerve are conformationally altered poliovirus (135S and 80S particles) and not 160S virion particles. However, the majority of poliovirus-related materials in the sciatic nerve showed a sedimentation coefficient of 160S26) (Fig. 9), and the 160S fraction was infectious in cultured cells of primate origin. It is not known, to date, why hPVR-mediated conformational change does not occur for poliovirus-related particles in the sciatic nerve. Possibly, a small number of hPVR per virion is not sufficient to result in viral conformational change but is able to induce endocytosis of the virus on the surface of synapses. Alternatively, a cellular factor(s) that inhibits viral uncoating could exist in the neural pathway of transgenic mice. If this is the case, the virus would require to be free from such a factor(s) before replicating in the neural cell body.
Fig. 9.
Analysis of virus-related materials in the sciatic nerve. Tg mice with sciatic nerve ligation were intramuscularly inoculated with [35S]methionine-labeled poliovirus Mahoney strain. Radioactive materials were recovered from the sciatic nerve and analyzed by sucrose density gradient centrifugation. (modified from ref. 26).
It has been reported that a human homolog (TCTEL1) of mouse Tctex-1, which is light chain-1 of the cytoplasmic dynein complex, binds the cytoplasmic domain of hPVRs (Fig. 5),32),33) and that treatment of the sciatic nerve with the micro-tubule-depolymerizing agent vinblastin results in slower retrograde transport of the virus to the spinal cord of transgenic mice.32) Taking into consideration all the observations mentioned above, a possible mechanism for the retrograde axonal transport is shown in Fig. 10. IM-inoculated poliovirus is possibly incorporated into cells by hPVR-mediated endocytosis at synapses without any hPVR-mediated conformational changes of the virion particle. The cytoplasmic domain of hPVR on the surface of endosomes that enclosed the poliovirion could interact with cytoplasmic dynein, and the endosomes could be retrogradely transported along microtubules through the axon to the neuron cell body where uncoating and replication of poliovirus occurs.
Fig. 10.
Possible mechanism for the retrograde axonal transport of poliovirus.
A reconstitution of the experimental system was established to support the above hypothesis, in which rat primary neurons were employed. Molecular imaging experiments indicated that endosomes carrying both poliovirus and hPVR move retrogradely through the axon of the primary neurons (S. Ohka et al., manuscript in preparation). However, much research is required to elucidate the mechanisms underlying the endocytosis of poliovirus at synapses and the manner and place where poliovirus replication begins in the neuron cell body. The efficiency of the neural dissemination of poliovirus has been compared between the virulent Mahoney and the attenuated Sabin 1 strains. The data suggest that the difference in efficiency is not significant.26)
Neurovirulence determinant
The difference in efficiency in the 2 dissemination routes (BBB permeation and retrograde axonal transport) was not observed between the virulent and attenuated poliovirus strains. Thus, the efficiency cannot be a distinct determinant for the expression of strain-specific poliovirus neuro-virulence. Thus, strain-specific neurovirulence levels appear to depend primarily on the replication ability of the virus in the CNS.
Comparative sequence analysis between the attenuated Sabin 3 and its neurovirulent revertants revealed an effective mutation of neurovirulence phenotype at nucleotide position 472.34) Molecular genetic analysis employing reverse genetics of poliovirus type 1 revealed that a relatively strong determinant of neurovirulence resides in the 5′ non-coding sequence of the viral RNA, particularly at nucleotide position 48035),36) (Fig. 11). A neurovirulence determinant in the poliovirus type 2 genome has also been investigated, and nucleotide position 481 has been found to be an important nucleotide for this phenotype.
Fig. 11.
Genome structure of recombinant type 1 polioviruses. The expected genome structures of the recombinant viruses are shown as a combination of the Sabin 1 (open boxes) and Mahoney (closed boxes) sequences. The numbers over the genome RNAs are the nucleotide positions from the 5′ end of the genome. Nucleotide differences between the Mahoney and Sabin 1 strains are shown at the bottom of the figure. A small protein VPg is indicated by closed circles at the 5′ end of the genomes. Lesion scores obtained from monkey neurovirulence tests are shown on the right of the corresponding genome RNAs. (modified from refs. 35 and 36).
These nucleotide positions exist within the region corresponding to the IRES.11) These results suggest that the neurovirulence levels of individual poliovirus strains reflect their IRES activities; that is, the capability of translation initiation activity in the CNS. Although there are multiple neuro-virulence determinants on the poliovirus RNA genome, it is interesting that the IRES regions regulate the CNS specificity of the virus. The cumulative evidences offered by the data obtained thus far38)–40) led us to the notion of “IRES-dependent virus tropism.” In fact, a chimeric poliovirus in which poliovirus IRES is replaced by hepatitis C virus IRES lost the ability to propagate in the brain of transgenic mice but it can grow in the liver.41) Thus, the structure and function of the poliovirus IRES should be elucidated for a better understanding of poliovirus pathogenesis.
Anti-poliovirus response of neural cells
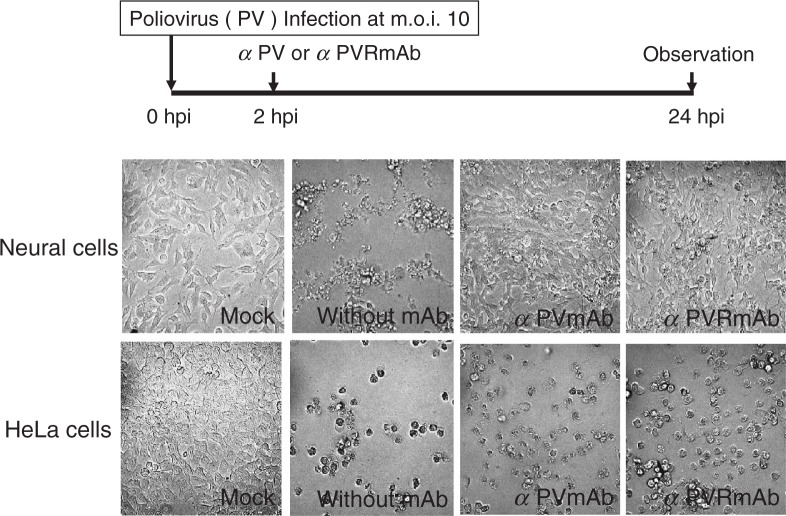
It has been reported that post-infection treatment with anti-poliovirus serum results in the survival of neural cells productively infected with virulent poliovirus.42) This phenomenon is specific for neural cells but has not been observed in other cells such as HeLa cells. Therefore, this observation might partially reflect in vivo pathogenesis of poliovirus specific to neural cells. This phenomenon has been reproduced using mAbs to poliovirus and hPVR instead of anti-poliovirus serum43) (Fig. 12). These results indicate that a second infection of neural cells with progeny poliovirus is required for the cells to express cytopathic effects. The reason why cytopathic effects in neural cells are blocked by prevention of the second infection is presently unknown. However, it is possible that the response of neural cells to the first poliovirus infection includes the expression of some cellular factors that inhibit poliovirus IRES activity. In fact, in the presence of mAbs to poliovirus or hPVR, poliovirus-specific protein synthesis was detected by 5 hours post infection (hpi) but not 7 hpi.43) In any event, this observation in infected neural cells in vitro is consistent with observations in an early study conducted by Bodian.44) Moreover, the reason why the second infection induces cytopathic effects in neural cells is totally unknown. Much research remains to be done to gain insights into the mechanisms of poliovirus neurovirulence.
Fig. 12.
Inhibition of the poliovirus-induced cytopathic effect in neural cells by mAb against poliovirus or hPVR. Neural cells (upper panels) or HeLa cells (lower panels) were infected with poliovirus type 1 Mahoney strain at an m.o.i. of 10. At 2 hpi, the cells were washed three times. Then, medium with or without mAb against poliovirus or hPVR was added to the culture. Poliovirus-infected or mock-infected cells were observed 24 hpi by microscope. (modified from ref. 43).
Acknowledgments
This review is a summary of my scientific achievements on molecular pathogenesis of polio-virus that have been carried out in Kitasato University, Tokyo Metropolitan Institute of Medical Science, and The University of Tokyo. I am grateful to so many collaborators on this work since 1977. I thank Etsuko Suzuki in preparing the manuscript and Seii Ohka in drawing Figures.
This work was supported in part by Grants-in-Aid for Specially Promoted Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Abbreviations:
- PVR
poliovirus receptor
- hPVR
human poliovirus receptor
- CNS
central nervous system
- IRES
internal ribosome entry site
- OPV
oral poliovaccine
- BBB
blood-brain barrier
- mAb
monoclonal antibody
- hpi
hours post infection
Profile
Akio Nomoto was born in 1946 and started his research career in 1969 with studies on the transfer RNA from baker’s yeast, in the Faculty of Pharmaceutical Science, the University of Tokyo, after his graduation from the same faculty. He first encountered poliovirus when he joined Prof. Eckard Wimmer’s laboratory, the State University of New York at Stony Brook, as a post-doctoral fellow in 1974. He worked on VPg, which is a small protein attached to the 5′ terminus of all the poliovirus RNAs except for its mRNA. He came back to Japan in 1977 and started as an associate professor at the School of Pharmaceutical Science, Kitasato University, and determined the total genome sequences of all three serotypes of poliovirus Sabin strains. After his moving to the Faculty of Medicine, the University of Tokyo as an associate professor in 1982, he identified the attenuating mutation on the RNA of the Sabin 1 vaccine strain of poliovirus using reverse genetics. This work opened a new avenue for elucidating the molecular mechanisms of the poliovirus pathogenesis. By using the knowledge obtained from above work, he constructed new candidates of type 2 and type 3 oral live vaccine strains by recombinant DNA technology. In 1987, he moved to Department of Microbiology, the Tokyo Metropolitan Institute of Medical Science as a Department Director, and another pioneering work was performed in this Institute, that is, discovery of human poliovirus receptor (hPVR) which determines species-specificity of poliovirus. He and his coworkers generated poliovirus-sensitive mice which were transgenic for hPVR. He was promoted to Professor at the University of Tokyo in 1991 and became Head of the Department of Microbiology at the Institute of Medical Science. Using the transgenic mice, he and his coworkers investigated poliovirus dissemination pathway in a whole body at molecular level, and proved that the transgenic mouse was a very useful animal model for studying molecular pathogenesis of poliovirus. He moved to the Department of Microbiology, Graduate School of Medicine, the University of Tokyo in 2000, where he continues to study poliovirus pathogenesis and is educating many students in the field of microbiology. He was awarded the Hideyo Noguchi Memorial Award for Medical Sciences in 1987, the Naito Foundation Research Prize in 1998, the Takeda Prize for Medical Science in 2002, and the Japan Academy Prize in 2004.

References
- 1.Mendelsohn, C.L., Wimmer, E. and Racaniello, V.R. (1989) Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell 56, 855–865 [DOI] [PubMed] [Google Scholar]
- 2.Koike, S., Horie, H., Ise, I., Okitsu, A., Yoshida, M., Iiizuka, N., Takeuchi, K., Takegami, T. and Nomoto, A. (1990) The poliovirus receptor protein is produced both as membrane-bound and secreted forms. EMBO J. 9, 3217–3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ren, R., Constantini, F., Gorgacz, E.J., Lee, J.J. and Racaniello, V.R. (1990) Transgenic mice expressing a human poliovirus receptor: a new model for poliomyelitis. Cell 63, 353–362 [DOI] [PubMed] [Google Scholar]
- 4.Koike, S., Taya, C., Kurata, T., Abe, S., Ise, I., Yonekawa, H. and Nomoto, A. (1991) Transgenic mice susceptible to poliovirus. Proc. Natl. Acad. Sci. USA 88, 951–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodian, D. (1955) Emerging concept of poliomyelitis infection. Science 122, 105–108 [DOI] [PubMed] [Google Scholar]
- 6.Sabin, A.B. (1956) Pathogenesis of poliomyelitis: reappraisal in the light of new data. Science 123, 1151–1157 [DOI] [PubMed] [Google Scholar]
- 7.Hogle, J.M., Chow, M. and Filman, D.J. (1985) Three-dimensional structure of poliovirus at 2.9 Å resolution. Science 229, 1358–1365 [DOI] [PubMed] [Google Scholar]
- 8.Belnap, D.M., McDermott, B.M.Jr., Filman, D.J., Cheng, N., Trus, B.L., Zuccola, H.J., Racaniello, V.R., Hogle, J.M. and Steven, A.C. (2000) Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc. Natl. Acad. Sci. USA 97, 73–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He, Y., Bowman, V.D., Mueller, S., Bator, C.M., Bella, J., Peng, X., Baker, T.S., Wimmer, E., Kuhn, R.J. and Rossmann, M.G. (2000) Interaction of the poliovirus receptor with poliovirus. Proc. Natl. Acad. Sci. USA 97, 79–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xing, L., Tjarnlund, K., Lindqvist, B., Kaplan, G.G., Feigelstock, D., Cheng, R.H. and Casasnovas, J.M. (2000) Distinct cellular receptor interactions in poliovirus and rhinoviruses. EMBO J. 19, 1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pelletier, J. and Sonenberg, N. (1988) Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334, 320–325 [DOI] [PubMed] [Google Scholar]
- 12.Sabin, A.B. and Boulger, L.R. (1973) History of Sabin attenuated poliovirus oral live vaccine strains. J. Biol. Stand. 1, 115–118 [Google Scholar]
- 13.Koike, S., Ise, I. and Nomoto, A. (1991) Functional domains of the poliovirus receptor. Proc. Natl. Acad. Sci. USA 88, 4104–4108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Selinca, H.-C., Zibert, A. and Wimmer, E. (1991) Poliovirus can enter and infect mammalian cells by way of an intercellular adhesion molecule 1 pathway. Proc. Natl. Acad. Sci. USA 88, 3598–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morrison, M.E., He, Y.-J., Wien, M.W., Hogle, J.M. and Racaniello, V.R. (1994) Homolog-scanning mutagenesis reveals poliovirus receptor residues important for virus binding and replication. J. Virol. 68, 2578–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aoki, J., Koike, S., Ise, I., Sato-Yoshida, Y. and Nomoto, A. (1994) Amino acid residues on human poliovirus receptor involved in interaction with poliovirus. J. Biol. Chem. 269, 8431–8438 [PubMed] [Google Scholar]
- 17.Bottino, C., Castriconi, R., Pende, D., Rivera, P., Nanni, M., Carnemolla, B., Cantoni, C., Grassi, J., Marcenaro, S., Reymond, N.et al. (2003) Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J. Exp. Med. 198, 557–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs, A., Cella, M., Giurisato, E., Shaw, A.S. and Colonna, M. (2004) Cutting edge: CD96 (tactile) promotes NK cell-target cell adhesion by interacting with the poliovirus receptor (CD155). J. Immunol. 172, 3994–3998 [DOI] [PubMed] [Google Scholar]
- 19.Arita, M., Koike, S., Aoki, J., Horie, H. and Nomoto, A. (1998) Interaction of poliovirus with its purified receptor and conformational alteration in the virion. J. Virol. 72, 3578–3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomez-Yafal, A., Kaplan, G., Racaniello, V.R. and Hogle, J.M. (1993) Characterization of poliovirus conformational alteration mediated by soluble cell receptors. Virology 197, 501–505 [DOI] [PubMed] [Google Scholar]
- 21.Gromeier, M. and Wetz, K. (1990) Kinetics of poliovirus uncoating in HeLa cells in a nonacidic environment. J. Virol. 64, 3590–3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren, R. and Racaniello, V.R. (1992) Human poliovirus receptor gene expression and poliovirus tissue tropism in transgenic mice. J. Virol. 66, 296–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abe, S., Ota, Y., Koike, S., Kurata, T., Horie, H., Nomura, T., Hashizume, S. and Nomoto, A. (1995) Neurovirulence test for oral live poliovaccines using poliovirus-sensitive transgenic mice. Virology 206, 1075–1083 [DOI] [PubMed] [Google Scholar]
- 24.Horie, H., Koike, S., Kurata, T., Sato-Yoshida, Y., Ise, I., Ota, Y., Abe, S., Hioki, K., Kato, H., Taya, C.et al. (1994) Transgenic mice carrying the human poliovirus receptor: new animal model for study of poliovirus neurovirulence. J. Virol. 68, 681–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang, W.-X., Terasaki, T., Shiroki, K., Ohka, S., Aoki, J., Tanabe, S., Nomura, T., Terada, E., Sugiyama, Y. and Nomoto, A. (1997) Efficient delivery of circulating poliovirus to the central nervous system independently of poliovirus receptor. Virology 229, 421–428 [DOI] [PubMed] [Google Scholar]
- 26.Ohka, S., Yang, W.-X., Terada, E., Iwasaki, K. and Nomoto, A. (1998) Retrograde transport of intact poliovirus through the axon via the fast transport system. Virology 250, 67–75 [DOI] [PubMed] [Google Scholar]
- 27.Ohka, S., Igarashi, H., Nagata, N., Sakai, M., Koike, S., Nochi, T., Kiyono, H. and Nomoto, A. (2007) Establishment of a poliovirus oral infection system in human poliovirus receptor-expressing transgenic mice that are deficient in alpha/beta interferon receptor. J. Virol. 81, 7902–7912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nathanson, N. and Langmuir, A.D. (1963) The Cutter incident: poliomyelitis following from aldehyde-inactivated poliovirus vaccination in the United States during the spring of 1955. III. Comparison of the clinical character of vaccinated and contact cases occurring after use of high rate lots of Cutter vaccine. Am. J. Hyg. 78, 61–81 [Google Scholar]
- 29.Ren, R. and Racaniello, V.R. (1992) Poliovirus spreads from muscle to the central nervous system by neural pathways. J. Infect. Dis. 166, 747–752 [DOI] [PubMed] [Google Scholar]
- 30.Gromeier, M. and Wimmer, E. (1998) Mechanism of injury-provoked poliomyelitis. J. Virol. 72, 5056–5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brady, S.T. (1991) Molecular motors in the nervous system. Neuron 7, 521–533 [DOI] [PubMed] [Google Scholar]
- 32.Ohka, S., Matsuda, N., Tohyama, K., Oda, T., Morikawa, M., Kuge, S. and Nomoto, A. (2004) Receptor (CD155)-dependent endocytosis of poliovirus and retrograde axonal transport of the endosome. J. Virol. 78, 7186–7198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mueller, S., Cao, X., Welker, R. and Wimmer, E. (2002) Interaction of poliovirus receptor CD155 with the dynein light chain Tctex-1 and its implication for poliovirus pathogenesis. J. Biol. Chem. 277, 7897–7904 [DOI] [PubMed] [Google Scholar]
- 34.Evans, D.M., Dunn, G., Minor, P.D., Schild, G.C., Cann, A.J., Stanway, G., Almond, J.W., Currey, K., Maizel, J.V.Jr. (1985) Increased neuro-virulence associated with a single nucleotide change in a noncoding region of the Sabin type 3 poliovaccine genome. Nature 314, 548–550 [DOI] [PubMed] [Google Scholar]
- 35.Omata, T., Kohara, M., Kuge, S., Komatsu, T., Abe, S., Semler, B.L., Kameda, A., Itoh, H., Arita, M., Wimmer, E. and Nomoto, A. (1986) Genetic analysis of the attenuation phenotype of poliovirus type 1. J. Virol. 58, 348–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawamura, N., Kohara, M., Abe, S., Komatsu, T., Tago, K., Arita, M. and Nomoto, A. (1989) Determinants in the 5′ noncoding region of poliovirus Sabin 1 RNA that influence the attenuation phenotype. J. Virol. 63, 1302–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macadam, A.J., Pollard, S.R., Ferguson, G., Dunn, G., Skuce, R., Almond, J.W. and Minor, P.D. (1991) The 5′ noncoding region of the type 2 poliovirus vaccine strain contains determinants of attenuation and temperature sensitivity. Virology 181, 451–458 [DOI] [PubMed] [Google Scholar]
- 38.Shiroki, K., Ishii, T., Aoki, T., Ota, Y., Yang, W.-X., Komatsu, T., Ami, Y., Arita, M., Abe, S., Hashizume, S. and Nomoto, A. (1997) Host range phenotype induced by mutations in the internal ribosomal entry site of poliovirus RNA. J. Virol. 71, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamoshita, N., Tsukiyama-Kohara, K., Kohara, M. and Nomoto, A. (1997) Genetic analysis of internal ribosomal entry site on hepatitis C virus RNA: implication for involvement of the highly ordered structure and cell type-specific transacting factors. Virology 233, 9–18 [DOI] [PubMed] [Google Scholar]
- 40.Isoyama, T., Kamoshita, N., Yasui, K., Iwai, A., Shiroki, K., Toyoda, H., Yamada, A., Takasaki, Y. and Nomoto, A. (1999) Lower concentration of La protein required for internal ribosome entry on hepatitis C virus RNA than on poliovirus RNA. J. Gen. Virol. 80, 2319–2327 [DOI] [PubMed] [Google Scholar]
- 41.Yanagiya, A., Ohka, S., Hashida, N., Okamaura, M., Taya, C., Kamoshita, N., Iwasaki, K., Sasaki, Y., Yonekawa, H. and Nomoto, A. (2003) Tissue specific replicating capacity of a chimeric poliovirus that carries the internal ribosome entry site of hepatitis C virus in a new mouse model transgenic for human poliovirus receptor. J. Virol. 77, 10479–10487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tolskaya, E.A., Ivannikova, T.A., Kolesnikova, M.S., Drozdov, S.G. and Agol, V.I. (1992) Postinfection treatment with antiviral serum results in survival of neural cells productively infected with virulent poliovirus. J. Virol. 66, 5152–5156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yanagiya, A., Jia, Q., Ohka, S., Horie, H. and Nomoto, A. (2005) Blockade of the poliovirus-induced cytopathic effect in neural cells by monoclonal antibody against poliovirus or the human poliovirus receptor. J. Virol. 79, 1523–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bodian, D. (1949) Histopathologic basis of clinical findings in poliomyelitis. Am. J. Med. 6, 563–578 [DOI] [PubMed] [Google Scholar]