Abstract
Whole genome duplication (WGD) is a major factor in the evolution of multicellular eukaryotes, yet by doubling the number of homologs, WGD severely challenges reliable chromosome segregation [1, 2, 3], a process conserved across kingdoms [4]. Despite this, numerous genome-duplicated (polyploid) species persist in nature, indicating early problems can be overcome [1, 2]. Little is known about which genes are involved – only one has been molecularly characterized [5]. To gain new insights into the molecular basis of adaptation to polyploidy, we investigated genome-wide patterns of differentiation between natural diploids and tetraploids of Arabidopsis arenosa, an outcrossing relative of A. thaliana [6, 7]. We first show that diploids are not preadapted to polyploid meiosis. We then use a genome scanning approach to show that while polymorphism is extensively shared across ploidy levels, there is strong ploidy-specific differentiation in 39 regions spanning 44 genes. These are discrete, mostly single-gene peaks of sharply elevated differentiation. Among these peaks are eight meiosis genes whose encoded proteins coordinate a specific subset of early meiotic functions, suggesting these genes comprise a polygenic solution to WGD-associated chromosome segregation challenges. Our findings indicate that even conserved meiotic processes can be capable of nimble evolutionary shifts when required.
Meiotic chromosome behavior in tetraploid A. arenosa
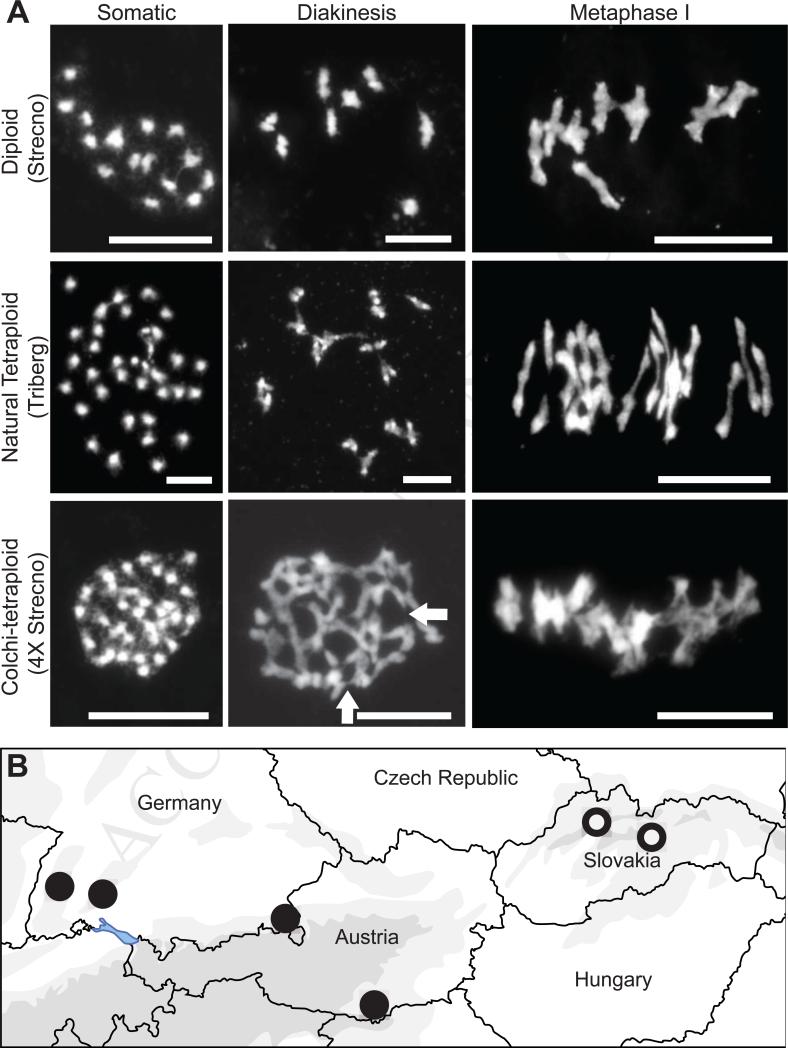
At least initially, WGD is commonly associated with deleterious chromosome mis-segregation arising from multivalent associations among available homologs [e.g. 1-3, 8-11]. This is especially challenging for autopolyploids, which arise from within-species duplication and have multiple approximately equally homologous chromosomes. We asked if for autotetraploid A. arenosa (1) the tetraploid material we are working with has diploid-like chromosome behavior, and (2) the diploid genome we are comparing to is not preadapted for polyploid meiosis, as has been seen in some species [e.g. 12]. Though bivalent formation among homologs appears to be random and inheritance tetrasomic in natural autotetraploid A. arenosa [13], metaphase I chromosomes associate predominantly as bivalents like in diploids ([14] and Fig. 1A). There are structural differences, however: tetraploids have significantly more rod bivalents (and fewer ring bivalents) than diploids, which indicates natural tetraploid A. arenosa averages fewer chiasmata per bivalent than diploid A. arenosa (Table S1). A reduction in chiasma number to one per bivalent has previously been suggested as a possible mechanism for meiotic diploidization in autopolyploids because limiting crossovers to one per chromosome prevents multivalent associations [e.g. 10, 11].
Figure 1. Chromosome spreads and map locations.
(A) DAPI-stained meiotic chromosome spreads. Left column shows chromosome counts, middle column, diakinesis, and right column, metaphase I. Top row shows diploid A. arenosa. Somatic chromosome counts are as expected (2N=16) and associations are bivalents. Second row shows a natural tetraploid. Chromosome count (2N=32) in somatic cells (left). Middle and right panels show bivalent associations. Bottom row shows neo-tetraploid A. arenosa. Somatic chromosome counts (left) confirmed tetraploidy (2N=32). Extensive multivalent formation and fine ectopic inter-chromosomal connections (examples indicated by arrows) are evident at diakinesis and metaphase I. (B) Map of populations. Tetraploid populations are indicated with closed circles and diploids with open circles.
We induced WGD in two diploid A. arenosa genotypes using colchicine and examined chromosome behavior of confirmed neotetraploids in diakinesis and metaphase I, when multivalents are readily discernable. Unlike natural autotetraploids, synthetic neotetraploids exhibit extensive multivalent formation and ectopic connections between the chromosomes (Fig. 1A; Table S1). The cytological abnormalities in the neotetraploid lines correlate with sharply reduced pollen viability: The two colchicine-doubled lines had only 3% and 5% pollen viability, in contrast to two natural autotetraploid lines that had 91% and 92% pollen viability. Thus diploid A. arenosa provides an “unevolved” comparison for the natural tetraploid. Bivalent associations and reduced estimated chiasma frequency in natural autotetraploids, and the aberrant meiosis of neotetraploids, are consistent with data from many other autopolyploids [e.g. 8-11], suggesting A. arenosa is a representative model for studying the molecular basis of adaptation to autopolyploid meiosis.
Evidence of polygenic selection in autotetraploid A. arenosa
Because of its connection to fertility [1, 2], selection for meiotic stability immediately following WGD should be intense. Thus we reasoned that alleles contributing to stable chromosome segregation in the autopolyploid should show reduced allelic diversity and excess differentiation between autotetraploids and diploids. High genetic diversity suggests A. arenosa autotetraploids did not undergo a severe recent bottleneck associated with WGD [7, 13] and/or have ongoing gene flow with diploids [15]. We have previously shown evidence that autotetraploid A. arenosa has undergone selective sweeps [13], but since diploids were not included, it remained unknown whether top outliers reflect adaptation to polyploidy, or species-wide patterns shared with diploids.
We used a genome scanning approach to compare the genomes of diploid and tetraploid A. arenosa. We short-read sequenced whole genomes from 16 natural autotetraploid and 8 diploid individuals from six natural populations (Fig. 1B; Table S2). We aligned reads to the closely related A. lyrata genome [16]. Over 46 million sites had coverage in all 24 individuals, of which about 5.6 million are polymorphic relative to the A. lyrata reference (Table 1). There is extensive shared variation between diploids and autotetraploids (>1.7 million sites), and remarkably few fixed differences (26 genome-wide; Table 1).
Table 1.
Genetic differentiation between diploid and tetraploid A. arenosa
| Description | Number |
|---|---|
| Total sites with coverage in all 24 individuals | 46,254,812 |
| Total polymorphic relative to A. lyrata reference | 5,577,375 |
| Fixed polymorphisms relative to A. lyrata reference | 120,576 |
| Shared polymorphisms between diploid and tetraploid A. arenosa | 1,701,318 |
| Private polymorphism among 8 diploid A. arenosa | 533,850 |
| Private polymorphism among 16 tetraploid A. arenosa | 3,221,605 |
| Fixed differences between diploids and tetraploids | 26 |
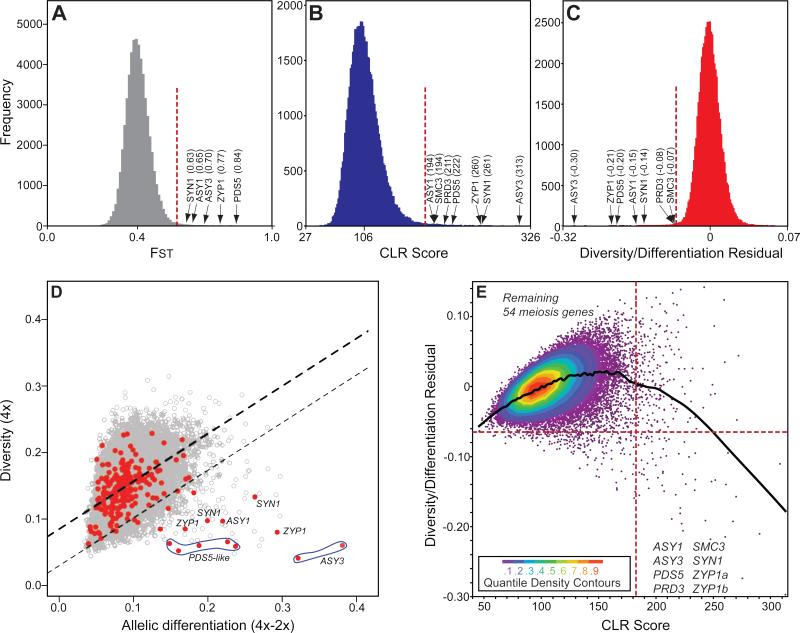
We scanned for signatures suggestive of selective sweeps by analyzing consecutive windows of 100 polymorphic sites (55,769 windows total) for 0.5% outliers in the distributions of three metrics: FST [17] (Fig. 2A), the two-dimensional site frequency spectrum (2dSFS) [18] (Fig. 2B), and the 0.5% most negative values of linear regression residuals from the relationship between diversity and differentiation. Outlier values for this “residuals” metric indicate excess differentiation for a given level of diversity (Fig. 2C, D). All 0.5% outlier windows for all three tests are given in Table S3. We generated an overlap list of windows found both among 0.5% outliers for 2dSFS and the residuals (Table S4). Though both the residuals and FST quantify genetic differentiation, we favored the former since it accounts for the positive relationship between differentiation and diversity (e.g. see Fig 2D).
Figure 2. Diversity and differentiation of meiosis genes relative to genome-wide patterns.
Genome-wide values for 100 SNP windows for FST (A), CLR Score (B), and Diversity/Differentiation residuals (C). X-axes are linear, indicate means, and outlier meiosis genes are labeled. (D) Nucleotide diversity of 100 SNP windows in tetraploids plotted against differentiation between ploidies. Heavy line shows linear regression and lighter line, 1% cutoff. Red dots represent 100 SNP windows in meiosis genes with extreme outliers labeled. Note: each gene can have multiple hits as it can have multiple 100 SNP windows. (E) CLR Score vs Diversity/Differentiation Residual for all windows. Dotted lines indicate 0.5% cutoffs. Meiosis genes are indicated in respective quadrants.
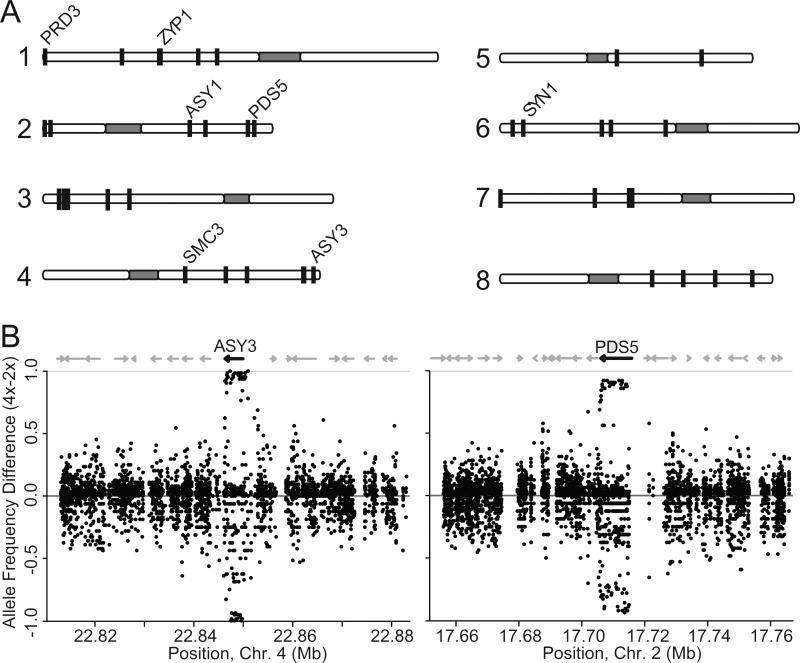
The overlap list contains 39 distinct differentiated regions spanning 44 genes; most contain only a single gene (Fig. 3A,B; Table S4), with rapid decay to background (e.g. Fig. 3B). Using paired end information and de novo assemblies aligned to A. lyrata, we verified gene order in these regions (see supplemental methods). This analysis showed that neighboring loci in these regions are syntenous between A. arenosa and A. lyrata, confirming that the rapid decay of differentiation reflects low linkage disequilibrium, not an alignment artifact. Low levels of linkage disequilibrium are likely due to the large effective population size of A. arenosa [6]. Six of the 44 genes overlap with our previous scan, even though the analyses used different methods and sample sets [11].
Figure 3. Most differentiated regions and examples of differentiation in two sweep candidates.
(A) Differentiated regions (vertical lines), with meiosis genes labeled. ZYP1 consists of tandem duplicates, ZYP1a and ZYP1b. (B) Two example differentiated regions in meiosis genes. Dots represent polymorphic SNPs. X-axis gives chromosome location. Y-axis shows degree of differentiation calculated by subtracting diploid from tetraploid allele frequency. Short gaps are regions in which reads did not align due to repeat masking, high intergenic polymorphism, or deletions in A. arenosa relative to A. lyrata. These were verified with alignment of an A. arenosa de novo assembly and paired end read information.
Meiosis genes are over-represented among genome scan outliers
Eight meiosis-related genes were on our overlap list of 39 regions and 44 genes (Fig. 2,3; Table S4). In GO category analysis, meiosis was the only significantly overrepresented functional category. However, there is some ambiguity in the GO category designation for meiosis genes (the GO designation contains 219 genes, many of which have no known role in meiosis), thus we generated a new list by searching A. thaliana gene descriptions (TAIR10; www.arabidopsis.org) to identify 71 (out of ~25,550) genes that are clearly annotated as having a role in meiosis. Of these 71 genes, 62 have good read alignment in A. arenosa (Table S5). A random list of 44 genes would not be expected to contain any meiosis genes on average (the probability is ~0.1).
We next asked whether meiosis genes as an overall class have consistently high differentiation, which could indicate they are under selection as a group even if most do not meet stringent 0.5% cutoffs. We compared differentiation of 100 SNP windows mapping within meiosis genes (Table S5) to windows in the rest of the genome using the residuals metric. Aside from the eight outliers, the distribution of values for windows falling in the remaining 54 well-aligned meiosis genes were not distinguishable from the genome-wide distribution (Fig. 2D,E; t-test p = 0.60). Thus, meiosis-related genes show no gene set enrichment for differentiation apart from the eight outliers. This result suggests the eight meiosis-associated genes with strong differentiation in A. arenosa represent a polygenic, naturally evolved solution to WGD-associated challenges. Among these eight, three were represented in a previous scan (ASY1, SMC3, PDS5)[13], while three othersdid not align in our previous study and were thus not included (ZYP1a, ZYP1b, ASY3).
Functional implications of identified meiosis genes
In all eight meiosis genes, sites with excess derived allele frequency encode predicted amino acid substitutions, though these are more common in tetraploids (Table S6). ZYP1a harbors 16 high frequency derived (relative to the A. lyrata reference) substitutions in the tetraploid that encode predicted coding changes, but none in the diploid. ASY3, however, shows highly divergent polymorphism in both ploidies.
The eight meiosis genes in our outlier set are not a random sample: Selection appears to have acted on multiple unlinked loci to shift the allelic landscape of coordinated events in early prophase I. All eight genes encode proteins crucial for the organization of chromosome structure, alignment, and synapsis of homologous chromosomes, and the controlled formation of crossovers [19-21]. First, PRD3 participates in the early initiation of homologous recombination [22, 23]. Coordination of subsequent events in recombination is dependent on the interplay between the recombination machinery and the chromosome axes. In yeast, this involves Red1, Hop1 and Rec8 [24] whose functional homologs in A. thaliana are ASY3, ASY1 and SYN1 [20, 25-29]. Their roles appear to be largely conserved [20, 25-29], and all are differentiated between A. arenosa ploidies. At zygotene aligned homologous chromosomes are brought into close apposition by the formation of the synaptonemal complex (SC) [30] cross-linked by a transverse filament protein, Zip1 [31], which also affects crossover fate [32]. In A. thaliana the SC transverse filament is encoded by tandem duplicates, ZYP1a and ZYP1b [33], both of which lie under a strong peak of ploidy-differentiation in A. arenosa. Two other differentiated genes are SMC3 and scaffold_202722.1 (At1g77600 in A. thaliana). The function of At1g77600 is unknown, but the encoded protein has high homology to PDS5/SPO76, which is required in fungi and animals for sister chromatid cohesion and regulation of SC formation in cooperation with cohesins, including SMC3 [34].
All of the meiosis proteins we identified are involved in coordinated processes that contribute to chromosome juxtaposition and chiasma formation. ASY1 and ASY3 proteins directly interact and their localization to the chromosome axis requires the presence of SYN1 [25]. SYN1 in turn has been shown by mass-spectrometry to co-precipitate with ASY1, ASY3, and ZYP1 (K. Osman and F. C. H. Franklin, unpublished data). The finding of differentiation interacting proteins suggests that adaptation to WGD-associated meiotic chromosome segregation challenges might have been multigenic; whether this reflects co-evolution or additive contributions to phenotype remains to be tested.
Conclusions
Understanding the genetic basis of naturally-evolved solutions to chromosome segregation with extra homologous copies is relevant to a range of WGD contexts, including crop improvement, polyploid human cancers, and our basic understanding of an evolutionarily important phenomenon. The genes that are sharply differentiated between diploid and tetraploid A. arenosa encode proteins that affect the initial juxtaposition and alignment of homologous chromosomes, formation of the SC and the controlled maturation of recombination intermediates into crossovers or non-crossovers [19-34]. Altering these processes can ultimately affect the number and distribution of crossover events [e.g. 19, 24-29, 32, 33]. Some cytological studies have found evidence that established polyploids can have reduced crossover frequencies relative to neotetraploids or diploid relatives, and this has been hypothesized as a mechanism of suppressing multivalent formation and thereby stabilizing polyploid meiosis [e.g. 10, 11]. Our cytological results are consistent with this, and our genome scan results provide a candidate set of genes that could mediate this outcome. It merits mention that an alternative possibility is that some of these alleles may promote unreduced gamete formation in diploids and thus directly contribute to polyploid formation.
There is evidence of parallels with other systems. For example, we observed strong differentiation in ASY1, whose homolog has been implicated in meiotic stability in allopolyploid wheat. The wheat gene Ph1, the only “diploidization gene” molecularly characterized to date [5, 35], promotes bivalent formation by solidifying similarity-based pairing fidelity. In the absence of Ph1, transcription of the wheat homolog of ASY1 is increased and its localization is affected, while decreased ASY1 activity in transgenic lines caused homeologous pairing [36]. Though the genes themselves are not homologs, there are functional similarities among the genes we identified and those critical to tetraploid, but not diploid yeast cells, which include genes involved in homologous recombination and sister chromatid cohesion [37]. Finally, in humans cancer cells are often polyploid [3]. Though they divide mitotically, a suite of meiosis genes, including a vertebrate homolog of ASY1 (HORMAD1), as well as homologs of ZYP1 and SYN1/REC8, are over-expressed in at least some cancers, where they may contribute to genomic instability and show promise as therapeutic targets [e.g. 3, 38, 39]. With ours, these studies indicate parallels between kingdoms in processes that affect chromosome segregation after WGD, while our work shows that this conserved process can make evolutionary shifts when necessary.
Data
All genomic sequencing reads are available from the NCBI SRA database under bioproject number SRP021057.
Experimental Procedures
Plant material
Plant growth and DNA preparation were previously described [13]. To generate neotetraploids, diploid SN seeds were treated with 0.1 % colchicine for 24 hours and confirmed tetraploidy with chromosome spreads. We assayed pollen viability (n=90-120 grains / line) using Alexander's stain [40].
Cytological procedures
We fixed inflorescences in 3:1 ethanol: acetic acid. Anthers were isolated and prepared as previously described [41]. Chromosomes were stained with 4, 6 diamidino-2-phenylindole (DAPI), mounted in Vectashield (Vector Lab. Burlingame, CA, USA) and visualized using a Nikon 90i Eclipse fluorescent microscope with NIS elements software.
Genome sequencing
Sequencing libraries were prepared using the Illumina Genomic Sample Preparation Kit and sequenced on an Illumina HiSeq2000. Reads were mapped to the repeatmasked Lyrata1.0 genome [16] using bowtie2 [42] and bam files were processed with Samtools [43] and Picard (http://picard.sourceforge.net/). We used GATK [44, 45] for indel realignment, duplicate removal, SNP discovery and genotyping using standard parameters for diploids and the ‘–ploidy 4’ option for tetraploids. See supplemental methods for diploid de novo assembly.
Genomic analysis
For details see supplemental methods. Sites with coverage in all 24 individuals were binned into 55,570 100-SNP sliding windows. We calculated FST between diploids and tetraploids following [17, 46]. We also used a composite likelihood ratio test of the diploid-tetraploid two-dimensional Site Frequency Spectrum (2dSFS) [18], and tested for regions with excess allelic differentiation between diploids and tetraploids, for a given diversity within tetraploids. Our final set of differentiated regions was defined as the overlap between these latter two tests
Supplementary Material
Highlights.
Natural autotetraploids accurately segregate chromosomes contrary to neotetraploids
Autotetraploids have fewer chiasmata per bivalent than diploids
39 genomic regions are sharply differentiated between diploids and tetraploids
Meiosis genes are over-represented among putative autopolyploidy adaptation loci
Acknowledgements
We thank Nancy Kleckner and Dan Hartl for critical reading of the manuscript and thoughtful discussions, Ben Hunter for technical assistance, and members of the Bomblies and Kleckner labs for helpful discussions. This work was supported through a Harvard University Milton Fund Award to KB, Ruth L. Kirschstein National Research Service Awards from the National Institutes of Health to LY (1 F32 GM096699) and KMW (1 F32 GM105293), and work in the FCHF lab is funded by the BBSRC, UK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Otto SP. The evolutionary consequences of polyploidy. Cell. 2007;131:452–462. doi: 10.1016/j.cell.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 2.Comai L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005;6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- 3.Storchova Z, Pellman D. From polyploidy to aneuploidy, genome instability and cancer. Nat. Rev. Mol. Cell Biol. 2004;5:45–54. doi: 10.1038/nrm1276. [DOI] [PubMed] [Google Scholar]
- 4.Gerton J, Hawley R. Homologous chromosome interactions in meiosis: diversity amidst conservation. Nat. Rev. Genet. 2005;6:477–487. doi: 10.1038/nrg1614. [DOI] [PubMed] [Google Scholar]
- 5.Griffiths S, Sharp R, Foote TN, Bertin I, Wanous M, Reader S, Colas I, Moore G. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature. 2006;439:749–752. doi: 10.1038/nature04434. [DOI] [PubMed] [Google Scholar]
- 6.Koch MA, Matschinger M. Evolution and genetic differentiation among relatives of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 2007;104:6272–6277. doi: 10.1073/pnas.0701338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmickl R, Paule J, Klein J, Marhold K, Koch MA. The evolutionary history of the Arabidopsis arenosa complex: Diverse tetraploids mask the Western Carpathian center of species and genetic diversity. PLoS One. 2012;7:e42691. doi: 10.1371/journal.pone.0042691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hazarika MH, Rees H. Genotypic control of chromosome behaviour in rye. X. Chromosome pairing and fertility in autotetraploids. Heredity. 1967;22:317–332. [Google Scholar]
- 9.Santos JL, Alfaro D, Sanchez-Moran E, Armstrong SJ, Franklin FCH, Jones GH. Partial diploidization of meiosis in autotetraploid Arabidopsis thaliana. Genetics. 2003;165:1533–1540. doi: 10.1093/genetics/165.3.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cifuentes M, Grandont L, Moore G, Chevre AM, Jenczewski E. Genetic regulation of meiosis in polyploid species: new insights into an old question. New Phytol. 2010;186:29–36. doi: 10.1111/j.1469-8137.2009.03084.x. [DOI] [PubMed] [Google Scholar]
- 11.Le Comber SC, Ainouche ML, Kovarik A, Leitch AR. Making a functional diploid: from polysomic to disomic inheritance. New Phytol. 2010;186:113–122. doi: 10.1111/j.1469-8137.2009.03117.x. [DOI] [PubMed] [Google Scholar]
- 12.Lavania UC. High bivalent frequencies in artificial autopolyploids of Hyoscyamus muticus L. Can. J. Genet. Cytol. 1985;28:7–11. [Google Scholar]
- 13.Hollister JD, Arnold BJ, Svedin E, Xue KS, Cilkes BP, Bomblies K. Genetic Adaptation Associated with Genome-Doubling in Autotetraploid Arabidopsis arenosa. PLoS Genetics. 2012;8:e1003093. doi: 10.1371/journal.pgen.1003093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carvalho A, Delgado M, Barao A, Frescatada M, Ribeiro E, Pikaard CS, Viegas W, Neves N. Chromosome and DNA methylation dynamics during meiosis in the autotetraploid Arabidopsis arenosa. Sex. Plant Reprod. 2010;23:29–37. doi: 10.1007/s00497-009-0115-2. [DOI] [PubMed] [Google Scholar]
- 15.Jørgensen MH, Ehrich D, Schmickl R, Koch MA, Brysting AK. Interspecific and interploidal gene flow in central European Arabidopsis (Brassicaceae). BMC Evol. Biol. 2011;11:346. doi: 10.1186/1471-2148-11-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nature Genet. 2011;43:476–481. doi: 10.1038/ng.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weir BS. Genetic Data Analysis II. Sinauer Associates; Sunderland, MA: 1996. [Google Scholar]
- 18.Nielsen R, Hubisz MJ, Hellmann I, Torgerson D, Andrés AM, Albrechtsen A, Gutenkunst R, Adams MD, Cargill M, Boyko A, et al. Darwinian and demographic forces affecting human protein coding genes. Genome Res. 2009;19:838–849. doi: 10.1101/gr.088336.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleckner N. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma. 2006;115:175–194. doi: 10.1007/s00412-006-0055-7. [DOI] [PubMed] [Google Scholar]
- 20.Osman K, Higgins JD, Sanchez-Moran E, Armstrong SJ, Franklin FCH. Pathways to meiotic recombination in Arabidopsis thaliana. New Phytologist. 2011;190:523–544. doi: 10.1111/j.1469-8137.2011.03665.x. [DOI] [PubMed] [Google Scholar]
- 21.Blat Y, Protacio RU, Hunter N, Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002;111:791–802. doi: 10.1016/s0092-8674(02)01167-4. [DOI] [PubMed] [Google Scholar]
- 22.De Muyt A, Pereira L, Chelysheva L, Gendrot G, Chambon A, Lainé-Choinard S, Pelletier G, Mercier R, Nogué F, Grelon M. A high throughput genetic screen identifies new early meiotic recombination functions in Arabidopsis thaliana. PLoS Genet. 2009;5:e1000654. doi: 10.1371/journal.pgen.1000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atcheson CL, DiDomenico B, Frackman S, Esposito RE, Elder RT. Isolation, DNA sequence, and regulation of a meiosis-specific eukaryotic recombination gene. Proc. Natl. Acad. Sci. U.S.A. 1987;84:8035–8039. doi: 10.1073/pnas.84.22.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim KP, Weiner BM, Zhang LR, Jordan A, Dekker J, Kleckner N. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell. 2010;143:924–937. doi: 10.1016/j.cell.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferdous M, Higgins JD, Osman K, Lambing C, Roitinger E, Mechtler K, Armstrong SJ, Perry R, Pradillo M, Cunado N, Franklin FCH. Inter-homolog crossing-over and synapsis in Arabidopsis meiosis are dependent on the chromosome axis protein AtASY3. PLoS Genet. 2012;8:e1002507. doi: 10.1371/journal.pgen.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Armstrong SJ, Caryl AP, Jones GH, Franklin FCH. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 2002;115:3645–3655. doi: 10.1242/jcs.00048. [DOI] [PubMed] [Google Scholar]
- 27.Bhatt AM, Lister C, Page T, Fransz P, Findlay K, Jones GH, Dickinson HG, Dean C. The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J. 1999;19:463–472. doi: 10.1046/j.1365-313x.1999.00548.x. [DOI] [PubMed] [Google Scholar]
- 28.Bai X, Peirson BN, Dong F, Xue C, Makaroff CA. Isolation and characterization of SYN1, a RAD21-like gene essential for meiosis in Arabidopsis. Plant Cell. 1999;11:417–430. doi: 10.1105/tpc.11.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamant O, Ma H, Cande WZ. Genetics of Meiotic Prophase I in Plants. Annu. Rev. Plant Biol. 2006;57:267–302. doi: 10.1146/annurev.arplant.57.032905.105255. [DOI] [PubMed] [Google Scholar]
- 30.Page SL, Hawley RS. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 2004;20:525–558. doi: 10.1146/annurev.cellbio.19.111301.155141. [DOI] [PubMed] [Google Scholar]
- 31.Sym M, Engebrecht J, Roeder GS. Zip1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- 32.Boerner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- 33.Higgins JD, Sanchez-Moran E, Armstrong SJ, Jones GH, Franklin FCH. The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Devel. 2005;19:2488–2500. doi: 10.1101/gad.354705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panizza S, Tanaka T, Hochwagen A, Eisenhaber F, Nasmyth K. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr. Biol. 2000;10:1557–1564. doi: 10.1016/s0960-9822(00)00854-x. [DOI] [PubMed] [Google Scholar]
- 35.Al-Kaff N, Knight E, Bertin I, Foote T, Hart N, Griffiths S, Moore G. Detailed dissection of the chromosomal region containing the Ph1 locus in wheat Triticum aestivum: with deletion mutants and expression profiling. Ann. Bot. 2008;105:863–872. doi: 10.1093/aob/mcm252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boden SA, Langridge P, Spangenberg G, Able JA. TaASY1 promotes homologous chromosome interactions and is affected by deletion of Ph1. Plant J. 2009;57:487–497. doi: 10.1111/j.1365-313X.2008.03701.x. [DOI] [PubMed] [Google Scholar]
- 37.Storchova Z, Breneman A, Cande J, Dunn J, Burbank K, O'Toole E, Pellman D. Genome-wide genetic analysis of polyploidy in yeast. Nature. 2006;443:541–547. doi: 10.1038/nature05178. [DOI] [PubMed] [Google Scholar]
- 38.Shahzad MM, Shin YH, Matsuo K, Lu C, Nishimura M, Shen DY, Kang Y, Hu W, Mora EM, Rodriguez-Aguayo C, et al. Biological significance of HORMA domain containing protein 1 (HORMAD1) in epithelial ovarian carcinoma. Cancer Lett. 2013;330:123–129. doi: 10.1016/j.canlet.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalejs M, Ivanov A, Plakhins G, Cragg MS, Emzinsh D, Illidge TM, Erenpreisa J. Upregulation of meiosis-specific genes in lymphoma cell lines following genotoxic insult and induction of mitotic catastrophe. BMC Cancer. 2006;6:6. doi: 10.1186/1471-2407-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alexander MP. Differential staining of aborted and nonaborted pollen. Biotechnic Histochem. 1969;44:117–122. doi: 10.3109/10520296909063335. [DOI] [PubMed] [Google Scholar]
- 41.Armstrong S. Spreading and fluorescence in situ hybridization of male and female meiocyte chromosomes from Arabidopsis thaliana for cytogenetical analysis. Methods Mol. Biol. 2013;990:3–11. doi: 10.1007/978-1-62703-333-6_1. [DOI] [PubMed] [Google Scholar]
- 42.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Meth. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DePristo M, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics. 2013;43:491–497. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altschuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ross-Ibarra J, Wright SI, Foxe JP, Kawabe A, DeRose-Wilson L, Gos G, Charlesworth D, Gaut BS. Patterns of polymorphism and demographic history in natural populations of Arabidopsis lyrata. PLoS ONE. 2008;3(6):e2411. doi: 10.1371/journal.pone.0002411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.