Abstract
To understand the epigenetic regulation required for germ cell-specific gene expression in the mouse, we analysed DNA methylation profiles of developing germ cells using a microarray-based assay adapted for a small number of cells. The analysis revealed differentially methylated sites between cell types tested. Here, we focused on a group of genomic sequences hypomethylated specifically in germline cells as candidate regions involved in the epigenetic regulation of germline gene expression. These hypomethylated sequences tend to be clustered, forming large (10 kb to ∼9 Mb) genomic domains, particularly on the X chromosome of male germ cells. Most of these regions, designated here as large hypomethylated domains (LoDs), correspond to segmentally duplicated regions that contain gene families showing germ cell- or testis-specific expression, including cancer testis antigen genes. We found an inverse correlation between DNA methylation level and expression of genes in these domains. Most LoDs appear to be enriched with H3 lysine 9 dimethylation, usually regarded as a repressive histone modification, although some LoD genes can be expressed in male germ cells. It thus appears that such a unique epigenomic state associated with the LoDs may constitute a basis for the specific expression of genes contained in these genomic domains.
Keywords: DNA methylation, primordial germ cell, epigenome, reprogramming, cancer testis antigen
1. Introduction
Among the many cells that constitute an organism, only the germ cell can transmit its genetic information to the next generation. To achieve this vital function, germ cells must possess a distinctive gene transcription programme and epigenomic features unique to this cell lineage. It is accepted that, during development, germ cells undergo marked changes in their global epigenetic status, termed ‘epigenetic reprogramming’.1,2 However, details of the epigenetic changes and their relationship to the establishment of germ cell-specific gene expression have been poorly characterized, possibly for technical reasons. The number of developing primordial germ cells (PGCs) in embryos is limited,3 restricting the conventional analysis of epigenetic status. Most of the genome-scale studies on DNA methylation have focused on CpG islands (CGIs) or gene promoters (e.g. Borgel et al.4). Currently, the importance of DNA methylation of other parts of genes, non-genic or intergenic regions for establishing gene expression and the nuclear organization of chromatin is increasingly recognized.5–7 Although recent research has advanced our understanding of the PGC epigenome,8–11 further studies are still required to gain more detailed information on epigenomic features of germline cells and their involvement in defining germ cell-specific gene expression.
In this study, we used a proven method of DNA methylation analysis called the HELP assay (HpaII tiny fragment enrichment by ligation-mediated polymerase chain reaction (PCR)).12–14 Because this method uses linker-mediated PCR, it can be adapted for the small-scale analysis of developing germ cells. Oda et al.14 developed an improved version of the method, nanoHELP. Here, we have fine-tuned the protocol further. This modified nanoHELP method provides a global analysis of the DNA methylation status of CCGG sites using only a subnanogram (≥0.5 ng) quantity of genomic DNA. The custom-made genomic microarray used in this study is unusual in that the CCGG sites of the intergenic regions as well as the promoters and gene bodies of the RefSeq genes could be tested. This HELP microarray may provide new information about previously unexplored parts of the germ cell epigenome. We applied this method to analyse DNA methylation in the mouse X chromosome. We reasoned that epigenomic features specific to germ cells can be found by focusing on the X chromosome, because the X chromosome carries many germ cell-expressed genes15,16 and undergoes major epigenetic changes (e.g. X chromosome reactivation) during germ cell development.17 In this analysis, we found for the first time a group of sequences that are specifically hypomethylated on the X chromosome of male germline cells. These sequences form relatively large genomic domains that harbour gene families displaying specific expression in germ cells. We term these regions as large hypomethylated domains (LoDs). LoDs have not been detected in previous studies, including recent whole-genome bisulphite sequence analyses,10 probably because mapping of bisulphite-converted short sequence reads onto locally duplicated regions such as LoDs is technically challenging. In contrast, the experimental design of the HELP assay, which involved removal of the potentially confounding effect of copy number difference12 and inclusion of a probe design that selects unique sequences for hybridization, was effective in finding LoDs. Interestingly, many genes with homology to human cancer testis antigen (CTA) genes are contained within the LoDs. CTA genes are normally expressed only in the germline, and are also expressed in some tumour cell types.18 The results presented in this study may shed light on the epigenetic basis for the germline gene expression programme and its relationship with oncogenesis.
2. Materials and methods
2.1. Sample preparation and purification of DNA and RNA
TMA5 cells are male embryonic stem (ES) cells derived from the 129/Sv mouse.19 The female ES#5 line was from F1 hybrid mice between TgN(deGFP)20Imeg (RBRC No. 00822) and MSM/Ms (RBRC No. 00209). The embryonic germ (EG) cell lines used in this study were TMA55G (male) and TMA58G (female).19 These ES and EG cell lines were cultured, as described previously.20 The Oct3/4–GFP transgenic mouse line TgN(deGFP)18Imeg (RBRC No. 00821)21 was used to collect PGCs from developing mouse embryos, as described previously.20 Germline stem (GS) cells were obtained from the RIKEN Cell Bank (RCB1968) and were cultured on a feeder layer as described.22 Germ cells expressing the Venus reporter were purified by fluorescence-activated cell sorting (FACS) from adult testis cells of the Mvh-Venus bacterial artificial chromosome transgenic mouse line, Tg(Mvh-Venus)1Rbrc (Mise and Abe, unpublished results).23 All animal experiments were approved by the Institutional Animal Experiment Committee of the RIKEN BioResource Center. DNA and RNA were extracted simultaneously from the same samples using an AllPrep DNA/RNA Micro Kit (Qiagen, Hilden, Germany). The quality of RNA samples was checked using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
2.2. Gene expression profiling
A 44K custom microarray24 was used for gene expression profiling throughout this study. This custom array covers all the known protein-coding genes as well as expression sequence tags derived from PGC cDNA libraries (Abe, unpublished) and was manufactured by Agilent Technologies. Total RNA was labelled with Cy3-CTP with a Quick Amp Labeling Kit (Agilent Technologies). Hybridization was performed according to the protocol suggested by the supplier. Hybridized slides were scanned using a microarray scanner (Agilent Technologies), and the signals were processed with the Feature Extraction software ver. 10.5.1.1 (Agilent Technologies). The processed signal data were normalized and analysed by the Gene Spring GX11.5 software (Agilent Technologies). The microarray experiments were conducted using biologically duplicated samples.
2.3. Modified nanoHELP: linker-mediated amplification and hybridization
The nanoHELP assay, a microarray-based DNA methylation analysis, was performed according to our previous reports12,14 with modifications. Briefly, genomic DNA (0.5–2 ng) was digested by HpaII or MspI in 100 μl of reaction mixture at 37°C overnight. This was followed by DNA purification with the MinElute Reaction Cleanup Kit (Qiagen), and the digested DNA was ligated to linker adapters, NHpaII12/NhpaII24 and JHpaII12/JhpaII2414 overnight at 16°C. After removing the linker adapters, the ligated DNA was added to a total of 50-μl PCR reaction mixture containing 1.5 μl each of 20 μM primer (NHpaII24, 5′-GCAACTGTGCTATCCGAGGGAAGC-3′; JHpaII24, 5′-CGACGTCGACTATCCATGAACAGC-3′), 10 μl of 5 M betaine (Sigma-Aldrich), 200 μM of dNTPs and 2.5 units of ExTaq DNA polymerase (TaKaRa Bio, Inc., Otsu, Japan) in a buffer supplied by the manufacturer. The mixture was heated at 72°C for 10 min and subjected to PCR amplification with the following parameters: 15 cycles at 95°C for 30 s and 72°C for 3 min, with a final extension at 72°C for 10 min. After the first round of amplification, one-tenth of the volume of the reaction was added to a fresh PCR reaction mix containing the same primers, and amplified for an additional 10–15 cycles14,25 with the same PCR parameters as described above. The PCR products were purified using the MinElute Kit (Qiagen). An additional column washing step with 750 μl of 35% guanidine hydrochloride (Nacalai Tesque, Inc., Kyoto, Japan) solution was performed to remove the residual primer–adapters. The amplified DNA originally digested with HpaII was labelled with Cy5-labelled Random 9-mers (TriLink Biotechnologies, San Diego, CA, USA), and the MspI-digested DNA was labelled with Cy3-Random 9-mers (TriLink Biotechnologies). The labelled DNAs were mixed and hybridized with a custom microarray (Roche NimbleGen, Madison, WI, USA) using a NimbleGen Array Hybridization Kit (http://www.nimblegen.com/products/lit/lit.html). After washing with the NimbleGen Array wash kit, the microarrays were scanned on an Agilent Technologies Scanner G2505C with a setting of 5-μm resolution. The HELP array experiments were performed using biological replicates. The raw data were processed using the NimbleScan 2.4 data extraction software (NimbleGen) to obtain the processed log2 (Cy5/Cy3) ratio data.
Our previously published ChIP-on-chip data for cumulus cells26 were reanalysed and used. The ChIP-on-chip experiment using GS cells was performed as described in the same paper.
2.4. Microarray design
The microarrays were designed to represent restriction fragments with 5′-CCGG restriction sites in a size range of 200–2000 bp (=CCGG segments) on mouse Chromosome 7 and the X chromosome. Ten 50-mer oligonucleotide probes were designed from unique sequences in each CCGG segment, avoiding repeat-masked regions and sequence ambiguities. Probe sequences were selected using a score-based selection algorithm, as described.12 Detailed information for the coverage of genomic regions on each chromosome and annotations of the CCGG segments are described in Supplementary Table S1. Information about the positions of probes, M-values obtained from different samples and k-means cluster number are described in Supplementary Table S2, and gff files of the HELP array data are available at our web site (http://www.brc.riken.go.jp/lab/mcd/mcd2/protocol/nanoHELP.html).
2.5. Data analysis
The steps in the HELP data analysis are shown schematically in Supplementary Fig. S1. Briefly, hybridization signal noise is first removed from the processed data by cutting off the values in the range of random sequence probes. In our microarray, 10 oligonucleotide probes are normally assigned to each CCGG segment. The median signal intensity of the 10 probes is calculated and used to define the segment's signal intensity. Using the median signal values, the HpaII/MspI ratio is then calculated for each CCGG segment and converted to a log2 value to obtain the M-value. After normalization of the microarray ratio data, hypomethylated and hypermethylated segments are distinguished using an R script (http://www.r-project.org/) that determines the threshold values based on a binarization method.27 The marginal width of the threshold is calculated using the Mahalanobis distance.28 The log2 value at the threshold is set as 0, so that unmethylated segments have a value of ≥0 and methylated segments have a negative value (<0). For interarray normalization of the HpaII/MspI ratio, the threshold value of each array data set is scaled to 0. All the bioinformatic analysis described here is based on the UCSC mm8 genome assembly (http://genome.ucsc.edu/).
2.6. Accession number
Gene expression microarray data and the HELP array data are available at the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) (Accession number GSE39895).
3. Results
3.1. DNA methylation analysis with subnanogram amounts of genomic DNA
For epigenomic analyses of developing cells, materials can be very limited in quantity, precluding conventional analytical techniques. One of our goals was to describe comprehensively the epigenomic changes during the development of early embryos and germ cells in the mouse. Towards this goal, we use the HELP assay,12 a proven, microarray-based method, for the analysis of DNA methylation.13 The original HELP protocol requires 10 μg of genomic DNA as the starting material,12 but Oda et al.14 established an improved version of the method, nanoHELP, for the analysis of a limited amount of starting DNA. Here, we have fine-tuned the protocol further for the analysis of 0.5–2 ng of starting material. The details of this method are described in Section 2, and the flow of the data analysis is presented in Supplementary Fig. S1.
The HELP assay is a microarray-based method that detects the subset of unmethylated HpaII fragments in the genome and uses the corresponding, methylation-insensitive MspI representations as a control. The M-value, an index of the methylation level, is calculated as log2(HpaII signal/MspI signal) as described in Section 2: unmethylated segment has a value of ≥0 and methylated segment has a negative value of <0. As shown in Supplementary Fig. S2A–D, the modified nanoHELP assay generated data having good correlations with the data obtained by the original protocol. To validate these results, bisulphite pyrosequencing analysis of six CpG sites was performed as described.12 The results showed that the modified nanoHELP assay could generate reliable data (Supplementary Fig. S2E).
3.2. Custom HELP microarray used in this study
The number of restriction sites for HpaII, 5′-CCGG, in the mouse genome (UCSC mm8) is 1 588 546, covering ∼7.5% of the total CpG dinucleotides in the genome (Supplementary Table S1A). The CCGG sites are distributed almost evenly over the mouse genome and do not show an apparent bias to a particular genomic context. Thus, the use of CCGG sites is suitable for obtaining a chromosome-wide view of CpG methylation profiles.
In this experiment, we designed a custom microarray harbouring 382 018 oligoprobes. We first selected HpaII or MspI fragments (designated here as CCGG segments) with a size range of 200–2000 bp, mostly from mouse Chromosomes 7 and X, and designed 10 unique sequence probes of 50 nucleotides per CCGG segment. The custom microarray can detect 22 128 CCGG segments on Chromosome 7 and 14 472 on the X chromosome, which correspond to 46 and 39% of the total segments on each chromosome, respectively. The CCGG segments were selected in an unbiased fashion except for a part of the Chromosome 7 and a small number of segments associated with some germ cell-related genes on Chromosomes 6, 11 and 12 (Supplementary Table S1B). Supplementary Table S1C and D describes the categorization of the CCGG segments based on the genome annotations. About 47% of the segments map to intergenic regions, 5% to promoter regions and 46% to bodies of RefSeq genes (Supplementary Table S1C). About 75% of the X-linked RefSeq genes are covered by this HELP microarray (Supplementary Table S1D). Because most of the CCGG segments in CGIs are <200 bp, ∼1% of CGIs annotated in the UCSC mm8 genome assembly can be assayed by this array (Supplementary Table S1E).
CGIs and gene promoter regions have been the main targets in most DNA methylation studies. However, the importance of DNA methylation in genomic regions outside the promoters is becoming increasingly apparent.5,6 It is expected that this microarray method should be appropriate for the analysis of previously unexplored and potentially informative parts of the genome.
3.3. Analysis of DNA methylation profiles of stem cells and germline cells
We performed DNA methylation profiling of the following samples: ES cells from male and female blastocysts, male and female EG cells established from PGCs at embryonic day 12.5 (E12.5), GS cells derived from spermatogonia22 and male and female PGCs purified from Oct3/4–GFP transgenic embryos20,21 in various stages by FACS. PGCs were isolated from male and female E10.5, E13.5 and E17.5 embryos. PGCs have not entered the gonads at E10.5, and that are colonized within the gonads in E13.5 embryos. At E17.5, PGCs are subjected to mitotic arrest in male gonads, and female PGCs are arrested in the early phase of meiosis.3 We isolated germ cells from newborn ovary and testis. Whole adult testis, thymus and brain were isolated from male mice and used for the analysis. Germ cells in the adult testis were purified by FACS from Mvh (mouse Vasa homolog)-Venus transgenic mouse (Mise and Abe, unpublished results).23 Gene expression profiling of all samples was conducted using our custom 44K microarray.
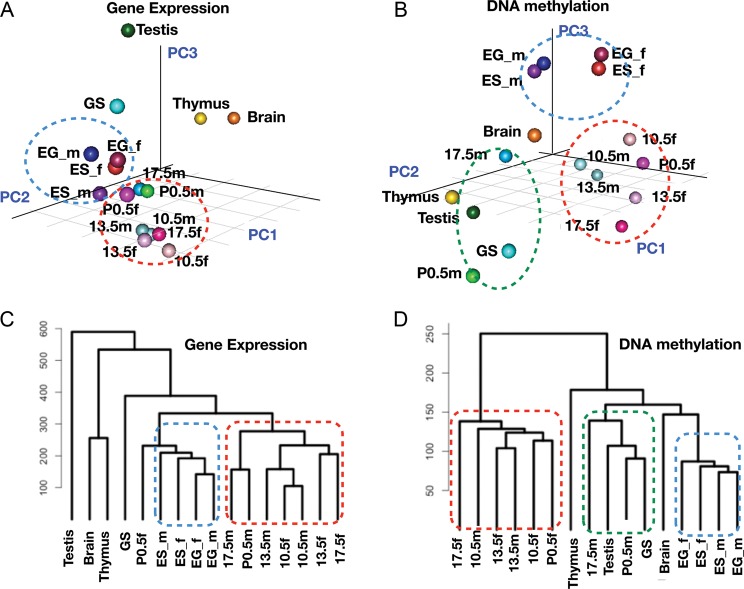
Figure 1 shows the results of principal component analysis (PCA) and hierarchical cluster analysis of the DNA methylation profiles and the gene expression profiles. In this comparison, pluripotent stem cells (i.e. ES and EG cells) and PGCs from various stages show similar but distinct expression profiles; ES and EG cells are positioned more closely (blue circle) relative to PGCs (red circle) (Fig. 1A and C). This result confirms our previous findings that PGCs possess a distinct transcription programme from ES cells, although both share the expression of common ‘signature genes’.20 In contrast, analysis of the DNA methylation profiles showed the differences between samples more clearly. PGC samples could be classified into two groups: one comprising female PGCs and early male PGCs (i.e. E10.5 and E13.5 in the red circle) and E17.5 and P0.5 male germ cells that formed a cluster together with GS cells and testis (green circle) (Fig. 1B and D). Male PGCs in different stages appeared to be more distantly related to each other than to female PGCs, suggesting that the DNA methylation profiles change more drastically during male PGC development. These results suggest that cell types can be classified by their DNA methylation profiles and that, in some cases, DNA methylation profiling can display differences in the cellular state more effectively.
Figure 1.
Profiling of gene expression and DNA methylation in germ cells and stem cells. (A) PCA of the expression profiles of germ cells, stem cells and adult organs. ES_m (male ES cells), ES_f (female ES cells), EG_m (male EG cells), EG_f (female EG cells), 10.5m (PGCs from male E10.5 embryos), 10.5f (female E10.5 PGCs), 13.5m (male E13.5 PGCs), 13.5f (female E13.5 PGCs), 17.5m (male E17.5 PGCs), 17.5f (female E17.5 PGCs), P0.5m (spermatogonia from P0.5 neonates), P0.5f (oocytes from P0.5 neonates) and GS cells. Testis, thymus and brain were isolated from male adult mice. (B) PCA analysis of DNA methylation profiles of germ cells, stem cells and adult organs. (C) Hierarchical clustering of gene expression profiles. (D) Hierarchical clustering of DNA methylation profiles.
3.4. k-means cluster analysis of DNA methylation profiles to visualize cell type-specific differentially methylated regions
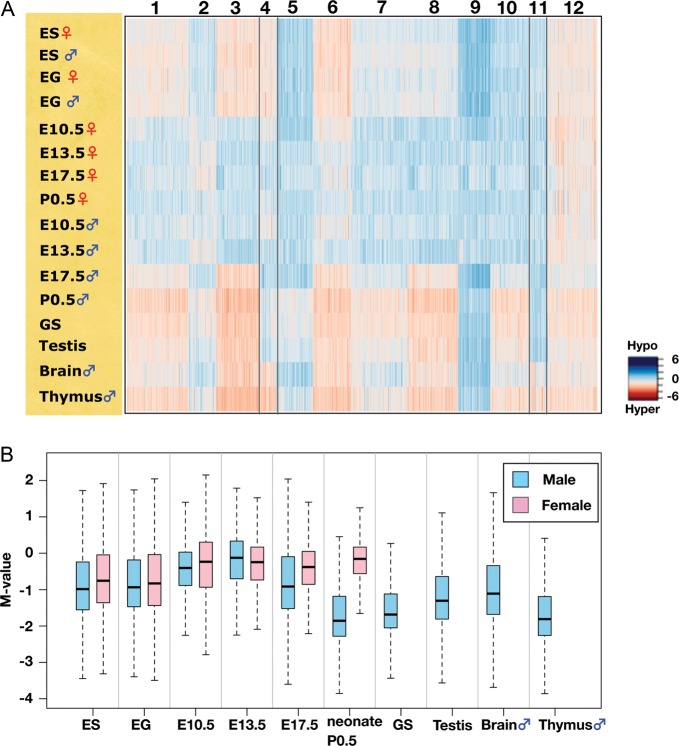
To visualize differences in the DNA methylation profiles of the samples tested, non-hierarchical k-means analysis (k = 12) was performed using the data obtained from the 28 217 informative CCGG segments; the result is shown as a heat map in Fig. 2A. One of the most conspicuous trends was that the genomes of the PGCs examined are mostly hypomethylated except for male E17.5 PGCs. A box plot of the M-value for each sample is shown in Fig. 2B. Global levels of DNA methylation are lower in the E10.5 PGC genome than in ES and EG cells, and the levels are even lower at E13.5. At E17.5, the methylation level of male PGCs is increased, whereas female PGCs maintain a hypomethylated status similar to that at E10.5 or E13.5. The difference in the methylation level between male and female germ cells is most prominent in neonates: male spermatogonia have a highly methylated genome. GS cells possess a similar DNA methylation profile to that of P0.5 spermatogonia (R = 0.80). Epigenetic features of GS cells have not been reported to date, and this result suggests that GS cells should provide a valuable in vitro model for epigenetic studies of spermatogonial cells. Adult testis, which comprises both germ and somatic cells, has a slightly lower DNA methylation level relative to spermatogonia (P0.5 male) and GS cells (Fig. 2B).
Figure 2.
DNA methylation dynamics during germ cell development. (A) k-means clustering of DNA methylation profiles of germ cells, stem cells and adult organs. DNA methylation levels of the CCGG segments are represented as a heat map (unmethylated segments in dark blue, M-value = 6.00; highly methylated segments in dark red, M-value = –6.00). (B) Changes in global DNA methylation levels during PGC development. The M-value was calculated for each sample and is shown as a box plot. The bottom and top of the boxes are the 25th and 75th percentile, respectively.
Although most of the PGC genomes are hypomethylated, genomic regions classified as Cluster 12 remain methylated at a level similar to that in the other cell types examined. The rest of the clusters showed some cell or tissue specificities in DNA methylation.
3.5. Characterization of germline-specific hypomethylated CCGG segments on the X chromosome
The clusters were then characterized by examining their tissue specificities in DNA methylation patterns. We noticed that Cluster 4 comprises the segments hypomethylated only in PGCs, GS cells and the testis, whereas these segments are hypermethylated in ES and EG cells, and somatic organs. These putative germline-specific hypomethylated segments were characterized further. Although no particular GO terms are enriched, Cluster 4 contains genes expressed in the germ cells of testis such as Xmr (Xlr-related, meiosis regulated)29,30 or CTA genes; e.g. the Mage (melanoma antigen) gene family.31 The number of Cluster 4 CCGG segments mapped onto the X chromosome is disproportionately high. There are 1004 segments in Cluster 4, and 715 (71.2%) are on the X chromosome, which has 14 472 CCGG segments in total. In contrast, 219 of Cluster 4 segments (21.8%) are on Chromosome 7, which carries 22 128 segments. Because genes expressed in germ cells or testis are known to be enriched on the X chromosome,15,16 we focused on the Cluster 4 segments of the X chromosome as candidates involved in the epigenetic regulation of germ cell-specific gene expression.
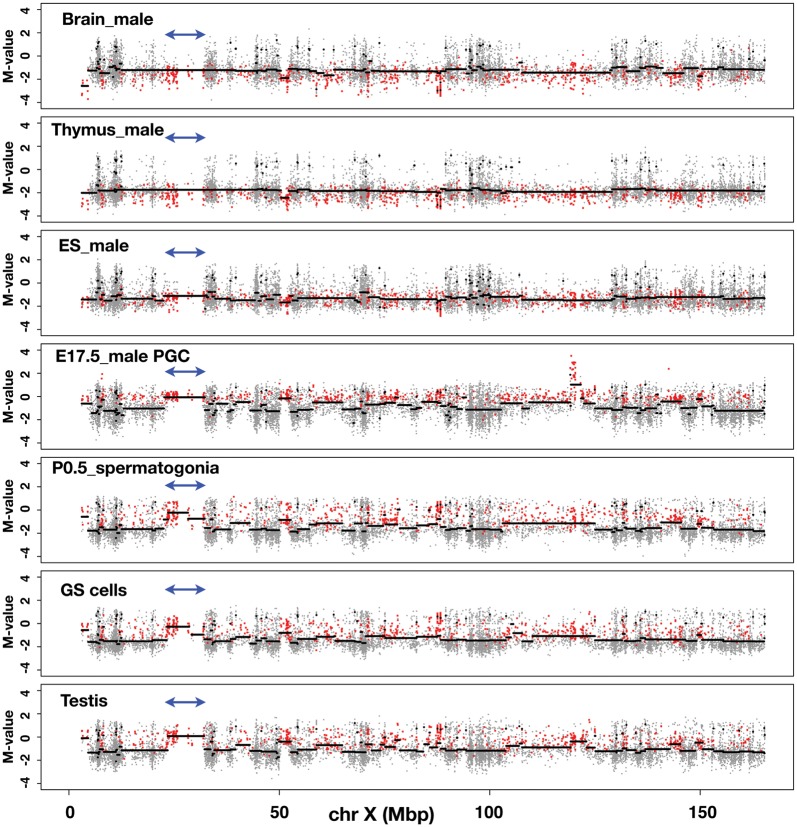
As shown in Fig. 3 and Supplementary Fig. S3A–C, we plotted the M-values of all the CCGG segments along the X chromosome using the data obtained from each sample (grey dots). To translate the M-value measurements into regions of equal M-value, we used a circular binary segmentation programme, which is used normally for comparative genomic hybridization analysis.32 Using this programme, we drew lines (black horizontal lines) to show regions of equal M-value. By tracing the line, we could identify the genomic regions in which the M-value changes significantly from the flanking regions. The M-values of the segments belonging to Cluster 4 are overlaid as red dots. The distributions of the M-values in the DNA of somatic cells (i.e. brain and thymus) along the entire X chromosome are similar to each other: the average M-value is less than –1, with some local exceptions. The Cluster 4 dots are mapped even below the average line, indicating that, as expected, Cluster 4 segments are hypermethylated in both brain and thymus (Fig. 3). In ES and EG cells (Fig. 3 and Supplementary Fig. S3A), the average M-values of the Cluster 4 segments do not change significantly along the X chromosome and are positioned below –1, suggesting that Cluster 4 segments are largely hypermethylated in the genomes of ES or EG cells. In sharp contrast, in E17.5 male PGC DNA, it appears that the average M-value line is often discontinuous, and that hypomethylated CCGG segments exist over relatively large, contiguous genomic regions (Fig. 3). For example, the average M-value of the segments within the ∼9 Mb genomic region harbouring the Xmr gene cluster (double-headed arrows) is close to 0, and Cluster 4 segments are enriched in this region.
Figure 3.
Discovery of large, contiguous genomic regions with low DNA methylation modifications in male germ cells. The methylation profiles of genomic DNA along the mouse X chromosome. Samples used for the analysis are indicated in each figure. The M-value of each CCGG segment obtained from the analysis of each sample DNA is plotted on the mouse X chromosome (grey dots). The y-axis represents the M-value; log2(HpaII/MspI). The black line was drawn using DNAcopy, a circular binary segmentation programme obtained from http://www.bioconductor.org/packages/2.3/bioc/html/DNAcopy.html.32 Red dots represent CCGG segments belonging to Cluster 4. Double-headed arrows indicate the position of the Xmr gene cluster.
It is obvious that the distribution of the Cluster 4 segments is not uniform, and that these Cluster 4 segments form ‘hypomethylated domains’ compared with their flanking regions. These trends persist in P0.5 spermatogonia and GS cells derived from spermatogonia (Fig. 3), with a few cell type-specific differences. In testis DNA, the overall methylation pattern of the Cluster 4 segments is essentially similar to that found in the male germline cells, described above (Fig. 3). We also examined earlier stages of male PGCs (Supplementary Fig. S3B). In E10.5 male PGCs, formation of hypomethylated domains, e.g. Xmr region, is not as obvious as seen in E17.5 male PGCs. In E13.5 male PGC DNA, the distribution of the Cluster 4 segments is similar to that found in E17.5 male PGCs. It thus appears that clustering of hypomethylated DNA segments become increasingly evident on the X chromosome during the development of male germ cells.
3.6. Discovery of large genomic regions hypomethylated specifically in male germline cells
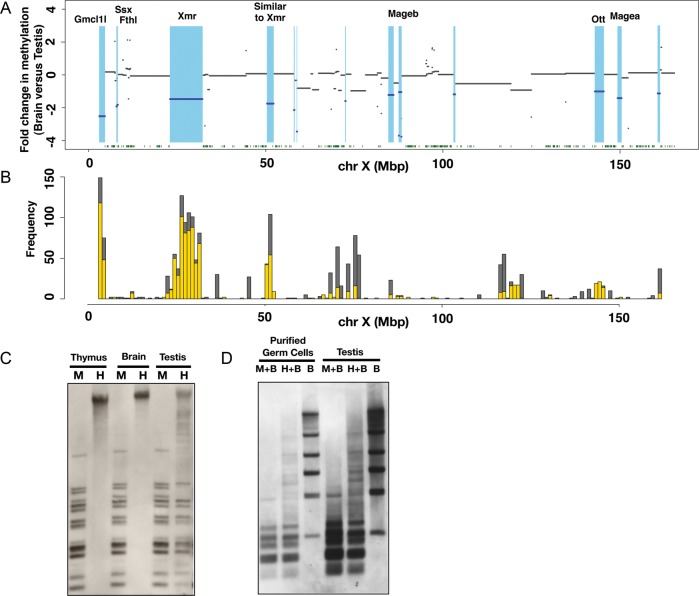
Although it is clear that segment DNAs possess generally lower methylation levels in female PGCs than in cells of somatic organs, the formation of hypomethylated DNA regions as seen in male PGCs is not evident in female PGCs (Supplementary Fig. S3B and C). We therefore decided to focus on the male germline-specific hypomethylated DNA regions comprised of Cluster 4 segments. To visualize the hypomethylated DNA regions in the male germline from a different viewpoint, we plotted fold differences in the methylation level between somatic and male germ cell DNA along the X chromosome (Fig. 4A). Because methylation patterns of Cluster 4 segments are essentially similar in testis, E17.5 and P0.5 male PGCs, the testis was chosen for this analysis. Brain was also used as somatic tissue for this analysis. As the data were plotted with a log2 scale, a negative value indicates the lower level of DNA methylation in the testis than in the brain. The plot revealed broad domains with lower methylation levels in the testis and therefore, in male germ cells of late stages (coloured light blue in Fig. 4A). These broad domains of hypomethylated DNA described above are distinct from CGIs, which are generally located within or near a promoter and have a typical length of 300–3000 bp.33 The broad and hypomethylated domains identified here are often much larger than CGIs and do not show preferential localization at promoter regions. Thus, these broad domains do not correspond to the known hypomethylated regions and may represent a hitherto unknown epigenomic entity. For convenience of discussion, we here designate such a broad and hypomethylated domain as an LoD. By definition, a LoD has a size of >10 kb and shows more than a 2-fold difference in M-value between germline and somatic cells (testis and brain in this case). Each LoD should also have at least one Cluster 4 segment.
Figure 4.
Demonstration of LoDs on the mouse X chromosome. (A) Differences in methylation levels between brain and testis genomic DNA along the mouse X chromosome. Fold changes in the methylation level, i.e. brain M-value versus testis M-value, were calculated for each CCGG segment, and plotted using the log2 scale along the X chromosome. The blue lines indicate genomic regions showing more than a 2-fold difference between the brain M-value and testis M-value. Light blue represents hypomethylated regions in the testis relative to the brain. Green dots at the bottom represent the positions of CGIs. (B) The positions of segmentally duplicated regions along the mouse X chromosome. Segmentally duplicated regions >1000 bases with >98% similarity are counted, and the frequencies of duplications (y-axis) are shown. (The data were from UCSC Genome Browser.) Grey bars represent duplications occurring on the X and other chromosomes, and yellow bars represent the frequencies of duplications mapped only on the X chromosome. (C) Methylation analysis of LoDs 10 and 12 by Southern blot hybridization. Genomic DNAs of the male thymus, male brain and testis were digested by either methylation-sensitive HpaII (H) or methylation-insensitive isoschizomer, MspI (M). The Southern blot was hybridized with a probe targeted to LoDs 10 and 12. A primer pair (FW: 5′-GCTGGGTCCAGCTTCCCTGG-3′, RV: 5′-TGGCACCCCTCCTGCCTGAT-3′) was used to amplify a 807-bp sequence using testis cDNA for generation of the probe. The 807-bp probe contains locally repeated sequences and corresponds to both LoDs 10 and 12 located upstream of Mageb1/b2 genes. (D) Methylation analysis of LoDs 10 and 12 in purified germ cells. Germ cells expressing the Mvh-Venus reporter were purified from adult testis by FACS. DNAs from purified germ cells and whole testis were digested with MspI plus BamHI (M + B), HpaII plus BamHI (H + B) or BamHI only (B). A Southern blot was made using these DNAs and hybridized with the same probe as used in Fig. 4C.
3.7. Overlap of LoDs with segmentally duplicated regions
Using the definition described above, we list the LoDs of the X chromosome in Table 1. There are 16 LoDs on the X chromosome (Table 1), and their sizes are generally large: 11 of the 16 LoDs are >100 kb (mean: 1 219 252 bp), and six of the large LoDs are >1 Mb. The mammalian genome is replete with segmentally duplicated regions.34 Although segmental duplications can be found on every chromosome, they are particularly abundant on the sex chromosomes. Because LoDs are generally large and contain gene families such as Xmr, we asked whether LoDs overlap with segmentally duplicated regions. As shown in Fig. 4A and B and Table 1, all LoDs on the X chromosome are found to contain segmentally duplicated regions. The use of the MspI control represents an unusual strength of the HELP assay to remove the potentially confounding effect of copy number variation.12 Combined with a probe design that selects only unique sequences for hybridization, these aspects ensure that the DNA methylation readout from regions of constitutive segmental duplication accurately reflects the underlying DNA methylation and is not influenced by DNA copy number.
Table 1.
List of LoDs on the mouse X chromosome
| LoD no. | Position | Length (bp) | Segment number on HELP array | Methylation |
Gene families | Description | Number of duplications |
Human CTA genes | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Brain M-value/testis M-value | CGI | Seg Dups | Total | Within LoD | Outside LoD on chrX | |||||||
| 1 | chrX:3035387-4561626 | 1 526 239 | 12 | −2.52 | 1344 | Gmcl1l | Germ cell-less protein-like 1 | 11 | 8 | 3 | ||
| 2 | chrX:7857031-7924410 | 67 379 | 5 | −1.924 | 88 | Ssx9 | Synovial sarcoma, X breakpoint 9 | 11 | 4 | 7 | CTA | |
| 3 | chrX:8116622-8236784 | 120 162 | 19 | −1.82 | 45 | Fthl17 | Ferritin, heavy polypeptide-like 17 | 7 | 6 | 1 | CTA | |
| 4 | chrX:22991291-32117922 | 9 126 631 | 100 | −1.4745 | 10 697 | Xmr | XMR protein (Xlr-related, meiosis regulated) | 32 | 28 | 4 | ||
| Gmcl1l | Germ cell-less homologue 1 (Drosophila)-like | 11 | 2 | 9 | ||||||||
| LOC236749 | Hypothetical protein LOC236749 | 1 | 1 | 0 | ||||||||
| 5 | chrX:50380769-52184422 | 1 803 653 | 48 | −1.75 | 1 | 1971 | Similar to Xmr protein | Adult male testis cDNA, RIKEN full-length enriched library, clone: 4930527E24 product: weakly similar to XMR PROTEIN | 30 | 16 | 14 | |
| 6 | chrX:57970833-58090999 | 120 166 | 5 | −2.14 | 23 | Ldoc1 | Leucine zipper, down-regulated in cancer 1 | 1 | 1 | 0 | ||
| 7 | chrX:58775958-58815172 | 39 214 | 13 | −3.45 | 10 | a1700019B21Rik | Mus musculus adult male testis cDNA, M. musculus RIKEN cDNA 1700019B21 gene (1700019B21Rik), transcript variant 1, non-coding RNA | 1 | 1 | 0 | ||
| 8 | chrX:72448732-72544743 | 96 011 | 4 | −1.59 | 67 | LOC238829 | Hypothetical protein LOC238829 (AK133378—M. musculus adult male testis cDNA, RIKEN full-length enriched library, clone: 4933402E19 product: hypothetical protein, full insert sequence) | 4 | 1 | 3 | ||
| 9 | chrX:84663998-86056286 | 1 392 288 | 23 | −1.2207 | 334 | Pet2 | Plasmacytoma expressed transcript 2 | 1 | 1 | 0 | ||
| 4932429P05Rik | M. musculus RIKEN cDNA 4932429P05 gene (4932429P05Rik), mRNA. (M. musculus adult male testis cDNA) SMEK homologue 3, putative | 1 | 1 | 0 | ||||||||
| 10 | chrX:87564706-87579103 | 14 397 | 20 | −3.70 | 1 | (up site of Mageb1, Mageb2) | ||||||
| 11 | chrX:87598220-88271067 | 672 847 | 15 | −1.0483 | 43 | Mageb5 | Melanoma antigen, family B, 5 | 2 | 2 | 0 | CTA | |
| Mageb1 | Melanoma antigen, family B, 1 | 2 | 1 | 1 | CTA | |||||||
| 12 | chrX:88271564-88285701 | 14 137 | 21 | −3.76 | 1 | (up site of Mageb1,Mageb2) | ||||||
| 13 | chrX:103040719-103447968 | 407 249 | 10 | −1.186 | 158 | A630033H20Rik | Hypothetical protein LOC213438 (product: similar to putative purinergic receptor P2Y10) | 1 | 1 | 0 | ||
| Gpr23 | G protein-coupled receptor 23 | 1 | 1 | 0 | ||||||||
| P2ry10 | Purinergic receptor P2Y, G-protein coupled 10 | 1 | 1 | 0 | ||||||||
| Zcchc5 | Zinc finger, CCHC domain containing 5 | 1 | 1 | 0 | ||||||||
| 14 | chrX:142924085-145405975 | 2 481 890 | 112 | −1.0089 | 2 | 1666 | Ott | Ovary testis transcribed | 17 | 16 | 1 | CTA |
| 15 | chrX:149333243-150403389 | 1 070 146 | 41 | −1.4149 | 211 | Magea | Melanoma antigen family A 2, 3, 5, 6, 8 | 7 | 5 | 2 | CTA | |
| Samt4 | M. musculus spermatogenesis associated multi-pass transmembrane protein 4 (Samt4), mRNA (hypothetical protein LOC75185) | 1 | 1 | 0 | ||||||||
| 16 | chrX:160673416-161229042 | 555 626 | 13 | −1.13 | 274 | |||||||
aNon-coding RNA.
Hypomethylation of two such domains, LoD 10 and 12, was confirmed by Southern blot analysis (Fig. 4C and D). Since LoD 10 and 12 contain homologous, locally repeated sequences, a hybridization probe can be used to assess the methylation status of both regions. The genomic DNAs of the thymus, brain and testis were digested by either methylation-sensitive HpaII or the methylation-insensitive MspI. In the HpaII digests of thymus and brain DNA, no bands were detected except for a hybridization signal in the unresolved part of the lanes, indicating that the genomic region is hypermethylated in somatic organs. In the HpaII digest of testis DNA, many bands were detected, and the band pattern was essentially the same as that found in the MspI digest, clearly indicating that this region is largely unmethylated in the testis. Given that the testis comprises both germ and somatic cells, we asked whether LoDs are hypomethylated in germ cells. We used an Mvh35-Venus reporter transgenic mouse line, in which germ cells are marked by Venus fluorescence protein (Mise and Abe, unpublished results). We also used FACS to purify the Venus-positive germ cells from the adult testis and performed Southern analysis. The results indicated that the genomic regions in the purified germ cells are indeed hypomethylated (Fig. 4C and D).
3.8. Predominance of genes expressed in male germ cells or in the testis in LoDs
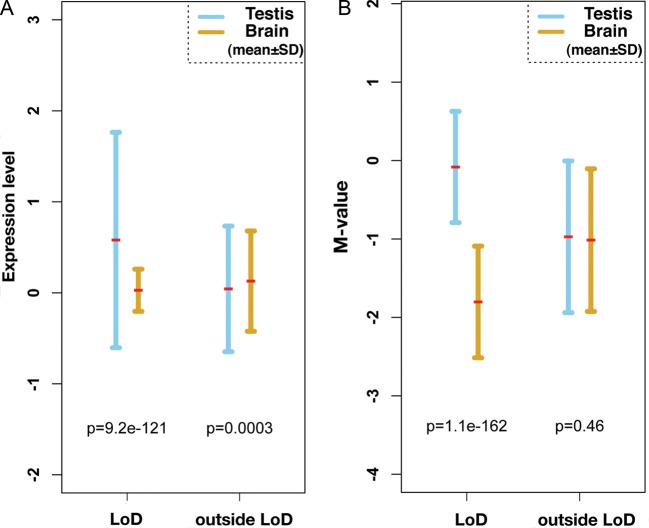
We noticed that most LoDs contain genes that are expressed in the testis. For example, Gmcl1l (germ cell-less protein-like 1-like), Ssx9, Fthl17, Xmr, Mageb, Ott, Samt4 and Magea are expressed in the testis and are included in LoDs 1, 2, 3, 4, 11, 14 and 15, respectively (Table 1). Expression of these genes are also detected in germ cells purified from adult testes (data not shown). If we omit LoDs 10, 12 and 16, which do not carry known genes, only LoDs 6 and 13 do not contain genes predominantly expressed in germ cells (Table 1 and Supplementary Table S3). The mean expression levels of genes contained in LoDs are shown in Fig. 5A. Genes within LoDs show significantly higher expression in the testis than in the brain. Figure 5B shows the mean levels of DNA methylation within and outside LoDs on the mouse X chromosome. Figure 5 indicates that there is an inverse correlation between the level of DNA methylation and the expression of genes in LoDs.
Figure 5.
Inverse relationship between DNA methylation and expression of genes within the LoDs. (A) A plot of the expression levels of genes contained in LoDs, and regions outside LoDs on the mouse X chromosome. Gene expression data were obtained from the Affymetrix Exon array data set.43 The expression values of exons contained in LoDs were averaged and plotted, and the data points from regions outside LoDs were similarly averaged. For statistical analysis of the data from regions outside LoDs, the same number of data points as those used to analyse within LoDs were randomly selected and used. (B) Mean methylation levels of genomic DNA within LoDs. The mean M-values from the CCGG segments contained in LoDs or in regions outside LoDs are shown. Statistical significance was tested by Wilcoxon t-test, and P-values are shown within the figures.
3.9. Genomic structures of LoDs: Xmr/Slx and Mageb regions
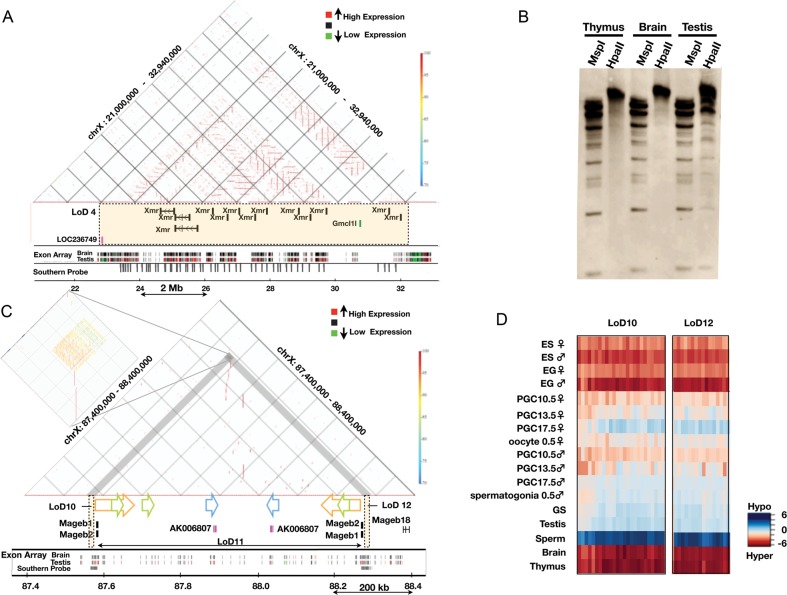
In addition, we demonstrate the detailed structures of two LoD regions (Fig. 6). LoD 4 is ∼9.1 Mb in size (chrX: 22 991 291–32 117 922) and contains three distinct genes/gene families, all of which are expressed specifically in the testis. Because Xmr is a synonymous gene with Slx,29,30 we call this gene/gene family either Xmr or Xmr/Slx in this study. Xmr/Slx is known to be expressed in spermatids, where it encodes a protein, SLX/XMR, normally localized in cytoplasm.29,30 Xmr/Slx represents a locally duplicated multigene family, whose copy number is at least 28 in LoD 4. Gmcl1l and LOC236749 are included in the same LoD, and both are expressed in the testis and in purified male germ cells (Fig. 6A; data not shown).
Figure 6.
Genomic structures of LoDs: Xmr and Mageb. (A) Genomic structure of LoD 4 (Xmr). The top section represents a similarity dot plot44 of LoD 4 and flanking regions (chrX: 21 000 000–32 940 000; UCSC mm8). Similarities of the sequences are colour-coded as shown by the colour bar on the right (high similarity in dark red, 100% similarity; low similarity in blue, 70% similarity). The horizontal lines represent direct repeats, and the vertical lines indicate IRs. The middle section depicts the locations of genes contained in LoD 4 (coloured rectangle). Exon array expression data for the brain and testis43 are shown at the bottom (high gene expression in red and low expression in green). The positions of sequences with homology to the Xmr cDNA probe are also shown by grey vertical bars (Southern probe). (B) Southern blot probed with the Xmr cDNA. Genomic DNAs from three organs were digested with either MspI or HpaII. Xmr cDNA probe (708 bp) was amplified from testis cDNA using primers, Xmr FW: 5′-AAGGGTGCAGTTGTGAAGGT-3′, Xmr Rv: 5′-TGTTGGTCTCCATGTTCATCA-3′. The hybridization signals in the unresolved part of the testis DNA blot are likely to reflect non-specific cross-hybridization. Southern blot analysis data using DNA doubly digested by BamHI plus either HpaII or MspI confirmed this notion (data not shown). (C) Genomic structure of LoDs 10, 11 and 12 (Mageb1/b2). Top: similarity dot plot. The vertical lines and coloured arrows indicate positions of IRs. The colouring of the similarities is the same as found in Fig. 6A. Homologous repeats are represented by the same colour. A magnified view of the IRs contained in LoDs 10 and 12 is presented on the left. Gene expression data for the brain and testis are shown at the bottom. The positions of sequences with homology to a probe used for Southern blot analysis (Fig. 4C and 4D) are shown by grey bars (Southern probe). (D) A DNA methylation heat map of LoDs 10 and 12. DNA methylation levels of the CCGG segments are represented as a heat map (unmethylated segments in dark blue, M-value = 6.00; highly methylated segments in dark red, M-value = –6.00).
The LoD 4 region represents one of the largest segmentally duplicated regions on the mouse X chromosome (Katsura and Satta, personal communication) and can be divided into four subregions (Supple-mentary Fig. S4). Subregion I spans ∼3 Mb and harbours tandemly repeated Xmr genes. Subregion II spans ∼3.8 Mb and comprises both tandem and inverted repeats (IRs) of Xmr genes. Subregion III contains tandem and IRs of Gmcl1l genes, which are duplicated on two distant sites on the X chromosome; the other site is also classified as LoD 1 (Table 1). Subregion IV is <1 Mb and contains tandem repeats of Xmr genes. One hundred and forty-one CCGG segments are mapped within LoD 4, and the fold difference in methylation level (brain versus testis) of these segments were calculated as described in Fig. 4A. The mean value is –1.4745, suggesting that the CCGG segments in this region are generally hypomethylated in the testis genome (Table 1 and Figs 3 and 4).
Because of the repetitive nature of the LoD region, HELP probes cannot be assigned for most of the subregions II and IV. To examine DNA methylation in these regions, we performed Southern analysis of the testis and brain DNA digested with either HpaII or MspI, and hybridized with an Xmr cDNA probe. As shown in Fig. 6A and Supplementary Fig. S4, the Xmr cDNA probe should be able to assess the methylation status of 161 restriction fragments. These fragments are distributed evenly within subregions I, II and IV, and fill the gaps of information provided by the nanoHELP assay, which tests only unique sequences. The results of the Southern blot analysis shown in Fig. 6B demonstrate that the Xmr region is highly methylated in the brain and liver, whereas a considerable proportion of the restriction fragments appear unmethylated in the testis.
It has been suggested that transcriptionally active genes are hypomethylated in their promoter region, while their gene bodies tend to be hypermethylated.6 However, a magnified view of the LoD 4 region (Supplementary Fig. S5) indicates that all CCGG segments in this region are hypomethylated in the testis and male PGCs regardless of their positions with respect to the Xmr genes. Both the probes positioned near the transcription start sites and the probes positioned at introns or even at intergenic regions are unmethylated in the testis, GS and male PGCs. The Southern blot analysis data suggest that the CCGG segments containing exons of the Xmr genes appear to be relatively hypomethylated in the testis (Fig. 6B). These results imply that methylation of the whole LoD 4 is subjected to a region-wide regulation. This feature is shared by other LoDs not described here.
Mageb belongs to the Mage (melanoma antigen) gene family, which is expressed in spermatogenic cells and in some cancer cells.31 Figure 6C shows a genomic region spanning ∼1 Mb that contains Mageb1 and Mageb2 genes. This region represents a large IR with arms of ∼400 kb in length. At the ends of both arms, LoDs 10 and 12 are located 4–2 kb upstream of the transcription start sites of Mageb1 and Mageb2, respectively. These two LoDs do not contain the Mageb locus itself (Supplementary Fig. S6). Both LoDs are highly homologous and ∼14 kb long, and comprise repeat sequences with a unit size of ∼3 kb. These sequences are both tandem and IRs (Fig. 6C; magnified part), are found only in these LoD regions and are clearly hypomethylated only in germ cells (Supplementary Fig. S6). Hypomethylation of LoDs 10 and 12 was confirmed by Southern blot analysis as described (Fig. 4C and D).
3.10. Developmental changes in the methylation levels of LoDs
LoDs are hypomethylated in the testis, GS cells and male PGCs. The methylation heat maps of LoDs 10 and 12 shown in Fig. 6D illustrate how the DNA methylation of LoDs changes during germ cell development. During development, the PGC genome undergoes global DNA demethylation, which is known to be completed between E11.5 and E13.5.2 In E10.5 PGCs, the LoDs tested here are not unmethylated completely, whereas demethylation of LoD DNA progresses in PGCs by E13.5. At E17.5, the LoD regions are largely unmethylated in both male and female PGCs. This trend persists in later stages of male germ cells, whereas the methylation levels of the LoDs appear to increase in newborn oocytes. The results together with the results shown in Supplementary Fig. S3 suggest that, in general, LoDs begin to form between E10.5 and E13.5, and distinct hypomethylated domains are established around E13.5 in the male germline. Despite the global increase in DNA methylation at later stages of male germline development (Fig. 2B), hypomethylation of LoD DNAs is maintained in male germ cells. Although LoDs 10 and 12 are shared by male and female PGCs, the overall DNA methylation patterns are not identical, suggesting that a distinct epigenomic status is generated in male and female germlines (Supplementary Fig. S3).
3.11. Coincidence of most LoDs with broad domains of the repressive histone mark, H3K9 dimethylation
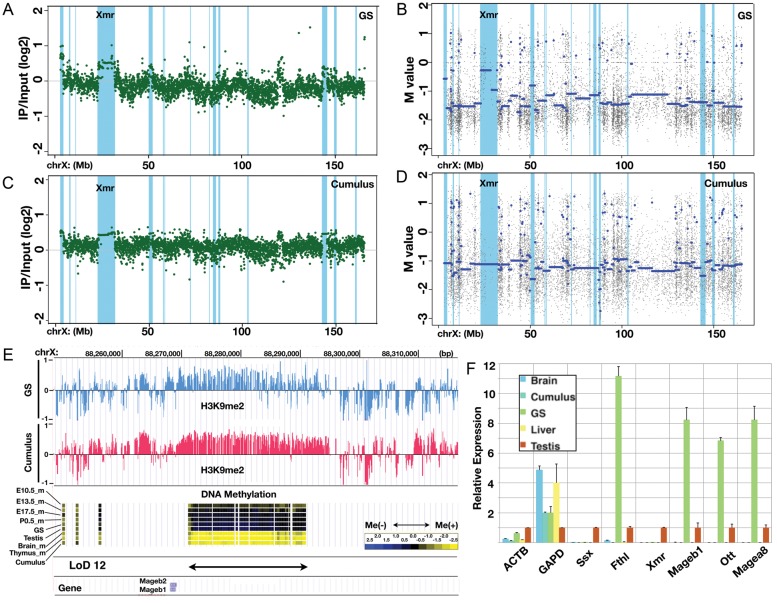
We have shown that most LoDs are broad genomic domains with low DNA methylation levels that form boundaries between the LoDs and other methylated parts of the genome. The mammalian genome can be divided into broad domains of distinct histone modifications.36,37 For example, LOCKs (large organized chromatin K9 modifications) are genomic domains with histone H3 lysine 9 dimethylation (H3K9me2) modification thought to be involved in region-wide gene repression.37 To investigate the relationship between LoDs and the repressive histone mark, we performed ChIP-on-chip analysis to detect H3K9me2 enrichment in GS cells as a representative of germ cells in this test and in cumulus (somatic cells in the ovary) cells as a somatic cell control.26 Figure 7 shows the H3K9me2 modification patterns on the X chromosome in both GS and cumulus cells. The overall pattern of H3K9me2 modifications along the X chromosome in GS cells is essentially similar to that in cumulus cells (Fig. 7A and C). Enrichment of the modifications along the LoD regions (coloured light blue) is seen in both GS and cumulus cells (Fig. 7A and C). In contrast, as expected, DNA methylation levels in the LoD regions are high in cumulus cells and low in GS cells (Fig. 7B and D). Figure 7E shows a magnified view of LoD 12, indicating that the hypomethylated region has the H3K9me2 mark. A significant enrichment of H3K9me2 is found in most (11 of 16) LoDs in GS cells (Supplementary Fig. S7).
Figure 7.
Large stretches of H3K9 dimethyl modifications overlapping LoDs. (A) ChIP-on-chip data for the H3K9me2 modification along the X chromosome in GS cells. The green dots represent individual probe data. The positions of LoDs are shown in light blue. (B) DNA methylation profile of GS cells within the X chromosome. The blue line denotes the average M-values. The positions of LoDs are shown in light blue. (C) ChIP-on-chip data for H3K9me2 modification along the X chromosome in cumulus cells. (D) The DNA methylation profile of cumulus cells within the X chromosome. (E) A magnified view of the LoD 12 region. Top, the H3K9me2 modification in GS cells (blue) and in cumulus cells (pink). Middle, DNA methylation data for E10.5 male PGCs, E13.5 male PGCs, E17.5 male PGCs, P0.5 spermatogonia, GS cells, testis, brain, thymus and cumulus cells. Bottom, positions of LoD and Mageb1/b2 genes. (F) Quantitative reverse transcription polymerase chain reaction (RT-PCR) expression analysis of genes in LoD regions. The Ssx9 (Ssx), Fthl17 (Fthl), Xmr, Mageb1, Ott and Magea8 genes are located within LoDs on the X chromosome. Beta-actin (ACTB) and Gapdh (GAPD) genes were also examined. The expression level in the testis is set at 1.0, and the expression levels in other cells and tissues (brain, cumulus cells, GS cells and liver) relative to that in the testis are shown. Primers used for this RT-PCR analysis are listed in Supplementary Table S4A.
Expression of six genes contained in the LoDs was examined in cumulus, GS, testis and two other somatic cell types by quantitative reverse transcription polymerase chain reaction (RT-PCR) analysis (Fig. 7F). Fthl17, Ott, Mageb and Magea are included in the LoDs and are expressed in the testis; the expression level of these genes is much higher in GS cells, but only negligible expression is detected in somatic cells. This result indicates that genes in hypomethylated LoDs can be expressed even though the same region has continuous H3K9me2 modifications (Fig. 7A and C and Supplementary Fig. S7), demonstrating peculiar epigenomic features of LoD regions. It is reasonable to expect that Ssx and Xmr are barely detectable in GS cells, because these genes become active in post-meiotic stages,16 whereas GS cells are derived from pre-meiotic spermatogonia. It is probable that, in GS cells, other factors required for the expression of post-meiotic genes (e.g. transcription factors) are lacking. These results suggest that DNA hypomethylation in LoDs may not be sufficient by itself, but is a prerequisite for the expression of LoD genes.
4. Discussion
In the present study, we have analysed the DNA methylation profiles of developing germ cells using the modified nanoHELP method, which requires only a limited amount of DNA. Recent studies by Guibert et al.9 using the methylated DNA immunoprecipitation analysis of a promoter array and Seisenberger et al.10 using a whole-genome bisulphite sequencing suggest that DNA demethylation of the PGC genome is initiated earlier than previously thought.1,2 Our finding that the PGC genome is substantially hypomethylated already at E10.5 is consistent with the result of Seisenberger et al.,10 confirming the technical reliability of our method. Our data from developing germ cells revealed for the first time the presence of large, hypomethylated DNA domains on the X chromosome of male germline cells in mice.
4.1. Discovery of large hypomethylated domains of epigenomic organization
Traditionally, epigenetic studies have focused on modifications of genes or elements adjacent to genes. However, with the development of genome-wide assays, recent studies have revealed marked clustering of particular histone modifications over relatively large genomic regions; e.g. LOCKs and BLOCs (broad local enrichments) enriched with the histone marks H3K9me2 and H3K27me3, respectively.36,37 These large epigenetic marks, LOCKs in particular, are thought to be involved in gene silencing. DNA methylation is found throughout the mammalian genome except for short unmethylated regions, CGIs, which typically occur around the transcription start sites of genes.6 The LoDs described in this work are also hypomethylated genomic regions, but are distinct from CGIs in terms of their size, tissue specificity and genomic structure. To our knowledge, large differentially methylated DNA regions showing germ cell specificities, such as LoDs, have not been previously reported. This may be because previous studies have focused only on methylation of gene promoters and not broader genomic contexts in germ cell samples. In contrast, our custom HELP chip method could assess the DNA methylation status of both genic and intergenic regions using the meager amounts of DNA that could be sampled from germ cell genomes in this study.
Seisenberger et al.10 recently reported the results of whole-genome bisulphite sequencing analysis of the mouse PGC genome. We analysed their data on E16.5 male PGCs to determine whether LoDs could be found at the single-nucleotide level and found that the number of sequence reads mapped to LoDs were significantly lower than that mapped to the flanking regions (data not shown). Given that mapping of bisulphite-converted short sequence reads onto locally duplicated regions is technically challenging, the probability of finding LoDs using the bisulphite sequencing data currently available seems low. In contrast, the HELP assay uses an MspI control to remove the potentially confounding effect of copy number variation12 along with a probe design that selects unique sequences for hybridization. These ensure that the DNA methylation readout from regions of segmental duplication is genuinely reflective of the underlying DNA methylation. Oda et al.38 reported that CGI methylation of an X-linked homeobox gene cluster spanning ∼1 Mb is under long-range regulation in a tissue-specific manner. Therefore, widespread changes in DNA methylation could occur depending on the cellular phenotype or differentiation status.
4.2. Peculiar epigenomic features of LoDs
LoDs have been detected based on arbitrary criteria, but most share common features. Most LoDs represent segmental duplications that harbour germline-expressed genes and overlap with large H3K9me2-enriched domains. Wen et al.37 described large H3K9me2-enriched chromatin blocks, LOCKs, in the human and mouse. The occurrence of LOCKs is differentiation specific: there are more LOCKs in differentiated cells, and genes contained in the LOCKs tend to be repressed during differentiation. Because LOCKs substantially overlap with lamin B-associated domains, a gene-silencing mechanism based on three-dimensional subnuclear organization has been proposed.37 We found that most LoDs are enriched with H3K9me2 modifications, and that at least four LoDs—1, 2, 3 and 4—correspond to the LOCKs described by Wen et al.37 (data not shown). Overlaps of other LoDs with LOCKs cannot be checked because LOCKs data are not available for the rest of the mouse X chromosome. Overlap of LoDs with LOCKs is counterintuitive because LOCKs are supposed to repress gene expression, whereas genes can be highly expressed within LoDs. This may be reconciled if we assume that gene silencing in LoDs is complete when both DNA methylation and H3K9me2 marks are established, but is derepressed in the absence of DNA methylation. Consistent with this idea, somatic cells such as cumulus cells, which have both marks, do not express the LoD genes, although germ cell genes can be active in DNA-hypomethylated but H3K9-dimethylated LoDs. The H3K9me2 histone methyltransferases, G9a and GLP, are required for DNA methylation in ES cells, but not in cancer cells.39 It is thus likely that DNA methylation and the H3K9me2 modification are not always interdependent, and that they can be regulated independently in the LoD regions of male germ cells and cancer cells.
4.3. Segmental duplication, hypomethylation and gene expression in germ cells and cancer cells
Through the analysis of germ cell-specific hypomethylated regions, we found that LoDs overlap with large segmentally duplicated regions, within which germ cell-expressed genes are commonly found. Some of these genes, such as Xmr, are found only in rodents. In contrast, the Mage gene family genes, Ssx and Fthl17, are conserved in the human genome and are known as CTA genes, which are expressed specifically in germ cells and in some tumour cell types. More than 260 CTA genes have been detected in the human (http://www.cta.lncc.br/), and half of them are on the X chromosome. Most of the X-linked CTA genes are organized as multicopy gene families.18 Warburton et al.40 searched the IR structures in the human genome and found that the X chromosome is replete with large IRs harbouring testis genes, most of which encode CTA genes. More than 40% of large IRs found in the mouse genome are on the X chromosome, and Ssx, Fthl17 and the Xmr loci are contained in such regions. Thus, three kinds of studies with different starting points reached the same conclusion: the X chromosome is abundant with duplicated regions containing germ cell-expressed genes, including CTA genes. To this, we add the new observation that these regions also have unique epigenomic features, i.e. widespread DNA hypomethylation and H3K9me2 enrichment. The epigenomic features of the LoDs could account for the finding that CTA genes can be activated by inhibition of DNA methylation but not by a reduction in H3K9 dimethylation,39,41 and suggest that DNA methylation is the key epigenetic mechanism involved with the regulation of LoD–CTA genes. It is not fully understood how DNA methylation regulates the coordinated expression of CTA genes in a cell type-specific manner. It is also necessary to clarify whether CTA gene expression contributes directly to oncogenesis or whether it simply reflects global chromatin changes that occur during tumour formation. Simpson et al.42 postulated an intriguing hypothesis that the aberrant expression of germline genes in cancer reflects the activation of the gametogenic programme, which is normally silenced in somatic cells. The gametogenic programme is normally repressed because germline-specific products would be harmful for normal somatic cells, whereas they would be advantageous for cancer cells. To test this hypothesis, it will be essential to elucidate the activation mechanism for the germline gene expression programme, as well as the epigenetic and chromatin status required for the operation of this programme. As shown in this study, widespread DNA hypomethylation may be a prerequisite for the activation of LoD genes, including CTA genes. In addition to DNA methylation, the nuclear chromatin environments within germ cells and/or tumour cells may also be important for long-range transcriptional control over large genomic regions, because LOCKs,37 LoDs and the partially methylated domains found in colorectal cancer7 are correlated with nuclear lamina-associated domains. Therefore, further studies of epigenomic features and the nuclear architecture of LoDs may shed light on the germline gene expression programme and its relationship to oncogenesis.42
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported, in part, by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan to K.A.
Supplementary Material
Acknowledgements
We wish to thank Drs T. Suzuki for help with bisulphite pyrosequencing, S. Matoba and A. Ogura for cumulus samples, and Y. Katsura and Y. Satta for information and discussion on segmentally duplicated regions of the X chromosome. We also thank Ms Y. Koga for help with the experiments.
Footnotes
Edited by Dr Toshihiko Shiroishi
References
- 1.Hajkova P., Erhardt S., Lane N., et al. Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 2.Hajkova P., Ancelin K., Waldmann T., et al. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–81. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McLaren A. Primordial germ cells in the mouse. Dev. Biol. 2003;262:1–15. doi: 10.1016/s0012-1606(03)00214-8. [DOI] [PubMed] [Google Scholar]
- 4.Borgel J., Guibert S., Li Y., et al. Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 2010;42:1093–100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 5.Meissner A., Mikkelsen T.S., Gu H., et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki M.M., Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–76. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 7.Berman B.P., Weisenberger D.J., Aman J.F., et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat. Genet. 2012;44:40–6. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Popp C., Dean W., Feng S., et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–05. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guibert S., Forné T., Weber M. Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res. 2012;22:633–41. doi: 10.1101/gr.130997.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seisenberger S., Andrews S., Krueger F., et al. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell. 2012;48:1–14. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hackett J.A., Sengupta R., Zylicz J.J., et al. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339:448–52. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khulan B., Thompson R.F., Ye K., Fazzari M.J., et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16:1046–55. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irizarry R.A., Ladd-Acosta C., Carvalho B., Wu H., Brandenburg S.A., Wen B., Feinberg A.P. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:780–90. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oda M., Glass J.L., Thompson R.F., et al. High-resolution genome-wide cytosine methylation profiling with simultaneous copy number analysis and optimization for limited cell numbers. Nucleic Acids Res. 2009;37:3829–39. doi: 10.1093/nar/gkp260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang P.J., McCarrey J.R., Yang F., Page D.C. An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 2001;27:422–6. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- 16.Mueller J.L., Mahadevaiah S.K., Park P.J., Warburton P.E., Page D.C., Turner J.M.A. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat. Genet. 2008;40:794–9. doi: 10.1038/ng.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugimoto M., Abe K. X chromosome reactivation initiates in nascent primordial germ cells in mice. PLoS Genet. 2007;3:e116. doi: 10.1371/journal.pgen.0030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caballero O.L., Chen Y.-T. Cancer/testis (CT) antigens: potential targets for immunotherapy. Cancer Sci. 2009;100:2014–21. doi: 10.1111/j.1349-7006.2009.01303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tada T., Tada M., Hilton K., et al. Epigenotype switching of imprintable loci in embryonic germ cells. Dev. Genes Evol. 1998;207:551–61. doi: 10.1007/s004270050146. [DOI] [PubMed] [Google Scholar]
- 20.Mise N., Fuchikami T., Sugimoto M., et al. Differences and similarities in the developmental status of embryo-derived stem cells and primordial revealed by global expression profiling. Genes Cells. 2008;13:863–77. doi: 10.1111/j.1365-2443.2008.01211.x. [DOI] [PubMed] [Google Scholar]
- 21.Ohbo K., Yoshida S., Ohmura M., et al. Identifi-cation and characterization of stem cells in prepubertal spermatogenesis in mice. Dev. Biol. 2003;258:209–25. doi: 10.1016/s0012-1606(03)00111-8. [DOI] [PubMed] [Google Scholar]
- 22.Kanatsu-Shinohara M., Ogonuki N., Inoue K., Miki H., Ogura A., Toyokuni S., Shinohara T. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol. Reprod. 2003;69:612–6. doi: 10.1095/biolreprod.103.017012. [DOI] [PubMed] [Google Scholar]
- 23.Ogonuki N., Inoue K., Hirose M., et al. A high-speed congenic strategy using first-wave male germ cells. PLoS One. 2009;4:e4943. doi: 10.1371/journal.pone.0004943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi S., Fujihara Y., Mise N., et al. The X-linked imprinted gene family Fthl17 shows predominantly female expression following the two-cell stage in mouse embryos. Nucleic Acids Res. 2010;38:3672–81. doi: 10.1093/nar/gkq113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ko M., Ko S., Takahashi N., Nishiguchi K., Abe K. Unbiased amplification of a highly complex mixture of DNA fragments by ‘lone linker’-tagged PCR. Nucleic Acids Res. 1990;18:4293–4. doi: 10.1093/nar/18.14.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inoue K., Kohda T., Sugimoto M., et al. Impeding Xist expression from the active X chromosome improves mouse somatic cell cloning. Science. 2010;330:496–9. doi: 10.1126/science.1194174. [DOI] [PubMed] [Google Scholar]
- 27.Otsu N. Threshold selection method from gray-level histograms. IEEE Trans. Syst. Man. Cybern. 1979;9:62–6. [Google Scholar]
- 28.Mahalanobis P.C. On the generalized distance in statistics. Proc. Natl Inst. Sci. India. 1936;2:49. [Google Scholar]
- 29.Reynard L.N., Turner J.M.A., Cocquet J., et al. Expression analysis of the mouse multi-copy X-linked gene, Xlr-related, meiosis-regulated (Xmr), reveals that Xmr encodes a spermatid-expressed cytoplasmic protein, SLX/XMR. Biol. Reprod. 2007;77:329–35. doi: 10.1095/biolreprod.107.061101. [DOI] [PubMed] [Google Scholar]
- 30.Cocquet J., Ellis P.J.I., Mahadevaiah S.K., Affara N.A., Vaiman D., Burgoyne P.S. A genetic basis for a postmeiotic X versus Y chromosome intragenomic conflict in the mouse. PLoS Genet. 2012;8:e1002900. doi: 10.1371/journal.pgen.1002900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clotman F., De Backer O., De Plaen E., Boon T., Picard J. Cell- and stage-specific expression of mage genes during mouse spermatogenesis. Mammal. Genome. 2000;11:696–9. doi: 10.1007/s003350010116. [DOI] [PubMed] [Google Scholar]
- 32.Olshen A., Venkatraman E., Lucito R., Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–72. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 33.Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–82. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 34.Bailey J., Yavor A., Massa H., Trask B., Eichler E. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–17. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujiwara Y., Komiya T., Kawabata H., et al. Isolation of a DEAD-family protein gene that encodes a murine homolog of Drosophila vasa and its specific expression in germ cell lineage. Proc. Natl Acad. Sci. USA. 1994;91:12258–62. doi: 10.1073/pnas.91.25.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pauler F.M., Sloane M.A., Huang R., et al. H3K27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome. Genome Res. 2009;19:221–33. doi: 10.1101/gr.080861.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wen B., Wu H., Shinkai Y., Irizarry R.A., Feinberg A.P. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat. Genet. 2009;41:246–50. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oda M., Yamagiwa A., Yamamoto S., et al. DNA methylation regulates long-range gene silencing of an X-linked homeobox gene cluster in a lineage-specific manner. Genes Dev. 2006;20:3382–94. doi: 10.1101/gad.1470906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Link P.A., Gangisetty O., James S.R., et al. Distinct roles for histone methyltransferases G9a and GLP in cancer germ-line antigen gene regulation in human cancer cells and murine embryonic stem cells. Mol. Cancer Res. 2009;7:851–62. doi: 10.1158/1541-7786.MCR-08-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warburton P., Giordano J., Cheung F., Gelfand Y., Benson G. Inverted repeat structure of the human genome: the X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome Res. 2004;14:1861–9. doi: 10.1101/gr.2542904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karimi M.M., Goyal P., Maksakova I.A., et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell. 2011;8:676–87. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simpson A.J.G., Caballero O.L., Jungbluth A., Chen Y.-T., Old L.J. Cancer/testis antigens, gametogenesis and cancer. Nat. Rev. Cancer. 2005;5:615–25. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 43.Pohl A., Sugnet C., Clark T., Smith K., Fujita P., Cline M.S. Affy exon tissues: exon levels in normal tissues in human, mouse and rat. Bioinformatics. 2009;25:2442–3. doi: 10.1093/bioinformatics/btp414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohtsubo Y., Ikeda-Ohtsubo W., Nagata Y., Tsuda M. GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinformatics. 2008;9:376. doi: 10.1186/1471-2105-9-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.