Abstract
Nonsyndromic oral clefts (NSOC) are the most common craniofacial birth defects in humans. The etiology of NSOC is complex, involving both genetic and environmental factors. Several genes that play a role in cellular proliferation, differentiation, and apoptosis have been associated with clefting. For example, variations in the homeobox gene family member MSX1, including a CA repeat located within its single intron, may play a role in clefting. The aim of this study was to investigate the association between MSX1 CA repeat polymorphism and NSOC in a Southern Brazilian population using a case-parent triad design. We studied 182 nuclear families with NSOC recruited from the Hospital de Clínicas de Porto Alegre in Southern Brazil. The polymorphic region was amplified by the polymerase chain reaction and analyzed by using an automated sequencer. Among the 182 families studied, four different alleles were observed, at frequencies of 0.057 (175 bp), 0.169 (173 bp), 0.096 (171 bp) and 0.67 (169 bp). A transmission disequilibrium test with a family-based association test (FBAT) software program was used for analysis. FBAT analysis showed overtransmission of the 169 bp allele in NSOC (P=0.0005). These results suggest that the CA repeat polymorphism of the MSX1 gene may play a role in risk of NSOC in populations from Southern Brazil.
Keywords: MSX1, Cleft lip and palate, Oral clefts
Introduction
Nonsyndromic oral clefts (NSOC) are the most common craniofacial birth defects in humans. The overall prevalence is approximately 1 in 700 live births, with variation among ethnic groups and geographic regions. Affected individuals need multidisciplinary care from birth until adulthood (1). Forms of NSOC include cleft lip with or without cleft palate (CL/P) and cleft palate only (CPO) (2).
Normal development of the lip and palate involves a series of closely coordinated events during development of the facial primordia that includes cell migration, growth, differentiation, and apoptosis (1). Perturbations in these events via genetic or environmental factors may affect proper morphogenesis of facial structures (2). The identification of genetic factors affecting NSOC has been the subject of intensive research in the last decade. Several oral cleft loci have been mapped, and the list of candidate genes includes IRF6, TGFA, MSX1, TGFB3, and FOXE1, among others (3). Recent epidemiological and genetic studies have shown that the phenotypes of NSOC have different etiologies. IRF6 plays a major role in cleft lip only (CLO) and FOXE1 in CPO (4).
The Msh homeobox 1 (MSX1) gene is strongly expressed in cranial neural crest (CNC) cells. CNC cells are pluripotent cells derived from the lateral ridges of the neural plate. These cells migrate ventrolaterally to the frontonasal prominence and the branchial arches and give rise to the jaws and the anterior skull (5). MSX1 mutations are found in 2% of cases of oral cleft (6). MSX1 has been mapped to chromosome 4p16. It consists of two exons and one intron spanning fewer than 4,300 bp. Build 129 of dbSNP reports 78 known single nucleotide polymorphisms (SNPs) in or near this gene, including 6 in the coding sequence (7). A CA repeat is located within the single intron in MSX1, and there is no apparent functional role for the allele size variation. However, there is evidence that an MSX1 antisense transcript, which encompasses the CA repeat, may be involved in craniofacial development (8). The aim of the present study was to investigate the association between MSX1 CA repeat polymorphisms and NSOC in a Southern Brazilian population.
Patients and Methods
A total of 182 nuclear pedigrees were enrolled. All affected individuals had CL/P or CPO and were recruited at the Hospital de Clínicas de Porto Alegre in Southern Brazil. All cases were nonsyndromic, and individuals did not present with any other birth defect, syndrome, or intellectual disability. This research protocol was approved by the Research Ethics Committee of the Hospital de Clínicas de Porto Alegre, and all individuals provided written informed consent.
DNA was extracted from blood samples according to the manufacturer's instructions (Gentra Puregene, Qiagen Inc., USA). The CA repeat alleles were identified by polymerase chain reaction (PCR) and fragment size analysis, as described by Hwang et al. (9). The primer sequences were: forward 5′-6-FAM-GGG CAT GTT GAT GTC TGC TGAC-3′ and reverse 5′-TTA GAT TGT CAT CAG TCCTC-3′.
PCR was performed in a reaction volume of 20 µL containing 20.0 µmol of each primer, 6.0 µM deoxyribonucleotide triphosphates (dNTPs; GeneAmp¯ dNTP, Applied Biosystems, USA), 0.5 units DNA polymerase (AmpliTaq¯ Gold, Applied Biosystems), and 2.0 ng DNA. The PCR conditions consisted of an initial denaturation at 94°C for 6 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 59°C for 30 s, and extension at 72°C for 1 min. The amplified fragments were analyzed with an ABI 3130X automated sequencer (Applied Biosystems) and with the Genescan and Genotyper software packages (Applied Biosystems) to distinguish the four CA repeat alleles (175, 173, 171, and 169 bp).
Statistical analysis was performed with the transmission disequilibrium test (TDT) in a family-based association test software program (FBAT; version 2.0.3: http://www.biostat.harvard.edu/fbat/fbat.htm) (10). TDT is a family-based study that compares the frequencies of alleles transmitted to the affected child to the alleles not transmitted by a heterozygous parent. It is a powerful test for association of alleles in the presence of population admixture (11).
Results
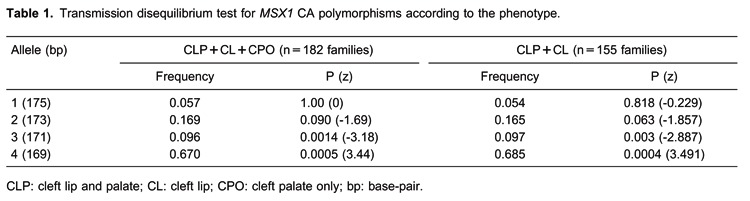
We studied 182 nuclear families, including 83 complete case-parent triads, totaling 462 individuals. Of the 182 probands, 94 (52%) were male and 88 (48%) were female. CL/P was more frequent in males (56%) and CPO was more frequent in females (73%). Among all of the individuals studied, most cases had CL/P (156 cases; 85.7%) and the rest had CPO (26 cases; 14.3%). Table 1 shows the TDT results. We observed overtransmission of the 169-bp allele in overall NSOC (P=0.0005). Analysis of the association of the CA repeat according to the phenotype was performed. We observed overtransmission of the 169-bp allele in cleft lip with or without cleft palate (P=0.0004). The CPO and CL groups could not be evaluated alone due to small sample size.
Discussion
Homeobox genes are highly conserved among various species and function as transcriptional regulators that control cellular proliferation, differentiation, and apoptosis, which are important for balanced cell growth and morphogenesis during normal embryonic development (5). MSX1 first emerged as a candidate gene for clefting in the Msx1 knockout mouse that had an abnormal phenotype, which included a foreshortened maxilla and CL/P (12). In humans, the first MSX1 mutation was identified in a family with autosomal dominant tooth agenesis (13). A subsequent study in a Dutch family with tooth agenesis and various combinations of CL/P and CPO found a nonsense mutation (Ser104stop) in exon 1 (14).
Another nonsense mutation (Q189X) in the homeodomain of MSX1, first identified in a Chinese family with a phenotype including tooth agenesis with cleft lip, was found to cause dramatically reduced mRNA expression compared to wild-type MSX1 mRNA. The authors suggested that most MSX1 mutations in the homeodomain cause isolated tooth agenesis, and mutations outside the homeodomain are associated with CL/P (15).
Jezewski et al. (6) performed complete sequencing of MSX1 in over 900 subjects with NSOC and identified several novel variants that may be directly causal. They also found three intronic variants that were associated with NSOC in Asian and Caucasian populations (6). Several studies have identified an association between the MSX1 gene and CL/P and CPO in different ethnic backgrounds (16- 22). However, a Chinese study found no association between three MSX1 variants and NSOC (23), similar to a recent study in the northeast of Brazil that found no association between six polymorphisms in the MSX1 gene and CL/P (24).
MSX1 antisense RNA is synthesized starting from the MSX1 3′-UTR to the middle of the intron and encompasses the CA repeat. A heterozygote for the CA marker could have a mismatch when the endogenous antisense RNA is binding to its corresponding sense, causing mRNA degradation and loss of function, thereby increasing the risk for clefting (16).
Previous case-parent trio studies of South American (Argentina, Bolivia, Brazil, Chile, Ecuador, Paraguay, Uruguay, and Venezuela) families with NSOC reported significant associations between the 169-bp allele of the MSX1 CA repeat polymorphism and CL/P (16,25). A case-control study in a Colombian population found an association between the 171-bp allele of the CA repeat and CL/P (26). These different results may be due to variations in the degree of admixture across the South American continent.
This study evaluated the association between the CA repeat polymorphism in the MSX1 gene and NSOC in a Southern Brazilian population, using a case-parent trio design. For the analysis, we used TDT, which allowed for the inclusion of cases of different ethnic origins and avoided the complications of mixed ancestry that can arise in case-control studies. This aspect is important in our study, because the Brazilian population represents an ethnic admixture of Europeans, Africans, and Amerindians, making it difficult to determine the ethnicity of cases and controls in our sample population (27,28).
The TDT results showed an association between the 169-bp allele and CL/P in our population of 182 familial trios. We found no association between any CA repeat polymorphism and CPO or CL due to the small sample size of our study. Because this result differs from previous studies in populations with different ethnic backgrounds, further studies are needed to confirm this association and to elucidate the role of the MSX1 intron region in NSOC.
Acknowledgments
Research supported by Fundo de Incentivo à Pesquisa e Eventos (FIPE), Hospital de Clínicas de Porto Alegre, and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Footnotes
First published online July 16, 2013.
References
- 1.Mossey PA, Little J, Munger RG, Dixon MJ, Shaw WC. Cleft lip and palate. Lancet. 2009;374:1773–1785. doi: 10.1016/S0140-6736(09)60695-4. [DOI] [PubMed] [Google Scholar]
- 2.Butali A, Mossey PA, Adeyemo WL, Jezewski PA, Onwuamah CK, Ogunlewe MO, et al. Genetic studies in the Nigerian population implicate an MSX1 mutation in complex oral facial clefting disorders. Cleft Palate Craniofac J. 2011;48:646–653. doi: 10.1597/10-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jugessur A, Murray JC. Orofacial clefting: recent insights into a complex trait. Curr Opin Genet Dev. 2005;15:270–278. doi: 10.1016/j.gde.2005.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixon MJ, Marazita ML, Beaty TH, Murray JC. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 2011;12:167–178. doi: 10.1038/nrg2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han J, Ishii M, Bringas P, Jr, Maas RL, Maxson RE, Jr, Chai Y. Concerted action of Msx1 and Msx2 in regulating cranial neural crest cell differentiation during frontal bone development. Mech Dev. 2007;124:729–745. doi: 10.1016/j.mod.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jezewski PA, Vieira AR, Nishimura C, Ludwig B, Johnson M, O'Brien SE, et al. Complete sequencing shows a role for MSX1 in non-syndromic cleft lip and palate. J Med Genet. 2003;40:399–407. doi: 10.1136/jmg.40.6.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingersoll RG, Hetmanski J, Park JW, Fallin MD, McIntosh I, Wu-Chou YH, et al. Association between genes on chromosome 4p16 and non-syndromic oral clefts in four populations. Eur J Hum Genet. 2010;18:726–732. doi: 10.1038/ejhg.2009.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blin-Wakkach C, Lezot F, Ghoul-Mazgar S, Hotton D, Monteiro S, Teillaud C, et al. Endogenous Msx1 antisense transcript: in vivo and in vitro evidences, structure, and potential involvement in skeleton development in mammals. Proc Natl Acad Sci U S A. 2001;98:7336–7341. doi: 10.1073/pnas.131497098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang SJ, Beaty TH, McIntosh I, Hefferon T, Panny SR. Association between homeobox-containing gene MSX1 and the occurrence of limb deficiency. Am J Med Genet. 1998;75:419–423. doi: 10.1002/(SICI)1096-8628(19980203)75:4<419::AID-AJMG14>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 10.Rabinowitz D, Laird N. A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum Hered. 2000;50:211–223. doi: 10.1159/000022918. [DOI] [PubMed] [Google Scholar]
- 11.Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- 12.Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 1994;6:348–356. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- 13.Vastardis H, Karimbux N, Guthua SW, Seidman JG, Seidman CE. A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat Genet. 1996;13:417–421. doi: 10.1038/ng0896-417. [DOI] [PubMed] [Google Scholar]
- 14.van den Boogaard MJ, Dorland M, Beemer FA, van Amstel HK. MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat Genet. 2000;24:342–343. doi: 10.1038/74155. [DOI] [PubMed] [Google Scholar]
- 15.Liang J, Zhu L, Meng L, Chen D, Bian Z. Novel nonsense mutation in MSX1 causes tooth agenesis with cleft lip in a Chinese family. Eur J Oral Sci. 2012;120:278–282. doi: 10.1111/j.1600-0722.2012.00965.x. [DOI] [PubMed] [Google Scholar]
- 16.Vieira AR, Orioli IM, Castilla EE, Cooper ME, Marazita ML, Murray JC. MSX1 and TGFB3 contribute to clefting in South America. J Dent Res. 2003;82:289–292. doi: 10.1177/154405910308200409. [DOI] [PubMed] [Google Scholar]
- 17.Lidral AC, Romitti PA, Basart AM, Doetschman T, Leysens NJ, Daack-Hirsch S, et al. Association of MSX1 and TGFB3 with nonsyndromic clefting in humans. Am J Hum Genet. 1998;63:557–568. doi: 10.1086/301956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanco R, Chakraborty R, Barton SA, Carreno H, Paredes M, Jara L, et al. Evidence of a sex-dependent association between the MSX1 locus and nonsyndromic cleft lip with or without cleft palate in the Chilean population. Hum Biol. 2001;73:81–89. doi: 10.1353/hub.2001.0002. [DOI] [PubMed] [Google Scholar]
- 19.Jugessur A, Lie RT, Wilcox AJ, Murray JC, Taylor JA, Saugstad OD, et al. Variants of developmental genes (TGFA, TGFB3, and MSX1) and their associations with orofacial clefts: a case-parent triad analysis. Genet Epidemiol. 2003;24:230–239. doi: 10.1002/gepi.10223. [DOI] [PubMed] [Google Scholar]
- 20.Park J, Park BY, Kim HS, Lee JE, Suh I, Nam CM, et al. MSX1 polymorphism associated with risk of oral cleft in Korea: evidence from case-parent trio and case-control studies. Yonsei Med J. 2007;48:101–108. doi: 10.3349/ymj.2007.48.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu PA, Li YL, Fan GZ, Wang K. [Association study on microsatellite polymorphisms of MSX1 gene and nonsyndromic cleft lip and palate] Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2007;24:325–327. [PubMed] [Google Scholar]
- 22.Vieira AR, McHenry TG, Daack-Hirsch S, Murray JC, Marazita ML. A genome wide linkage scan for cleft lip and palate and dental anomalies. Am J Med Genet A. 2008;146A:1406–1413. doi: 10.1002/ajmg.a.32295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang YQ, Ma J, Ma M, Deng Y, Li YD, Ren HW, et al. Association between MSX1 variants and oral clefts in Han Chinese in western China. DNA Cell Biol. 2011;30:1057–1061. doi: 10.1089/dna.2010.1208. [DOI] [PubMed] [Google Scholar]
- 24.Cardoso M, Bezerra J, Oliveira G, Soares C, Oliveira S, de Souza K, et al. MSX1 gene polymorphisms in non-syndromic cleft lip and/or palate. Oral Dis. 2013;19:507–512. doi: 10.1111/odi.12033. [DOI] [PubMed] [Google Scholar]
- 25.Suazo J, Santos JL, Carreno H, Jara L, Blanco R. Linkage disequilibrium between MSX1 and non-syndromic cleft lip/palate in the Chilean population. J Dent Res. 2004;83:782–785. doi: 10.1177/154405910408301009. [DOI] [PubMed] [Google Scholar]
- 26.Otero L, Gutierrez S, Chaves M, Vargas C, Bermudez L. Association of MSX1 with nonsyndromic cleft lip and palate in a Colombian population. Cleft Palate Craniofac J. 2007;44:653–656. doi: 10.1597/06-097.1. [DOI] [PubMed] [Google Scholar]
- 27.Alves-Silva J, da Silva Santos M, Guimaraes PE, Ferreira AC, Bandelt HJ, Pena SD, et al. The ancestry of Brazilian mtDNA lineages. Am J Hum Genet. 2000;67:444–461. doi: 10.1086/303004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carvalho-Silva DR, Santos FR, Rocha J, Pena SD. The phylogeography of Brazilian Y-chromosome lineages. Am J Hum Genet. 2001;68:281–286. doi: 10.1086/316931. [DOI] [PMC free article] [PubMed] [Google Scholar]


