Abstract
Yellow mosaic disease (YMD) is one of the major diseases affecting mungbean (Vigna radiata (L.) Wilczek). In this study, we report the mapping of the quantitative trait locus (QTL) for mungbean yellow mosaic India virus (MYMIV) resistance in mungbean. An F8 recombinant inbred line (RIL) mapping population was generated in Thailand from a cross between NM10-12-1 (MYMIV resistance) and KPS2 (MYMIV susceptible). One hundred and twenty-two RILs and their parents were evaluated for MYMIV resistance in infested fields in India and Pakistan. A genetic linkage map was developed for the RIL population using simple sequence repeat (SSR) markers. Composite interval mapping identified five QTLs for MYMIV resistance: three QTLs for India (qYMIV1, qYMIV2 and qYMIV3) and two QTLs for Pakistan (qYMIV4 and qYMIV5). qYMIV1, qYMIV2, qYMIV3, qYMIV4 and qYMIV5 explained 9.33%, 10.61%, 12.55%, 21.93% and 6.24% of variation in disease responses, respectively. qYMIV1 and qYMIV4 appeared to be the same locus and were common to a major QTL for MYMIV resistance in India identified previously using a different resistant mungbean.
Keywords: mungbean, mungbean yellow mosaic virus, MYMIV, QTL, SSR
Introduction
Mungbean (Vigna radiata (L.) Wilczek) is an important legume crop of South and Southeast Asia. The total planting area of mungbean is about 6 M ha. Mungbean is relatively tolerant to drought and can be harvested within 60–75 days, so it can be grown as a sole crop or as a component in various cropping systems for its dry seeds, which are rich in proteins, amino acids and minerals. Several foods are made from mungbean seeds. Apart from the seeds, mungbean sprouts are very popular in Asian cuisine.
One of the most destructive and important diseases of mungbean is yellow mosaic disease (YMD). The disease mainly occurs in South Asian countries, viz. India, Sri Lanka, Pakistan and Bangladesh. The disease can reduce mungbean seed yield by up to 100% or even kill a plant infected at an early vegetative stage. YMD is caused by geminivirus (genus Begomovirus, family Geminiviridae), which has bipartite genomes (DNA A and DNA B). Begomoviruses cause mungbean yellow mosaic virus (MYMV), mungbean yellow mosaic India virus (MYMIV), Dolichos yellow mosaic virus (DoYMV) and horsegram yellow mosaic virus (HgYMV), which have spread throughout South Asia. MYMV and MYIMV are the main pathogens causing YMD in mungbean grown in India (Hussain et al. 2004, Pant et al. 2001), while only MYIMV is responsible for YMD in Pakistan (Ilyas et al. 2010). The viruses are transmitted by whitefly (Bemisia tabaci) and can infect mungbean at all growth stages. Disease symptoms after virus infection depend on the host species and susceptibility of each plant. Disease symptoms vary from a few small yellow specks or spots on a few leaves, to yellowing or chlorosis of all leaves of the whole plant followed by necrosis. In highly susceptible plants, symptoms includes shortening of internodes, severe stunting of plants with no yield or few flowers and deformed pods producing small, immature and shriveled seeds.
Chemical control of the virus vector, whitefly, is ineffective and is detrimental to the environment. Using resistant cultivars is the best option to manage YMD. Improved resistance to YMD is now the major goal in mungbean breeding programs in several mungbean production countries. Resistance to YMD in mungbean was reported to be controlled by a single major recessive gene with modifiers (Malik et al. 1986, Thakur et al. 1997), or two recessive genes (Dhole and Reddy 2012) or complementary recessive genes (Shukla and Pandya 1985). However, a major difficulty in breeding YMD-resistant mungbean is field screening for the disease, which is hampered by non-uniform development of the disease due to fluctuation of the whitefly population in different locations and different seasons. Marker-assisted selection (MAS) is a useful tool for genetic improvement of crops. Selection based on molecular markers associated with the target trait helps in decreasing the number of phenotypic evaluations and thus reducing time and cost and increasing gain from selection. So far, there are few reports on DNA markers associated with YMD resistance in mungbean or blackgram, a related species of mungbean and all except one were based on gene tagging. Maiti et al. (2011) reported that resistance gene analog (RGA) markers, YR4 and CYR1, were associated with resistance to MYMIV in blackgram (Vigna mungo (L.) Hepper). CYR1 cosegregated with the resistance in F2 and F3 populations from the cross between resistant and susceptible blackgram and was completely linked with the MYMIV-resistant blackgram germplasm. They further demonstrated that marker CYR1 was also associated with resistance in the mungbean germplasm. Recently, Dhole and Reddy (2013) reported that SCAR marker MYMVR-583 is associated with MYMV resistance in mungbean. The marker is 6.8 cM from the resistance gene. Gupta et al. (2013) reported that CEDG180 is linked to MYMIV resistance in blackgram at a distance of 12.9 cM. In a recent study, which is the only QTL mapping study of YMD resistance in mungbean, Chen et al. (2013) identified four major QTLs on three different linkage groups for MYMIV resistance using AFLP and SSR markers.
We report quantitative trait locus (QTL) mapping of MYMIV resistance in mungbean using SSR markers from mungbean and other related legumes in this paper. The objective of this study was to locate and compare QTLs for MYMIV resistance in India and Pakistan.
Materials and Methods
Plant materials and DNA extraction
A population of 122 recombinant inbred lines (RILs) was developed from a cross between Kamphaeng Saen 2 (KPS2) (female parent) and NM10-12-1 (male parent). KPS2 is a popular high-yielding mungbean variety grown in Thailand but is susceptible to MYMIV, while NM10-12-1 is a breeding line from Pakistan resistant to MYMIV. The RILs were developed at Kasetsart University, Kamphaeng Saen, Thailand by the single seed descent method from individual F2 plants until F8 generation. This population is the same as that used by Prathet et al. (2012) for mapping QTLs conferring resistance to iron-deficient chlorosis caused by calcareous soil.
MYMIV evaluation
YMD resistance was evaluated in India and Pakistan. In India, the RIL population and parents were evaluated for resistance under field conditions with a high population of whitefly and severe incidence of MYMIV at Punjab Agricultural University, Ludhiana, Punjab, India during August to October 2012. This location is the same site as that used for MYMIV evaluation, reported recently by Chen et al. (2013). The RILs and parents were sown in a randomized complete block design (RCBD) with three replications. Each replicate had 15 plants in a row of 3 m in length with 40-cm inter-row distance. A local susceptible check variety was sown after every four rows of the tested entries. Thirty days after planting (DAP), infection and disease severity were visually scored for MYMIV. In each replication, 10 to 15 plants (depending on germination) of each entry were scored separately by two trained staff. The plants were scored using the following scale of 1 (highly resistant) to 5 (highly susceptible) (Table 1).
Table 1.
Disease scales for scoring of mungbean yellow mosaic India virus in India and Pakistan
| India | |||
|---|---|---|---|
|
| |||
| Score | Symptoms | Disease reaction | |
| 0 | No visible symptoms on leaves or very minute yellow specks (1–2) on leaves | Highly resistant | |
| 1 | Small yellow specks with restricted spread covering up to 5 % leaf area | Resistant | |
| 2 | Yellow mottling covering up to 15% leaf area | Moderately resistant | |
| 3 | Yellow mottling and discoloration of up to 30% leaf area | Moderately susceptible | |
| 4 | Pronounced yellow mottling and discoloration of leaves and pods, reduction in leaf size and stunting of plants covering up to 75% of foliage | Susceptible | |
| 5 | Severe yellow discoloration of leaves covering >75% of foliage, stunting of plants and reduction in pod size | Highly susceptible | |
|
| |||
| Pakistan | |||
|
| |||
| Score | Symptoms | Percentage of disease indexa | Disease reaction |
|
| |||
| 0 | Complete absence of symptoms | 0 | Immune |
| 1 | Few small yellow specks or spots on few leaves seen | 0.01–10 | Highly resistant |
| 2 | Bright yellow specks or spots common on leaves, easily observed and some coalesced | 10.01–25 | Resistant |
| 3 | Mostly coalesced bright yellow specks or spots common on leaves, but no or minor reduction in yield | 25.01–40 | Moderately resistant |
| 4 | Plants showing coalesced bright yellow specks or spots on all leaves, with no or minor stunting and set fewer normal pods | 40.01–60 | Susceptible |
| 5 | Yellowing or chlorosis of all leaves on whole plant followed by necrosis, shortening of internode, severe stunning of plants with no yield or few flowers and produce deformed pods | >60.01 | Highly susceptible |
Calculated as (sum of all disease ratings/total number of plants scored) × 20.
For YMD evaluation in Pakistan, the parents and the RIL population were evaluated for resistance at the Nuclear Institute for Agricultural Biology, Faisalabad, Pakistan in July 2008. They were evaluated under field conditions with a high incidence of whiteflies and MYMIV. Individual RIL and each parent were arranged in a RCBD with three replications. Each replication had 24 to 28 plants in a row of 4 m in length with 40 cm inter-row distance. The parents were sown as checks after every two tested entries. Infection and disease severity were visually scored for MYMIV reactions at 30 DAP. The scoring system was different from that used in India. Experimental units (each plant of each RIL) were scored using scale of 1 (immune) to 5 (highly susceptible) (Table 1). Then, the score was used to calculate the percentage disease index for each line following Akhkar et al. (2009).
Genetic linkage map and analysis of new markers
A genetic linkage map previously developed for the same RIL population to locate QTLs controlling resistance to calcareous soil (Prathet et al. 2012) was used in this study. In brief, the population was analyzed with 62 polymorphic SSR markers screened from 1,203 SSR markers derived from mungbean, azuki bean (Vigna angularis), cowpea (Vigna unguiculata) and common bean (Phaseolus vulgaris), together with 8 polymorphic AFLP markers generated from 4 primer combinations. The linkage map was constructed using software JoinMap 3.0 (Van Ooijen and Voorrips 2001). The polymorphic markers were assigned to linkage groups (LGs) using a minimum logarithm of an odds (LOD) score of 4.0 and maximum recombination frequency (r) of 0.5. The Kosambi mapping function (Kosambi 1944) was used to calculate the genetic distance between markers. Linkage groups were named following the azuki bean linkage map (Han et al. 2005). The analysis assigned 54 polymorphic markers to 12 linkage groups (LG) with a total genetic distance of 991.98 cM, while 16 markers were unlinked (Prathet et al. 2012). The shortest and the longest LGs were 23.66 cM and 139.45 cM, respectively, with the average total distance per LG of 82.66 cM. The number of markers per LG was 2 (LG 9B) to 8 (LG 5), with an average of 4.5.
To confirm the association between the markers, CYR1, YR4 and MYMIV resistance in mungbean (Maiti et al. 2011), these markers were screened for polymorphism between the parents. Total genomic DNA was isolated from young leaves of NM10-12-1 and KPS2 using the CTAB method described by Lodhi et al. (1994) with a slight modification. The concentration of DNA was determined by comparing against lamda DNA and diluted to 1 ng/μl for marker analysis. PCR was performed using a total volume of 10 μl containing 1 ng genomic DNA, 1x Taq buffer, 2 mM MgCl2, 0.2 mM dNTPs, 1 U Taq DNA polymerase (Fermentas) and 5 pmol each of forward and reverse primers. The DNA was amplified in a GeneAmp PCR System 9700 (Applied Biosystems) under the following conditions: 94°C for 2 min followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1 min, with a final extension step of 72°C for 10 min. The PCR products were separated on 5% denaturing polyacrylamide gel and visualized by silver staining.
QTL analysis
Markers associated with QTLs for MYMIV resistance were determined by single regression analysis at P = 0.001 using R-program 2.10.0. To locate the position of the QTLs on the linkage map, composite interval mapping (CIM) was performed with software WinQTL Cartographer 2.5 (Wang et al. 2010). Significant LOD thresholds value for declaring QTLs were computed by a 1,000 run of permutation tests at P = 0.05, P = 0.01 and P = 0.001.
Results
MYMIV resistance in the RIL population in India and Pakistan
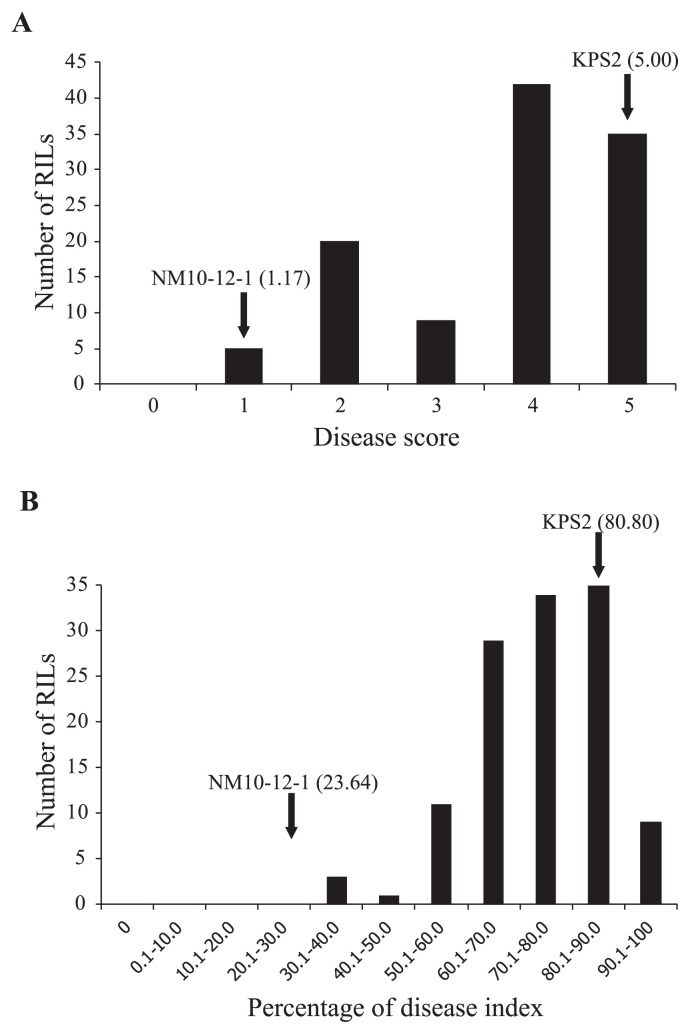
Resistance to MYMIV in the RILs and parents was evaluated under field conditions in India and Pakistan using different scoring systems. In India, 11 lines did not germinate and one of the three replicates had to be discarded due to poor stand from soil-borne disease infection in several plots. KPS2 and NM10-12-1 showed highly susceptible and resistant reactions with scores of 5.00 and 1.17, respectively. The disease scores among the RILs varied from 0.71 to 5.00 with an average of 3.73 (Supplemental Table 1). The correlation coefficient (r) of the disease score between the two replications was moderate, being 0.64 and significant (P < 0.0001, df = 108). Frequency distribution of the disease scores in the RILs showed skewness towards the susceptible parent (Fig. 1A). Of the 111 RILs, none were highly resistant, 5 were resistant, 20 were moderately resistant, 9 were moderately susceptible, 42 were susceptible and 35 were highly susceptible.
Fig. 1.
Frequency distribution of disease scores and percentages of the disease index in the mungbean RIL population derived from cross KPS2 × NM10-12-1 evaluated for resistance to mungbean yellow mosaic India virus in 2012 in Punjab, India (A) and in 2008 in Faisalabad, Pakistan (B).
For the evaluation in Pakistan, KPS2 and NM10-12-1 were highly susceptible and highly resistant with percentage disease indices of 80.80% and 23.64%, respectively (Supplemental Table 1). The RILs showed percentage disease indices between 31.89% and 100%, with an average of 74.11%. Correlation of the percentage disease index between replications was very high and significant (r = 0.97 to 0.98, P < 0.0001, df = 120). Frequency distribution of the percentage of disease index in the population demonstrated skewness towards KPS2 (Fig. 1B).
There was a significant correlation between disease responses in Pakistan and India (r = 0.37, df = 109, P = 0.0001).
QTLs conditioning MYMIV resistance
QTL analysis by single marker analysis revealed that only one marker, cp02662 on LG2, was associated with MYMIV resistance in India. All seven markers on LG2, except CEDG244, were associated with MYMIV resistance in Pakistan. The coefficient of determination (R2) of the significant marker in India was 8.37%. R2 of the significant markers in Pakistan ranged from 9.88% for CEDG108 to 25.10% for cp03715.
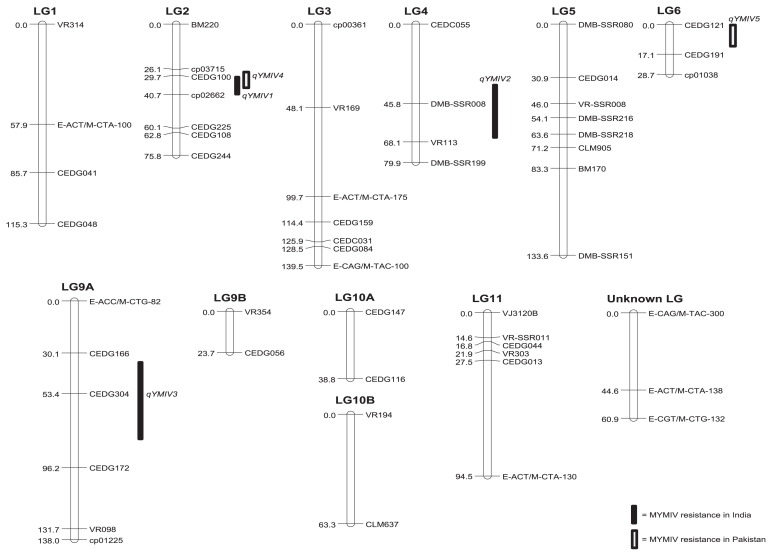
CIM was employed to locate QTL for MYMIV resistance on the linkage map. The LOD score threshold for MYMIV resistance in India and Pakistan was determined by a permutation test. Three QTLs were identified for the MYMIV in India (Fig. 2 and Table 2). The QTLs were each on LGs 2, 4 and 9A and were designated as qMYMIV1, qMYMIV2 and qMYMIV3, respectively. qMYMIV1 was located between markers CEDG100 and cp02662, showing an LOD score of 2.62, which was significant at P = 0.05. This QTL had an additive effect of 0.35 and explained 9.33% of the variation in disease reaction. qMYMIV2 was localized between markers DMB-SSR008 and VR113, showing LOD score of 2.54, which was significant at P = 0.05. This QTL showed an additive effect of −0.50 and accounted for 10.67% of the variation in the disease score. qMYMIV3 was located between markers CEDG166 and CEDG304 and showed significance at P = 0.01 with an LOD score of 3.42. It had an additive effect of 0.42 and accounted for 12.55% of the disease score variation. qMYMIV1 and qMYMIV3 alleles from NM10-12-1 increased the resistance, while qMYMIV2 alleles from NM10-12-1 decreased the resistance.
Fig. 2.
An SSR-based linkage map of the mungbean RIL population derived from the cross KPS2 × NM10-12-1 and position of the QTLs conditioning yellow mosaic India virus resistance, as detected by composite interval mapping.
Table 2.
Quantitative trait loci conditioning resistance to mungbean yellow mosaic India virus in F8 mungbean RIL populations from KPS2 and NM10-12-1 in India and Pakistan
| Location | QTL name | Linkage group | Marker interval | Position (cM)a | LOD score | PVE (%)b | Additive effect |
|---|---|---|---|---|---|---|---|
| India | qYMIV1 | 2 | CEDG100–cp02662 | 38.8 | 2.62 | 9.33 | 0.35 |
| qYMIV2 | 4 | DMB-SSR008–VR113 | 51.8 | 2.54 | 10.67 | −0.50 | |
| qYMIV3 | 9A | CEDG166–CEDG304 | 50.2 | 3.42 | 12.55 | 0.42 | |
| Pakistan | qYMIV4 | 2 | CEDG100–cp02662 | 29.7 | 10.00 | 27.93 | 6.41 |
| qYMIV5 | 6 | CEDG121–CEDG191 | 0.1 | 2.55 | 6.24 | 3.04 |
Position on the linkage group.
Percentage of phenotypic variance explained by the QTL.
Two QTLs, qMYMIV4 and qMYMIV5, were detected by CIM for disease resistance in Pakistan. qMYMIV4 had an LOD score of 10.00 and was significant at P = 0.001. It is located on LG2 between markers CEDG100 and cp02662 (Fig. 2). qMYMIV4 had an additive effect of 6.41% and explained 27.93% of the variation in the percentages of the disease index. qMYMIV5 was detected on LG6 between markers CEDG121 and CEDG191. This QTL showed a LOD score of 2.55 and was significant at P = 0.05. It had an additive effect of 3.04% and explained 6.24% of the percentage of the disease index. At both QTLs, alleles from NM10-12-1 increased resistance at this locus. It is important to note that another QTL detected on LG9A almost reached significance at P = 0.05 (LOD threshold = 2.35) (Supplemental Fig. 1). This locus was located between markers CEDG166 and CEDG304 with an LOD score of 2.27.
Discussion
In this study, we located QTLs for resistance to MYMIV in two different countries, India and Pakistan. CIM identified five QTLs for resistance: three QTLs in India (qMYMIV1, qMYMIV2 and qMYMIV3) and two QTLs in Pakistan (qMYMIV4 and qMYMIV5) (Table 2 and Fig. 2). Although the disease evaluation was conducted in different locations and years and with different scoring systems, a common QTL was identified. qMYMIV1 detected in India and qMYMIV4 identified in Pakistan are in the same marker interval and can be considered as the same QTL. In addition, qMYMIV3 detected in India showed the highest genetic effect and was almost significant in Pakistan. Therefore, two out of the five QTLs detected in India and Pakistan appeared to be common. Moreover, qMYMIV1 and qMYMIV4 detected in our study and the major QTL reported by Chen et al. (2013) appeared to be the same, although only one SSR marker (DMB-SSR151) was common between the two genetic linkage maps. The location for evaluation for the resistance in India in our study (Punjab Agricultural University, Ludhiana) was the same as that reported by Chen et al. (2013). Based on the mungbean genetic map reported by Kajonphol et al. (2012), SSR marker DMB-SSR158, which links to the major QTL controlling MYMIV resistance, MYMIVr9_25, identified by Chen et al. (2013), was on LG2 between markers CEDG100 and CEDG108. Our linkage groups and those of Kajonphol et al. (2012) are the same and CEDG100 and CEDG108 were located on LG2 of both maps. A QTL for the resistance in both India and Pakistan was located between the two markers in our study. Therefore, MYMIVr9_25, qMYMIV1 and qMYMIV4 are common QTLs conferring MYMIV resistance. This locus was detected in different locations and years, with different sources of resistance and scoring systems; thus, it is highly robust and can be used in marker-assisted selection. DMB-SSR151, which is linked to MYMIVr8_48.8, a QTL controlling MYMIV identified by Chen et al. (2013), was mapped to LG5 of our map, but no QTL was detected in this genome region in our study. The resistance parent used in our study, NM10-12-1 and that of Chen et al. (2013), NM92 (a derivative of NM36 × VC2768B), were both developed by the Nuclear Institute for Agriculture and Biology (NIAB), Pakistan (Ali et al. 1997). NM36 is derived from a cross between accession 6601 (MYIMV tolerant) and VC1973A (MYIMV susceptible) (Ali et al. 1997), while NM10-12-1 is a mutant line from gamma irradiation whose original parent is not known. However, based on the pedigree provided by Ali et al. (1997), NM10-12-1 is possibly derived from 6601.
In blackgram, resistance gene analog markers YR4 and CYR1 were reported to be completely linked with resistance to MYMIV (Maiti et al. 2011). Marker CYR1 was also associated with resistance in mungbean (amplifies DNA band in resistant mungbean but not in susceptible mungbean). CYR1 is proposed as part of the candidate disease resistance (R) gene (Maiti et al. 2011). Recently, the R gene CYR1 was fully isolated from blackgram (Maiti et al. 2012). Protein CYR1 may act as a signaling molecule to protect blackgram from MYMIV and be involved in recognizing the effector molecule of the pathosystem to contribute to incompatible interactions (Maiti et al. 2012). In the present study, markers YR4 and CYR1 were used in a polymorphism survey of the parents, but none showed polymorphism. Although CRY1 showed no polymorphism between KPS2 and NM10-12-1, the association between CYR1 and MYMIV resistance in mungbean cannot be ruled out. The contrast between our result and that of Maiti et al. (2011) may possibly be due to the difference in genotypes of mungbean. DNA B of a MYMV isolate is an important determinant of host range between mungbean and blackgram. Balaji et al. (2004) showed that agroinoculation of mungbean and blackgram with two different DNA B components together with DNA A of a blackgram isolate of MYMV-Vig resulted in distinctly different symptoms between the two crops. Additional research is required to clarify whether CYR1 is involved in MYMIV resistance in mungbean. In addition, SSR marker CEDG180, which was reportedly associated with a major gene controlling MYMIV resistance in blackgram (Gupta et al. 2013), showed no polymorphism between our mapping parents. Based on the linkage map of mungbean reported by Isemura et al. (2012), CEDG180 is mapped to LG10. This suggests that the genetics of resistance to MYMIV in mungbean and blackgram are different.
Supplementary Materials
Acknowledgements
This research was supported by the Thailand Graduate Institute of Science and Technology (TGIST) of the National Science and Technology Development Agency (NSTDA), Thailand and by the Center of Advanced Studies for Agriculture and Food, Kasetsart University, under the National Research University Program of the Office of the Higher Education Commission, Ministry of Education, Thailand.
Literature Cited
- Akhtar, K.P., Kitsanachandee, R., Srinives, P., Abbas, G., Asghar, M.J., Shah, T.M., Atta, B.M., Chatchawankanphanich, O., Sarwar, G., Ahmad, M.et al. (2009) Field evaluation of mungbean recombinant inbred lines against mungbean yellow mosaic disease using new disease scale in Thailand. Plant Pathol. J. 25: 422–428 [Google Scholar]
- Ali, M., Malik, I.A., Sabir, H.M. and Ahmad, B. (1997) The mungbean green revolution in Pakistan. Asian Vegetable Research and Development Center, Shanhua, Taiwan [Google Scholar]
- Balaji, V., Vanitharani, R., Karthikeyan, A.S., Anbalagan, S. and Veluthambi, K. (2004) Infectivity analysis of two variable DNA B components of Mungbean yellow mosaic virus-Vigna in Vigna mungo and Vigna radiata. J. Biosci. 29: 297–308 [DOI] [PubMed] [Google Scholar]
- Chen, H.M., Ku, H.S., Schafleitner, R., Bains, T.S., Kuo, G.C., Liu, C.A. and Nair, R.M. (2013) The major quantitative trait locus for mungbean yellow mosaic Indian virus resistance is tightly linked in repulsion phase to the major bruchid resistance locus in a cross between mungbean [Vigna radiata (L.) Wilczek] and its wild relative Vigna radiata ssp. sublobata. Euphytica 192: 205–216 [Google Scholar]
- Dhole, V.J. and Reddy, K.S. (2012) Genetic analysis of resistance to mungbean yellow mosaic virus in mungbean (Vigna radiata). Plant Breed. 131: 414–417 [Google Scholar]
- Dhole, V.J. and Reddy, K.S. (2013) Development of a SCAR marker linked with a to MYMV resistance gene in mungbean (Vigna radiata). Plant Breed. 132: 127–132 [Google Scholar]
- Gupta, S., Gupta, D.S., Anjum, T.K., Pratap, A. and Kumar, J. (2013) Inheritance and molecular tagging of MYMIV resistance gene in blackgram (Vigna mungo L. Hepper). Euphytica 193: 27–37 [Google Scholar]
- Han, O.K., Kaga, A., Isemura, T., Wang, X.W., Tomooka, N. and Vaughan, D.A. (2005) A genetic linkage map for azuki bean (Vigna angularis (Willd.) Ohwi & Ohashi). Theor. Appl. Genet. 111: 1288–1299 [DOI] [PubMed] [Google Scholar]
- Hussain, M., Qazi, J., Mansoor, S., Iram, S., Bashir, M. and Zafar, Y. (2004) First Report of Mungbean yellow mosaic India virus on mungbean in Pakistan. New Disease Rep. 9: 13 [Google Scholar]
- Ilyas, M., Qazi, J., Mansoor, S. and Briddon, R.W. (2010) Genetic diversity and phylogeography of begomoviruses infecting legumes in Pakistan. J. Gen. Virol. 91: 2091–2101 [DOI] [PubMed] [Google Scholar]
- Isemura, T., Kaga, A., Tabata, S., Somta, P., Srinives, P., Shimizu, T., Jo, U., Vaughan, D.A. and Tomooka, N. (2012) Construction of a genetic linkage map and genetic analysis of domestication related traits in mungbean (Vigna radiata). PLoS One 7(8): e41304. 10.1371/journal.pone.0041304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajonphol, T., Sangsiri, C., Somta, P., Toojinda, T. and Srinives, P. (2012) SSR map construction and quantitative trait loci (QTL) identification of major agronomic traits in mungbean (Vigna radiata (L.) Wilczek). SABRAO J. Breed. Genet. 44: 71–86 [Google Scholar]
- Kosambi, D.D. (1944) The estimation of map distances from recombination values. Ann. Eugen. 12: 172–175 [Google Scholar]
- Lodhi, M.A., Ye, G.N., Weeden, N.F. and Reisch, B.I. (1994) A simple and efficient method for DNA extraction from grapevine cultivars and Vitis species. Plant Mol. Biol. Rep. 12: 6–13 [Google Scholar]
- Maiti, S., Basak, J., Kundagrami, S., Kundu, A. and Pal, A. (2011) Molecular marker-assisted genotyping of mungbean yellow mosaic India virus resistant germplasms of mungbean and urdbean. Mol. Biotechnol. 47: 95–104 [DOI] [PubMed] [Google Scholar]
- Maiti, S., Paul, S. and Pal, A. (2012) Isolation, characterization, and structure analysis of a non-TIR-NBS-LRR encoding candidate gene from MYMIV-resistant Vigna mungo. Mol. Biotechnol. 52: 217–233 [DOI] [PubMed] [Google Scholar]
- Malik, L.A., Sarwar, G. and Ali, Y. (1986) Genetic studies in mungbean (Vigna radiata (L.) Wilczek). Inheritance of tolerance to mungbean yellow mosaic virus and some morphological characters. Pak. J. Botany 18: 189–198 [Google Scholar]
- Pant, V., Gupta, D., Choudhury, N.R., Malathi, V.G., Varma, A. and Mukherjee, S.K. (2001) Molecular characterization of the Rep protein of the blackgram isolate of Indian mungbean yellow mosaic virus. J. Gen. Virol. 82: 2559–2567 [DOI] [PubMed] [Google Scholar]
- Prathet, P., Somta, P. and Srinives, P. (2012) Mapping QTL conferring resistance to iron deficiency chlorosis in mungbean [Vigna radiata (L.) Wilczek]. Field Crops Res. 137: 230–236 [Google Scholar]
- Shukla, G.P. and Pandya, B.P. (1985) Resistance to yellow mosaic in greengram. SABRAO J. Genet. Breed 17: 165–171 [Google Scholar]
- Thakur, R.P., Patel, P.N. and Verma, J.P. (1997) Genetical relationships between reactions to bacterial leaf spot, yellow mosaic and Cercospora leaf spot diseases in mungbean (Vigna radiata). Euphytica 26: 765–774 [Google Scholar]
- Van Ooijen, J.W. and Voorrips, R.E. (2001) JoinMap 3.0: software for the calculation of genetic linkage maps. Plant Research International, Wageningen, the Netherlands [Google Scholar]
- Wang, S., Basten, C.J. and Zeng, Z.B. (2010) Windows QTL Cartographer 2.5. Department of statistics, North Carolina State University, Raleigh [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.