Abstract
In soybean, the I gene inhibits pigmentation over the entire seed coat, resulting in yellow seeds. It is thought that this suppression of seed coat pigmentation is due to naturally occurring RNA silencing of chalcone synthase genes (CHS silencing). Fully pigmented seeds can be found among harvested yellow seeds at a very low percentage. These seed coat pigmented (scp) mutants are generated from yellow soybeans by spontaneous recessive mutation of the I gene. A candidate for the I gene, GmIRCHS, contains a perfect inverted repeat (IR) of a CHS pseudogene (pseudoCHS3) and transcripts of GmIRCHS form a double-stranded CHS RNA that potentially triggers CHS silencing. One CHS gene, ICHS1, is located 680 bp downstream of GmIRCHS. Here, the GmIRCHS–ICHS1 cluster was compared in scp mutants of various origins. In these mutants, sequence divergence in the cluster resulted in complete or partial loss of GmIRCHS in at least the pseudoCHS3 region. This result is consistent with the notion that the IR of pseudoCHS3 is sufficient to induce CHS silencing, and further supports that GmIRCHS is the I gene.
Keywords: CHS genes, inverted repeat, mutant, RNA silencing, seed coat pigmentation, soybean
Introduction
In soybean (Glycine max), the I (inhibitor) locus determines the spatial distribution of pigments in the epidermal layer of the seed coat. The I locus has four alleles (I, ii, ik and i) and the dominance relationships are I > ii > ik > i. The i allele leads to a self-pigmented seed coat, i.e., the entire seed coat surface is pigmented. The I allele inhibits the production and accumulation of pigments over the entire seed coat. The ii and ik alleles inhibit pigmentation except in the hilum and the saddle-shaped region (the hilum and a small surrounding region), respectively. All yellow soybean cultivars carry the I allele for a nonpigmented hilum or the ii allele for a pigmented hilum. Inhibition of seed coat pigmentation by the I locus, at least for the I and ii alleles, has been suggested to be the result of naturally occurring RNA silencing of chalcone synthase (CHS) genes, hereafter referred to simply as CHS silencing (Kanazawa 2008, Nagamatsu et al. 2007, Senda et al. 2004, 2012, Tuteja et al. 2004). Fully pigmented seeds are found among the harvested seeds of yellow soybean cultivars, although the frequency is usually quite low (Bernard and Weiss 1973). This seed coat pigmentation phenomenon in yellow soybean occurs via spontaneous mutation from either the I or ii allele to the i allele; CHS silencing does not occur in pigmented soybeans with the i/i genotype (Kasai et al. 2004, Tuteja et al. 2004). The aim of our study was to elucidate the molecular mechanism of the seed coat pigment mutation from the I allele to the i allele, which hereafter we simply call “scp mutation” in this paper.
Regardless of the I locus genotype, a CHS1-specific probe commonly detects a single HindIII fragment in which CHS3 and CHS1 are clustered and this fragment is not affected by the scp mutation (Akada and Dube 1995, Senda et al. 2002a, 2002b, Todd and Vodkin 1996). Interestingly, in yellow soybeans with the I allele, an extra HindIII fragment is also detected using a CHS1-specific probe and this fragment is affected by the scp mutation (Kasai et al. 2007, Senda et al. 2002a, 2002b, Todd and Vodkin 1996). CHS1 in the extra HindIII fragment tightly linked to the I allele was regarded as a duplicated CHS1 (designated dCHS1) (Todd and Vodkin 1996) and was later designated ICHS1 (I-linked CHS1) to distinguish it from CHS1 in the CHS3–CHS1 cluster (Senda et al. 2002a). A candidate for the I allele, GmIRCHS (Glycine max inverted repeat of CHS pseudogene), is located 680 bp upstream of ICHS1 (Kasai et al. 2007). GmIRCHS is composed of a 5′-portion of GmJ1 (including the promoter region) and a perfect inverted repeat (IR) of a CHS pseudogene (pseudoCHS3). GmJ1 encodes a type III DnaJ-like protein, but its function is still unknown (Kasai et al. 2007, Miernyk 2001). Soybean CHS genes consist of two exons (exon1 and exon2) split by an intron. PseudoCHS3 is missing the 5′-portion (exon1, the intron and a small part of exon2) of CHS3. The IR of pseudoCHS3 includes pseudoCHS3 and its complementary sequence; it was suggested that transcription of GmIRCHS leads to the formation of double-stranded RNA (dsRNA) of the CHS pseudogene (Kasai et al. 2007, Kurauchi et al. 2011). In general, RNA silencing is triggered by a dsRNA structure of the target gene; therefore, the IR structure of pseudoCHS3 in GmIRCHS is likely to be important for inducing CHS silencing (Senda et al. 2012).
We previously compared the GmIRCHS–ICHS1 cluster in three scp mutants (i/i genotype), each of which was found in a different yellow soybean cultivar (Miyagi shirome, Toyohomare) or strain (Karikei 584) with the I/I genotype in Japan (Kasai et al. 2007, Senda et al. 2002b). The IR structure of pseudoCHS3 in GmIRCHS was missing in all the three scp mutants, supporting that the IR region of pseudoCHS3 may be essential for the function of the I allele; more noteworthy was that the patterns of structural changes in the GmIRCHS–ICHS1 cluster were not identical to one another (Kasai et al. 2007, Senda et al. 2002b). However, only three scp mutants were used for the analysis and greater numbers of scp mutants are required to confirm the importance of the IR of pseudoCHS3 and to further characterize the patterns of structural changes in the GmIRCHS–ICHS1 cluster (Senda et al. 2012). In the current study, we compared the regions corresponding to the GmIRCHS–ICHS1 cluster in 22 scp mutants, including the three that were previously analyzed.
Materials and Methods
Plant materials
Twenty-two scp mutants with the i/i genotype were found in Japanese yellow soybean cultivars or strains with the I/I genotype (Table 1). These scp mutants were collected in northern Japan (the Hokkaido and Tohoku areas). Seven scp mutants (EnM1–EnM7) derived from a single cultivar (cv. Enrei) were isolated in different fields. The following scp mutants were also isolated from the same cultivar: THM1 and THM2 from cv. Toyohomare, and YHM1 and YHM2 from cv. Yukihomare.
Table 1.
List of scp mutants used in previous studies and the current work
| Name of scp mutant | Origin (cultivar/strain) | Size of mutant-specific polymorphic fragment (kb) | Scp mutant type | Size of insertion at the divergence point | Accession number of DDBJ/EMBL/GenBank databases | |
|---|---|---|---|---|---|---|
|
| ||||||
| CHS probe | CHS1 probe | |||||
| C127M | strain Chukei127 | 5.1 Ha | 5.1 H | I | 13 bp | AB822565 |
| EnM1 | cv. Enrei | 7.7 Ea | 7.7 E | NAb | NA | |
| EnM2 | cv. Enrei | 1.6 E | NDb | I | AB822566 | |
| EnM3 | cv. Enrei | 3.7 H | 3.7 H | I | 36 bp | AB822567 |
| EnM4 | cv. Enrei | 2.2 E | ND | II | AB822568 | |
| EnM5 | cv. Enrei | 5.1 E | 5.1 E | I | 15 bp | AB822569 |
| EnM6 | cv. Enrei | 2.8 H | ND | I | 5 bp | AB822570 |
| EnM7 | cv. Enrei | 4.7 E | 4.7 E | II | AB822571 | |
| K557M | strain Karikei557 | 2.2 E | ND | II | AB822572 | |
| K584M | strain Karikei584 | 6.2 E | 6.2 E | I | AB083125 | |
| K629M | strain Karikei629 | 6.2 E | 6.2 E | I | AB822573 | |
| K699M | strain Karikei699 | 3.7 E | ND | I | AB822574 | |
| MSM | cv. Miyagi shirome | 7.7 E | ND | I | 3 bp | AB083126 |
| OSM | cv. Osuzu | 2.5 H | ND | I | AB822575 | |
| SKM | cv. Suzukari | 2.2 E | ND | II | AB822576 | |
| THM1 | cv. Toyohomare | 2.2 E | ND | II | AB264312 | |
| THM2 | cv. Toyohomare | 2.9 H | ND | I | 2 bp | AB822577 |
| TUM | cv. Tamaurara | 1.4 H | ND | I | 2 bp | AB822578 |
| TYM | cv. Tachiyutaka | 2.5 E | ND | II | 27 bp | AB822579 |
| YHM1 | cv. Yukihomare | 2.7 H | ND | I | 23 bp | AB822580 |
| YHM2 | cv. Yukihomare | 2.2 E | ND | II | AB822581 | |
| YSM | cv. Yukishizuka | 4.9 H | 4.9 H | I | 5 bp | AB822582 |
H: HindIII fragment, E: EcoRI fragment.
ND: not detected, NA: not analyzed.
Genomic DNA and seed coat RNA extraction
Soybean genomic DNA and seed coat RNA was extracted as described by Kasai et al. (2007).
Southern blot and RNA gel blot analyses
Southern blot and RNA gel blot analyses were carried out as described previously (Kasai et al. 2007, Senda et al. 2002b). DNA fragments for a CHS probe to detect all CHS gene members, a CHS1 probe to detect only CHS1 members and a DnaJ probe to detect GmJ1 were amplified by PCR; a 530-bp DNA fragment for the CHS probe was amplified using the primers CHSFP and CHSRP, a 680-bp fragment for the CHS1 probe was amplified using primers CHS1FP and CHS1RP and a 460-bp DNA fragment was amplified using primers DnaJ FP and DnaJ RP (Kasai et al. 2007). The sequences of these primers are listed in Table 2.
Table 2.
Sequences of primers used in PCR analyses
| Name | RE Site Additiona | Sequenceb |
|---|---|---|
| CHSFP | None | 5′-AGGCAAGACATGGTGGT-3′ |
| CHSRP | None | 5′-GGAACATCCTTGAGGAG-3′ |
| CHS1FP | None | 5′-GCAAAAACTTAAAGTTGGAATAAAATTTGGC-3′ |
| CHS1RP | None | 5′-CATCCTAGCTGGTTAAGAAAAGAATGGA-3′ |
| DnaJ FP | None | 5′-AAAACGACAGCTAATCACGC-3′ |
| DnaJ RP | None | 5′-TCTGGAAGGATCAGACAGGG-3′ |
| 1 | None | 5′-CACAATACGTTTTTTCAAACCGG-3′ |
| 2 | None | 5′-TTCCCCCTGCCCTGCAAATGCTTC-3′ |
| 3 | EcoRI | 5′-GCCGAATTCGAAACACAATACGTTTTTTCAAACCGGCCAGCC-3′ |
| 4 | EcoRI | 5′-GCCGAATTCCCTGCAAATGCTTCTTTTTGTATACCAG-3′ |
| 5 | None | 5′-TCCAGGGTGATCCTATGGAAGGACTGACCC-3′ |
| 6 | EcoRI | 5′-GCCGAATTCCATTATGCATTGCAATAAGATGGGGTCAGG-3′ |
| 7 | None | 5′-ATGCGTTCTTATGGCTTAACCG-3′ |
Restriction enzyme is abbreviated as RE.
Extra restriction enzyme sites added are underlined.
Inverse PCR
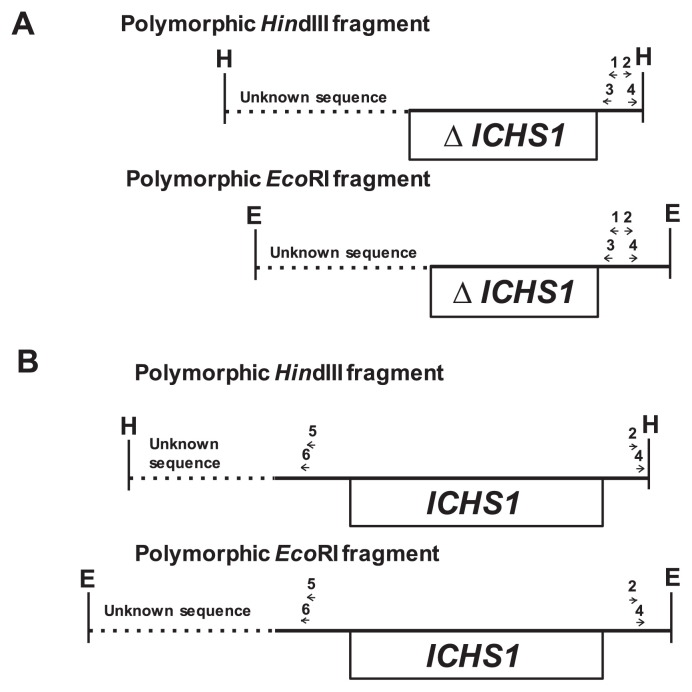
Inverse PCR (IPCR) was performed as described by Kasai et al. (2007). The positions of the primers used for IPCRs are shown in Fig. 1. For IPCR of scp mutants in which a polymorphic EcoRI or HindIII fragment was hybridized with the CHS probe but not the CHS1 probe, two pairs of primers (primer set, 1 and 2; nested primer set, 3 and 4) were designed to anneal with the 3′-downstream region of ICHS1 or its complementary region (Fig. 1A). In other scp mutants in which a polymorphic EcoRI or HindIII fragment was detected with both the CHS and CHS1 probes, two pairs of primers (primer set, 2 and 5; nested primer set, 4 and 6) were designed for IPCR; primer 5 and its nested primer 6 annealed with the 5′-upstream region of ICHS1 (Fig. 1B). The sequences of primers 1–6 used for IPCRs are listed in Table 2.
Fig. 1.
Schematic representation of the locations and orientations of primers (horizontal arrows) used for IPCR in two groups of scp mutants. (A) Scp mutants in which a polymorphic EcoRI or HindIII fragment hybridized to the CHS probe but not the CHS1 probe. (B) Scp mutants in which a polymorphic EcoRI or HindIII fragment was detected with both the CHS and CHS1 probes. The sequences of these primers are described in Table 2. Unknown sequence in HindIII or EcoRI polymorphic fragments is shown by a dotted line. The open boxes indicate deleted (denoted by ΔICHS1) or complete ICHS1 regions. The restriction sites are: E, EcoRI; H, HindIII.
Cloning and DNA sequencing
Cloning and DNA sequencing were performed as described previously (Kasai et al. 2007).
Results
RFLP analysis between yellow soybean and scp mutants
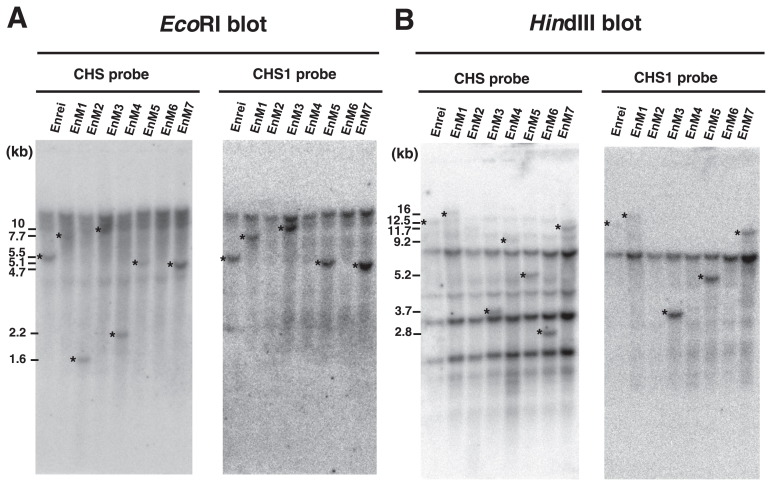
CHS and CHS1 probes were used to detect all CHS family members and CHS1 genes specifically. In yellow soybeans with the I/I genotype, both of these probes hybridize 5.5-kb EcoRI and 12.5-kb HindIII fragments, in which the GmIRCHS–ICHS1 cluster is located (Kasai et al. 2007, Senda et al. 2002a, 2002b) (Figs. 2, 3). We previously reported that these restriction fragments were shifted in size or lost in three scp mutants (K584M, MSM and THM1), resulting in different polymorphic patterns (Kasai et al. 2007, Senda et al. 2002a, 2002b). Each of these three scp mutants was isolated from a different yellow soybean cultivar (Miyagi shirome or Toyohomare) or strain (Karikei 584) with the I/I genotype (Table 1). This raised the question of whether patterns of structural changes in the GmIRCHS–ICHS1 cluster are specific to cultivars/strains. To address this question, we performed restriction fragment length polymorphism (RFLP) analysis among seven Enrei-derived scp mutants isolated from different fields. A CHS probe to detect all CHS gene members was hybridized to a Southern blot of EcoRI- or HindIII-digested genomic DNA from Enrei (I/I genotype) and its seven scp mutants, EnM1–EnM7 (i/i genotype). As shown in Fig. 3, RFLPs were found between Enrei and its scp mutants and also found among the seven scp mutants. Enrei displayed 5.5-kb EcoRI and 12.5-kb HindIII fragments harboring the GmIRCHS–ICHS1 cluster. These fragments were not present in any of the scp mutants, which instead displayed polymorphic restriction fragments of varying sizes (Fig. 3 and Table 1). After removing the CHS probe, the CHS1 probe detecting only CHS1 genes was rehybridized to the same blot. An additional polymorphism was noted: as with the CHS probe, the CHS1 probe also hybridized to a 5.5-kb EcoRI fragment and a 12.5-kb HindIII fragment in Enrei and a polymorphic EcoRI and/or HindIII fragment in EnM1, EnM3, EnM5 and EnM7, whereas it did not hybridize to the polymorphic EcoRI and/or HindIII fragments detected by the CHS probe in EnM2, EnM4 or EnM6 (Fig. 3 and Table 1). As shown in Fig. 2, the CHS1 probe hybridizes to the 5′-upstream region of ICHS1, indicating that at least this region was deleted in EnM2, EnM4 and EnM6, while the entirety or a part of this region was retained in EnM1, EnM3, EnM5 and EnM7. Similarly to the Enrei-derived scp mutants, RFLPs were also found between the two pairs of scp mutants (THM1/THM2 and YHM1/THM2) each derived from the same cultivar (Toyohomare and Yukihomare, respectively) (Table 1). These results indicated that the patterns of structural changes in the I → i mutations of the GmIRCHS–ICHS1 cluster were not specific to cultivars/strains. We also identified a polymorphic EcoRI or HindIII fragment in other scp mutants (i/i genotype) isolated from different yellow soybean cultivars/strains with the I/I genotype. The various sizes of polymorphic restriction fragments in all 22 scp mutants used in previous studies and this work are summarized in Table 1.
Fig. 2.
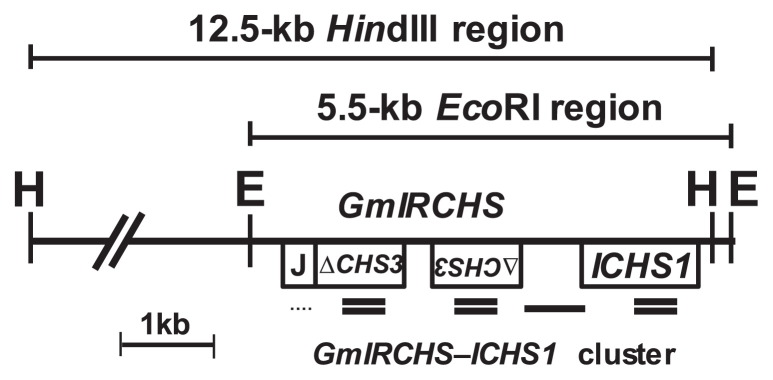
Schematic representation of the GmIRCHS–ICHS1 cluster region. In GmIRCHS, the GmJ1 part and pseudoCHS3 are denoted by J and ΔCHS3, respectively. The locations of CHS, CHS1 and DnaJ probe hybridization are shown by double lines, a single line and a dotted line, respectively. The sizes of EcoRI and HindIII fragments containing the GmIRCHS–ICHS1 cluster are indicated.
Fig. 3.
RFLP analysis of a yellow soybean cultivar (cv. Enrei) and its derived scp mutants (EnM1–EnM7). The CHS (left panel) and CHS1 (right panel) probes were hybridized to EcoRI (A) and HindIII (B) blots. The polymorphic bands are indicated by asterisks. Sizes of the polymorphic bands are shown in kb.
Nucleotide sequence analysis of polymorphic restriction fragments in scp mutants
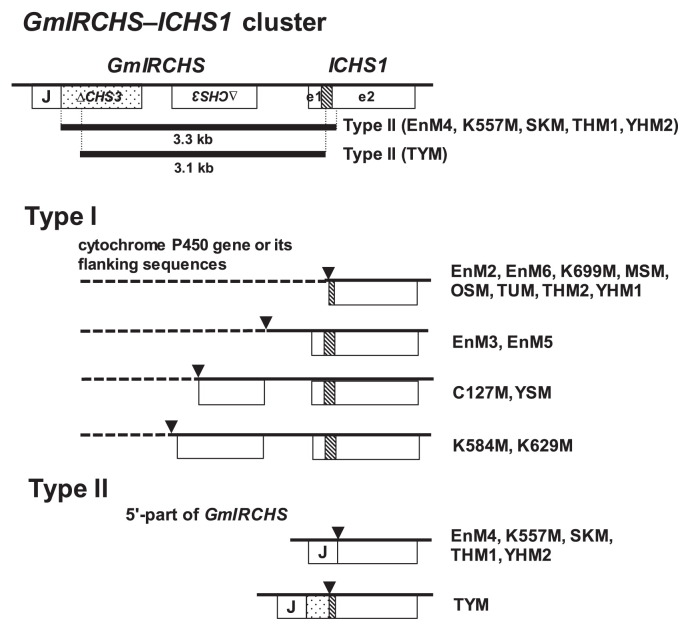
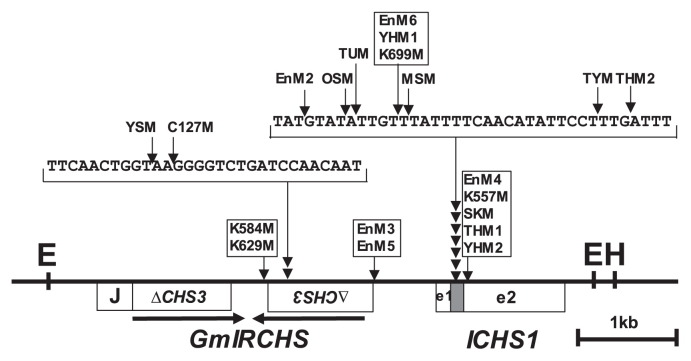
In previous studies, we found that the downstream sequences of ICHS1 were not changed by the mutation from I to i (Kasai et al. 2007, Senda et al. 2002b). Using IPCR as described in the Materials and Methods, we amplified part of the polymorphic EcoRI or HindIII fragments in the scp mutants (Fig. 1). Each amplified fragment was cloned and sequenced. However, part of the polymorphic EcoRI fragments detected in EnM1 and EnM7 were not amplified, probably due to large size and/or the secondary structure of the fragment. The nucleotide sequence data reported are available in the DDBJ/EMBL/GenBank databases under the accession numbers AB822565–AB822582 (Table 1). Sequence comparison with the GmIRCHS–ICHS1 cluster (accession number AB264311) revealed sequence divergence in which the upstream sequence from the point of divergence was replaced with different sequences (Fig. 4). The points of divergence were located within the GmIRCHS–ICHS1 cluster and consequently the IR structure of pseudoCHS3 in GmIRCHS was missing in all analyzed scp mutants (Fig. 5). We also searched for sequences homologous to the sequences upstream of the points of divergence. As shown in Fig. 4, the matching sequences in the database were divided into two types: type I, a cytochrome P450 gene or its flanking sequences; type II, the 5′-part of GmIRCHS. Of the three previously reported scp mutants, K584M (accession number AB083125) and MSM (accession number AB083126) belonged to type I (Senda et al. 2002b), while THM1 (accession number AB264312) belonged to type II (Kasai et al. 2007). In several scp mutants belonging to type I (C127M, EnM3, EnM5, EnM6, MSM, THM2, TUM, YHM1, YSM) or II (TYM), small insertions from 2 to 36 bp were found at the divergence points (Table 1).
Fig. 4.
Schematic representation of the GmIRCHS–ICHS1 cluster in yellow soybean with the I/I genotype and the corresponding region in scp mutants (types I and II). Deleted regions in type II mutants are shown by bold lines. The stippled box indicates most of the exon2 region of pseudoCHS3. The shaded box, e1 and e2 denote an intron, exon1 and exon2 of ICHS1, respectively. Points of divergence are indicated by triangles. A cytochrome P450 gene or its flanking regions are indicated by dotted lines.
Fig. 5.
Identification of divergence points in scp mutants by comparison with the GmIRCHS–ICHS1 cluster sequence. The positions and relative orientations of IRs of pseudoCHS3 are shown by horizontal arrows. The divergence points, from which the 5′-upstream sequences are changed, are indicated by vertical arrows. Scp mutants with the same divergence point are boxed.
Type I scp mutants
The cytochrome P450 gene (Glyma08g11570) consists of two exons (exon1 and exon2) split by an intron, although the detailed function of its translational products is unknown. The sequence of its homolog has also been registered in the database (the accession number AX196297), although its origin (i.e., soybean cultivar/strain) is unknown. Sequence comparison between Glyma08g11570 and its homolog revealed that the sequence difference was located in the intron; the 5′- and 3′-flanking sequences were identical (data not shown). We mapped the points of divergence in the type I scp mutants by comparing with the cytochrome P450 gene and its flanking sequences (Fig. 6). Divergent points were located at various sites in and around the cytochrome P450 geneas well as in the GmIRCHS–ICHS1 cluster (Figs. 5, 6), leading to polymorphism among the type I scp mutants (Table 1).
Fig. 6.
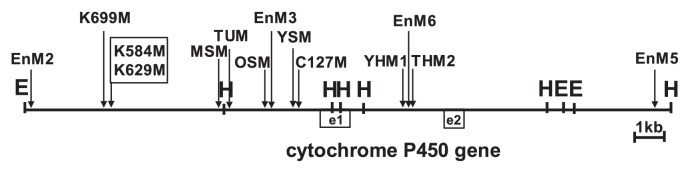
Identification of divergence points in type I scp mutants by comparison with the cytochrome P450 gene and its flanking sequences. The restriction map was constructed based on the registered sequence under accession number AX196297. The divergence points, from which the 3′-downstream sequences are changed to sequences within the GmIRCHS–ICHS1 cluster, are indicated by vertical arrows. The open boxes labeled e1 and e2 denote exon1 and exon2 of the cytochrome P450 gene. Scp mutants with the same divergence point are boxed.
Type II scp mutants
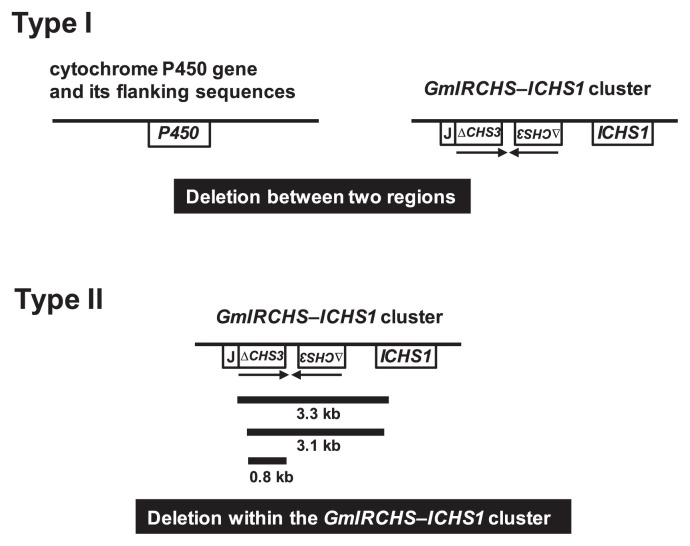
It has been suggested that the polymorphic 2.2-kb EcoRI region specific to THM1 (Table 1) is generated via a 3.3-kb deletion from the GmIRCHS–ICHS1 cluster in the 5.5-kb EcoRI region (Fig. 4) (Kasai et al. 2007). Sequences of the polymorphic 2.2-kb EcoRI regions detected in EnM4, K557M, SKM and YHM2 were completely identical to that of THM1, indicating that the 3.3-kb deletion occurred within the GmIRCHS–ICHS1 cluster (Fig. 4). The sequence of the polymorphic 2.5-kb EcoRI fragment specific to TYM (Table 1) was analyzed. Comparison with the GmIRCHS–ICHS1 cluster revealed that a 3.1-kb deletion had occurred within the GmIRCHS–ICHS1 cluster (Fig. 4).
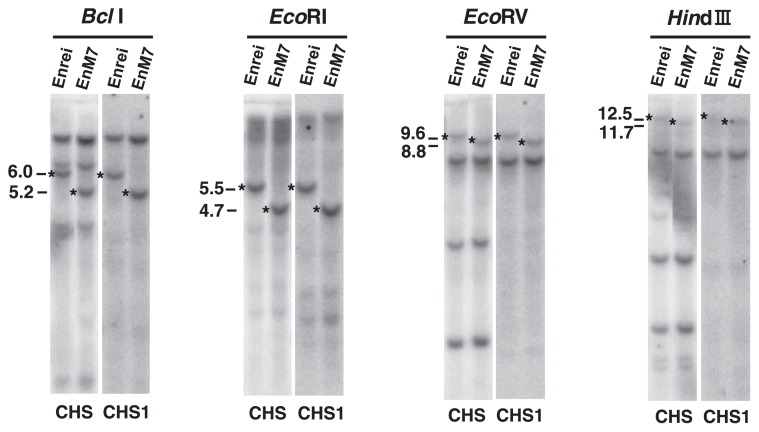
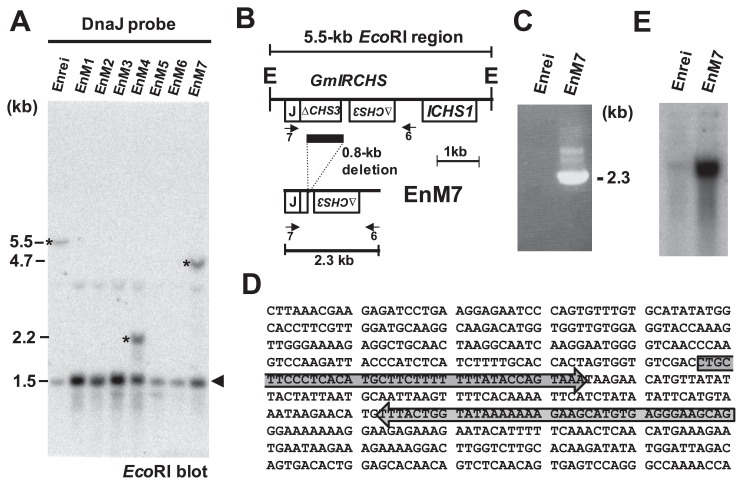
We next analyzed EnM7, although we were not able to amplify part of the 4.7-kb polymorphic EcoRI fragment (Table 1) by the IPCR methods described above. According to the RFLP analysis with the CHS and CHS1 probes, the polymorphic BclI, EcoRI, EcoRV and HindIII fragments in EnM7 were 0.8 kb smaller than those in the WT (cv. Enrei), suggesting that a 0.8-kb deletion may have occurred in the GmIRCHS–ICHS1 cluster (Fig. 7). In addition, the DnaJ probe to detect GmJ1 hybridized to the polymorphic 4.7-kb EcoRI fragment in EnM7 (Fig. 8A), indicating that the DnaJ and CHS1 probe-hybridizing regions in the GmIRCHS–ICHS1 cluster remained, at least in part. We designed primer 7 for the GmJ1 part of GmIRCHS, and PCR was carried out with primers 7 and 6 (Fig. 8B). In the WT, PCR amplification was impossible, because the IR of pseudoCHS3 inhibited the primer annealing step of the PCR reaction by intra-strand annealing between pseudoCHS3 and its complementary sequence (Kasai et al. 2007). In contrast, a 2.3-kb fragment was amplified specifically in EnM7 (Fig. 8C). Sequence analysis and comparison with the GmIRCHS sequence revealed that a 0.8-kb region containing a large part of pseudoCHS3 was deleted in EnM7 (Fig. 8B). As a result of the 0.8-kb deletion, an IR of only 38-bp remained in EnM7 (Fig. 8D).
Fig. 7.
RFLP analysis of Enrei and EnM7. The CHS (left panel) and CHS1 (right panel) probes were hybridized to BclI, EcoRI, EcoRV and HindIII blots. The polymorphic bands are indicated by asterisks. Sizes of the polymorphic bands are shown in kb.
Fig. 8.
0.8-kb deleted region of GmIRCHS identified in EnM7. (A) RFLP analysis of Enrei and its derived scp mutants (EnM1–EnM7) with the DnaJ probe. The polymorphic bands are indicated by asterisks. A 1.5-kb EcoRI fragment containing the 5′-portion of GmJ1 is denoted by an arrowhead. Sizes of the detected polymorphic bands are shown in kb. (B) Schematic representation of the GmIRCHS deletion in EnM7. For comparison, the GmIRCHS–ICHS1 cluster is also presented. The 0.8-kb deleted region is denoted by a black bar. The locations of primers 6 and 7 used for PCR analysis in Fig. 8C are indicated by horizontal arrows. E, EcoRI restriction site. (C) PCR analysis using primers 6 and 7 in Enrei and EnM7. (D) The remaining 38-bp IR sequences in EnM7 are indicated by gray arrows. (E) RNA gel blot analysis of CHS transcripts in the seed coat.
Discussion
In soybean, CHS is encoded by a multigene family composed of at least nine members, CHS1–CHS9, which are classified into two subfamilies based on sequence similarities in the ORFs, CHS1–CHS6/CHS9 and CHS7/CHS8 (Kurauchi et al. 2009, Tuteja and Vodkin 2008). In the seed coat tissues of pigmented soybeans with the i/i genotype, in which CHS silencing does not occur, CHS7 and CHS8 account for the majority of CHS transcripts, while the other CHS genes including CHS3 are in the minority (Kasai et al. 2004, Tuteja et al. 2004). In GmIRCHS, the size of pseudoCHS3 forming the IR is 1087 bp, consisting of a 955-bp sequence corresponding to most of exon2 and 132 bp of the 3′-untranslated region (UTR). RNase protection assay showed that the IR of pseudoCHS3 is completely transcribed and a 1087-bp intramolecular dsRNA is formed (Kurauchi et al. 2011). It is anticipated that this dsRNA could be processed into primary short interfering RNAs (siRNAs) by a Dicer-like protein (DCL). Production of primary CHS siRNA sequences from the 1087-bp intramolecular dsRNA is limited in both strands to most of exon2 and the 3′-UTR of CHS3 so cleavage sites in CHS mRNAs for RNA-induced silencing complexes (RISCs) guided by the primary CHS siRNAs are restricted to these locations. It was suggested that an RNA-dependent RNA polymerase (RdRP) could produce dsRNAs from CHS7/CHS8 mRNAs cleaved by RISCs guided by the primary CHS siRNAs, generating aberrant dsRNAs that could be processed into CHS7/CHS8 siRNAs by a DCL (Kurauchi et al. 2009, Tuteja et al. 2009). Therefore, for the production of CHS7/CHS8 siRNAs and the occurrence of CHS silencing, cleavage of CHS7/CHS8 mRNAs led by the primary CHS siRNAs may be required. The levels of nucleotide identity between the 955-bp sequence (most of exon2) of pseudoCHS3 and the corresponding regions of CHS7 and CHS8 are high (82% identity), suggesting that the primary CHS siRNA-directed cleavage sites are likely to be present in the exon2 region of CHS7/CHS8 mRNAs (Kurauchi et al. 2009). However, nucleotide sequence alignment of the 132-bp 3′-UTR sequence of pseudoCHS3 to the corresponding regions in CHS7 and CHS8 was not possible, indicating that primary CHS siRNA-directed cleavage sites are not present in the 3′-UTRs of CHS7 and CHS8 (Kurauchi et al. 2009).
In this study, we compared the GmIRCHS–ICHS1 cluster in 22 scp mutants including three previously analyzed mutants (Kasai et al. 2007, Senda et al. 2002b). In one scp mutant (EnM1), although RFLP analysis suggested that sequence divergence occurred in the GmIRCHS–ICHS1 cluster, we were not able to perform further analysis because part of the polymorphic EcoRI fragment was not amplified by IPCR (Table 1). Therefore, we used the remaining 21 scp mutants for further analysis. In 20 of the 21 scp mutants, loss of the IR structure of pseudoCHS3 was observed, suggesting that the IR of pseudoCHS3 may be essential for the function of the I gene. The only exception was the EnM7 mutant, in which an IR of only 38 bp still remained despite a 0.8-kb deletion. The 38-bp sequence forming an IR in EnM7 is located 95 bp downstream from the TAA stop codon, i.e. in the 3′-UTR of CHS3. In EnM7, even if the IR of the 38-bp sequence is transcribed and the resulting 38-bp dsRNA is processed into primary siRNAs by a DCL, they are unlikely to guide RISCs to CHS7 or CHS8 mRNAs for cleavage, resulting in abrogation of CHS silencing. In fact, in RNA gel blot analysis, a strong signal of CHS transcripts was detected in the EnM7 seed coat, indicating that CHS silencing was abrogated by the 0.8-kb deletion (Fig. 8E).
Based on the patterns of structural changes in the GmIRCHS–ICHS1 cluster, the 21 scp mutants were classified into two types (I and II) (Fig. 9 and Table 1). In type II, a region ranging from 0.8 kb to 3.3 kb within the cluster was deleted. There is a closely-spaced IR of pseudoCHS3 in GmIRCHS. IRs are hotspots of genomic instability associated with deletion events in a wide range of organisms (Gebow et al. 2000, Gordenin et al. 1993, Kramer et al. 1996, Leach 1994, Lobachev et al. 1998, 2000). In type I, deletion may also have occurred, between the cytochrome P450 gene (including its flanking sequences) and the GmIRCHS–ICHS1 cluster; at least, complete or partial deletion of GmIRCHS was suggested by Southern blot analysis.
Fig. 9.
Mechanisms of i allele generation in scp mutants. Sizes and locations of deleted regions in type II scp mutants are shown. The positions and orientations of the IR of pseudoCHS3 in GmIRCHS are indicated by horizontal arrows.
In type I, if some or all of GmIRCHS is not deleted but remains elsewhere, an additional polymorphic restriction fragment should be detected with a CHS probe. RFLP analysis with a CHS probe between the WT and scp mutants showed a single polymorphic restriction band (Table 1), suggesting that complete or partial deletion occurred in GmIRCHS. The physical locations of the cytochrome P450 gene and the GmIRCHS–ICHS1 cluster are uncertain in yellow soybean genomes with the I/I genotype; exactly how far apart these regions are located is unclear. The Glyma08g11570 sequence is included in the registered sequence (accession number EF623854), which is the complete sequence of a genomic bacterial artificial chromosome (BAC) clone (BAC77G7-a) isolated from the soybean BAC library of cultivar Williams 82 with the ii/ii genotype (Tuteja and Vodkin 2008). Notably, BAC77G7-a harbors a candidate for the ii allele, located ca. 63 kb away from Glyma08g11570, suggesting that Glyma08g11570 is linked to the I locus. Why is the cytochrome P450 gene and its flanking sequences a hot spot for deletion of the GmIRCHS? Determination of both the relative orientation and the distance between the cytochrome P450 gene and the GmIRCHS–ICHS1 cluster would be needed to elucidate the molecular mechanism of type I scp mutation in more detail.
Finally, comparison of the GmIRCHS–ICHS1 cluster in many scp mutants suggested that loss of the IR of pseudoCHS3 in GmIRCHS is likely to lead to seed coat pigmentation in yellow soybean. We previously reported that GmIRCHS of a Japanese yellow soybean cultivar, Toyoharuka, has a structural difference: the 5′-portion of GmJ1 extends to the middle of the intron and this extended region replaces pseudoCHS3, the result of which is that the IR structure of pseudoCHS3 characteristic of GmIRCHS is missing and only a complementary sequence of pseudoCHS3 remains (Kasai et al. 2009, Ohnishi et al. 2011). This new type was designated GmASCHS (Glycine max antisense CHS pseudogene) to distinguish it from GmIRCHS, and was also found in another yellow soybean strain, 0518BW-8 (Kasai et al. 2009, Rodriguez et al. 2013). The structure of GmASCHS suggested that antisense RNA of pseudoCHS3 may be transcribed and such antisense RNA was actually detected by RT-PCR (Kasai et al. 2009). It is possible that the antisense pseudoCHS3 RNA also forms a CHS dsRNA structure by hybridization with endogenous CHS transcripts or by the action of RdRP, leading to induction of CHS silencing. Thus, if antisense pseudoCHS3 RNA can be transcribed, the loss of IR structure in GmIRCHS may lead to a yellow seed phenotype, not the scp phenotype.
Acknowledgments
This work was supported in part by JSPS KAKENHI Grant Number 24580002 and a Grant for Hirosaki University Institutional Research.
Literature Cited
- Akada, S. and Dube, S.K. (1995) Organization of soybean chalcone synthase gene clusters and characterization of a new member of the family. Plant Mol. Biol. 29: 189–199 [DOI] [PubMed] [Google Scholar]
- Bernard, R.L. and Weiss, M.G. (1973) Qualitative genetics. In: Caldwell, B.E. (ed.) Soybean: Improvement, Production, and Uses. American Society of Agronomy, Madison, WI, pp. 117–154 [Google Scholar]
- Gebow, D., Miselis, N. and Liber, H.L. (2000) Homologous and non-homologous recombination resulting deletion: Effects of p53 status, microhomology, and repetitive DNA length and orientation. Mol. Cell. Biol. 20: 4028–4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordenin, D.A., Lobachev, K.S., Degtyareva, N.P., Malkova, A.L., Perkins, E. and Resnick, M.A. (1993) Inverted DNA repeats: a source of eukaryotic genome instability. Mol. Cell. Biol. 13: 5315–5322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa, A. (2008) RNA silencing manifested as visibly altered phenotypes in plants. Plant Biotechnol. 25: 423–435 [Google Scholar]
- Kasai, A., Watarai, M., Yumoto, S., Akada, S., Ishikawa, R., Harada, T., Niizeki, M. and Senda, M. (2004) Influence of PTGS on chalcone synthase gene family in yellow soybean seed coat. Breed. Sci. 54: 355–360 [Google Scholar]
- Kasai, A., Kasai, K., Yumoto, S. and Senda, M. (2007) Structural features of GmIRCHS, candidate of the I gene inhibiting seed coat pigmentation in soybean: implications for inducing endogenous RNA silencing of chalcone synthase genes. Plant Mol. Biol. 64: 467–479 [DOI] [PubMed] [Google Scholar]
- Kasai, A., Ohnishi, S., Yamazaki, H., Funatsuki, H., Kurauchi, T., Matsumoto, T., Yumoto, S. and Senda, M. (2009) Molecular mechanism of seed coat discoloration induced by low temperature in yellow soybean. Plant Cell Physiol. 50: 1090–1098 [DOI] [PubMed] [Google Scholar]
- Kramer, P.R., Stringer, J.R. and Sinden, R.R. (1996) Stability of an inverted repeat in a human fibrosarcoma cell. Nucleic Acids Res. 24: 4234–4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurauchi, T., Matsumoto, T., Taneda, A., Sano, T. and Senda, M. (2009) Endogenous short interfering RNAs of chalcone synthase genes associated with inhibition of seed coat pigmentation in soybean. Breed. Sci. 59: 419–426 [Google Scholar]
- Kurauchi, T., Kasai, A., Tougou, M. and Senda, M. (2011) Endogenous RNA interference of chalcone synthase genes in soybean: Formation of double-stranded RNA of GmIRCHS transcripts and structure of the 5′ and 3′ ends of short interfering RNAs. J. Plant Physiol. 168: 1264–1270 [DOI] [PubMed] [Google Scholar]
- Leach, D.R. (1994) Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays 16: 893–900 [DOI] [PubMed] [Google Scholar]
- Lobachev, K.S., Shor, B.M., Tran, H.T., Taylor, W., Keen, J.D., Resnick, M.A. and Gordenin, D.A. (1998) Factors affecting inverted repeat stimulation of recombination and deletion in Saccharomyces cerevisiae. Genetics 148: 1507–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev, K.S., Stenger, J.E., Kozyreva, O.G., Jurka, J., Gordenin, D.A. and Resnick, M.A. (2000) Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J. 19: 3822–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk, J.A. (2001) The J-domain proteins of Arabidopsis thaliana: an unexpectedly large and diverse family of chaperones. Cell Stress Chaperones 6: 209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamatsu, A., Masuta, C., Senda, M., Matsuura, H., Kasai, A., Hong, J.S., Kitamura, K., Abe, J. and Kanazawa, A. (2007) Functional analysis of soybean genes involved in flavonoid biosynthesis by virus-induced gene silencing. Plant Biotechnol. J. 5: 778–790 [DOI] [PubMed] [Google Scholar]
- Ohnishi, S., Funatsuki, H., Kasai, A., Kurauchi, T., Yamaguchi, N., Takeuchi, T., Yamazaki, H., Kurosaki, H., Shirai, S., Miyoshi, T.et al. (2011) Variation of GmIRCHS (Glycine max inverted repeat CHS pseudogene) is related to tolerance of low temperature-induced seed coat discoloration in yellow soybean. Theor. Appl. Genet. 122: 633–642 [DOI] [PubMed] [Google Scholar]
- Rodriguez, T.O., Rodas, F.R., Oyoo, M.E., Senda, M. and Takahashi, R. (2013) Inverted repeat of chalcone synthase 3 pseudogene is associated with seed coat discoloration in soybean. Crop Sci. 53: 518–523 [Google Scholar]
- Senda, M., Jumonji, A., Yumoto, S., Ishikawa, R., Harada, T., Niizeki, M. and Akada, S. (2002a) Analysis of the duplicated CHS1 gene related to the suppression of the seed coat pigmentation in yellow soybeans. Theor. Appl. Genet. 104: 1086–1091 [DOI] [PubMed] [Google Scholar]
- Senda, M., Kasai, A., Yumoto, S., Akada, S., Ishikawa, R., Harada, T. and Niizeki, M. (2002b) Sequence divergence at chalcone synthase gene in pigmented seed coat soybean mutants of the Inhibitor locus. Genes Genet. Syst. 77: 341–350 [DOI] [PubMed] [Google Scholar]
- Senda, M., Masuta, C., Ohnishi, S., Goto, K., Kasai, A., Sano, T., Hong, J.S. and MacFarlane, S. (2004) Patterning of virus-infected Glycine max seed coat is associated with suppression of endogenous silencing of chalcone synthase genes. Plant Cell 16: 807–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senda, M., Kurauchi, T., Kasai, A. and Ohnishi, S. (2012) Suppressive mechanism of seed coat pigmentation in yellow soybean. Breed. Sci. 61: 523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd, J.J. and Vodkin, L.O. (1996) Duplications that suppress and deletions that restore expression from a chalcone synthase multigene family. Plant Cell 8: 687–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja, J.H., Clough, S.J., Chan, W.-C. and Vodkin, L.O. (2004) Tissue-specific gene silencing mediated by a naturally occurring chalcone synthase gene cluster in Glycine max. Plant Cell 16: 819–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja, J.H. and Vodkin, L.O. (2008) Structural features of the endogenous CHS silencing and target loci in the soybean genome. Crop Sci. 48: S-49–S-68 [Google Scholar]
- Tuteja, J.H., Zabala, G., Varala, K., Hudson, M. and Vodkin, L.O. (2009) Endogenous tissue-specific short interfering RNAs silence the chalcone synthase gene family in Glycine max seed coats. Plant Cell 21: 3063–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]