Abstract
Brain aging is associated with diminished circadian clock output and decreased expression of the core clock proteins, which regulate many aspects of cellular biochemistry and metabolism. The genes encoding clock proteins are expressed throughout the brain, though it is unknown whether these proteins modulate brain homeostasis. We observed that deletion of circadian clock transcriptional activators aryl hydrocarbon receptor nuclear translocator–like (Bmal1) alone, or circadian locomotor output cycles kaput (Clock) in combination with neuronal PAS domain protein 2 (Npas2), induced severe age-dependent astrogliosis in the cortex and hippocampus. Mice lacking the clock gene repressors period circadian clock 1 (Per1) and period circadian clock 2 (Per2) had no observed astrogliosis. Bmal1 deletion caused the degeneration of synaptic terminals and impaired cortical functional connectivity, as well as neuronal oxidative damage and impaired expression of several redox defense genes. Targeted deletion of Bmal1 in neurons and glia caused similar neuropathology, despite the retention of intact circadian behavioral and sleep-wake rhythms. Reduction of Bmal1 expression promoted neuronal death in primary cultures and in mice treated with a chemical inducer of oxidative injury and striatal neurodegeneration. Our findings indicate that BMAL1 in a complex with CLOCK or NPAS2 regulates cerebral redox homeostasis and connects impaired clock gene function to neurodegeneration.
Introduction
Circadian rhythms are controlled on a molecular level by cell-autonomous core clock machinery that is present in most cells in the body (1, 2). Circadian output from the suprachiasmatic nucleus (SCN) in the hypothalamus synchronizes tissue-specific cellular clocks to the light-dark cycle. The core circadian clock consists of a set of interacting transcriptional activators and repressors. The activators, or “positive limb” components BMAL1 and its binding partners CLOCK or NPAS2 heterodimerize, bind E-box motifs, and regulate the transcription of a wide variety of genes (3, 4). These positive limb proteins drive the transcription of circadian repressors, or “negative limb” components, including period (PER1-3) and cryptochrome (CRY1 and 2), which in turn inhibit the transcriptional activity of the BMAL1:CLOCK/NPAS2 heterodimers. This cell-autonomous clock machinery serves to synchronize intracellular gene expression to external cues such as light and to align physiologic oscillations in cells and tissues throughout the body. Furthermore, each core clock gene performs unique cellular functions that are distinct from its role in maintaining circadian oscillation, implying that clock genes might control key cellular processes via circadian or noncircadian mechanisms (5).
In peripheral tissues, clock genes serve as critical regulators of cellular metabolism and redox homeostasis and have been implicated in the aging process (6–9). Mice with targeted deletion of Bmal1 display loss of behavioral and physiologic circadian rhythms and develop increased systemic oxidative stress and signs of accelerated aging (9, 10). Conversely, aging is associated with diminished expression of positive-limb clock genes in mouse brain, and impaired circadian oscillation and oxidative injury are associated with brain aging and age-related neurodegenerative conditions in humans, suggesting a possible link between circadian clock dysfunction, oxidative stress, and age-related neurodegeneration (11–15). However, it is unknown whether core clock genes play any role in maintaining neuronal health or if these genes influence neurodegeneration.
Core clock genes are expressed throughout the brain (11, 16), though their function and importance in brain regions other than the SCN are poorly understood. BMAL1 has been implicated in hippocampal and astrocytic function (17–20). In Drosophila, deletion of the Period gene exacerbates brain pathology in neurodegeneration-prone mutants (21). In mice, Bmal1 deletion is associated with impairments in learning and memory as well as subtle increases in brain ROS (22), though no connection between clock genes and neurodegeneration has been clearly established in vertebrates. Thus, we hypothesized that core circadian clock function might regulate redox homeostasis in the mouse brain and that genetic disruption of circadian function might facilitate neuronal injury and neurodegeneration.
Results
Oscillation of circadian clock genes is controlled by Bmal1 in cerebral cortex.
As circadian clock genes expressed in non-SCN brain regions might influence neuronal homeostasis, we examined the expression of selected core clock genes in cerebral cortex samples from young WT mice. Bmal1 and its transcriptional targets Per2 and Dbp all demonstrated circadian oscillation with phases that were similar to those observed in pituitary tissue from a previous experiment (23), as well as to those described in rat cortex (ref. 17 and Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI70317DS1). Expression of Dbp, a PAR bZIP transcription factor that is directly regulated by BMAL1/CLOCK-mediated transcription and serves as a marker of BMAL1/CLOCK transcriptional output (24), decreased by 86% in Bmal1 KO cortex, while expression of RevErba (Nr1d1), another BMAL1 target gene, decreased by 83% (Supplemental Figure 1C). Notably, Per2 mRNA increased by an average of 46%, perhaps due to loss of transcriptional repression of Per2 by RevErba, as previously described (25). Thus, core clock genes are expressed and oscillate with appropriate phase in cerebral cortex, and deletion of Bmal1 elicits transcriptional changes in non-SCN regions similar to those seen in peripheral tissues.
Bmal1 deletion causes age-dependent neuropathology and synaptic degeneration.
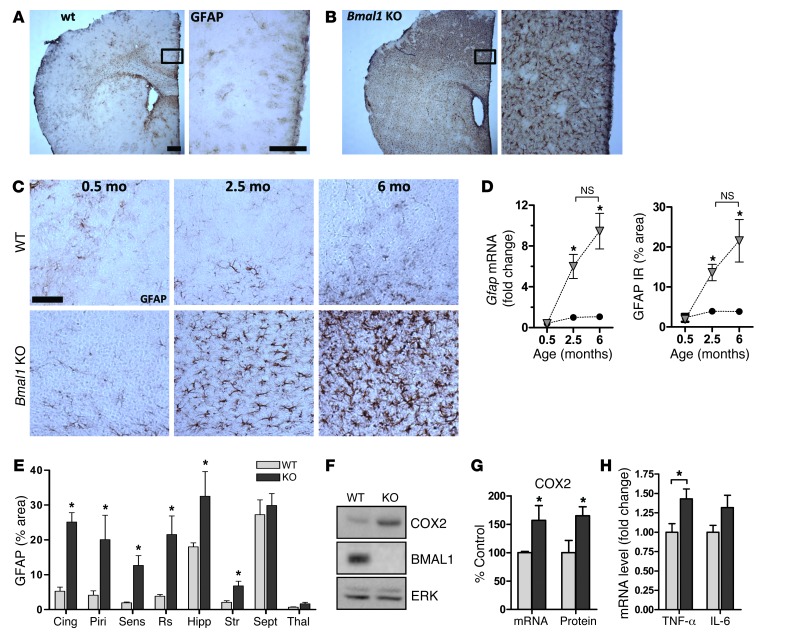
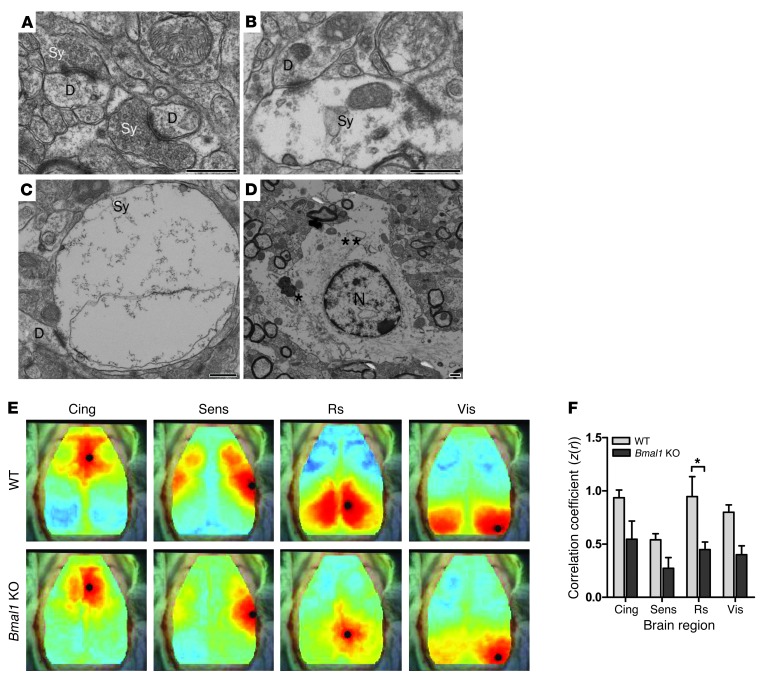
Global Bmal1 KO mice lack circadian rhythmicity in gene transcription and behavior and develop a variety of pathologies reminiscent of accelerated aging (9). Thus, we examined brain pathology in Bmal1 KO mice at 4 to 6 months of age, before peripheral pathologies become severe (9). Brains from 6-month-old Bmal1 KO mice appeared structurally normal with no developmental malformations, and cresyl violet staining demonstrated appropriate neural architecture with no significant thinning of hippocampal cell layers (Supplemental Figure 2). However, immunostaining for glial fibrillary acidic protein (GFAP) revealed striking astrocyte activation, a general marker of brain and neuronal injury that was most severe in cortical regions and hippocampus and less evident in thalamus and brainstem (Figure 1, A, B, and E). Astrocytosis was age dependent, as we did not observe it in 2-week-old Bmal1 KO mice, but it was clearly evident by 2.5 months of age and progressed by 6 months, as quantified by both Gfap mRNA levels and GFAP immunoreactivity (Figure 1, C and D). Astrocyte activation is often associated with increased expression of proinflammatory cytokines, including cyclooxygenase-2 (COX2, Ptghs2) and TNF-α (Tnfa), which exacerbate neurodegeneration (26, 27). We found that Ptghs2 and Tnfa mRNAs and COX2 protein were significantly increased in Bmal1 KO cortex, consistent with chronic neuroinflammation (Figure 1, F–H). Microglial activation was generally subtle in 6-month-old Bmal1 KO brain, though we observed substantial activation in isolated mice that lived until 8 months of age (Supplemental Figure 3). Fluoro-Jade C staining of Bmal1 KO cortex labeled numerous cellular processes in the dentate gyrus of the hippocampus and septal nuclei, which presumably represent injured neuronal processes (Supplemental Figure 2E). We performed ultrastructural analysis of the Bmal1 KO retrosplenial cortex with transmission electron microscopy, which showed many activated astrocytes throughout the tissue (Figure 2, A–D). Neuronal cell bodies and subcellular organelles appeared normal, as did dendritic structures. However, we noted frequent abnormal axonal synaptic terminals that were not seen in the control tissue. These presynaptic terminals were swollen and devoid of normal synaptic vesicles and cytoskeletal architecture, suggesting degeneration (Figure 2, B and C). Interestingly, the synaptic cleft itself appeared intact, as did the postsynaptic terminal. The axons themselves were structurally normal with intact myelination. Thus, Bmal1 deletion leads to widespread astrocytosis as well as discrete degeneration of presynaptic axonal terminals.
Figure 1. Marked age-dependent cerebral astroglial activation in Bmal1 KO mice.
GFAP staining of sections from 6-month-old WT (A) and Bmal1 KO (B) mice shows severe astrogliosis throughout the brain of KO mice and most severe in cortex. Scale bars: 200 μm. (C) GFAP staining of retrosplenial cortex sections from WT and KO mice at 2 weeks of age (0.5 months), 2.5 months, and 6 months of age demonstrates age-dependent astrogliosis, which is present by age 2.5 months. Scale bar: 100 μm (D) Quantification of Gfap mRNA by qPCR and GFAP immunoreaction (IR; % area) by immunostaining of cortex samples shows age-dependent increases in astrogliosis. qPCR was normalized to 18S mRNA levels and is expressed as fold change compared with 2.5-month-old WT control mice. Black circles represent WT mice, and gray triangles represent KO mice (n = 3 mice/point). *P < 0.05 by 1-way ANOVA versus 2-week-old WT mice. (E) Region-specific astrocyte activation in Bmal1 KO brain. Ten mice per genotype were stained for GFAP, while 3 mice per genotype were quantified. Cing, cingulate cortex; Piri, piriform cortex; Sens, sensory cortex; Rs, retrosplenial cortex; Hipp, hippocampus; Str, striatum; Sept, septum; Thal, thalamus. (F) Increased COX2 protein (F and G) and Ptghs2 mRNA (G) in 6-month-old Bmal1 KO cortex. (H) Increased Tnfa mRNA by qPCR in Bmal1 KO cortex. n = 4 mice/genotype (G and H). *P < 0.05 versus WT by 2-way ANOVA with Bonferroni’s post test.
Figure 2. Synaptic degeneration and impaired functional connectivity in Bmal1 KO cortex.
(A–C). Electron micrographs showing presynaptic terminals (Sy) in 6-month-old WT (A) and Bmal1 KO (B and C) retrosplenial cortex. Note that in the Bmal1 KO cortex, the synaptic terminals are swollen and relatively devoid of synaptic vesicles, while the presynaptic and postsynaptic membranes, synaptic cleft, and dendritic spine (D) have normal morphology. Bmal1 KO mice showed both normal and abnormal terminals. (D) An activated astrocyte with a prominent Golgi complex (*) and islands of rough ER (**) around the nucleus (N). This cell is recognized by its abundance of intermediate filaments and cytoplasm with a lucent matrix. Activated astrocytes and numerous organelle-rich astrocytic processes were seen throughout the Bmal1 KO cortical tissue. Scale bars: 500 nm. (E) Composite functional connectivity maps from all mice generated using fcOIS. Shown are the seed locations (black circle) and the map of connectivity with that region (red indicates a positive correlation; blue indicates a negative correlation). (F) Connectivity (correlation coefficient z score) between corresponding contralateral cortical regions (n = 5 mice/genotype, all 6 months of age). Cing, cingulate; Sens, sensory; Rs, retrosplenial; Vis, visual. *P < 0.05 by 2-way ANOVA with Bonferroni’s post test.
Functional connectivity is impaired in Bmal1 KO brain.
We next sought to characterize the functional consequences of the neuropathological changes using optical intrinsic signal functional connectivity imaging (fcOIS) (28). This approach visualizes alterations in regional cortical blood flow in anesthetized mice and generates maps of resting-state functional connectivity between contralateral brain regions. fcOIS is highly sensitive to neuritic pathology in the absence of overt neuronal loss in mouse models of brain aging and Alzheimer disease (AD) (29). As shown in Figure 2, E and F, 5- to 6-month-old Bmal1 KO mice exhibited diminished functional connectivity throughout the cortex, most significantly in the retrosplenial cortex, a region that is severely affected in mouse models of brain aging and AD (29) and that showed severe astrogliosis in Bmal1 KO mice (see Figure 1E). Thus, the neuropathology seen in Bmal1 KO mice coincides in space and time with impaired neuronal network functional connectivity.
Brain-specific deletion of Bmal1 replicates neuropathology despite intact behavioral rhythms and sleep-wake cycle.
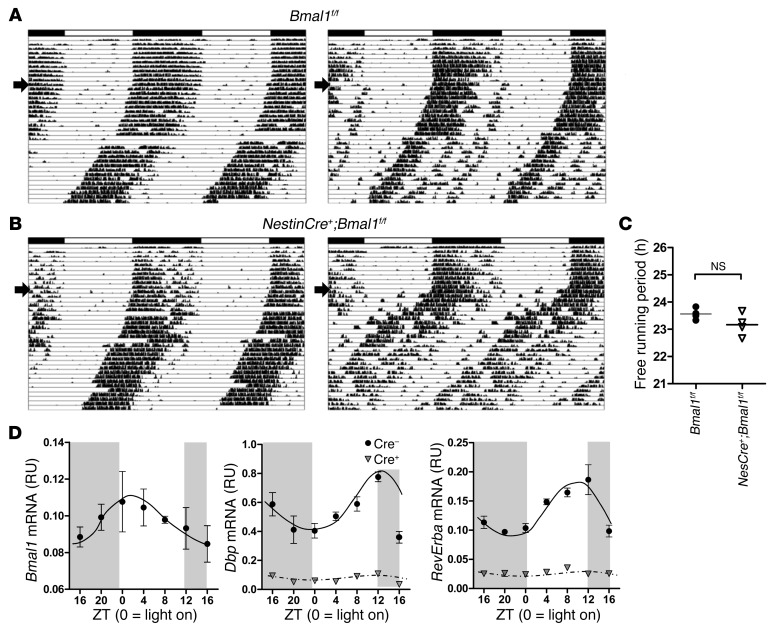
Mice with global deletion of Bmal1 have a variety of peripheral pathologies and a lack of circadian oscillation in the sleep-wake cycle, which could potentially contribute to brain pathology (9, 10, 30). To address this, we generated NestinCre+:Bmal1flox/flox mice (referred to herein as NestinCre+;Bmal1f/f) in which Bmal1 is deleted in most neurons, astrocytes, and oligodendrocytes, with residual Bmal1 expression in microglia (31). These mice do not display gross peripheral pathologies. Similar mice were previously reported to have partially intact Bmal1 expression in the SCN, rescuing the circadian regulation of locomotor activity and sleep (32). Locomotor circadian rhythms were largely retained in our NestinCre+;Bmal1f/f mice as compared with mice lacking Cre, with no genotype difference in the free-running period, although there was a trend toward a slightly shorter free-running period, similar to that reported previously (ref. 32, Figure 3, A–C, and Supplemental Figure 4A). Using EEG, we saw that grossly normal sleep-wake oscillation was retained in NestinCre+;Bmal1f/f mice in 12-hour light/12-hour dark housing conditions (Supplemental Figure 4, B and C). Despite relatively normal activity and sleep rhythms, the expression and oscillation of clock genes were disrupted in cortex tissue from NestinCre+;Bmal1f/f. As shown in Figure 3D, Bmal1f/f control mice entrained to a 12-hour light/12-hour dark schedule that were then placed in constant darkness for 24 hours showed circadian oscillation of Bmal1, Dbp, and RevErba in cerebral cortex, similar to that seen in WT mice and in phase with rhythms observed in liver in previous experiments (see Supplemental Figure 1). However, expression of Dbp and RevErba, both BMAL1 target genes, declined by approximately 90% and became arrhythmic in NestinCre+;Bmal1f/f littermates (Figure 3D).
Figure 3. Brain-specific deletion of Bmal1 disrupts circadian transcriptional regulation in cortex despite intact behavioral circadian rhythms.
(A and B) Actograms showing wheel-running activity in 3- to 4-month-old Bmal1flox/flox control mice (A) and NestinCre+;Bmal1f/f mice (B). Each panel shows data from a representative mouse, recorded for 10 days in a 12-hour light/12-hour dark cycle, then for 30 days in constant darkness (start of constant darkness denoted by arrow). (C) Free-running time for all mice analyzed in A (n = 4/genotype). There was no statistical difference between groups (mean = 23.56 hours for control and 23.17 hours for Nestin-Bmal1 mice; P = 0.14 by 2-tailed Student’s t test). (D) Circadian clock gene expression in cerebral cortex tissue from control (Bmal1f/f, Cre–) and brain-specific Bmal1 KO mice (NestinCre;Bmal1f/f, Cre+; gray triangles). Mice were housed in constant darkness for 24 hours, then harvested every 4 hours. mRNA levels were quantified by qPCR and were normalized to 18S rRNA (n = 2–4 mice/genotype/time point). RU, relative units.
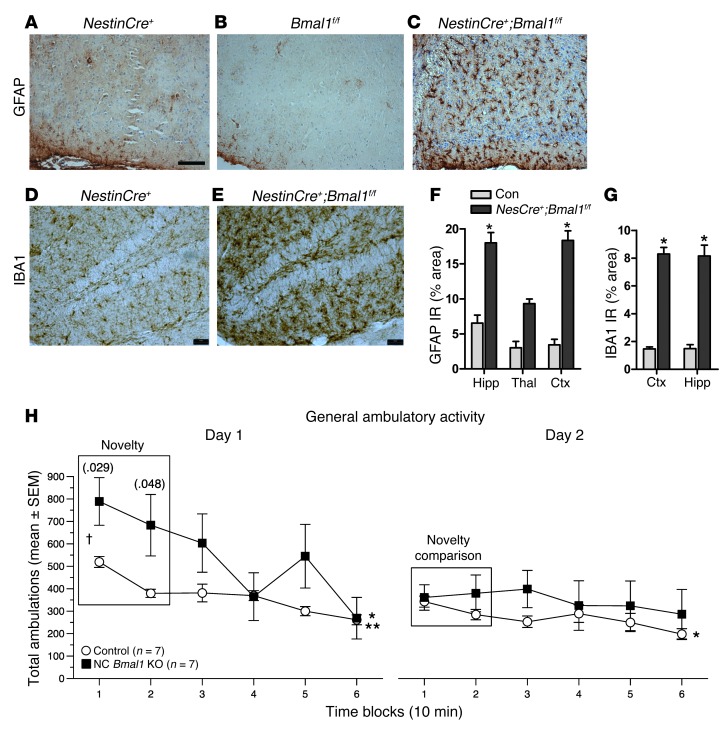
Despite intact behavioral circadian rhythms and rhythmic sleep-wake, NestinCre+;Bmal1f/f mice showed the same severe age-dependent astrogliosis we saw in global Bmal1 KO mice, as well as widespread microglial activation, which was more severe than that observed in global Bmal1 KO mice (Figure 4, A–G). We conclude that the brain phenotype seen in global Bmal1 KO mice is due to local loss of BMAL1 function within neurons/glia, and not to peripheral pathologies, changes in the sleep-wake cycle, or loss of peripheral circadian rhythms.
Figure 4. Brain-specific deletion of Bmal1 causes neuropathology and behavioral abnormalities.
GFAP staining shows marked astrocyte activation in the retrosplenial cortex of NestinCre+:Bmal1f/f mice (C), but not in NestinCre+ (A) or Bmal1f/f controls (B). Hippocampal microglial activation assessed by IBA1 immunoreactivity in a representative Cre+ control (D) and NestinCre+;Bmal1f/f mice (E). Scale bars: 200 μm. Quantification of GFAP (F) and IBA1 (G) immunoreactivity by percentage of area (n = 4 mice/genotype). *P < 0.05 versus control by 2-way ANOVA with Bonferroni’s post test. Ctx, cortex. (H) One-hour locomotor behavioral test reveals a significantly abnormal response to a novel environment in NestinCre+;Bmal1f/f mice (black squares) as compared with Bmal1f/f controls. Data for total ambulations are shown; similar data for vertical rearings are shown in Supplemental Figure 5. n = 7 mice/genotype. P values from repeated-measures ANOVA are displayed for novelty analysis. P < 0.05 for habituation analysis for both genotypes on day 1, but only for Bmal1f/f on day 2; *Bmal1f/f; **NestinCre+;Bmal1f/f.
Since global Bmal1 KO mice exhibit behavioral abnormalities, including novelty-induced hyperactivity (22), we examined the performance of NestinCre;Bmal1f/f mice in a 1-hour locomotor behavioral test. Like Bmal1 global KOs, NestinCre+;Bmal1f/f mice displayed significant abnormalities in their response to a novel environment in 1-hour locomotor testing, as a repeated measures ANOVA of the total ambulation data yielded significant genotype by time (F statistic[5,60] = 3.06, P = 0.040), and genotype by time by test day (F[5,60] = 3.70, P = 0.011) interactions, while the vertical rearing data yielded a significant genotype by time by test day (F[5,60] = 3.58, P = 0.007) interaction (Figure 4H and Supplemental Figure 5). A planned analysis of performance over the first two time blocks to assess novelty on day 1 yielded a significant genotype effect for total ambulations (F[1,12] = 6.60, P = 0.025) and vertical rearing (F[1,12] = 6.75, P = 0.023), with NestinCre+;Bmal1f/f mice showing increased total activity (block 1 P = 0.029, block 2 P = 0.048) and number of rearings (block 1 P = 0.028) compared with WT controls. We found that all mice showed evidence of habituation on day 1 (P = 0.0004 and P < 0.00005, respectively), although only the control mice showed habituation on day 2 (P = 0.022). However, for rearing frequencies, only the NestinCre+;Bmal1f/f mice showed habituation on day 1 (P = 0.012), while neither group showed habituation to rearing on day 2. Thus, unlike the previous report with global Bmal1 KO mice, it was not clear that NestinCre+;Bmal1f/f mice showed impaired habituation. Ultimately, these data are consistent with the hypothesis that NestinCre+;Bmal1f/f mice respond abnormally to novelty. This may represent a behavioral consequence of the observed neuropathology.
Bmal1 regulates redox gene expression and oxidative stress in the brain.
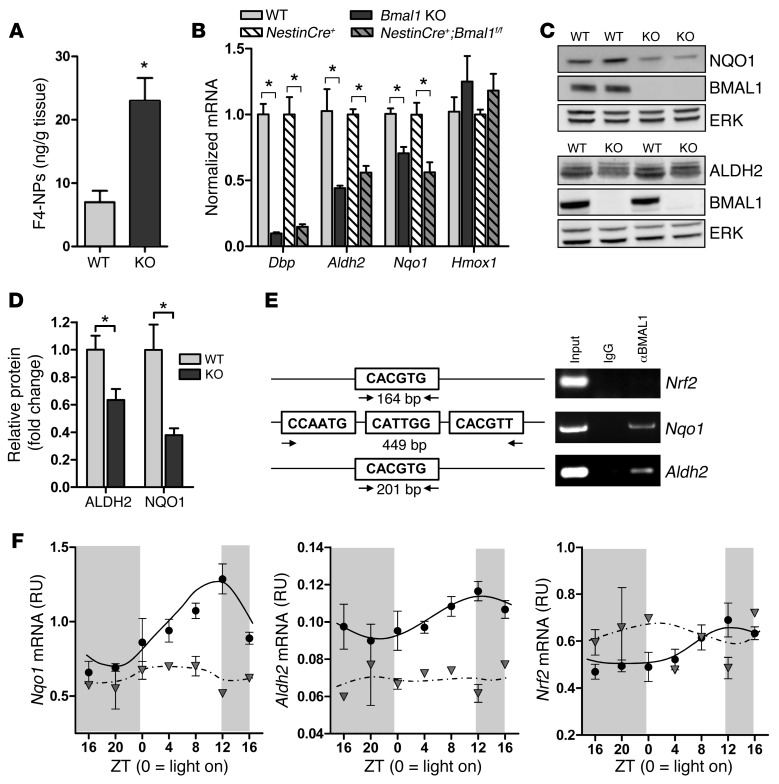
Circadian clock genes have been implicated in the regulation of oxidative stress in several organs (9). Using mass spectrometry, we observed that cortical F4-neuroprostanes (F4-NPs), markers of neuronal membrane lipid peroxidation (33), were increased 3-fold in 6-month-old Bmal1 KO mice, reflective of neuronal oxidative damage (Figure 5A). A similar, albeit nonsignificant, trend was evident for F2-isoprostanes (F2-IPs) (Supplemental Figure 6A), general markers of lipid peroxidation in all cell types. Marked increases in 4-hydroxynonenal Michael adducts, markers of lipid peroxidation, were also observed by immunohistochemistry throughout the Bmal1 KO brain (Supplemental Figure 6, B and C). To investigate the regulation of oxidative stress by the circadian clock, we examined the circadian oscillation of 89 candidate genes (see Supplemental Table 1) known to be involved in the regulation of oxidative stress in several tissues using the CircaDB Circadian Expression Profiles database ( http://bioinf.itmat.upenn.edu/circa), which queries microarray data from several published circadian microarray experiments from non-brain tissues. Circadian oscillation was detected using the JTK_CYCLE nonparametric algorithm (34), with a P value cutoff of 0.05. We also examined the expression level of this same panel of genes in WT and Bmal1 KO pituitary tissue using previously published microarray data (ref. 35; GEO accession number GSE29664). From this, we identified a set of candidate redox genes, each of which have been previously implicated in neurodegeneration, including Ucp2, Sod2, Prdx6, Aldh2, Nfe2l2 (NRF2), and the NRF2 target genes Nqo1 and Hmox1. We examined the expression of these genes in WT and Bmal1 KO cortex at a single time point (Zeitgeber time [ZT] 6, Figure 5B and Supplemental Figure 7A) and in NestinCre+;Bmal1f/f brain tissue at ZT 0, 6, and 12 (Figure 5B and Supplemental Figure 7, A and B). Of these, the expression of Nqo1 and Aldh2 was significantly reduced in both Bmal1 KO and Nestin+;Bmal1f/f cortex as compared with controls at ZT 6. We thus examined the regulation of Nqo1 and Aldh2 in more detail.
Figure 5. Bmal1 deletion induces oxidative stress and redox defense gene dysregulation.
(A) Increased F4-NP levels as quantified by liquid chromatography tandem mass spectrometry (LC-MS/MS) in 6-month-old Bmal1 KO cortex, indicative of neuronal membrane lipid peroxidation (n = 5 mice/genotype). (B) Quantification of Dbp and redox gene expression in Bmal1 KO and NestinCre+;Bmal1f/f cortex versus controls at ZT 6 (n = 5–6 mice/genotype). Bmal1 KO values were normalized and compared with WT cortex, while NestinCre+;Bmal1f/f values were normalized and compared with NestinCre+ controls. (C) Representative Western blots showing decreased NQO1 (upper blots) and ALDH2 (lower blots) protein in Bmal1 KO brain at ZT 6. ERK is shown as a loading control. (D) Quantification of ALDH2 and NQO1 protein (n = 5 mice/genotype). Shown is the mean + SEM for all graphs. *P < 0.05 versus control by Student’s t test (A) or 2-way ANOVA with Bonferroni’s post test (B and D). (E) ChIP assay in WT mice at ZT 6 demonstrating that BMAL1 does not bind to a canonical E-box in the Nrf2 promoter, but does bind a noncanonical E-box in the Nqo1 promoter and a canonical E-box in the Aldh2 promoter. Total lysate (input, positive control) and immunoprecipitates prepared using nonspecific IgG (negative control) are shown. (F) BMAL1 regulates cortical expression of Nqo1 and Aldh2, but not Nrf2. Frontal cortex samples were collected every 4 hours from Bmal1f/f control mice (black circles) or NestinCre+;Bmal1f/f mice (gray triangles) as in Figure 3D, and redox genes were quantified by qPCR (n = 2–3 mice/time point/genotype).
ALDH2 scavenges reactive aldehydes generated during mitochondrial respiration and protects neurons against oxidative stress–induced neurodegeneration (36, 37). In our Bmal1 KO cortex samples, Aldh2 mRNA levels were significantly decreased on average by 58% and protein by 37% at ZT 6 (Figure 5, B–D). Nqo1 encodes NADPH dehydrogenase (quinone 1), a critical redox defense enzyme that reduces toxic quinones, suppresses oxidative damage, and may prevent neurodegeneration (38). Nqo1 mRNA and protein levels were also significantly decreased in Bmal1 KO brain, while mRNA was diminished by approximately 50% in NestinCre;Bmal1f/f cortex (Figure 5, B–D). We performed ChIP assays from WT mouse cortex to determine whether BMAL1 directly regulates the transcription of these genes. BMAL1 binds to noncanonical E-box motifs in the Nqo1 promoter (Figure 5E). ChIP also showed that BMAL1 binds to a canonical E-box in the Aldh2 promoter, but does not bind a canonical E-box in the Nrf2 promoter. Accordingly, the NRF2 target genes heme oxygenase 1 (Hmox1) and glutamate-cysteine ligase catalytic subunit (Gclc) showed nonsignificant trends toward increased expression at all time points in both Bmal1 KO and NestinCre+;Bmal1f/f cortex (Figure 5B and Supplemental Figure 7B), suggesting a normal NRF2-mediated response to increased oxidative stress, and showing that suppressed Nqo1 and Aldh2 expression in Bmal1 KO cortex is not due to general inhibition of NRF2 signaling. We examined the circadian expression of these redox genes in cortex of Bmal1f/f and NestinCre+;Bmal1f/f mice and observed that Nqo1 oscillated in phase with other BMAL1 targets such as Dbp and RevErba (see Figure 3 for comparison), while Aldh2 and Nrf2 showed minimal oscillation (Figure 5F). Nqo1 and Aldh2 mRNA levels decreased at all time points and were arrhythmic in NestinCre+;Bmal1f/f cortex, further demonstrating transcriptional regulation by BMAL1. Thus, neuronal oxidative damage is evident in Bmal1 KO brain, and numerous redox-related transcripts exhibit circadian oscillation. Moreover, BMAL1 regulates the transcription of key redox response genes, including Nqo1 and Aldh2.
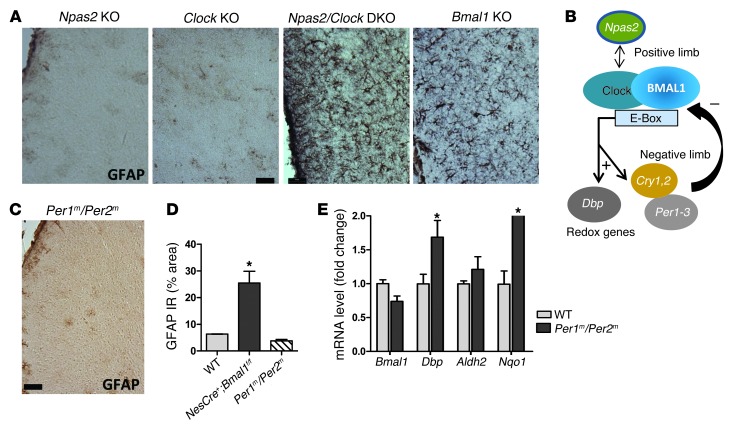
Dual deletion of Clock and Npas2 phenocopies Bmal1 deletion, while dual Per1 and Per2 mutation does not.
Next, we sought to determine whether the neuropathology was specific to deletion of Bmal1 or common to other core clock genes. BMAL1 can heterodimerize with either CLOCK or NPAS2 to drive gene transcription. Because CLOCK and NPAS2 serve redundant roles in core clock function in the SCN (39) and are both expressed in the brain, we examined brains from Npas2 KO, Clock KO, and Npas2/Clock double-KO (DKO) mice for neuropathology. While Npas2 and Clock single-gene KOs resembled WT mice, Npas2/Clock DKO mice recapitulated the Bmal1 KO phenotype, showing marked astroglial activation throughout the brain, but was most severe in cortex (Figure 6A). Thus, disruption of the heterodimeric positive-limb transcriptional complex, either via deletion of Bmal1 or of both of its binding partners, leads to neuropathology. The negative limb of the core clock consists of several BMAL1 targets, including Period 1-3 (Per1-3, Figure 6B). Per1m/Per2m double-mutant mice have disrupted circadian clock function and become immediately arrhythmic in constant darkness (40, 41), but have intact Bmal1 expression. In contrast to positive limb disruption, there was no evidence of increased astrogliosis in age-matched Per1m/Per2m DKO mice (Figure 6, C and D). Per1m/Per2m cortex also showed an opposite transcriptional pattern from that seen in Bmal1 KO mice, with increased levels of Dbp (demonstrating derepression of Bmal1 transcriptional activity) and Nqo1 and a trend toward increased Aldh2 (Figure 6E). Thus, deletion of negative limb repressors (Per1/2) enhances BMAL1-mediated transcription of target genes such as Dbp and expression of Nqo1/Aldh2. Taken together, these findings show that transcriptional regulation specifically by positive-limb BMAL1:NPAS2/CLOCK heterodimers is required to prevent neuropathology in the brain.
Figure 6. Genetic disruption specifically of the positive limb of the circadian clock causes neuropathology.
(A) GFAP staining of retrosplenial cortex from 4-month-old Npas2 KO, Clock KO, Npas2/Clock DKO, and Bmal1 KO mice shows that complete disruption of the positive limb of the core clock (depicted in B) is required to elicit neuropathology. (C) Absence of astrogliosis in retrosplenial cortex from 4-month-old Per1m/Per2m mice, which lack negative limb clock function and have dysfunctional circadian oscillation. (D) Quantification of GFAP immunoreactivity in Per1m/Per2m mice (% area). NestinCre+;Bmal1f/f cortex is shown for comparison. (E) qPCR data from 4-month-old Per1m/Per2m cortex show a transcriptional profile opposite that of Bmal1 KO brain (see Figure 1). (D and E) n = 4 mice/genotype. *P < 0.05 versus control by 2-way ANOVA with Bonferroni’s post test. Scale bars: 50 μm (A and C).
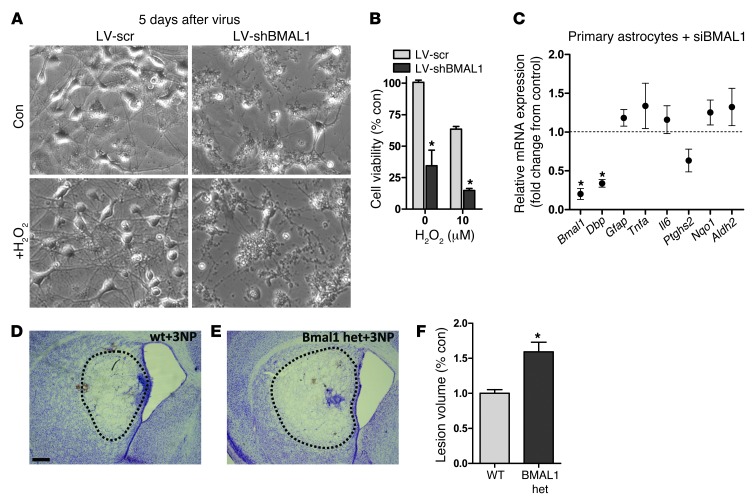
Diminished Bmal1 expression exacerbates neurodegeneration in vitro and in vivo.
In primary mouse neuron–enriched cultures, infection with a lentiviral shRNA (LV-shBMAL1) achieved an approximately 50% decrease in Bmal1 and an approximately 60% decrease in Dbp mRNA when compared with sister cultures treated with an identical lentivirus expressing a scrambled shRNA (LV-shSCR) (Supplemental Figure 8). By day 5 after lentiviral exposure, LV-shBMAL1 cultures exhibited spontaneous neurite degeneration and cell death, while LV-shSCR–treated cells continued to appear healthy (Figure 7, A and B). Treatment with a low concentration of hydrogen peroxide (H2O2) exacerbated cell death even further in LV-shBMAL1 cultures. Knockdown of Bmal1 in Neuro2a neuroblastoma cells using siRNA targeted at a distinct sequence from the lentiviral shRNA did not induce spontaneous cell death, but did increase cell death caused by the mitochondrial toxin rotenone (Supplemental Figure 9, B and C), suggesting that the toxic effects of Bmal1 knockdown in primary neurons are not due to off-target effects of the shRNA. Conversely, siRNA-mediated knockdown of Bmal1 (~85% decrease) in primary astrocyte cultures had no effect on cell viability (Supplemental Figure 9A), did not induce significant astrocyte activation (as assessed by Gfap mRNA upregulation), did not suppress Aldh2 or Nqo1 expression, and did not induce inflammatory gene expression (Tnfa, Ptghs2, Il6), as compared with scrambled siRNA-treated cells (Figure 7C). This suggests that loss of Bmal1 expression in neurons, not astrocytes, is the primary driver of pathology. To address the hypothesis that gene dosage of Bmal1 might modulate oxidative neurodegeneration in vivo, we used the mitochondrial complex III inhibitor 3-nitroproprionic acid (3-NP), which induces oxidative injury and striatal neurodegeneration (42). We treated WT and Bmal1 hemizygous mice (Bmal1+/–), which show no spontaneous neuropathologic phenotype in the age range we examined, with intrastriatal stereotactic injection of 3-NP, and 3 days later measured striatal lesion size by cresyl violet and Fluoro-Jade C staining (see Supplemental Figure 10). As shown in Figure 7, D–F, Bmal1+/– mice had significantly larger lesions than WT mice, supporting the notion that Bmal1 expression levels play a critical role in neuronal redox homeostasis and neurodegeneration.
Figure 7. Diminished Bmal1 expression enhances neurodegeneration.
(A and B). Bmal1 knockdown in primary neuronal cultures induces neuronal death. (A) Representative phase-contrast photomicrographs from DIV 7 mouse cortical neuron–enriched cultures 5 days after treatment with LV expressing scrambled shRNA (LV-scr) or BMAL1 shRNA (LV-shBMAL1) and after 24 hours of treatment with vehicle (Con) or H2O2. Original magnification, ×10. (B) Quantification of cell viability as assessed by MTT assay. Data are representative of three independent experiments. (C) siRNA-mediated knockdown of Bmal1 in primary astrocyte cultures does not induce astrocyte activation or suppress Nqo1 or Aldh2 transcription. mRNA levels quantified by qPCR and normalized to 18S rRNA. Data are expressed as fold change from sister cultures transfected with scrambled siRNA. The mean ± SEM of four separate experiments is shown. *P < 0.05 by 1-way ANOVA with Bonferroni’s post test. (D and E) Representative photomicrographs showing area of striatal neurodegeneration (dotted line) in WT (D) and Bmal1 hemizygous (E) mice 3 days after intrastriatal injection of 3-NP. Scale bar: 250 μm. (F) Quantification of 3-NP striatal lesion volume as assessed by cresyl violet staining. *P < 0.05 by 2-way ANOVA with a Bonferroni’s post test, as compared with control condition. het, heterodimer.
Discussion
It is now evident that core clock genes regulate critical aspects of cellular biology in many organs and that disruption of normal core clock function may precipitate disease pathology. Indeed, impaired clock gene expression in mice can cause a variety of pathologies, including diabetes, vascular disease, obesity, and accelerated aging (6, 8, 9, 43). Despite these findings, relatively little is known about the role of clock genes in other brain regions. Our results show that disruption of the positive limb of the circadian clock in the brain, either via deletion of Bmal1 (either globally on in a brain-specific manner) or Npas2 and Clock, induces oxidative stress, widespread astrocyte activation, axonal terminal degeneration, and disrupted resting-state functional connectivity. This neuropathology is not due to a disruption in peripheral physiology, sleep disruption, or systemic circadian mechanisms and is not recapitulated by mutation of the negative-limb genes Per1 and Per2. The degree of astrogliosis and oxidative stress do not perfectly correlate, as thalamus shows minimal astrogliosis but significant increases in 4-HNE, suggesting that the regionality of brain pathology in Bmal1 KO mice is driven both by the degree of oxidative damage and the inherent susceptibility of a given region. The expression levels of Bmal1 appear to be critical, as Bmal1+/– mice, though not exhibiting overt pathology or circadian phenotype, are more sensitive than WT mice to oxidative neurodegeneration caused by the mitochondrial inhibitor 3-NP, while even partial knockdown of Bmal1 in primary neurons induces spontaneous neurodegeneration. These data reveal a critical role for the circadian clock positive-limb transcriptional complex (BMAL1:CLOCK/NPAS2) in neuronal redox homeostasis and protection from neurodegeneration.
BMAL1:NPAS2/CLOCK heterodimers regulate the transcription of many circadian and noncircadian genes, and both sets of transcripts are likely relevant to neuronal function (5). In our studies, the neuropathology in Bmal1 KO and Clock/Npas2 DKO mice does not appear to be due to loss of systemic circadian rhythms or sleep-wake disturbance, since (a) NestinCre+;Bmal1f/f mice develop neuropathology despite normal behavioral rhythms and sleep-wake oscillation; (b) Per1m/Per2m mice, which have impaired circadian rhythms, do not develop pathology; and (c) Bmal1+/– mice, which exhibit normal circadian oscillation, have enhanced sensitivity to 3-NP–induced neurodegeneration. It thus appears that the levels of Bmal1 and/or Clock and Npas2 are the important factors, rather than the circadian oscillation of transcripts. However, the levels and activity of Bmal1, Npas2, and Clock are intimately tied to circadian oscillation, and conditions that disrupt oscillation can suppress Bmal1 expression or transcriptional activity. These include cellular senescence, sleep deprivation, and pulsed-light exposure (44–46). Therefore, these circadian “stressors” may predispose individuals to age-related neurodegeneration by suppressing positive-limb clock gene expression and activity. Circadian dysfunction is observed in several neurodegenerative diseases, including AD and Parkinson disease (12, 13, 47), suggesting that disease-related loss of clock gene function might impact pathogenesis and disease progression. Accordingly, epidemiologic data show that diminished circadian function in humans imparts an increased risk of developing future dementia (48). Our findings suggest that further study of the regulation of circadian clock genes in non-SCN brain regions in aging and neurodegenerative diseases is warranted and that therapies targeted at bolstering positive-limb clock gene expression in the brain might have neuroprotective effects.
Aging is a major risk factor for neurodegenerative diseases, and the core clock is intimately intertwined with the aging process, as aging impairs the expression of Bmal1 and Clock (11), while disruption of Bmal1 recapitulates many aspects of age-related pathology (9). In vascular smooth muscle cells, senescence is associated with a marked decline in Bmal1 expression (44), while the expression of Bmal1 and Clock in cerebral cortex is substantially diminished in aged mice (11). Our results predict that aging-related declines in Bmal1 expression in the brain could impair redox defense gene expression, exacerbate oxidative stress, and facilitate neurodegeneration. Indeed, aged brain shares many characteristics with Bmal1 KO brain, including increased oxidative damage, diminished redox defense gene expression, increased Ptghs2 expression, and impaired retrosplenial functional connectivity. Thus, declining positive-limb core clock activity in non-SCN brain may be part of a feed-forward cycle with aging, which may exacerbate specific age-related pathogenic events that contribute to neurodegeneration (49–52).
Our findings, as well as previous studies, suggest that the core clock plays an important role in redox homeostasis. Bmal1 KO mice exhibit increased ROS levels in spleen, kidney, and brain, and treatment with the glutathione precursor N-acetyl cysteine extends lifespan in these mice (9, 22, 53). The cellular redox state shows circadian oscillation, which is dependent on Bmal1 expression both in cultured fibroblasts and the mouse SCN (54, 55). Furthermore, the acetylation of multiple critical mitochondrial proteins shows circadian oscillation, suggesting clock-mediated control of the mitochondrial redox state (56). Conversely, clock function is modulated by the redox state of the cell (7, 57, 58). In the brain, our observation of impaired expression of Aldh2 and Nqo1, despite ongoing oxidative stress, suggests that the core clock is required for appropriate protective responses to oxidant injury, illustrating a new aspect of this relationship between the clock and ROS. Aldh2 and Nqo1 are directly regulated by BMAL1, and their expression is diminished when Bmal1 is deleted. However, the expression of both genes increases in parallel with Dbp (a marker of positive-limb transcriptional activity) in Per1m/Per2m DKO brain, suggesting that alleviation of Per1/2-mediated repression of BMAL1 transcriptional activity enhances Aldh2 and Nqo1 expression. Both NQO1 and ALDH2 are critical mediators of the cellular antioxidant response and are closely linked to neurodegeneration. ALDH2 scavenges reactive electrophiles within mitochondria, and deletion of Aldh2 in mice causes oxidative damage and neuronal death, while impaired ALDH2 activity has been implicated as a cause of dopaminergic neurodegeneration in Parkinson disease (36, 37). NQO1 catalyzes the reduction of reactive quinones and oxidized proteins, prevents ROS-mediated cytotoxicity, and is upregulated in vulnerable brain regions in AD (38, 59). Thus, impaired Aldh2 and Nqo1 expression may contribute to neuronal pathology in Bmal1 KO brain.
In summary, our findings draw a novel link between the core circadian clock, brain oxidative stress, and neurodegeneration. This relationship has many potential implications for age-related neurodegenerative diseases and suggests that further study of the regulation and function of core clock genes in non-SCN brain regions in health and disease is warranted.
Methods
Reagents.
The following antibodies were used: ALDH2 and NQO1 monoclonals (Epitomics); BMAL1 polyclonal (Bethyl Laboratories, Inc.); COX2 polyclonal (Cayman Chemical); ERK (Cell Signaling Technology); GFAP (Dako); IBA1 (Wako); and 4-HNE Michael Adduct (EMD Millipore). Cell culture media and reagents (neurobasal media, B27 supplement) and TaqMan quantitative PCR (qPCR) primer sets were obtained from Invitrogen. 3-Nitropropionic acid was purchased from Sigma-Aldrich.
Mice.
Bmal1+/–, NestinCre+, and Bmal1f/f mice were obtained from The Jackson Laboratory and were bred at Washington University. A second strain of NestinCre+;Bmal1f/f mice was bred at the University of Pennsylvania and was used in some experiments. A second strain of Bmal1+/– mice was originally obtained from C. Bradfield (University of Wisconsin, Madison, Wisconsin, USA) and bred at the University of Pennsylvania. Npas2 KO (originally provided by S. McKnight, UT Southwestern Medical School, Dallas, Texas, USA), Clock KO, and Npas2/Clock DKO mice were bred and housed at the University of Massachusetts Medical School. Per1ldc/Per2ldc (referred to herein as Per1m/Per2m) were originally provided by S. Reppert (University of Massachusetts, Worcester, Massachusetts, USA) and were bred at Washington University. All mice were maintained on a C57Bl6 background. Mice were housed under 12-hour light/12-hour dark conditions.
Optical imaging of functional connectivity.
OIS imaging of resting-state functional connectivity in mice was performed as previously described (28, 29). Briefly, mice were anesthetized with an i.p. ketamine/xylazine mixture (86.9 mg/kg ketamine, 13.4 mg/kg xylazine) and allowed 30 minutes for anesthetic transition. Once induced, each animal was placed on a heating pad maintained at 37°C (mTCII; Cell MicroControls) and its head secured in a stereotactic frame. Next, the scalp was reflected, exposing approximately 1 cm2 of the skull. Sequential illumination of the skull surface was provided at four wavelengths by a ring of light-emitting diodes placed approximately 10 cm above the mouse’s head. Images were captured using a cooled, frame-transfer EMCCD camera (iXon 897; Andor Technology), which was time synchronized and controlled via computer using custom-written software (MATLAB; MathWorks). Images were acquired at a frame rate of 120 Hz, and 7–9 five-minute imaging sessions were performed per mouse.
Western blotting.
Tissue samples were homogenized by sonication on ice in radioimmunoprecipitation (RIPA) buffer (Pierce, Thermo Scientific) containing cOmplete protease inhibitors and PhosSTOP phosphatase inhibitors (Roche). PAGE and Western blotting were performed using Invitrogen Novex gels and reagents and an iBlot transfer device per the manufacturer’s instructions. Bands were visualized using Lumigen TMA-6 chemiluminescence reagents on a Syngene GBOX imaging system. Band intensity was quantified using ImageJ software (NIH) and was normalized to the ERK loading control.
qPCR.
For RNA isolation, brain tissue was homogenized by trituration through a 23-gauge needle in TRIzol (Invitrogen). Chloroform (1:5) was added, samples were agitated then centrifuged at 13,000 g for 15 minutes at 4°C, and the chloroform layer was removed, diluted 1:1 in 70% ethanol, then purified using RNeasy columns and reagents (QIAGEN). RNA concentration was measured using a NanoDrop spectrophotometer, and reverse transcription was performed using a high-capacity RNA-cDNA kit (Applied Biosystems [ABI]) with 1 μg RNA per 20 μl reaction. Real-time qPCR was performed using ABI TaqMan primers and reagents on an ABI Prizm 7500 thermocycler according to the manufacturer’s instructions. All mRNA measurements were normalized to 18S rRNA or β-actin mRNA levels.
Primary neuronal and astrocyte cultures.
Primary cultures were prepared from E17 C57/Bl6 WT cerebral cortices, which were dissected and incubated in 5 ml HBSS containing 3 mg bovine pancreatic trypsin (Sigma-Aldrich) at 37°C for 15 minutes. Cortices were then gently triturated in warm HBSS plus 10% FBS, diluted in warmed MEM with 10% FBS to a concentration of 275,000 cells/ml, and then added to 24- or 96-well plates precoated with poly-D-lysine (50 μg/ml; Sigma-Aldrich) and laminin (25 μg/ml; Invitrogen). After 24 hours, 60% of the media was replaced with neurobasal media plus 1X B27 supplement and 5 mM L-glutamine. Lentivirus was added to neurons on days in vitro (DIV) 2 at a concentration of 1 × 106 trophic units (TU) per milliliter for all viruses (2.5 TU/cell). On DIV 4, lentivirus-containing media were removed and replaced with neurobasal plus B27 and glutamine. On DIV 6 (4 days after lentiviral exposure), the media were changed to neurobasal plus B27 without antioxidants, with or without 10 μM hydrogen peroxide. Twenty-four hours later, the cells were harvested for RNA extraction, or cell viability was determined by MTT assay. For astrocyte cultures, P1 mice were dissected as described above. Cells were diluted in DMEM with F-12 supplement (Invitrogen) and 15% FBS and 10 ng/ml epidermal growth factor (Sigma-Aldrich).
Immunohistochemistry.
Immunostaining was performed as previously described (29). Briefly, mice were anesthetized via i.p. pentobarbital, then perfused for 3 minutes with ice-cold Dulbecco’s modified PBS (DPBS) containing 3 g/l heparin. One hemisphere was fixed in 4% paraformaldehyde for 24 hours (4°C), then cryoprotected with 30% sucrose in PBS (4°C) for 48 hours, and 50-micron serial coronal sections were cut on a freezing sliding microtome. Sections were incubated in 0.3% hydrogen peroxide for 10 minutes, blocked for 30 minutes in TBS containing 3% serum and 0.25% Triton X-100, then incubated overnight in TBS plus 0.25% Triton X-100 (Sigma-Aldrich) with primary antibody and 1% serum at 4°C. Sections were incubated for 1 hour with biotinylated secondary antibody, then washed and incubated with 1:400 dilution of streptavidin-conjugated HRP (VECTASTAIN ABC Elite; Vector Laboratories), then with diaminobenzidine substrate with hydrogen peroxide and nickel chloride.
Quantification of immunoreactivity.
Images from a given brain region from three sections per mouse that were 150 microns apart were converted to grayscale, thresholded such that all GFAP immunoreactivity was included, then the percentage of area was calculated using ImageJ software. Sections from all mice included in a given analysis were stained in a single batch, and threshold values were held constant for all mice in that batch. The mean percentage of the area for each region from each mouse was compiled for statistical analysis.
LV preparation. shRNA sequences targeting Bmal1 (Arntl, clone NM_007489.1-2418s1c1, CCATTGATACAAGTCAATCTA) or a scrambled sequence were cloned into a lentiviral vector containing a U6 promoter (ubiquitous expression) upstream of the shRNA as well as a phosphoglycerate kinase (Pgk) promoter driving a GFP tag.
3-NP treatment.
Sterile 3-NP (100 nmol/μl) in PBS (pH 7.4) was injected with a 30-gauge Hamilton syringe needle at the following coordinates relative to the bregma: +0.98 mm anterior, +1.5 mm lateral, and depth of 2.6 mm. The needle was left in place for 5 minutes, then 0.5 μl (total of 50 nmol) of 3-NP was injected over a 5-minute period. Three days later, the mice were perfused as described above.
siRNA experiments.
siRNAs (scrambled or Bmal1 targeted) were obtained from Thermo Scientific (Dharmacon SMARTpool siRNAs, which consists of a pool of 5 siRNA targeting Bmal1). Neuro2a neuroblastoma cells (ATCC) were cultured in DMEM plus 10% FBS and were plated at a density of 3 × 105 cells/ml (0.5 ml per well in a 24-well plate). One microliter of 20 μM siRNA was added to each well. Neuro2a cells were transfected with Lipofectamine 2000 reagent, while astrocytes were transfected with RNAiMax reagent (both from Invitrogen), according to the manufacturer’s instructions.
Additional methods, including behavioral testing, electron microscopy, ChIP, mass spectrometry, and Fluoro-Jade staining, are available in the Supplemental Methods.
Statistics.
A 2-tailed Student’s t test was used when a single variable was compared between two genotypes, and 2-way ANOVA with Bonferroni’s post test was used when multiple variables were compared between genotypes. The cutoff for significance was P < 0.05. In all figures, the graphs depict the mean ± SEM.
Study approval.
All animal studies were performed in accordance with protocols approved by the Animal Studies Committees of Washington University and the University of Pennsylvania.
Supplementary Material
Acknowledgments
This study was supported by NIH grants K08NS079405 and R25NS065745 (to E.S. Musiek), HL097800 (to G.A. FitzGerald and J.B. Hogenesch), P01NS074969 (to D.M. Holtzman), P30NS057105 (to D.M. Holtzman), NS056125 (to D.R. Weaver), an Ellison Medical Foundation Senior Scholar Award (to D.M. Holtzman), the Cure Alzheimer’s Fund (to D.M. Holtzman), and an AAN Clinical Research Training Fellowship (to J.H. Roh). G.A. FitzGerald is a McNeill Professor of Translational Medicine and Therapeutics. The Hope Center Viral Vector Core is supported by NIH grant P30 NS057105. The authors thank Krystal Emmer, Floy Stewart, and Tom Mahan for histological assistance, Michael Adam for assistance with mouse breeding, Ronald Perez and the Hope Center Surgical Core at Washington University for performing stereotactic injections, and Mingjie Li for lentivirus preparation.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(12):5389–5400. doi:10.1172/JCI70317.
Jee Hoon Roh’s present address is: Department of Neurology, Asan Medical Center, Seoul, Republic of Korea.
See the related Commentary beginning on page 4994.
References
- 1. Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001. 63: 647 676 [DOI] [PubMed] [Google Scholar]
- 2.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hogenesch JB, Gu YZ, Jain S, Bradfield CA. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci U S A. 1998;95(10):5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gekakis N, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280(5369):1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 5.Yu EA, Weaver DR. Disrupting the circadian clock: gene-specific effects on aging, cancer, and other phenotypes. Aging. 2011;3(5):479–493. doi: 10.18632/aging.100323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330(6009):1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293(5529):510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 8.Rudic RD, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2(11):e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20(14):1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bunger MK, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103(7):1009–1017. doi: 10.1016/S0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyse CA, Coogan AN. Impact of aging on diurnal expression patterns of CLOCK and BMAL1 in the mouse brain. Brain Res. 2010;1337:21–31. doi: 10.1016/j.brainres.2010.03.113. [DOI] [PubMed] [Google Scholar]
- 12.Witting W, Kwa IH, Eikelenboom P, Mirmiran M, Swaab DF. Alterations in the circadian rest-activity rhythm in aging and Alzheimer’s disease. Biol Psychiatry. 1990;27(6):563–572. doi: 10.1016/0006-3223(90)90523-5. [DOI] [PubMed] [Google Scholar]
- 13.Hu K, Van Someren EJ, Shea SA, Scheer FA. Reduction of scale invariance of activity fluctuations with aging and Alzheimer’s disease: Involvement of the circadian pacemaker. Proc Natl Acad Sci U S A. 2009;106(8):2490–2494. doi: 10.1073/pnas.0806087106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farajnia S, et al. Evidence for neuronal desynchrony in the aged suprachiasmatic nucleus clock. J Neurosci. 2012;32(17):5891–5899. doi: 10.1523/JNEUROSCI.0469-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondratova AA, Kondratov RV. The circadian clock and pathology of the ageing brain. Nat Rev Neurosci. 2012;13(5):325–335. doi: 10.1038/nrn3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abraham U, Prior JL, Granados-Fuentes D, Piwnica-Worms DR, Herzog ED. Independent circadian oscillations of Period1 in specific brain areas in vivo and in vitro. J Neurosci. 2005;25(38):8620–8626. doi: 10.1523/JNEUROSCI.2225-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rath MF, Rohde K, Fahrenkrug J, Moller M. Circadian clock components in the rat neocortex: daily dynamics, localization and regulation. Brain Struct Funct. 2012;218(2):551–562. doi: 10.1007/s00429-012-0415-4. [DOI] [PubMed] [Google Scholar]
- 18.Jilg A, et al. Temporal dynamics of mouse hippocampal clock gene expression support memory processing. Hippocampus. 2010;20(3):377–388. doi: 10.1002/hipo.20637. [DOI] [PubMed] [Google Scholar]
- 19.Valnegri P, et al. A circadian clock in hippocampus is regulated by interaction between oligophrenin-1 and Rev-erbalpha. Nat Neurosci. 2011;14(10):1293–1301. doi: 10.1038/nn.2911. [DOI] [PubMed] [Google Scholar]
- 20.Marpegan L, et al. Circadian regulation of ATP release in astrocytes. J Neurosci. 2011;31(23):8342–8350. doi: 10.1523/JNEUROSCI.6537-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krishnan N, Rakshit K, Chow ES, Wentzell JS, Kretzschmar D, Giebultowicz JM. Loss of circadian clock accelerates aging in neurodegeneration-prone mutants. Neurobiol Dis. 2012;45(3):1129–1135. doi: 10.1016/j.nbd.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondratova AA, Dubrovsky YV, Antoch MP, Kondratov RV. Circadian clock proteins control adaptation to novel environment and memory formation. Aging. 2010;2(5):285–297. doi: 10.18632/aging.100142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes ME, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5(4):e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gachon F, et al. The loss of circadian PAR bZip transcription factors results in epilepsy. Genes Dev. 2004;18(12):1397–1412. doi: 10.1101/gad.301404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baggs JE, Price TS, DiTacchio L, Panda S, Fitzgerald GA, Hogenesch JB. Network features of the mammalian circadian clock. PLoS Biol. 2009;7(3):e52. doi: 10.1371/journal.pbio.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Candelario-Jalil E, et al. Assessment of the relative contribution of COX-1 and COX-2 isoforms to ischemia-induced oxidative damage and neurodegeneration following transient global cerebral ischemia. J Neurochem. 2003;86(3):545–555. doi: 10.1046/j.1471-4159.2003.01812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teismann P, et al. Cyclooxygenase-2 is instrumental in Parkinson’s disease neurodegeneration. Proc Natl Acad Sci U S A. 2003;100(9):5473–5478. doi: 10.1073/pnas.0837397100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White BR, Bauer AQ, Snyder AZ, Schlaggar BL, Lee JM, Culver JP. Imaging of functional connectivity in the mouse brain. PLoS One. 2011;6(1):e16322. doi: 10.1371/journal.pone.0016322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bero AW, et al. Bidirectional relationship between functional connectivity and amyloid-β deposition in mouse brain. J Neurosci. 2012;32(13):4334–4340. doi: 10.1523/JNEUROSCI.5845-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laposky A, Easton A, Dugovic C, Walisser J, Bradfield C, Turek F. Deletion of the mammalian circadian clock gene BMAL1/Mop3 alters baseline sleep architecture and the response to sleep deprivation. Sleep. 2005;28(4):395–409. doi: 10.1093/sleep/28.4.395. [DOI] [PubMed] [Google Scholar]
- 31.Tronche F, et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23(1):99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 32.Mieda M, Sakurai T. Bmal1 in the nervous system is essential for normal adaptation of circadian locomotor activity and food intake to periodic feeding. J Neurosci. 2011;31(43):15391–15396. doi: 10.1523/JNEUROSCI.2801-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts LJ 2nd, et al. Formation of isoprostane-like compounds (neuroprostanes) in vivo from docosahexaenoic acid. J Biol Chem. 1998;273(22):13605–13612. doi: 10.1074/jbc.273.22.13605. [DOI] [PubMed] [Google Scholar]
- 34.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25(5):372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guillaumond F, et al. DNA microarray analysis and functional profile of pituitary transcriptome under core-clock protein BMAL1 control. Chronobiol Int. 2012;29(2):103–130. doi: 10.3109/07420528.2011.645707. [DOI] [PubMed] [Google Scholar]
- 36.Ohsawa I, Nishimaki K, Murakami Y, Suzuki Y, Ishikawa M, Ohta S. Age-dependent neurodegeneration accompanying memory loss in transgenic mice defective in mitochondrial aldehyde dehydrogenase 2 activity. J Neurosci. 2008;28(24):6239–6249. doi: 10.1523/JNEUROSCI.4956-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fitzmaurice AG, et al. Aldehyde dehydrogenase inhibition as a pathogenic mechanism in Parkinson disease. Proc Natl Acad Sci U S A. 2013;110(2):636–641. doi: 10.1073/pnas.1220399110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dinkova-Kostova AT, Talalay P. NAD(P)H:quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Arch Biochem Biophys. 2010;501(1):116–123. doi: 10.1016/j.abb.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeBruyne JP, Weaver DR, Reppert SM. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci. 2007;10(5):543–545. doi: 10.1038/nn1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30(2):525–536. doi: 10.1016/S0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 41.Zheng B, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105(5):683–694. doi: 10.1016/S0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- 42.Beal MF, et al. 3-Nitropropionic acid neurotoxicity is attenuated in copper/zinc superoxide dismutase transgenic mice. J Neurochem. 1995;65(2):919–922. doi: 10.1046/j.1471-4159.1995.65020919.x. [DOI] [PubMed] [Google Scholar]
- 43.Paschos GK, et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med. 2013;18(12):1768–1777. doi: 10.1038/nm.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kunieda T, et al. Cellular senescence impairs circadian expression of clock genes in vitro and in vivo. Circ Res. 2006;98(4):532–539. doi: 10.1161/01.RES.0000204504.25798.a8. [DOI] [PubMed] [Google Scholar]
- 45.Mongrain V, La Spada F, Curie T, Franken P. Sleep loss reduces the DNA-binding of BMAL1, CLOCK, and NPAS2 to specific clock genes in the mouse cerebral cortex. PLoS One. 2011;6(10):e26622. doi: 10.1371/journal.pone.0026622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grone BP, et al. Acute light exposure suppresses circadian rhythms in clock gene expression. J Biol Rhythms. 2011;26(1):78–81. doi: 10.1177/0748730410388404. [DOI] [PubMed] [Google Scholar]
- 47.Morton AJ, Wood NI, Hastings MH, Hurelbrink C, Barker RA, Maywood ES. Disintegration of the sleep-wake cycle and circadian timing in Huntington’s disease. J Neurosci. 2005;25(1):157–163. doi: 10.1523/JNEUROSCI.3842-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tranah GJ, et al. Circadian activity rhythms and risk of incident dementia and mild cognitive impairment in older women. Ann Neurol. 2011;70(5):722–732. doi: 10.1002/ana.22468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Montine TJ, et al. Lipid peroxidation in aging brain and Alzheimer’s disease. Free Radic Biol Med. 2002;33(5):620–626. doi: 10.1016/S0891-5849(02)00807-9. [DOI] [PubMed] [Google Scholar]
- 50.Suh JH, et al. Decline in transcriptional activity of Nrf2 causes age-related loss of glutathione synthesis, which is reversible with lipoic acid. Proc Natl Acad Sci U S A. 2004;101(10):3381–3386. doi: 10.1073/pnas.0400282101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vlassenko AG, et al. Spatial correlation between brain aerobic glycolysis and amyloid-β (Aβ) deposition. Proc Natl Acad Sci U S A. 2010;107(41):17763–17767. doi: 10.1073/pnas.1010461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ho L, Pieroni C, Winger D, Purohit DP, Aisen PS, Pasinetti GM. Regional distribution of cyclooxygenase-2 in the hippocampal formation in Alzheimer’s disease. J Neurosci Res. 1999;57(3):295–303. doi: 10.1002/(SICI)1097-4547(19990801)57:3<295::AID-JNR1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 53.Kondratov RV, Vykhovanets O, Kondratova AA, Antoch MP. Antioxidant N-acetyl-L-cysteine ameliorates symptoms of premature aging associated with the deficiency of the circadian protein BMAL1. Aging (Albany NY). 2009;1(12):979–987. doi: 10.18632/aging.100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khapre RV, Kondratova AA, Susova O, Kondratov RV. Circadian clock protein BMAL1 regulates cellular senescence in vivo. Cell Cycle. 2011;10(23):4162–4169. doi: 10.4161/cc.10.23.18381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang TA, et al. Circadian rhythm of redox state regulates excitability in suprachiasmatic nucleus neurons. Science. 2012;337(6096):839–842. doi: 10.1126/science.1222826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Masri S, et al. Circadian acetylome reveals regulation of mitochondrial metabolic pathways. Proc Natl Acad Sci U S A. 2013;110(9):3339–3344. doi: 10.1073/pnas.1217632110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hirayama J, Cho S, Sassone-Corsi P. Circadian control by the reduction/oxidation pathway: catalase represses light-dependent clock gene expression in the zebrafish. Proc Natl Acad Sci U S A. 2007;104(40):15747–15752. doi: 10.1073/pnas.0705614104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng X, Yang Z, Yue Z, Alvarez JD, Sehgal A. FOXO and insulin signaling regulate sensitivity of the circadian clock to oxidative stress. Proc Natl Acad Sci U S A. 2007;104(40):15899–15904. doi: 10.1073/pnas.0701599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.SantaCruz KS, Yazlovitskaya E, Collins J, Johnson J, DeCarli C. Regional NAD(P)H:quinone oxidoreductase activity in Alzheimer’s disease. Neurobiol Aging. 2004;25(1):63–69. doi: 10.1016/S0197-4580(03)00117-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.