Abstract
Huntington’s disease (HD) is the result of expression of a mutated Huntingtin protein (mtHtt), and is associated with a variety of cellular dysfunctions including excessive mitochondrial fission. Here, we tested whether inhibition of excessive mitochondrial fission prevents mtHtt-induced pathology. We developed a selective inhibitor (P110-TAT) of the mitochondrial fission protein dynamin-related protein 1 (DRP1). We found that P110-TAT inhibited mtHtt-induced excessive mitochondrial fragmentation, improved mitochondrial function, and increased cell viability in HD cell culture models. P110-TAT treatment of fibroblasts from patients with HD and patients with HD with iPS cell–derived neurons reduced mitochondrial fragmentation and corrected mitochondrial dysfunction. P110-TAT treatment also reduced the extent of neurite shortening and cell death in iPS cell–derived neurons in patients with HD. Moreover, treatment of HD transgenic mice with P110-TAT reduced mitochondrial dysfunction, motor deficits, neuropathology, and mortality. We found that p53, a stress gene involved in HD pathogenesis, binds to DRP1 and mediates DRP1-induced mitochondrial and neuronal damage. Furthermore, P110-TAT treatment suppressed mtHtt-induced association of p53 with mitochondria in multiple HD models. These data indicate that inhibition of DRP1-dependent excessive mitochondrial fission with a P110-TAT–like inhibitor may prevent or slow the progression of HD.
Introduction
Huntington’s disease (HD) is an inherited autosomal dominant neurodegenerative disorder characterized by adult onset of motor dysfunctions, psychiatric disturbances, and intellectual decline (1). HD is caused by abnormal expansion of a CAG trinucleotide repeat found in the first exon of the huntingtin gene (Htt), which is translated into a long stretch of glutamines in a 348-kDa protein with unclear function (2). Although the HD mutation has been identified, the molecular and cellular basis of HD is far less known, and treatment for this disease remains a challenge.
Some studies suggest that disrupted mitochondrial integrity and function may contribute to the mechanism of progressive neurodegeneration in HD (3–7), and a recent review argues for focusing new drug discovery effort for HD that will correct mitochondrial structural defects (8). Mitochondria are a highly dynamic network whose size is a result of 2 opposing processes of fusion and fission. Fusion and fission are regulated by a set of conserved GTPases. Optical atrophy 1 (OPA1) with mitofusin 1 and 2 (MFN1 and -2) regulate fusion, whereas dynamin-related protein 1 (Drp1) regulates fission (9, 10).
Drp1 is located mainly in the cytosol. Upon activation, Drp1 oligomerizes and is recruited to punctuate spots on the mitochondrial surface (11–13). Drp1 binds to mitochondrial adaptors, such as Fis1, Mff, and MiD49/51, thus assembling future fission sites and severing mitochondrial membranes in a GTP hydrolysis-dependent fission (14–17).
A potential causal role of impaired mitochondrial fission in neuronal damage of HD has recently been suggested. 3-Nitropropionic acid (3-NP), an irreversible inhibitor of the mitochondrial respiratory complex II and a neurotoxin, causes excessive mitochondrial fission, neuronal cell death and HD-like symptoms in primates and rodents (18). Cells expressing mutant Htt (mtHtt) protein are more sensitive to oxidative stress-induced mitochondrial fragmentation (19), fibroblasts harvested from HD patients exhibit mitochondrial fragmentation, and neurons with mtHtt displayed defects in anterograde and retrograde mitochondrial transport along axons and increased cell death (3, 4, 20). These defects can be rescued by overexpression of Drp1K38A, a dominant-negative mutant of Drp1 (3, 4, 20). These lines of evidence suggest that the increase in cytotoxicity induced by mtHtt protein containing expanded polyglutamine tracts is mediated, at least in part, by increased mitochondrial fragmentation. Furthermore, mtHtt was reported to interact with pro-fission protein Drp1 in HD transgenic mouse models and in brains of HD patients (4, 20). The interaction of mtHtt with Drp1 leads to increased Drp1 GTPase activity and results in defective mitochondrial movement and synaptic defects (4, 20). Thus, Drp1 hyperactivation might play an important role in the progression of HD. However, how Drp1 mediates mitochondrial dysfunction in HD and whether pharmacological inhibition of Drp1 activation is sufficient to reduce mtHtt-induced neurotoxicity and neurodegeneration are not known.
p53 is a central stress sensor responding to a myriad of insults, including DNA damage, senescence, and oxidative stress, to orchestrate apoptosis, autophagy-associated cell death, and necrosis via transcription-dependent and -independent mechanisms (21, 22). Upon stress, a cytoplasmic pool of p53 mainly translocates to the mitochondrial fraction, an event that precedes its effect on nuclear functions (23–26). Accumulation of p53 on the mitochondria is observed in animal models of HD and in cell cultures, and is associated with mitochondrial depolarization and mitochondrial complex IV inactivity (27, 28). Suppression and/or deletion of p53 prevents mitochondrial dysfunction and reduces neurodegeneration in both Drosophila and transgenic mouse models of HD (27). Thus, p53 may be an important mediator in mtHtt-induced mitochondrial toxicity and the resulting neurodegeneration.
We recently developed the Drp1-selective peptide inhibitor P110-TAT, which inhibits excessive mitochondrial fission and dysfunction without affecting basal mitochondrial structure and functions (29). Here we use P110-TAT to identify the molecular basis of mtHtt-induced pathology and to determine whether inhibition of excessive mitochondrial fission in several culture models of HD, in the R6/2 mouse HD model in vivo and in HD patient-derived cells, is sufficient to inhibit the pathology induced by mtHtt.
Results
Inhibition of Drp1 hyperactivation by P110-TAT in HD cell culture models.
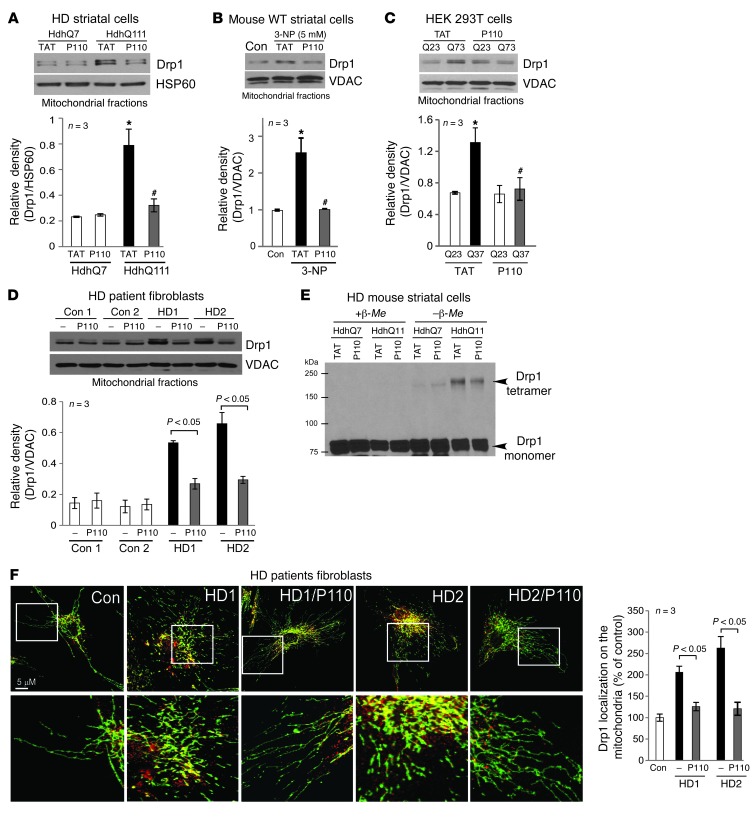
We first examined Drp1 association with the mitochondria, an essential step in initiating fission (9, 10). We used a striatal cell line from HD knock-in mice (HdhQ111), which express a full-length mtHtt containing 111 CAG repeats, as a cell culture model of HD (28). Relative to striatal cells carrying 7 CAG repeats (HdhQ7; wild-type), striatal HdhQ111 cells exhibited a 3- to 4-fold increase in Drp1 association with the mitochondria (Figure 1A). Treatment of HdhQ111 cells with P110-TAT, an inhibitor of Drp1 translocation to the mitochondria (29), abolished the increased translocation of Drp1 to the mitochondria relative to that in cells treated with the control peptide (TAT, Figure 1A; 1 μM each). A similar P110-TAT–induced inhibition of Drp1 association with the mitochondria was observed in wild-type striatal cells exposed to 3-NP, a neurotoxin that causes HD-like symptoms in vivo (ref. 18 and Figure 1B) and in HEK293T cells expressing the fragment of Htt gene containing 73 polyglutamine repeats (73Q, mutant, Figure 1C). A 3- to 4-fold increase in mitochondrial association of Drp1 was observed also in 2 lines of dermal fibroblasts obtained from HD patients relative to fibroblasts from control subjects and this increased Drp1 association with the mitochondria was corrected by treatment with P110-TAT (Figure 1D). Note that P110-TAT treatment in striatal HdhQ7 cells (wild-type), in cells expressing a fragment of Htt carrying 23 polyglutamine repeats (23Q, wild-type), and in fibroblasts from normal subjects had minimal effect on mitochondrial Drp1 levels (Figure 1, A, C, and D). These data were consistent with our recent findings that P110-TAT treatment mainly blocks stress-induced accumulation of Drp1 on the mitochondria (29). In addition, P110-TAT treatment had no effect on the total protein levels of Drp1 and Htt as well as on components of mitochondrial fusion machinery (Mfn1/2 and OPA1) (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI70911DS1). Thus, P110-TAT treatment suppressed Drp1 accumulation on the mitochondria in response to genetic or chemical stresses that model HD.
Figure 1. Treatment with P110-TAT blocked Drp1 association with the mitochondria in HD cell cultures.
(A–D) Drp1 association with the mitochondria and the effect of peptide treatment on this association was determined in 4 culture models of HD: (A) cultured mouse striatal cells expressing normal (HdhQ7) and mtHtt protein (HdhQ111) treated with peptide P110-TAT (P110, 1 μM) for 3 days; (B) mouse wild-type striatal cells incubated with P110-TAT (1 μM) for 30 minutes prior to 4-hour 3-NP exposure; (C) HEK293T cells transfected with Htt containing 23Q and 73Q repeats, followed by 2-day treatment with P110-TAT (1 μM); and (D) HD patient fibroblasts and normal fibroblasts treated with P110-TAT (1 μM) for 3 days. Western blot analysis of mitochondrial fractions was determined with anti-Drp1 antibodies (loading controls, VDAC and HSP60). Data are mean ± SEM, 3 independent experiments. (E) Total lysates of HD striatal cells were subjected to Western blot analysis using reducing or non-reducing gels and monomeric (~75-kDa) and tetrameric (~200-kDa) Drp1 levels determined; representative of 3 independent experiments. (F) Confocal microscopy of stained normal and HD patient fibroblasts with anti-Drp1 (1:250) and anti-Tom20 (mitochondria marker; 1:500). Bottom panels show enlargement of the boxed areas. Supplemental Figure 2A provides images of multiple cells. Drp1/Tom20 colocalization was determined using confocal microscopy (Pearson’s co-efficiency); data are mean ± SEM, 3 independent experiments. At least 100 cells/group were counted by an observer blinded to experimental conditions. *P < 0.05 vs. control cells; #P < 0.05 vs. cells treated with TAT.
Drp1 activation is mediated, at least in part, by its polymerization (30, 31). In HdhQ111 striatal cells, Drp1 tetramer (~200 kDa) levels increased relative to that in HdhQ7 wild-type cells and were reduced by P110-TAT treatment, as compared with TAT control treatment (Figure 1E). Furthermore, confocal microscopy analysis revealed greater localization of Drp1 on mitochondria in 2 lines of fibroblasts derived from HD patients as compared with fibroblasts obtained from control subjects (Figure 1F, single cell; Supplemental Figure 2A, multiple cells). Again, P110-TAT treatment in these HD patient–derived fibroblasts dramatically reduced mitochondrial localization of Drp1 (from 210% to 125% in fibroblasts derived from patient HD1 and from 260% to 120% in fibroblasts derived from HD2, relative to fibroblasts of a control subject; Figure 1F). Taken together, these data demonstrated that treatment with peptide P110-TAT inhibited Drp1 hyperactivation induced by mtHtt in 3 cell models of HD and in fibroblasts derived from HD patients, but not in control cells.
P110-TAT treatment reduced mitochondrial dysfunction and improved cell viability in HD cell culture models.
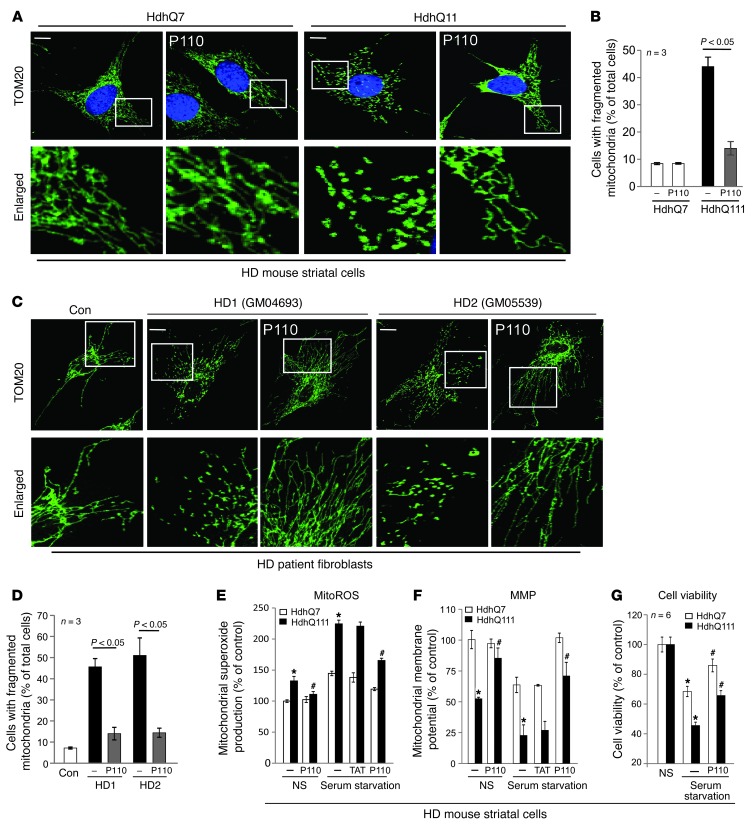
Next we determined the changes in mitochondrial morphology in both cultured HdhQ111 mouse striatal cells and in fibroblasts derived from HD patients. Mitochondria in these cells exhibited dot- or sphere-like clusters, indicating mitochondrial fragmentation (Figure 2, A and C, and Supplemental Figure 2, B and C, show a large field with multiple cells per field). In contrast, treatment with P110-TAT corrected mitochondrial morphology; the number of cells with fragmented mitochondria was reduced from 45% to 15% in HdhQ111 cells relative to that in HdhQ7 cells and from 47% and 51% to 14% and 15% in 2 fibroblasts derived from patients with HD (see Figure 2, B and D, for quantitation). Similar to our recent finding (29), P110-TAT treatment in wild-type striatal cells appears to have no effect on mitochondrial morphology (e.g., Figure 2A and Supplemental Figure 2B). We previously showed that the benefit of P110-TAT depends on the presence of Drp1 (29); in HdhQ111 cells treated with Drp1 silence RNA (Drp1-siRNA), P110-TAT had no effect on mitochondrial fragmentation (Supplemental Figure 2D), further confirming the specificity of P110-TAT for Drp1.
Figure 2. P110-TAT treatment reduced mitochondrial fragmentation and corrected mitochondrial dysfunction.
(A–D) Cultured HD mouse striatal cells (A and B) and human fibroblasts from control and HD patients (C and D) were treated with P110-TAT (1 μM) for 3 days. The cells were then stained with anti-Tom20 antibody (green) and Hoechst stain (scale bars: 5 μm). Mitochondrial morphology was analyzed using a ×60 oil lens. The boxed area in each upper micrograph is enlarged in the panels below. Images of multiple cells are provided in Supplemental Figure 2, B and C. Histogram in B and D provides quantitation of the percentage of cells with fragmented mitochondria relative to the total number of cells; data are the mean ± SEM of 3 independent experiments, counted by an observer blinded to the experimental conditions. At least 100 cells per group were counted. (E–G) Twenty hours of serum starvation was performed. (E) Mitochondrial superoxide production was determined using the mitochondrial superoxide indicator, MitoSOX red, in the indicated groups, and red fluorescence from 3 independent experiments was quantified. (F) Mitochondrial membrane potential was determined using Tetramethylrhodamine, methyl ester (TMRM). Fluorescence was quantitated using a fluorescence reader. (G) Cell viability was measured using an MTT assay. The data are presented as mean ± SEM of percentage relative to control mitochondria from 3 independent experiments. *P < 0.05 vs. wild-type cells under normal or serum starvation conditions; #P < 0.05 vs. HD mutant cells under normal or serum starvation conditions.
Drp1-induced excessive mitochondrial fission resulted in mitochondrial damage, which has been proposed to be a potential mechanism contributing to the pathogenesis of HD (4, 20, 32). Thus, we next determined whether inhibition of Drp1 hyperactivation by P110-TAT treatment (Figures 1 And 2A) blocked mitochondrial dysfunction induced by mtHtt. We first determined the production of mitochondrial superoxide (O2–), a major source of mitochondrial ROS (mitoROS). The levels of mitoROS were significantly elevated in HdhQ111 striatal cells, and this increase was abolished by treatment with P110-TAT (Figure 2E) when cultured in 10% serum (P = 0.05). We also found that HdhQ111 striatal cells were much more sensitive to stress when cultured in the absence of serum as compared with cells cultured in 10% serum. Again, P110-TAT treatment significantly reduced O2– production (~70% reduction; Figure 2E). Further, a decrease in mitochondrial membrane potential (MMP) in HdhQ111 cells cultured under the same conditions as above was corrected by treatment with P110-TAT (Figure 2F). Finally, P110-TAT treatment increased cell viability in these HD mouse striatal cells subjected to serum starvation (Figure 2G). Again, P110-TAT treatment had minimal effects on mitochondrial function and cell viability in wild-type striatal cells under normal cultured conditions (Figure 2, E–G). Thus, inhibition of Drp1 by P110-TAT treatment reduced mitochondrial dysfunction and cellular damage in a HD rodent cell culture model.
Drp1 and p53 interdependently induce mitochondrial damage in HD cell culture model.
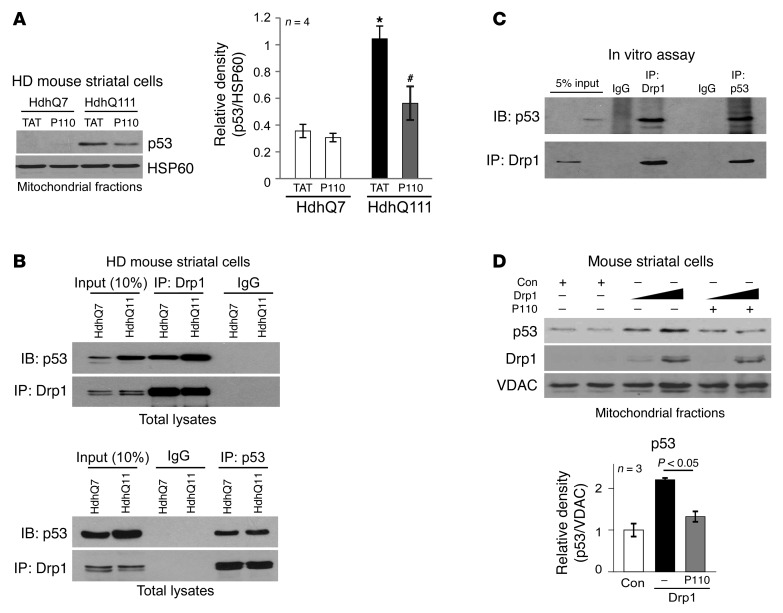
We next examined further the molecular basis of mtHtt-induced mitochondrial dysfunction. A number of studies, including ours, demonstrated that hyper-activation of Drp1 caused mitochondrial damage and led to increased apoptosis, necrosis and autophagy-associated cell death (29, 33–37). However, the mechanism by which Drp1 mediates mitochondrial dysregulation and cell death is poorly understood. The levels of p53, a tumor suppressor gene, were elevated in various HD models and p53 accumulation on the mitochondria had been suggested to play a role in the pathology associated with HD (27, 28). Recent studies reported that p53 participates in Drp1-related cell death in cardiomyocytes exposed to hydrogen peroxide (H2O2) (38) and in neuronal cells in response to camptothecin (CPT), an inducer of DNA damage (39). Based on these lines of evidence, we hypothesized that p53 might be involved in Drp1-induced mitochondrial dysfunction in HD models. We observed an approximately 3-fold increase in p53 association with mitochondria isolated from HdhQ111 striatal cells (Figure 3A). Treatment with P110-TAT reduced this association as compared with that in HdhQ7 striatal cells treated with the control peptide, TAT (Figure 3A), suggesting that P110-TAT treatment suppresses p53 mitochondrial localization in HD cell cultures. P110-TAT treatment had no effect on the total protein level of p53 in either HdhQ7 or HdhQ111 cells (Supplemental Figure 2E). P110-TAT peptide corresponds to a short sequence within Drp1 (amino acids 49–55) and is designed to block the interaction between Drp1 and its mitochondrial adaptor Fis1 (29). We found no sequence similarity between p53 and peptide P110-TAT or between p53 and Fis1 (data not shown), ruling out the possibility that P110-TAT directly competes with p53 binding to 1 of these 2 proteins.
Figure 3. Drp1 bound to p53.
(A) HD striatal cells were treated with P110-TAT or control peptide TAT (1 μM each) for 3 days, and p53 association with the mitochondria was determined by Western blot analysis (loading control, HSP60). Histogram: the data represent mean ± SEM of 3–4 independent experiments. *P < 0.05 vs. wild-type cell treated with TAT; #P < 0.05 vs. HD striatal cells treated with TAT. (B) Top: Total lysates of HD striatal cells were subjected to immunoprecipitation with anti-Drp1 antibody, and immunoprecipitates were analyzed by immunoblotting with anti-p53 and anti-Drp1 antibodies. Data are representative of 3 independent experiments. Bottom: Immunoprecipitates obtained using anti-p53 antibodies were analyzed by immunoblotting using anti-Drp1 and anti-p53 antibodies. Data are representative of 2 independent experiments. (C) GST-Drp1 recombinant protein (500 ng) was incubated with p53 recombinant protein (500 ng). Immunoprecipitates with anti-Drp1 antibodies or anti-p53 antibodies were analyzed by immunoblotting with the indicated antibodies. Data are representative of 2 independent experiments. (D) Mouse wild-type striatal cells were transfected with either control vector or Drp1 plasmid (5 μg and 10 μg, respectively) for 36 hours. In the presence or absence of P110-TAT (1 μM), protein levels of p53 and Drp1 on the mitochondria were analyzed by Western blotting (loading control, VDAC). Quantification of p53 mitochondrial level in cells with Drp1 (10 μg) is presented as mean SEM of 3 independent experiments in the histogram.
Given that Drp1 and p53 translocated to the mitochondria in HD cell culture (Figure 1, A–D, and Figure 3A), and that blocking Drp1 recruitment to the mitochondria reduced p53 accumulation in this organelle (Figure 3A), we next examined the possibility that Drp1 and p53 form a complex. Analysis of anti-Drp1 immunoprecipitates by immunoblotting with anti-p53 antibodies demonstrated the association of Drp1 with p53 in HD striatal cells (Figure 3B, top panel). In reciprocal experiments, immunoblot analysis of anti-p53 immunoprecipitates with anti-Drp1 antibodies confirmed the interaction (Figure 3B, bottom panel). In an in vitro assay, we further demonstrated that recombinant protein Drp1 interacted with recombinant protein p53, suggesting a direct interaction of the 2 proteins (Figure 3C).
We expressed Drp1 in wild-type striatal cells and determined p53 association with the mitochondria. Increase in Drp1 levels caused a proportional increase in p53 accumulation in the mitochondrial fraction. Importantly, P110-TAT treatment significantly reduced p53 mitochondrial association when compared with that in cells in the control group (Figure 3D). (Note that Drp1 expression had no effect on either p53 total protein or mRNA levels; Supplemental Figure 3A). In contrast, expression of p53 in either mouse striatal cells or p53-null H1299 cells did not increase Drp1 association with the mitochondria (Supplemental Figure 3B) and had no effect on Drp1 cellular levels (Supplemental Figure 3C). These data collectively suggest that Drp1 influences p53 mitochondrial localization, but that p53 is not necessary for Drp1 association with this organelle. In addition, treatment with pifithrin-α (PFTα), an inhibitor of p53 transactivation (40), had no effect on Drp1 and p53 levels in the mitochondrial fraction isolated from mouse HD striatal cells HdhQ111 (Supplemental Figure 3D). Taken together, these data indicate that Drp1, and not p53, determines translocation of the complex to the mitochondria under conditions relevant to HD.
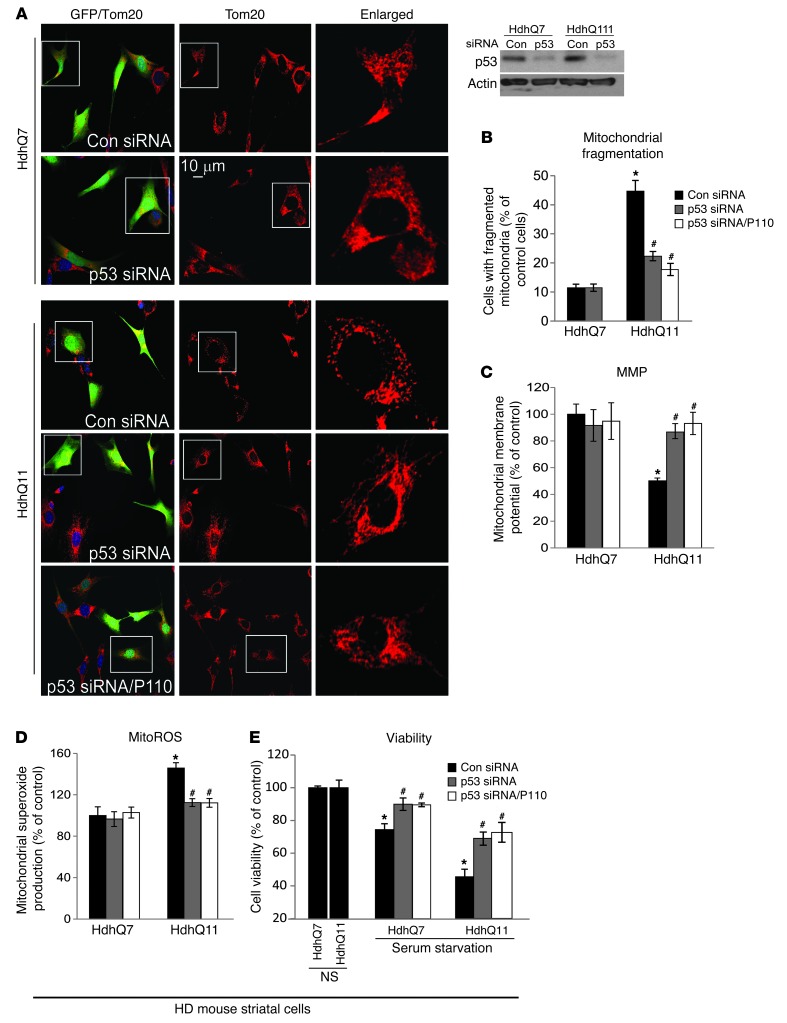
To examine the functional importance of p53 in Drp1-mediated mitochondrial damage in HD, we determined mitochondrial morphology, function, and cell survival in HdhQ111 mouse striatal cells after p53 levels were reduced using p53 siRNA (psiRNA-mp53, GFP tagged) (Figure 4A, insert). Silencing p53 in HdhQ111 cells (see GFP-labeled cells) reduced the number of cells with fragmented mitochondria from 43% to 19% relative to that in HdhQ7 cells treated with control siRNA (psiRNA-con, GFP labeling; Figure 4, A and B). Silencing p53 in HdhQ111 cells also reduced increased mitochondrial ROS production and the loss of MMP and promoted cell viability when compared with those in HdhQ111 cells treated with control siRNA (Figure 4, C–E). Note that quantification only accounts for GFP-labeled cells, which were those transfected with either p53 siRNA or control siRNA (Figure 4, B–E). Importantly, the effects of p53 silencing on mitochondria and cell survival are similar to those in HD striatal cells treated with P110-TAT (see Figure 2). Furthermore, treatment with P110-TAT following p53 silencing under the same conditions above had no additive improvement on mitochondrial morphology, mitochondrial function and cell viability (Figure 4, A–E), suggesting that Drp1 and p53 are part of the same signaling pathway in HD. Thus, mtHtt-induced accumulation of p53 on mitochondria may be a key mediator of Drp1 hyperactivation-induced mitochondrial impairment.
Figure 4. p53 is required for Drp1-induced mitochondrial dysfunction in HD striatal cells.
HD striatal cells were transfected with either control siRNA (psiRNA-GFP) or p53 siRNA (psiRNA-mp53-GFP) for 2 days in the presence or absence of P110-TAT (P110, 1 μM). (A) Cells were stained with Tom20 antibody (red), and mitochondrial morphology was analyzed in GFP-labeled cells using a ×40 oil lens. Scale bars: 10 μm. A Western blot shows p53 levels after siRNA treatments. (B) GFP-positive cells with fragmented mitochondria as percentage of total number of GFP-positive cells. (C) Mitochondrial membrane potential was determined using TMRM. The red fluorescence was quantitated in GFP-positive cells. (D) Mitochondrial superoxide production was determined using the mitochondrial superoxide indicator, MitoSOX red, in the indicated groups. Red fluorescence was quantitated in GFP-positive cells. Data are the mean ± SEM of 3 independent experiments, counted by an observer blinded to the experimental conditions. At least 100 cells were counted per treatment group. (E) Cell viability was measured using an MTT assay after 20 hours of serum starvation. Data are presented as mean ± SEM of 3 independent experiments. *P < 0.05 vs. HdhQ7 and #P < 0.05 vs. HhdQ111 cells with control siRNA under normal or serum starvation conditions.
P110-TAT treatment reduces mitochondrial dysfunction and neurite shortening in neurons derived from HD patient induced pluripotent stem cells (iPS cells).
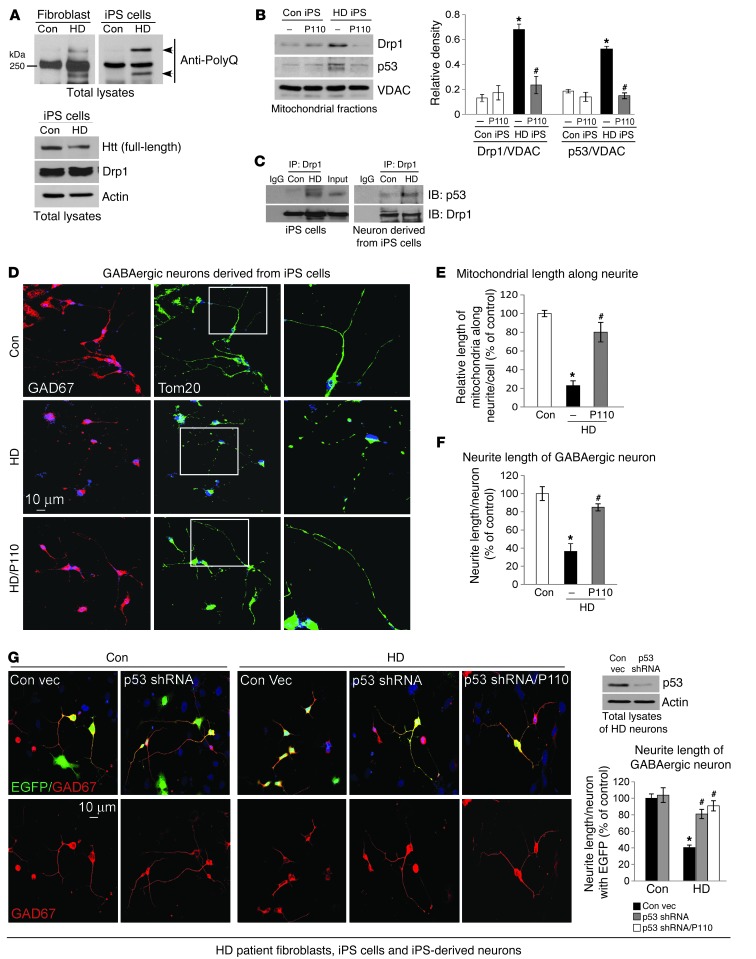
iPS cells generated from HD patient fibroblasts have been recently used as a model to study the pathology of HD. Neurons derived from HD patient iPS cells conserved the same number of CAG repeats in patients (41–43) and exhibited some HD-associated pathology in neuronal morphology and function (41–44) as well as sensitivity to stressors (41, 45). Thus, these patient-derived neuronal cells represent a unique human disease-specific cellular system to elucidate disease mechanisms underlying HD and provide a platform of patient cells for screening therapeutic candidates (46–49). Using the technique as described in refs. 50 and 51, we generated iPS cells lines from fibroblasts of 2 HD patients (adult patient GM04693 and juvenile patient GM05539; 5 lines/patient). We also derived iPS cells from fibroblasts of healthy subjects (Con 1 and Con 2; 3 lines/subject). Of those, 1 line from normal fibroblast Con 2 (Con-iPS cells) and 1 line from HD patient GM05539 (HD-iPS cells) were further characterized and were shown to be fully reprogrammed, as judged by colony morphology, sustained growth for long-term passaging (30 passages), and karyotype stability (Supplemental Figure 4). The HD-derived iPS cells expressed antigens of embryonic stem cells including OCT3/4, SOX2, SSEA-4, TRA1–60, and TRA1–81 and were positive in alkaline phosphatase (AP) staining (Supplemental Figure 4). After 15 days of spontaneous differentiation, embryonic bodies (EB) formed from these lines can give rise to cells immunopositive for 3 main embryo germ layer markers, α-SMA (mesoderm), β-tubulin (ectoderm), and GATA-4 (endoderm), showing in vitro pluripotent differentiation ability (Supplemental Figure 4). The expression of mtHtt in patient fibroblasts was conserved in HD patient iPS cells (Figure 5A), and total levels of mitochondrial dynamic-related proteins, including Drp1, were not significantly different between Con-iPS cells and HD-iPS cells (Figure 5A and Supplemental Figure 5A).
Figure 5. P110-TAT treatment normalized mitochondrial and neurite length in GABAergic neuronal cells derived from HD patient-iPScells.
(A) Left: Expression of polyglutamine-expansion disease marker (PolyQ) was determined using Western blot of total lysates in fibroblasts or Con-iPS or HD-iPS human cells. Right: Htt protein levels and Drp1 were determined using Western blot (loading control, actin). Data represent 2 independent experiments. (B) Con-iPS and HD-iPS cells were treated with P110-TAT (1 μM) for 3 days, and mitochondrial levels of Drp1 and p53 were determined with Western blot (loading control, VDAC). Data represent mean ± SEM, 3 independent experiments. (C) Immunoprecipitation analysis at the indicated groups confirmed the interaction between Drp1 and p53. Data represent 2 independent experiments. (D) GABAergic neurons were derived from Con-iPS or HD-iPS human cells and then stained with anti-Tom20 and anti-GAD67. Right panels show enlargements of boxed areas. (E and F) Quantitation of mitochondrial length along neurites of GABAergic neurons (E) and neurite length of GABAergic neurons (F). Data represent 3 independent experiments. (G) Neurons were transduced by lentiviral particles containing p53 shRNA or control vector (EGFP-tagged), then cells were stained with anti-GAD67 antibodies (red). Neurite length of GABAergic neurons with EGFP labeling is quantified, representative of 3 independent experiments. Western blot confirmed p53 silencing in HD neurons (actin, loading control). In F and G, quantification was performed by an observer blinded to experimental conditions. At least 30 neurons/group were analyzed. Data are mean ± SEM. *P < 0.05 vs. Con; #P < 0.05 vs. HD.
We first isolated mitochondrial fractions from Con- and HD-iPS cells and determined the levels of Drp1 and p53 in the mitochondrial fraction. The levels of both mitochondria-associated Drp1 and p53 in HD-iPS cells were increased 3- to 4-fold relative to Con-iPS cells and these increases were abolished by treatment with P110-TAT for 5 days (Figure 5B). Again, P110-TAT had no significant effect on the total protein levels of Drp1 and p53 in these experimental groups (Supplemental Figure 5B). These data are consistent with our above findings (Figures 1 and 3, Supplemental Figure 1). Furthermore, in both the undifferentiated HD-iPS cells (Figure 5C, left panel) and the neurons derived from HD-iPS cells (Figure 5C, right panel), we observed interaction between Drp1 and p53.
We differentiated Con- and HD-iPS cells into GABAergic neurons and medium spiny neurons (MSNs), which are susceptible to degeneration in HD (45, 52, 53), using the protocol described in Methods. Generation of these neurons was tested by co-staining neuronal marker β-tubulin (TUJ1) or MAP2 with GAD67 (GABAergic neuronal marker), calbindin (striatal neuronal marker), and DARPP-32 (MSN neuronal marker), respectively (Supplemental Figure 5). Thus, similar to previous studies (41, 44, 45), both Con- and HD-iPS cells can be differentiated into GABAergic striatal neurons.
Next we determined the benefit of P110-TAT treatment on this human-derived cultured neuronal model of HD. Twenty days after differentiation, we started to treat neuronal cells derived from Con-iPS or HD-iPS cells with the Drp1 peptide inhibitor, P110-TAT (1 μM/day for 5 consecutive days). In HD patient–derived cells immunopositive for anti-GAD67 (a GABAergic neuronal marker), we observed that the mitochondria (green) were extensively fragmented along the neurites, when compared with those in cells derived from Con-iPS cells (Figure 5D; compare middle and top panels). This fragmented mitochondrial morphology was corrected by P110-TAT treatment (Figure 5D, bottom; see Figure 5E for quantification by an observer blinded to the experimental conditions). We also measured the length of the neurites of the GABAergic neurons. The analyses showed significantly longer neurites in GABAergic neurons from Con-iPS cells as compared with neurons derived from HD-iPS cells (Figure 5D; see Figure 5F for quantitation by an observer blinded to the experimental conditions). Importantly, P110-TAT treatment improved the morphology of GABAergic neurons derived from HD-iPS cells; the neurite length increased from 37% to 84% relative to that in neurons derived from Con-iPS cells (Figure 5F). Further, downregulating p53 by p53 shRNA (EGFP tagged) also increased the neurite length of GABAergic neurons (Figure 5G); the effect was similar to that in neurons treated with either P110-TAT alone (Figure 5, D and F) or P110-TAT in the presence of p53 shRNA (Figure 5G).
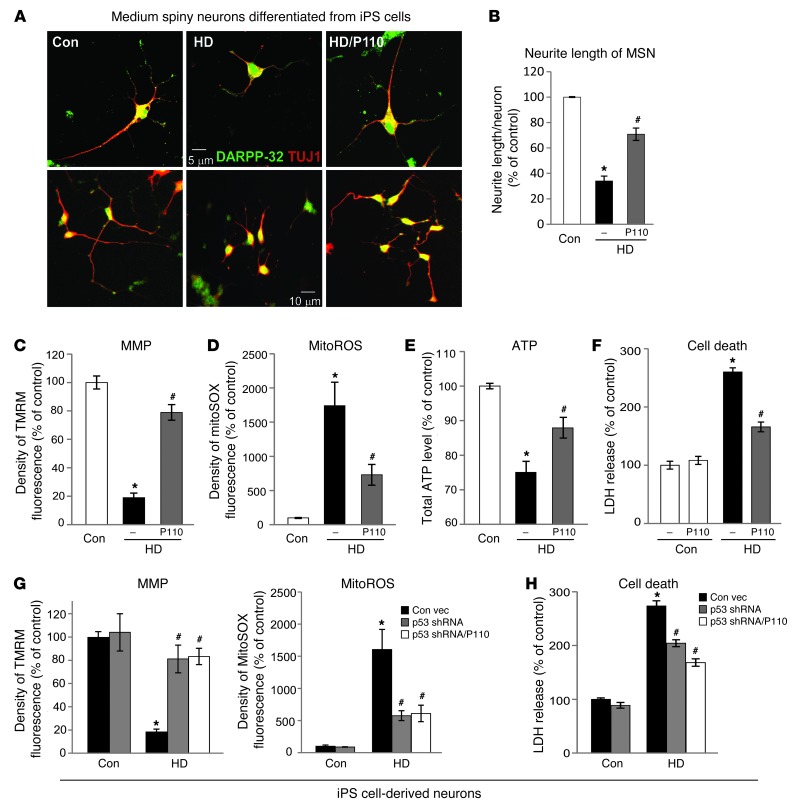
Among GABAergic striatal neurons, medium spiny neurons (MSNs) are the most affected in HD patients and are largely degenerated in the late stage of the disease (54–57). MSNs derived from HD-iPS cells exhibited morphological abnormalities, including fewer and shorter neurites as compared with those from Con-iPS–derived MSNs (Figure 6A; see Figure 6B for quantification by an observer blinded to the experimental conditions). Importantly, P110-TAT treatment of HD-derived MSNs doubled neurite length, from 34% to 70%, relative to that in neurons derived from Con-iPS–derived MSNs (Figure 6, A and B). Moreover, in these HD patient–derived neurons, a loss of MMP, an increase in mitochondrial ROS formation and decreased ATP levels were also observed (Figure 6, C–E), which were greatly corrected by 5 days of treatment with P110-TAT. Further, consistent with a previous study (41), increased cell death was observed in HD patient iPS cell–derived neurons by removing the growth factor, BDNF, and P110-TAT treatment reduced cell death by 60% (Figure 6F). In parallel, reducing the levels of p53 by p53 shRNA suppressed mitochondrial ROS production and the loss of MMP as well as reduced cell death in HD patient–derived neurons, which was not further affected by P110-TAT treatment, suggesting that p53 and Drp1 affect the same molecular event (Figure 6, G and H). Collectively, these data above demonstrate that P110-TAT treatment corrected the neuronal structure abnormalities of 2 types of neurons as well as the mitochondrial structure and function abnormalities seen in human cells derived from HD patients.
Figure 6. P110-TAT treatment reduced neurite shortening of MSNs from HD-iPS cells and corrected mitochondrial damage and cell death in patient neurons.
(A) Representative imaging on MSNs derived from Con- and HD-iPS cells. Top: A single neuron from each experimental group (original magnification, ×40). Bottom: A cluster of neurons at lower magnification (original magnification, ×20). (B) Data represent 3 independent experiments. At least 50 neurons/group were analyzed by an observed blinded to the experimental conditions. (C) Neuronal cells were labeled with TMRM fluorescence dye to indicate MMP. Density of TMRM red fluorescence only in cells with a neuronal-like morphology (multipolar cell bodies with at least 2 processes) was further quantitated as described in ref. 91. (D) Neuronal cells were stained with MitoSOX red, and density was analyzed in cells immunopositive for anti–DARPP-32. (E) Total ATP levels were measured using total lysates of mixed neuronal cells. Data represent 3 independent experiments. At least 50 neurons per group were analyzed. (F) After removal of BDNF for 48 hours, neuronal cell death was determined by the release of lactate dehydrogenase (LDH). Data represent 3 independent experiments. (G) Fluorescence density of EGFP-positive cells from neurons transduced by lentiviral particles containing p53 shRNA or control vector. Data represent 3 independent experiments. At least 50 EGFP-positive neurons per group were analyzed. (H) Neuronal cell death was determined by LDH release 2 days after removal of BNDF. Data are mean ± SEM. *P < 0.05 vs. Con; #P < 0.05 vs. HD.
P110-TAT treatment reduced neurological defects in R6/2 HD transgenic mice.
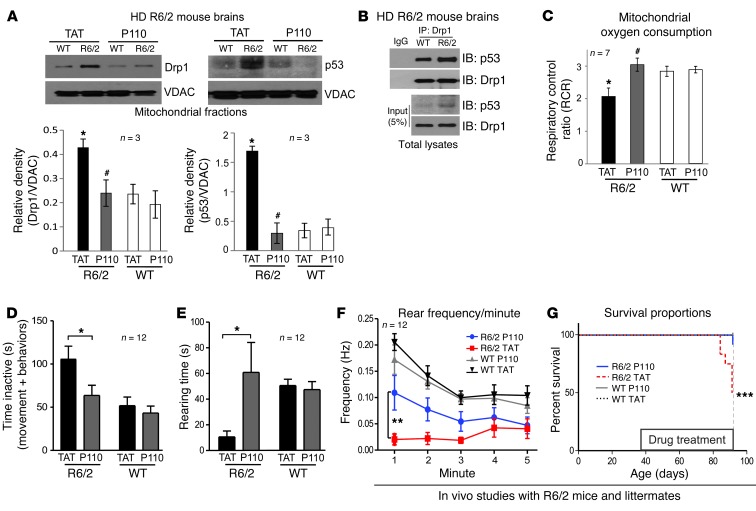
We next asked whether treatment with P110-TAT is protective in in vivo HD animal model. R6/2 mice, expressing exon 1 of the human HD gene carrying more than 120 CAG repeats, exhibit progressive neurological phenotype that mimics the features of HD in humans. The mice develop progressive neurological phenotypes gradually with mild phenotype (e.g., resting tremor) as early as 5 weeks of age and severe symptoms (including reduced mobility and seizures) at 9–11 weeks, with many of the mice dying by 14 weeks (58). To determine whether treatment of mtHtt-induced neurological deficits is due to Drp1 activation, R6/2 HD transgenic mice were treated with the Drp1 inhibitor, P110-TAT, using Alzet osmotic mini-pumps (delivering 3 mg/kg/day) from age 5 weeks to 13 weeks (the pumps were replaced once, after 28 days). Note that the peptide was stable in the pump used in vivo during the 28 days of delivery, as determined by an analytical HPLC with UV detection using a RP-C18 column (Supplemental Figure 6B). Moreover, a FITC-positive signal was observed in neurons of mouse brain after 1 and 14 days of continual systemic delivery of FITC-conjugated P110-TAT peptide (Supplemental Figure 6A). Combined with the efficacy studies, these data confirm that the P110-TAT peptide is delivered to the neurons in vivo. In addition, 8-week treatment with P110-TAT at 3 mg/kg/day is well-tolerated in naive wild-type mice (see information in the figure legend of Supplemental Figure 6C). As shown in Figure 7A, R6/2 HD transgenic mice exhibited 2-fold and 4-fold increase in Drp1 and p53 association with brain mitochondria, respectively, as compared with mitochondria isolated from wild-type littermates, and treatment with P110-TAT for 8 weeks (using Alzet pumps; 3 mg/kg/day; from age 5 to 13 weeks) reduced mitochondrial association of both Drp1 and p53 to wild-type levels. Moreover, the interaction between Drp1 and p53 was confirmed in brains of the HD R6/2 mice by co-immunoprecipitation (Figure 7B).
Figure 7. P110-TAT treatment reduced neurological defects and improved mitochondria function in HD R6/2 mice.
(A) Mitochondria were isolated from brains of R6/2 mice after 8 weeks of TAT or P110-TAT treatment (3 mg/kg/day). Drp1 and p53 levels in the mitochondrial fraction were determined using Western blot analysis (loading control, VDAC). Data represent 3 mice. (B) Immunoprecipitates obtained from HD R6/2 mice brains using anti-Drp1 antibodies were analyzed by immunoblotting using anti-Drp1 and anti-p53. (C) Mitochondrial oxygen consumption of brain mitochondria from wild-type and R6/2 mice was analyzed after 8 weeks of TAT or P110-TAT treatment, and the RCR was calculated. *P < 0.05 vs. wild-type; #P < 0.01 vs. R6/2; n = 7/group). (D) P110-TAT treatment improved the mobility of R6/2 mice relative to TAT-treated R6/2 mice, to levels similar to wild-type mice treated with either peptide (R6/2 TAT, n = 9; R6/2 P110-TAT, n = 11; WT TAT, n = 12; WT P110-TAT, n = 12; *P < 0.05). (E and F) Rearing time (E) and frequency (F) were significantly lower in R6/2 mice treated with TAT, compared with R6/2 mice treated with P110-TAT and wild-type mice treated with either peptide (**P < 0.01). (G) Survival of R6/2 mice treated with TAT was significantly lower than for R6/2 mice treated with P110-TAT or wild-type mice (Mantel Cox log-rank test, χ2 = 17.27; 3 degrees of freedom; ***P = 0.0006; n = 12 per group). Data represent mean ± SEM.
We next measured the respiratory control rate (RCR; the ratio between the state 3 respiratory rate and state 4 respiratory rate, which is an indicator of the integrity of the inner mitochondrial membrane and a sensitive indicator of oxidative phosphorylation defects in isolated mitochondria. RCR is considered to be a useful general measure of function in isolated mitochondria (59–61). This ratio, which indicates oxygen consumption, was reduced in brain mitochondria isolated from HD R6/2 mice relative to wild-type littermates (Figure 7C). In vivo treatment with P110-TAT corrected this mitochondrial dysfunction (Figure 7C).
Deterioration in motor and cognitive function is the first manifestation of HD progression. We first set up a number of behavioral assessments to study motor and cognitive function in these mice. Rotor-rod and mobility in an activity chamber were used for assessment of motor function, and the Y-maze was used for assessment of working memory. As seen in Supplemental Figure 7, there was an apparent functional deficit in vehicle-treated mice, and P110-TAT treatment led to improvement in this deficit. In cohorts A and B tested at different experimental periods, P110-TAT increased the total distance moved in 10 weeks old R6/2 mice (motor activity) and ameliorated the cognitive deficits at 9 weeks of age in the Y-maze test (methods previously described in refs. 62, 63; Supplemental Figure 7).
We also noted that, independent of the treatment, the animals were very fragile, and stress mediated by repetitive testing and handling led to seizure and death. We therefore assessed behavioral function in an independent cohort of mice at a single time point (13 weeks old) using a simple home-cage assessment test, to minimize handling and stress (see Methods). In a double-blind behavior analysis, we found that R6/2 mice treated with the control peptide TAT were much less active overall (Figure 7D); their period of immobility lasted almost twice as long as that of wild-type littermates. The R6/2 mice treated with TAT also exhibited a 5-fold reduction in their ability to stand on their rear limbs (Figure 7E) and a 10-fold reduction in rearing frequency (Figure 7F), all indicative of major motor deficits. In contrast, the group of R6/2 mice treated with P110-TAT for 8 weeks did not display the above motor deficits compared to wild-type mice, suggesting that P110-TAT can prevent the progression of disease: R6/2 mice treated with P110-TAT displayed inactivity levels (Figure 7D) and rearing time (Figure 7E) comparable to those of wild-type mice and showed greatly increased rearing frequency compared with the mice treated with TAT (Figure 7F). P110-TAT treatment in wild-type littermates had no significant effects on any of these behaviors (Figure 7). Thus, P110-TAT treatment reduced the progression of motor deficits in R6/2 mice. Furthermore, we found that P110-TAT treatment increased survival of R6/2 mice (Figure 7G). By the age of 13 weeks (before brain samples were collected for biochemical and immunohistochemical analyses), 6 of the 12 mice treated with TAT died, whereas only 1 of the 12 mice treated with P110-TAT died. Again, P110-TAT had no effect on mortality of wild-type littermates.
P110-TAT treatment reduced neuropathology in R6/2 HD transgenic mice.
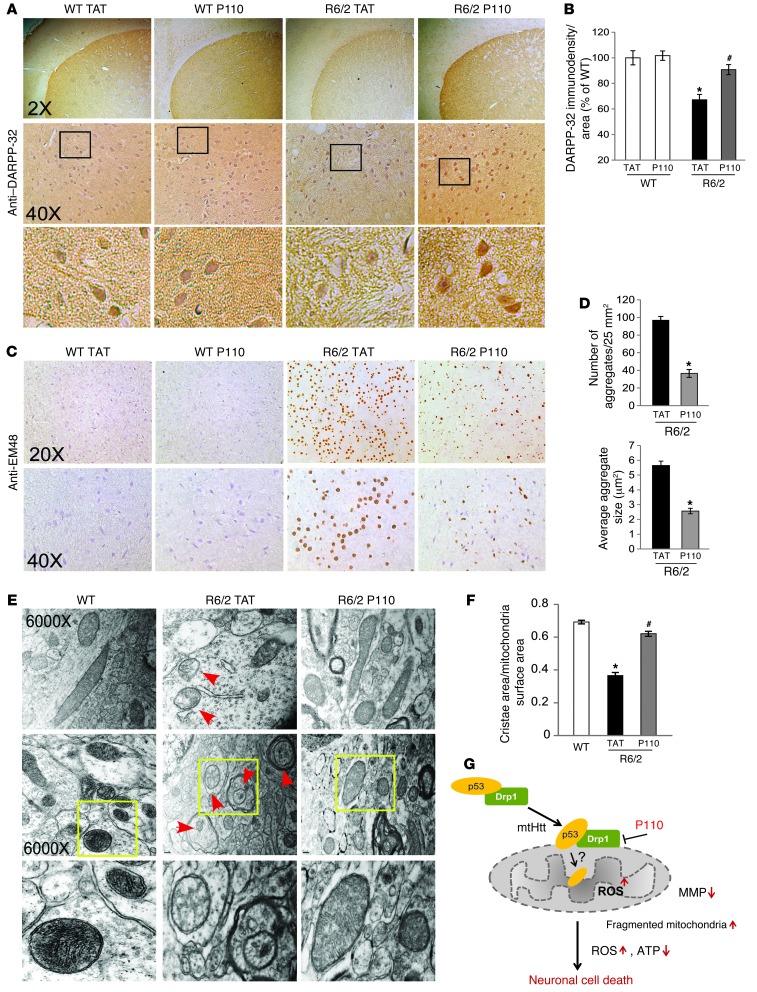
We next determined the effects of P110-TAT on striatal neuronal damage in R6/2 mice, measured by the loss of DARPP-32 immunoreactivity. DARPP-32 is expressed in 96% of the striatal medium-sized spiny neurons and is substantially reduced in HD transgenic mice (64–67). Consistent with previous reports (64, 67), DARPP-32 immunostaining was 33% lower in striatum of 13-week-old R6/2 mice as compared with their WT littermates (Figure 8, A and B). P110-TAT treatment of R6/2 mice increased the immunodensity of DARPP-32 from 67% to 90% relative to TAT-treated WT littermates. Thus, P110-TAT treatment suppresses striatal neuronal loss in HD R6/2 mice.
Figure 8. P110-TAT reduces neuropathology in HD R6/2 mice.
(A and C) Coronal sections of mouse brains were stained with anti-DARPP-32 Abs (A) or anti-EM48 Abs (C). Photomicrographs of DARPP-32 or EM-48 immunostaining were obtained of the dorsolateral striatum of TAT-treated or P110-TAT-treated mice. Bottom panels show magnification of boxed areas. (B) Quantification of DARPP-32 immunodensity (see Methods) by an observer blinded to the experimental conditions. (D) Quantification of the number (left) and average size (right) of Htt aggregates recognized by anti-EM48 by an observer blinded to the experimental conditions. Data represent 3 mice. (E) Transmission electron microscopy images of striatum samples from 13-week-old wild-type and R6/2 mice. Arrows indicate mitochondria with a loss of cristae. Original magnification, ×6,000. Top and middle panels show different section orientations; bottom panels show magnification of boxed areas. Representative images are shown. (F) Quantification of ratio between cristae surface area and mitochondrial surface area. At least 40 mitochondria were analyzed in each group by an observer blinded to the experimental conditions. (G) Drp1 and p53 interact in the cytoplasm and translocate together to the mitochondria in a Drp1-dependent manner in the presence of mtHtt. P110-TAT, a selective peptide inhibitor of Drp1, blocked Drp1 association with the mitochondria, which in turn inhibited Drp1/p53 accumulation on the mitochondria and suppressed the subsequent p53-induced mitochondrial and neuronal damage incurred under conditions relevant to HD. *P < 0.05 vs. wild-type mice treated with TAT; #P < 0.05 vs. R6/2 mice treated with TAT.
Intraneuronal Htt aggregates are a hallmark pathology of HD (68, 69), although their role in disease progression is not clear (70–72). Previous studies showed that Htt aggregates are not present in wild-type mice but are prominent features of the R6/2 striatum (58, 68). Using EM48 antibodies, which specifically recognize the aggregated form of Htt (69), we assessed the effect of P110-TAT treatment. Striatum of R6/2 mice showed extensive EM48-positive aggregates (Figure 8C). Fewer and smaller Htt aggregates were observed in striatum of R6/2 mice treated with P110-TAT; there was a 60% reduction in the number of Htt aggregates relative to R6/2 mice treated with TAT (Figure 8, C and D). Increased oxidative stress in HD brains has been associated with formation of Htt aggregates (5, 73). Thus, it is possible that P110-TAT treatment inhibits Drp1 hyperactivation-induced mitochondrial oxidative stress which, in turn, reduces formation of Htt aggregates in R6/2 mice.
Consistent with previous studies showing mitochondrial cristae disorganization in models of HD (3, 4), electron microscopy analysis showed a 50% reduction in cristae in brains of HD R6/2 mice, relative to that in wild-type mice (Figure 8, E and F). P110-TAT treatment corrected the disruption in mitochondrial morphology (Figure 8, E and F). Cristae disruption is downstream of mitochondrial fragmentation (3, 4, 74). Thus, the data suggest that inhibition of Drp1 association with the mitochondria by P110-TAT, which reduced mitochondrial fragmentation, may have caused the improved organization of the mitochondrial cristae.
Taken together, the above data in cell culture models of HD and in a transgenic mouse HD model demonstrated that mtHtt-induced pathology can be greatly reduced by inhibition of Drp1 hyperactivation using P110-TAT treatment. Importantly, P110-TAT treatment greatly improved the motor activity and survival of HD mice and reduced neuropathology induced by mtHtt in these animals. Therefore, our data suggest that excessive mitochondrial fission might be the main cause of mtHtt-induced pathologies in these models.
Discussion
Increased mitochondrial fragmentation in HD cell cultures and HD animal models was recently documented (3, 4, 18, 20, 32). Two recent studies demonstrated that inhibition of Drp1 by a Drp1-dominant negative mutant, Drp1K38A, reduced mitochondrial fragmentation and cristae disorganization, which in turn led to a reduction of mitochondrial dysfunction and cell death in a number of HD cell culture models (3, 4). These studies highlighted a critical role of Drp1 dysfunction in the pathogenesis of HD. In this study we used 6 models of HD to determine whether inhibition of Drp1-induced excessive mitochondrial fission is sufficient to inhibit mtHtt-induced pathology. These include mouse striatal cell cultures expressing a full-length mtHtt, mice expressing mtHtt (R6/2), cultured HD patient–derived primary fibroblasts, iPS cells, and 2 different types of neurons derived from these patients. In all these models, consistent with previous studies (3, 4), Drp1 association with the mitochondria was much greater relative to cells that do not express mtHtt. Importantly, the Drp1 inhibitor, P110-TAT, which inhibits only excessive mitochondrial fission (29), inhibited mtHtt-induced Drp1 association with the mitochondria, and inhibited the increase in mitochondrial fragmentation, dysfunction, and cell damage in all these HD models. Thus, these results demonstrate that inhibiting Drp1 hyperactivation is sufficient to inhibit HD-related neuropathology (see schematic in Figure 8G).
Mitochondrial fragmentation is caused by either excessive mitochondrial fission or by a defect in mitochondrial fusion. Overexpression of OPA1, a mitochondrial fusion-related protein, has similar effects on correcting mitochondrial fragmentation and cristae disruption as Drp1 inhibition by Drp1K38A in cultured HD cells derived from either HD mice or HD patients (3). In our study, P110-TAT treatment had no effect on OPA1 protein levels in various HD cell culture models (Supplemental Figures 1 and 5). Thus, P110-TAT treatment likely has a protective role distinct from that of OPA1 in these HD model cells.
We found that Drp1 and p53 physically interact, as evidenced by in vivo and in vitro co-immunoprecipitation studies, and that Drp1 is required for p53 translocation to the mitochondria; blocking Drp1 association with the mitochondria by P110-TAT inhibits this event. Thus, translocation of p53 to the mitochondria may reflect the translocation of Drp1/p53 complex to this compartment, which is likely an initiating event in the subsequent mitochondrial damage and cell death in the context of HD. Mutant Htt has been shown to interact with and activate Drp1 (4, 20) and p53 (27). Thus, it is possible that mtHtt forms complexes with both Drp1 and p53 on the mitochondria in a process that leads to mitochondrial pathology in HD.
Mitochondria-associated p53 induces multiple types of damage to this organelle. For example, p53 can induce the expression of some downstream targets, such as Bax and Puma, which lead to apoptosis in HD cell models (75). p53 can also enter the mitochondrial matrix to induce oxidative stress through interaction with p66SHC and MnSOD (76, 77). Further, p53 activation inhibits mitochondrial complex IV activity by suppression of SCO2 (synthesis of cytochrome c oxidase isoform 2, a core component of complex IV), which in turn disrupts the integrity of the mitochondrial respiratory chain and suppresses ATP production (78). In addition, p53 can be imported into the mitochondrial inner membrane compartment, where it interacts with cyclophilin D to induce necrosis in response to oxidative stress (79). Together, these disturbances of mitochondrial integrity caused by p53 are potential mechanisms of mtHtt-induced Drp1/p53 complex–mediated mitochondrial dysfunction in models of HD. Indeed, in this study, we found that either inhibition of Drp1 by P110-TAT treatment or silencing p53 by siRNA or shRNA has similar effects on mitochondrial morphology and function and on cell viability in both HD mouse striatal cells and in neurons derived from HD patient-iPS cells. Interestingly, the protective effect of P110-TAT was abolished when p53 was knocked down, further supporting our hypothesis that Drp1 and p53 form a complex and interdependently induce mitochondrial damage in HD.
In addition to rodent cell culture models of HD, we also compared neurons derived from HD patient iPS cells to those from normal subjects. In agreement with previous studies (41, 44, 45), the HD patient neurons exhibited morphological abnormalities, thus providing a unique model to study HD neuropathology in the context of patient genotype. Treatment with peptide inhibitor P110-TAT improved neuronal morphology as evidenced by increased neurite length of patient GABAergic striatal neurons. Many of the mitochondrial dysfunctions were also corrected in HD patient–derived neurons. These mitochondrial dysfunctions were associated with the Drp1/p53 interaction, which is also found in neurons derived from HD patient iPS cells, relative to neurons derived from control subjects. This p53/Drp1 association may provide further explanation of mitochondrial dysfunction-associated mtHtt-induced neurodegeneration in HD.
The use of the R6/2 mouse model of HD allowed us to further determine the benefit of treatment with P110-TAT in vivo. We first confirmed that P110-TAT is well tolerated in normal mice; there was no change in body weight or gross pathological changes (see information in the figure legend of Supplemental Figure 6C), and there was no change in mitochondrial functions or behaviors in wild-type mice treated with P110-TAT for 8 weeks, as compared with control mice or mice treated with the control peptide carrier, TAT. We found that the motor activity (as measured by exploration of the home cage and spontaneous rearing activity) was greatly impaired in the HD mouse model relative to wild-type littermates. Importantly, sustained treatment of these HD mice with P110-TAT using an Alzet pump implanted subcutaneously, resulted in a significant improvement in these mice and reduced overall mortality during the treatment period. Moreover, P110-TAT treatment reduced neuropathology in R6/2 mice as demonstrated by an increase in the density of medium spiny neurons and a reduction of Htt aggregate formation. Thus, our finding in R6/2 mice provides the first evidence of preclinical usefulness of P110-TAT for treatment of HD. R6/2 HD mice expressing a fragment of mHtt exhibit early-onset and rapidly progressing behavioral and neuropathological phenotypes associated with early death. This model is a suitable first screening model of drug candidates for HD and has already been used to make significant contributions in identifying promising preclinical leads in HD (80–82). However, this model does not replicate the development of chronic pathology seen in human HD (80–82). Further studies using mice expressing full-length mtHtt, such as YAC128 mice (83), may help address that aspect of the disease.
Together, our studies in HD cell cultures, including patient neurons derived from HD-iPS cell lines, and in an in vivo mouse model of HD provide new insight into the mechanism of Drp1 hyperactivation-induced mitochondrial impairment and neurodegeneration in HD. It suggests that inhibiting Drp1 hyperactivation may be sufficient to inhibit mtHtt-induced pathologies. We propose that a Drp1 inhibitor, such as P110-TAT, may be useful in preventing or slowing the progression of HD in humans.
Methods
Antibodies and reagents.
3-NP and protease inhibitor cocktail were purchased from Sigma-Aldrich. PFTα was from Tocris Bioscience. Recombinant protein Drp1 was from Abnova, and p53 recombinant protein was from ProSpec. Antibodies for Tom20, p53, and HSP60 were from Santa Cruz Biotechnology, Inc., Drp1 (DLP1) and OPA1 were from BD Biosciences. Antibody for Fis1 was from Proteintech, VDAC from MitoScience, full-length Htt (MAB2166) and anti-PolyQ (MAB1574) from Millipore, pan-actin from Sigma-Aldrich, and Mfn1 from Abcam. Anti-mouse IgG and anti-rabbit IgG, peroxidase-linked, species-specific antibodies were from Thermo-Scientific and Abcam (only recognize light chain for immunoprecipitation in Figure 3B, Figure 5C, and Figure 7B). The Drp1/Fis1 peptides were synthesized in the lab of Daria Mochly-Rosen at Stanford University and conjugated to TAT-carrier peptide (amino acids 47–57) for transmembrane delivery. Note that TAT47–57–based delivery was used in culture and in vivo and was found to be safe and efficacious in delivery peptide cargoes to all cells and also across the blood-brain barrier (84–86).
Constructs and transfection.
Full-length Drp1 and p53 plasmids were obtained from Addgene. Httex1Q23 and Q73 constructs were obtained from the CHDI Foundation. Cells were transfected with Lipofectamine 2000 (Invitrogen) following the manufacturer’s protocol.
Cell culture.
Immortalized striatal cell lines HdhQ111 mutant and HdhQ7 wild-type that were derived from striatal cells from HdhQ111/111 and HdhQ7/Q7 knock-in transgenic mice (expressing 111 and 7 glutamine repeats, respectively) were obtained from the CHDI Foundation. Cells were cultured in DMEM supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 400 μg/ml G418. Cells were grown at 33°C in a 5% CO2 incubator. Cells with fewer than 14 passages than 14 were used in all experiments. TAT or P110-TAT peptides were added once daily for the duration of the experiment at 1 μM final concentration.
HD patient fibroblasts (HD1: GM04693 from a 33-year-old male patient; HD2: GM05539 from a 10-year-old male patient; purchased from Coriell Institute, USA) and normal fibroblasts (Con 1, fibroblasts from adult, HDFa; Con 2, fibroblasts from juvenile, HDFn; purchased from Invitrogen) were maintained in MEM supplemented with 15% (vol/vol) heat-inactivated fetal calf serum and 1% (vol/vol) penicillin/streptomycin. All of the above cells were maintained at 37°C in 5% CO2-95% air. TAT or P110-TAT peptides were added once daily for the duration of the experiment at 1 μM final concentration.
RNA interference.
For silencing p53 in HD striatal cells, mouse p53 siRNA (psiRNA-mp53, GFP conjugated) and control psiRNA plasmid were purchased from InvivoGen. HdhQ7 and HdhQ111 cells were transfected either with psiRNA-mp53 or control psiRNA using TransIT-2020 Transfection Reagent (Mirus Bio LLC), according to the manufacturer’s instructions.
In neurons derived from HD patient iPS cells, we used lentiviral transduction to downregulate p53. Briefly, 1 day prior to transfection, confluent 10-cm plates of 293T cells were seeded at 2 × 106 cells/plate. The expression plasmid pLVUHshp53 (p53 shRNA, EGFP tagged) and psPAX2 (packaging vector) and pMD2.G (enveloping vector) were obtained from Addgene. For each 10-cm dish, 6 μg expression vector, 6 μg psPAX2, and 0.75 μg pMD2.G (8:8:1) were transfected using TransIT-2020 Transfection Reagent (Mirus Bio, LLC). A day after transfection, the medium was replaced with fresh neuronal culture media. Culture media was subsequently collected 48 and 72 hours after transfection. Virus-containing medium was filtered through 0.45-μm pore filters, and 4 μg/ml polybrene was then added. Transduction of neurons was conducted by incubating the cells with the virus-containing media overnight at 37°C, in 5% CO2. Cultured neurons were then washed, fresh neuronal medium was added and neurons were cultured for an additional 5 days.
Isolation of mitochondrial-enriched fraction and lysate preparation.
Cells were washed with cold PBS and incubated on ice in lysis buffer (250 mM sucrose, 20 mM HEPES-NaOH, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, protease inhibitor cocktail, phosphatase inhibitor cocktail) for 30 minutes on ice. Cells were scraped and then disrupted 10 times by repeated aspiration through a 25-gauge needle, followed by a 30-gauge needle. Mice brains were minced and homogenized in isolation buffer (300 mM sucrose, 10 mM HEPES, 2 mM EGTA, pH 7.2, at 4°C) containing 0.1 mg/ml of type I protease (bovine pancreas) to release mitochondria and later washed in the same buffer in the presence of 1 mg/ml bovine serum albumin. The homogenates were spun at 800 g for 10 minutes at 4°C, and the resulting supernatants were spun at 10,000 g for 20 minutes at 4°C. The pellets were washed with lysis buffer and spun at 10,000 g again for 20 minutes at 4°C. The final pellets were suspended in lysis buffer containing 1% Triton X-100 and were mitochondrial-rich lysate fractions. The mitochondrial membrane protein VDAC or HSP60 was used as a marker and loading control.
Immunoprecipitation.
Cells were lysed in total cell lysate buffer (50 mM Tris-HCl, pH 7.5, containing 150 mM NaCl, 1% Triton X-100, and protease inhibitor). Soluble protein was incubated with the indicated antibody overnight at 4°C and protein A/G beads for 1 hour. Immunoprecipitates were washed 3 times with cell lysate buffer and were analyzed by SDS-PAGE and immunoblotting with antibodies.
Immunocytochemistry.
Cells cultured on coverslips were washed with cold PBS, fixed in 4% formaldehyde, and permeabilized with 0.1% Triton X-100. After incubation with 2% normal goat serum (to block nonspecific staining), fixed cells were incubated overnight at 4°C with indicated antibodies. Cells were washed with PBS and incubated for 60 minutes with FITC-conjugated or rhodamine-conjugated secondary antibody, followed by incubation with Hoechst dye (1:10,000; Invitrogen) for 10 minutes. Coverslips were mounted, and slides were imaged by confocal microscopy (Fluoview FV100; Olympus). To determine mitochondrial superoxide production in cultures, cells were incubated with 5 μM MitoSOX red mitochondrial superoxide indicator (Invitrogen) for 10 minutes at 37°C. To measure the membrane potential of mitochondria in cultures, cells were incubated with 0.25 μM tetra-methyl rhodamine (TMRM) (Invitrogen) for 20 minutes at 37°C. Neurons were transduced by lentiviral particles containing p53 shRNA or control vector (both EGFP-tagged) for 5 days in the presence or absence of P110-TAT (1 μM/day for 5 days), then cells were stained with fluorescence probes of TMRM to indicate MMP or with mitoSOX to indicate mitoROS . Fluorescence density was quantitated in EGFP positive cells. The images were visualized by microscope and quantification was carried out using NIH ImageJ software.
Determination of total ATP level.
The cellular ATP concentration was measured using an ATP Colorimetric/Fluorometric Assay Kit (BioVision). Cells were lysed in 50 μl of ATP assay buffer, and total ATP level was determined at 570 nm using a microplate reader, according to the manufacturer’s instructions.
Measurement of cell viability.
Striatal cell lines from HD knock-in mice were treated with the Drp1 peptide inhibitor P110-TAT or a control peptide, TAT (at 1 μM each) in FBS-free DMEM medium or in 10% serum for 24 hours. Cell viability was measured using an in vitro toxicology assay, MTT-based kit (Sigma-Aldrich), according to the manufacturer’s instructions. Neurons derived from iPS cells were subjected to withdrawal of the growth factor BDNF for 48 hours. Cell death was determined by measuring LDH release to the culture medium, as described previously (87).
Generation of iPS cell lines.
HD iPS cell lines were generated in the Case Western Reserve University (CWRU) Pluripotent Stem Cell Facility. In brief, fibroblasts were plated at 1 × 104 cells/cm2 and infected with VSV-g pseudotyped hSTEMCCA lentivirus containing a polycistronic coding sequence for the human genes OCT3/4, SOX2, KLF4, and MYC (50). Cells were transitioned to human pluripotent stem cell media consisting of DMEM-F12 supplemented with 20% knockout serum replacement (KSR), 20 ng/ml FGF2, 2 mM Glutamax, 1× non-essential amino acids, and 0.1 mM 2-mercaptoethanol. Nascent iPS cell colonies were individually picked and expanded around day 30. Established lines were passaged as small clusters every 4–5 days with enzymes (1 mg/ml collagenase type IV and 1 mg/ml dispase). Irradiated mouse embryonic fibroblasts (MEFs) served as a feeder layer for iPS cell derivation and subsequent expansion. iPS cell lines were plated on Matrigel-coated (BD Bioscience) 6-well plates in feeder-free medium (Applied Stem Cells Inc.). For characterization of validated iPS cells, alkaline phosphatase staining was carried out using the stem cell characterization kit (Millipore). Immunostaining was performed to immunolabel nuclear, cytoplasmic, and surface markers of stem cells, including Oct3/4 (1:500; Santa Cruz Biotechnology), SOX2 (1:200; Abgent), SSEA4 (1:50), TRA1–60 (1:50), and TRA1–81 (1:50) (Millipore). DNA fingerprinting and karyotyping were performed by Cell Line Genetics. The HD-derived iPS cells expressed antigens of embryonic stem cells including OCT3/4, SOX2, SSEA-4, TRA1–60, and TRA1–81 and were positive in alkaline phosphatase (AP) staining (Supplemental Figure 4).
Neuronal differentiation.
We followed the differentiation protocols as shown in (41, 45, 51) with some modifications. Briefly, iPS cells were dissociated with Accutase (Invitrogen), plated onto 6-well plates precoated with 2.5% Matrigel and allowed to reach 90% confluence in feeder-free medium. For the first 10 days, cells were treated with SB431542 (10 μM; Tocris Bioscience) and Noggin (100 ng/ml) in neural medium (NM) with FGF2 (20 ng/μl) and EGF (20 ng/μl). NM contained neurobasal and DMEM (1:1), B-27 supplement minus vitamin A (×50; Invitrogen), N2 supplement (×100; Invitrogen), GlutaMax (×100; Invitrogen), 100 U/ml penicillin, and 100 μg/ml streptomycin (Fisher Scientific). For the next 10 days, cells were treated with human recombinant Sonic hedgehog (SHH, 200 ng/ml), DKK1 (100 ng/ml), BDNF (20 ng/ml), and 10 μM Y27632 (Sigma-Aldrich) in neuronal differentiation medium. Neuronal differentiation medium contained Neurobasal and DMEM (1:3), B27, N2, GlutaMax, and PS. Cells were then switched to treatment with BDNF (20 ng/ml), ascorbic acid (200 μM, Sigma-Aldrich), cAMP (0.5 mM; Sigma-Aldrich), and Y27632 (10 μM) in neuronal differentiation medium for the following 5 days. The medium was changed every other day. All growth factors were purchased from PeproTech. After 20-day differentiation, cells were treated with P110-TAT (1 μM/day for 5 days). GABAergic neuron was confirmed by staining with anti-GAD67 antibody (a GABAergic neuronal marker). Medium spiny neuron (MSN) identity was confirmed by co-staining with anti-Tuj-1 antibody (a neuronal marker, red) and anti-DARPP-32 antibody (a marker of MSN, green).
Consistent with previous reports on differentiation to other neuronal lineages (44, 87), we observed the variability in the numbers of GABAergic striatal neurons between batches of differentiation. Then, we stained cells with both markers of mitochondria (anti-TOM20) and markers for GABAergic striatal neurons (GAD67 or DARPP-32) to ensure the observation of mitochondria in these neurons.
For spontaneous embryoid body (EB) differentiation, iPS cells were seeded into ultra low attachment hydrogel (Corning) plates containing DMEM: F12 supplemented with 20% FBS, 2 mM L-glutamine, 0.1 mM non-essential amino acids, and 0.1 mM 2-mercaptoethanol differentiation medium. After 8 days of growth in the suspension, the EBs were transferred to gelatin-coated dishes containing the same medium to allow the cells to expand. Seven days later, the cells were fixed and stained with α-SMA (1:500; Santa Cruz Biotechnology), GATA-4 (1:200; Millipore), and TUJ1 (1:500; Covance).
For immunostaining, neurons were passed onto 12-mm poly-L-ornithine/laminin-coated coverslips (Fisher) and grown in 24-well plates. Cells were fixed with 4% formaldehyde in PBS for 20 minutes at room temperature. The coverslips were washed twice in PBS and incubated in 0.1% Triton solution followed by blocking solution (0.05% Triton, 2% normal goat serum). The samples were incubated with the indicated primary antibodies overnight at 4°C and then washed 3 times in PBS at room temperature before incubation with secondary antibodies (Alexa Fluor 488–conjugated anti-mouse/rabbit IgG [1:500] and/or Alexa Fluor 586–conjugated anti-mouse/rabbit IgG [1:500]) for 1 hour at room temperature in blocking solution. The samples were then washed 5 times with PBS and incubated for 10 minutes with Hoechst 33258 (1:10,000; Invitrogen). The coverslips were mounted with AquaMount on glass microscope slides and imaged by microscopy. The following primary antibodies were used: Tom20 (1:500; Santa Cruz Biotechnology), Tuj1 (1:500; Covance), MAP2 (1:200; Cell Signaling), GAD67 (1:200; Millipore), Calbindin (1:100; R&D Systems) and DARPP-32 (1:100; Epitomics). The imaging was observed using an Olympus Fluoview FV100 microscope.
Animal model of HD.
Male R6/2 mice and their wild-type littermates were purchased from the Jackson Laboratory [B6CBA-TgN (HD exon 1)62; JAX stock number: 006494]. These mice are transgenic for the 5′ end of the human HD gene carrying 100–150 glutamine (CAG) repeats. R6/2 mice used in this study had an average of 131 ± 2.3 (mean ± SD) CAG repeats. Mice were maintained with a 12-hour light/12-hour dark cycle (on at 6:00 am, off at 6:00 pm).
Measurement of mitochondrial oxygen consumption in R6/2 HD mice.
Mitochondrial O2 consumption was monitored in 125 mM sucrose, 65 mM KCl, 10 mM HEPES, 2 mM KH2PO4, 100 mMEGTA, and 2 mM MgCl2 in the presence of 0.01% BSA at pH 7.2 containing 0.1 mg mitochondrial protein/ml using a computer-interfaced Clark-type electrode (OROBOROS Oxygraph-2k) operating with continuous stirring at 37°C. Succinate, malate, and glutamate (2 mM of each) were used as substrates, and ADP (1 mM) was added to induce state 3 respiratory rate. A subsequent addition of oligomycin (1 μg/ml) was used to determine the state 4 respiratory rate. RCR was calculated by dividing state 3 by state 4 oxygen consumption rates, which demonstrates the tightness of the coupling between mitochondrial respiration and phosphorylation.
Peptide treatment and behavior tests in R6/2 HD mice.
Hemizygous R6/2 mice (Tg) and their wild-type littermates were implanted with a 28-day osmotic pump (Alzet) containing TAT control peptide or P110-TAT peptide, which delivered peptides to the mice at a rate of 3 mg/kg/day. The first pump was implanted subcutaneously in the backs of 5-week-old mice between the shoulders and replaced once, after 4 weeks. At the end of the treatment, the 13-week-old mice were tested to assess motor function. The mice were placed in a clean cage with clean bedding for 5 minutes and were recorded using a Sony HandyCam full HD camcorder (model HDR-CX220/R). Distance traveled and behaviors including grooming, rearing, digging, and seizures were analyzed. The overall survival during the study period was recorded and the remaining mice were sacrificed at 13 weeks of age for sample harvest. All behavior and survival tests and analyses were conducted by an experimenter who was blind to genotypes and drug groups.
Immunohistochemistry in HD R6/2 mice.
Mice were deeply anesthetized and transcardially perfused with 4% paraformaldehyde in 0.1 M phosphate buffer (PB; pH 7.4) at 13 weeks of age. Brains were processed for paraffin embedment. Brain sections (7 μm, coronal) were used for immunohistochemical localization of DARPP-32 (1:500; Epitomics) or Htt protein (1:200; clone EM48; Millipore) using the IHC Select HRP/DAB kit (Millipore).
For quantification of DARPP-32 immunostaining, striatal sections (2 sections per mouse in each hemisphere; therefore, 4 sections per mouse) were viewed and digitized using a Nikon microscope (magnification, ×20). Within the striatum, images of the 0.58 × 0.44 mm2 sample field (3 images/field for a total of 12 images per mouse) were collected from rostral to mid-striatal levels and dorsal striatum for all animals, since HD pathology progresses from dorsal to ventral, anterior to posterior, and medial to lateral within this brain area (88). The immunodensity was quantitated using NIH ImageJ software with plug-ins of color deconvolution, with the method described in ref. 89.
To quantify Htt aggregates, we followed the method as described in (90) with some modification. Briefly, color digital images of Htt aggregates were obtained with a ×40 objective lens from the dorsal lateral striatum and converted to grayscale images using the ImageJ software. The threshold of the grayscale images were then adjusted until staining of soluble Htt disappeared. The same parameters were used to take and adjust all images. We used the particle analysis function of ImageJ to measure the number and size of Htt aggregates larger than 0.1 μm2. Five images from each mouse were analyzed.
The same image exposure times and threshold settings were used for sections from all treatment groups. Quantification was conducted by an observer blinded to the study conditions.
Electron microscopy.
Thin sections of respective brain tissue were fixed in 2.5% glutaraldehyde in 0.1 mol/l cacodylate buffer, pH 7.4. The fixed material was sectioned at the Stanford Electron Microscopy Facility. Sections of 75 to 80 nm were taken, picked up on Formvar/carbon-coated 75 mesh Ni grids and stained for 20 seconds in 1:1 saturated Uranyl acetate (~7.7%) in acetone followed by staining in 0.2% lead citrate for 3 to 4 minutes for contrast. Mitochondrial samples were observed in a JEOL 1230 transmission electron microscope at 80 kV and photos were taken using a Gatan Multiscan 791 digital camera.
Western blot analysis.
Protein concentrations were determined by Bradford assay. Then, protein was resuspended in Laemmli buffer, loaded on SDS-PAGE and transferred onto nitrocellulose membranes. Membranes were probed with the indicated antibody, followed by visualization with ECL.
Statistics.
Data are expressed as means ± SEM. Statistical analysis was assessed by unpaired Student’s t test and 1-way or 2-way ANOVA. Whenever significant F values were obtained, Tukey adjustment was used for multiple comparison purposes. The standard Mantel Cox log-rank test was used to assess survival. Significance of changes in neurological symptoms was analyzed with the Fisher’s exact test. Statistical significance was considered achieved when the P value was less than 0.05.
Study approval.
All experiments in animals were in accordance with protocols approved by the Institutional Animal Care and Use Committee of Stanford University and were performed based on the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Sufficient actions were considered for reducing pain or discomfort of subjects during the experiments.
Supplementary Material
Acknowledgments
The authors thank Case Western Reserve University Pluripotent Stem Cell Facility, directed by Paul Tesar, for generating HD-iPS cells and iPS cell characterization. They also thank Anna Sato for technical assistance in the study of the R6/2 mice. The work was supported in part by a Clinical and Translational Science Collaborative core facility grant at Case Western Reserve University (to X. Qi), by Sanofi BioX Stanford grant and NIH HL52141 (to D. Mochly-Rosen), and by NINDS P30 center core grant (NS069375-01A1) (to M. Monbureau and M. Shamloo).
Footnotes
Conflict of interest: A patent on the design and application of mitochondrial fission peptide inhibitor has been filed.
Citation for this article: J Clin Invest. 2013;123(12):5371–5388. doi:10.1172/JCI70911.
References
- 1.Ross CA, Tabrizi SJ. Huntington’s disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10(1):83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- 2.[No authors listed]. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993;72(6):971–983. doi: 10.1016/0092-8674(93)90585-E. [DOI] [PubMed] [Google Scholar]
- 3.Costa V, et al. Mitochondrial fission and cristae disruption increase the response of cell models of Huntington’s disease to apoptotic stimuli. EMBO Mol Med. 2010;2(12):490–503. doi: 10.1002/emmm.201000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song W, et al. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat Med. 2011;17(3):377–382. doi: 10.1038/nm.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Browne SE, Ferrante RJ, Beal MF. Oxidative stress in Huntington’s disease. Brain Pathol. 1999;9(1):147–163. doi: 10.1111/j.1750-3639.1999.tb00216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oliveira JM. Nature and cause of mitochondrial dysfunction in Huntington’s disease: focusing on huntingtin and the striatum. J Neurochem. 2010;114(1):1–12. doi: 10.1111/j.1471-4159.2010.06741.x. [DOI] [PubMed] [Google Scholar]
- 7.Oettinghaus B, Licci M, Scorrano L, Frank S. Less than perfect divorces: dysregulated mitochondrial fission and neurodegeneration. Acta Neuropathol. 2012;123(2):189–203. doi: 10.1007/s00401-011-0930-z. [DOI] [PubMed] [Google Scholar]
- 8.Costa V, Scorrano L. Shaping the role of mitochondria in the pathogenesis of Huntington’s disease. EMBO J. 2012;31(8):1853–1864. doi: 10.1038/emboj.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125(7):1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 10.Knott AB, Perkins G, Schwarzenbacher R, Bossy-Wetzel E. Mitochondrial fragmentation in neurodegeneration. Nat Rev Neurosci. 2008;9(7):505–518. doi: 10.1038/nrn2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan DC. Mitochondrial fusion and fission in mammals. Annu Rev Cell Dev Biol. 2006;22:79–99. doi: 10.1146/annurev.cellbio.22.010305.104638. [DOI] [PubMed] [Google Scholar]
- 12.Chang CR, Blackstone C. Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Ann N Y Acad Sci. 2010;1201:34–39. doi: 10.1111/j.1749-6632.2010.05629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fannjiang Y, et al. Mitochondrial fission proteins regulate programmed cell death in yeast. Genes Dev. 2004;18(22):2785–2797. doi: 10.1101/gad.1247904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.James DI, Parone PA, Mattenberger Y, Martinou JC. hFis1, a novel component of the mammalian mitochondrial fission machinery. J Biol Chem. 2003;278(38):36373–36379. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- 15.Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, Ryan MT. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 2011;12(6):565–573. doi: 10.1038/embor.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao J, et al. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J. 2011;30(14):2762–2778. doi: 10.1038/emboj.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otera H, et al. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol. 2010;191(6):1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liot G, Bossy B, Lubitz S, Kushnareva Y, Sejbuk N, Bossy-Wetzel E. Complex II inhibition by 3-NP causes mitochondrial fragmentation and neuronal cell death via an NMDA- and ROS-dependent pathway. Cell Death Differ. 2009;16(6):899–909. doi: 10.1038/cdd.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, Lim PJ, Karbowski M, Monteiro MJ. Effects of overexpression of huntingtin proteins on mitochondrial integrity. Hum Mol Genet. 2009;18(4):737–752. doi: 10.1093/hmg/ddn404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shirendeb UP, et al. Mutant huntingtin’s interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington’s disease. Hum Mol Genet. 2012;21(2):406–420. doi: 10.1093/hmg/ddr475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458(7242):1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moll UM, Wolff S, Speidel D, Deppert W. Transcription-independent pro-apoptotic functions of p53. Curr Opin Cell Biol. 2005;17(6):631–636. doi: 10.1016/j.ceb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Vaseva AV, Moll UM. The mitochondrial p53 pathway. Biochim Biophys Acta. 2009;1787(5):414–420. doi: 10.1016/j.bbabio.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchenko ND, Wolff S, Erster S, Becker K, Moll UM. Monoubiquitylation promotes mitochondrial p53 translocation. EMBO J. 2007;26(4):923–934. doi: 10.1038/sj.emboj.7601560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y, et al. p53 translocation to mitochondria precedes its nuclear translocation and targets mitochondrial oxidative defense protein-manganese superoxide dismutase. Cancer Res. 2005;65(9):3745–3750. doi: 10.1158/0008-5472.CAN-04-3835. [DOI] [PubMed] [Google Scholar]
- 26.Galluzzi L, Morselli E, Kepp O, Vitale I, Pinti M, Kroemer G. Mitochondrial liaisons of p53. Antioxid Redox Signal. 2011;15(6):1691–1714. doi: 10.1089/ars.2010.3504. [DOI] [PubMed] [Google Scholar]
- 27.Bae BI, et al. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington’s disease. Neuron. 2005;47(1):29–41. doi: 10.1016/j.neuron.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Trettel F, et al. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet. 2000;9(19):2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- 29.Qi X, Qvit N, Su YC, Mochly-Rosen D. A novel Drp1 inhibitor diminishes aberrant mitochondrial fission and neurotoxicity. J Cell Sci. 2013;126(pt 3):789–802. doi: 10.1242/jcs.114439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mears JA, Lackner LL, Fang S, Ingerman E, Nunnari J, Hinshaw JE. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat Struct Mol Biol. 2011;18(1):20–26. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu PP, Patterson A, Stadler J, Seeburg DP, Sheng M, Blackstone C. Intra- and intermolecular domain interactions of the C-terminal GTPase effector domain of the multimeric dynamin-like GTPase Drp1. J Biol Chem. 2004;279(34):35967–35974. doi: 10.1074/jbc.M404105200. [DOI] [PubMed] [Google Scholar]
- 32.Reddy PH, Shirendeb UP. Mutant huntingtin, abnormal mitochondrial dynamics, defective axonal transport of mitochondria, and selective synaptic degeneration in Huntington’s disease. Biochim Biophys Acta. 2012;1822(2):101–110. doi: 10.1016/j.bbadis.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Jiang H, Chen S, Du F, Wang X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell. 2012;148(1–2):228–243. doi: 10.1016/j.cell.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 34.Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27(2):433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frank S, et al. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1(4):515–525. doi: 10.1016/S1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 36.Reddy PH, Reddy TP, Manczak M, Calkins MJ, Shirendeb U, Mao P. Dynamin-related protein 1 and mitochondrial fragmentation in neurodegenerative diseases. Brain Res Rev. 2011;67(1–2):103–118. doi: 10.1016/j.brainresrev.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su YC, Qi X. Inhibition of excessive mitochondrial fission reduced aberrant autophagy and neuronal damage caused by LRRK2 G2019S mutation. Hum Mol Genet. 2013;22(22):4546–4561. doi: 10.1093/hmg/ddt301. [DOI] [PubMed] [Google Scholar]
- 38.Li J, Donath S, Li Y, Qin D, Prabhakar BS, Li P. miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genet. 2010;6(1):e1000795. doi: 10.1371/journal.pgen.1000795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang DB, et al. Declines in drp1 and parkin expression underlie DNA damage-induced changes in mitochondrial length and neuronal death. J Neurosci. 2013;33(4):1357–1365. doi: 10.1523/JNEUROSCI.3365-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Komarov PG, et al. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285(5434):1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- 41.HD iPSC Consortium. Induced pluripotent stem cells from patients with Huntington’s disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell. 2012;11(2):264–278. doi: 10.1016/j.stem.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castiglioni V, Onorati M, Rochon C, Cattaneo E. Induced pluripotent stem cell lines from Huntington’s disease mice undergo neuronal differentiation while showing alterations in the lysosomal pathway. Neurobiol Dis. 2012;46(1):30–40. doi: 10.1016/j.nbd.2011.12.032. [DOI] [PubMed] [Google Scholar]
- 43.Camnasio S, et al. The first reported generation of several induced pluripotent stem cell lines from homozygous and heterozygous Huntington’s disease patients demonstrates mutation related enhanced lysosomal activity. Neurobiol Dis. 2012;46(1):41–51. doi: 10.1016/j.nbd.2011.12.042. [DOI] [PubMed] [Google Scholar]
- 44.Jeon I, et al. Neuronal properties, in vivo effects, and pathology of a Huntington’s disease patient-derived induced pluripotent stem cells. Stem Cells. 2012;30(9):2054–2062. doi: 10.1002/stem.1135. [DOI] [PubMed] [Google Scholar]
- 45.Zhang N, An MC, Montoro D, Ellerby LM. Characterization of human Huntington’s disease cell model from induced pluripotent stem cells. PLoS Curr. 2010;2:RRN1193. doi: 10.1371/currents.RRN1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gunaseeli I, Doss MX, Antzelevitch C, Hescheler J, Sachinidis A. Induced pluripotent stem cells as a model for accelerated patient- and disease-specific drug discovery. Curr Med Chem. 2010;17(8):759–766. doi: 10.2174/092986710790514480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benraiss A, Goldman SA. Cellular therapy and induced neuronal replacement for Huntington’s disease. Neurotherapeutics. 2011;8(4):577–590. doi: 10.1007/s13311-011-0075-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kiskinis E, Eggan K. Progress toward the clinical application of patient-specific pluripotent stem cells. J Clin Invest. 2010;120(1):51–59. doi: 10.1172/JCI40553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481(7381):295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Somers A, et al. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells. 2010;28(10):1728–1740. doi: 10.1002/stem.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yazawa M, et al. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature. 2011;471(7337):230–234. doi: 10.1038/nature09855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maksimovic ID, et al. Oxidative damage and metabolic dysfunction in experimental Huntington’s disease: selective vulnerability of the striatum and hippocampus. Vojnosanit Pregl. 2001;58(3):237–242. [PubMed] [Google Scholar]
- 53.Han I, You Y, Kordower JH, Brady ST, Morfini GA. Differential vulnerability of neurons in Huntington’s disease: the role of cell type-specific features. J Neurochem. 2010;113(5):1073–1091. doi: 10.1111/j.1471-4159.2010.06672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Graveland GA, Williams RS, DiFiglia M. Evidence for degenerative and regenerative changes in neostriatal spiny neurons in Huntington’s disease. Science. 1985;227(4688):770–773. doi: 10.1126/science.3155875. [DOI] [PubMed] [Google Scholar]
- 55.Tang TS, et al. Disturbed Ca2+ signaling and apoptosis of medium spiny neurons in Huntington’s disease. Proc Natl Acad Sci U S A. 2005;102(7):2602–2607. doi: 10.1073/pnas.0409402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Melone MA, Jori FP, Peluso G. Huntington’s disease: new frontiers for molecular and cell therapy. Curr Drug Targets. 2005;6(1):43–56. doi: 10.2174/1389450053344975. [DOI] [PubMed] [Google Scholar]
- 57.Kelly CM, Dunnett SB, Rosser AE. Medium spiny neurons for transplantation in Huntington’s disease. Biochem Soc Trans. 2009;37(pt 1):323–328. doi: 10.1042/BST0370323. [DOI] [PubMed] [Google Scholar]
- 58.Mangiarini L, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87(3):493–506. doi: 10.1016/S0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 59.Zischka H, et al. Liver mitochondrial membrane crosslinking and destruction in a rat model of Wilson disease. J Clin Invest. 2011;121(4):1508–1518. doi: 10.1172/JCI45401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brand MD, Nicholls DG. Assessing mitochondrial dysfunction in cells. Biochem J. 2011;435(2):297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gottlieb E, Armour SM, Thompson CB. Mitochondrial respiratory control is lost during growth factor deprivation. Proc Natl Acad Sci U S A. 2002;99(20):12801–12806. doi: 10.1073/pnas.202477599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Faizi M, et al. Thy1-hAPP(Lond/Swe+) mouse model of Alzheimer’s disease displays broad behavioral deficits in sensorimotor, cognitive and social function. Brain Behav. 2012;2(2):142–154. doi: 10.1002/brb3.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Coutellier L, Beraki S, Ardestani PM, Saw NL, Shamloo M. Npas4: a neuronal transcription factor with a key role in social and cognitive functions relevant to developmental disorders. PLoS One. 2012;7(9):e46604. doi: 10.1371/journal.pone.0046604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bibb JA, et al. Severe deficiencies in dopamine signaling in presymptomatic Huntington’s disease mice. Proc Natl Acad Sci U S A. 2000;97(12):6809–6814. doi: 10.1073/pnas.120166397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Canals JM, et al. Brain-derived neurotrophic factor regulates the onset and severity of motor dysfunction associated with enkephalinergic neuronal degeneration in Huntington’s disease. J Neurosci. 2004;24(35):7727–7739. doi: 10.1523/JNEUROSCI.1197-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Almeida LP, Ross CA, Zala D, Aebischer P, Deglon N. Lentiviral-mediated delivery of mutant huntingtin in the striatum of rats induces a selective neuropathology modulated by polyglutamine repeat size, huntingtin expression levels, and protein length. J Neurosci. 2002;22(9):3473–3483. doi: 10.1523/JNEUROSCI.22-09-03473.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simmons DA, Mehta RA, Lauterborn JC, Gall CM, Lynch G. Brief ampakine treatments slow the progression of Huntington’s disease phenotypes in R6/2 mice. Neurobiol Dis. 2011;41(2):436–444. doi: 10.1016/j.nbd.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davies SW, et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90(3):537–548. doi: 10.1016/S0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 69.DiFiglia M, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277(5334):1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 70.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431(7010):805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 71.Gutekunst CA, et al. Nuclear and neuropil aggregates in Huntington’s disease: relationship to neuropathology. J Neurosci. 1999;19(7):2522–2534. doi: 10.1523/JNEUROSCI.19-07-02522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hatters DM. Protein misfolding inside cells: the case of huntingtin and Huntington’s disease. IUBMB Life. 2008;60(11):724–728. doi: 10.1002/iub.111. [DOI] [PubMed] [Google Scholar]
- 73.Tsunemi T, et al. PGC-1α rescues Huntington’s disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci Transl Med. 2012;4(142):142ra97. doi: 10.1126/scitranslmed.3003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Germain M, Mathai JP, McBride HM, Shore GC. Endoplasmic reticulum BIK initiates DRP1-regulated remodelling of mitochondrial cristae during apoptosis. EMBO J. 2005;24(8):1546–1556. doi: 10.1038/sj.emboj.7600592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 76.Sharpless NE, DePinho RA. p53: good cop/bad cop. Cell. 2002;110(1):9–12. doi: 10.1016/S0092-8674(02)00818-8. [DOI] [PubMed] [Google Scholar]
- 77.Pani G, Galeotti T. Role of MnSOD and p66shc in mitochondrial response to p53. Antioxid Redox Signal. 2011;15(6):1715–1727. doi: 10.1089/ars.2010.3499. [DOI] [PubMed] [Google Scholar]
- 78.Matoba S, et al. p53 regulates mitochondrial respiration. Science. 2006;312(5780):1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 79.Vaseva AV, Marchenko ND, Ji K, Tsirka SE, Holzmann S, Moll UM. p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell. 2012;149(7):1536–1548. doi: 10.1016/j.cell.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang WX, Gray M. Neurobiology of Huntington’s Disease: Applications to Drug Discovery. Mouse models for validating preclinical candidates for Huntington’s disease. In: Lo DC, Hughes RE, eds. Boca Raton, Florida, USA: CRC Press; 2011:Chapter 7. [Google Scholar]
- 81.Switonski PM, Szlachcic WJ, Gabka A, Krzyzosiak WJ, Figiel M. Mouse models of polyglutamine diseases in therapeutic approaches: review and data table. Part II. Mol Neurobiol. 2012;46(2):430–466. doi: 10.1007/s12035-012-8316-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Trancikova A, Ramonet D, Moore DJ. Genetic mouse models of neurodegenerative diseases. Prog Mol Biol Transl Sci. 2011;100:419–482. doi: 10.1016/B978-0-12-384878-9.00012-1. [DOI] [PubMed] [Google Scholar]
- 83.Slow EJ, et al. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum Mol Genet. 2003;12(13):1555–1567. doi: 10.1093/hmg/ddg169. [DOI] [PubMed] [Google Scholar]
- 84.Qi X, Inagaki K, Sobel RA, Mochly-Rosen D. Sustained pharmacological inhibition of deltaPKC protects against hypertensive encephalopathy through prevention of blood-brain barrier breakdown in rats. J Clin Invest. 2008;118(1):173–182. doi: 10.1172/JCI32636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bright R, et al. Protein kinase C delta mediates cerebral reperfusion injury in vivo. J Neurosci. 2004;24(31):6880–6888. doi: 10.1523/JNEUROSCI.4474-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Qi X, Disatnik MH, Shen N, Sobel RA, Mochly-Rosen D. Aberrant mitochondrial fission in neurons induced by protein kinase Cδ under oxidative stress conditions in vivo. Mol Biol Cell. 2011;22(2):256–265. doi: 10.1091/mbc.E10-06-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qi X, Mochly-Rosen D. The PKCδ-Abl complex communicates ER stress to the mitochondria — an essential step in subsequent apoptosis. J Cell Sci. 2008;121(pt 6):804–813. doi: 10.1242/jcs.024653. [DOI] [PubMed] [Google Scholar]
- 88.Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP. Neuropathological classification of Huntington’s disease. J Neuropathol Exp Neurol. 1985;44(6):559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 89.Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2001;23(4):291–299. [PubMed] [Google Scholar]
- 90.Gharami K, Xie Y, An JJ, Tonegawa S, Xu B. Brain-derived neurotrophic factor over-expression in the forebrain ameliorates Huntington’s disease phenotypes in mice. J Neurochem. 2008;105(2):369–379. doi: 10.1111/j.1471-4159.2007.05137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cooper O, et al. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson’s disease. Sci Transl Med. 2012;4(141):141ra90. doi: 10.1126/scitranslmed.3003985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.