Abstract
Basal autophagy is a crucial mechanism in cellular homeostasis, underlying both normal cellular recycling and the clearance of damaged or misfolded proteins, organelles and aggregates. We showed here that enhanced levels of autophagy induced by either autophagic gene overexpression or voluntary exercise ameliorated desmin-related cardiomyopathy (DRC). To increase levels of basal autophagy, we generated an inducible Tg mouse expressing autophagy-related 7 (Atg7), a critical and rate-limiting autophagy protein. Hearts from these mice had enhanced autophagy, but normal morphology and function. We crossed these mice with CryABR120G mice, a model of DRC in which autophagy is significantly attenuated in the heart, to test the functional significance of autophagy activation in a proteotoxic model of heart failure. Sustained Atg7-induced autophagy in the CryABR120G hearts decreased interstitial fibrosis, ameliorated ventricular dysfunction, decreased cardiac hypertrophy, reduced intracellular aggregates and prolonged survival. To determine whether different methods of autophagy upregulation have additive or even synergistic benefits, we subjected the autophagy-deficient CryABR120G mice and the Atg7-crossed CryABR120G mice to voluntary exercise, which also upregulates autophagy. The entire exercised Atg7-crossed CryABR120G cohort survived to 7 months. These findings suggest that activating autophagy may be a viable therapeutic strategy for improving cardiac performance under proteotoxic conditions.
Introduction
Autophagy is a conserved catabolic process that is essential for maintaining cellular homeostasis. Normal autophagic processes degrade long-lived proteins and cytoplasmic organelles, providing their components for recycling (1, 2). More recently, autophagy has been identified as being a major contributor in the clearance of misfolded and aggregated proteins in the cytosol of mammalian cells (3, 4). The biological importance of basal autophagy in the clearance of misfolded proteins under physiological conditions can be inferred from conditional gene ablation for some of the essential autophagy genes: Atg7 deletion in hepatocytes, Atg5 and Atg7 deletion in neurons, and Atg5 deletion in cardiomyocytes led to the accumulation of ubiquitin-positive protein aggregates in inclusion bodies (5–8). Impairment of autophagy leads to decreased cytosolic protein turnover and increased aggregate protein accumulation under normal metabolic conditions (9).
Recent studies have shown that a primary failure in autophagy could be responsible for the accumulation and aggregation of misfolded proteins in neurodegenerative disorders, such as Alzheimer’s disease, transmissible spongiform encephalopathy, Parkinson’s disease, and Huntington’s disease (10). Induction of autophagy using pharmacological intervention (11), autophagic gene overexpression (12), and voluntary exercise (13–15) protected cells against the toxic insults of aggregate-prone proteins associated with neurodegeneration by promoting their clearance. Similar to the cytoplasmic phenomena observed in neurodegenerative diseases, desmin-related cardiomyopathy (DRC) is characterized by the accumulation of cytotoxic misfolded proteins in the form of preamyloid oligomers and protein aggregates (16). Earlier studies from our laboratory showed that Tg mice with cardiomyocyte-specific expression of mutated αB-crystallin (CryABR120G mice; ref. 17) accurately recapitulated the human DRC phenotype, with dilated cardiomyopathy and heart failure (18–21). Ultrastructural observations (18) and autophagic flux assays in CryABR120G-expressing cardiomyocytes revealed reduced autophagic function, which we hypothesized contributed to failed clearance of the misfolded proteins and aggregates (22).
To determine whether morbidity and mortality could be improved in the CryABR120G model by restoration of autophagy, we generated Tg mice overexpressing Atg7 in cardiomyocytes, in an attempt to increase autophagic flux in the hearts. ATG7 is a noncanonical E1-like enzyme that uniquely recognizes 2 distantly related ubiquitin-like proteins, ATG8 and ATG12, directing them to their respective E2 partners, ATG3 and ATG10. They are then typically conjugated to the substrates phosphatidylethanolamine and ATG5, respectively (23–26). Genetic deletion of Atg7 by conditional knockdown impaired autophagosome formation and constitutive turnover of cytoplasmic components, which suggests that ATG7 is necessary for autophagosome formation (5).
Herein, we demonstrated that increased Atg7 expression was sufficient to increase autophagy in the heart and that this increase did not cause any detectable pathology or compromise cardiac function in normal mice. When these mice were crossed with CryABR120G mice, the probability of survival was increased, and morbidity decreased significantly.
Results
Generation of Tg mice in which enhanced levels of Atg7 can be induced.
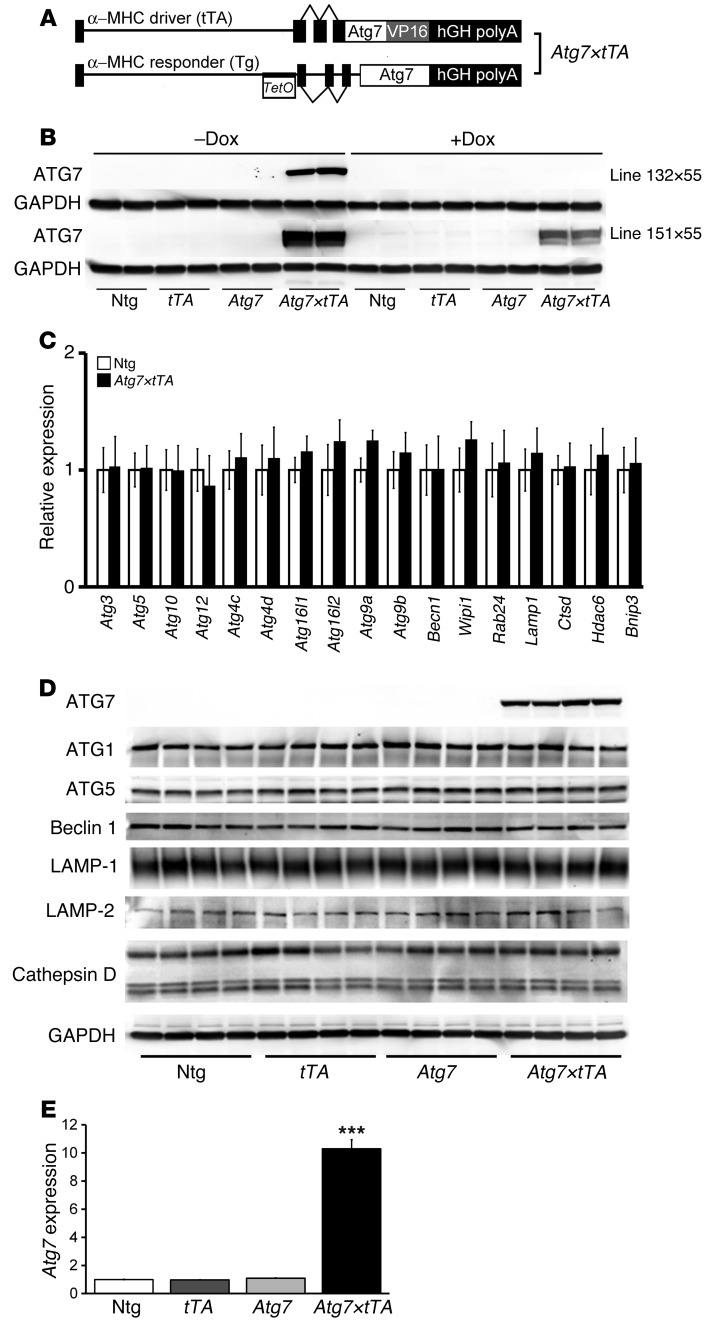
In an attempt to induce high levels of basal autophagy, we generated an inducible Tg mouse (17) containing the ATG7 cDNA behind a modified myosin heavy chain (α-MHC) promoter cassette to generate responder Atg7-Tg mice (27). We hypothesized that overexpression of a single, potentially rate-limiting protein might result in increased autophagic activity. The responder Atg7-Tg mice were crossed with doxycycline-controlled (Dox-controlled) transcriptional activator mice (referred to herein as tTA-Tg mice; ref. 28) to generate double-Tg Atg7×tTA mice (Figure 1A). Typical expression patterns for 2 lines (line 132 and line 151) showed that Dox treatment effectively inhibited transgene expression (Figure 1B). As the transgene in line 132 was very tightly regulated by Dox treatment, we selected this line for further study. We titrated other transcripts associated with autophagy in Atg7×tTA mice and noted no significant upregulation of those RNAs compared with control nontransgenic (Ntg) mice (Figure 1C). We validated these data using selective Western blot analyses and confirmed that upregulation of Atg7 expression did not lead to increased steady-state levels of other autophagy- and lysosomal-associated proteins (Figure 1D). Quantitation of transgene expression via Western blotting confirmed 10-fold overexpression of Atg7 in the hearts of Atg7×tTA mice relative to control Ntg, Atg7-Tg, or tTA-Tg hearts (Figure 1E). We also observed Atg7 expression–dependent regulation of p62 in the 2 Atg7×tTA lines, line 132 and line 151, as Dox treatment effectively inhibited p62 expression (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI70877DS1). Quantitation of p62 expression showed 4.5-fold overexpression in Atg7×tTA compared with Ntg, Atg7-Tg, and tTA-Tg hearts in line 132 (Supplemental Figure 1, B and C).
Figure 1. Generation and characterization of cardiomyocyte-specific inducible Atg7-overexpressing mice (Atg7×tTA).
(A) Construct design of the binary Tg system regulated by Dox to inducibly overexpress Atg7 in the heart. (B) Representative Western blot showing ATG7 cardiac protein from 2 lines (line 132 and line 151) of 3-month-old Ntg, tTA-Tg, Atg7-Tg, and Atg7×tTA mice without or with Dox (17). (C) Autophagy-related transcript analyses in Atg7×tTA hearts. Shown is a direct groupwise comparison of fold change in mRNA levels in male Atg7×tTA and Ntg hearts at 5 months (n = 3 per group). (D) Western blot showing ATG7 expression and expression levels of autophagy-related proteins in the hearts of 3-month-old Ntg, tTA-Tg, Atg7-Tg, and Atg7×tTA (line 132×55) mice. (E) Quantitation of Atg7 expression for line 132. ***P < 0.001 versus all other groups, Tukey’s post-hoc test.
Elevated Atg7 levels are sufficient to trigger increased cardiac autophagy in Tg mice.
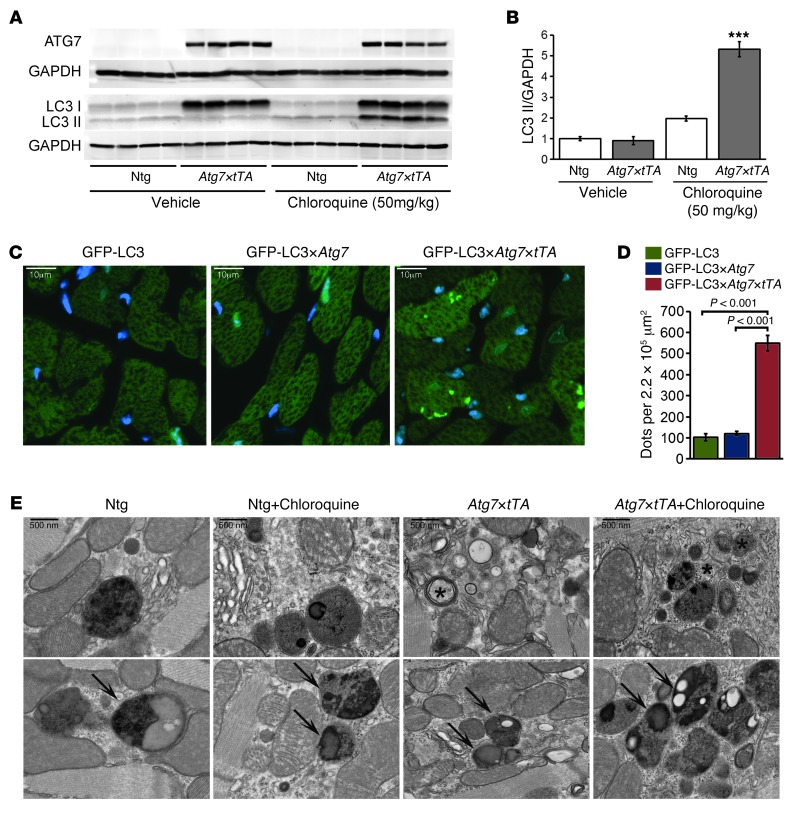
To evaluate whether autophagy was induced by Atg7 overexpression, we carried out an autophagic flux assay (28, 29). Mice were treated with chloroquine (50 mg/kg) for 5 days to block lysosomal function. The assay showed increased levels of the auophagosome membrane–associated lipidated protein LC3-II in Atg7×tTA hearts (Figure 2, A and B). Quantitation of p62 expression showed no significant changes after chloroquine treatment in both Atg7×tTA and Ntg hearts (Supplemental Figure 1, D and E). We also crossed Atg7-Tg and Atg7×tTA mice with reporter mice expressing a GFP-labeled marker of autophagosomes, GFP-LC3, to evaluate autophagic flux in vivo. The number of GFP-LC3 puncta was significantly increased by Atg7 overexpression in the GFP-LC3×Atg7×tTA mice (Figure 2, C and D). Evaluation of the ultrastructure of Atg7×tTA hearts after lysosomal inhibition by chloroquine treatment also showed marked upregulation of the number of intracellular autophagic structures. Most notably, Atg7×tTA hearts showed a large number of amphisomes (Figure 2E), an intermediate structure that develops after an autophagosome engulfs its cargo and fuses with other vesicular structures such as multivesicular endosomes.
Figure 2. Autophagy in Atg7×tTA mouse hearts.
(A) Autophagic flux assay (see Methods) showed increased LC3-II levels by Western blot analysis in Ntg and Atg7×tTA hearts (n = 4 per treatment). (B) Densitometry analysis of LC3-II expression relative to GAPDH. ***P < 0.001 vs. Ntg, Tukey’s post-hoc test. (C) Representative images of GFP-LC3 puncta (autophagosomes) in hearts from male GFP-LC3 control, GFP-LC3×Atg7, and GFP-LC3×Atg7×tTA mice. (D) Quantification of GFP puncta per microscopic field (220,000 μm2) in LV. (E) Ultrastructural analyses confirmed an increase in autophagic structures in Atg7×tTA hearts. Arrows denote amphisomes; asterisks denote multilamellar bodies. Scale bars: 10 μm (C); 500 nm (E).
Normal heart function and morphology are preserved when autophagy is upregulated.
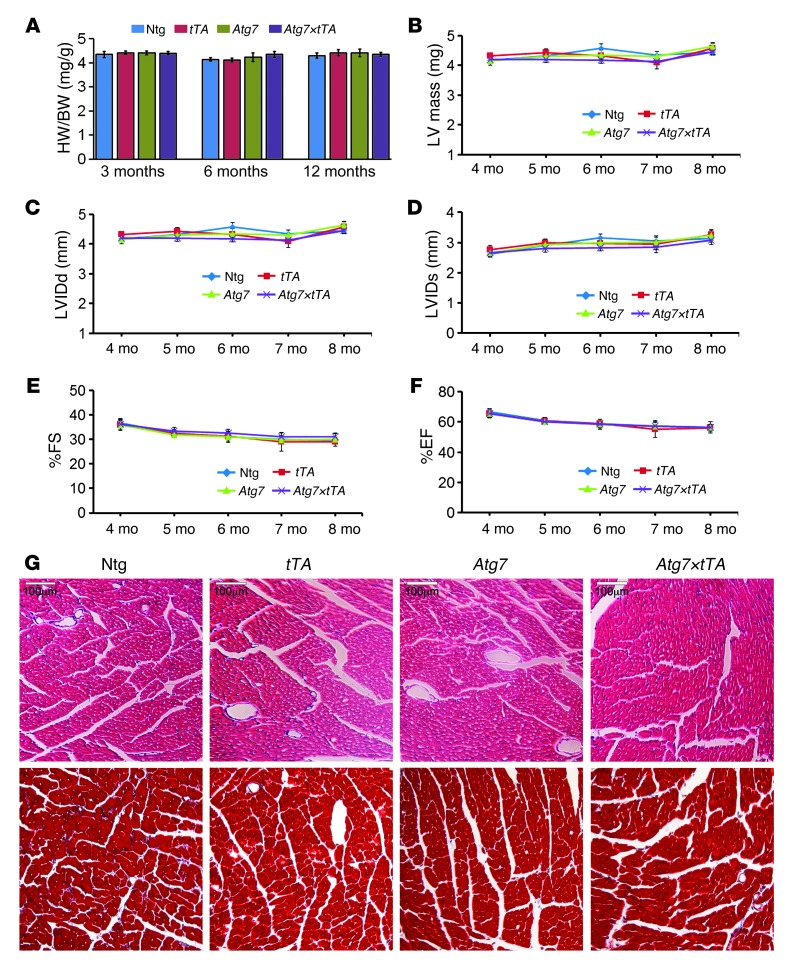
We anticipated that upregulation of autophagy for prolonged periods under basal conditions might lead to cardiac disease. Heart weight/body weight (HW/BW) ratios at 3, 6, and 12 months of age in Ntg, Atg7-Tg, tTA-Tg, and Atg7×tTA mice showed no significant differences (Figure 3A). Serial changes in LV mass index by echocardiography at 4–8 months across genotypes also showed normal heart size in Atg7×tTA mice (Figure 3B). Atg7×tTA hearts displayed normal LV dimensions, as measured by LV internal diameters in end-diastole and end-systole (LVIDd and LVIDs, respectively; Figure 3, C and D). LV function, as measured by percent fractional shortening (%FS) and ejection fraction (%EF), was also unaffected by increased autophagic flux (Figure 3, E and F). H&E- and Masson’s trichrome–stained histological sections showed no pathological features in the hearts of Atg7×tTA mice compared with tTA-Tg, Atg7-Tg, and Ntg littermates (Figure 3G). Despite our expectations that increased autophagy might be pathologic in healthy hearts, we concluded that Atg7×tTA animals lack any notable cardiac pathology and have normal anatomy and function through at least mid-adulthood.
Figure 3. Cardiac function and histology of Atg7×tTA mouse hearts.
(A) HW/BW ratios at 3, 6, and 12 months in Ntg, tTA-Tg, Atg7-Tg, and Atg7×tTA mice (n ≥ 6 per group). (B) Serial changes in LV mass index by echocardiography across genotypes, indicative of normal heart size. (C–F) Effect of Atg7 overexpression on echocardiography indices of LV end-diastolic diameter, measured by (C) LVIDd and (D) LVIDs, and LV function, measured by (E) %FS and (F) %EF (n ≥ 10 per group). (G) H&E (top) and Masson’s trichrome (bottom) staining of cardiac histological sections from 6-month-old mice showed no differences or overt pathology with Atg7 overexpression. Scale bars: 100 μm.
Atg7-induced autophagy has a beneficial effect in a proteotoxic DRC mouse model.
To test the hypothesis that upregulating basal autophagy in a proteotoxic model of cardiomyopathy is beneficial, we crossed Atg7×tTA mice with CryABR120G animals. Previously, we extensively characterized the CryABR120G model, which accurately recapitulates human DRC: the hearts display protein aggregation, myofibrillar disarray, and dysfunction and fail by 6–8 months of age (18–21). We previously reported reduced autophagic activity in mutant CryABR120G mouse hearts, as shown both by ultrastructural observation (18) and by decreased autophagic flux in CryABR120G-expressing cardiomyocytes (22). In the present study, we analyzed autophagic gene expression in more detail in CryABR120G hearts. As expected on the basis of our previous data, direct groupwise comparison of CryABR120G and control hearts at 5 months revealed significant downregulation of autophagy-related and -regulatory genes in CryABR120G versus control mice (Supplemental Figure 2A). We confirmed the transcript data at the protein level by Western blot analyses of selected markers of autophagy, including ATG10, ATG5, ATG5-ATG12 complex, Beclin 1, and BNIP3 (Supplemental Figure 2B).
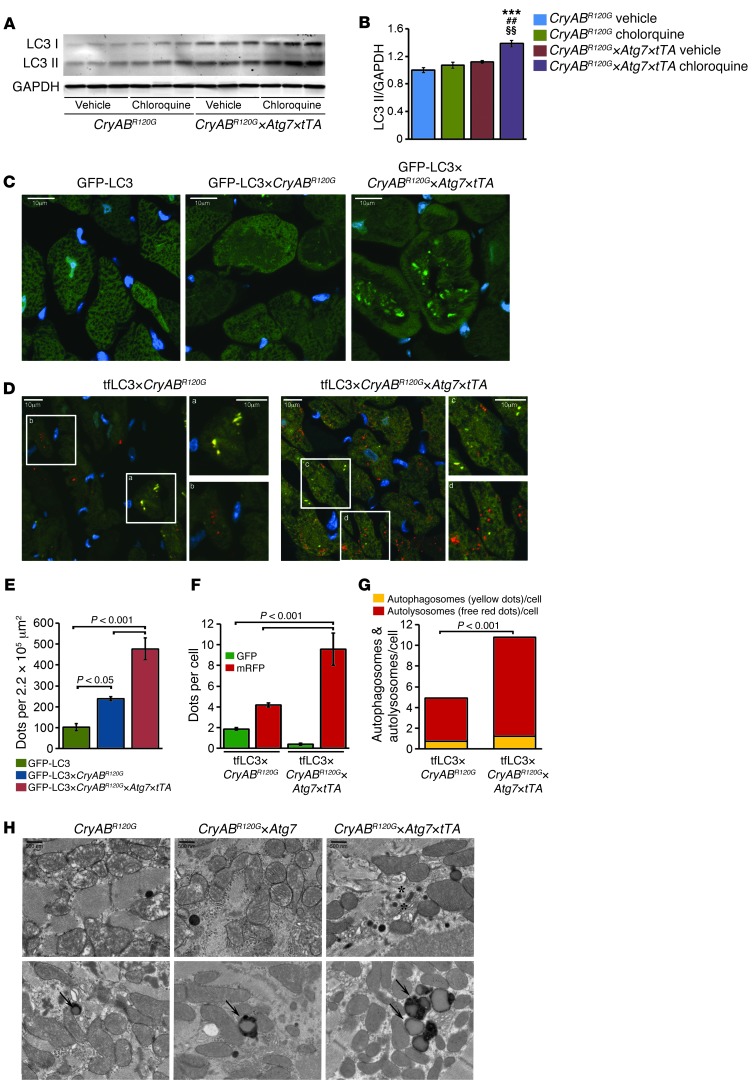
To evaluate whether autophagy is induced by Atg7 overexpression in CryABR120G hearts, we carried out an autophagic flux assay (28, 29) and treated the mice with chloroquine (50 mg/kg) for 5 days to block lysosomal function. The assay showed increased LC3-II levels in hearts of the induced Atg7-crossed CryABR120G triple-Tg mice (referred to herein as CryABR120G×Atg7×tTA mice; Figure 4, A and B). Quantitation of p62 expression showed significantly increased expression of p62 in CryABR120G×Atg7×tTA hearts (Supplemental Figure 3, A and B), and no changes were observed after chloroquine treatment in CryABR120G×Atg7×tTA or CryABR120G×tTA hearts (Supplemental Figure 1, C and D). We also used reporter mice to measure relative levels of autophagic activity. CryABR120G×Atg7×tTA mice were crossed with the GFP-LC3 reporter mouse (30). The resulting induced quadruple-Tg GFP-LC3×CryABR120G×Atg7×tTA mice showed significantly increased numbers of GFP-LC3 green puncta compared with the different control cohorts (Figure 4, C and E).
Figure 4. Autophagy flux analysis in CryABR120G×Atg7×tTA mouse hearts.
(A) Autophagic flux assay showed increased LC3-II levels in CryABR120G×Atg7 hearts (n = 3 per treatment). (B) Densitometry analysis showed increased LC3-II relative to GAPDH in CryABR120G×Atg7 hearts. ***P < 0.001 vs. vehicle-treated CryABR120G; ##P < 0.001 vs. chloroquine-treated CryABR120G; §§P < 0.01 vs. vehicle-treated CryABR120G×Atg7×tTA, Tukey’s post-hoc test. (C) Representative images of GFP-LC3 puncta (autophagosomes) in hearts from male GFP-LC3 control, GFP-LC3×CryABR120G, and GFP-LC3×CryABR120G×Atg7×tTA mouse hearts. Immunofluorescence analysis of heart sections showing punctate LC3 staining in 4-month-old mice. DAPI (blue) was used to identify nuclei. Original magnification, ×60. (D) Representative images of autophagosomes (yellow puncta) (mRFP-GFP-LC3) and their maturation into autolysosomes (red puncta) in hearts from male tf-LC3×CryABR120G and tf-LC3×CryABR120G×Atg7×tTA mice. Boxed regions are shown enlarged at right. (E) Quantification of GFP puncta per microscopic field (220,000 μm2) in LV. (F) Mean numbers of GFP green and mRFP red puncta per cell. (G) Mean numbers of autophagosomes and autolysosomes (represented by yellow and red puncta, respectively, in D) per cell. (H) Representative TEM of CryABR120G, CryABR120G×Atg7, and CryABR120G×Atg7×tTA hearts showing increased membranous structures (asterisks) and autophagosomes (arrows) in CryABR120G×Atg7×tTA hearts. Scale bars: 10 μm (C and D); 500 nm (H).
Recently, Sadoshima’s group developed reporter mice that transgenically overexpress a tandem fluorescent mRFP-GFP-LC3 (tf-LC3) in a cardiac-specific manner and used these animals to evaluate autophagic flux in vivo (31). These tf-LC3 mice can be used to measure autophagosome and autolysosome formation simultaneously, as LC3 puncta labeled with both GFP and mRFP represent autophagosomes, whereas those labeled with mRFP alone represent autolysosomes (31). We crossed tf-LC3 reporter mice with CryABR120G×Atg7×tTA mice. In hearts of the resulting tf-LC3×CryABR120G×Atg7×tTA mice, Atg7 overexpression increased autophagic flux, as shown by significantly increased autophagosome and autolysosome levels (Figure 4, D, F, and G). Ultrastructural analyses confirmed the increased numbers of autolysosomes: their accumulation was widespread and easily observed (Figure 4H).
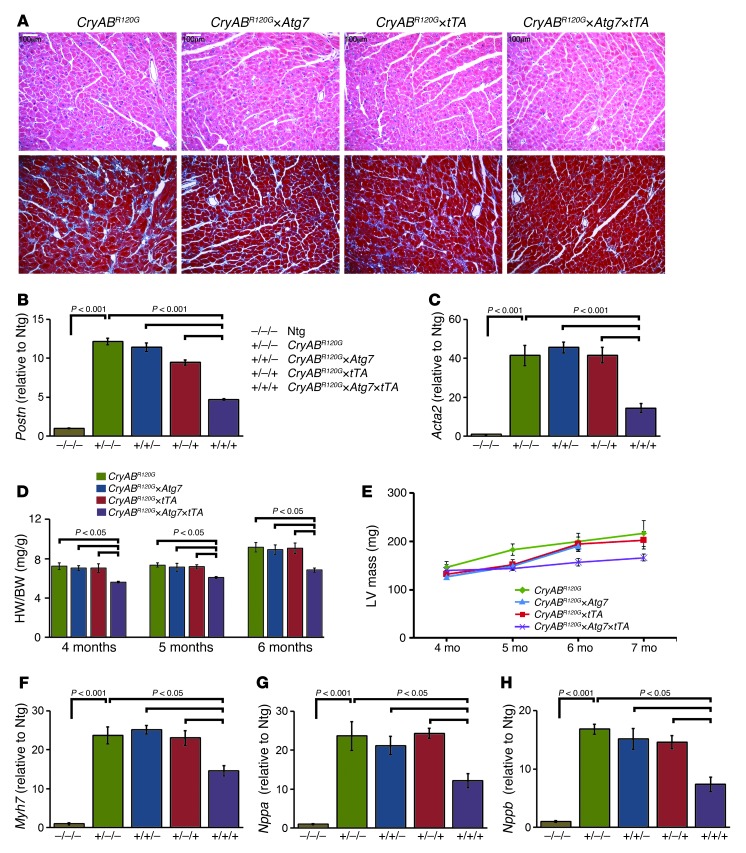
We next sought to determine whether enhanced autophagy affects the developing pathology of the CryABR120G mouse heart. Hearts from CryABR120G mice at 4 months displayed pathological remodeling, characterized by cardiomyocyte disarray and extensive interstitial fibrosis (Figure 5A). Markers for genetic activation of enhanced fibrosis, including Postn (encoding periostin) and Acta2 (encoding SMA), were significantly elevated in CryABR120G versus Ntg hearts (Figure 5, B and C). HW/BW ratios at 4, 5, and 6 months of age and serial echocardiographic determination of the LV mass index at 4–7 months showed significant reductions in CryABR120G×Atg7×tTA hearts (Figure 5, D and E). Expression of 3 genetic markers of cardiac distress and hypertrophy in the mouse — β-MHC (Myh7), atrial natriuretic factor (Nppa), and B-type natriuretic peptide (Nppb) — were also significantly reduced at 4 months compared with CryABR120G, CryABR120G×Atg7, and CryABR120G×tTA mice (Figure 5, F–H).
Figure 5. Histology and morphometry of CryABR120G×Atg7×tTA mouse hearts.
(A) H&E (top) and Masson’s trichrome (bottom) staining of cardiac sections from 4-month-old CryABR120G, CryABR120G×Atg7, CryABR120G×tTA, and CryABR120G×Atg7×tTA mice (n ≥ 6 per group). (B and C) mRNA expression of Postn (B) and Acta2 (C). Values are expressed as fold change versus Ntg (n = 3 per group). (D) HW/BW ratio at 4, 5, and 6 months of age. (E) Serial changes in LV mass index by echocardiography across genotypes (n ≥ 10 mice per group). (F–H) mRNA expression of Myh7 (F), Nppa (G), and Nppb (H). Values are expressed as fold change versus Ntg control (n = 6 per group). P values were determined by Tukey’s post-hoc test. Scale bars: 100 μm.
Induction of autophagy reduces CryABR120G-induced aggregate formation.
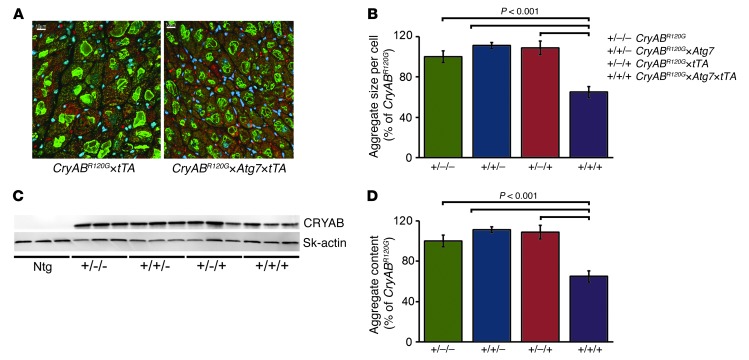
CryABR120G-mediated DRC is a protein misfolding disease characterized by deposition of insoluble proteins, whose accumulation can have both toxic (14) and protective functions (16). Immunofluorescent staining for CRYAB was used to quantitate aggregate accumulation on a per-cell-area basis. Although both CryABR120G×tTA and CryABR120G×Atg7×tTA hearts contained protein aggregates, aggregate size per cell was significantly reduced in the latter (Figure 6, A and B). We also evaluated the effect of induced autophagy on the amount of CRYAB protein accumulation in aggregates, compared with soluble CRYAB, by Western blot analysis of soluble and insoluble proteins fractionated from CryABR120G, CryABR120G×Atg7, CryABR120G×tTA, and CryABR120G×Atg7×tTA hearts. As expected, these analyses showed significant aggregate accumulations in CryABR120G hearts compared with Ntg controls, while CryABR120G×Atg7×tTA hearts showed decreased aggregate levels (Figure 6, C and D). These results suggest that autophagy can effectively prevent and/or help clear the proteotoxic aggregates in cardiomyocytes.
Figure 6. Overexpression of Atg7 significantly reduced misfolded protein accumulation in CryABR120G hearts.
(A) Immunofluorescence staining for CRYAB (green) with troponin I counterstaining (red) showed decreased aggregate levels in CryABR120G×Atg7×tTA cardiomyocytes. (B) Quantification of CryAB aggregates showed decreased aggregate accumulation in CryABR120G×Atg7×tTA mice. (C) Representative Western blot showing aggregated CRYAB protein levels present in the insoluble fractions. (D) Densitometry analysis showed decreased aggregate in the insoluble protein fraction in CryABR120G×Atg7×tTA mice (n = 6 per group). P values were determined by Tukey’s post-hoc test. Scale bars: 10 μm.
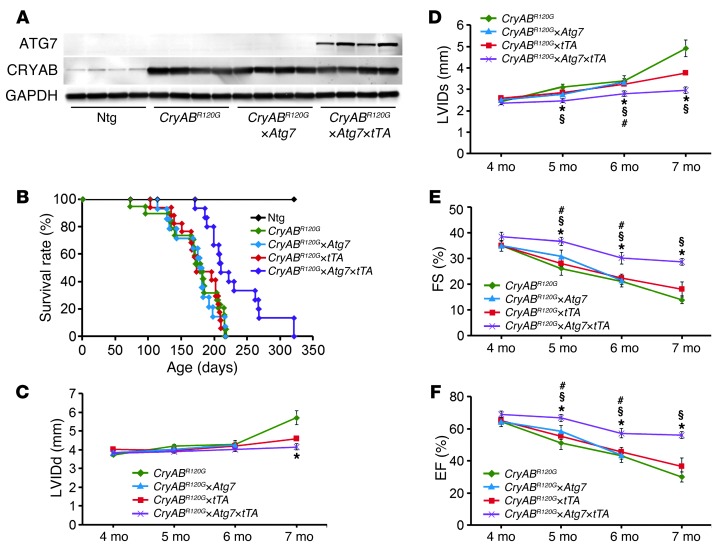
CRYAB levels were examined in the different cohorts in order to determine whether the Atg7 overexpression simply downregulates steady-state levels in CryABR120G×Atg7×tTA animals; this did not appear to be the case (Figure 7A). The sustained Atg7-induced autophagy in CryABR120G×Atg7×tTA hearts prolonged survival by 40%, although the mice did die prematurely, with no survivors by 10.5 months of age (Figure 7B). Upregulation of autophagy also had beneficial effects on conserving LV dimension and performance (Figure 7, C–F).
Figure 7. Cardiac hemodynamics in CryABR120G×Atg7×tTA mice.
(A) Western blot showing ATG7 and CRYAB expression in Ntg, CryABR120G, CryABR120G×Atg7, and CryABR120G×Atg7×tTA mice. (B) Kaplan-Meier curves of CryABR120G (n = 20), CryABR120G×tTA (n = 17), CryABR120G×Atg7 (n = 14), and CryABR120G×Atg7×tTA (n = 15) mice. (C–F) Echocardiography indices of LV end-diastolic diameter, measured by (C) LVIDd and (D) LVIDs, and LV function, measured by (E) %FS and (F) %EF (n ≥ 10 mice per group). *P < 0.05 vs. CryABR120G; #P < 0.05 vs. CryABR120G×tTA; §P < 0.05 vs. CryABR120G×Atg7, Tukey’s post-hoc test.
Effect of voluntary exercise on survival and cardiac function in CryABR120G×Atg7×tTA mice.
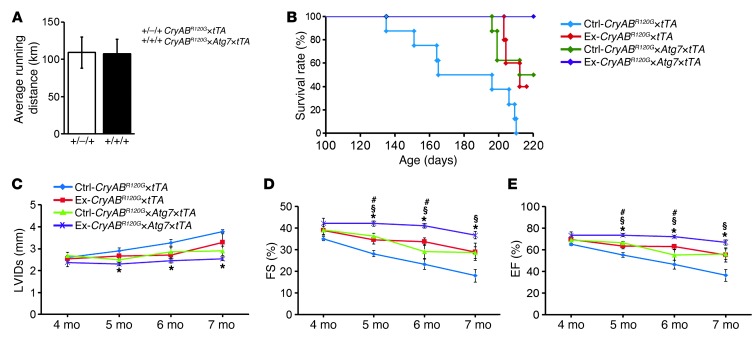
Exercise can increase autophagy levels in both normal and diseased tissues (32–35). We previously showed that exercise could prolong survival in the CryABR120G model (17), and sought to determine whether the effects of exercise and autophagic induction by Atg7 expression were additive or even synergistic. We therefore subjected mice to long-term voluntary wheel exercise: 32 1-month-old male CryABR120G×tTA and CryABR120G×Atg7×tTA mice were housed in regular cages (nonexercised control; n = 8 per genotype) or in cages equipped with voluntary running wheels (exercised group; n = 8 per genotype), with 1 mouse per cage. There were no significant differences between the CryABR120G×tTA and CryABR120G×Atg7×tTA mice with respect to average total running distance (Figure 8A). All nonexercised CryABR120G×tTA mice died from heart failure by 7 months, whereas 50% of exercised CryABR120G×tTA mice were alive at this time (Figure 8B). Induction of autophagy via Atg7 overexpression in the CryABR120G×Atg7×tTA group resulted in additive benefits, with 100% survival at 7 months (Figure 8B). This group also showed conserved LV function, as measured by LVIDs, %FS, and %EF (Figure 8, C–E).
Figure 8. Effect of voluntary exercise on CryABR120G×Atg7×tTA mice.
(A) Average running distance traveled by mice subjected to voluntary exercise for 7 months. (B) Kaplan-Meier survival curves of CryABR120G×tTA (n = 8) and CryABR120G×Atg7×tTA (n = 9) mice. (C–E) Echocardiography indices of (C) LV end-diastolic diameter, measured by LVIDs function, and LV function, measured by (D) %FS and (E) %EF, in male nonexercised and exercised mice (n = 8 per group). *P < 0.05 vs. nonexercised CryABR120G×tTA; #P < 0.05 vs. exercised CryABR120G×tTA; §P < 0.05 vs. nonexercised CryABR120G×Atg7×tTA, Tukey’s post-hoc test.
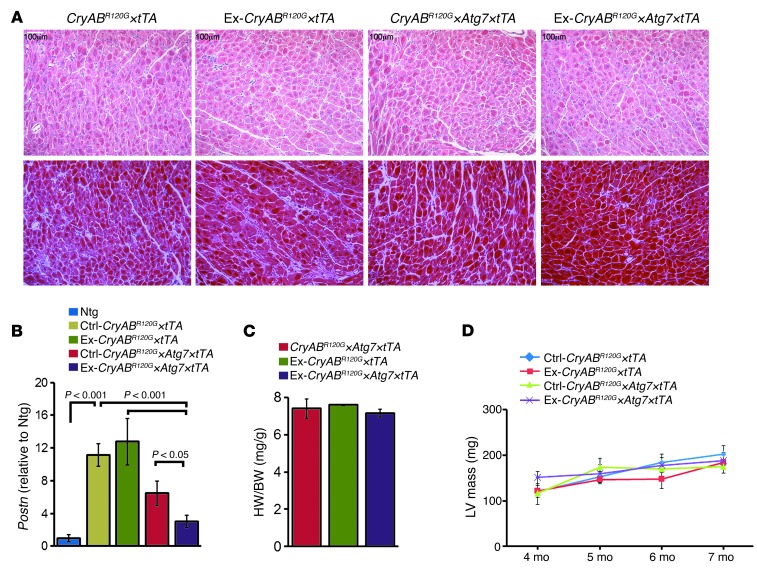
Exercise had no effect on the induction of fibrosis or HW/BW ratios in CryABR120G×tTA hearts (Figure 9, A and C). As noted above, Atg7 overexpression and the resulting increase in autophagy did effectively decrease the fibrotic response, and exercised CryABR120G×Atg7×tTA mice showed further decreases, as demonstrated by decreased Postn mRNA levels and reduction of extensive interstitial fibrosis (Figure 9, A and B). However, exercise had no effect on any of the experimental groups in terms of either HW/BW ratio or LV mass as assessed by echocardiography (Figure 9, C and D).
Figure 9. Histology and morphometry of exercised CryABR120G×Atg7×tTA mouse hearts.
(A) H&E (top) and Masson’s trichrome (bottom) staining of cardiac sections from 7-month-old nonexercised and exercised CryABR120G×tTA and CryABR120G×Atg7×tTA mice. (B) Postn mRNA levels, expressed as fold change relative to Ntg (n = 3 per group). (C) HW/BW ratios at 7 months of age (n ≥ 6 per group). (D) Serial changes in LV mass index by echocardiography across genotypes (n = 8 per group). P values were determined by Tukey’s post-hoc test. Scale bars: 100 μm.
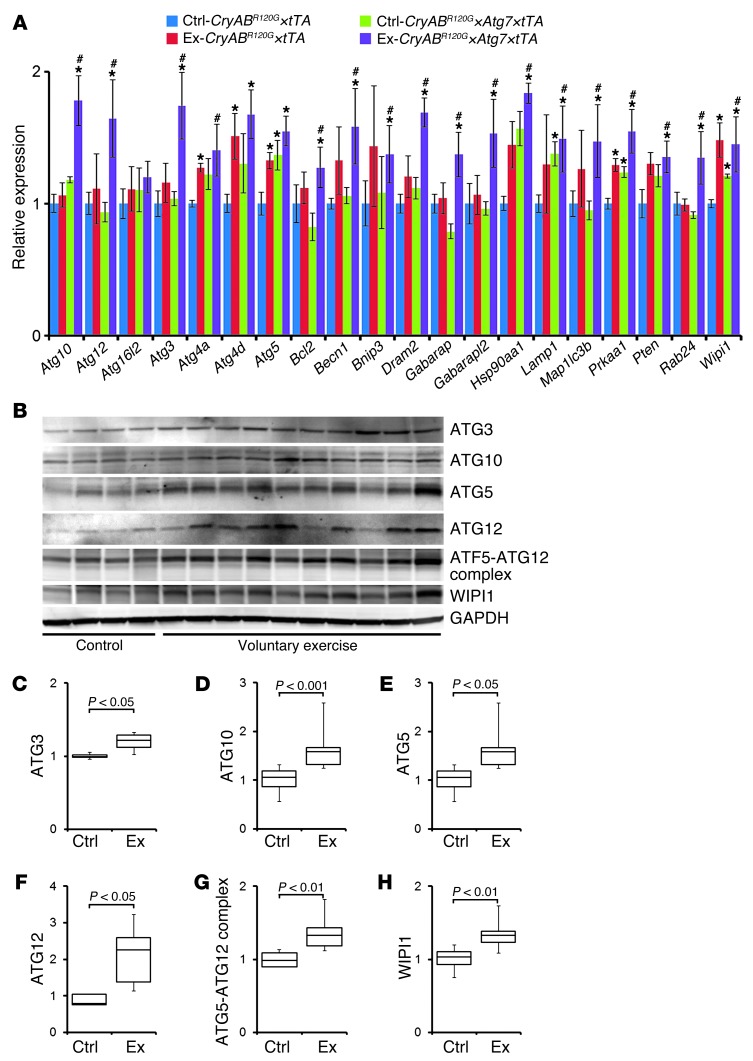
The additive beneficial effects of exercise in the CryABR120G×Atg7×tTA cohorts implied that exercise might be affecting autophagy in a manner different from simple Atg7 overexpression. Exercise can induce autophagy-related and -regulatory genes in both human cardiac and skeletal muscle (36). To better define the autophagy response in exercised CryABR120G×Atg7×tTA hearts, we again titrated transcripts involved in the autophagy-lysosomal pathways that might be regulated by voluntary exercise (Figure 10). Interestingly, in contrast with the data obtained in Atg7×tTA hearts, exercised CryABR120G×Atg7×tTA hearts showed significant upregulation in essentially all analyzed transcripts encoding proteins involved in autophagic vacuole formation and protein transport (Atg3, Atg10, Atg12, Map1lc3b, Rab24, Gabarapl2, Becn1, and Wipi1) and protein targeting to membranes/vacuoles and linking autophagosomes to lysosomes (Gabarap), compared with both nonexercised control groups (Figure 10A). Exercised mice also showed significantly increased levels of transcripts that mediate autophagy in response to other, intracellular signals (e.g., Dram2), as well as transcripts encoding proteins that function as coregulators of autophagy and apoptosis (Bcl2, Prkaa1, and Pten). We confirmed changes at the cognate protein levels as well for selected peptides by Western blot analyses (Figure 10B). Quantitation of these data showed significant upregulation of the autophagy-related proteins ATG3, ATG10, ATG5, ATG12, ATG5-ATG12 complex, and WIPI1 in exercised versus nonexercised CryABR120G×Atg7×tTA mice (Figure 10, C–H). Coimmunoprecipitation experiments showed that voluntary exercise decreased the BCL2/Beclin 1 interaction (Supplemental Figure 4), indicative of increased autophagy by voluntary exercise.
Figure 10. Autophagy response in exercised CryABR120G×Atg7×tTA mouse hearts.
(A) Autophagy PCR array. Direct groupwise comparisons of mRNA levels in male exercised and nonexercised CryABR120G×Atg7×tTA and CryABR120G×tTA hearts at 5 months of age, expressed as fold change versus nonexercised CryABR120G×tTA (n = 3 per group). *P < 0.05 vs. nonexercised CryABR120G×tTA; #P < 0.05 vs. nonexercised CryABR120G×Atg7×tTA, 2-tailed Student’s t test. (B–H) Western blot demonstrating protein level changes of some of the upregulated genes identified by PCR array in CryABR120G×Atg7×tTA hearts. The obtained PCR array results were validated by Western blot (B). Densitometry analysis showed upregulation of autophagy-related proteins ATG3 (C), ATG10 (D), ATG5 (E), ATG12 (F), ATG5-ATG12 complex (G), and WIPI1 (H) in exercised (n = 10 per group) versus nonexercised (n = 4 per group) CryABR120G×Atg7×tTA hearts. Boxes represent interquartile ranges; lines represent medians; whiskers represent ranges. P values were determined by Tukey’s post-hoc test.
We performed autophagic flux assays to confirm increased autophagy in the hearts of exercised CryABR120G×Atg7×tTA mice and found increased levels of LC3-II compared with nonexercised control hearts (Supplemental Figure 5, A and B). Immunostaining with LC3 antibody also showed increased levels of LC3 in exercised CryABR120G×Atg7×tTA hearts (Supplemental Figure 5, C–E). Ultrastructural analyses confirmed high numbers of autolysosomes, relative to those seen in control sections, in the exercised CryABR120G×Atg7×tTA cohort (Supplemental Figure 5F). Although these data clearly showed that autophagy was upregulated, we wished to determine whether other pathways, such as the ubiquitin proteasome system and ER stress responses, were also activated, as these might each or both play an important role in the observed response. However, we did not find any marked changes in the expression of proteins involved in the ubiquitin proteasome system (19S, 20S, and 26S) or ER stress response (Atf6α, XBP1, pelF2α, and GRP78) after voluntary exercise (Supplemental Figure 6).
Discussion
The normal housekeeping functions of basal or constitutional autophagy are important or even essential for intracellular clearance of cellular waste, including protein aggregates, damaged or senescent organelles, and misfolded proteins. This key protective mechanism is crucial for maintaining healthy cells, as evidenced by studies of mice lacking the autophagy genes Atg7 and Atg5. Those animals develop symptoms of neurodegeneration and neuronal cell death even in the absence of any overtly harmful gene products (6, 7, 37). Accumulation of protein aggregates and aggresomes are also detected in human hearts with DRC (38, 39), idiopathic or ischemic cardiomyopathies (40, 41), and load-induced heart failure (42), all of which can ultimately lead to contractile dysfunction and heart failure. Basal autophagy is particularly vital in cardiomyocytes, as these cells are normally terminally differentiated and cannot decrease toxic concentrations of waste by cell division. By generating mice with cardiac-specific, inducible Atg7 expression, we attempted to define the in vivo physiological and pathological role, if any, of induced autophagy, and addressed a hypothesized beneficial role for upregulation of autophagy in a proteotoxic heart failure model of DRC.
Cardiac-specific Atg7 overexpression induces basal autophagy.
ATG7 is an essential protein for basal autophagy, as its ablation resulted in complete loss of autophagy, accumulation of abnormal organelles, and ubiquitinated protein aggregates, with death occurring within 1 day after birth (5). Although numerous in vitro (30, 43, 44) and in vivo (30, 45) genetic models have been developed with increased cellular autophagy, they were limited to preconditioning by starvation. In the present study, using inducible, cardiac-specific expression of Atg7, our data showed that even in the absence of starvation or some other stimulus, high Atg7 expression enhanced autophagic flux. Strikingly, cardiomyocyte-specific expression of Atg7 resulted in normal cardiac morphometry and functionality despite increased autophagic flux. This novel model thus allows for studying the chronic effect of constitutively high levels of autophagy in a normal organ system, challenging the concept that significantly elevated levels of autophagy are necessarily detrimental to an otherwise healthy organ system. The lack of any increase in the other autophagic proteins (Figure 1C) is somewhat surprising and implies that ATG7 is a key rate-limiting component for autophagy in this model. However, this may be somewhat misleading, as we did not undertake a comprehensive analysis of all the potential key players in the pathway. Further studies using comprehensive array analyses for the different mice used in this study will be necessary to fully understand the sufficiency of ATG7’s role in the overall regulation of cardiomyocyte autophagy.
Induced autophagy in the heart is protective in a model of cardiac proteinopathy.
Accumulation of intracellular aggregates has been reported in different genetic models of cardiac disease induced by the expression of Tg Mst1, truncated Mybpc3, DesΔ7, PQ83, and CryABR120G, but the prevalence of autophagic deficiency in a majority of these models is unclear (16, 39, 46–48). Clearance of aggregation-prone proteins dependent upon autophagy has been reported for different cell-based models, in which either autophagy inhibitors were added (49, 50) or Atg gene expression was decreased by siRNA transfection (51). Autophagic inhibition resulted in delayed or compromised clearance of the protein aggregates, whereas induction of autophagy enhanced clearance (49, 50). Prior studies from our laboratory using the CryABR120G model of cardiac proteotoxicity reported reduced autophagic activity (22). Notably, in the present study, we found that increasing autophagy in CryABR120G hearts by Atg7 overexpression attenuated and/or delayed the onset of the morphological and functional pathologies and significantly increased the probability of survival into mid-adulthood. We also observed significant reductions in activation of the gene programs normally associated with developing cardiac pathology, such as activation of the fetal gene program seen during the development of pathological hypertrophy (52) and cardiac fibrosis (17).
Physical exercise has numerous health benefits, decreasing the probabilities of developing cardiovascular disease, diabetes, cancer, and neurodegenerative disease (53). The beneficial protective effects of voluntary exercise are evident from different mouse models of neurodegenerative disease which, upon exercise, show delayed onset of neurological deficits (14), decreased amyloid load (13), and significantly decreased levels of neural amyloid deposits (15). Moreover, evidence from observational and randomized trials also demonstrates that regular exercise contributes to primary, secondary, and tertiary prevention of cardiovascular disease (54, 55). In the present study, we sought to determine whether the effects of exercise and autophagic induction by ATG7 expression were additive or even synergistic. We chose the voluntary cage wheel as the exercise intervention because it avoids the physical and psychological stressors associated with forced exercise paradigms (20). Voluntary exercise is relatively mild in nature, compared with more intense forced exercise models (56), and had little or no effect on LV mass across genotypes, as assessed by echocardiography (Figure 8, C–E). As we previously showed (20), long-term voluntary exercise increased the survival probability of CryABR120G×tTA mice to 50%, whereas the genetically identical nonexercised control group died from heart failure by 7 months. The data from this group closely matched that obtained with the nonexercised CryABR120G×Atg7×tTA control cohort; however, induction of autophagy by voluntary exercise in this group resulted in additive benefits, with 100% survival at 7 months (Figure 8B). Whether this is simply due to even higher levels of autophagic flux is currently being explored, but we believe a more likely hypothesis is that multiple pathways able to augment survival are being activated. Consistent with the observed effects on survival probability, exercise resulted in significant improvements in maintaining cardiac function, as assessed by echocardiography, and these beneficial effects were increased in the presence of increased Atg7 overexpression (Figure 8, C–E).
In contrast to the Atg7-overexpressors, assessment of autophagy-related gene expression in the mice subjected to voluntary exercise showed elevated levels of virtually all test transcripts involved in a variety processes associated with autophagy, including autophagic vacuole formation, protein ubiquitination, and protein transport. These data are consistent with previous reports defining the effects of physical exercise on autophagy-related and -regulatory genes such as ATG12, MAP1LC3B, GABARAPL1, and ATG4B in human muscle (34, 35). Interestingly, voluntary exercise significantly induced Bcl2 and Becn1 in CryABR120G×Atg7×tTA mice. A recent study also showed that exercise induces autophagy by regulating BCL2/Beclin 1 signaling in skeletal and cardiac muscle of fed mice, but not in autophagy-deficient mice (36).
The observed increase in p62 expression in CryABR120G×Atg7×tTA mice is somewhat surprising, as earlier studies showed marked accumulation of p62 in the brain and liver of Atg7-deficient mice (57). Although total p62 expression can be inversely correlated with autophagic activity (58), p62 levels were unchanged after chloroquine treatment in Atg7×tTA mice. Moreover, CryABR120G×Atg7×tTA mice also showed increased p62, along with increases in autophagic flux. However, it is not yet clear whether p62 is degraded solely through the autophagy pathway, and it can be transcriptionally regulated during autophagy (59), which may confound the interpretation of p62 levels as an overall indicator of autophagic flux.
In summary, the present study demonstrated that basal autophagy cleared cytosolic misfolded and aggregated proteins by engulfing them in autophagic vesicles that ultimately fused with the lysosome for degradation of their content. CryABR120G hearts showed extensive aggregate formation. Isolated regions in some of these misfolded proteins are able to assume a β-pleated sheet structure and interact with each other to form a series of intermediate but stable structures, resulting in cytotoxic soluble preamyloid oligomers (16). Further upregulation of autophagy via the different mechanisms associated with voluntary exercise had additional beneficial effects, which implies that induced autophagy by various means may be therapeutically beneficial.
Methods
Animals.
Atg7×tTA mice (cardiomyocyte-specific inducible Atg7-overexpressing mice) were generated by first inserting ATG7 cDNA into the αMHC promoter cassette to generate responder Atg7-Tg mice (27). Tg mice were identified by PCR analysis of genomic DNA isolated from ear clips. The responder Atg7-Tg mice were crossed with tTA-Tg (tet-controlled transcriptional activator) mice to generate the double-Tg Atg7×tTA mice. 2 mouse lines, line 132 and line 152, were treated with 1% (w/v) Dox in drinking water supplemented with 2% sucrose to confirm inducible transgene expression by Dox. Male CryABR120G mice (FVB/N mice with cardiac-specific overexpression of CryAB containing the R120G missense mutation) have been described previously (39).
Autophagic flux.
For the assessment of autophagic flux in the heart, mice were subjected to intraperitoneal injection with chloroquine diphosphate (50 mg/kg body weight; Sigma-Aldrich) (60) every 24 hour for 5 days. Hearts were harvested, and LC3-II protein levels were determined by Western blot analysis. For in vivo determination of autophagic flux, mice were crossed with GFP-LC3 and tf-LC3 Tg mice, each of which provides an efficient in vivo reporter for autophagy (31, 61). To avoid autophagic induction during sample collection, hearts were perfused with 4% paraformaldehyde in cardioplegic buffer (0.1 M PBS, pH 7.4, 50–100 mM KCl, and 5% dextrose). Tissues were harvested and further fixed with 4% paraformaldehyde in PBS overnight (7–12 hours), followed by treatment with 15% sucrose in PBS for 4 hours and then with 30% sucrose solution overnight. Tissue samples were embedded in Tissue-Tek OCT (Sakura Finetechnical Co. Ltd.) and stored at –80°C. Frozen tissue samples were then sectioned at 5 μm thickness, air dried for 30 minutes, rehydrated in PBS for 5 minutes, mounted using a reagent containing DAPI, and viewed under a fluorescence microscope. The number of GFP and mRFP dots was determined by manual counting of fluorescent puncta in 5 fields from 3 different myocyte preparations using a ×60 objective as described previously (31, 61).
Autophagy-related transcript analyses.
The mouse Autophagy RT2 Profiler PCR Array (SABiosciences) was used to study autophagy-specific gene expression profiles in accordance with the manufacturer’s recommendations. Briefly, total RNA was isolated with TRI reagent (Molecular Research Center) and further purified using the RNeasy Mini Kit (SABiosciences). DNA contamination was removed from the samples by treatment with RNase-free DNase, and RNA concentration and purity was determined with a NanoDrop ND-1000 (Thermo Scientific). The first-strand cDNA was synthesized from total RNA, and real-time PCR was performed with the SuperArray PCR master mix using SYBR green mastermixes (SABiosciences) and a CFX-96 real-time cycler (Bio-Rad Laboratories). All data were normalized to Gapdh, and the amplification data (fold change in Ct of all genes) were analyzed by the ΔΔCt method.
Echocardiography.
Cardiac ultrasound was performed on isoflurane-anesthetized mice from 3 to 7 months of age with a VisualSonics Vevo 770 Imaging System using a 30-MHz transducer. 2-dimensional directed M-mode echocardiographic images along the parasternal short axis were recorded to determine LV systolic function and expressed as %FS. M-mode measurements of LV end-diastolic and end-systolic chamber size were calculated as (LVIDd – LVIDs)/LVIDd and expressed as a percentage.
Histological analysis.
Hearts were collected, fixed in 10% buffered formalin, and embedded in paraffin. Serial 5-μm heart sections from each group were stained with H&E or Masson’s trichrome. Interstitial fibrotic regions were quantified using Metamorph analysis of the percentage of blue area in the Masson’s trichrome–stained sections.
Immunofluorescence microscopy.
Paraffin-embedded heart sections were used for immunofluorescence analyses as described previously (39). The following primary antibodies were used: rabbit anti-ATG7 (1:100; Novus), rabbit anti-LC3 (1:50; Cell Signaling Technology Inc.), mouse anti–troponin I (1:1,000; Millipore), rabbit anti-CRYAB (1:400; Assay Designs). Alexa Fluor 488– or Alexa Fluor 568–conjugated secondary antibody (Molecular Probes) directed against mouse or rabbit IgG was used, and DAPI (Invitrogen) was used to identify nuclei. Immunofluorescent staining for CRYAB was used to quantitate the cytoplasmic area occupied by the aggresomes, as described previously (18, 39). Other antibodies used in these studies included anti-ATG3, anti-ATG10, anti-ATG12, and pelF2α (1:1,000; Cell Signaling); anti-ATG10 (1:1,000; Millipore); anti-ATG5, anti-WIPI1, and anti-GRP78 (1:200; Santa Cruz Biotechnology); anti-p62 (1:1,000; ProGen); anti-BNIP3, anti-BCL2, anti–proteasome 19S, anti–proteasome 20S, and anti–proteasome 26S (1:1,000; AbCam); anti-ATF6α (1:1,000; Imgenex); and anti-XIBP1 (1:1,000; LifeSpan BioSciences).
Electron microscopy.
Hearts were perfused with 1% paraformaldehyde and 2% glutaraldehyde in cardioplegic buffer, then with 1% paraformaldehyde and 2% glutaraldehyde in 0.1M cacodylate buffer (pH 7.2), postfixed in 1% OsO4, and processed for thin sectioning. Sections were counterstained with uranium, and lead salts and examined with a Hitachi 7600 transmission electron microscope. Images were acquired with an AMT digital camera.
RNA isolation and quantitative real-time PCR analysis.
Total RNA was isolated with TRI reagent (Molecular Research Center) according to the manufacturer’s protocol. Quantitative real-time PCR (qRT-PCR) was performed with a CFX-96 instrument (Bio-Rad) using Taqman probes (Applied Biosystems) for Postn (Applied Biosystems), Acta2 (Applied Biosystems), and cardiac fetal genes (Myh7, Nppa, and Nppb; Applied Biosystems). All data were normalized to Gapdh (Applied Biosystems) content and expressed as fold increase over the control group.
Cell fractionation, SDS-PAGE, and immunoblotting.
To prepare soluble and insoluble fractions, hearts were harvested in cold PBS (pH 7.4) containing 1% Triton-X100, 2.5 mM EDTA, 0.5 mM PMSF, and a complete protease inhibitor mixture and then vortexed for 30 seconds. The heart extracts were centrifuged at 12,000 g for 15 minutes, and the supernatants were collected (soluble fraction). The pellets were dissolved in DNAase (1 mg/kl in 10 mM Tris, 15 mM MgCl2; Roche) and sonicated on ice, and the protein was quantitated with a modified Bradford assay. The insoluble protein was then diluted in RIPA buffer, and 3 μg resuspended insoluble protein was used for subsequent immunoblotting with appropriate antibodies.
Protein lysates were separated on SDS-PAGE using precast 7.5%–15% Criterion Gels (BioRad) and transferred to PVDF membranes (BioRad). Membranes were blocked for 1 hour in 5% nonfat dried milk and exposed to primary antibodies overnight. The following primary antibodies were used for immunoblotting: anti-ATG7 (1:1,000; ProSci); anti-ATG1 (1:1,000; Sigma-Aldrich); anti–ATG5-ATG12 (1:1,000; Cosmo Bio Co.); anti–Beclin 1, anti–cathepsin D, anti-LAMP1, and anti-LAMP2 (1:200; Santa Cruz Biotechnology); anti-LC3 (1:1,000; Cell Signaling Technology Inc.); anti-CRYAB (1:5,000; Assay Designs); and anti-GAPDH (1:7,500; Chemicon). Membranes were then washed, incubated with alkaline phosphatase–conjugated secondary antibodies (Santa Cruz Biotechnology), and exposed with ECF reagent (Amersham), and the resultant signal was quantitated on a STORM 820 fluorescent scanner (Molecular Dynamics) using ImageQuant version 5.2 (Molecular Dynamics).
Voluntary exercise studies.
We used 32 1-month-old male CryABR120G×tTA and CryABR120G×Atg7×tTA mice randomly divided into 4 groups (n = 8 per group): nonexercised CryABR120G×tTA, exercised CryABR120G×tTA, nonexercised CryABR120G×Atg7×tTA, and exercised CryABR120G×Atg7×tTA. Mice were housed in regular cages or in cages equipped with voluntary running wheels, with 1 mouse per cage.
Statistics.
Data are expressed as mean ± SEM. All statistical tests were done with SigmaPlot 9.0 software. Statistical analyses between 2 groups were analyzed by 2-tailed Student’s t test. Groups of 3 or more were analyzed with 1-way ANOVA, followed by Tukey’s post-hoc test. A P value less than 0.05 was considered statistically significant.
Study approval.
Animals were handled in accordance with the principles and procedures of the Guide for the Care and Use of Laboratory Animals. The Institutional Animal Care and Use Committee at Cincinnati Children’s Hospital approved all experimental procedures.
Supplementary Material
Acknowledgments
This work was supported by NIH grants P01HL69779, P01HL059408, R01HL05924, and R011062927 and a Trans-Atlantic Network of Excellence grant from Le Fondation Leducq (to J. Robbins) as well as by American Heart Association Postdoctoral Fellowship grants (to S. Bhuiyan).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(12):5284–5297. doi:10.1172/JCI70877.
References
- 1.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443(7113):780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 4.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34(3):259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 5.Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169(3):425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441(7095):885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 7.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441(7095):880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 8.Nakai A, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13(5):619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 9.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 10.Martinez-Vicente M, Cuervo AM. Autophagy and neurodegeneration: when the cleaning crew goes on strike. Lancet Neurol. 2007;6(4):352–361. doi: 10.1016/S1474-4422(07)70076-5. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Arencibia M, Hochfeld WE, Toh PP, Rubinsztein DC. Autophagy, a guardian against neurodegeneration. Semin Cell Dev Biol. 2010;21(7):691–698. doi: 10.1016/j.semcdb.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pickford F, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118(6):2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazarov O, et al. Environmental enrichment reduces Abeta levels and amyloid deposition in transgenic mice. Cell. 2005;120(5):701–713. doi: 10.1016/j.cell.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 14.van Dellen A, Blakemore C, Deacon R, York D, Hannan AJ. Delaying the onset of Huntington’s in mice. Nature. 2000;404(6779):721–722. doi: 10.1038/35008142. [DOI] [PubMed] [Google Scholar]
- 15.Adlard PA, Perreau VM, Pop V, Cotman CW. Voluntary exercise decreases amyloid load in a transgenic model of Alzheimer’s disease. J Neurosci. 2005;25(17):4217–4221. doi: 10.1523/JNEUROSCI.0496-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanbe A, et al. Desmin-related cardiomyopathy in transgenic mice: a cardiac amyloidosis. Proc Natl Acad Sci U S A. 2004;101(27):10132–10136. doi: 10.1073/pnas.0401900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8(4):445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maloyan A, Sayegh J, Osinska H, Chua BH, Robbins J. Manipulation of death pathways in desmin-related cardiomyopathy. Circ Res. 2010;106(9):1524–1532. doi: 10.1161/CIRCRESAHA.109.212639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maloyan A, et al. Biochemical and mechanical dysfunction in a mouse model of desmin-related myopathy. Circ Res. 2009;104(8):1021–1028. doi: 10.1161/CIRCRESAHA.108.193516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maloyan A, Gulick J, Glabe CG, Kayed R, Robbins J. Exercise reverses preamyloid oligomer and prolongs survival in alphaB-crystallin-based desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2007;104(14):5995–6000. doi: 10.1073/pnas.0609202104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maloyan A, et al. Mitochondrial dysfunction and apoptosis underlie the pathogenic process in alpha-B-crystallin desmin-related cardiomyopathy. Circulation. 2005;112(22):3451–3461. doi: 10.1161/CIRCULATIONAHA.105.572552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pattison JS, Osinska H, Robbins J. Atg7 induces basal autophagy and rescues autophagic deficiency in CryABR120G cardiomyocytes. Circ Res. 2011;109(2):151–160. doi: 10.1161/CIRCRESAHA.110.237339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taherbhoy AM, et al. Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway. Mol Cell. 2011;44(3):451–461. doi: 10.1016/j.molcel.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noda NN, et al. Structural basis of Atg8 activation by a homodimeric E1, Atg7. Mol Cell. 2011;44(3):462–475. doi: 10.1016/j.molcel.2011.08.035. [DOI] [PubMed] [Google Scholar]
- 25.Yamaguchi M, et al. Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7. Nat Struct Mol Biol. 2012;19(12):1250–1256. doi: 10.1038/nsmb.2451. [DOI] [PubMed] [Google Scholar]
- 26.Kaiser SE, et al. Noncanonical E2 recruitment by the autophagy E1 revealed by Atg7-Atg3 and Atg7-Atg10 structures. Nat Struct Mol Biol. 2012;19(12):1242–1249. doi: 10.1038/nsmb.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanbe A, Gulick J, Hanks MC, Liang Q, Osinska H, Robbins J. Reengineering inducible cardiac-specific transgenesis with an attenuated myosin heavy chain promoter. Circ Res. 2003;92(6):609–616. doi: 10.1161/01.RES.0000065442.64694.9F. [DOI] [PubMed] [Google Scholar]
- 28.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4(2):151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rubinsztein DC, et al. In search of an “autophagomometer”. Autophagy. 2009;5(5):585–589. doi: 10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
- 30.Zhu H, et al. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007;117(7):1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hariharan N, Zhai P, Sadoshima J. Oxidative stress stimulates autophagic flux during ischemia/reperfusion. Antioxid Redox Signal. 2011;14(11):2179–2190. doi: 10.1089/ars.2010.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giannuzzi P, Temporelli PL, Corra U, Tavazzi L. Antiremodeling effect of long-term exercise training in patients with stable chronic heart failure: results of the Exercise in Left Ventricular Dysfunction and Chronic Heart Failure (ELVD-CHF) Trial. Circulation. 2003;108(5):554–559. doi: 10.1161/01.CIR.0000081780.38477.FA. [DOI] [PubMed] [Google Scholar]
- 33.Haykowsky MJ, Liang Y, Pechter D, Jones LW, McAlister FA, Clark AM. A meta-analysis of the effect of exercise training on left ventricular remodeling in heart failure patients: the benefit depends on the type of training performed. J Am Coll Cardiol. 2007;49(24):2329–2336. doi: 10.1016/j.jacc.2007.02.055. [DOI] [PubMed] [Google Scholar]
- 34.Jamart C, Benoit N, Raymackers JM, Kim HJ, Kim CK, Francaux M. Autophagy-related and autophagy-regulatory genes are induced in human muscle after ultraendurance exercise. Eur J Appl Physiol. 2012;112(8):3173–3177. doi: 10.1007/s00421-011-2287-3. [DOI] [PubMed] [Google Scholar]
- 35.Jamart C, Francaux M, Millet GY, Deldicque L, Frere D, Feasson L. Modulation of autophagy and ubiquitin-proteasome pathways during ultra-endurance running. J Appl Physiol. 2012;112(9):1529–1537. doi: 10.1152/japplphysiol.00952.2011. [DOI] [PubMed] [Google Scholar]
- 36.He C, et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481(7382):511–515. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klionsky DJ. Neurodegeneration: good riddance to bad rubbish. Nature. 2006;441(7095):819–820. doi: 10.1038/441819a. [DOI] [PubMed] [Google Scholar]
- 38.Vicart P, et al. A missense mutation in the alphaB-crystallin chaperone gene causes a desmin-related myopathy. Nat Genet. 1998;20(1):92–95. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- 39.Wang X, et al. Expression of R120G-alphaB-crystallin causes aberrant desmin and alphaB-crystallin aggregation and cardiomyopathy in mice. Circ Res. 2001;89(1):84–91. doi: 10.1161/hh1301.092688. [DOI] [PubMed] [Google Scholar]
- 40.Heling A, et al. Increased expression of cytoskeletal, linkage, and extracellular proteins in failing human myocardium. Circ Res. 2000;86(8):846–853. doi: 10.1161/01.RES.86.8.846. [DOI] [PubMed] [Google Scholar]
- 41.Kostin S, et al. Myocytes die by multiple mechanisms in failing human hearts. Circ Res. 2003;92(7):715–724. doi: 10.1161/01.RES.0000067471.95890.5C. [DOI] [PubMed] [Google Scholar]
- 42.Tannous P, et al. Intracellular protein aggregation is a proximal trigger of cardiomyocyte autophagy. Circulation. 2008;117(24):3070–3078. doi: 10.1161/CIRCULATIONAHA.107.763870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sengupta A, Molkentin JD, Yutzey KE. FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem. 2009;284(41):28319–28331. doi: 10.1074/jbc.M109.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J. Deacetylation of FoxO by Sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circ Res. 2010;107(12):1470–1482. doi: 10.1161/CIRCRESAHA.110.227371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arsov I, et al. BAC-mediated transgenic expression of fluorescent autophagic protein Beclin 1 reveals a role for Beclin 1 in lymphocyte development. Cell Death Differ. 2008;15(9):1385–1395. doi: 10.1038/cdd.2008.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanbe A, et al. Reversal of amyloid-induced heart disease in desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2005;102(38):13592–13597. doi: 10.1073/pnas.0503324102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pattison JS, Sanbe A, Maloyan A, Osinska H, Klevitsky R, Robbins J. Cardiomyocyte expression of a polyglutamine preamyloid oligomer causes heart failure. Circulation. 2008;117(21):2743–2751. doi: 10.1161/CIRCULATIONAHA.107.750232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarikas A, et al. Impairment of the ubiquitin-proteasome system by truncated cardiac myosin binding protein C mutants. Cardiovasc Res. 2005;66(1):33–44. doi: 10.1016/j.cardiores.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 49.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11(9):1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 50.Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem. 2003;278(27):25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 51.Iwata A, et al. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc Natl Acad Sci U S A. 2005;102(37):13135–13140. doi: 10.1073/pnas.0505801102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vikstrom KL, Bohlmeyer T, Factor SM, Leinwand LA. Hypertrophy, pathology, and molecular markers of cardiac pathogenesis. Circ Res. 1998;82(7):773–778. doi: 10.1161/01.RES.82.7.773. [DOI] [PubMed] [Google Scholar]
- 53.Handschin C, Spiegelman BM. The role of exercise and PGC1alpha in inflammation and chronic disease. Nature. 2008;454(7203):463–469. doi: 10.1038/nature07206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sofi F, Capalbo A, Cesari F, Abbate R, Gensini GF. Physical activity during leisure time and primary prevention of coronary heart disease: an updated meta-analysis of cohort studies. Eur J Cardiovasc Prev Rehabil. 2008;15(3):247–257. doi: 10.1097/HJR.0b013e3282f232ac. [DOI] [PubMed] [Google Scholar]
- 55.Golbidi S, Laher I. Molecular mechanisms in exercise-induced cardioprotection. Cardiol Res Pract. 2011;2011:972807. doi: 10.4061/2011/972807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fewell JG, et al. A treadmill exercise regimen for identifying cardiovascular phenotypes in transgenic mice. Am J Physiol. 1997;273(3 pt 2):H1595–H1605. doi: 10.1152/ajpheart.1997.273.3.H1595. [DOI] [PubMed] [Google Scholar]
- 57.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131(6):1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 58.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakaso K, et al. Transcriptional activation of p62/A170/ZIP during the formation of the aggregates: possible mechanisms and the role in Lewy body formation in Parkinson’s disease. Brain Res. 2004;1012(1–2):42–51. doi: 10.1016/j.brainres.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 60.Grumati P, et al. Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat Med. 2010;16(11):1313–1320. doi: 10.1038/nm.2247. [DOI] [PubMed] [Google Scholar]
- 61.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15(3):1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.