Abstract
More than 80% of the aggressive alveolar rhabdomyosarcoma (ARMSs) harbor a PAX3-FKHR fusion transcription factor, which regulates cell motility and promotes metastasis. Our hypothesis is that PAX3-FKHR regulates cell motility by regulating the expression of its transcriptional targets that are also its downstream effectors, which if identified, may lead to novel therapeutic approaches for treating ARMS. Here we report that PAX3-FKHR regulates the expression of pleiotrophin (PTN) by binding specifically to a paired-box domain binding-site in the PTN promoter, indicating that PTN is a transcriptional target of PAX3-FKHR. Significantly, we show that PTN regulates ARMS cell motility. Taken together, we have identified PTN as a novel transcriptional target of PAX3-FKHR that promotes ARMS cell motility. PTN may be a novel therapeutic target for the treatment of ARMS.
Keywords: rhabdomyosarcoma, PAX3-FKHR, PTN, motility, metastasis
1. Introduction
Rhabdomyosarcoma (RMS) is the most common soft tissue sarcoma in children. Two subtypes of RMS, embryonal (ERMS) and alveolar (ARMS), have been identified. More than 80% of the more aggressive ARMSs harbor a PAX3-FKHR fusion transcription factor. The unique expression, function, and subcellular location of PAX3-FKHR contribute to its oncogenic behavior by modifying cell growth, differentiation, and migration [1]. Various evidence suggests that PAX3-FKHR regulates cell migration, contributing to the high propensity of ARMS to metastasize [2–4].
It is possible to prevent ARMS metastasis by downregulating PAX3-FKHR. However, no pharmacologic inhibitor of PAX3-FKHR is currently available; identification of druggable transcription targets of PAX3-FKHR that are also downstream effectors of PAX3-FKHR–mediated cell migration and metastasis may lead to novel therapeutic approaches for treating ARMS [1]. Significant effort has been made to identify transcription targets of PAX3-FKHR, and several transcription targets of PAX3-FKHR that are involved in ARMS cell migration have been reported [1, 5–7].
Here we report that pleiotrophin (PTN) is a transcriptional target of PAX3-FKHR and, for the first time, show that PTN regulates cell motility in ARMS cells. Therefore, PTN is a transcription target of PAX3-FKHR and a downstream effector of PAX3-FKHR–mediated cell migration and metastasis. Our findings uncover a novel function of PTN in ARMS tumorigenesis, providing supporting evidence for our hypothesis that PAX3-FKHR regulates cell motility by regulating the expression of its transcriptional targets that are also its downstream effectors, and suggesting that PTN may be a novel therapeutic target for the treatment of ARMS.
2. Materials and methods
2.1. Cell culture
Rh30, Rh41, RD, HEK293T, NIH3T3 cells, and the establishment of PAX3-FKHR knockdown (KD1) and control (CON) stable clones of Rh30 have been described previously [5–7]. Phenol red–free Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA) was used for all luminescence assays.
2.2. Real-time reverse transcription polymerase chain reaction
We used an iCycler iQ real-time polymerase chain reaction (PCR) detection system (Bio-Rad, Hercules, CA) to perform real-time reverse transcription PCR for the measurement of mRNA levels. Extraction of total RNA, preparation of cDNA, PCR primers for PAX3-FKHR and glucose-6-phosphate dehydrogenase (GAPDH), the conditions for PCR, and the quantitation of mRNA levels have been previously described [5, 7, 8]. Briefly, the cycle threshold (Ct) values of each gene of interest and of GAPDH were calculated for each sample. The normalized value was derived by subtracting the Ct value of GAPDH from that of the gene of interest (ΔCt). Data are shown as mRNA fold change (2−ΔΔCt) relative to the mRNA level of the corresponding transcript in the control samples as indicated. PTN sense primer: ATGCAGGCTCAACAGTACCAG. Anti-sense primer: GCTTGCACTCAGCTCCAGT.
2.3. Western blot
Lysates (20 µg/lane) were prepared and processed as previously described [7]. Anti-FKHR antibodies (H-128; sc-11350) and anti-GFP antibodies (sc-9996) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA); anti-actin antibodies (A5441) were obtained from Sigma (St. Louis, MO).
2.4. Transient transfections of siRNAs and plasmids
A PAX3-FKHR–specific siRNA (PF siRNA) and control non-targeting siRNA (NT siRNA) were synthesized as described previously [5]. Pooled PTN siRNA (ON-TARGETplus SMARTpool L-018793-00-0005), individual PTN siRNA 1 (J-018793-07-0005), and individual PTN siRNA2 (J-018793-08-0005) were obtained from Thermo Scientific (Chicago, IL). Cells were plated in a 24-well plate 24 hours prior to transfection with siRNAs (10 nM) by using Lipofectamine RNAiMAX reagent (Invitrogen) or plasmids by using FuGENE 6 (Invitrogen). Wound healing assays were performed 24 hours after transfection. For evaluation of expression levels, cells were harvested 48 hour after transfection. The IncuCyte system was used to monitor and quantify cell motility in the scratch wound-healing assay, as previously described [7].
2.5. Cell invasion assay
Cell invasion assay was performed by using the CytoSelect 96-well cell invasion assay (Cell Biolabs, San Diego, CA) [9]. Briefly, 100 µL of Rh30 cells at 1 × 106/mL in a serum-free RPMI medium with or without recombinant PTN protein (R&D Systems, Minneapolis, MN) were added to the top chamber, while the bottom wells were filled with RPMI supplemented with 10% fetal bovine serum. Cells on the upper surface of the membrane were removed after 24 hours of incubation. The invasive cells at the bottom surface of the membrane were dislodged by the cell detachment solution, lysed, and quantified with CyQuant GR Dye using an EnVision plate reader (PerkinElmer, Waltham, MA). Results were expressed as percentage of fluorescence intensity relative to the no-treatment control (% cell invasion).
2.6. Luciferase assay
PTN-luc, a luciferase reporter in which the expression of luciferase is controlled by the PTN promoter containing a PAX3-FKHR binding site, 5’-GTGCTTTTCCATTCTGAATATCATTA-3’, was obtained from SwitchGear (Menlo Park, CA). PTNm-luc was created by mutating the paired-box domain (PD)–binding site, GTGCT, to ATTTA. NIH3T3 cells were co-transfected with pcDNA3 or pcDNA3-PAX3-FKHR [5], PTN-luc or PTNm-luc, and a constitutively expressed Renilla luciferase reporter (PRL-TK, a control for transfection efficiency) (Promega) by using FuGENE 6. Twenty-four hours after transfection, 10,000 cells were plated in each well of a 96-well culture plate and grown for an additional 24 hours before luciferase assay using the Dual-Glo Luciferase Assay System (Promega). PTN promoter activity was expressed in relative luciferase units (RLU), derived by normalizing firefly luciferase activity with Renilla luciferase activity.
2.7. Electrophoretic mobility shift assay
Electrophoretic mobility shift assay (EMSA) was performed as previously described [7]. Briefly, 3 µL of nuclear extract or in vitro–translated protein was incubated at room temperature for 20 min in a 20 µL volume containing 2.5% glycerol, 5 mM MgCl2, 50 ng/µL Poly (dI.dC), 0.05% NP-40, and biotin end-labeled PTN probe (GGGTGGAAGAAGAGGCTGGTTTGAAGAGAGTGCTTTTCCATTCTGAATATCATTAAAGG), with or without unlabeled PTN or PTN_m (GGGTGGAAGAAGAGGCTGGTTTGAAGAGAATTTATTTCCATTCTGAATATCATTAAAGG) as indicated.
2.8. Statistical analysis
Results are expressed as the mean ± SD of at least 3 independent experiments. Student’s t-test was used to determine the statistical significance of the difference between the paired samples. Differences were considered significant if p ≤ 0.05 (*), 0.01 (**), or 0.001 (***) and non-significant (ns) if p > 0.05. Where applicable, sample pairs are noted in brackets.
Results and discussion
3.1. PAX3-FKHR regulates the expression of PTN
To identify genes whose expression is directly affected by the levels of PAX3-FKHR in an ARMS cellular context, we developed a clone of ARMS cell line Rh30 (KD1) in which PAX3-FKHR is stably downregulated and showed that downregulating PAX3-FKHR significantly decreased the cells’ motility [7]. We used microarray analysis and identified genes whose expression decreased when PAX3-FKHR was downregulated. Among these genes are those previously reported as PAX3-FKHR target genes [1, 10], and novel target genes.
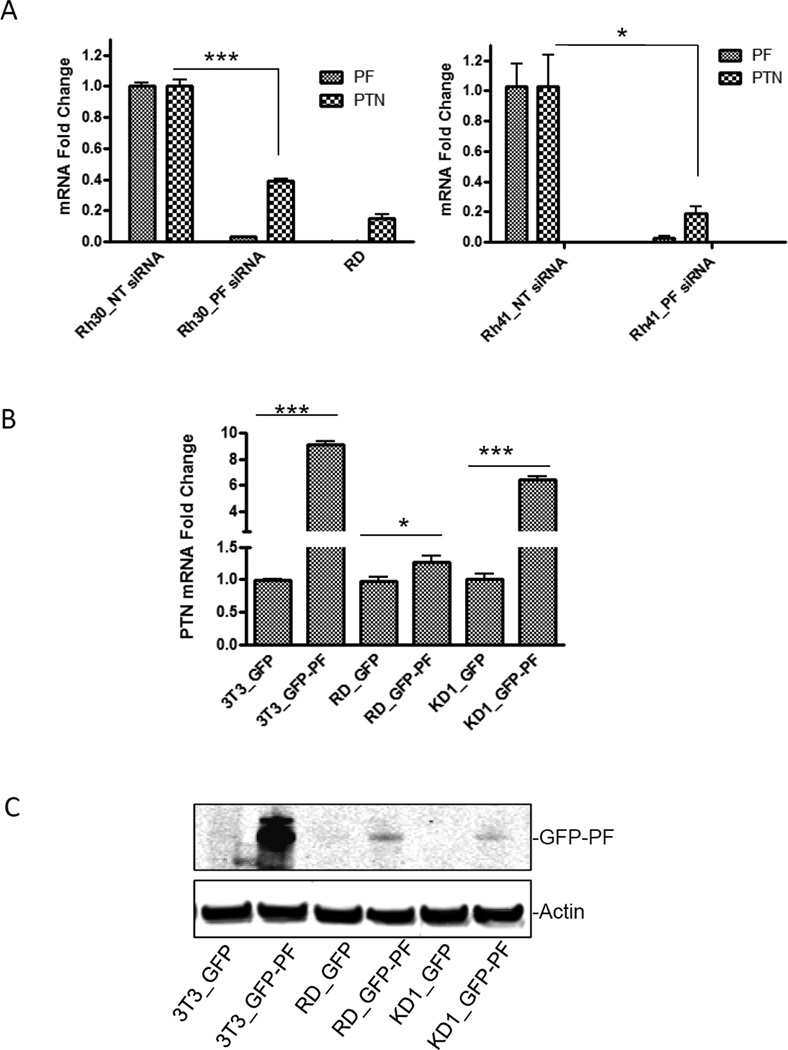
PTN was identified as a gene whose expression is significantly downregulated (an average of 7.7 fold change) in KD1 clone [7] (Table S1). To confirm that the reduced PTN expression was correlated to the decreased PAX3-FKHR levels, Rh30 cells were transiently transfected with siRNA specifically targeting PAX3-FKHR (PF siRNA). Downregulation of PAX3-FKHR (97% reduction at mRNA level) led to a downregulation of PTN mRNA (61% reduction) (Fig. 1A, left panel). Similar results were observed in another ARMS cell line, Rh41 (Fig. 1A, right panel). Additionally, the mRNA level of PTN was very low in RD (Fig. 1A), a PAX3-FKHR–negative ERMS cell line. When PAX3-FKHR was transfected into RD, NIH3T3 or KD1, PTN levels were elevated (Fig. 1B). The PAX3-FKHR-mediated elevated PTN level was most significant in the KD1 clone, even though the expression of PAX3-FKHR was moderately low (Fig. 1C). In contrast, the elevated level of PTN in response to PAX3-FKHR overexpression in RD cells was marginal, suggesting that a permissive cellular environment is required for PAX3-FKHR to induce the expression of PTN. Thus, the expression of PTN correlates with the level of PAX3-FKHR in a permissive cellular environment, suggesting that PTN might be a transcriptional target of PAX3-FKHR. These findings also highlight the importance of choosing an appropriate cellular model to study the regulation of PAX3-FKHR target genes.
Fig. 1. PAX3-FKHR regulates the expression of PTN.
(A) The mRNA levels of PAX3-FKHR (PF) or PTN in Rh30 (left panel) or Rh41 cells (right panel) transiently transfected with NT siRNA (Rh30_NT siRNA and Rh41_NT siRNA) were set as 1, and the mRNA levels of PF or PTN in cells transiently transfected with PF siRNA (Rh30_PF siRNA and Rh41_PF siRNA) or in RD were expressed as “mRNA fold change” by normalizing to the corresponding control. (B) RD, NIH3T3 (3T3), and KD1 cells were transiently transfected with either GFP empty vector or GFP-PAX3-FKHR (GFP-PF). The mRNA levels of PTN in 3T3_GFP, RD_GFP, or KD1_GFP were arbitrarily set as 1. (C) Protein levels of GFP-PF in cells (3T3, RD, or KD1) transiently transfected with either GFP or GFP-PF, detected by using an anti-GFP antibody. Actin was an equal loading control.
3.2. Induction of PTN by PAX3-FKHR involved a PD binding site–dependent transactivation
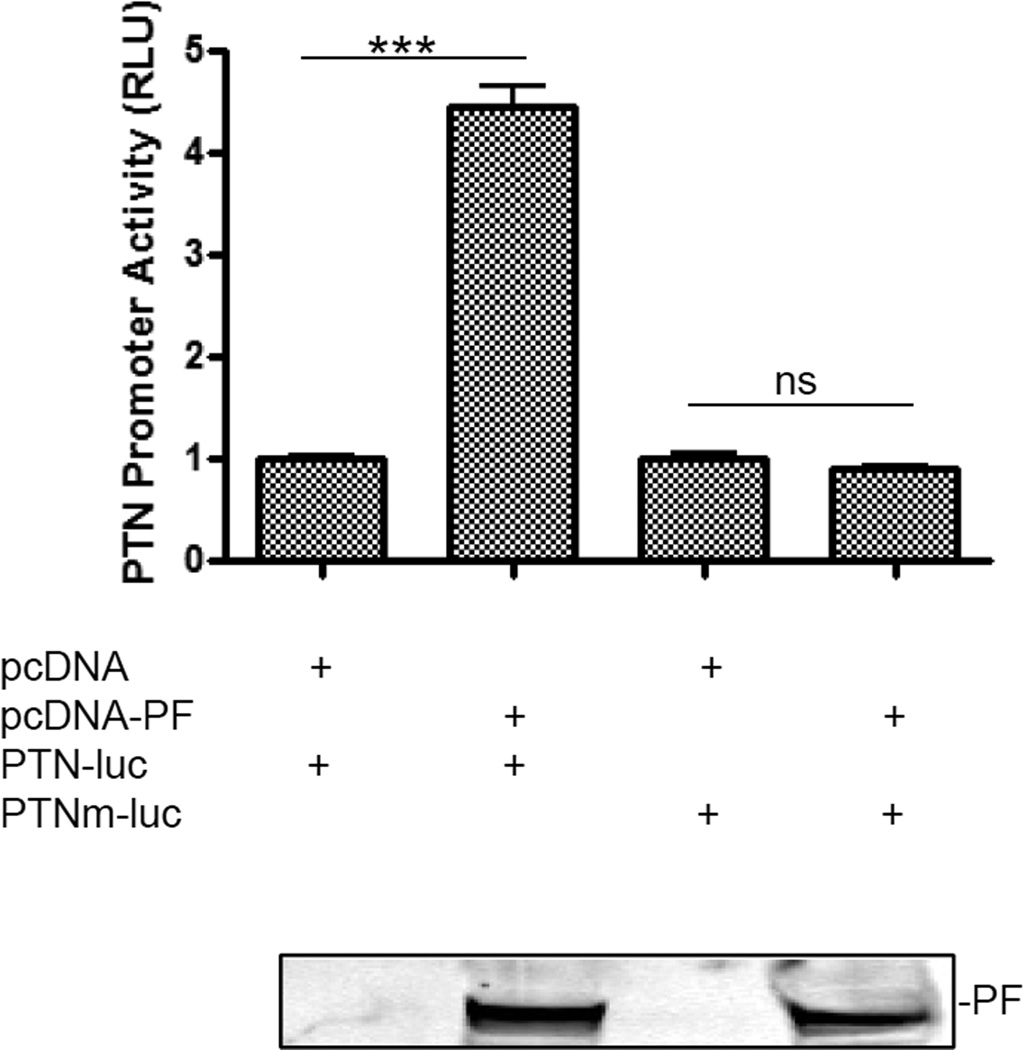
To investigate whether PAX3-FKHR controls PTN transcription by binding to the PTN promoter, we analyzed the promoter of PTN and identified a putative PAX3-FKHR binding site, 5’-GTGCTTTTCCATTCTGAATATCATTA-3’ (−288 to −263 from the transcription start site). To evaluate the contribution of this putative PAX3-FKHR binding site to the transactivation of PTN, PTN-luc was used. In NIH3T3 cells, a PAX3-FKHR–negative cell line commonly used to evaluate the function of PAX3-FKHR, expression of PAX3-FKHR significantly transactivated PTN-luc (Fig. 2). To confirm this observation, we mutated the putative PD binding site in PTN-luc to generate PTNm-luc. As shown in Fig. 2, mutating the PD-binding site of the PTN promoter abolished its transactivation by PAX3-FKHR. The requirement of an intact PD binding site for PAX3-FKHR–mediated transactivation was consistent with previous reports [10–12]. Our data suggest that the promoter of PTN contains a binding site for PAX3-FKHR, which is required for PAX3-FKHR–mediated transactivation.
Fig. 2. PAX3-FKHR induces PTN in a PD binding site–dependent manner.
Activity with pcDNA transfection was set as 1. PAX3-FKHR (PF) protein level is shown below the bar graph by using Anti-FKHR antibodies in a Western-blotting analysis.
3.3. PAX3-FKHR participates in regulating PTN gene transcription
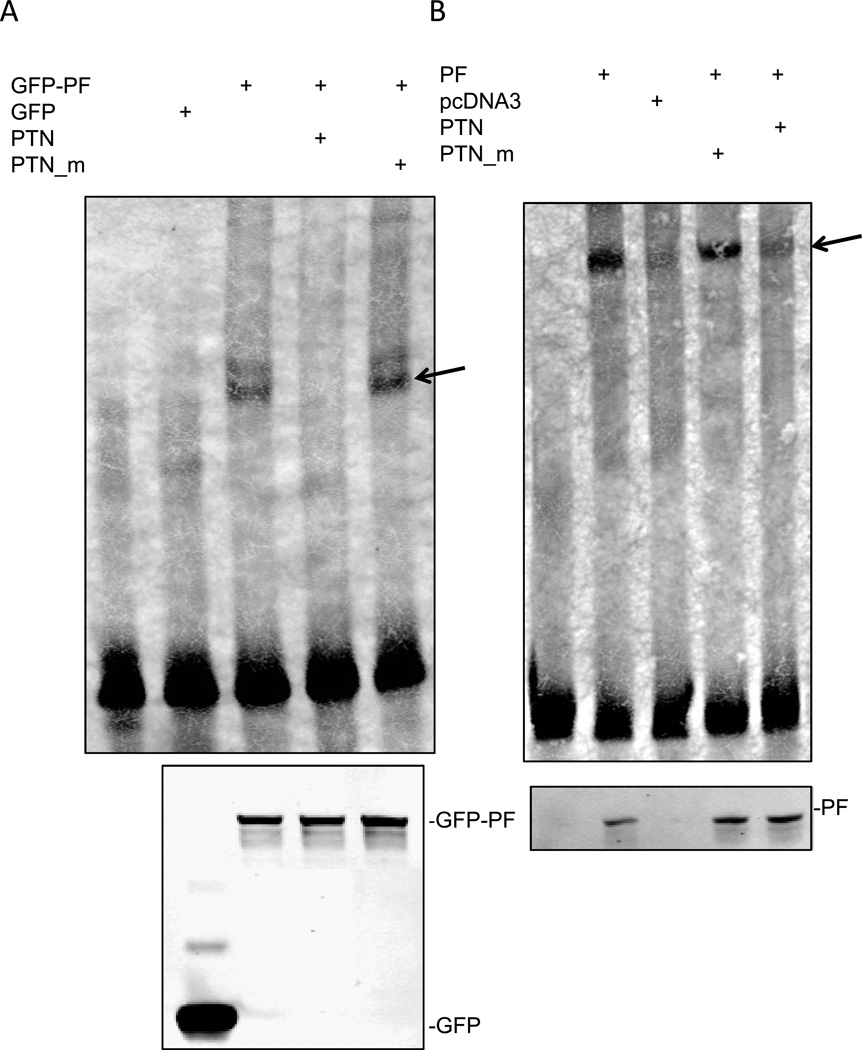
We next investigated whether PAX3-FKHR interacted with the promoter of PTN by performing an EMSA. As shown in Fig. 3A, biotin-labeled wild-type PTN probe (lane 1) was shifted by the addition of nuclear extract from HEK293T cells transfected with GFP-PAX3-FKHR (lane 3), but not by nuclear extract from HEK293T cells transfected with a control vector (GFP only) (lane 2), indicating the formation of a specific complex between GFP–PAX3-FKHR and the biotin-labeled wild-type PTN probe. The specific DNA-protein complex was disrupted by the inclusion of a wild-type unlabeled PTN (lane 4), but not by a mutated unlabeled PTN_m (lane 5). The amount of GFP–PAX3-FKHR and GFP used in the EMSA assay is shown in Fig. 3A (lower panel). Similar results were obtained when in vitro–translated PAX3-FKHR was used (Fig. 3B). These results indicate that PAX3-FKHR binds to the promoter of PTN in a PD binding site–dependent manner, confirming that PTN is a transcriptional target of PAX3-FKHR.
Fig. 3. PAX3-FKHR binds to the PTN promoter.
(A) EMSA analysis of GFP-PAX3-FKHR binding to PTN promoter. All lanes contain labeled PTN probe in addition to other components as indicated. A 100-fold excess of unlabeled wild-type (PTN) or PD binding site mutant (PTN_m) was used to compete with the labeled PTN DNA probe for GFP-PAX3-FKHR binding. GFP-PF, nuclear extract prepared from HEK293T cells transfected with GFP-PAX3-FKHR; GFP, nuclear extract prepared from HEK293T cells transfected with GFP vector only. Arrow indicates the PAX-FKHR-DNA complexes. The amount of GFP-PF and GFP used for the EMSA is shown in the lower panel, using an anti-GFP antibody. (B) EMSA analysis of PAX3-FKHR binding to PTN promoter using in vitro translated PAX3-FKHR (PF) or vector control (pcDNA). The amount of PF used for the EMSA is shown in the lower panel by using anti-FKHR antibodies.
3.4. PTN promotes cell motility in ARMS
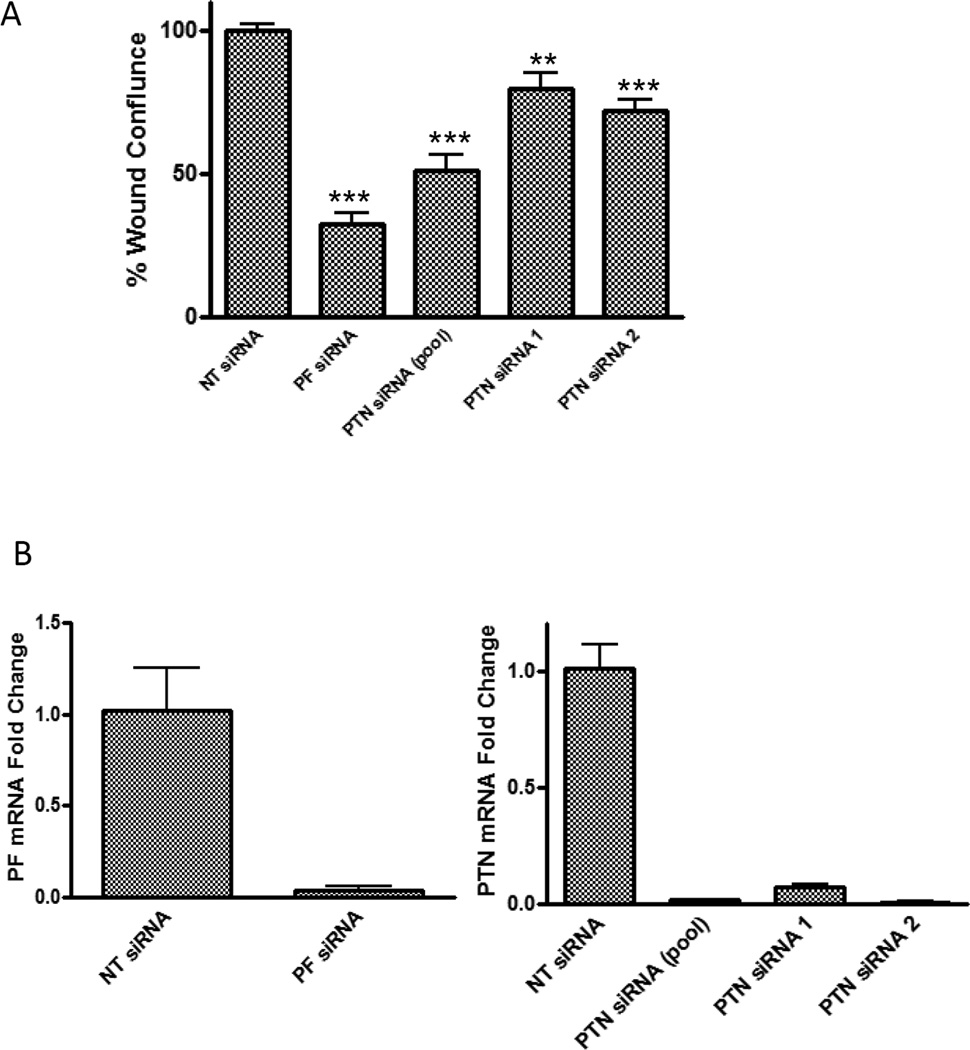
Downregulation of PAX3-FKHR decreases cell motility of Rh30 [7]. To determine whether PAX3-FKHR’s regulation of motility in ARMS cells is mediated by PTN, we first measured the effect of downregulating PTN on Rh30 cell motility in a wound-healing assay. As shown in Fig. 4, pooled PTN siRNA reduced Rh30 cell motility by 49%, whereas PF siRNA reduced the cell motility by 67%. PTN siRNA has less effect on wound healing than PF siRNA does, possibly because other transcriptional targets of PAX3-FKHR also play roles in regulating cell motility. The high efficiency of siRNA-mediated PTN and PAX3-FKHR downregulation is shown in Figure 4B. The effect of PTN siRNA on cell motility was confirmed by using two individual PTN siRNAs, PTN siRNA 1 and PTN siRNA 2 (Figure 4A).
Fig. 4. Downregulation of PTN decreases cell motility in Rh30 cells.
(A) Rh30 cells were transfected as indicated. The data, expressed as percentage of wound confluence, represent the quantitation of wound healing 30 hours after the scratch was created. The wound confluence of NT siRNA was set as 100%. (B) The knockdown efficiency was confirmed by using real-time reverse transcription PCR.
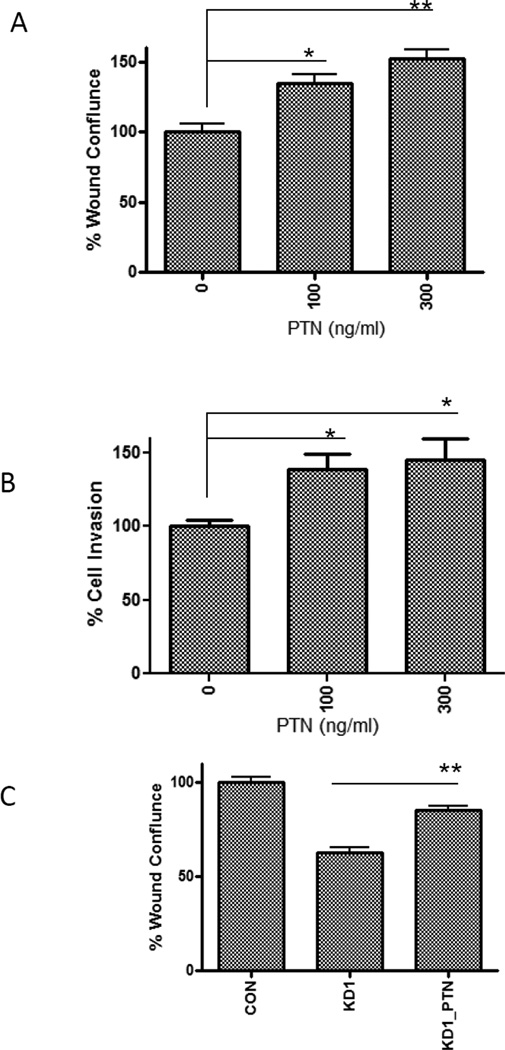
To further investigate the effect of PTN levels on cell motility, we showed that recombinant PTN protein enhanced wound healing (Fig. 5A) and cell invasion (Fig. 5B) of Rh30 and rescued the reduced wound healing in the KD1 clone (Fig. 5C). These results point to a possible role of PTN in the invasive and metastatic properties of ARMS cells.
Fig. 5. Recombinant PTN promotes cell motility and invasion.
(A) PTN enhances Rh30 cell motility in a dose-dependent manner. Wound healing assay was performed as described in Fig. 4. The wound confluence of Rh30 cells without PTN was set as 100%. (B) PTN enhanced Rh30 cell invasion in a dose-dependent manner. The cell invasion without PTN was set as 100%. (C) PTN rescued the reduced cell motility in the KD1 clone. PTN at 100 ng/ml was used (KD1_PTN). The wound confluence of control Rh30 clone (CON) was set as 100%.
Multiple transcriptional targets of PAX3-FKHR have been identified [1,7]. We chose PTN for further investigation because PTN is known to regulate cell migration, and the purpose of our research is to identify the transcriptional targets of PAX3-FKHR that are also its downstream effectors in regulating cell migration. PTN, a heparin-binding growth factor highly expressed in various human cancers [13–16], activates its cell surface receptors to regulate cellular functions, including cell adhesion, proliferation, and migration [17,18], and plays an important role in tumor promotion. A high level of PTN significantly advances the “aggressiveness” of breast cancer cells in vivo [13]. However, how the expression of PTN is regulated in ARMS and whether PTN plays a role in ARMS tumorigenesis or contributes to the aggressive metastatic behavior of ARMS have not yet been investigated. The rationale of our study is that if the transcription targets of PAX3-FKHR also mediate the function of PAX3-FKHR in regulating ARMS tumorigenesis, then these PAX3-FKHR transcription targets would be novel therapeutic targets for ARMS. In this study, we have established that PTN is a transcriptional target of PAX3-FKHR and, for the first time, shown that PTN regulates cell motility in ARMS cells. PTN could be a novel therapeutic target for ARMS.
Supplementary Material
Acknowledgements
We thank Yong-Dong Wang, Jing Wu, Jimmy Cui, Granger Ridout, and Ruoning Wang for technical assistance; other members of the Chen group for valuable discussions, and David Galloway of the Department of Scientific Editing for editing the manuscript. This work was supported by the American Lebanese Syrian Associated Charities (ALSAC), St. Jude Children’s Research Hospital, National Cancer Institute grant P30CA027165, and the St. Baldrick’s Foundation (Award Number: 179692).
Abbreviations
- PTN
pleiotrophin
- RMS
rhabdomyosarcoma
- ARMS
alveolar rhabdomyosarcoma
- ERMS
embryonal rhabdomyosarcoma
- PF
PAX3-FKHR
- PD
paired-box domain
- EMSA
electrophoretic mobility shift assay
References
- 1.Linardic CM. PAX3-FOXO1 fusion gene in rhabdomyosarcoma. Cancer Lett. 2008;270:10–18. doi: 10.1016/j.canlet.2008.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kikuchi K, Tsuchiya K, Otabe O, Gotoh T, Tamura S, Katsumi Y, Yagyu S, Tsubai-Shimizu S, Miyachi M, Iehara T, Hosoi H. Effects of PAX3-FKHR on malignant phenotypes in alveolar rhabdomyosarcoma. Biochem Biophys Res Commun. 2008;365:568–574. doi: 10.1016/j.bbrc.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 3.Ebauer M, Wachtel M, Niggli FK, Schafer BW. Comparative expression profiling identifies an in vivo target gene signature with TFAP2B as a mediator of the survival function of PAX3/FKHR. Oncogene. 2007;26:7267–7281. doi: 10.1038/sj.onc.1210525. [DOI] [PubMed] [Google Scholar]
- 4.Xia SJ, Holder DD, Pawel BR, Zhang C, Barr FG. High expression of the PAX3-FKHR oncoprotein is required to promote tumorigenesis of human myoblasts. Am J Pathol. 2009;175:2600–2608. doi: 10.2353/ajpath.2009.090192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeng FY, Cui J, Liu L, Chen T. PAX3-FKHR sensitizes human alveolar rhabdomyosarcoma cells to camptothecin-mediated growth inhibition and apoptosis. Cancer Lett. 2009;284:157–164. doi: 10.1016/j.canlet.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng FY, Dong H, Cui J, Liu L, Chen T. Glycogen synthase kinase 3 regulates PAX3-FKHR-mediated cell proliferation in human alveolar rhabdomyosarcoma cells. Biochem Biophys Res Commun. 2010;391:1049–1055. doi: 10.1016/j.bbrc.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu L, Wang YD, Wu J, Cui J, Chen T. Carnitine palmitoyltransferase 1A (CPT1A): a transcriptional target of PAX3-FKHR and mediates PAX3-FKHR-dependent motility in alveolar rhabdomyosarcoma cells. BMC cancer. 2012;12:154. doi: 10.1186/1471-2407-12-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pondugula SR, Tong AA, Wu J, Cui J, Chen T. Protein phosphatase 2Cbetal regulates human pregnane X receptor-mediated CYP3A4 gene expression in HepG2 liver carcinoma cells. Drug Metab Dispos. 2010;38:1411–1416. doi: 10.1124/dmd.110.032128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lam KK, Chiu PC, Chung MK, Lee CL, Lee KF, Koistinen R, Koistinen H, Seppala M, Ho PC, Yeung WS. Glycodelin-A as a modulator of trophoblast invasion. Hum Reprod. 2009;24:2093–2103. doi: 10.1093/humrep/dep205. [DOI] [PubMed] [Google Scholar]
- 10.Cao L, Yu Y, Bilke S, Walker RL, Mayeenuddin LH, Azorsa DO, Yang F, Pineda M, Helman LJ, Meltzer PS. Genome-wide identification of PAX3-FKHR binding sites in rhabdomyosarcoma reveals candidate target genes important for development and cancer. Cancer research. 2010;70:6497–6508. doi: 10.1158/0008-5472.CAN-10-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, Wang C. Identification of a new class of PAX3-FKHR target promoters: a role of the Pax3 paired box DNA binding domain. Oncogene. 2007;26:1595–1605. doi: 10.1038/sj.onc.1209958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Wang C. Nephroblastoma overexpressed (NOV/CCN3) gene: a paired-domain-specific PAX3-FKHR transcription target that promotes survival and motility in alveolar rhabdomyosarcoma cells. Oncogene. 2011;30 doi: 10.1038/onc.2011.69. 3549-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deuel TF, Zhang N, Yeh HJ, Silos-Santiago I, Wang ZY. Pleiotrophin: a cytokine with diverse functions and a novel signaling pathway. Archives of biochemistry and biophysics. 2002;397:162–171. doi: 10.1006/abbi.2001.2705. [DOI] [PubMed] [Google Scholar]
- 14.Perez-Pinera P, Chang Y, Deuel TF. Pleiotrophin, a multifunctional tumor promoter through induction of tumor angiogenesis, remodeling of the tumor microenvironment, and activation of stromal fibroblasts. Cell Cycle. 2007;6:2877–2883. doi: 10.4161/cc.6.23.5090. [DOI] [PubMed] [Google Scholar]
- 15.Lu KV, Jong KA, Kim GY, Singh J, Dia EQ, Yoshimoto K, Wang MY, Cloughesy TF, Nelson SF, Mischel PS. Differential induction of glioblastoma migration and growth by two forms of pleiotrophin. The Journal of biological chemistry. 2005;280:26953–26964. doi: 10.1074/jbc.M502614200. [DOI] [PubMed] [Google Scholar]
- 16.Jager R, List B, Knabbe C, Souttou B, Raulais D, Zeiler T, Wellstein A, Aigner A, Neubauer A, Zugmaier G. Serum levels of the angiogenic factor pleiotrophin in relation to disease stage in lung cancer patients. British journal of cancer. 2002;86:858–863. doi: 10.1038/sj.bjc.6600202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng ZJ, Gao SB, Wu Y, Xu XF, Hua X, Jin GH. Lung cancer cell migration is regulated via repressing growth factor PTN/RPTP beta/zeta signaling by menin. Oncogene. 2010;29:5416–5426. doi: 10.1038/onc.2010.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mikelis C, Sfaelou E, Koutsioumpa M, Kieffer N, Papadimitriou E. Integrin alpha(v)beta(3) is a pleiotrophin receptor required for pleiotrophin-induced endothelial cell migration through receptor protein tyrosine phosphatase beta/zeta. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2009;23:1459–1469. doi: 10.1096/fj.08-117564. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.