Abstract
Brucella quorum sensing has been described as an important regulatory system controlling crucial virulence determinants such as the VirB type IV secretion system and the flagellar genes. However, the basis of quorum sensing, namely the production of autoinducers in Brucella has been questioned. Here, we report data obtained from the use of a genetic tool allowing the in situ detection of long-chain N-acyl-homoserine lactones (AHL) activity at single bacterium level in Brucella melitensis. These data are consistent with an intrinsic production of AHL by B. melitensis in low concentration both during in vitro growth and macrophage infection. Moreover, we identified a protein, named AibP, which is homologous to the AHL-acylases of various bacterial species. In vitro and during infection, expression of aibP coincided with a decrease in endogenous AHL activity within B. melitensis, suggesting that AibP could efficiently impair AHL accumulation. Furthermore, we showed that deletion of aibP in B. melitensis resulted in enhanced virB genes expression and VirB8 production as well as in a reduced flagellar genes expression and production of FlgE (hook protein) and FliC (flagellin) in vitro. Altogether, these results suggest that AHL-dependent quorum sensing and AHL-quorum quenching coexist in Brucella, at least to regulate its virulence.
Introduction
Quorum sensing (QS), in its broadest sense, is a regulatory system that allows genetic reprogramming in response to small diffusible signalling molecules called autoinducers that are produced and released by bacteria [1]. Once a threshold concentration is reached, autoinducers modulate the activity of a target sensor kinase or a transcriptional regulator, leading to the induction or repression of target genes. This threshold level is usually reached at high bacterial population density, and QS allows individual cells to coordinate gene expression at the level of the whole population in a cell-density-dependent manner [2].However, production of autoinducers by only few bacteria in a restricted environment with limited diffusion rate can be sufficient to reach the threshold concentration [3], and even a single bacterium in an enclosed environment is able to engage QS [4]. Thus QS signal molecules allow bacteria not only to sense quorum per se, but also environmental factors such as diffusion and confinement [1]. QS is used by many bacterial species to regulate numerous functions, including virulence, symbiosis, biofilm formation, swarming motility, antibiotic resistance and plasmid conjugation [5], [6].
In Gram-negative bacteria, N-acyl homoserine lactones (AHLs) are commonly used as the chemical cues of QS. They are synthesized by LuxI-type AHL-synthases from acyl-acyl carrier proteins and S-adenosylmethionine [7], and they differ one from another on the length of their acyl side chains that additionally may contain C3 substitutions (either a hydroxyl or a carbonyl group). The amphiphilic nature of AHLs allows them to freely diffuse across bacterial membranes, although an efflux pump is required in some cases [8]. Once the threshold intracellular concentration is reached, AHLs bind to their cognate LuxR-type transcriptional regulator(s) and modulate their activity [5], [6].
The Gram-negative bacteria Brucella spp. are the etiologic agent of brucellosis, a chronic disease affecting wild and domestic animals but also humans worldwide [9]. Although details on their infectious cycle within natural mammalian hosts are lacking, it is commonly assumed that successful establishment of persistent infections by Brucella spp. is linked to its ability to survive and replicate within host phagocytic cells, while remaining inconspicuous and avoiding a strong inflammatory response at the onset of infection [10], [11].
In previous studies, we have reported the identification of QS components in Brucellamelitensis. First, we demonstrated in vitro a low level of production of AHLs, namely N-dodecanoyl-homoserine lactone (C12-HSL) and probably N-3-oxo-dodecanoyl-homoserine lactone (3-oxo-C12-HSL), although Brucella lacks a classical AHL-synthase [12]. Secondly, we identified two conserved LuxR-type regulators named VjbR and BabR (also known as BlxR) that contain a predicted N-terminal AHL-binding domain and a C-terminal HTH DNA-binding domain [13], [14]. Moreover, several works by independent groups have suggested that (i) VjbR and BabR are global transcriptional regulators controlling genes involved in Brucella virulence, stress response and metabolism, some of them being identified as direct VjbR targets [13]–[16]; and (ii) that C12-HSL modifies the transcriptional activity of VjbR and BabR [12]–[17].
The virB operon, encoding proteins of the VirB type IV secretion system (T4SS), and flagellar genes were the first identified QS targets in Brucella, and both are induced by VjbR [13]. The T4SS is one of Brucella’s major virulence factors, as it is essential for the control of Brucella-containing vacuole (BCV) trafficking towards a replication permissive organelle [18]. Expression of the virB operon requires VjbR and its deletion causes strong attenuation of Brucella in cellular infection models as well as in vivo [19], [20]. While non-motile, Brucella spp. possess 31 genes encoding flagellar proteins [21]. VjbR is a transcriptional activator of the gene encoding the flagellar master regulator FtcR [22]. Flagellar genes are required for the establishment of chronic infections, as all B. melitensis flagellar mutants tested so far, except for the ▵fliC and ▵flbT strains, are attenuated in vivo [20], [21], [23], [24].
The role of Brucella QS components has also been evaluated during infection. Deletion of vjbR was shown to cause a strong attenuation of B. melitensis virulence both in cellular and murine infection models [13]. Conversely, it was proposed that C12-HSL negatively influences Brucella virulence as its exogenous addition on early infected macrophages impairs B. melitensis intracellular replication, an effect that has been suggested to occur through the alleviation of VjbR-mediated induction of the virB operon expression [16]. Thus, VjbR would promote Brucella virulence, whereas C12-HSL would dampen it.
Despite these observations, it is still not known whether C12-HSL, 3-oxo-C12-HSL, VjbR and BabR function in a bona fide QS regulatory system in Brucella and what would be the environmental factor monitored by AHLs. Moreover, as Brucella AHL synthase remains elusive [25] and, as only low concentration of AHLs has been detected in culture supernatants, their intrinsic production by Brucella was recently questioned [17].
In the current study, we used a genetic tool that provided results consistent with an intrinsic production of long-chain AHLs within B. melitensis both in culture in vitro and during cellular infection. In both cases, we found that the concentration of QS signals within B. melitensis was not population-density-dependent. In vitro, this was largely due to the expression of aibP, which encodes an AHL-inactivating protein homologous to the AHL-acylases of various bacterial species. Therefore, in addition to be capable of QS, we propose that Brucella is also capable of self “quorum quenching” [26]. Similarly to QS, AibP-mediated quorum quenching was found to regulate both virB and flagellar genes expression, although it acted on them in opposite ways.
Results
In situ detection of long-chain AHL activity in B. melitensis
Brucella has been described to produce long-chain AHLs, namely C12-HSL and probably 3-oxo-C12-HSL. However, they were only detected at low levels in supernatants of B. melitensis cultures in stationary phase of growth [12]. We hypothesized that this low extracellular concentration could be due to the control of AHL synthesis by Brucella and/or to the limited diffusion of these hydrophobic molecules across bacterial membranes [8]. For these reasons, we investigated the AHL production kinetics of B. melitensis in vitro by using a sensitive AHL-responsive reporter system rather than by monitoring and quantifying their appearance in culture medium. The AHL reporter system developed by Hentzer et al. [27], which is based on the production of the unstable GFP(ASV) following activation of the Pseudomonas aeruginosa LuxR-type regulator LasR (Figure S1A), appeared to be suited for the detection of endogenous AHL activity in B. melitensis. Indeed, although 3-oxo-C12-HSL is the cognate signal molecule of LasR, the regulator responds to AHLs with chain length above C10 [27]. Moreover, the instability of GFP(ASV) makes it an interesting reporter protein to quantify rapid changes in AHL concentrations. In order to use it in Brucella with a high sensitivity, this genetic tool was cloned on the medium copy and broad host range vector pBBRMCSI (Figure S1A). A control plasmid that does not contain lasR was used to ensure that no endogenous regulator could induce gfp(ASV) in Brucella (Figure S1A).
Prior to its use in Brucella, we tested the sensitivity and confirmed the specificity of the QS reporter plasmid in Escherichia coli, in which QS is not based on endogenous AHL production [28]. Analysis of GFP(ASV) production has been performed by flow cytometry after a 4h incubation with various concentrations of exogenous synthetic AHLs and fixation of bacteria. As expected, gfp(ASV) was not induced in the absence of AHL (Figure S1B). Moreover, the system is specific for long-chain AHLs, as C12-HSL and 3-oxo-C12-HSL, but not C4-HSL triggered a dose-dependent response (Figure S1B). As expected, the cognate signal molecule of LasR, i.e. 3-oxo-C12-HSL, was the most efficient inducer of the system (Figure S1B).
The QS reporter plasmid and the control plasmid were then separately introduced into B. melitensis 16M by conjugation to give rise to B. melitensis QS reporter and B. melitensis control strains, respectively. Fluorescence microscopy and flow cytometry analysis were used to validate both strains. Similar to what has been observed with E. coli, the B. melitensis QS reporter strain but not the control strain responded to exogenously supplied C12-HSL and 3-oxo-C12-HSL in a concentration-dependent manner. Indeed, both the number of GFP(ASV)-positive bacteria and the GFP(ASV) fluorescence intensity increased proportionally to signal molecules concentration (Figure 1). The absence of GFP(ASV) signal in the control strain incubated with C12-HSL suggested that no Brucella regulator could take place of LasR to induce the reporter gene. The QS reporter vector was thus adapted to our purpose, as activation of heterologously expressed LasR, and the subsequent induction of gfp(ASV) reporter gene in B. melitensis was expected to reflect the presence of one or several long-chain AHLs within this species.
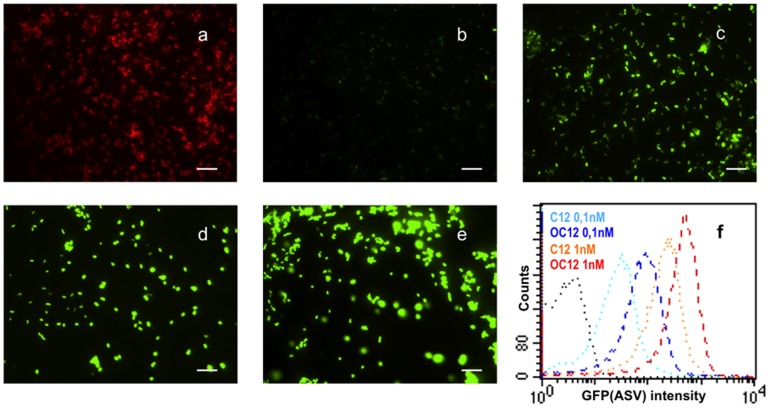
Figure 1. Validation of the specificity and sensitivity of the QS reporter system in Brucella melitensis 16M.
(a) Immunofluorescence of the B. melitensis control strain incubated 4 hours with C12-HSL (1 µM) and labelled with monoclonal A76-12G12 anti-LPS antibody (red). No GFP(ASV) signal is detected. (from b to e) Observation of GFP(ASV) production by the B. melitensis reporter strain after a 4h incubation with various concentrations of synthetic C12-HSL; (b) 1nM, (c) 10nM, (d) 100nM, (e) 1 µM; scale bar 5 µm. (f) Measurement of GFP(ASV) fluorescence intensity by flow cytometry (5×104 events acquired) in the B. melitensis QS reporter strain fixed after a 4h-incubation with 0.1nM or 1nM of C12-HSL or 3-oxo-C12HSL. The B. melitensis control strain was used as a negative control (black dotted line). The results are representative of at least two independent experiments.
Endogenous AHL activity within B. melitensis does not increase proportionally to bacterial density
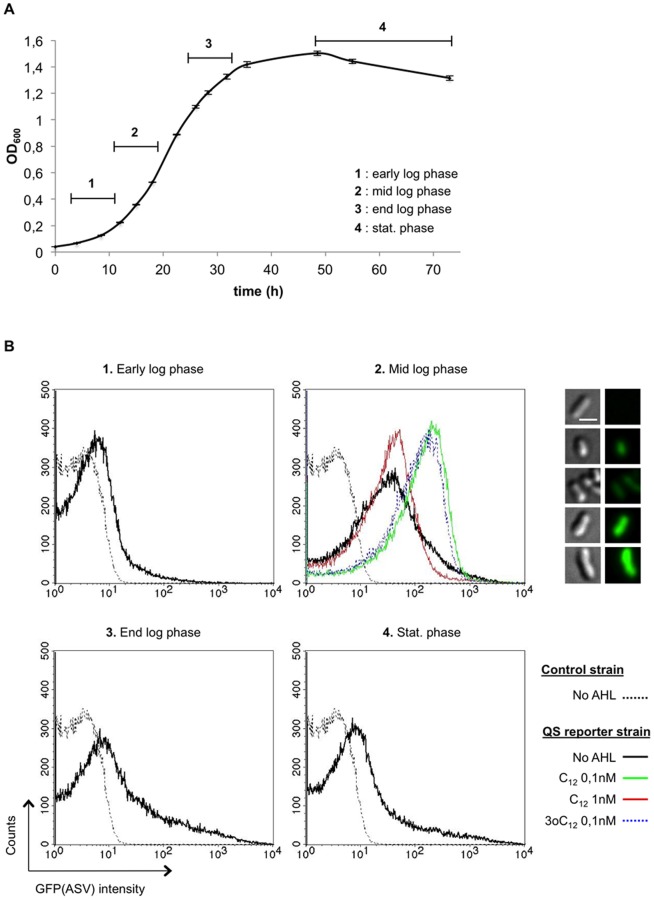
As the LasR-based reporter system allows for semi-quantitative detection of long-chain AHLs activity, we used it in B. melitensis 16M to follow its own AHLs production during in vitro growth. The B. melitensis QS reporter and control strains were grown in 2YT rich medium. At different times post-inoculation, bacterial samples were fixed and analysed by flow cytometry for GFP(ASV) production. Results representative for each of the different 4 growth phases determined by growth rate calculation are presented in Figure 2. GFP(ASV) signal was not observed for the control strain, whatever the growth phase considered. Conversely, we could detect a GFP(ASV) signal at each time of bacterial growth when using the QS reporter strain (Figure 2B). Detecting an endogenous long-chain AHL activity in B. melitensis is consistent with the previously reported production of C12-HSL and 3-oxo-C12-HSL in vitro [12]. Interestingly, a peak of GFP(ASV) production was observed during the mid-log phase, i.e. between 12 and 18h of growth, which corresponded to an OD600 between 0.3 and 0.6 (Figure 2A and 2B). It was followed by a decrease at the end exponential and stationary phases at which bacterial density is the highest (Figure 2A and 2B). Moreover, the measurement of GFP(ASV) fluorescence intensity after the addition of synthetic AHLs on the B. melitensis QS reporter strain harvested in the early log phase (corresponding to a low intrinsic production of AHL) indicated that maximal intrabacterial concentration of endogenous long-chain AHL in the tested conditions would not be higher than 1nM of C12-HSL or 0.1nM of 3-oxo-C12-HSL (Figure 2B panel 2).
Figure 2. Transient production of long-chain AHLs by B. melitensis in liquid culture.
The B. melitensis QS reporter strain was grown in 2YT and cell density was determined when fluorescence intensity of GFP(ASV) was assessed by flow cytometry (5×104 events acquired). (A) Growth curve of the B. melitensis QS reporter strain. Numbers represent the 4 distinct phases of growth. OD600, optical density at 600nm. (B) Histograms of GFP(ASV) fluorescence intensity representative of the growth phases represented in A. The B. melitensis control strain was used as a negative control. In (B2), the peak of GFP(ASV) fluorescence intensity due to endogenous AHLs was compared with results obtained after a 4h-incubation of the B. melitensis QS reporter strain with synthetic C12-HSL or 3-oxo-C12-HSL (bacteria from the early log phase were used). The insets show differential interference contrast (DIC) and FITC fluorescence microscopy of (from top to bottom) the negative control strain, the QS reporter strain in the absence of exogenous AHL and incubated with C12-HSL 0.1nM, with C12-HSL 1nM, and with 3-oxo-C12-HSL 0.1 nM respectively. Results are representative of three independent experiments.
Identification of a conserved putative AHL-acylase in Brucella
We hypothesized that the decrease in intrabacterial AHL activity observed during vegetative growth could reflect a decrease in AHL synthesis, and/or could be due to the degradation of signal molecules. As the mechanisms of AHL synthesis in Brucella remains unknown, the first hypothesis could not be further tested. However, in order to determine whether AHL production in Brucella could be regulated by self-quorum quenching activity, a bioinformatic screen of Brucella spp. genomes was carried out to identify homologs of the two classes of AHL-degrading enzymes described to date, i.e. AHL-lactonases and AHL-acylases. No AHL-lactonase homologous to Bacillus spp. AiiA [26], Rhodococcus erythropolis QsdA [29], or Ochrobactrum sp. strain T63 AidH [30] were found. However, BLASTP searches allowed us to find a hypothetical protein in Brucella sharing 17–28% identity with characterized or predicted AHL-acylases from various species (e value from 1e-04 to 3e-86) (Figure S2). This 761 aa-long protein was named AibP (AHL-inactivating Brucella protein) and was found in all of the sequenced Brucella genomes, except for that of B. ovis. In B. melitensis, a frameshift mutation due to a 1 bp deletion is responsible for the interruption of aibP, which then covers two ORFs, i.e. BMEII0212 and BMEII0211 (Figure S2). No homologs were found in close phylogenetic relatives of Brucella, such as Ochrobactrum anthropi, Sinorhizobium spp., Mesorhizobium loti, A. tumefaciens, Rickettsia spp. or Bartonella spp. As part of the N-terminal nucleophile (Ntn) hydrolases superfamily, AHL-acylases are produced into a single inactive precursor that consists of a signal peptide followed by a α-subunit, a spacer sequence and a ß-subunit [31]–[34]. Further in silico analysis of AibP showed that conserved regions are equally observed in the predicted α- and ß-subunits. Unlike most AHL-acylases, Brucella spp. homolog is not predicted to have a N-terminal signal peptide. More important is the conservation in AibP of amino acids known to be part of the active site of characterized Ntn hydrolases and that are essential to their post-translational processing and/or their enzymatic activity (Figure S2) [35]–[39]. Among them, the glycine-serine pair, which represents the cleavage site between the spacer peptide and the ß-subunit, generating a free N-terminal nucleophile serine that has been demonstrated to be essential to the activity of Ralstonia AHL acylase [31].
The decrease in AHL activity within Brucella is correlated with aibP expression
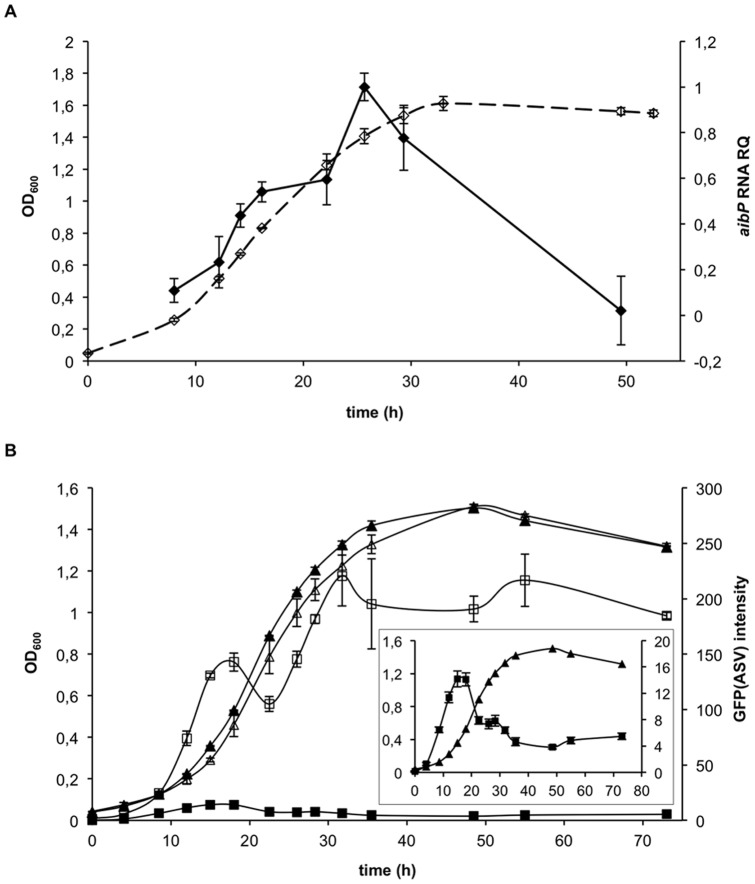
In order to know whether we could link the decrease in AHL activity observed when B. melitensis 16M entered the end log phase of growth in 2YT with the expression of aibP, quantitative real-time PCR (qRT-PCR) was performed to follow variations in the relative level of its transcript. We found that aibP mRNA level increased during exponential growth up to ten-fold, and was followed by a decline upon entry into stationary phase (Figure 3A). As the peak of aibP mRNA level coincided with the decrease in AHL activity (Figure 2B), it was tempting to speculate that both events are directly linked. In order to further test this hypothesis, we constructed an aibP non-polar mutant of B. melitensis (▵aibP). This mutant produced smooth LPS, and bacterial and colony morphology were not different from the wt strain (data not shown). Moreover, the aggregative phenotype previously reported for some B. melitensis QS-mutants [16], [40] was not observed (data not shown). By using the QS reporter plasmid described above, we could first suggest that AibP inactivates synthetic C12-HSL and 3-oxo-C12-HSL supplied to B. melitensis cultures, as the mean GFP(ASV) fluorescence intensity measured by flow cytometry after incubation of the wt or the ▵aibP strain with these AHL was higher in the mutant than in the wt (Figure S3). More significantly, we could also suggest an activity of AibP toward self-generated AHL. Indeed, by monitoring the endogenous long-chain AHL activity in the ▵aibP mutant (i.e. in the absence of any added AHL) we found that the mean fluorescence intensity of GFP(ASV) in the mutant strain population was higher than in the isogenic wt strain population throughout all bacterial growth phases (Figure 3B). This suggests that the mutant accumulated higher levels of long-chain AHL during in vitro growth: while their concentration decreased during the log phase of B. melitensis wt growth, bacteria showed the typical population density-dependent accumulation of AHL in the absence of aibP (Figure 3B). This result strongly suggests that AibP is an efficient enzyme with an activity against physiological concentrations of long-chain AHLs produced by Brucella, and that it plays a critical role in the regulation of QS signals concentration in vitro.
Figure 3. The decrease in Brucella AHLs activity during in vitro growth is linked to aibP expression.
(A) The relative amount of aibP mRNA (black forms) during vegetative growth of B. melitensis (open forms) in 2YT medium was determined by qRT-PCR. Result is representative of three independent experiments. Error bars represent standard deviation from biological triplicates. OD600, optical density at 600nm; RQ, relative quantity. (B) The activity of endogenous long-chain AHLs was followed using the QS reporter system. B. melitensis wt (black forms) and ▵aibP (open forms) QS reporter strains were grown in 2YT and cell density was determined (triangle) for each culture when GFP(ASV) fluorescence intensity was evaluated (square). The activity of endogenous long-chain AHL(s) at each time is expressed as the geometric mean of GFP(ASV) fluorescence intensity measured by flow cytometry (5×104 events acquired). The B. melitensis control strain was used as a negative control (not shown). Insert: the scale of the graph was modified for analysis in the wt strain. Results are representative of three independent experiments. Error bars represent standard deviation from biological triplicates.
AHL synthesis and degradation occur within B. melitensis during macrophage infection
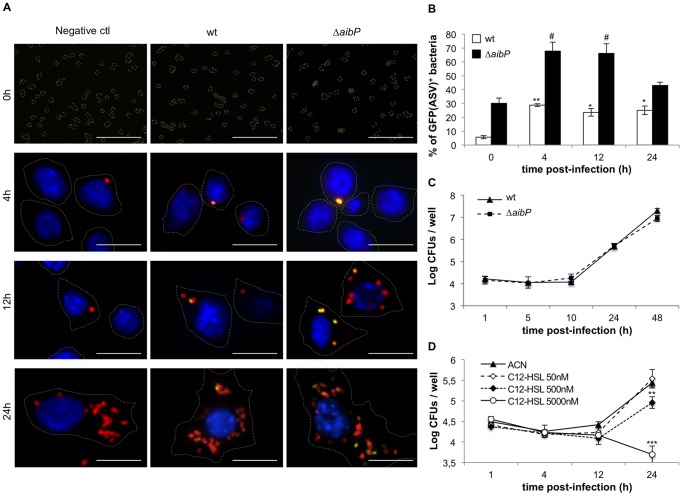
Brucella is usually referred to as a “facultatively extracellular intracellular parasite” [41] to emphasize its impressive ability to survive and replicate in phagocytic cells, which are thought to be the primary niche of the bacteria within the host. We took advantage of the QS reporter tool to determine whether Brucella produces AHL during its intracellular life. The murine macrophage line RAW264.7 was infected with B. melitensis wt or ▵aibP strain each bearing either the control or the QS reporter plasmid and grown to stationary phase in RPMI medium supplemented with erythritol. In this condition, the GFP(ASV) signal of both wt and ▵aibP strains was found drastically reduced compared to bacteria grown in 2YT (Figure S4). Having an inoculum containing mostly GFP(ASV)-negative bacteria was important in order to highlight a potential de novo synthesis of AHLs during macrophage infection. Prior to infection (0h) as well as at 4-, 12- and 24 hours post-infection (p.i.), cells were fixed and examined by fluorescence microscopy in order to measure the mean intensity of GFP(ASV) fluorescence in individual bacteria (Figure 4A). The number of GFP(ASV)-positive bacteria (Figure 4B) was determined as described in the Material and Methods section. Compared to the inoculum (0h), we observed a reproducible increase (minimum 5-fold) in the number of GFP(ASV)-positive B. melitensis wt bacteria 4h p.i., suggesting an induction of AHL production following infection (Figures 4A and 4B). This percentage was not found to increase at later time p.i. even when bacteria entered the replication phase (Figures 4A and 4B). Thus, AHL production by B. melitensis did not seem to depend on the bacterial number in infected cells. Similar to what has been observed in vitro, both the number of GFP(ASV)-positive bacteria and the fluorescence intensity of GFP(ASV) were higher when the ▵aibP strain was used to infect RAW 264.7 macrophages (Figures 4A and 4B), indicating that AibP-mediated inactivation of self-produced AHL occurred during cell infection.
Figure 4. B. melitensis both produces and degrades long-chain AHLs during macrophage infection.
(A and B) B. melitensis wt and ▵aibP QS reporter strains were used to infect monolayers of RAW264.7 murine macrophages. Prior to (0h) and during infection, bacteria or cells were fixed, bacteria were labelled with a monoclonal A76-12G12 anti-LPS antibody and DNA was labelled with DAPI. (A) Immunofluorescence micrographs are representative from at least two independent experiments. Bacteria LPS appears in red, DNA in blue. Scale bar, 10 µm. (B) The percentage of GFP(ASV)-positive bacteria at different times post-infection was determined as described in the Material and Methods section. Error bars represent the standard deviation from two independent experiments. Data have been analyzed by ANOVA I after testing the homogeneity of variance (Bartlett). * and ** denote significant differences (P < 0.05 and P < 0.01) in relation to wt bacteria prior to infection (0h) while # denotes a significant difference (P < 0.05) in relation to ▵aibP bacteria prior to infection. (C) Intracellular replication of B. melitensis wt and ▵aibP strains in RAW264.7 murine macrophages. At indicated times, cells were lysed and intracellular colony forming units (CFUs) were determined. Error bars represent the standard deviation of triplicates in one representative experiment out of three. (D) RAW264.7 macrophages were infected with B. melitensis wt in the presence of C12-HSL or ACN (negative control) and treated as described (C). ** and *** denote significant (P < 0.01 and P < 0.001 respectively) differences in relation to infection by wt bacteria in the presence of CAN (Bartlett and ANOVA I analysis).
It is noteworthy that, despite its apparent inability to control AHL concentration as the wt does, the ▵aibP mutant strain did not display any defect in its capacity to enter, survive and replicate in various cell lines, as indicated by CFUs recovery (see Figure 4C for RAW264.7 infection; data not shown for bovine SV40 macrophages and HeLa cells). This observation contrasts with the effect of exogenous C12-HSL addition on early infected cell that has been previously reported to impair bacterial replication [16]. However this inhibitory effect was found to occur mostly when very high concentrations of C12-HSL (5 µM) were used (Figure 4D).
AibP-mediated quorum quenching regulates expression of virulence genes
Since QS has been shown to play a role in the control of virulence genes expression in Brucella, we investigated whether its regulation by AHL inactivation through AibP could also impact it. We chose to assess the expression of the virB operon and flagellar genes in the ▵aibP mutant as these genes were the first identified QS targets required for full virulence of Brucella.
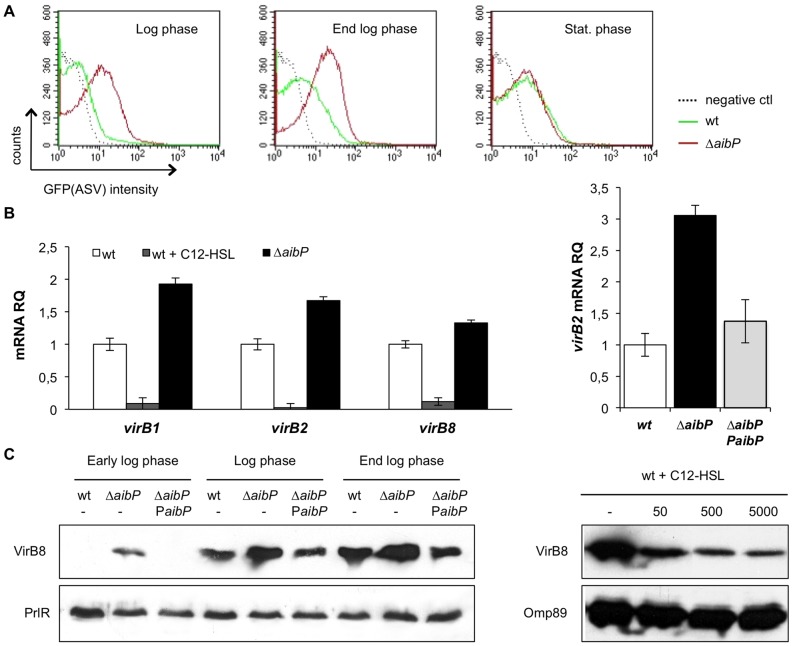
First, we compared the activity of the virB operon promoter (PvirB) in the wt and ▵aibP strains during their growth in 2YT medium. A fusion of the B. melitensis virB1 upstream region to the promoterless gfp(ASV) was constructed and the resulting plasmid pBBR-PvirB-gfp(ASV) was introduced into both strains. Bacteria carrying the pBBR-PvirB-gfp(ASV) vector were fixed at different times of the growth curve and GFP(ASV) production was measured by flow cytometry. Results representative of each growth phase are shown in Figure 5A. In agreement with the previously reported growth phase-dependent regulation of virB operon expression in rich medium [13], [42], we noted that PvirB was barely active in B. melitensis wt strain during the exponential growth, while it was induced at the end log phase and reached its maximal activity in the stationary phase (Figure 5A). Interestingly, an earlier induction of PvirB in the log phase was observed for the ▵aibP mutant (Figure 5A). Moreover, PvirB activity at the end log phase of growth remained higher in the mutant, while it returned to the wt level in the stationary phase (Figure 5A). The earlier induction of virB operon in the ▵aibP mutant was confirmed at the mRNA and protein levels respectively by qRT-PCR on virB1, virB2 and virB8 mRNAs (Figure 5B, left panel), and by Western blot analysis of VirB8 production in cultured bacteria (Figure 5C). Overexpression of virB2 (Figure 5B, right panel) and overproduction of VirB8 (Figure 5C) were both compensated by the expression of aibP from a replicative vector, consistent with complementation of the aibP deletion. This phenotype of the ▵aibP strain was in striking contrast with the downregulation of virB genes expression and VirB8 production in wt B. melitensis grown in the presence of C12-HSL (Figures 5B and 5C). However, the absence of a negative effect of long-chain AHL accumulation in the ▵aibP strain is in agreement with its ability to replicate at the wt level intracellularly (Figure 4C).
Figure 5. Self-quorum quenching regulates virB genes expression.
(A) B. melitensis wt and ▵aibP strains both carrying the pBBR PvirB-gfp(ASV) plasmid were grown in 2YT and GFP(ASV) fluorescence intensity was measured at indicated phases of growth by flow cytometry (5×104 events acquired). Results are representative of two independent experiments. (B) The relative abundance of virB1, virB2, and virB8 mRNAs was determined by qRT-PCR on RNA isolated from bacteria harvested at the early exponential phase of growth in 2YT supplemented or not with exogenous C12-HSL (5 µM). Deletion of aibP results in significant upregulation of virB genes (P<0.001 in Student’s t test), whereas exogenous C12-HSL significantly downregulates their expression (P<0.001 in Student’s t test). ▵aibP PaibP on the right panel is the complemented strain. Results are representative of two independent experiments. Error bars represent standard deviation from biological triplicates. (C) (Left panel) Western Blot analysis of VirB8 production performed on whole protein lysates of bacteria harvested at the indicated phases of growth in 2YT. ▵aibP PaibP is the complemented strain. (Right panel) Bacteria were harvested in the log phase of growth in 2YT in the absence (–) or in the presence of C12-HSL. The VirB8 protein was detected at its expected size (26,5 kDa). Detection of PrlR or Omp89 proteins was used to normalize total protein content.
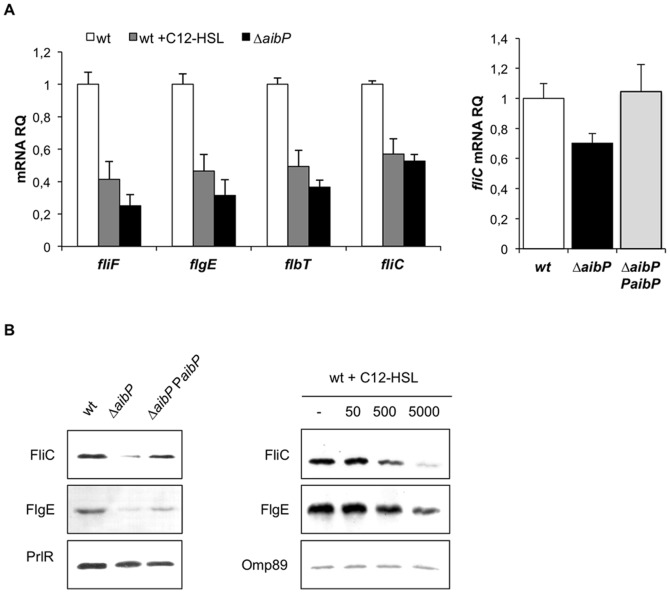
Flagellar genes are also targeted by QS regulation in Brucella [13], [22]. Expression of flagellar genes recently reported as class II and class III genes in the non-classical hierarchy of flagellum biogenesis in B. melitensis was monitored by qRT-PCR on bacteria harvested at the early exponential growth phase in 2YT medium. fliC is a class III gene that codes for the filament flagellin subunit; fliF, flgE and flbT are class II genes encoding the basal body MS-ring protein, the hook protein and a regulator needed for flagellin production, respectively [43]. Expression of all these genes was significantly downregulated in the ▵aibP mutant, and this effect was also observed in wt bacteria supplied with exogenous C12-HSL (Figure 6A). These results were confirmed by Western blot analysis of FlgE and FliC abundance in the wt and ▵aibP mutant strains as we observed that both hook protein and flagellin levels were reduced in the ▵aibP mutant or when wt was grown in 2YT supplemented with high concentrations of C12-HSL (Figure 6B). Moreover, the use of a replicative plasmid carrying the aibP gene allowed complementation of this phenotype (Figure 6A and 6B).
Figure 6. Self-quorum quenching regulates flagellar genes expression.
(A) The relative abundance of fliF, flgE, flbT, and fliC mRNAs was determined by qRT-PCR on RNA isolated from bacteria harvested at the early exponential growth phase in 2YT supplemented or not with exogenous C12-HSL (5 µM). Deletion of aibP and addition of exogenous C12-HSL on B. melitensis both result in significant downregulation of the expression of the tested genes (P<0.001 in Student’s t test). ▵aibP PaibP (right panel) is the complemented strain. Results are representative of two independent experiments. Error bars represent standard deviation from biological triplicates. (B) (Left panel) Western Blot analysis of FliC and FlgE production performed on whole protein lysates of bacteria harvested at the early log phase of growth in 2YT. ▵aibP PaibP is the complemented strain. (Right panel) Bacteria were harvested at the early log phase of growth in 2YT in the absence (–) or in the presence of C12-HSL. The FliC and FlgE proteins were detected at their expected size (29 kDa and 41 kDa respectively). Detection of the PrlR or Omp89 was used to normalize total protein content.
Discussion
Recent studies have identified and characterized components of a QS system in B. melitensis. These include C12-HSL and probably 3-oxo-C12-HSL as signal molecules [12], and VjbR and BabR/BlxR as responding LuxR-type regulators [13]–[16], [44]. However, the fact that Brucella lacks a classical AHL-synthase and that only low levels of AHLs were detected in the supernatant of B. melitensis stationary-phase cultures brought up questions about the basis of such a system in Brucella, namely the intrinsic production of QS cues. Here, we sought to confirm endogenous production of AHLs in Brucella by using a sensitive reporter system that allows specific in situ detection of long-chain AHLs activity at single bacterium level. In this genetic tool, the gfp(ASV) gene is transcriptionally controlled by the P. aeruginosa LuxR-type LasR (Figure S1A). Detecting the activation of heterologously expressed LasR within Brucella appeared to be suited for our purpose as C12-HSL and 3-oxo-C12-HSL are both potent activator of LasR (Figures S1B and Figure 1). Moreover, this system monitors AHL concentration directly within bacteria, allowing us to bypass the problem of slow diffusion of long chain AHLs across bacterial membrane. Finally, determining the amount of AHLs retained by bacteria rather than extracellular AHLs is particularly relevant as the former can be considered as the level of biologically active QS signals. In this respect, significant differences between the intracellular and extracellular pools of AHLs have been already observed in bacterial cultures [8], [45]. By using this QS reporter system, we observed a production of GFP(ASV) by B. melitensis during in vitro growth (Figure 2B) but also during its intracellular trafficking within infected macrophages (Figures 4A and 4B). These findings constitute the second data consistent with an intrinsic production of long-chain AHLs by Brucella in culture, and suggest for the first time that signal molecules are produced by B. melitensis in the relevant context of cell infection.
In several Proteobacteria, the QS system is apparently incomplete. For example, LuxR-type QS regulators that lack a cognate LuxI-like AHL synthase have been called “LuxR orphans” [46] or “solos” [47]. In some AHL-producing Proteobacteria, they could function as “extra” LuxR receptors that are connected to complete AHL-dependent QS systems [48]–[50]. However “solos” have also been found in non-AHL-producing bacteria where they detect AHLs produced by neighbouring bacteria [28], exogenous signals produced by eukaryotic hosts [51] or endogenously produced signalling molecules of different nature [52]. While we cannot rule out the possibility of a response of VjbR and BabR to host-derived molecules or to endogenous signals that are not AHLs, we report here that B. melitensis 16M produces signals that activate LasR and that can be inactivated by a protein homologous to AHL-acylases, therefore supporting the idea of an endogenous production of AHLs. The finding that mutations of VjbR in critical residues involved in AHL binding affect virB expression in the absence of exogenously supplied AHLs in vitro reinforces this idea [16]. It should be noted that the absence of an obvious classical AHL-synthase in a bacterial strain is not synonymous of absence of self-produced QS cues. As for Brucella, a Shewanella isolate was shown to produce AHLs whereas attempts to identify AHL-synthase(s) were unsuccessful, suggesting a novel family of AHL synthases that remain to be discovered [53].
QS was originally described as used by bacteria to coordinate gene expression at the population level and, thus, as a mean for bacteria to monitor population density [5], [6]. In vitro, we found that the GFP(ASV) fluorescence intensity measured within the B. melitensis QS reporter strain decreased at the end exponential and stationary phases of growth at which bacterial density is the highest (Figure 2). The QS circuitry in cultured Brucella is thus non-classical in that the intrabacterial AHLs concentration does not reflect the cell population density. We found that this was largely due to the expression of aibP, which encodes an AHL-inactivating protein homologous to the AHL-acylases of various bacterial species (Figure S2) [31]–[34], [54]–[57]. Indeed, when B. melitensis lacks aibP, accumulation of GFP(ASV) was observed throughout the growth of the QS reporter strain, suggesting that its deletion restores the typical population density-dependent accumulation of AHLs (Figure 3B).
The identification of a mechanism of signal molecules turnover in B. melitensis is an additional finding consistent with the hypothesis of a self-AHL production. Along with the production of AHL-antagonists and the enzymatic modification of AHLs, their degradation by AHL-lactonases or AHL-acylases is one of the processes of QS interference known as quorum quenching [26], [58], [59]. AHL-acylases are part of the N-terminal nucleophile (Ntn) hydrolases superfamily comprising the penicillin acylase family enzymes, cephalosporin acylases (CA) and aculeacin A acylases [31]. They hydrolyze the amide bond between the homoserine lactone (HSL) and the acyl-side chain of AHL via a nucleophilic attack, resulting in HSL and fatty acid products that cannot spontaneously reform a functional QS signal molecule [31], [60]. Although the identification of the products released upon AibP-mediated inactivation of AHLs would be required to clearly demonstrate it, the conservation of the Glyα, Ser1ß, His23ß, Tyr33ß and Asn244/269ß residues that are essential for the enzymatic activity of Ntn hydrolases suggests that AibP is an AHL-acylase (Figure S2).
Expression of aibP during the exponential growth phase could be the main reason why Taminiau et al. [12] found only low levels of C12-HSL and 3-oxo-C12-HSL in B. melitensis stationary-phase culture supernatants. However, our semi-quantitative analysis suggests that even in the absence of AibP, the concentration of AHLs would remain low within B. melitensis when compared with other bacteria. Indeed, at the peak of GFP(ASV) fluorescence intensity in vitro, we suggest that concentrations of C12-HSL and 3-oxo-C12-HSL within B. melitensis wt would not be higher than 1nM and 0.1nM, respectively (Figure 2B panel 2), while they would be approximately ten-fold higher in the ▵aibP mutant strain (Figure 3B). In contrast, AHLs reach micromolar concentrations in cultures fluids of bacteria such as Vibrio fischeri [61], A. tumefaciens [62] and P. aeruginosa [63]. A low concentration of endogenous long-chain AHLs and their possible intrabacterial sequestration due to their slow diffusion across membranes could account for our inability to detect AHL production by B. melitensis ▵aibP in cross-streak assays with biosensors strains, or by incubating the latter with culture supernatants of the former strain (data not shown).
A signal functions as a signal per se only if it appears but also disappears at a certain time. This also holds true for QS signals. The inactivation of self-produced AHLs by Gram-negative bacteria could be a regulatory mechanism by which they exit a quorum-sensing mode. Nevertheless, it remains unclear whether it takes place in other bacteria, as AHL-acylases from P. aeruginosa (PvdQ) and P. syringae (HacA and HacB) and an AHL-lactonase (AttM/BlcC) from A. tumefaciens have been suggested to be part of AHL-independent metabolic pathways by acting against endogenous substrates other than self-produced AHLs [32], [33], [45], [64], [65]. Although we cannot yet say for certain that AHLs are the natural substrates of Brucella AibP, several observations suggest that it mediates self-quorum quenching. First, the timing of aibP expression during in vitro growth is consistent with the kinetics of GFP(ASV) production in the B. melitensis QS reporter strain, as the peak of aibP mRNA level coincided with the decrease in GFP(ASV) intensity (Figure 3). Secondly, on the contrary to P. aeruginosa [32], P. syringae [33] and A. tumefaciens [45], in which deleting genes encoding AHL-inactivating enzymes does not affect the intracellular levels of AHLs, the higher GFP(ASV) fluorescence intensity of the ▵aibP strain compared to the wt strain (Figures 3B and 4B) suggests that endogenous expression of aibP in Brucella would allow degradation of self-produced AHLs and maintain them at low concentrations. Finally, the alterations in the ▵aibP mutant of virB and flagellar genes expression (Figures 5 and 6), which were the first identified targets of VjbR and BabR [13], [44] support the hypothesis that AHLs inactivation by AibP is effectively capable of quenching QS-dependent functions in B. melitensis.
Since its discovery in Vibrio fischeri [66], the conceptual role of QS has evolved, notably with the highlighting that bacteria can use autoinducers not only to sense quorum per se but potentially any environmental and biotic factor influencing their concentrations (for reviews, see [1], [3], [67]). By using the B. melitensis QS reporter strain, we could show an early production of AHLs during cell infection (1h, 4h and 8h p.i.), i.e. at a time at which infected macrophages contain only few bacteria (from 1 to 3 in average) that have not yet started replication. This suggests that a single Brucella individually enclosed in their vacuole could switch from a “non-quorate” to a “quorate” state, in the absence of neighbouring bacteria. Studies of single Staphylococcus aureus entrapped within an endosomal compartment in endothelial cells [68] or within a physical and chemical isolated nanostructure droplet [4] have demonstrated that confinement of an individual isolated bacterium can induce QS. In the case of Brucella, VjbR is required early in host cell infection, at least to induce virB whose expression reaches a peak at 5h p.i. in B. abortus during infection of J774 macrophages [69]. Thus, the expression of a QS regulator along with the production of QS cues at very early times p.i. provide single vacuole-enclosed Brucella with the possibility to engage QS. This could result in genetic reprogramming required for virulence genes expression but also for the metabolic adaptation of the bacteria to the host environment [14], [15], [70]. Production of QS cues could thus allow Brucella to sense confinement in the BCV rather than quorum.
It is interesting to note that aibP is part of the VjbR and C12-HSL regulons [15]. More precisely, it is induced by VjbR and repressed by the addition of exogenous C12-HSL, at least through a VjbR-independent mechanism, which could therefore involve BabR. aibP is thus part of the QS network of Brucella. Further supporting this idea is our finding that deletion of aibP in B. melitensis was found to impact the expression of QS-regulated genes (Figures 5 and 6). Interestingly, supplying B. melitensis wt cultures with C12-HSL or 3-oxo-C12-HSL (500nM or 5 µM) mimics the effect of aibP deletion on flagellar genes expression but not on virB expression, although increased concentrations of both AHLs are expected in the ▵aibP mutant. A similar observation was made in infected macrophages where the ▵aibP mutant replicated at the wt level, whereas addition of 5 µM C12-HSL or 3-oxo-C12-HSL on cells reduced the intracellular replication of B. melitensis (Figures 4C and 4D). This suggests that Brucella QS is far from being completely understood and that regulation of the virB operon is even more complex than anticipated. Since the identification of C12-HSL as the major signal molecule produced by B. melitensis [12], studies on Brucella QS were based on the use of synthetic C12-HSL either on (i) recombinant QS regulator VjbR [17], (ii) on bacteria during vegetative growth [12], [13] or (iii) on intracellular bacteria at different stages of macrophages infection [16]. They all conclude that VjbR functions as a transcriptional activator of virB only in the absence of C12-HSL in order to promote cellular infection. However, it should be noted that the concentration of synthetic signal molecules used to report an effect on VirB (and flagellar proteins) production and bacterial multiplication within cells were at least 500 fold higher than the concentration of endogenous C12-HSL or 3-oxo-C12-HSL estimated in the B. melitensis ▵aibP mutant (5 µM vs 10 nM). Here, we did not find B. melitensis to produce such concentrations of long chain AHLs, which could therefore be physiologically irrelevant. It is also noteworthy that the absence of AibP in B. melitensis could result not only in higher C12-HSL and 3-oxo-C12-HSL concentrations but also of other unidentified AHL(s) that would not have been detected by Taminiau et al. [12] because of the previously unsuspected AibP activity. These unidentified AHL(s) could have a positive effect on virB expression that would be consistent with its earlier induction in the ▵aibP mutant (Figure 5). Moreover, it is possible that AibP impacts Brucella virulence not only by regulating the AHLs level, but also by its involvement in yet-to-be discovered metabolic processes. As mentioned earlier, other substrates than AHLs have been either identified or suspected for several so-called quorum-quenching enzymes [65], [71]. The links between virulence and metabolism in Brucella become more and more appreciated, and it is now clear that regulatory mechanisms of Brucella virulence also regulates metabolism, which in turn impacts bacterial virulence [70]. Thus, if the deletion of aibP leads to changes in the metabolic state of Brucella, through the ability of AibP to modify (an) unknown substrate(s), this could indirectly result in alteration of Brucella virulence.
In conclusion we propose that, although its AHL synthase remains elusive, B. melitensis synthesizes low levels of long-chain AHLs both during in vitro growth and cell infection, and that it carefully controls their concentration through the expression of aibP that encodes an AHL inactivating protein. AHL-dependent quorum-sensing and quorum-quenching would thus coexist, and they both regulate expression of virulence genes, including the virB operon and flagellar genes.
Materials and Methods
Bacteria and growth conditions
The bacterial strains used in this study were E. coli DH10B (Gibco BRL), E. coli S17-1 [72] and smooth virulent B. melitensis 16M (Biotype1, ATCC 23456). E. coli and B. melitensis strains were grown with shaking at 37°C, respectively in Luria-Bertani and in 2YT medium (10% yeast extract, 10 g liter−1 tryptone, 5 g liter−1 NaCl) containing appropriate antibiotics. The B. melitensis QS reporter strains used for cellular infection were grown in RMPI 1640 (Gibco) supplemented with 2 g liter−1 erythritol.
Chloramphenicol, kanamycin and nalidixic acid were used at 20 µg/ml, 50 µg/ml and 25 µg/ml, respectively.
Synthetic C4-HSL, C12-HSL, and 3-oxo-C12-HSL were purchased from Sigma-Aldrich and prepared in acetonitrile (ACN). They were added to bacterial growth media at the indicated final concentrations. The same volume of ACN was used as a negative control.
B. melitensis growth curve in 2YT were performed as follows: a stationary-phase overnight culture (2YT, 10 ml) was back-diluted to obtain an optical density at 600nm (OD600) of 0.05. Growth was measured by reading OD600 at indicated times post-inoculation. 100-ml cultures were used to follow GFP(ASV) production from B. melitensis QS reporter strains and from B. melitensis harbouring the pBBR-PvirB-gfp(ASV) vector. 100-ml or 250-ml cultures were used to harvest bacteria for qRT-PCR analysis. 50-ml cultures were used to harvest bacteria for Western blot analysis. The growth rate constant µ was determined from the following equation: ln Nt – ln N0 = µ(t – t0) in which N is OD600.
Molecular techniques and plasmids
DNA manipulations were performed according to standard techniques [73]. Primers used are listed in Table 1.
Table 1. Primers used in this work.
| Primer | Sequence |
| FPaibP | GGGGTACCCCTTCCGAAATGGTTGGAAGG |
| RPaibP | GGAATTCCGAAGATCTTCTCCGATATAAGAATGGCCG |
| FTaibP | GAAGATCTTCGGAATTCCACACAGAAATCGGGGAGG |
| RTaibP | CGGGATCCCGCAACGTGTCGAGAAACGC |
| F-BHIaibP | CGGGATCCATGAACGTCGCGAGTGC |
| R-XbaIaibP | GCTCTAGATTAAGATGGCTGCATAATCAGG |
| gfpasv-F | CTCGAGATGCGTAAAGGAGAAGAAC |
| gfpasv-R | GGTACCTTAAACTGATGCAGCGTAG |
| pvirB-F | GGATCCGAAGTCCTTTCCGTCCTG |
| pvirb-R | CTCGAGGTCTCCTTCTCAGAGAATG |
| aibP-F | AACAATTGGGCGGTGGA |
| aibP-R | AATTTCATAGGCCCGATGC |
| flbT-F | AACTTCTGAACGATGCGACAT |
| flbT-R | AAAGCTGGCGCAGCG |
| flgE-F | TTCCGTGAACGCTGC |
| flgE-R | GAAACGAGATCGCCCGT |
| fliC-F | CTTCGTACAATCGTTCCGGT |
| fliC-R | CCATGGTCTTCGCATCAGT |
| fliF -F | CCTACGAGACGCTCTATGTCG |
| fliF-R | AAGGGAATGCCAGCTTCAC |
| virB1-F | ACGACAGCACAGTCACTGGAAG |
| virB1-R | TTCGGCAGATTGTACCTGTTGA |
| virB2-F | GCAAAAAGTGCTGGACTTGCTA |
| virB2-R | CCATCTTGTAACCGGACCAGAT |
| virB8-F | TGGATAAATACTGGCTCTCGCA |
| virB8-R | GGTTTCGTAGTCCTTTTGCAGC |
| 16S rRNA-F | ACGCCGTAAACGATGAATGTT |
| 16S rRNA-R | CCCAGGCGGAATGTTTAATG |
To obtain an AHL-monitor vector usable in Brucella, the previously described pMHLAS vector [27] was digested with NotI and the restriction fragment ends were filled in with Klenow DNA polymerase. The fragment containing the Plac-lasR and PlasB-gfp(ASV) was then inserted into the EcorV site of pBBR1 MCS-I [74] to give rise to the QS reporter plasmid used in this study (Figure S1A). The control vector was obtained by digestion of the previously described pMHLB [27] with BamHI and HindIII, and insertion of the fragment containing PlasB-gfp(ASV) into the corresponding sites of the pBBR1 MCS-I [74] (Figure S1A).
The B. melitensis 16M ▵aibP mutant was obtained by allelic replacement as previously described [75]. Briefly, upstream and downstream regions flanking the BMEII0212-BMEII0211 sequence were amplified by polymerase chain reaction (PCR) from B. melitensis 16M genomic DNA using the primer pairs (i) FPaibP and RPaibP, (ii) FTaibP and RTaibP. A second PCR was used to ligate the two PCR products by cohesive ends. The PCR product (aibP upstream/downstream) was inserted into the Asp718 and BamHI sites of pSKoriTcat to generate pSKoriTcat-▵aibP. The aphA4 non-polar deletion cassette [75] was excised from pUC4aphA4 with BamHI, and subsequently cloned into the BglII site of pSKoriTcat-▵aibP to generate the pSKoriTCat-▵aibP::aphA4 plasmid. The plasmid was transformed into E. coli strain S17-1 and introduced into B. melitensis 16M by conjugation. Clones for which a double recombination event occurred (Cms, Kanr) were selected. Gene replacement was confirmed by PCR.
In order to obtain the complementation vector pBBR1-aibP, the BMEII0212-BMEII0211 sequence was first amplified by PCR from B. melitensis 16M genomic DNA using the F-BHIaibP and R-XbaIaibP primers. The PCR product (BamHI-aibP-XbaI) was then cloned into the EcorV site of pGEM. In a last step, this fragment was excised using BamHI and XbaI, and inserted into the corresponding site of pBBR1 MCS-I [74] downstream the endogenous Plac. This final vector was transformed into E. coli S17-1 and introduced into B. melitensis 16M ▵aibP by conjugation.
The pBBR1-gfp(ASV) vector used to clone PvirB upstream the promoterless gfp(ASV) was generated as follows: the gfp(ASV) gene encoding an unstable variant of GFPmut3 was amplified by PCR from the pMHLAS vector [27] using the gfpasv-F and gfpasv-R primers, and the PCR product (XhoI-gfp(ASV)-Asp718) was inserted into the corresponding site of pBBR1-MCSI vector [74] in the opposite orientation to the Plac. The promoter of the virB operon was supposed to localize within a sequence that comprises the last 30bp of its upstream ORF and going until their predicted ribosome-binding site (RBS). It was amplified by PCR using the pvirB-F and pvirB-R primers. It was then cloned into the BamHI and XhoI sites of the pBBR1-gfp(ASV) vector in order to position the predicted RBS at -6 from the gfp(ASV) start codon.
RNA preparation
For analysis of the relative abundance of aibP, flagellar genes and virB genes, B. melitensis strains were grown as described above. Total RNA was extracted from B. melitensis strains as follows. At indicated phases of growth, culture samples (10 ml) were harvested, bacteria were resuspended in 100 µl SDS 10% and 20 µl proteinase K (20 mg ml−1) and incubated at 37°C with shaking for 1h. TriPure Isolation Reagent (Roche) (1 ml) was added and suspensions were vigorously shaken. After 10 min of incubation at 65°C, chloroform (300 µl) was added, the suspensions were shaken and incubated at room temperature for 5-10 min. Samples were centrifuged at 14 000 rpm for 15 min at 4°C. Then 500 µl of isopropanol was added and RNA was precipitated overnight at –20°C. After a centrifugation (13 000rpm, 30 min), the pellet was washed with ethanol 75%. RNA was dried at room temperature, resuspended in RNase-free water, and contaminating DNA was removed by DNase I treatment (Fermentas) following the manufacturer’s instructions. RNA samples were assessed for quality by the Agilent Bioanalyzer or by electrophoresis. RNA quantity was measured using a NanoDrop spectrophotometer (ND-1000, Thermo Fisher Scientific).
Relative quantification of mRNAs via the comparative cycle threshold method
1 µg of total RNA was reverse-transcribed into cDNA by using random hexamer primers and the Transcriptor First Strand cDNA synthesis kit (Roche) following manufacturer’s instructions. 250 ng of cDNA were then used as template for quantitative real-time PCR (qRT-PCR) in 96-well optical reaction plates using Power SYBR® Green PCR Master Mix (12.5 µl, Applied Biosystems) and primers (1 µl and 10 µM each, sequences are listed in Table 1) that were designed with PrimerExpress™ 2.0 (Applied Biosystems) and that allowed amplification of sequences from 80 to 100 bp long. Deionized RNase free water was added to make the total volume up to 25 µl/well. We used the thermal cycling conditions recommended by Applied Biosystems, which included 10min at 95°C for thermal activation of the AmpliTaq Gold ® DNA polymerase and 40 two-step cycles of denaturation for 15s at 95°C and annealing/extension for 1min at 60°C. Technical triplicates were performed. Relative quantification using the ▵▵Ct method was performed for each set of primer in an Applied Biosystems real-time PCR instrument. The samples from wt bacteria were used as a reference, and the 16S rRNA was used for normalization. A negative control used for each qRT-PCR reaction showed that no genomic DNA contamination occurred in the RNA samples (data not shown). Standard deviation was calculated on biological triplicates.
Western blot analysis
For analysis of FlgE, FliC and VirB8 production, B. melitensis strains were grown as described above. At indicated growth phases, bacterial samples were harvested. Bacteria were concentrated in PBS at an OD600 of 10 in 50 µl, prior to a 1h-inactivation at 80°C, and addition of 2x SDS-sample buffer. The proteins were resolved on a 12% polyacrylamide gel and transferred to Hybond ECL nitrocellulose membranes (Amersham). The immunodetection of proteins was performed using anti-FlgE (1/3000), anti-FliC (1/3000) or anti-VirB8 (1/1000) rabbit polyclonal sera [21], [76], and with anti-Omp89 mAb (A53/10B2) (1/1000), or polyclonal anti-PrlR antibodies (1/1000) [77]. The detection of primary antibodies was performed using donkey anti-rabbit (Amersham) horseradish peroxidase-conjugated secondary antibodies, and visualized using the ECL system (Amersham). The measured molecular masses of FlgE, FliC and VirB8 are 41, 29 and 26,5 kDa, respectively.
Flow cytometric analysis
Validation of the QS reporter strains was performed as follows. E. coli and B. melitensis control of QS reporter strains were grown overnight in the conditions described above, and were back-diluted to an OD600 of 0.2 in medium supplemented with AHLs at the indicated concentrations. After a 4h- or an 8h-incubation, bacteria were washed in PBS and fixed in 2% paraformaldehyde, pH 7.4, at 37°C for 15min. After an additional wash in PBS, bacteria were used for flow-cytometric analysis of GFP(ASV) production with a FACScalibur using CellQuest software (Becton Dickinson) as previously described [78]. Bacteria were gated according to size and scatter to eliminate debris from analysis. Then, 50,000 individual events were excited with a 488-nm argon-ion laser, and emission light was detected through a 530-nm bandpass filter. The same process of fixation, wash and flow cytometric analysis method was used to follow B. melitensis endogenous AHLs activity during in vitro growth.
Cellular infection and immunofluorescence
RAW264.7 murine macrophages were inoculated at a multiplicity of infection of 300:1. Plating serial dilutions of the inocula validated infectious doses. Briefly, bacteria were centrifuged onto cells at 400 g for 10 min at 4°C and then incubated 1h at 37°C with 5% CO2 atmosphere. Cells were washed twice with medium and then incubated in medium supplemented with 50 µg ml−1 gentamicin to kill extracellular bacteria. For bacterial counts, cells were washed twice with PBS and lysed in PBS/0.1% X-100 triton (Sigma). Serial dilutions in PBS were plated onto 2YT media plates, and CFUs were counted after 3 days of incubation at 37°C. For fluorescence microscopy analysis, cells were fixed in 2% paraformaldehyde, pH 7.4, at 37°C for 15min and then processed for immunofluorescence labeling with A76-12G12 anti-LPS monoclonal primary antibody (undiluted hybridoma culture supernatant) and a goat anti-mouse IgG secondary antibody coupled to Texas Red (1/500) (Invitrogen) as previously described [79]. DAPI (4,6-diamidino-2-phenylindole) was incubated along with the secondary antibody to label DNA. Fluorescence analysis was performed with a Nikon i80 microscope. The proportion of GFP(ASV)-positive bacteria after infection by QS reporter strains was determined as follows. At each time p.i., the GFP(ASV) mean fluorescence intensity of individual bacteria was determined as the mean pixel intensity of bacteria observed with the FITC filter by using the Nikon’s imaging software NIS-Elements. We subtracted from these values the green fluorescence background (autofluorescence) that was defined as the mean fluorescence intensity of bacteria bearing the control plasmid plus 2 standard deviations. After infection with the QS reporter strains, bacteria were considered as GFP(ASV)-positive if their fluorescence signal was clearly above the background, i.e. above a threshold of fluorescence intensity arbitrarily fixed to 100.
Supporting Information
The quorum sensing reporter system allows in situ semi-quantitative detection of long chain AHLs. (A) Schematic drawings of the QS reporter strain and its control. The plasmid carried by the QS reporter strain contains the divergently transcribed Plac-lasR and PlasB-gfp(ASV) fusions. The Plac drives constitutive expression of lasR, whose product induces gfp(ASV) by direct binding of the PlasB in the presence of long-chain AHLs. The plasmid carried by the control strain does not contain the Plac-lasR fusion and is used to control that no endogenous regulator in the strain interferes with the system. Solid lines and dashed lines indicate respectively expected and unexpected interactions, based on the literature. Dotted lines on right panel indicate unknown interactions that are controlled in this study. (B) Validation of the specificity and sensitivity of the QS reporter system in E. coli. The graphs represent measurement of GFP(ASV) fluorescence intensity in bacteria by flow cytometry (5×104 events acquired) after a 4h-incubation with various AHLs. The control strain incubated with 1 µM of C12-HSL was used as a negative control. The results are representative of two independent experiments.
(TIF)
Multiple sequence alignment of Brucella AibP with characterized or predicted Ntn hydrolases. In addition to B. suis 1330 AibP (BR_A1089) and B. melitensis AibP (BMEII0212 and BMEII0211), AHL-acylases from Pseudomonas aeruginosa (PvdQ, QuiP and the putative AHL-acylase PA1893), Pseudomonas syringae (HacA and HacB), Ralstonia eutropha (predicted AHL-acylase Reut_A1841), Ralstonia strain Xj12B (AiiD), Ralstonia solanacearum (Aac) and Streptomyces strain M664 (AhlM), as well as cephalosporin acylase (CAD) from Pseudomonas diminuta and penicillin G acylase (PGA) from E. coli were used in the alignment (ClustalW). Stretches of amino acids with no similarity have been collapsed into numbers that occur in the primary sequence. The ‘*’ symbol indicates identical residues, the ‘:’ symbol indicates conserved substitutions, the ‘.’ symbol indicates semi-conserved substitution. Yellow boxes indicate conserved residues of relevance to autoproteolysis and catalysis in characterized Ntn hydrolases.
(TIFF)
Stronger response of B. melitensis QS reporter strain to AHLs in the absence of aibP . B. melitensis wt and ▵aibP QS reporter strains grown to end log phase were incubated separately for 8 hours with C12-HSL (left panel) or 3-oxo-C12-HSL (right panel) (0.1 nM or 1 nM) prior to fixation and analysis of GFP(ASV) fluorescence intensity by flow cytometry (5×104 events acquired). The B. melitensis control strain was used as a negative control. Results are representative of at least two independent experiments.
(TIF)
AHLs activity of B. melitensis QS reporter strains is lower when grown in RPMI-erythritol medium. B. melitensis wt and ▵aibP QS reporter strains were grown in 2YT or in RPMI medium supplemented with erythritol (2gl−1) for 24h before fixation and fluorescence microscopy analysis in order to determine the percentage of GFP(ASV)-positive bacteria as described in the Material and Methods section.
(TIF)
Acknowledgments
M.T. would especially like to thank Christian Didembourg for his technical assistance with the flow cytometry experiments and Cécile Nicolas for helpful discussion. M.T. and JJ.L. are very grateful to Paul Williams and Miguel Cámara for helpful discussions about QS and Quorum quenching. C.D. and JJ.L. are grateful to Michael Givskov and Morten Hentzer for the pMHLAS vector.
Funding Statement
Part of this work was funded by an ARC Convention from the French community of Belgium (no. 08/13-015) and by the Interuniversity Attraction Poles Programme initiated by the Belgian Science Policy Office (http://www.belspo.be/belspo/iap/index_en.stm). AM, JL and CD were recipients of a specialization grant from FRIA (Fonds pour la Formation à la Recherche dans l’Industrie et l’Agriculture). MT has a PhD grant as “Aspirant du FNRS’’ (http://www.fnrs.be/fr/financer-les-chercheurs/introduction.html). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Platt TG, Fuqua C (2010) What's in a name? The semantics of quorum sensing. Trends Microbiol 18: 383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ng WL, Bassler BL (2009) Bacterial quorum-sensing network architectures. Annu Rev Genet 43: 197–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Redfield RJ (2002) Is quorum sensing a side effect of diffusion sensing? Trends Microbiol 10: 365–370. [DOI] [PubMed] [Google Scholar]
- 4. Carnes EC, Lopez DM, Donegan NP, Cheung A, Gresham H, et al. (2010) Confinement-induced quorum sensing of individual Staphylococcus aureus bacteria. Nat Chem Biol 6: 41–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fuqua C, Parsek MR, Greenberg EP (2001) Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet 35: 439–468. [DOI] [PubMed] [Google Scholar]
- 6. Williams P (2007) Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology 153: 3923–3938. [DOI] [PubMed] [Google Scholar]
- 7. Parsek MR, Val DL, Hanzelka BL, Cronan JE Jr Greenberg EP (1999) Acyl homoserine-lactone quorum-sensing signal generation. Proc Natl Acad Sci U S A 96: 4360–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pearson JP, Van Delden C, Iglewski BH (1999) Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J Bacteriol 181: 1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Godfroid J, Scholz HC, Barbier T, Nicolas C, Wattiau P, et al.. (2011) Brucellosis at the animal/ecosystem/human interface at the beginning of the 21st century. Prev Vet Med. [DOI] [PubMed]
- 10. Roop RM 2nd, Gaines JM, Anderson ES, Caswell CC, Martin DW (2009) Survival of the fittest: how Brucella strains adapt to their intracellular niche in the host. Med Microbiol Immunol 198: 221–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barquero-Calvo E, Chaves-Olarte E, Weiss DS, Guzman-Verri C, Chacon-Diaz C, et al. (2007) Brucella abortus uses a stealthy strategy to avoid activation of the innate immune system during the onset of infection. PLoS One 2: e631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Taminiau B, Daykin M, Swift S, Boschiroli ML, Tibor A, et al. (2002) Identification of a quorum-sensing signal molecule in the facultative intracellular pathogen Brucella melitensis . Infect Immun 70: 3004–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Delrue RM, Deschamps C, Leonard S, Nijskens C, Danese I, et al. (2005) A quorum-sensing regulator controls expression of both the type IV secretion system and the flagellar apparatus of Brucella melitensis . Cell Microbiol 7: 1151–1161. [DOI] [PubMed] [Google Scholar]
- 14. Uzureau S, Lemaire J, Delaive E, Dieu M, Gaigneaux A, et al. (2010) Global analysis of quorum sensing targets in the intracellular pathogen Brucella melitensis 16 M. J Proteome Res. 9: 3200–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weeks JN, Galindo CL, Drake KL, Adams GL, Garner HR, et al. (2010) Brucella melitensis VjbR and C12-HSL regulons: contributions of the N-dodecanoyl homoserine lactone signaling molecule and LuxR homologue VjbR to gene expression. BMC Microbiol 10: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Uzureau S, Godefroid M, Deschamps C, Lemaire J, De Bolle X, et al. (2007) Mutations of the quorum sensing-dependent regulator VjbR lead to drastic surface modifications in Brucella melitensis . J Bacteriol 189: 6035–6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Arocena GM, Sieira R, Comerci DJ, Ugalde RA (2010) Identification of the quorum-sensing target DNA sequence and N-Acyl homoserine lactone responsiveness of the Brucella abortus virB promoter. J Bacteriol 192: 3434–3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Celli J, de Chastellier C, Franchini DM, Pizarro-Cerda J, Moreno E, et al. (2003) Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J Exp Med 198: 545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. O'Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli ML, Bourg G, et al. (1999) A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis . Mol Microbiol 33: 1210–1220. [DOI] [PubMed] [Google Scholar]
- 20. Zygmunt MS, Hagius SD, Walker JV, Elzer PH (2006) Identification of Brucella melitensis 16M genes required for bacterial survival in the caprine host. Microbes Infect 8: 2849–2854. [DOI] [PubMed] [Google Scholar]
- 21. Fretin D, Fauconnier A, Kohler S, Halling S, Leonard S, et al. (2005) The sheathed flagellum of Brucella melitensis is involved in persistence in a murine model of infection. Cell Microbiol 7: 687–698. [DOI] [PubMed] [Google Scholar]
- 22. Leonard S, Ferooz J, Haine V, Danese I, Fretin D, et al. (2007) FtcR is a new master regulator of the flagellar system of Brucella melitensis 16M with homologs in Rhizobiaceae. J Bacteriol 189: 131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petersen E, Chaudhuri P, Gourley C, Harms J, Splitter G (2011) Brucella melitensis Cyclic-di-GMP Phosphodiesterase BpdA Controls Expression of Flagellar Genes. J Bacteriol. [DOI] [PMC free article] [PubMed]
- 24. Terwagne M, Ferooz J, Rolan HG, Sun YH, Atluri V, et al. (2013) Innate immune recognition of flagellin limits systemic persistence of Brucella . Cell Microbiol 15: 942–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terwagne M, Uzureau S, Letesson JJ (2012) Brucella quorum sensing: much more than sensing quorum. In: Lopez-Goni I, O'Callaghan D, editors. Brucella : molecular microbiology and genomics. United Kingdom: Caister Academic Press. pp. 163–178.
- 26. Dong YH, Wang LH, Xu JL, Zhang HB, Zhang XF, et al. (2001) Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411: 813–817. [DOI] [PubMed] [Google Scholar]
- 27. Hentzer M, Riedel K, Rasmussen TB, Heydorn A, Andersen JB, et al. (2002) Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 148: 87–102. [DOI] [PubMed] [Google Scholar]
- 28. Ahmer BM (2004) Cell-to-cell signalling in Escherichia coli and Salmonella enterica . Mol Microbiol 52: 933–945. [DOI] [PubMed] [Google Scholar]
- 29. Uroz S, Oger PM, Chapelle E, Adeline MT, Faure D, et al. (2008) A Rhodococcus qsdA-encoded enzyme defines a novel class of large-spectrum quorum-quenching lactonases. Appl Environ Microbiol 74: 1357–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mei GY, Yan XX, Turak A, Luo ZQ, Zhang LQ (2010) AidH, an alpha/beta-hydrolase fold family member from an Ochrobactrum sp. strain, is a novel N-acylhomoserine lactonase. Appl Environ Microbiol 76: 4933–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin YH, Xu JL, Hu J, Wang LH, Ong SL, et al. (2003) Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol Microbiol 47: 849–860. [DOI] [PubMed] [Google Scholar]
- 32. Huang JJ, Han JI, Zhang LH, Leadbetter JR (2003) Utilization of acyl-homoserine lactone quorum signals for growth by a soil pseudomonad and Pseudomonas aeruginosa PAO1. Appl Environ Microbiol 69: 5941–5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shepherd RW, Lindow SE (2009) Two dissimilar N-acyl-homoserine lactone acylases of Pseudomonas syringae influence colony and biofilm morphology. Appl Environ Microbiol 75: 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sio CF, Otten LG, Cool RH, Diggle SP, Braun PG, et al. (2006) Quorum quenching by an N-acyl-homoserine lactone acylase from Pseudomonas aeruginosa PAO1. Infect Immun 74: 1673–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Duggleby HJ, Tolley SP, Hill CP, Dodson EJ, Dodson G, et al. (1995) Penicillin acylase has a single-amino-acid catalytic centre. Nature 373: 264–268. [DOI] [PubMed] [Google Scholar]
- 36. McVey CE, Walsh MA, Dodson GG, Wilson KS, Brannigan JA (2001) Crystal structures of penicillin acylase enzyme-substrate complexes: structural insights into the catalytic mechanism. J Mol Biol 313: 139–150. [DOI] [PubMed] [Google Scholar]
- 37. Kim S, Kim Y (2001) Active site residues of cephalosporin acylase are critical not only for enzymatic catalysis but also for post-translational modification. J Biol Chem 276: 48376–48381. [DOI] [PubMed] [Google Scholar]
- 38. Kim Y, Yoon K, Khang Y, Turley S, Hol WG (2000) The 2.0 A crystal structure of cephalosporin acylase. Structure 8: 1059–1068. [DOI] [PubMed] [Google Scholar]
- 39. Li Y, Chen J, Jiang W, Mao X, Zhao G, et al. (1999) In vivo post-translational processing and subunit reconstitution of cephalosporin acylase from Pseudomonas sp. 130. Eur J Biochem 262: 713–719. [DOI] [PubMed] [Google Scholar]
- 40. Godefroid M, Svensson MV, Cambier P, Uzureau S, Mirabella A, et al. (2010) Brucella melitensis 16M produces a mannan and other extracellular matrix components typical of a biofilm. FEMS Immunol Med Microbiol 59: 364–377. [DOI] [PubMed] [Google Scholar]
- 41. Moreno E, Moriyon I (2002) Brucella melitensis: a nasty bug with hidden credentials for virulence. Proc Natl Acad Sci U S A 99: 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sieira R, Comerci DJ, Sanchez DO, Ugalde RA (2000) A homologue of an operon required for DNA transfer in Agrobacterium is required in Brucella abortus for virulence and intracellular multiplication. J Bacteriol 182: 4849–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ferooz J, Lemaire J, Letesson JJ (2011) Role of FlbT in flagellin production in Brucella melitensis . Microbiology 157: 1253–1262. [DOI] [PubMed] [Google Scholar]
- 44. Rambow-Larsen AA, Rajashekara G, Petersen E, Splitter G (2008) Putative quorum-sensing regulator BlxR of Brucella melitensis regulates virulence factors including the type IV secretion system and flagella. J Bacteriol 190: 3274–3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Khan SR, Farrand SK (2009) The BlcC (AttM) lactonase of Agrobacterium tumefaciens does not quench the quorum-sensing system that regulates Ti plasmid conjugative transfer. J Bacteriol 191: 1320–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Patankar AV, Gonzalez JE (2009) Orphan LuxR regulators of quorum sensing. FEMS Microbiol Rev 33: 739–756. [DOI] [PubMed] [Google Scholar]
- 47. Subramoni S, Venturi V (2009) LuxR-family 'solos': bachelor sensors/regulators of signalling molecules. Microbiology 155: 1377–1385. [DOI] [PubMed] [Google Scholar]
- 48. Danino VE, Wilkinson A, Edwards A, Downie JA (2003) Recipient-induced transfer of the symbiotic plasmid pRL1JI in Rhizobium leguminosarum bv. viciae is regulated by a quorum-sensing relay. Mol Microbiol 50: 511–525. [DOI] [PubMed] [Google Scholar]
- 49. Hoang HH, Becker A, Gonzalez JE (2004) The LuxR homolog ExpR, in combination with the Sin quorum sensing system, plays a central role in Sinorhizobium meliloti gene expression. J Bacteriol 186: 5460–5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee JH, Lequette Y, Greenberg EP (2006) Activity of purified QscR, a Pseudomonas aeruginosa orphan quorum-sensing transcription factor. Mol Microbiol 59: 602–609. [DOI] [PubMed] [Google Scholar]
- 51. Ferluga S, Bigirimana J, Hofte M, Venturi V (2007) A LuxR homologue of Xanthomonas oryzae pv. oryzae is required for optimal rice virulence. Mol Plant Pathol 8: 529–538. [DOI] [PubMed] [Google Scholar]
- 52.Brachmann AO, Brameyer S, Kresovic D, Hitkova I, Kopp Y, et al.. (2013) Pyrones as bacterial signaling molecules. Nat Chem Biol. [DOI] [PubMed]
- 53. Tait K, Williamson H, Atkinson S, Williams P, Camara M, et al. (2009) Turnover of quorum sensing signal molecules modulates cross-kingdom signalling. Environ Microbiol 11: 1792–1802. [DOI] [PubMed] [Google Scholar]
- 54. Chen CN, Chen CJ, Liao CT, Lee CY (2009) A probable aculeacin A acylase from the Ralstonia solanacearum GMI1000 is N-acyl-homoserine lactone acylase with quorum-quenching activity. BMC Microbiol 9: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huang JJ, Petersen A, Whiteley M, Leadbetter JR (2006) Identification of QuiP, the product of gene PA1032, as the second acyl-homoserine lactone acylase of Pseudomonas aeruginosa PAO1. Appl Environ Microbiol 72: 1190–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Morohoshi T, Nakazawa S, Ebata A, Kato N, Ikeda T (2008) Identification and characterization of N-acylhomoserine lactone-acylase from the fish intestinal Shewanella sp. strain MIB015. Biosci Biotechnol Biochem 72: 1887–1893. [DOI] [PubMed] [Google Scholar]
- 57. Romero M, Diggle SP, Heeb S, Camara M, Otero A (2008) Quorum quenching activity in Anabaena sp. PCC 7120: identification of AiiC, a novel AHL-acylase. FEMS Microbiol Lett 280: 73–80. [DOI] [PubMed] [Google Scholar]
- 58. Uroz S, Dessaux Y, Oger P (2009) Quorum sensing and quorum quenching: the yin and yang of bacterial communication. Chembiochem 10: 205–216. [DOI] [PubMed] [Google Scholar]
- 59. Dong YH, Wang LY, Zhang LH (2007) Quorum-quenching microbial infections: mechanisms and implications. Philos Trans R Soc Lond B Biol Sci 362: 1201–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Leadbetter JR, Greenberg EP (2000) Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus . J Bacteriol 182: 6921–6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Eberhard A, Burlingame AL, Eberhard C, Kenyon GL, Nealson KH, et al. (1981) Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 20: 2444–2449. [DOI] [PubMed] [Google Scholar]
- 62. Zhang HB, Wang LH, Zhang LH (2002) Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens . Proc Natl Acad Sci U S A 99: 4638–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pearson JP, Passador L, Iglewski BH, Greenberg EP (1995) A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa . Proc Natl Acad Sci U S A 92: 1490–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lamont IL, Martin LW (2003) Identification and characterization of novel pyoverdine synthesis genes in Pseudomonas aeruginosa . Microbiology 149: 833–842. [DOI] [PubMed] [Google Scholar]
- 65. White CE, Finan TM (2009) Quorum quenching in Agrobacterium tumefaciens: chance or necessity? J Bacteriol 191: 1123–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hastings JW, Nealson KH (1977) Bacterial bioluminescence. Annu Rev Microbiol 31: 549–595. [DOI] [PubMed] [Google Scholar]
- 67. Boyer M, Wisniewski-Dye F (2009) Cell-cell signalling in bacteria: not simply a matter of quorum. FEMS Microbiol Ecol 70: 1–19. [DOI] [PubMed] [Google Scholar]
- 68. Qazi SN, Counil E, Morrissey J, Rees CE, Cockayne A, et al. (2001) agr expression precedes escape of internalized Staphylococcus aureus from the host endosome. Infect Immun 69: 7074–7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sieira R, Comerci DJ, Pietrasanta LI, Ugalde RA (2004) Integration host factor is involved in transcriptional regulation of the Brucella abortus virB operon. Mol Microbiol 54: 808–822. [DOI] [PubMed] [Google Scholar]
- 70. Barbier T, Nicolas C, Letesson JJ (2011) Brucella adaptation and survival at the crossroad of metabolism and virulence. FEBS Lett 585: 2929–2934. [DOI] [PubMed] [Google Scholar]
- 71. Roche DM, Byers JT, Smith DS, Glansdorp FG, Spring DR, et al. (2004) Communications blackout? Do N-acylhomoserine-lactone-degrading enzymes have any role in quorum sensing? Microbiology 150: 2023–2028. [DOI] [PubMed] [Google Scholar]
- 72. Simon R, Priefer U, Pühler A (1983) A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology 10: 783–791. [Google Scholar]
- 73.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, et al.. (1991) Current protocols in Molecular Biology. New-York: John Wiley & Sons.
- 74. Kovach ME, Phillips RW, Elzer PH, Roop RM 2nd, Peterson KM (1994) pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16: 800–802. [PubMed] [Google Scholar]
- 75. Dozot M, Boigegrain RA, Delrue RM, Hallez R, Ouahrani-Bettache S, et al. (2006) The stringent response mediator Rsh is required for Brucella melitensis and Brucella suis virulence, and for expression of the type IV secretion system virB. Cell Microbiol 8: 1791–1802. [DOI] [PubMed] [Google Scholar]
- 76. Rouot B, Alvarez-Martinez MT, Marius C, Menanteau P, Guilloteau L, et al. (2003) Production of the type IV secretion system differs among Brucella species as revealed with VirB5- and VirB8-specific antisera. Infect Immun 71: 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mirabella A, Yanez Villanueva RM, Delrue RM, Uzureau S, Zygmunt MS, et al. (2012) The two-component system PrlS/PrlR of Brucella melitensis is required for persistence in mice and appears to respond to ionic strength. Microbiology 158: 2642–2651. [DOI] [PubMed] [Google Scholar]
- 78. Jubier-Maurin V, Rodrigue A, Ouahrani-Bettache S, Layssac M, Mandrand-Berthelot MA, et al. (2001) Identification of the nik gene cluster of Brucella suis: regulation and contribution to urease activity. J Bacteriol 183: 426–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pizarro-Cerda J, Meresse S, Parton RG, van der Goot G, Sola-Landa A, et al. (1998) Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect Immun 66: 5711–5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The quorum sensing reporter system allows in situ semi-quantitative detection of long chain AHLs. (A) Schematic drawings of the QS reporter strain and its control. The plasmid carried by the QS reporter strain contains the divergently transcribed Plac-lasR and PlasB-gfp(ASV) fusions. The Plac drives constitutive expression of lasR, whose product induces gfp(ASV) by direct binding of the PlasB in the presence of long-chain AHLs. The plasmid carried by the control strain does not contain the Plac-lasR fusion and is used to control that no endogenous regulator in the strain interferes with the system. Solid lines and dashed lines indicate respectively expected and unexpected interactions, based on the literature. Dotted lines on right panel indicate unknown interactions that are controlled in this study. (B) Validation of the specificity and sensitivity of the QS reporter system in E. coli. The graphs represent measurement of GFP(ASV) fluorescence intensity in bacteria by flow cytometry (5×104 events acquired) after a 4h-incubation with various AHLs. The control strain incubated with 1 µM of C12-HSL was used as a negative control. The results are representative of two independent experiments.
(TIF)
Multiple sequence alignment of Brucella AibP with characterized or predicted Ntn hydrolases. In addition to B. suis 1330 AibP (BR_A1089) and B. melitensis AibP (BMEII0212 and BMEII0211), AHL-acylases from Pseudomonas aeruginosa (PvdQ, QuiP and the putative AHL-acylase PA1893), Pseudomonas syringae (HacA and HacB), Ralstonia eutropha (predicted AHL-acylase Reut_A1841), Ralstonia strain Xj12B (AiiD), Ralstonia solanacearum (Aac) and Streptomyces strain M664 (AhlM), as well as cephalosporin acylase (CAD) from Pseudomonas diminuta and penicillin G acylase (PGA) from E. coli were used in the alignment (ClustalW). Stretches of amino acids with no similarity have been collapsed into numbers that occur in the primary sequence. The ‘*’ symbol indicates identical residues, the ‘:’ symbol indicates conserved substitutions, the ‘.’ symbol indicates semi-conserved substitution. Yellow boxes indicate conserved residues of relevance to autoproteolysis and catalysis in characterized Ntn hydrolases.
(TIFF)
Stronger response of B. melitensis QS reporter strain to AHLs in the absence of aibP . B. melitensis wt and ▵aibP QS reporter strains grown to end log phase were incubated separately for 8 hours with C12-HSL (left panel) or 3-oxo-C12-HSL (right panel) (0.1 nM or 1 nM) prior to fixation and analysis of GFP(ASV) fluorescence intensity by flow cytometry (5×104 events acquired). The B. melitensis control strain was used as a negative control. Results are representative of at least two independent experiments.
(TIF)
AHLs activity of B. melitensis QS reporter strains is lower when grown in RPMI-erythritol medium. B. melitensis wt and ▵aibP QS reporter strains were grown in 2YT or in RPMI medium supplemented with erythritol (2gl−1) for 24h before fixation and fluorescence microscopy analysis in order to determine the percentage of GFP(ASV)-positive bacteria as described in the Material and Methods section.
(TIF)