Abstract
Polymyxins are often considered as a last resort to treat multidrug resistant P. aeruginosa but polymyxin resistance has been increasingly reported worldwide in clinical isolates. Polymyxin resistance in P. aeruginosa is known to be associated with alterations in either PhoQ or PmrB. In this study, mutant strains of P. aeruginosa carrying amino acid substitution, a single and/or dual inactivation of PhoQ and PmrB were constructed to further understand the roles of PhoQ and PmrB in polymyxin susceptibility. Polymyxin B resistance was caused by both inactivation and/or amino acid substitutions in PhoQ but by only amino acid substitutions of PmrB. Alterations of both PhoQ and PmrB resulted in higher levels of polymyxin B resistance than alteration of either PhoQ or PmrB alone. These results were confirmed by time-killing assays suggesting that high-level polymyxin resistance in P. aeruginosa is caused by alterations of both PhoQ and PmrB.
Keywords: Pseudomonas aeruginosa, Polymyxin B Resistance, phoPQ, pmrAB
1. Introduction
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen that is a frequent cause of hospital-acquired infections [1–5]. P. aeruginosa accounts for 11% to 14% of all nosocomial infections and is a major problem for people hospitalized with cancer, cystic fibrosis, or burns [2]. Treatment usually involves the use of one or more antibiotics such as β-lactams, aminoglycosides, or quinolones. Combination therapy is usually recommended as it reduces the risk of antibiotic resistance and enhances the eradication rate. Despite the use of combination therapy, there are numerous reports of emergence of multidrug resistant P. aeruginosa. When multidrug P. aeruginosa occurs in critically ill patients, polymyxins are often the last resort for treatment of the infections [6,7]. However, polymyxin B associated multidrug resistant P. aeruginosa have been increasingly reported worldwide [8–11].
Polymyxins are a group of cationic polypeptide antibiotics consisting of five different compounds (A to E); only polymyxin B and E (colistin) have been used in clinical practice. Polymyxin B differs by only one amino acid from polymyxin E and both have essentially the same antibacterial spectrum, susceptibilities, and resistant mechanisms [12,13]. Polymyxins interacts with lipopolysaccharide (LPS) of the outer membrane of Gram-negative bacteria and displace the calcium and magnesium bridges that stabilize the LPS disrupting the outer membrane producing cell death [14]. Based on in vitro studies using P. aeruginosa PAO1, it is known that polymyxin B resistance can be induced by arnBCADTEF that is modulated through a two-component regulatory systems of phoPQ and pmrAB under culture conditions limiting magnesium (e.g., 20 μM) [15–18]. In clinical isolates using normal culture conditions (e.g., Luria-Bertani or Mueller Hinton) for P. aeruginosa, polymyxin B resistance has been associated with alterations in either PhoQ or PmrB [1,19]. Polymyxin B resistant P. aeruginosa PAK selected in vitro by increasing concentrations of polymyxin B was found to have amino acid substitutions in PmrB [20]. Other studies using P. aeruginosa PAO1 suggested cross-talk between the phoPQ and pmrAB systems [17,18]. Although involvement of PhoQ and PmrB in polymyxin B resistance is known in P. aeruginosa, molecular details of polymyxin susceptibility remain unclear including the degree of polymyxin susceptibility and the correlation with alterations in PhoQ and/or PmrB. In this study, polymyxin susceptibility was examined in P. aeruginosa harboring inactivation and/or amino acid substitutions in PhoQ and/or PmrB.
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
P. aeruginosa PAO1 and clinical isolates of P. aeruginosa (M38100B4, NY214, NY215, NY220) reported previously [1,19] were used for this study. E. coli strains (DH5α and SM10) were also used for general cloning experiments. All bacterial strains were routinely cultured in Luria-Bertani (LB) medium. When needed, antibiotics were supplemented in the LB medium as ampicillin (100 μg/mL), gentamicin (15 μg/mL), and tetracycline (10 μg/mL) for E. coli. Antibiotics were also added to LB medium to select P. aeruginosa gene-knockout strains using carbenicillin (100 μg/mL), gentamicin (80 μg/mL), and tetracycline (80 μg/mL). Divalent cation-adjusted Mueller-Hinton (MH) (Oxoid, Ogdensburg, New York) broth was used for antibiotic susceptibility testing. All antibiotics and other chemicals used in this study were purchased from Sigma (Sigma, St. Louis, MO). Solutions of these compounds were prepared by dissolving in sterile doubledistilled water or the solvent suggested by the manufacturer and sterilized by filtering through the 0.4 μm disposable membranes (Millipore, Billerica, Massa-chusetts). The final pH of prepared medium was ~7.0 after appropriate adjustments.
2.2. Antibiotic Susceptibility Testing
Antibiotic susceptibility was determined as minimal inhibitory concentrations (MICs) using the broth dilution method as described [25]. Briefly, polymyxin B was added to MH broth to achieve serial two-fold dilutions between 0.25 and 32 μg/mL using sterile 17 × 100 mm snapped-cap Falcon culture tubes (1 mL/tube; Fisher Scientific). Fresh overnight cultures of each bacterial strain were diluted in saline to an optical density at 600 nm of 0.08 to 0.12 (approximately 1 × 108 viable cells per mL). A portion of the adjusted cell suspension (5 μL for ~105–6 cells) was inoculated to each MH broth containing polymyxin B as indicated. The cell cultures were then incubated overnight (16 to 18 hrs) at 37°C without shaking. The MIC was defined as the lowest concentration that completely inhibited growth of the inoculums. MIC values were confirmed by three independent experiments.
2.3. Genomic DNA Extraction, PCR and DNA Sequence Analysis
Genomic DNA extraction, PCR amplification, and cloning experiment were performed as described [19]. PCR fragments were used to determine DNA sequences by a commercial DNA sequencing service (GENEWIZ, South Plainfield, NJ).
2.4. Construction of Gene-Knockout Strains
The gene phoQ was knocked-out by inserting a gentamicin resistance cassette (Gm) from pGMΩ1 [21] into NruI of the 3060-bp DNA fragment containing oprH- phoPQ. The gene pmrB was knocked-out by inserting a tetracycline resistance cassette (Tc) derived from Tn5 (Epicentre Biotechnologies, Madison, WI) into PstI of the 1437-bp DNA fragment containing pmrB. To delete the genes of oprH-phoPQ a 1906-bp PstI fragment from the 3060-bp fragment containing oprH-phoPQ was replaced by the Gm cassette. To delete the genes of pmrAB, a 1468-bp PstI fragment from the 3157-bp SalI/SacI fragment containing pmrAB was replaced by the Gm cassette. The DNA fragments carrying the gene-knockout or -deletion were inserted into the conjugative plasmid of pRTP1 [22] and the resulting plasmids were used to knockout the genes for P. aeruginosa by biparental conjugation methods as described [23]. The gene replacement by Gm or Tc was confirmed for authenticity by PCR methods as described [24].
2.5. Time-Killing Assay
P. aeruginosa strains were grown overnight in LB broth. The cells were diluted in saline to an optical density at 600 nm of 0.08 to 0.12 and 5 μL of each diluted cells (~105–6 cells) were inoculated into 1 mL of MH broth supplemented with appropriate amount of polymyxin B, following incubation at 37°C. Aliquots (100 μL) were withdrawn at specific time intervals (4, 8, and 12 hours) and spread on LB agar plates with further dilution before spreading on plain LB agar plates. After overnight incubation (16 to 18 hours) at 37°C, cells that survived were counted as colony forming units (CFU).
3. Results and Discussions
3.1. A Single Inactivation of PhoQ or PmrB in Polymyxin B Susceptibility
P. aeruginosa PAO1 and a clinical isolate of P. aeruginosa M38100B4 were used to study the effects of PhoQ- or PmrB-inactivation on polymyxin B susceptibility. P. aeruginosa PAO1 carrying PhoQ-inactivation showed an MIC of 4 μg/mL of polymyxin B whereas the parental PAO1 MIC was 0.5 μg/mL. Polymyxin B susceptibility of the PAO1 carrying the PhoQ-inactivation was fully restored by introducing an intact clone of oprH-PhoPQ from the parental PAO1. These results are consistent with the previous observation that truncated-PhoQ in the clinical isolate of P. aeruginosa yielded an MIC of 8 μg/mL of polymyxin B. The PAO1 carrying PmrB-inactivation showed no significant change of MIC (0.5 μg/mL of polymyxin B) and inactivation of both PhoQ and PmrB retained the effect of only inactivation of PhoQ (MIC 4 μg/mL), indicating that inactivation of PmrB did not affect polymyxin susceptibility. Previous reports however showed that a single amino acid substitution (M292T) in PmrB increased the MIC of polymyxin B to 8 μg/mL [1] and inactivation of PmrB in this isolate decreased the MIC to 0.5 μg/mL. The isolate carrying a single amino acid substitution in PmrB was used to inactivate PhoQ resulting in an MIC of 16 μg/mL with full complementation of susceptibility by an intact clone of oprH- phoPQ (Table 1). These results suggest that amino acid substitutions of PmrB rather than inactivation whereas either inactivation or amino acid substitutions of PhoQ results in polymyxin resistance. These results also suggest that although both PhoQ and PmrB play roles in polymyxin B resistance, their specific roles in polymyxin B susceptibility may be different.
Table 1.
MICs of P. aeruginosa strains to polymyxin B.
| Strains | MICs to polymyxins (μg/mL)a |
|---|---|
| PAO1 | 0.5 |
| PAO1 (phoQ::Gm) | 4 |
| PAO1 (phoQ::Gm)/pAU124b | 0.25 |
| PAO1 (pmrB::Tc) | 0.5 |
| PAO1 (phoQ::Gm/pmrB::Tc) | 4 |
| M38100B4c | 8 |
| M38100B4 (phoQ::Tc) | 16 |
| M38100B4 (phoQ::Tc)/pAU124 | 8 |
| M38100B4 (pmrB::Tc) | 0.5 |
MIC measurement repeated three times with identical results.
pAU124 carries a full length of oprH-phoPQ from P. aeruginosa PAO1.
M38100B4 carried a single amino acid substitution (M292T) in PmrB (1).
3.2. Dual Alterations of PhoQ and PmrB in Polymyxin B Susceptibility
Amino acid substitutions in either PhoQ or PmrB showed MIC’s of 4 to 8 μg/mL among clinical isolates of P. aeruginosa [1,19]. To examine the effects of alterations in both PhoQ and PmrB on polymyxin B susceptibility, P. aeruginosa PAO1 and the clinical P. aeruginosa isolate M38100B4 were used following alterations in both PhoQ and PmrB. P. aeruginosa PAO1 carrying an PhoQ-inactivation and deletion of a full length of pmrAB (phoQ:: Tc/ΔpmrAB::Gm) was constructed and this dual mutant strain was transformed by plasmids carrying amino acid substitutions in PmrB, M292T for pAU129 and A247T/ Y345H for pAU154 as reported previously [1,19]. The dual mutant strain retained an MIC of 4 μg/mL which was the same MIC as found with PhoQ-inactivation alone. However, the mutant strain carrying the plasmids (pAU129 or pAU154) showed an MIC of 8 μg/mL whereas the MIC of the same mutant strain carrying the cloning vector (pUCP18) was unchanged (4 μg/mL) (Table 2). Similarly, the polymyxin B resistant clinical isolate M38100B4 carrying M292T in PmrB was used to construct a dual mutant strain carrying a deletion of the full length of oprH-phoPQ. This mutant strain was transformed by plasmids carrying amino acid substitutions or inactivated-PhoQ, V260G for pAU146, truncated-PhoQ for pAU147, H223R for pAU148 reported previously [19]. The clinical isolate M38100B4 in which the full length of oprH-phoPQ was deleted retained its MIC of 8 μg/mL. However, the same mutant strain carrying the amino acid substitutions in PhoQ or truncated-PhoQ showed an MIC of 16 μg/mL; the cloning vector alone had no changed in MIC (Table 2). These results suggest that the alterations in both PhoQ and PmrB cause higher level resistance to polymyxin B than that of alterations in either PhoQ or PmrB alone.
Table 2.
MICs of mutant P. aeruginosa to polymyxin B.
| Strains | MICs to polymyxin B (μg/mL)a |
|---|---|
| PAO1 (phoQ::Tc/ΔpmrAB::Gm) | 4 |
| PAO1 (phoQ::Tc/ΔpmrAB::Gm)/pAU129 | 8 |
| PAO1 (phoQ::Tc/ΔpmrAB::Gm)/pAU154 | 8 |
| PAO1 (phoQ::Tc/ΔpmrAB::Gm)/pUCP18 (vector) | 4 |
| M38100B4 (ΔoprH-phoPQ::Gm) | 8 |
| M38100B4 (ΔoprH-phoPQ::Gm)/pAU146 | 16 |
| M38100B4 (ΔoprH-phoPQ::Gm)/pAU147 | 16 |
| M38100B4 (ΔoprH-phoPQ::Gm)/pAU148 | 16 |
| M38100B4 (ΔoprH-phoPQ::Gm)/pUCP18 (vector) | 8 |
MIC measurement repeated three times with identical results.
Note: pAU129, amino acid substitution (M292T) in PmrB; pAU154, amino acid substitutions (A247T; Y345H) in PmrB; pAU146, amino acid substitution (V260G) in PhoQ; pAU147, truncated-PhoQ; pAU148, amino acid substitution (H223R) in PhoQ (see details in the text).
The effect of alterations of both PhoQ and PmrB was confirmed by time-killing assays using the PAO1 mutant strain (phoQ::Tc/ΔpmrAB::Gm) carrying a plasmid pAU129 (M292T in PmrB), the same mutant strain carrying a cloning vector (pUCP18), and the parental strain PAO1. As shown in Figure 1(a), the PAO1 mutant strain carrying alterations in both PhoQ and PmrB survived with significantly higher numbers of cells than the same strain carrying a single alteration (PhoQ inactivation). In contrast, the parental PAO1 was killed within 12 hours in the presence of 4 μg/mL of polymyxin B. Similarly, the clinical isolate M38100B4 carrying an inactivated-PhoQ (i.e., alterations in both PhoQ and PmrB) also survived at significantly higher numbers of cells than that of the parental M38100B4 (Figure 1(b)). These results confirmed the above observations that P. aeruginosa carrying alterations in both PhoQ and PmrB resulted in higher levels of polymyxin B resistance than that of alterations in either PhoQ or PmrB alone.
Figure 1.
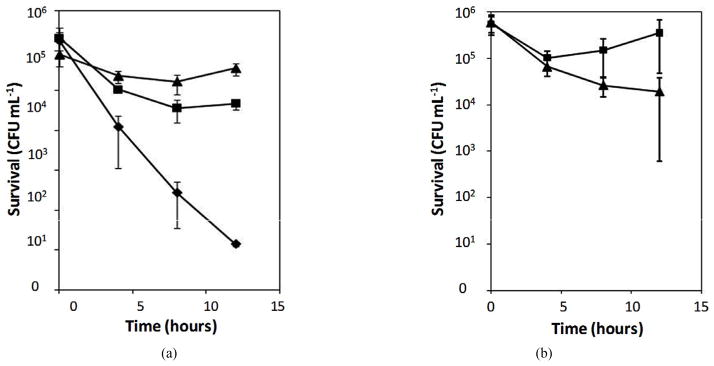
Time-killing assays of Pseudomonas aeruginosa. Time-killing assays were performed using 4 and 8 μg/mL of polymyxin B for PAO1 (a) and clinical isolate M38100B4 (b), respectively, as described in Materials and Methods. The clinical isolate M38100B4 carries an amino acid substitution (M292T) in PmrB causing polymyxin B resistance (MIC 8 μg/mL) as reported (1). Error bars were obtained from three independent assays. (a) Diamond, a strain PAO1; triangle, a strain PAO1 (phoQ::Gm/ΔpmrAB::Tc)/pAU129 (pmrAB carrying M292T in PmrB); square, a strain PAO1 (phoQ::Gm/ΔpmrAB::Tc) /pAU126 (intact pmrAB from PAO1); (b) Triangle, a strain M38100B4; squares, a strain M38100B4 carrying inactivated-PhoQ. The clones of pAU126 and pAU129 were used from the previous reports (1).
To further substantiate the effects of alterations in both PhoQ and PmrB 3 clinical isolates (NY214, NY215, and NY220) of P. aeruginosa reported previously (19) were used to induce high-level resistance to polymyxin B under polymyxin B selection pressure. The 3 clinical isolates were incubated in LB broth containing 4 μg/mL of polymyxin B for 24 hours and plated on LB agar plates containing 8 μg/mL of polymyxin B. A single colony from each culture was used for 3 additional passages on plain LB agar plates. Three colonies from the third passage of each culture showed high-level resistance to polymyxin B (MICs of 16 to 32 μg/mL). DNA sequence analysis revealed that the high-level resistant colonies harbored inactivation and/or amino acid substitutions of both PhoQ and PmrB (Table 3). These results clearly demonstrated that alterations in both PhoQ and PmrB resulted in high-level polymyxin B resistance among clinical P. aeruginosa isolates.
Table 3.
DNA sequence analysis of PhoQ and PmrB from polymyxin B resistant clinical isolates of P. aeruginosa and their MICs.
| P. aeruginosa isolatesa | Amino acid alterationb
|
MICs to polymyxin B (μg/mL)c | |
|---|---|---|---|
| PhoQ | PmrB | ||
| NY214R1 | Amino acid substitution at V260G | Amino acid substitution at M292T | 16 |
| NY214R2 | Amino acid substitution at V260G | Amino acid substitution at M292T | 16 |
| NY214R3 | Amino acid substitution at V260G | Amino acid substitution at M292T | 16 |
| NY215R1 | Frame-shift mutation by 17 nucleotides deletion from 1278818 with truncated-protein at position 152nd amino acid | Amino acid substitution at A247T | 16 |
| NY215R2 | Frame-shift mutation by 17 nucleotides deletion from 1278818 with truncated-protein at position 152nd amino acid | Amino acid substitution at A247T | 16 |
| NY215R3 | Frame-shift mutation by 17 nucleotides deletion from 1278818 with truncated-protein at position 152nd amino acid | Amino acid substitution at Y345H | 16 |
| NY220R1 | Amino acid substitution at V260G | Amino acid substitution at A247T and Y345H | 32 |
| NY220R2 | Amino acid substitution at V260G | Amino acid substitution at A247T and Y345H | 32 |
| NY220R3 | Amino acid substitution at H223R | Amino acid substitution at Y345H | 16 |
Polymyxin B resistant strains of NY214R, NY215R, and NY220R were induced from NY214, NY215, and NY220 reported previously (19); see details in the text.
A full length of PhoQ (1347-bp; 448 amino acids) and of PmrB (1434-bp; 477 amino acids) was compared to that of P. aeruginosa PAO1 (www.pseudomonas.com).
MIC measurement repeated three times with identical results.
In this study, three different approaches were used to understand the roles of PhoQ and PmrB alterations in relation to polymyxin B susceptibility. The clinical isolate M38100B4 had polymyxin B resistance (MIC 8 μg/mL) due to a single amino acid substitution in PmrB which increased MIC from 8 to 16 μg/mL after inactivation of PhoQ. The second approach was alterations of both PhoQ and PmrB, which showed higher levels of polymyxin B resistance than those with alterations of either PhoQ or PmrB alone. The third was that the time-killing rate of strains carrying alterations of both PhoQ and PmrB was much slower than that of the strains carrying alterations in either PhoQ or PmrB alone. Lastly, 3 clinical isolates were used to substantiate the roles of alterations in both PhoQ and PmrB in polymyxin B resistance. All of the observations demonstrate that the alterations in both PhoQ and PmrB result in higher levels of polymyxin B resistance than do alterations in either PhoQ or PmrB alone.
Acknowledgments
We are grateful to Dr. David Y. Graham for critical review of the manuscript. This study was supported by a grant from NIH (1 SC3 GM094053-01).
References
- 1.Abraham N, Kwon DH. A Single Amino Acid Substitution in PmrB Is Associated with Polymyxin B Resistance in Clinical Isolate of Pseudomonas aeruginosa. FEMS Microbiology Letters. 2009;298(2):249–254. doi: 10.1111/j.1574-6968.2009.01720.x. [DOI] [PubMed] [Google Scholar]
- 2.Driscoll JA, Brody SL, Kollef MH. The Epidemiology, Pathogenesis and Treatment of Pseudomonas aeruginosa Infections. Drugs. 2007;67(3):351–368. doi: 10.2165/00003495-200767030-00003. [DOI] [PubMed] [Google Scholar]
- 3.Lodise TP, Miller CD, Graves J, Furuno JP, McGregor JC, Lomaestro B, et al. Clinical Prediction Tool to Identify Patients with Pseudomonas aeruginosa Respiratory Tract Infections at Greatest Risk for Multidrug Resistance. Antimicrobial Agents and Chemotherapy. 2007;51(2):417–422. doi: 10.1128/AAC.00851-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McVay CS, Velasquez M, Fralick JA. Phage Therapy of Pseudomonas aeruginosa Infection in a Mouse Burn Wound Model. Antimicrobial Agents and Chemotherapy. 2007;51(6):1934–1938. doi: 10.1128/AAC.01028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mittal R, Khandwaha RK, Gupta V, Mittal PK, Harjai K. Phenotypic Characters of Urinary Isolates of Pseudomonas aeruginosa & Their Association with Mouse Renal Colonization. The Indian Journal of Medical Research. 2006;123(1):67–72. [PubMed] [Google Scholar]
- 6.Li J, Nation RL, Turnidge JD, Milne RW, Coulthard K, Rayner CR, et al. Colistin: The Re-Emerging Antibiotic for Multidrug-Resistant Gram-Negative Bacterial Infections. The Lancet Infectious Diseases. 2006;6(9):589–601. doi: 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- 7.Arnold TM, Forres GN, Messmer KJ. Polymyxin Antibiotics for Gram-Negative Infections. American Journal of Health-System Pharmacy. 2007;64(8):819–826. doi: 10.2146/ajhp060473. [DOI] [PubMed] [Google Scholar]
- 8.Falagas ME, Rafailidis PI, Matthaiou DK, Virtzili S, Nikita D, Michalopoulos A. Pandrug-Resistant Klebsiella pneumoniae, Pseudomonas aeruginosa and Acinetobacter baumannii Infections: Characteristics and Outcome in a Series of 28 Patients. International Journal of Antimicrobial Agents. 2008;32(5):450–454. doi: 10.1016/j.ijantimicag.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 9.Wang CY, Jerng JS, Chen KY, Lee LN, Yu CJ, Hsueh PR, et al. Pandrug-Resistant Pseudomonas aeruginosa among Hospitalised Patients: Clinical Features, Risk-Factors and Outcomes. Clinical Microbiology and Infection. 2006;12(1):63–68. doi: 10.1111/j.1469-0691.2005.01305.x. [DOI] [PubMed] [Google Scholar]
- 10.Falagas ME, Bliziotis IA. Pandrug-Resistant Gram-Negative Bacteria: The Dawn of the Post-Antibiotic Era? International Journal of Antimicrobial Agents. 2007;29(6):630–636. doi: 10.1016/j.ijantimicag.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 11.Landman D, Bratu S, Alam M, Quale J. Citywide Emergence of Pseudomonas aeruginosa Strains with Reduced Susceptibility to Polymyxin B. Journal of Antimicrobial Chemotherapy. 2005;55(6):954–957. doi: 10.1093/jac/dki153. [DOI] [PubMed] [Google Scholar]
- 12.Gupta S, Govil D, Kakar PN, Prakash O, Arora D, Das S, et al. Colistin and Polymyxin B: A Re-Emergence. Indian Journal of Critical Care Medicine. 2009;13(2):49–53. doi: 10.4103/0972-5229.56048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zavascki AP, Goldani LZ, Li J, Nation RL. Polymyxin B for the Treatment of Multidrug-Resistant Pathogens: A Critical Review. Journal of Antimicrobial Chemotherapy. 2007;60(6):1206–1215. doi: 10.1093/jac/dkm357. [DOI] [PubMed] [Google Scholar]
- 14.Yeaman MR, Yount NY. Mechanisms of Antimicrobial Peptide Action and Resistance. Pharmacological Reviews. 2003;55(1):27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 15.Macfarlane EL, Kwasnicka A, Hancock RE. Role of Pseudomonas aeruginosa PhoP-PhoQ in Resistance to Antimicrobial Cationic Peptides and Aminoglycosides. Microbiology. 2000;146(10):2543–2554. doi: 10.1099/00221287-146-10-2543. [DOI] [PubMed] [Google Scholar]
- 16.Macfarlane EL, Kwasnicka A, Ochs MM, Hancock RE. PhoP-PhoQ Homologues in Pseudomonas aeruginosa Regulate Expression of the Outer-Membrane Protein OprH and Polymyxin B Resistance. Molecular Microbiology. 1999;34(2):305–316. doi: 10.1046/j.1365-2958.1999.01600.x. [DOI] [PubMed] [Google Scholar]
- 17.McPhee JB, Bains M, Winsor G, Lewenza S, Kwasnicka A, Brazas MD, et al. Contribution of the PhoP-PhoQ and PmrA-PmrB Two-Component Regulatory Systems to Mg2+-Induced Gene Regulation in Pseudomonas aeruginosa,” Journal of. Bacteriology. 2006;188(11):3995–4006. doi: 10.1128/JB.00053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McPhee JB, Lewenza S, Hancock RE. Cationic Antimicrobial Peptides Activate a Two-Component Regulatory System, PmrA-PmrB, That Regulates Resistance to Polymyxin B and Cationic Antimicrobial Peptides in Pseudomonas aeruginosa. Molecular Microbiology. 2003;50(1):205–217. doi: 10.1046/j.1365-2958.2003.03673.x. [DOI] [PubMed] [Google Scholar]
- 19.Barrow K, Kwon DH. Alterations in Two-Component Regulatory Systems of phoPQ and pmrAB Are Associated with Polymyxin B Resistance in Clinical Isolates of Pseudomonas aeruginosa. Antimicrobial Agents and Chemotherapy. 2009;53(12):5150–5154. doi: 10.1128/AAC.00893-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moskowitz SM, Ernst RK, Miller SI. pmrAB, a Two-Component Regulatory System of Pseudomonas aeruginosa That Modulates Resistance to Cationic Antimicrobial Peptides and Addition of Aminoarabinose to Lipid A,” Journal of. Bacteriology. 2004;186(2):575–579. doi: 10.1128/JB.186.2.575-579.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park SM, Lu CD, Abdelal AT. Cloning and Characterization of argR, a Gene That Participates in Regulation of Arginine Biosynthesis and Catabolism in Pseudomonas aeruginosa PAO1,” Journal of. Bacteriology. 1997;179(17):5300–5308. doi: 10.1128/jb.179.17.5300-5308.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stibitz S, Black W, Falkow S. The Construction of a Cloning Vector Designed for Gene Replacement in Bordetella pertussis. Gene. 1986;50(1–3):133–140. doi: 10.1016/0378-1119(86)90318-5. [DOI] [PubMed] [Google Scholar]
- 23.Gambello MJ, Iglewski BH. Cloning and Characterization of the Pseudomonas aeruginosa lasR Gene, a Transcriptional Activator of Elastase Expression,” Journal of. Bacteriology. 1991;173(9):3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trieber CA, Taylor DE. Mutations in the 16S rRNA Genes of Helicobacter pylori Mediate Resistance to Tetracycline,” Journal of. Bacteriology. 2002;184(8):2131–2140. doi: 10.1128/JB.184.8.2131-2140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon DH, Lu CD. Polyamines Induce Resistance to Cationic Peptide, Aminoglycoside, and Quinolone Antibiotics in Pseudomonas aeruginosa PAO1. Antimicrobial Agents and Chemotherapy. 2006;50(5):1615–1622. doi: 10.1128/AAC.50.5.1615-1622.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]


