Abstract
Background
The mechanism(s) by which alcohol causes cell injury are still not clear but a major mechanism appears to be the role of lipid peroxidation and oxidative stress in alcohol toxicity. CYP2E1-generated ROS contributes to the ethanol-induced oxidant stress and inhibition of CYP2E1 activity decreases ethanol-induced fatty liver. The transcription factor Nrf2 regulates the expression of many cytoprotective enzymes which results in cellular protection against a variety of toxins.
Method
The current study was designed to evaluate the ability of sulforaphane, an activator of Nrf2, to blunt CYP2E1-dependent, ethanol-induced steatosis in vivo and in vitro.
Results
The sulforaphane treatment activated Nrf2, increased levels of the Nrf2 target heme oxygenase -1 and subsequently lowered oxidant stress as shown by the decline in lipid peroxidation and 3-Nitrotyrosine protein adducts and an increase in GSH levels after the acute ethanol treatment. It decreased ethanol-elevated liver levels of triglycerides and cholesterol and Oil Red O staining. Similar results were found in vitro as addition of sulforaphane to HepG2 E47 cells, which express CYP2E1, elevated Nrf2 levels and decreased the accumulation of lipid in cells cultured with ethanol. Sulforaphane treatment had no effect on levels of or activity of CYP2E1.
Conclusions
Sulforaphane proved to be an effective in vivo inhibitor of acute ethanol–induced fatty liver in mice.
General significance
The possible amelioration of liver injury which occurs under these conditions by chemical activators of Nrf2 is of clinical relevance and worthy of further study.
1. INTRODUCTION
The pathways by which alcohol causes cell injury are still not clear but a major mechanism appears to be the role of lipid peroxidation and oxidative stress in alcohol toxicity [1–4]. Fatty liver is a uniform and early response of the liver to ethanol consumption. Previously, fatty liver was considered benign, however, it is now known that fatty liver can increase sensitivity to hepatotoxins such as lipopolysaccharide [5] and high levels of fatty acids promote lipotoxicity [6,7]. Hence, there is a need to understand the mechanisms responsible for fatty liver production by ethanol. Early hypotheses for mechanisms responsible for fatty liver production by ethanol included redox state changes (elevated NADH) when ethanol was metabolized by alcohol dehydrogenase, elevated formation of acetyl CoA from ethanol/acetaldehyde oxidation and impairment of β-oxidation of fatty acids [8]. Recent studies [9–12] have identified mechanisms which regulate the synthesis and the oxidation of fatty acids as being central to how ethanol produces fatty liver. It appears that ethanol-induced steatosis may be related to oxidative stress as initially shown by Diluzio [13] who reported that antioxidants can prevent the ethanol-induced fatty liver, and studies showing that overexpression of the manganese superoxide dismutase can partially reduce the ethanol-induced steatosis [14]. CYP2E1-generated ROS contributes to the ethanol-induced oxidant stress [15–17] and inhibition of CYP2E1 activity decreases ethanol-induced fatty liver [18–20]. Both chronic and acute (binge) ethanol-induced fatty liver was blunted in CYP2E1 knockout mice but restored in humanized CYP2E1 knockin mice [20, 21]. Inhibition of CYP2E1 or CYP2E1-generated oxidative stress, by blunting ethanol-induced fatty liver, would likely be effective in preventing ethanol toxicity.
The transcription factor Nrf2 regulates the expression of many cytoprotective enzymes which results in cellular protection against a variety of insults produced by electrophilic and oxidative chemicals [22–24]. Nrf2 has been shown to be protective against a variety of drugs which can cause hepatotoxicity, lung injury, neurotoxicity, carcinogenesis and inflammation [25]. Exposure to many chemicals, oxidants and cellular stresses leads to increased production of Nrf2, disassociation of Nrf2 from the Kelch-like ECH associated protein Keap1 and subsequent entry of Nrf2 into the nucleus. In view of the importance of Nrf2 in upregulating many critical protective enzymes, there has been considerable interest in efforts to activate Nrf2 signaling by administration of low molecular weight molecules [26–28]. Sulforaphane is an isothiocyanate generated from the enzymatic cleavage of glucoraphanin, which is present in considerable amounts in brassica vegetables such as cabbage, kale, and broccoli [29]. It is thought to induce a phase II detoxification response by promoting the release of Nrf2 from the Keap 1-Nrf2 cytoplasmic complex. Sulforaphane has been shown to be protective against oxidative stress [30] via activation of Nrf2 [31–34]. The current study was designed to evaluate the ability of sulforaphane to blunt CYP2E1-dependent, ethanol-induced steatosis in vivo and in vitro.
2. MATERIALS and METHODS
2.1 IN VITRO MODEL
HepG2 E47 cells are HepG2 cells which were transfected with human CYP2E1 cDNA and constitutively express human CYP2E1 [35]. The cells were treated with or without 100 mM ethanol in the absence or presence of 6 uM sulforaphane. Cells treated with ethanol were cultured in a CO2 incubator which was saturated with 100 mM ethanol to minimize ethanol evaporation from the medium. Fresh medium with or without ethanol or sulforaphane was added daily. Lipid droplets were assayed by Oil Red O staining. Cells were transferred to cover slides after treatment and fixed with 10% buffered formalin for 10 min, then placed in absolute propylene glycol for 5 min and stained with pre-warmed Oil Red O solution for 10 min in a 60 degree water bath. Slides were differentiated in propylene glycol solution for 5 min and stained in Gill’s hematoxylin for 30 sec. After mounting with glycerol-PBS medium, the red stained lipid droplets were observed under the light microscope. Levels of triglycerides (TGs) and cholesterol were determined as described below.
2.2 IN VIVO MODEL
Male SV129 humanized CYP2E1 knockin (KI) mice [36,37], 20–30g (a gift from Dr. F. Gonzales, NCI, NIH), were bred and maintained in the Center of Laboratory Animal Sciences, Icahn School of Medicine at Mount Sinai (21). KI mice with high levels of CYP2E1 were used in this study since the effects of ethanol on oxidative stress and liver injury were more robust in these mice [21] thus making the possible protection by sulforaphane easier to evaluate. Mice were housed in temperature controlled animal facilities with 12-h light/12-h dark cycles and given access to water and Purina standard chow ad libitum. The mice received humane care, and experiments were carried out according to the criteria outlined in the Guide for the Care and Use of Laboratory Animals and with approval of our Institute’s Animal Care and Use Committee. Mice were gavaged twice a day with 30% ethanol at the dose of 3g/kg body weight for 5 days. Some mice were injected IP with sulforaphane at the dose of 0.05 g/Kg once a day for 5 days. Controls were gavaged with saline. Blood serum and liver were isolated for further analysis after pentobarbital administration at the end of the treatment.
2.3 WESTERN BLOTS
SDS-PAGE was carried out as previously described to determine levels of Nrf2, CYP2E1, and HO-1 [38,39]. Proteins were resolved using SDS-PAGE and were transferred to nitrocellulose membranes for blotting. The results were analyzed using an Odyssey fluorescence scanning system and the blots were quantified relative to the signals obtained for β-actin with ImageJ software from NIH.
CYP2E1 catalytic activity was assayed by the oxidation of p-nitrophenol to p-nitrocatechol [40]. Serum ALT and AST activities were assayed using Infinity kits from Thermo,
2.4 LIPID ACCUMULATION AND LIPID PEROXIDATION
Whole liver was removed and rapidly excised into fragments and washed with cold saline. One aliquot was placed in 10% formalin solution for paraffin blocking. Another aliquot was was placed in 50 mM sodium phosphate containing 18% sucrose at 4 degree C overnight and then frozen and cut into 10-um frozen sections for Oil Red O staining [20,21]. Liver homogenates were prepared in 150 mM KCL, pH 7.4 and stored at −80 degree C.
Steatosis was determined by Oil Red O staining as described above and H&E staining. TG and cholesterol content [20,21] in liver tissue were determined using kits from Pointe Scientific, INC (T7531-150, C7510-120 respectively). To assay lipid peroxidation, hepatic tissue or HepG2 cell extracts were placed in 1× PBS and homogenized in a polytron homogenizer for 10 strokes. 0.1 ml of the homogenate was incubated with 0.1 ml of a solution containing trichloroacetic acid [TCA; 15% (wt/vol)]-thiobarbituric acid [TBA; 0.375% (wt/vol)]-HCl solution (0.25 N)- butylated hydroxytoluene in a boiling water bath for 15 min. After being allowed to cool at room temperature, the samples were centrifuged at 10,000 rpm (8500×g) for 5 minutes. The resulting supernatant was used to determine the formation of TBA-reactive components by evaluating absorbance at 535 nm as previously described [20,21].
2.5 Glutathione Levels
Liver homogenate (20 ul), prepared as above was added to 20 ul of 10% W/V trichloroacetic acid and allowed to sit for 15 minutes at room temperature. Samples were then spun down at 10,000 rpm for 5 minutes. The resulting supernatant was added to 900 ul of buffer A (0.1 M sodium phosphate, pH 7.4, 5 µM EDTA, 0.6 mM 5,5'-Dithiobis(2-nitrobenzoic acid), 0.2 mM NADPH tetrasodium salt, pH 7.5) and 100 ul of buffer B (0.1 M sodium phosphate buffer, ~10 U/mL glutathione reductase pH 7.5). Levels of reduced GSH were measured by evaluating absorbance at 420 nm at 37°C for 2 minutes as previously described (20,21). Varying concentrations of reduced glutathione treated as above served as a standard.
2.6 3-NITROTYROSINE PROTEIN ADDUCTS
Liver tissue embedded in paraffin was sectioned, mounted onto microscope slides and deparffinized by immersing in the following for 3–5 minutes each in the given order: xylene, xylene, 100% ethanol, 95% ethanol, 80% ethanol, then briefly rinsing in tap water and then distilled water. The slides were stained using an Immunoperoxidase Secondary Detection System (DAB150, Millipore) with anti-3NT IgG as the primary antibody.
2.7 Nrf2 Activation
Nrf2 DNA binding activity in liver tissue was determined using the TransAM® Nrf2 Transcription Factor ELISA kit (50296) from Active Motif Inc. This kit contains a 96-well plate to which an oligonucleotide containing an antioxidant responsive element (ARE) which is the DNA binding site of Nrf2 has been immobilized. Briefly, liver tissue was homogenized and hepatocyte nuclear extract was isolated by using nuclear extract kit from Active Motif (Catalog #40010). Nrf2 contained in the nuclear extract then binds specifically to this oligonucleotide and is detected through use of and antibody directed against Nrf2. Addition of a second antibody conjugated to horseradish peroxidase (HRP) provides a colorimetic readout which can be quantified by spectrophotometer (41, 42).
Statistical Analysis
Statistical analysis was performed using one-way analysis of variance with subsequent post hoc comparisons by Scheffe. Values reflect means ± standard error. The number of experiments is indicated in the figure legends.
3 Results
3.1 Sulforaphane treatment induces Nrf2 in liver of CYP2E1 knockin mice
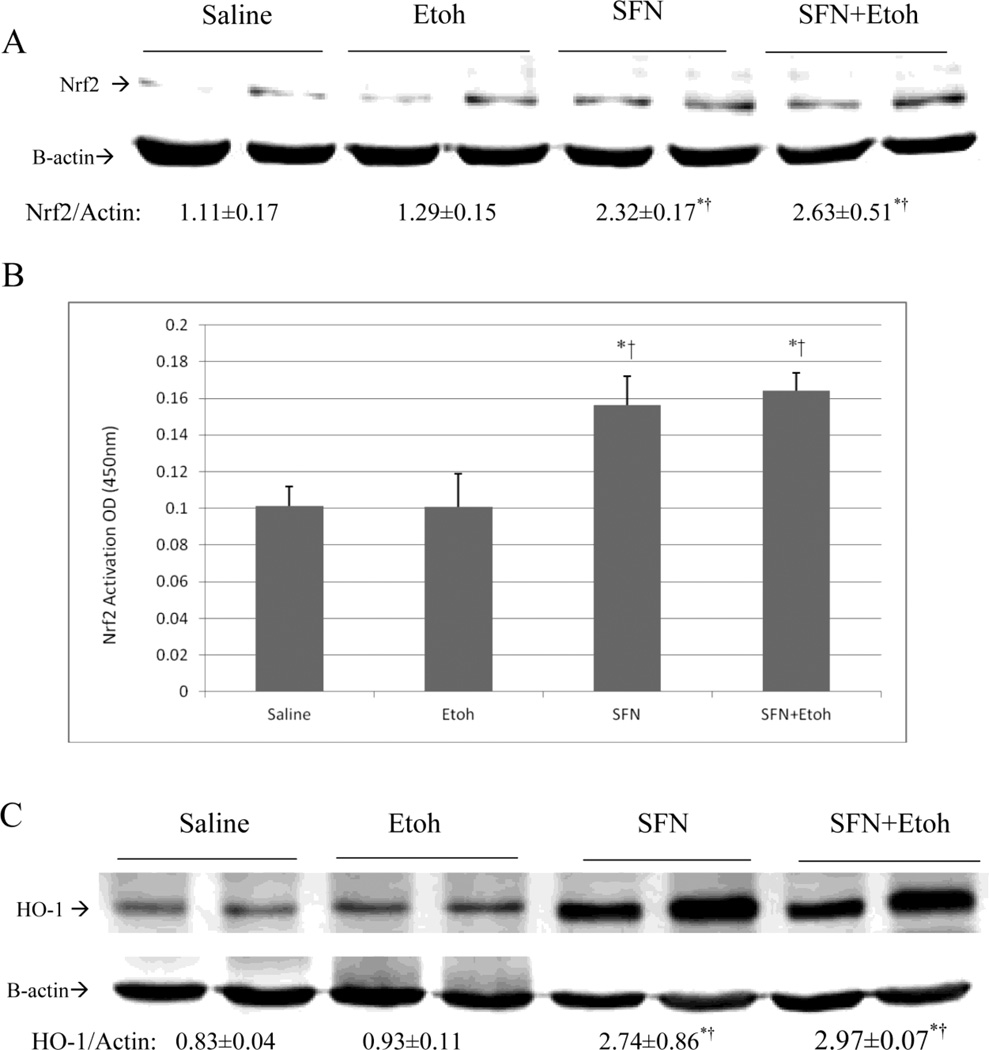
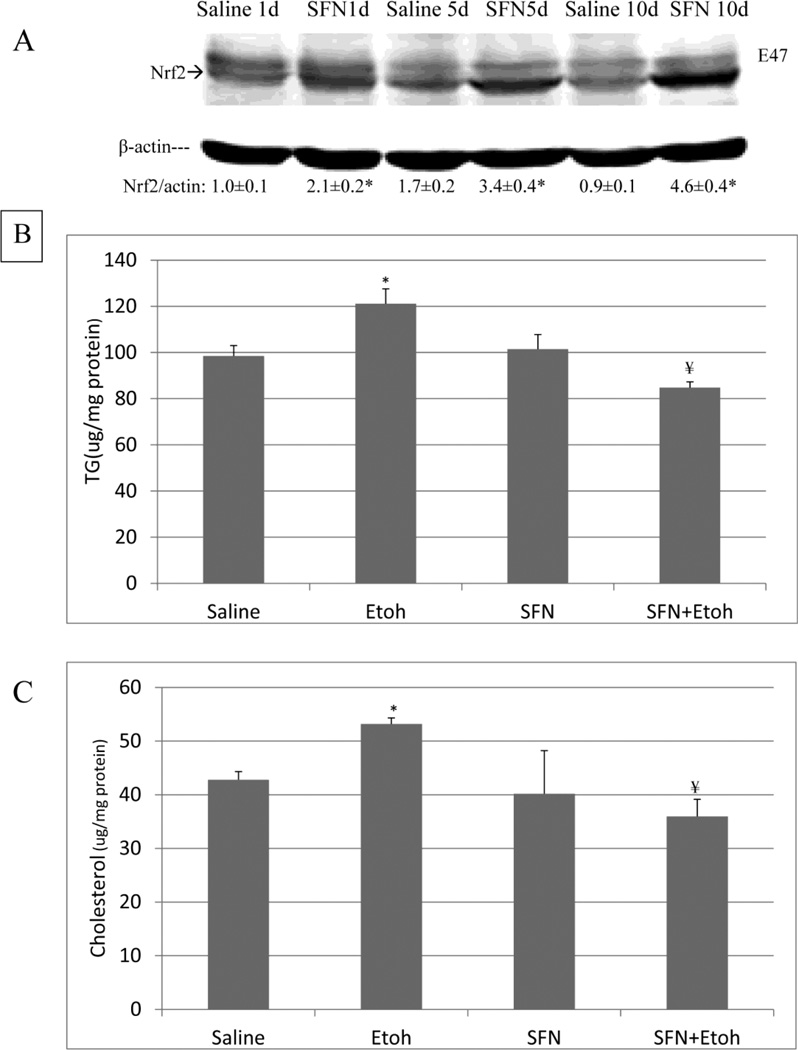
SV129 CYP2E1 KI mice were gavaged with 30% ethanol, 3g/kg body weight, or with saline twice a day in the absence or presence of 0.05 g/Kg sulforaphane once a day for a total of five days. Compared with 6g/kg body weight once a day, we separate the dose to 3g/kg body weitht twice a day, which may prevent mice to develop hypothermia. There was no significant weight loss or changes in food intake between groups (data not shown). ALT and AST levels were not significantly elevated by the binge ethanol treatment (data not shown). Ethanol treatment alone did not significantly increase levels of Nrf2 or promote activation of Nrf2 (Fig. 1(A, B)). CYP2E1 KI mice treated with sulforaphane exhibited increased protein levels of Nrf2 (Fig. 1(A)) as well as an increase in levels of activated Nrf2 (Fig. 1(B)) both in the absence and presence of ethanol compared to mice treated with saline or ethanol alone.
Fig. 1.
Sulforaphane treatment elevates Nrf2 and HO-1 levels and activates Nrf2. Humanized CYP2E1 knockin mice were gavaged with 30% ethanol at a dose of 3g/kg or saline twice a day for 5 days. Some mice were also treated with 0.05 g/Kg sulforaphane injected daily IP. A: levels of Ntf2. Results under the blots are expressed as the Nrf2/actin ratio. B: Nrf2 DNA binding activation was assayed using an ELISA kit as described in Materials and Methods. C: levels of HO-1. Results under the blots refer to the HO-1/ actin ratio. Results are from 3 experiments. *,¥ P<0.05 compared to saline or to ethanol- treated mice.
HO-1 is a Nrf2 inducible target [43]. To further validate that sulforaphane activates NRF2 in our in vivo experimental model, levels of HO-1 were determined. Induction of Nrf2 in sulforaphane treated CYP2E1 knockin mice resulted in about 3-fold increases in HO-1 levels in mice treated with sulforaphane alone or with sulforaphane plus ethanol (Fig. 1(C)). Ethanol treatment alone had no effect on HO-1 levels (Fig. 1(C)), analogous to its lack of effect on Nrf2 levels (Fig. 1 (A,B)).
3.2 Sulforaphane treatment protects against binge ethanol-induced steatosis
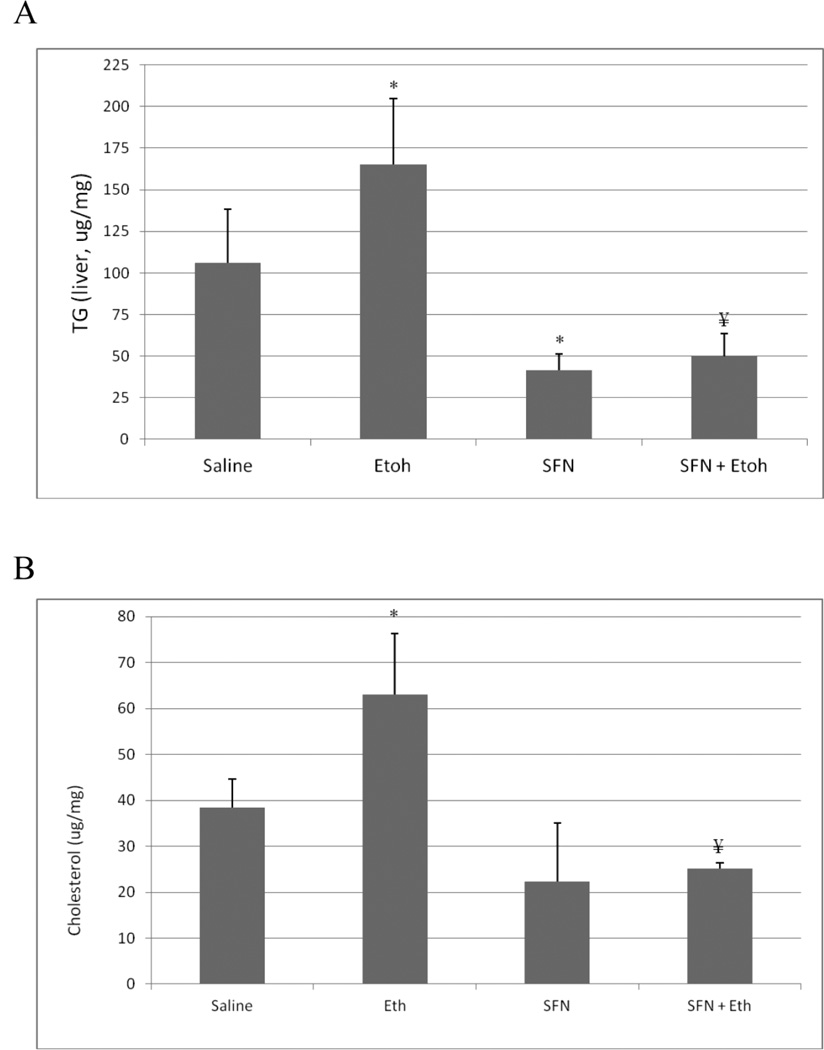
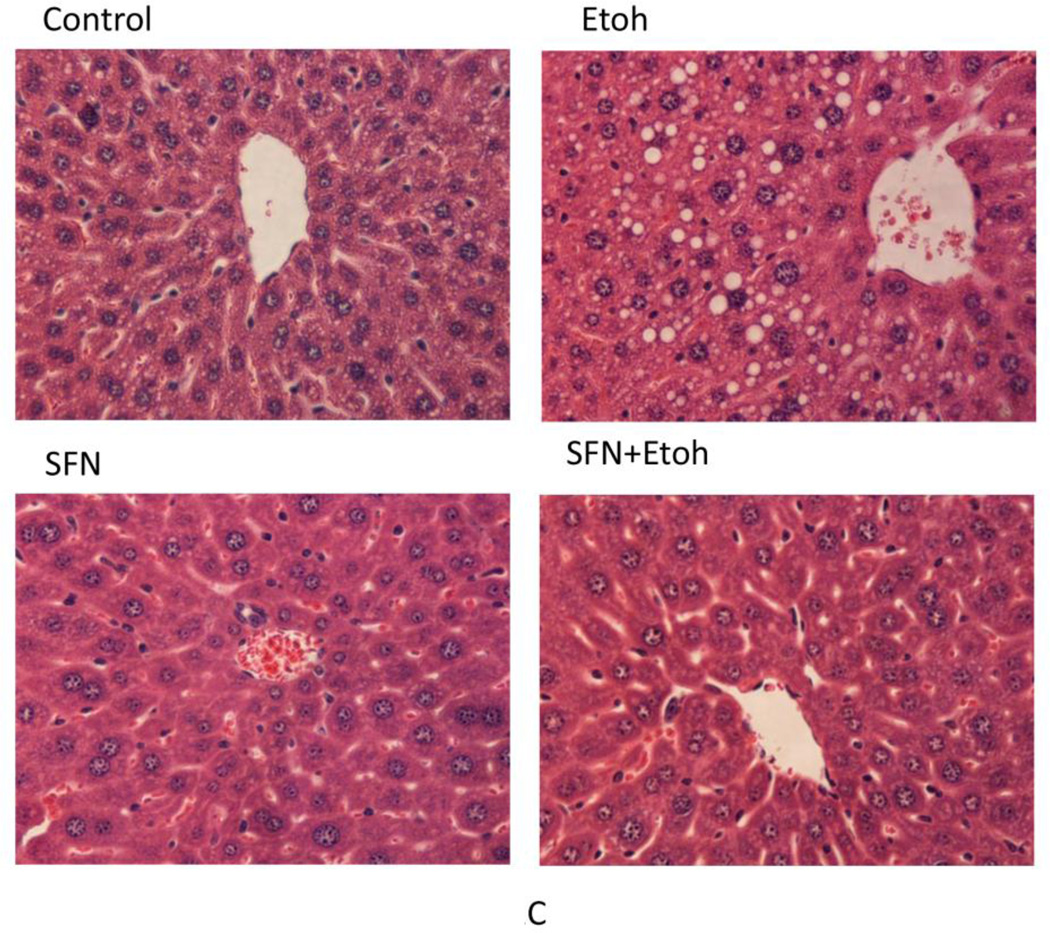
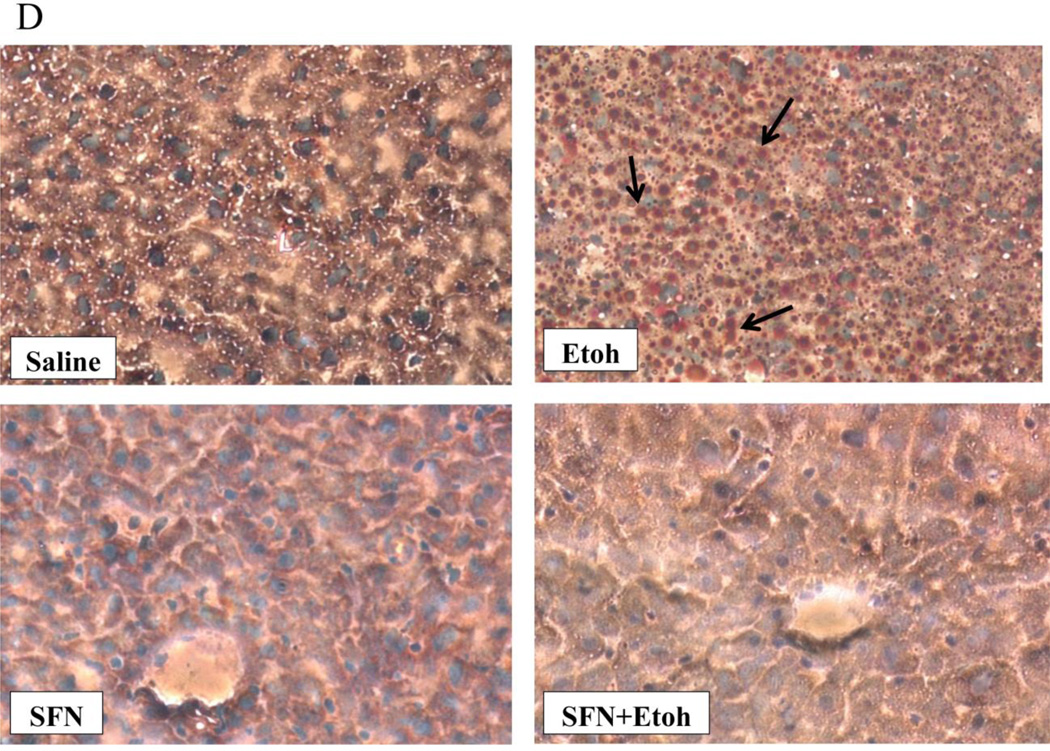
The binge ethanol treatment increased hepatic levels of TGs and cholesterol by about 50% (Fig. 2 (A,B)). The ethanol induced increases in hepatic TGs and cholesterol were reduced by treatment with sulforaphane (Fig. 2(A,B)). Interestingly, the sulforaphane treatment lowered the saline control levels of TGs and cholesterol as well as the ethanol-elevated levels. Histopathology confirmed the ethanol –induced steatosis and the prevention of lipid accumulation in the ethanol-treated mice by the sulforaphane treatment (Fig. 2(C)). Oil Red O staining was also reduced in sulforaphane treated mice compared to mice treated with ethanol alone (Fig. 2(D)).
Fig. 2.
Sulforaphane treatment prevents ethanol-induced steatosis. Mice were treated with ethanol in the absence or presence of sulforaphane as decribed in the legend to Fig 1. A; Hepatic triglyceride levels. B: Hepatic cholesterol levels. C: Liver H&E staining, 400× magnification. D: Oil Red O liver staining, 200× magnification. Results for A and B are from 3 experiments. * P< 0.05 for ethanol alone or sulforaphane alone as compared to saline controls. ¥ P< 0.05 for the effects of sulforaphane on levels of TG or cholesterol in livers from ethanol –treated mice compared to ethanol alone.
Several lipogenesis and lipolysis factors such PPARα, SCD, ACCα, or SREBP-1 regulate the synthesis and the oxidation of fatty acids, to determine whether SFN affects these factors, the immunoblots were carried out to determine the contents of these four proteins, no significant changes had found (data not showed).
3.3 Sulforaphane treatment protects against binge ethanol-induced oxidative stress
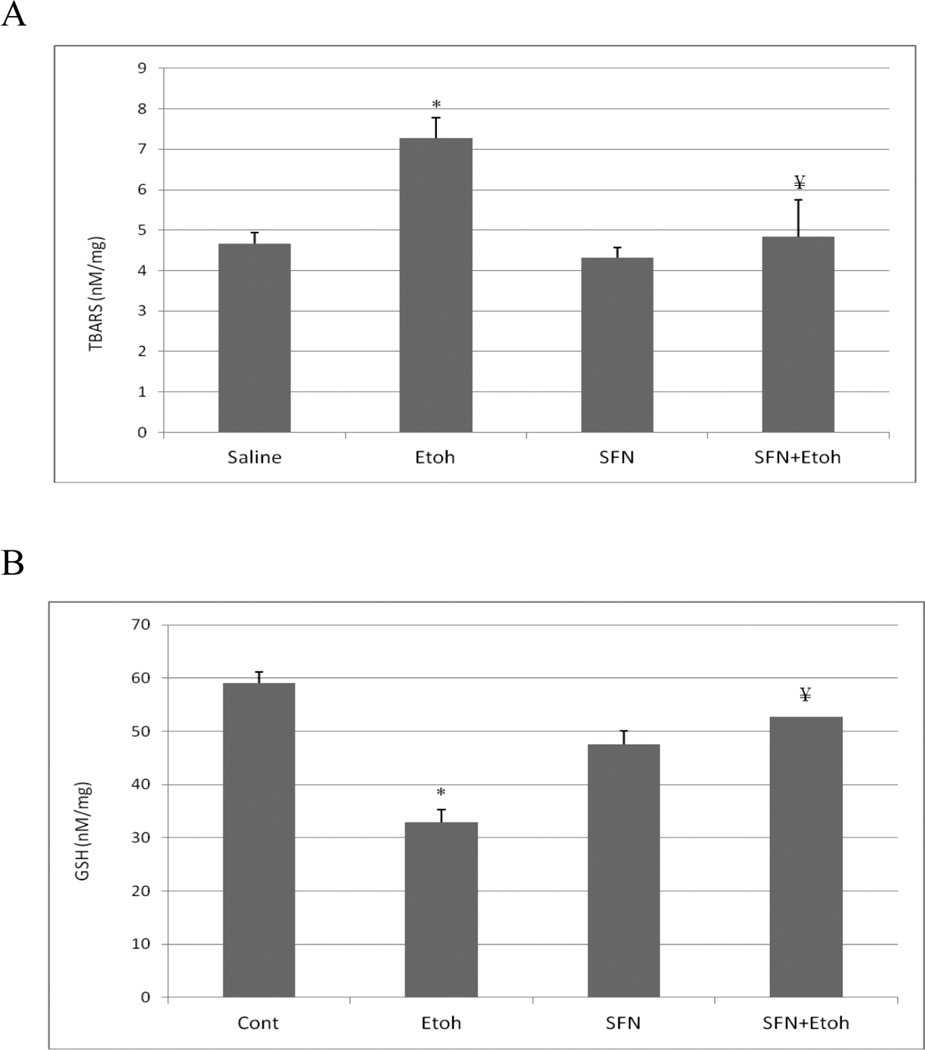
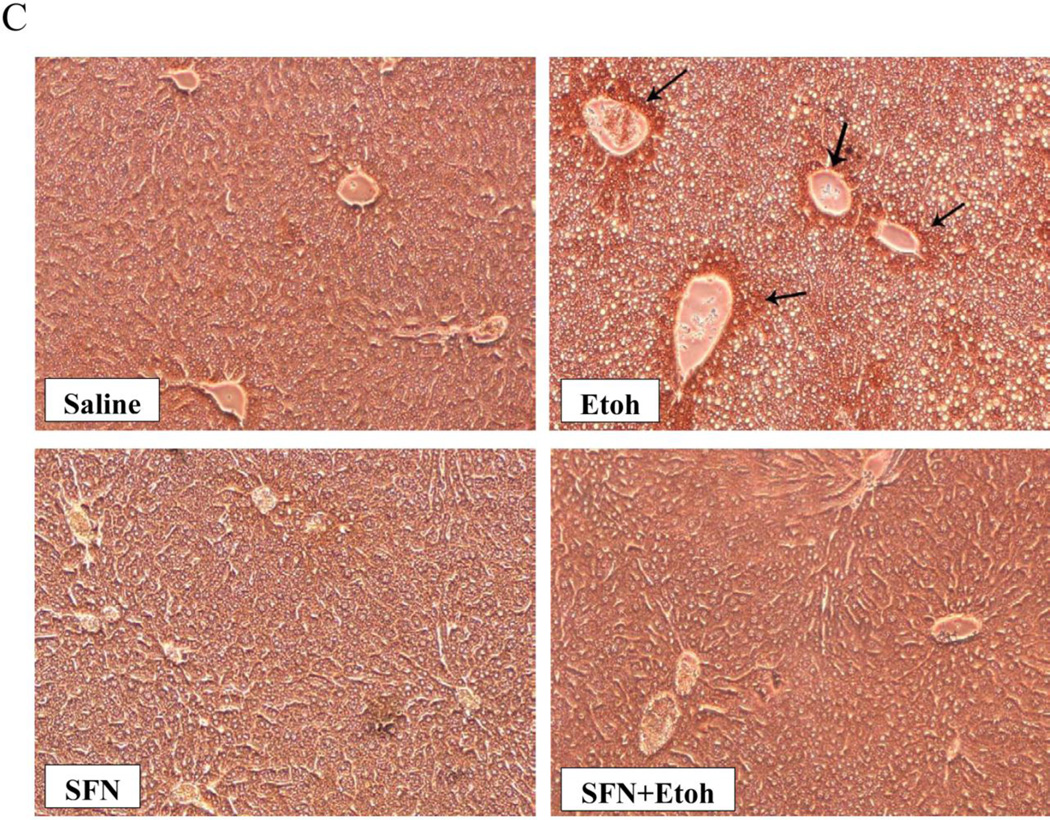
Hepatic TBARS levels, as a reflection of lipid peroxidation were elevated about 50% by the binge ethanol treatment (Fig. 3(A)). This ethanol-induced increase in lipid peroxidation was blunted by sulforaphane (Fig. 3(A)). Hepatic GSH levels were lowered about 50% by the ethanol treatment and this decline was prevented by sulforaphane (Fig. 3(B)). Immunohistochemical staining for the oxidative/nitrosative stress marker 3-nitrotyrosine (3NT) showed that ethanol treatment elevated 3-NT staining especially in the area around the central veins, the area with greatest CYP2E1 expression (Fig. 3(C)). There was a decrease in the intensity of 3-NT staining in mice treated with sulforaphane plus ethanol compared to mice treated with ethanol alone (Fig. 3(C)).
Fig. 3.
Sulforaphane treatment blunts binge-ethanol-induced oxidative stress. Mice were treated with ethanol in the absence or presence of sulforaphane as described in the legend to Fig. 1. A; Hepatic levels of TBARs as a reflection of lipid peroxidation. B; Hepatic levels of GSH. C: Immunohistochemical staining for 3-NT protein adducts in liver. Results for A and B are from 3 experiments. * P< 0.05 for the ethanol –induced increase in TBARs or decrease in GSH compared to saline. ¥ P<0.05 for the effect of sulforaphane on the ethanol-induced changes. Arrows in C point to the pericentral zone of the liver acinus.
3.4 Sulforaphane treatment does not affect CYP2E1 activity or content but increases autophagy
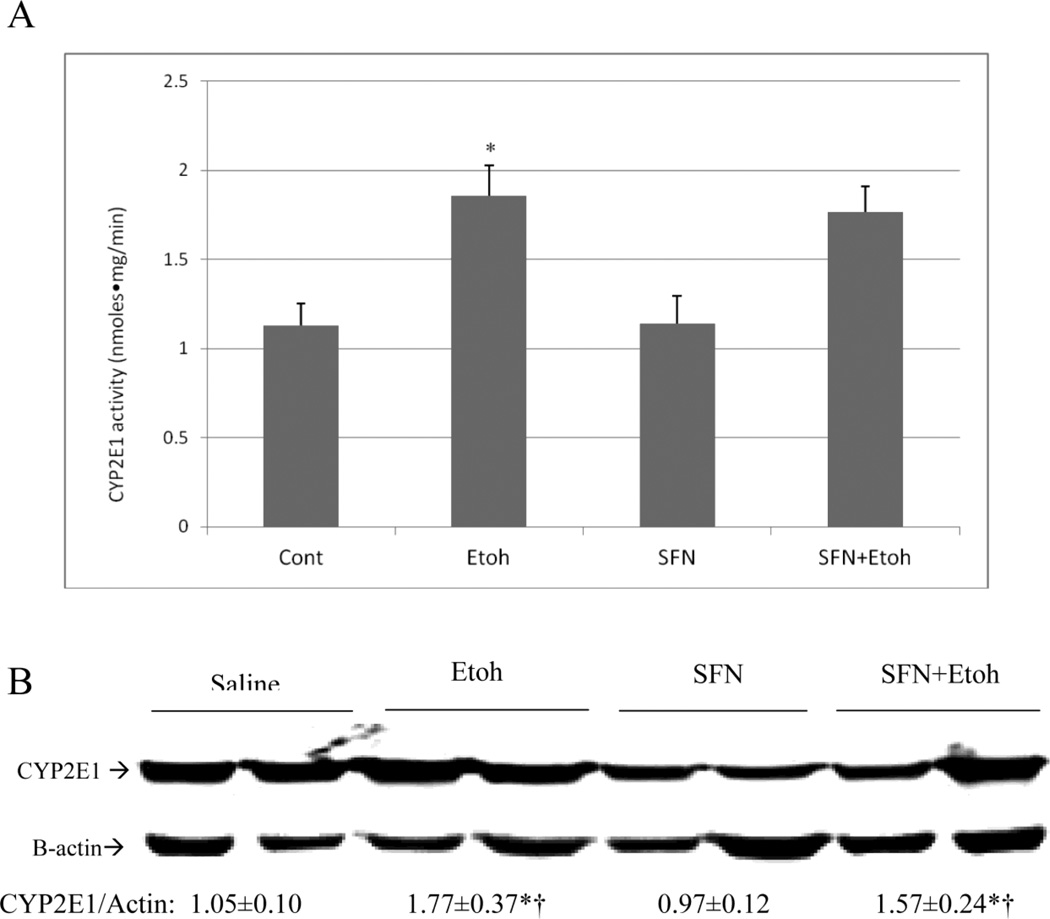
Treatment with sulforaphane did not affect the catalytic activity or protein levels of CYP2E1 in the saline controls (Fig. 4(A,B)). There was a 60–70% increase in CYP2E1 activity and protein levels after the binge ethanol treatment and these increases were not affected by sulroraphane (Fig. 4(A,B)).
Fig. 4.
Effect of sulforaphane on CYP2E1. A: CYP2E1 catalytic activity was assayed by the oxidation of para-nitrophenol by liver microsomes. B; CYP2E1 protein levels. Results are expressed as the CYP2E1/actin ratio and are shown under the blots. *,¥ P< 0.05 for ethanol alone or ethanol plus sulforaphane treatment as compared to saline alone or saline plus sulforaphane treatment. N= 3 experiments.
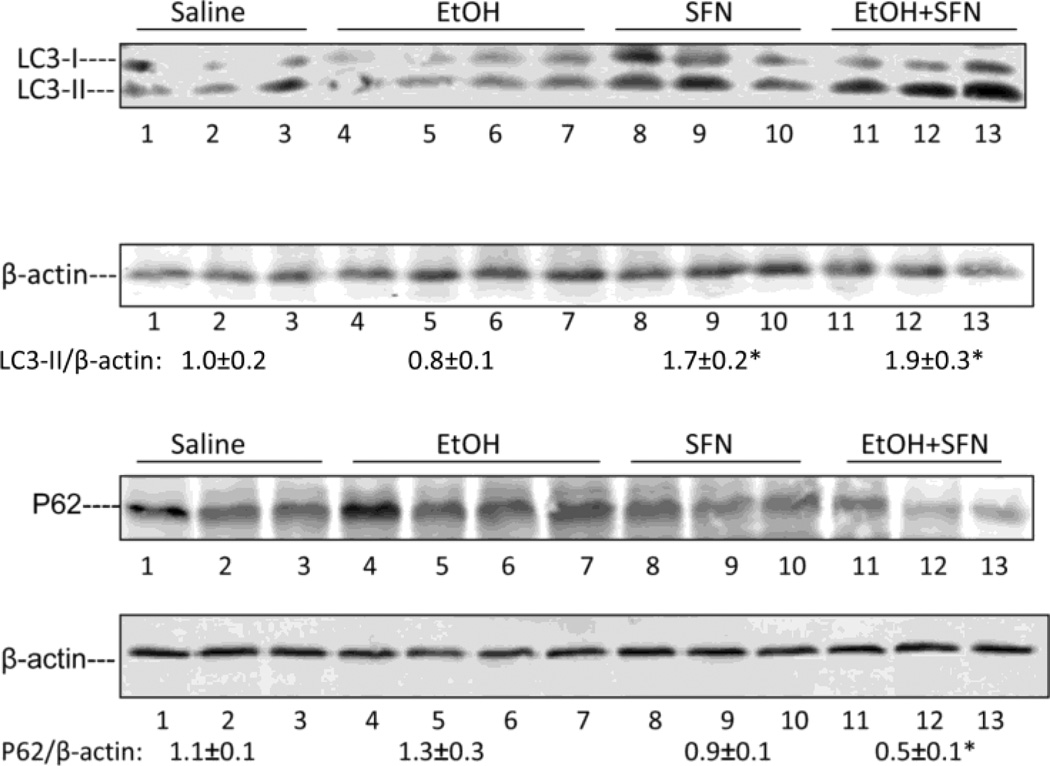
Autophagy is a degradative process involving delivery of cytoplasmic components to the lysosome for degradation. Autophagy was shown to promote lipid droplet breakdown in liver hepatocytes (lipophagy, [44]). The regulation and function of autophagy and lipid metabolism were shown to be reciprocally related to each other as inhibition of autophagy increased lipid accumulation while lipid accumulation inhibited autophagy [44, 47]. Autophagy may be involved in the regulation of alcohol-induced liver injury [46, 47]. The acute ethanol binge used in this study was previously shown to decrease autophagy as reflected by a 30% decrease in the hepatic LC3-II/LC3-I ratio [48]. Fig 5 shows that the binge ethanol treatment decreased the LC3-II/ LC3-I ratio about 20%, however, this ratio was elevated by combined treatment with ethanol plus sulforaphane (Fig. 5). P62/SQSTM1 is degraded by autophagy through direct interaction with LC3. An increase in autophagy leads to a decrease in the accumulation of P62 [49]. Levels of P62 were very low after the treatment with ethanol plus sulforaphane (Fig. 5), consistent with enhanced autophagy.
Fig. 5.
Ethanol plus sulforaphane treatment increased hepatic autophagy. The LC3-II/β-actin ratios and levels of p62 were measured by immunoblot analysis of liver extracts after treatment with saline (N=3), ethanol alone (N=4), sulforaphane alone (N=3) or combined treatment with ethanol plus sulforaphane (N=3). Densitometric results are shown under the blots. *, P<0.05 compared with saline group.
3.5 Sulforaphane prevents ethanol elevation of lipids in CP2E1-expressing HepG2 cells
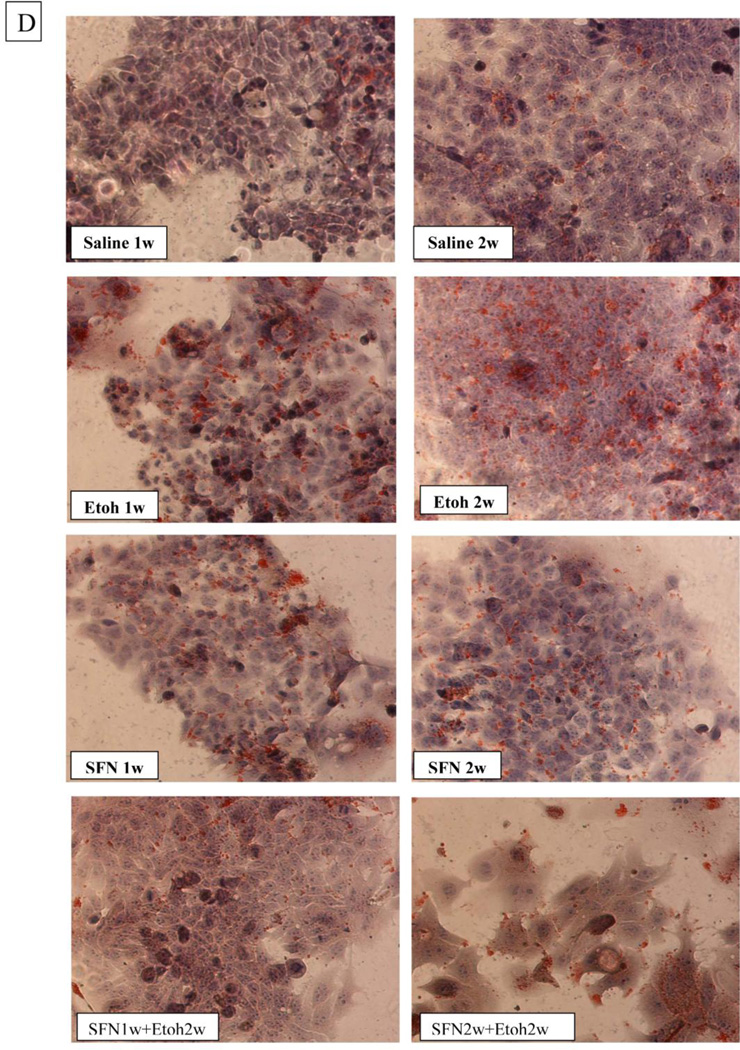
E47 HepG2 cells were used as an in vitro CYP2E1 expressing model to evaluate whether sulforaphane can protect against ethanol / CYP2E1 –induced steatosis. E47 cells were treated with 6 uM sulforaphene for 1 or 5 or 10 days. Levels of Nrf2 were elevated 2, 3, and 4.6 fold, respectively (Fig. 6(A)). Treatment with 100 mM ethanol for 7 days produced a 20–25 % increase in cellular levels of TG and cholesterol (Fig. 6(B)); no increase was found with HepG2 C34 cells which do not express CYP2E1 (data not shown: [48]). Sulforaphane completely prevented the increase in TG and cholesterol produced by ethanol in the E47 cells (Fig. 6(B)). One and two weeks of treatment of E47 cells with ethanol increased the intensity of Oil Red O staining (Fig. 6(C)). This increase was blunted by treatment with sulforaphane (Fig. 6(C)).
Fig. 6.
Sulforaphane prevents ethanol-induced increases in lipid accumulation in HepG2 E47 cells. A: Levels of Nrf2 in E47 cells incubated with 6 uM sulforaphane or saline for 1,5 or 10 days. Results are expressed as the Nrf2/actin ratio and are shown under the blots. * P< 0.05 compared to saline, N=3 expreiments. B: Levels of TGs and C levels of cholesterol in E47 cells incubated with 100 mM ethanol or saline in the absence or presence of 6 uM sulforaphane for 7 days. * P< 0.05 compared to saline; ¥ P< 0.05 compared to ethanol alone. N=3 experiments. D: Oil Red O staining of E47 cells after 1 or 2 weeks of incubation with buffer alone, 100 mM ethanol alone, 6 uM sulforaphane alone or ethanol plus sulforaphane. Note the increased staining after 1 or 2 weeks of incubation with ethanol and the decrease in this staining when ethanol was incubated in the presence of sulforaphane.
4 DISCUSSION
One major factor in ethanol-induced liver injury is the generation of reactive oxygen species and production of an imbalance between this generation and the cellular levels of anti-oxidant defense enzymes [1–4]. Since Nrf2 is a master transcription factor for regulation of levels of anti-oxidants, there has been increasing interest as to how ethanol alters Nrf2 activation and whether Nrf2 can modulate the effects of ethanol on the liver. Lamle et al [50] established a central role for Nrf2 in protecting mice against ethanol-induced liver injury as Nrf2 knockout mice fed ethanol for 4 to 7 days displayed increased mortality, marked steatosis and elevated liver pathology as compared to controls. The ethanol feeding to the Nrf2 knockouts caused decreases in total and in mitochondrial GSH levels and structural and functional damage to liver mitochondria. Somewhat surprising was the lack of significant increases in ethanol-induced oxidant stress in the Nrf2 knockouts [50]. Nrf2 was found to be protective against ethanol toxicity to mouse embryos as maternal ethanol treatment elevated Nrf2 protein and DNA binding in mouse embryos in association with increases in downstream Nrf2 target antioxidant genes [51]. Overexpression of Nrf2 by adenoviral delivery protected cerebral cortical neurons from ethanol-induced apoptotic death by increasing cellular GSH levels. Conversely, Nrf2 knockdown increased neuronal cell death [52]. Nrf2 was shown to protect against ethanol toxicity in vitro as the flavonoid quercetin protected human hepatocytes in culture against toxicity produced by 100 mM ethanol by a mechanism associated with activation of Nrf2 and enhanced production of HO-1 [53]. An interesting recent study showed that Nrf2, in graded amounts, could prevent against acute ethanol-induced oxidative stress and lipid accumulation in mice [54]. Ethanol was more toxic to Nrf2 knockout mice than controls and least toxic to Keap1 knockout and liver-specific Keap1 knockout mice with enhanced levels of Nrf2. Ethanol-induced oxidant stress and activation of SREBP and lipogenic genes was highest in the Nrf2 knockout and lowest in the Keap1 knockout mice [54]. Thus, Nrf2 protected against ethanol-induced oxidative stress and fat accumulation by upregulating antioxidant defense and decreasing lipogenic genes.
CYP2E1 contributes to chronic and acute ethanol-induced oxidative stress and fatty liver as shown with experiments using inhibitors of CYP2E1 [18–20] and CYP2E1 knockout and knocking mice [20, 21]. The ethanol-induced steatosis was associated with induction of CYP2E1 and increased generation of reactive oxygen species and could be blocked by antioxidants. Possible modulation of CYP2E1 toxicity and generation of ROS by Nrf2 was evaluated in previous studies. Chronic feeding of ethanol and acute administration of high doses (5–6 g/Kg) of ethanol increased Nrf2 protein and mRNA levels in association with increases in CYP2E1 protein [38,39,55]. Pyrazole, an inducer of CYP2E1, also increases Nrf2 levels [38, 56]. Basal levels of Nrf2 protein and mRNA, and Nrf2 binding to a consensus anti-oxidant response element probe by nuclear extracts were all higher in HepG2 E47 cells which express CYP2E1 than C34 cells which do not [38]. Associated with the increase in Nrf2 was an increase in Nrf2 target proteins such as HO-1 and the catalytic subunit of gamma glutamyl cysteine synthase in the E47 cells. These increases were prevented by SiRNA against Nrf2 [38]. The SiRNA against Nrf2 caused a loss of E47 cell viability without any effect on the C34 cells and sensitized the E47 cells to prooxidants such as arachidonic acid [39]. Overexpression of Nrf2 by plasmid delivery increased E47 cell levels of HO-1 and GSH and protected the E47 cells against oxidant-induced injury [39]. These studies suggested that Nrf2 is protective against CYP2E1-generated ROS and toxicity.
The goal of the current study was to evaluate whether Nrf2 was protective against ethanol-induced CYP2E1-dependent fat accumulation. Although previously considered benign, fatty liver caused by ethanol sensitizes the liver to other toxins and plays a role in the progression of ethanol injury. Chemical activation of Nrf2 by sulforaphane was studied in hopes of finding an effective pharmacological method as possible therapy against ethanol toxicity. The chemopreventive activites of sulforaphane have been recently reviewed [57]. Besides antioxidant activity largely via activation of Nrf2, sulforaphane can decrease phase 1 cytochrome P450 enzymes, increase phase II enzymes, decrease HIF alpha and COX 2 among other enzymes [57]. Sulforaphane has been shown to lower the pathology and decrease oxidative stress in livers of mice fed a methionine choline deficient diet in which steatohepatitiis occurs [58]. CYP2E1 plays an important role in models of chemical and high fat induced non-alcohol induced steatohepatitis and insulin resistance [59–61] and activation of Nrf2 has been shown to be protective against development of steatohepatitis [62]. Sulforaphane has been shown to inhibit CYP2E1 catalytic activity in isolated microsomes (oxidation of para nitrophenol) with a Ki of 37 uM and to block the genotoxicity of nitroso-dimethylamine, a procarcinogen substrate for oxidation by CYP2E1 [63,64]. Hence, the Nrf2 activation coupled to possible inhibition of CYP2E1 was thought to make sulforaphane an attractive chemical to blunt the toxic actions associated with CYP2E1. Sulforaphane proved to be an effective in vivo inhibitor of acute ethanol –induced fatty liver in mice. The sulforaphane treatment activated Nrf2 and subsequently lowered oxidant stress as shown by the decline in TBARS and 3-NT protein adducts and an increase in GSH levels after the acute ethanol treatment. The sulforaphane treatment had no effect on levels of or activity of CYP2E1 suggesting a major mechanism for its protection is likely activation of Nrf2 and induction of protective anti-oxidant enzymes. HO-1 was increased in mouse livers after sulforaphane treatment. The combined treatment with ethanol plus sulforaphane elevated the LC3-II/LC3-I ratio and decreased P62 levels, effects which may reflect increased autophagy. Increased autophagy may contribute to the decline in the ethanol-induced lipid accumulation. Effects of sulforaphane on lipogenic enzymes and transcription factors, cytokines such as TNF alpha, mitogen activated kinases such as JNK, mitochondrial dysfunction, and further studies on ethanol-induced autophagy are future directions planned to expand the current study. CYP2E1 activates many important toxicological relevant drugs such as acetaminophen, nitrosamines, halogenated hydrocarbons, benzene, etc and is activated under many pathophysiological conditions such as diabetes, obesity, fatty liver disease and alcohol toxicity. The possible amelioration of liver injury which occurs under these conditions by chemical activators of Nrf2 is of clinical relevance and worthy of further study as recently reviewed [63].
Highlight.
Activation of Nrf2 by sulforaphane was studied in CYP2E1 KI mice and HepG2 E47 cells which express CYP2E1.
The activation of Nrf2 by sulforaphane prevents binge-ethanol induced oxidative stress and steatosis in CYP2E1 KI mice.
Sulforaphane prevents ethanol-induced increases in lipid accumulation in HepG2 E47 cells.
Binge-ethanol plus sulforaphane increases hepatic autophagy.
Sulforaphane may be effective in lowering ethanol/CYP2E1-induced liver injury.
ACKNOWLEGEMEMTS
These studies were supported by USPHS grants RO1 AA018790 and R21 AA021362 from the National Institute on Alcohol Abuse and Alcoholism, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Nordman R, Riviere C, Rouach H. Implication of free radical mechanisms in ethanol-induced cellular injury. Free Rad. Biol. Med. 1992;12:219–240. doi: 10.1016/0891-5849(92)90030-k. [DOI] [PubMed] [Google Scholar]
- 2.Bondy SC. Ethanol toxicity and oxidative stress. Toxicol. Lett. 1992;63:231–242. doi: 10.1016/0378-4274(92)90086-y. [DOI] [PubMed] [Google Scholar]
- 3.Cederbaum AI. Introduction serial review: alcohol, oxidative stress and cell injury. Free Rad. Biol. Med. 2001;31:1524–1526. doi: 10.1016/s0891-5849(01)00741-9. [DOI] [PubMed] [Google Scholar]
- 4.Arteel GE. Oxidants and antioxidants in alcohol-induced liver disease. Gastroenterology. 2003;124:778–790. doi: 10.1053/gast.2003.50087. [DOI] [PubMed] [Google Scholar]
- 5.Yang SQ, Lin HZ, Lane MD, Clemens M, Diehl AM. Obesity increases sensitivity to endotoxin liver injury: implications for the pathogenesis of steatohepatitis. Proc. Natl. Acad. Sci. USA. 1997;94:2557–2562. doi: 10.1073/pnas.94.6.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang S, Lin H, Diehl AM. Fatty liver vulnerability to endotoxin-induced damage despite NF-kappaB induction and inhibited caspase 3 activation. Am. J. Physiol. Gastrointest. Liver Physiol. 2001;281:G382–G392. doi: 10.1152/ajpgi.2001.281.2.G382. [DOI] [PubMed] [Google Scholar]
- 7.Koteish A, Yang S, Lin H, Huang X, Diehl AM. Chronic ethanol exposure potentiates lipopolysaccharide liver injury despite inhibiting Jun N-terminal kinase and capase 3 activation. J. Biol. Chem. 2002;277:13037–13044. doi: 10.1074/jbc.M101632200. [DOI] [PubMed] [Google Scholar]
- 8.Purohit V, Gao B, Song BJ. Molecular mechanisms of alchoholic fatty liver. Alcoholism: Clin. Exp. Res. 2009;33:191–205. doi: 10.1111/j.1530-0277.2008.00827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.You M, Crabb DW. Recent advances in alcoholic liver disease II. Minireview: molecular mechanisms of alcoholic fatty liver. Am. J. Physiol. GI Liver Physiol. 2004;287:G1–G6. doi: 10.1152/ajpgi.00056.2004. [DOI] [PubMed] [Google Scholar]
- 10.You M, Crabb DW. Molecular mechanisms of alcoholic fatty liver: role of sterol regulatory element-binding proteins. Alcohol. 2004;34:39–43. doi: 10.1016/j.alcohol.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Crabb DW, Liangpunsaku S. Alcohol and lipid metabolism. J. Gastroent. Hepatol. 2006;21:S56–S60. doi: 10.1111/j.1440-1746.2006.04582.x. [DOI] [PubMed] [Google Scholar]
- 12.Sozio M, Crabb DW. Alcohol and lipid metabolism. Am. J. Physiol. Endocrinol. Metab. 2008;295:E10–E16. doi: 10.1152/ajpendo.00011.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiLuzio NR. Prevention of the acute ethanol-induced fatty liver by the simultaneous administration of antioxidants. Life Sci. 1964;3:113–118. doi: 10.1016/0024-3205(64)90189-4. [DOI] [PubMed] [Google Scholar]
- 14.Wheeler MD, Nakagami M, Bradford BU, Uesugi T, Mason RP, Connor HD, Dikalova A, Kadiiska M, Thurman RG. Overexpression of manganese superoxide dismutase prevents alcohol-induced liver injury in the rat. J Biol Chem. 2001;276:36664–36672. doi: 10.1074/jbc.M105352200. [DOI] [PubMed] [Google Scholar]
- 15.Caro AA, Cederbaum AI. Oxidative stress, Toxicology and Pharmacology of Cyp2E1. Annual Rev. Pharmacol. Toxicol. 2004;44:27–42. doi: 10.1146/annurev.pharmtox.44.101802.121704. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez FJ. Role of cytochromes P450 in chemical toxicity and oxidative stress: studies with CYP2E1. Mutat. Res. 2005;569:101–110. doi: 10.1016/j.mrfmmm.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 17.Butura A, Nilsson K, Morgan K, Morgan TR, French S, Johanssson I, Schuppe-Koistinen I, Ingelman-Sundberg M. The impact of CYP2E1 on the development of alcoholic liver disease as studied in a transgenic mouse model. J Hepatol. 2008;50:572–583. doi: 10.1016/j.jhep.2008.10.020. [DOI] [PubMed] [Google Scholar]
- 18.Morimoto M, Hagbjork AL, Wan YJ, Fu PC, Clot P, Albano E, Ingelman-Sundberg M, French SW. Modulation of experimental alcohol-induced liver disease by cytochrome P450 2E1 inhibitors. Hepatology. 1995;21:1610–1617. [PubMed] [Google Scholar]
- 19.Gouillon Z, Lucas D, Li J, Hagbjork AL, French BA, Fu P, Fang C, Ingelman-Sundberg M, Donohue TM, Jr, French SW. Inhibition of ethanol-induced liver disease in the intragastric feeding rat model by chlormethiazole. Proc. Soc. Exp. Biol. Med. 2000;224:302–308. doi: 10.1046/j.1525-1373.2000.22435.x. [DOI] [PubMed] [Google Scholar]
- 20.Lu Y, Zhuge J, Wang X, Bai J, Cederbaum AI. Cytochrome P4502E1 contributes to ethanol-induced fatty liver in mice. Hepatology. 2008;47:1483–1494. doi: 10.1002/hep.22222. [DOI] [PubMed] [Google Scholar]
- 21.Lu Y, Wu D, Wang X, Ward SC, Cederbaum AI. Chronic alcohol-induced liver injury and oxidant stress are decreased in cytochrome P4502E1 knockout mice and restored in cytochrome P4502E1 knockin mice. Free Radical Biol. Med. 2010;49:1406–1416. doi: 10.1016/j.freeradbiomed.2010.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev. Pharmacol. Toxicol. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen T, Yang CS, Pickett CB. The pathways and molecular mechanisms regulating Nrf2 activation in response to chemical stress. Free Radical. Biol. Med. 2004;37:433–441. doi: 10.1016/j.freeradbiomed.2004.04.033. [DOI] [PubMed] [Google Scholar]
- 24.Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Molec. Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 25.Osburn WO, Kensler TW. Nrf2 signaling: an adaptive response pathway for protection against environmental toxic insults. Mutation Res. 2008;659:31–39. doi: 10.1016/j.mrrev.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kensler TW, Egner P, Dolan P, Groopman JD, Roebuck BD. Mechanisms of protection against aflatoxin tumorigenicity in rats fed oltipraz and related 1,2 dithiol-3-thiones and 1,3 dithiol-3-one. Cancer Res. 1987;47:4271–4277. [PubMed] [Google Scholar]
- 27.Hu R, Xu C, Shen G, Jain MR, Khor TU, Gopalkrishnan A, Lin W, Reddy B, Chan Y, Kong ANT. Gene expression profiles induced by cancer chemopreventive isothiocyanate sulforaphane in the liver of C57BL/6J mice and C57B6/6J/Nrf2 (-1-) mice. Cancer Lett. 2006;243:170–192. doi: 10.1016/j.canlet.2005.11.050. [DOI] [PubMed] [Google Scholar]
- 28.Yates M, Kwak M, Egner P, Groopman JD, Bodreddigari S, Suttler TR, Baumgartner KJ, Roebuck BD, Liby KT, Yore MM, Honda T, Gibble GW, Sporn MB, Kensler TW. Protection against aflatoxin-induced tumorigenesis through induction of Nrf2-regulated pathways by the triterpenoid 1-[2-cyano-3-12-dioxouleana-1,9 (11)-diem-27-oyl]imidazole. Cancer Res. 2006;66:2488–2494. doi: 10.1158/0008-5472.CAN-05-3823. [DOI] [PubMed] [Google Scholar]
- 29.Lai RH, Keck AS, Walling MA, West LG, Jeffery EH. Evaluation of the safety and bioactivity of purified and semi-purifed glucoraphanin. Food Chem Toxicol. 2008;46:195–201. doi: 10.1016/j.fct.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 30.Guerrero-Beltran CE, Calderon-Oliver M, Pedriza-Chaverri J, Chirino YI. Protective effect of sulforaphane against oxidative stress: recent advances. Exp Toxicol Pathol. 2010;64:503–508. doi: 10.1016/j.etp.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Gan N, Mi L, Sun X, Dai G, Chung FL, Song L. Sulforaphane protects Microcystin –LR-induced toxicity through activation of the Nrf2-mediated defensive response. Toxicol. Appl. Pharmacol. 2010;247:129–137. doi: 10.1016/j.taap.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J, Shaikh ZA. Activation of Nrf2 by cadmium and its role in protection against cadmium-induced apoptosis in rat kidney cells. Toxicol. Appl. Pharmacol. 2009;241:81–89. doi: 10.1016/j.taap.2009.07.038. 2009. [DOI] [PubMed] [Google Scholar]
- 33.Wagner AE, Ernst I, Iori R, Desel C, Rimbach G. Sulforaphane but not ascorbigen, indole-3-carbinole and ascorbic acid activates the transcription factor Ntf2 and induces phase-2 and antioxidant enzymes in human keratinocytes in culture. Experimental Dermatology. 2010;19:137–144. doi: 10.1111/j.1600-0625.2009.00928.x. [DOI] [PubMed] [Google Scholar]
- 34.Greco T, Shafer J, Fiskum G. Sulforaphane inhibits mitochondrial permeability transition and oxidative stress. Free Radical Biol Med. 2011;51:2164–2171. doi: 10.1016/j.freeradbiomed.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu D, Cederbaum AI. Development and properties of HepG2 cells that constitutively express CYP2E1. In: Nagy L, editor. Alcohol: methods and Protocols. Humana Press; 2008. pp. 137–150. [DOI] [PubMed] [Google Scholar]
- 36.Cheung C, Yu AM, Ward JM, Krausz JW, Akiyama TE, Feigenbaum L, Gonzalez FJ. The CYP2E1-humanized transgenic mouse: role of CYP2E1 in acetaminophen hepatoxicity. Drug Metab Disp. 2005;33:449–457. doi: 10.1124/dmd.104.002402. [DOI] [PubMed] [Google Scholar]
- 37.Cheung C, Gonzalez FJ. Humanized mouse lines and their application for prediction of human drug metabolism and toxicological risk assessment. J Pharmacol Exp Ther. 2008;327:288–299. doi: 10.1124/jpet.108.141242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gong P, Cederbaum AI. Nrf2 is increased by CYP2E1 in rodent liver and HepG2 cells and protects against oxidative stress caused by CYP2E1. Hepatology. 2006;43:144–153. doi: 10.1002/hep.21004. [DOI] [PubMed] [Google Scholar]
- 39.Gong P, Cederbaum AI. Transcription factor Nrf2 protects HepG2 cells against CYP2E1 plus arachidonic acid-dependent toxicity. J. Biol. Chem. 2006;281:14573–14579. doi: 10.1074/jbc.M600613200. [DOI] [PubMed] [Google Scholar]
- 40.Reinke LA, Moyer MJ. P-Nitrophenol hydroxylation: a microsomal oxidation which is highly inducible by ethanol. Drug Metab Disp. 1985;13:548–552. [PubMed] [Google Scholar]
- 41.Zeng T, Zeng C, Song F, Zhao X, Yu L, Zhu Z, Xie K. The activation of HO-1/Nrf-2 contributes to the protective effects of diallyl disulfide (DADS) against ethanol-induced oxidative stress. Biochim Biophys Acta. 2013;1830:4848–4859. doi: 10.1016/j.bbagen.2013.06.028. [DOI] [PubMed] [Google Scholar]
- 42.Saidu NE, Touma R, Asalli IA, Jacob C, Montenarh M. Diallyl tetrasulfane activates both the elF2α and Nrf2/HO-1 pathways. Biochim Biophys Acta. 2013;1830:2214–2225. doi: 10.1016/j.bbagen.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 43.Wu KC, Liu JJ, Klaassen CD. Nrf2 activation prevents cadmium-induced acute liver injury. Toxicol. Appl. Pharmacol. 2012;263:14–20. doi: 10.1016/j.taap.2012.05.017. [DOI] [PubMed] [Google Scholar]
- 44.Dong H, Czaja MJ. Regulation of lipid droplets by autophagy. Trends Endocrin. Metab. 2011;22:234–240. doi: 10.1016/j.tem.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amir M, Czaja MJ. Autophagy in nonalcoholic steatohepatitis. Expert. Review Gastrorenterol. Hepatology. 2011;52:159–166. doi: 10.1586/egh.11.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding WX, Li M, Chen X, Ni HN, Lin CW, Gao W, Lu B, Stolz DB, Clemens DL, Yin XM. Autophagy reduces acute ethanol –induced hepatotoxicty and steatosis in mice. Gastroenterol. 2010;139:1740–1752. doi: 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding WX, Manley S, Ni HN. The emerging role of autophagy in alcoholic liver disease. Exp. Biol. Med. 2011;236:546–556. doi: 10.1258/ebm.2011.010360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu D, Wang X, Zhou R, Yang L, Cederbaum AI. Alcohol steatosis and cytotoxicity: the role of cytochrome P4502E1 and autophagy. Free Radical Biol. Med. 2012;53:1346–1357. doi: 10.1016/j.freeradbiomed.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lamle J, Marhenke S, Borlak J, Marhenke S, Borlak J, von-Wasielewski R, Eriksson CJ, Geffers R, Manns MP, Yamamoto M. Nuclear factor-erthyoid 2-related factor 2 prevents Alcohol-induced fulminant liver injury. Gastroenterology. 2008;134:1159–1168. doi: 10.1053/j.gastro.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 51.Dong J, Sulik KK, Chen S. Nrf2-mediated transcriptional induction of antioxidant response in mouse embryos exposed to ethanol in vivo. Antioxid. Redox Signal. 2008;10:2023–2033. doi: 10.1089/ars.2007.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Narasimhan M, Mahimainathan L, Rathinam ML, Riar AK, Henderson GI. Overexpression of Nrf2 protects cerebral cortical neurons from ethanol-induced apoptotic death. Molec. Pharmacol. 2011;80:988–999. doi: 10.1124/mol.111.073262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao P, Nussler A, Liu, Hao L, Song F, Schirmeier H, Nussler N. Quercetin protects human hepatocytes from ethanol-derived oxidative stress by inducing heme oxygenase-1 via the MAPK/Nrf2 pathways. J. Hepatol. 2007;47:253–261. doi: 10.1016/j.jhep.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 54.Wu KC, Liu JJ, Klaassen CD. Role of Nrf2 in preventing ethanol-induced oxidative stress and lipid accumulation. Toxicol Appl. Pharmacol. 2012;262:321–329. doi: 10.1016/j.taap.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 55.Lu Y, Zhang XH, Cederbaum AI. Ethanol induction of CYP2A5: role of CYP2E1-ROS-Nrf2 pathway. Toxicol Sci. 2012;128:427–438. doi: 10.1093/toxsci/kfs164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gilmore WJ, Hartmann G, Piquette-Miller M, Marriott J, Kirby GM. Effects of lipopolysaccharide-stimulated inflammation and pyrazole-mediated hepatocellular injury on mouse hepatic CYP2A5 expression. Toxicology. 2003;184:211–226. doi: 10.1016/s0300-483x(02)00581-4. [DOI] [PubMed] [Google Scholar]
- 57.Sharma R, Sharma A, Chaudhary P, Sahu M, Jaiswal S, Awashti S, Awashti YC. Role of 4-hydroxynonenal in chemopreventive activities of sulforaphane. Free Rad Biol Med. 2012;52:2177–2185. doi: 10.1016/j.freeradbiomed.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Okada K, Warabi E, Sugimoto H, Horie M, Tokushige K, Ueda T, Harada N, Taguchi K, Hashimoto E, Itoh K, Ishii T, Utsunomiya H, Yamamoto M, Shoda J. Ntf2 inhibits iron accumulation and counteracts oxidative stress-induced liver injury in nutritional steatohepatitis. J. Gastroenterology. 2012;47:924–935. doi: 10.1007/s00535-012-0552-9. [DOI] [PubMed] [Google Scholar]
- 59.Abdelmegeed MA, Banerjee A, Yoo SH, Jang S, Gonzalez FJ, Song BJ. Critical role of cytochrome P4502E1 (CYP2E1) in the development of high fat-induced non-alcoholic steatohepatitis. J. Hepatology. 2012;57:860–866. doi: 10.1016/j.jhep.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zong H, Armoni M, Harel C, Karnieli E, Pessin JE. Cytochrome P4502E1 CYP2e1 knockout mice are protected against high-fat diet-induced obesity and insulin resistance. Am. J. Physiol. Endocrinol Metab. 2012;302:E532–E539. doi: 10.1152/ajpendo.00258.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aubert J, Begriche K, Knockaert L, Robin MA, Fromenty B. Increased expression of cytochrome P450 2E1 in nonalcoholic fatty liver disease: mechanisms and pathophysiological role. Clinics and Research in Hepatology and Gastroenterology. 2011;35:630–737. doi: 10.1016/j.clinre.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 62.Zhang YKJ, Wu KC, Liu JJ, Klaassen CD. Nrf2 deficiency improves glucose tolerance in mice fed a high fat diet. Toxicol. Appl. Pharmacol. 2012;264:305–314. doi: 10.1016/j.taap.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barcelo S, Gardiner JM, Gescher A, Chipman JK. CYP2E1-mediated mechanism of anti-genotoxicity of the broccoli constituent sulforaphane. Carcinogenesis. 1996;17:277–282. doi: 10.1093/carcin/17.2.277. [DOI] [PubMed] [Google Scholar]
- 64.Barcelo S, Mace K, Pfeifer AMA, Chipman JK. Production of DNA strand breaks by N-nitrosodimethylamine and 2-amino-3-methylimidazol [4,5-f] quinoline in THLE cells expressing human CYP isoenzymes and inhibiton by sulforaphane. Mutation Res. 1998;402:111–120. doi: 10.1016/s0027-5107(97)00288-1. [DOI] [PubMed] [Google Scholar]
- 65.Bataille AM, Manautou JE. Nrf2: A potential target for new therapeutics in liver disease. Clin. Pharmacol.Ther. 2012;92:340–348. doi: 10.1038/clpt.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]