Abstract
In vitro models that accurately mimic the microenvironment of invading glioblastoma multiform (GBM) cells will provide a high-throughput system for testing potential anti-invasion therapies. Here, we investigate the ability of chitosan-polycaprolactone polyblend nanofibers to promote a migratory phenotype in human GBM cells by altering the nanotopography of the nanofiber membranes. Fibers were prepared with diameters of 200 nm, 400 nm, and 1.1 µm, and were either randomly oriented or aligned to produce six distinct nanotopographies. Human U-87 MG GBM cells, a model cell line commonly used for invasion assays, were cultured on the various nanofibrous substrates. Cells showed elongation and alignment along the orientation of aligned fibers as early as 24 hrs and up to 120 hrs of culture. After 24 hrs of culture, human GBM cells cultured on aligned 200 nm and 400 nm fibers showed marked upregulation of invasion-related genes including β-catenin, Snail, STAT3, TGF-β, and Twist, suggesting a mesenchymal change in these invading cells. After 120 hrs, there was only a slight upregulation of these genes in human GBM cells cultured on 400 nm and 1.1 µm aligned and random fibers, indicating slower, less pronounced transformation. The decrease in expression of these genes from 24 hrs to 120 hrs on 200 nm and 400 nm aligned fibers was attributed to asymmetric cell division, a hallmark of mesenchymal-like cells. Therefore, small (200 and 400 nm) diameter aligned nanofibers induce the greatest degree of phenotype change indicative of cell invasion behavior in human GBM. Additionally, cells cultured on 400 nm aligned fibers showed similar migration profiles as those reported in vivo, and thus these nanofibers should provide a unique high-throughput in vitro culture substrate for developing anti-invasion therapies for the treatment of GBM.
Keywords: migration, in vitro model, microenvironment, cancer, EMT, nanofiber
1. Introduction
Glioblastoma multiforme (GBM) is an aggressive brain cancer characterized by its ability to invade into surrounding brain tissue allowing it to evade therapy and recur.[1] Some therapies such as antiangiogenic approaches actually increase GBM cell invasion,[2] which results in disproportionately high patient morbidity and mortality, and underscores the importance of inhibiting invasion to effectively treat the disease.[3] Preventing the formation of a diffuse tumor through inhibition of cell invasion would allow effective treatment with radiation and chemo therapies and complete removal with neurosurgery. Unfortunately, the lack of in vitro models that accurately mimic the invading GBM cell microenvironment has greatly stalled research in anti-invasion treatments.
The major routes of GBM cell invasion are extracellular matrix (ECM)-mediated cell mobility on blood vessel surfaces and down white matter tracts.[3b,4] As a result, this selective invasion pattern indicates the ECM plays a decisive role in mediating migration pathways. Therefore, in vitro models must accurately mimic the physicochemical structure of the ECM encountered by invading cells. The chemical structure of neural ECM in which GBM cells invade is mainly hyaluronic acid (HA) and proteoglycans.[3b,5] The architecture of the invading GBM cell ECM is nanosized, high-aspect ratio fibers.[6] Recent studies have shown that cell surface receptor expression (EGFR versus PDGFR) may play a key role in determining whether GBM invade white versus gray matter in patients.[7] This unique topography of white matter ECM, as compared to gray matter, is thought to promote GBM cell invasion.[8] Indeed, it is well established that the tumor microenvironment plays a critical role in cancer cell progression and survival.[9]
The most common in vitro models for studying cell invasion and migration are hydrogels consisting of ECM molecules such as collagen, fibronectin, and laminin (e.g. Matrigel matrix). However, the brain ECM has a very low content of these fibrous proteins, limiting the utility of these models for brain tumors.[10] Although collagen is located in the basal lamina of brain blood vessels where GBM cells invade, it is not degraded by invading cells in vivo.[11] The requirement for collagen degradation in these in vitro invasion models, such as transwell assays, could lead to divergent results between in vitro and in vivo trials. More appropriate are hydrogels of HA that better mimic the chemical structure of the brain ECM;[10,12] however, these isotropic hydrogels lack the directional mechanical stimuli present in the brain ECM. The most representative model for studying GBM cell invasion in vitro is the organotypic slice assay where cancer cells are seeded on live slices of brain in appropriate culture medium.[13] This allows for the cancer cells to be subjected to the natural microenvironment in which they invade. However, the brain slice assay can produce discordant results due to dying brain tissue that cannot survive in vitro.
Electrospun nanofibers offer a unique tool for modeling the brain ECM for GBM cell migration. Their anisotropic elongated structures and nanotopography more accurately reflect the mechanical and structural cues present in the brain ECM, and their engineerability allows for fine-tuning their properties and confers ease in reproducibility. Electrospun nanofibers of polycaprolactone (PCL) with average diameters around 1 µm have been used to study GBM cell migration.[8,14] These studies found that migration distance and velocity were dependent on the alignment of the fibers, and that migration was highly dependent on STAT3 signaling. While these studies more accurately mimicked the mechanical and structural properties of the brain ECM, the PCL used to synthesize the fibers is synthetic and lacks appropriate biological properties.
To generate a more relevant in vitro model for brain ECM to study GBM cell migration, we electrospun polyblend nanofibers of chitosan and PCL. These nanofibers combine the biological properties conferred by the natural polymer, chitosan, with the aqueous stability provided by the synthetic polymer, PCL.[15] Chitosan, a polysaccharide derived from the exoskeletons of crustaceans shares structural similarity with HA, a major component of brain ECM. Therefore, the chemical structure of these nanofibers better represents the ECM of invading GBM cells, and may provide the proper biological stimuli. We prepared the fibers with diameters ranging from 200 nm to 1.1 µm to identify the nanotopography that best promotes cell invasion. GBM cells seeded on various nanofibers were imaged to identify cell morphology and assayed using PCR to determine the extent of overexpression of invasion related genes. This study should identify key morphological aspects of the brain ECM which promote GBM cell migration to provide a model for studying effective anti-migration treatments.
2. Results and Discussion
2.1. Nanofiber Synthesis and Characterization
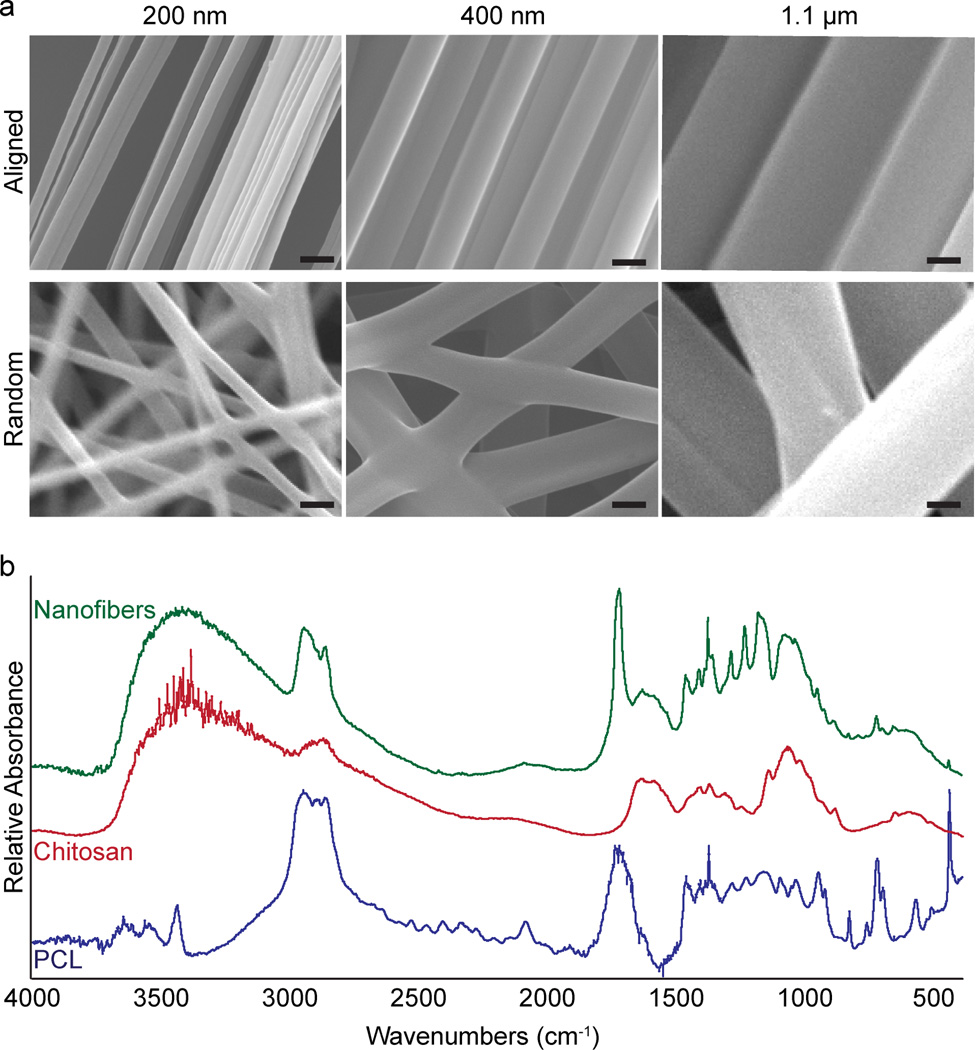
Chitosan-PCL polyblend nanofibers are synthesized by electrospinning solutions of chitosan and PCL prepared in TFA. The diameter of the fibers can be easily tuned by adjusting the viscosity of the solution, where more viscous solutions yield larger fiber diameters. The alignment of nanofibers is achieved using the parallel-electrode technique. The electrospun fibers are collected into 3D mats on glass cover slips. Figure 1a shows the SEM images of the fibrous mats with either randomly-oriented (bottom row) or aligned (top row) fibers of 200 nm, 400 nm, and 1.1 µm diameters. The fibers are highly aligned with uniform fiber diameters (Figure 1a, top row). FTIR analysis confirmed the presence of both chitosan and PCL in the nanofibers (Figure 1b). The previous quantification of nanofiber diameters and alignment have shown high uniformity and marked alignment.[15b,15c] The uniformity of the fiber diameters ensures accurate conclusions can be drawn from changes in cell behavior due to nanotopography. Furthermore, the highly aligned fibers allow cells to maintain an invasive phenotype along the white matter tract mimicking fibers rather than changing directions at kinks or fiber cross sections.[8]
Figure 1.
Structural and chemical analysis of chitosan-PCL fibers. a) SEM images showing aligned (top row) and random-oriented (bottom row) fibers of different diameters. Scale bars correspond to 400 nm. b) FTIR analysis confirming the presence of chitosan and PCL in the nanofibers.
2.2. Imaging cell morphologies and alignment on various nanofiber substrates
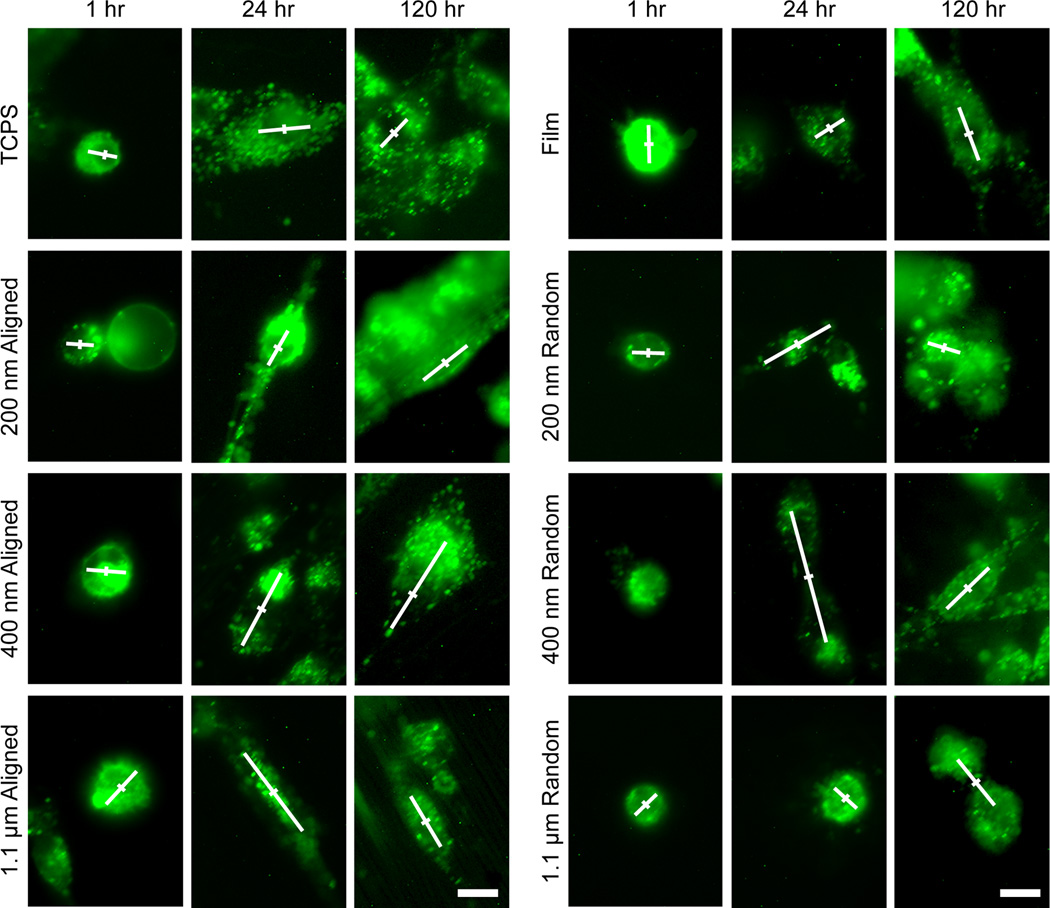
The cell behavior on nanofibers was first assessed through imaging of cell morphologies using bright field microscopy, fluorescence microscopy, and SEM imaging. U-87 MG human GBM cells were chosen as the model cell line for testing the nanofibers since they are a representative cell line commonly used for migration and invasion studies.[16] Cells were seeded on randomly oriented and aligned nanofibers of 200 nm, 400 nm, and 1.1 µm diameter, 2D films of chitosan-PCL, and tissue culture polystyrene (TCPS) and imaged after 24 and 120 hrs. The 2D films served to identify whether the surface chemistry of chitosan and PCL alone induces invasive changes in GBM cells.
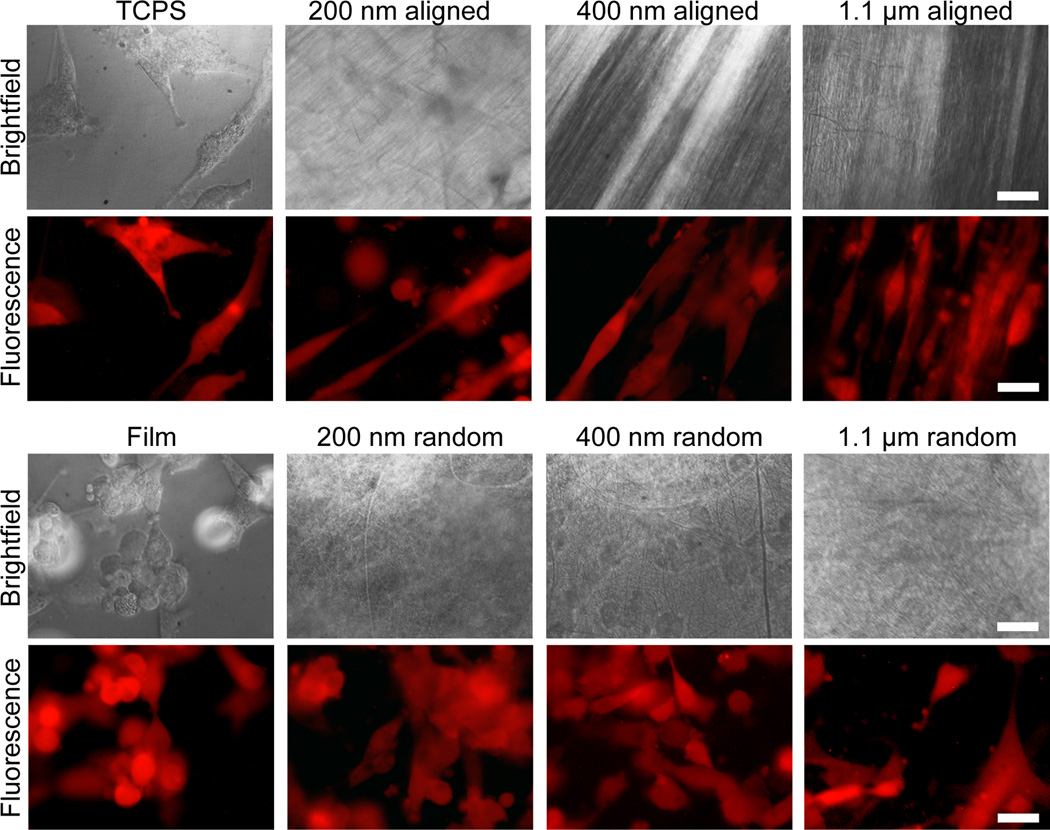
After 24 hrs of culture on various substrates, U-87 MG cells became elongated and aligned on aligned fibers (Figure 2), similar to the invasive morphology observed in vivo, a common observation for aligned nanotopographies.[17] Cells showed less spreading and no alignment on the isotropic randomly oriented fibers and film. The small gaps between the randomly oriented fibers reduced the contact area for the cells and may have prevented their elongation. These gaps between the fibers could also hinder cell migration by requiring the cell to interrupt its forward motion to find the closest nanofiber substrate to attach to. This is a structural advantage of the aligned nanofibers for promoting cell migration where long, uninterrupted tracts are present to maintain cell migration.[8] Cells on TCPS showed normal spreading and morphology for standard tissue culture.
Figure 2.
Cells cultured on TCPS and chitosan-PCL films and fibers for 24 hrs. RFP expressing U-87 MG GBM cells showed alignment along fiber orientation on aligned fibers (fiber orientations are shown in the corresponding brightfield images where cells cannot be well seen due to the opacity of the fibers), with 200 nm aligned fibers showing the greatest cell elongation. The scale bars correspond to 20 µm.
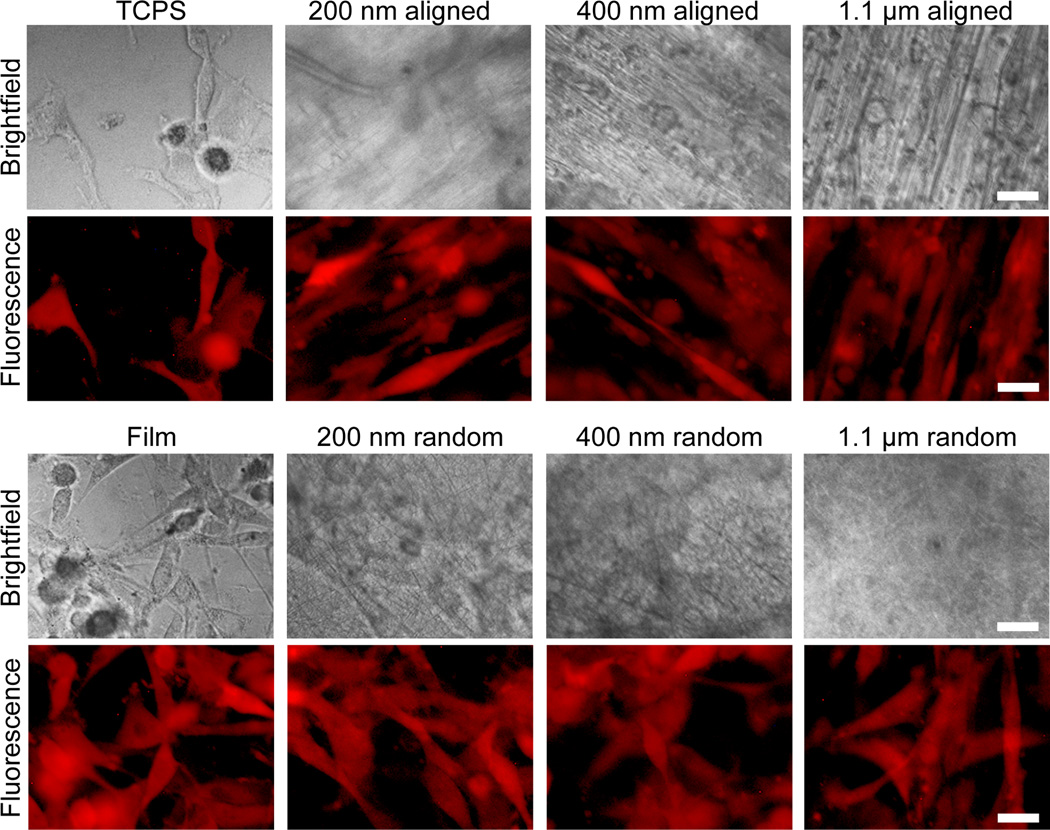
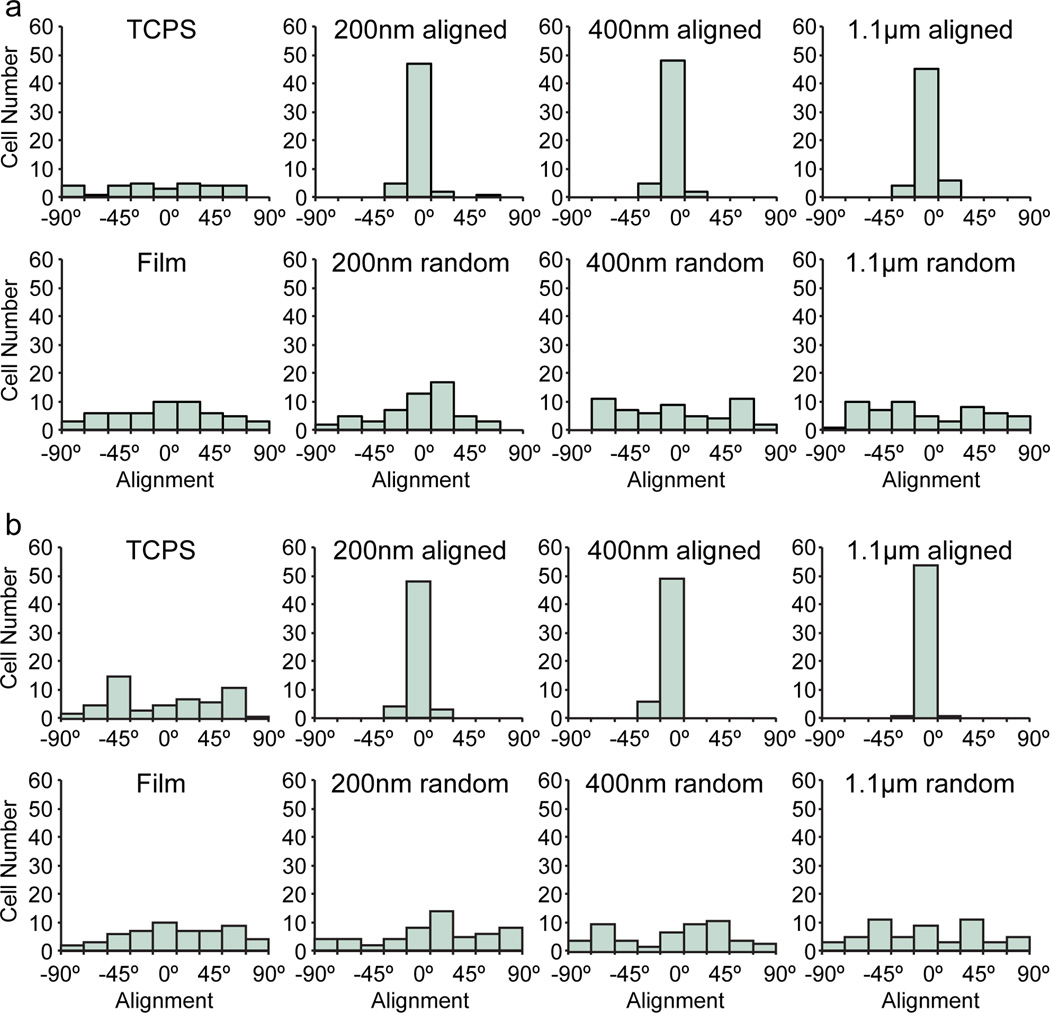
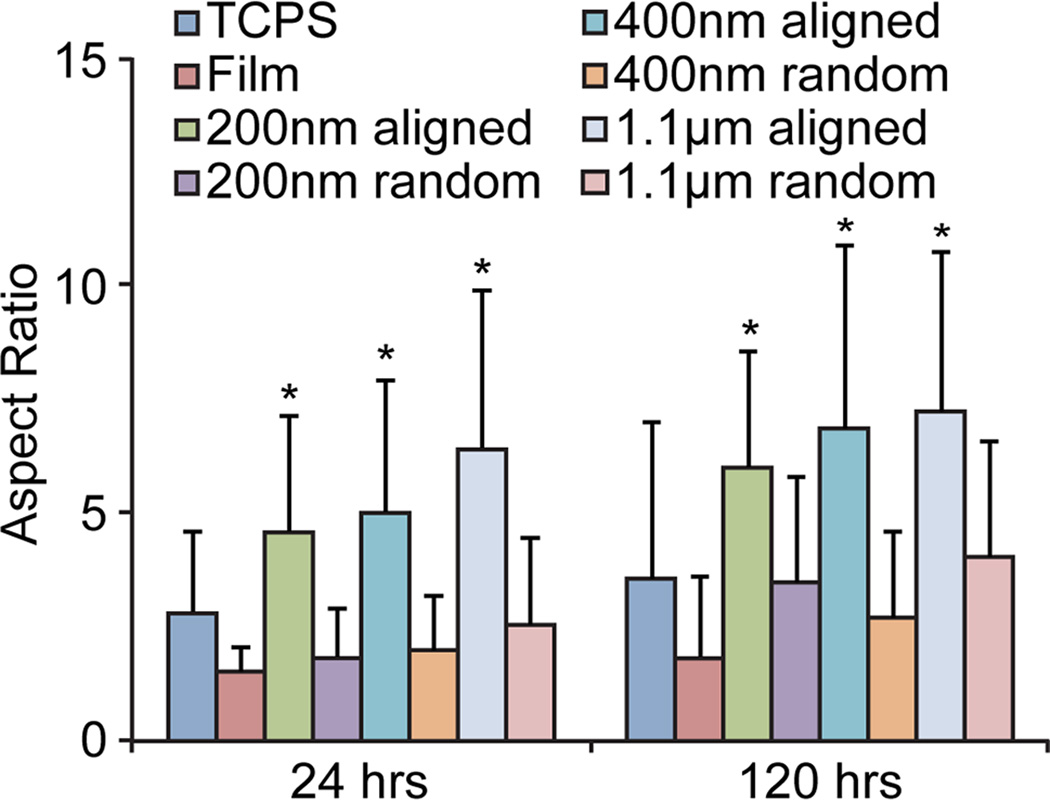
After 120 hrs of culture, cells were elongated and aligned on all aligned fibers of different diameters (Figure 3). Quantification using ImageJ confirmed the high degree of alignment on the aligned nanofibers as compared to the randomly oriented fibers, film, and TCPS (Figure 4) for both 24 hr and 120 hr time points. Furthermore, cells on the aligned fibers had a more elongated morphology as compared to those on TCPS, film, and randomly oriented fibers as quantified through aspect ratio measurements (Figure 5). However, there was no difference in elongation between fibers of different diameters at 24 or 120 hrs. Although the aspect ratio seems to increase with increasing fiber diameter, these increases were not statistically significant (p = 0.16 for 24 hrs and p = 0.48 for 120 hrs by ANOVA). This indicates the differences in curvatures between 200 nm, 400 nm, and 1.1 µm fibers were not enough to promote differences in elongation. This mesenchymal-like morphology suggests cells on aligned fibers may be undergoing a mesenchymal change to more malignant and invasive cancer cells. No mesenchymal-like morphology was observed in the randomly oriented fibers or 2D film samples.
Figure 3.
Cells cultured on various substrates for 120 hrs. RFP expressing U-87 MG GBM cells showed alignment along the orientation of aligned fibers (the fiber orientations are shown in the brightfield images where cells cannot be well seen due to the opacity of the fibers), with 200 nm and 400 nm aligned fibers showing similar elongation. The scale bars correspond to 20 µm.
Figure 4.
Alignment quantification of cells cultured on various substrates. Cells that were cultured for a) 24 hrs and b) 120 hrs on aligned fibers showed much greater alignment than those cultured on randomly-oriented fibers, films, and TCPS. Histograms are binned in 20° intervals. Fifty-five cells were counted for each condition.
Figure 5.
Aspect ratios of cells cultured on various substrates for 24 and 120 hrs. Cells cultured on aligned fibers were more elongated (larger aspect ratio) than those cultured on TCPS, films, or randomly-oriented fibers. * indicates a statistical significance (p < 0.01) as compared to TCPS, films, and randomly-oriented fibers as determined by an unpaired t test. Between 12 and 26 cells were measured for each condition.
2.3. Cell migration on various nanofibers substrates
In order to show that cells migrate along the nanofibers, red fluorescent protein (RFP) expressing cells were seeded on TCPS, 200 nm, 400 nm, and 1.1 µm aligned fibers and allowed to attach for a day prior to time-course imaging. Images were acquired every 10 min for 24 hrs and cells tracked manually using ImageJ (Figure 6a). Cells cultured in TCPS moved around randomly and did not migrate far from their original location. On the other hand, cells cultured on the aligned fibers migrated in linear paths corresponding to the direction of the fiber orientation. This directional persistence indicates the cells do, in fact, migrate as well as align on the fibers regardless of fiber diameter.
Figure 6.
Cell migration on nanofibers. a) Traces of cell migration over 15–24 hrs on TCPS and 200 nm aligned, 400 nm aligned, and 1.1 µm aligned nanofibers. b) Migration velocity over time in representative cells cultured on various substrates. c) Maximum migration velocity of cells cultured on various substrates. At least six cells were measured for each condition. d) Median migration velocity of cells cultured on various substrates. At least six cells were measured for each condition. * and # in panels c–d indicate statistical significance (p < 0.01) compared to TCPS and 400 nm fibers, respectively, as determined by Student’s t test. e) Effective cell migration velocities on nanofibers. At least six cells were measured for each condition. * in panel e indicates a statistical significance (p < 0.01) as compared to TCPS, 200 nm, and 1.1 µm fibers as determined by Student’s t test.
To see if the migratory behavior of the cells were different on fibers of different diameters, cell velocity was calculated by measuring the distance the cell traveled between images and plotted over time (Figure 6b). These representative plots showed differences in migration profiles between cells cultured on fibers of different diameters. Interestingly, cells on 200 nm aligned fibers had large spikes in cell velocity. These spikes are also observed in vivo and correspond to the migration of daughter cells immediately after cell division.[8,18] The spikes were also present in cells cultured on the larger diameter fibers and TCPS. The maximum cell velocities (Figure 6c) were highest for cells cultured on 200 nm fibers at 87 ± 33 µm hr−1, significantly larger than TCPS (35 ± 14 µm hr−1) and 400 nm fibers (40 ± 12 µm hr−1), with cells on 1.1 µm fibers in the middle (60 ± 15 µm hr−1). While the cells cultured on 200 nm fibers had larger maximum velocities, similar to those recorded in vivo,[18b] the velocity of the cells away from these peaks was much lower than 400 nm fibers but similar to TCPS and 1.1 µm fibers. Therefore, we calculated the median cell velocity on the different substrates (Figure 6d). Median cell velocity was highest in cells cultured on 400 nm fibers (13 ± 3 µm hr-−1), similar to that on brain slice cultures,[18b] with cells on 200 nm fibers, 1.1 µm fibers, and TCPS having similar median velocities. Again, the median velocities were similar to those observed in vivo.[18]
These migration data show that there is a marked difference in migratory behavior of GBM cells cultured on fibers of different diameters, which may provide insight into cell invasion in the brain. Cells seemed to prefer migrating along the 400 nm fibers with a constant higher velocity and fewer and smaller bursts of rapid movement, which may reflect the ability of this nanotopography to mimic the ECM of invading GBM cells in vivo. Indeed, in vivo analyses have revealed that GBM cells migrate faster and more efficiently on the abluminal side of blood vessels.[18a] This is better shown from the effective cell velocity, which takes into account the net distance the cell traveled from its starting point (Figure 6e). The effective velocity is thought to better represent invasion in the brain.[8] The effective velocity of cells was highest on 400 nm fibers at 2.5 ± 1 µm hr−1, very similar to that reported for cells invading along microvessels in vivo.[18a] This was significantly larger than on the other substrates indicating the cells migrated further and faster along the fiber axis on the 400 nm fibers. Therefore, the distinct nanotopography of the 400 nm aligned nanofibers provided a substrate that effectively mimicked in vivo cell migration.
2.4. Differential gene expression on various nanofiber substrates
The change in expression profile can provide clues into which fiber nanotopographies induced the greatest degree of invasion, and thus which best mimic the microenvironment of invading GBM cells. To assess the effect of nanotopography on the invasive phenotype of the cells, polymerase chain reaction (PCR) was run on invasion related genes, including Twist, Snail, TGF-β, STAT3, and β-catenin, after 24 hrs (Figure 7a) and 120 hrs (Figure 7b) of cell culture. First, Twist expression promotes invasion and has been linked to mesenchymal change in human GBM.[19] Twist expression was slightly elevated after 24 hrs of culture on 200 nm and 400 nm aligned fibers as compared to TCPS culture. After 120 hrs, Twist expression decreased to the same level as TCPS on 200 nm fibers and remained slightly elevated on 400 nm aligned fibers.
Figure 7.

Gene expression in GBM cells cultured on various substrates after 24 hrs of cell seeding. The expression levels of β-catenin, Snail, STAT3, TGF- β, and Twist were determined in U-87 MG cells after a) 24 hrs and b) 120 hrs of culture by PCR using GAPDH as the reference gene and TCPS as the control.
Twist activates the expression of other invasion and mesenchymal genes such as Snail. Snail is also involved in cancer cell invasion and its expression in GBM is highly correlated with histological grade and invasive phenotype.[20] After 24 hrs, Snail was highly overexpressed in cells cultured on 200 nm and 400 nm aligned fibers showing a 59-fold and 29-fold higher expression, respectively, compared to TCPS. High expression levels relative to their isotropic and 2D film controls were also observed on 200 nm and 400 nm random fibers, but to a much lower extent than on aligned fibers. After 120 hrs, overexpression shifted toward the larger diameter fibers, but was not specific to aligned fibers. A similar trend was observed with TGF-β expression. Like Snail, TGF-β is correlated with poor prognosis of GBM patients and is involved in cell invasion.[21] After 24 hrs, TGF-β was highly overexpressed on 200 nm aligned fibers which shifted to larger diameter 400 nm and 1.1 µm fibers with no selectivity towards alignment after 120 hrs, which may be a result of increased cell density and cell-cell signaling. Both Snail and TGF-β induce a mesenchymal morphology change,[22] which was observed on the aligned 200 nm and 400 nm fibers.
Lastly, STAT3 and β-catenin expression profiles showed similar trends to Snail and TGF-β. STAT3 is highly expressed in GBM stem cells,[23] and has been shown to be critical for glioblastoma cell migration on nanofibers,[14] and β-catenin enhances self-renewal,[24] a hallmark of cancer stem cells.[9,25] Cells cultured on 200 nm fibers for 24 hrs showed slightly higher expressions of STAT3 and β-catenin, which then shifted towards larger diameter 400 nm and 1.1 µm fibers with no selectivity towards alignment after 120 hrs. Cells cultured on the 200 nm aligned fibers showed the highest STAT3 expression after 24 hrs (3.7-fold higher than TCPS), which may explain the higher migration velocity of these cells on fibers of this size.
Therefore, overexpression of these invasion related genes was observed in cells cultured on smaller diameter 200 nm and 400 nm aligned fibers after 24 hrs. After 120 hrs of culture, the gene expression profiles shifted to larger diameter fibers with no specificity to alignment, and much lower expression levels. This shows the specific nanotopography of the nanofibers is critical for inducing a mesenchymal change to a more invasive phenotype at an early time point for drug treatment trials. The microenvironment provided by the aligned fibers induced a morphological change in GBM cells, which promoted a mesenchymal transition and overexpression of invasion and cancer stem cell related genes. This selective response of the GBM cells to the smaller diameter fibers suggests that the higher curvature of these nanofibers promotes the migratory behavior. Over time, the expression of the invasion related genes decreased which may be a result of asymmetric cell division.
2.5. Asymmetric cell division on aligned nanofibers
Endocytosis and membrane recycling play a large role in cell signaling and promote the progression of cancer. In fact, cancer cells regulate the distributions of endosomes and cell signaling proteins to control cell migration and division.[26] During cell division, the selective distribution, or polarization, of key signaling factors leads to asymmetric cell division which is thought to maintain cancer stem cells, or mesenchymal-like cells.[27] Since cell invasion is a mesenchymal feature of GBM,[19a,28] we assessed cell polarity by staining cell membranes with DiO oil. As the cells divide, the membrane stain is passed to daughter cells. Furthermore, newly formed endosomes also contain dye as they bud off from the stained membrane. The distributions of these endosomes in migrating and dividing cells can provide clues into the mesenchymal-like nature and invasive potential of these cells.
DiO stained cells were seeded on the nanofibers and imaged 1hr, 24 hrs, and 120 hrs after seeding (Figure 8). After 24 hrs and 120 hrs, DiO was incorporated into endosomes during cell signaling and trafficking. In TCPS, film, and randomly oriented fiber cultured cells, the distribution of DiO fluorescence was similar throughout the cells. However, in aligned fiber cultured cells, and most notably 400 nm aligned fiber cultured cells, the distribution of DiO fluorescence was asymmetric around the cell center. This asymmetric endosomal trafficking and membrane recycling was observed in 200 nm and 400 nm aligned fibers at the 24 hr time point and is an indicator of asymmetric cell division. This asymmetric cell division provides further evidence of a mesenchymal-like phenotype in cells cultured on these diameter aligned fibers. It further explains the decrease in gene expression over time in 200 nm and 400 nm fibers since mesenchymal-like cells can give rise to non-mesenchymal progeny.
Figure 8.
DiO membrane stained U-87 MG cells cultured on various substrates. Immediately after seeding, cells membranes were completely stained with DiO oil. The white lines in the images indicate the distribution of endosomes with the center of the cell marked with a perpendicular line for reference. Scale bars correspond to 10 µm.
3. Conclusion
We have examined the ability of chitosan-PCL polyblend nanofibers to promote a migratory phenotype in human GBM cells in vitro based on specific nanotopographies. An in vitro model of GBM cell migration will provide researchers with a reliable and cost-effective system for discovering anti-migration therapies. Human GBM cells cultured on small diameter 200 nm and 400 nm aligned fibers showed the greatest overexpression of invasion related genes after 24 hrs, which was attributed to a mesenchymal change. A decrease in expression of these genes after 120 hrs was a result of asymmetric cell division, a hallmark of mesenchymal-like cells. Additionally, 400 nm aligned fibers promoted similar cell migration profiles as those observed in vivo. These polyblend nanofibers provide significant advantages over currently used in vitro models for cell migration in that they are more reproducible and robust than organotypic brain slice models and better mimic the extracellular matrix of invading GBM cells than other nanofibers due to their unique morphology and chemical structure. These nanofibers will hopefully provide a better in vitro model for studying cell invasion to accelerate research in anti-migration therapies for more effective treatment of GBM.
4. Experimental Section
Materials
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise specified. Chitosan and PCL were used as received. Dulbecco’s modified Eagle’s medium (DMEM), antibiotic-antimycotic, and Dulbecco’s phosphate buffered saline (D-PBS) were purchased from Invitrogen (Carlsbad, CA). Fetal bovine serum (FBS) was purchased from Atlanta Biologicals (Atlanta, GA). U-87 MG human GBM cell lines and Minimum Essential Media (MEM) were purchased from American Type Culture Collection (ATCC, Manassas, VA). Cells were maintained according to manufacturer’s instructions in fully supplemented MEM (MEM with 10% FBS and 1% antibiotic-antimycotic) at 37° C and 5% CO2 in a fully humidified incubator.
Nanofiber synthesis
Chitosan-PCL polyblend nanofibers were synthesized according to previously published procedures.[15] Briefly, solutions of chitosan (5 wt%) and PCL (10 wt%) were prepared in trifluoroacetic acid (TFA) and mixed at a chitosan solution to PCL solution ratio of 2:3. Approximately 2 mL of the chitosan-PCL solution was loaded into a 3 mL disposable syringe with a 0.5 mm diameter tip. Randomly oriented fibers were prepared by electrospinning onto a grounded rotating drum and aligned fibers were prepared by electrospinning onto a pair of grounded parallel electrodes separated by approximately 2 cm. Fiber diameters were adjusted by changing the concentration of the chitosan-PCL solution and thus the viscosity. Fibers were then collected onto 12 mm glass cover slips. Chitosan-PCL films were prepared by spin coating the solution onto cover slips. Samples were neutralized, washed, and sterilized before cell experiments.
Generation of stably expressing RFP cells
U-87 MG cells were transfected with the pDsRed-Max-N1 plasmid (Addgene plasmid 21718, Addgene Inc., Cambridge, MA)[29] using Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA). Transfected cells were selected for neomycin resistance with 1 mg mL−1 G418 and sorted on a BD FACS Aria flow cytometer (Becton, Dickinson and Company, Franklin Lakes, NJ) to obtain pure populations of U-87 MG-RFP cells. Cells were maintained in fully supplemented DMEM containing 1 mg mL−1 G418.
Cell seeding and live imaging
Cells were seeded on sterilized nanofibers in 24-well plates at 10,000 cells per sample in 1 mL fully supplemented culture medium for SEM imaging and PCR analyses, or in 6-well plates at 50,000 cells per sample in 7 mL fully supplemented culture medium for bright field and fluorescence microscopy. Bright field and fluorescence images were obtained 24 hrs and 120 hrs after cell seeding to observe short- and long-term morphological changes using a 40× or 20× immersion objective (Nikon Instruments, Melville, NY) on an upright fluorescence microscope (Nikon Instruments, Melville, NY) using appropriate filters and Ri1 Color Cooled Camera System (Nikon Instruments, Melville, NY), and the NIS Elements software package (Nikon Instruments, Melville, NY).
Quantification of cell aspect ratio and alignment
Cell aspect ratio was quantified in images obtained with the 20× immersion objective using Photoshop (Adobe, San Jose, CA) through manual measurements of the long and short axes of the cell. At least 12 cells were measured in each image. Cell alignment was quantified in images obtained with the 20× objective using ImageJ (NIH, Bethesda, MD) through manual measurement of the long axis of the cell. Fifty-five cells were measured in each image and the average angle of the long axis of the cells was calculated. The angle of each cell away from the average angle was calculated and plotted as a histogram with binning of 20° (i.e., −90 – −70, −70 – −50, −50 – −30, −30 – −10, −10 – 10, 10 – 30, 30 – 50, 50 – 70, and 70 – 90).
Time-course imaging
U-87 MG-RFP cells were seeded on the various substrates and allowed to attach for 24 hrs before imaging. Time-course images were acquired every 10 min using a 20× immersion objective. Image sequences were analyzed using ImageJ and the MTrackJ plugin. Cell migration was determined by manually selecting cell location over the entire imaging sequence. Cell velocity was calculated by measuring the distance the cell traveled between imaging time points (10 min), and effective cell velocity was calculated by measuring the distance the cell traveled from the first to the last imaging time point.
PCR analysis
Cells cultured on nanofibers were detached using Versene (Invitrogen, Carlsbad, CA), pelleted through centrifugation at 500 rcf, and stored at −80°C before analysis. To ensure only cells on the fibers were collected, fibers were removed from the glass cover slips using forceps and placed into Versene. Cell pellets were processed to purify mRNA using the RNeasy kit (Qiagen, Valencia, CA) and cDNA generated with RT-PCR using the iScript cDNA synthesis kit (BioRad, Hercules, CA). Primers were designed using Primer-BLAST (NCBI, Bethesda, MD). Expression of β-catenin, Snail, STAT3, TGF- β, and Twist were probed using the iTaq SYBR Green Supermix (BioRad, Hercules, CA) with GAPDH as the reference gene.
Cell membrane staining
Cell membranes were stained with DiO oil (Invitrogen, Carlsbad, CA) immediately prior to seeding on nanofibers. Cells were imaged 1 hr, 24 hrs, and 120 hrs after seeding using a 40× immersion objective (Nikon Instruments, Melville, NY) on an upright fluorescence microscope (Nikon Instruments, Melville, NY) using appropriate filters and Ri1 Color Cooled Camera System (Nikon Instruments, Melville, NY). Asymmetric cell division was assessed by manually identifying the center of the cell using Photoshop and comparing the fluorescence intensity at opposite ends of the cell.
Acknowledgements
This work was supported in part by NIH grants R01CA134213 and R01EB006043 and a Kyocera Professorship Endowment. FK acknowledges support from the Ruth L. Kirschstein NIH Training grant T32CA138312. We acknowledge the use of the SEM at the Department of Materials Science and Engineering. We thank Prof. Benjamin S. Glick for submitting the pDsRed-Max-N1 plasmid to Addgene.
Contributor Information
Forrest M. Kievit, Department of Materials Science and Engineering, University of Washington, Seattle, WA, 98195, USA Department of Neurological Surgery, University of Washington, Seattle, WA, 98195, USA.
Ashleigh Cooper, Department of Materials Science and Engineering, University of Washington, Seattle, WA, 98195, USA.
Soumen Jana, Department of Materials Science and Engineering, University of Washington, Seattle, WA, 98195, USA.
Matthew C. Leung, Department of Materials Science and Engineering, University of Washington, Seattle, WA, 98195, USA
Kui Wang, Department of Materials Science and Engineering, University of Washington, Seattle, WA, 98195, USA.
Dennis Edmondson, Department of Materials Science and Engineering, University of Washington, Seattle, WA, 98195, USA.
David Wood, Department of Materials Science and Engineering, University of Washington, Seattle, WA, 98195, USA.
Jerry S. H. Lee, Department of Chemical and Biomolecular Engineering, Johns Hopkins University, Baltimore, MD, 21218, USA Center for Strategic Scientific Initiatives, National Cancer Institute, National Institutes of Health, Bethesda, MD, 20892, USA.
Richard G. Ellenbogen, Department of Neurological Surgery, University of Washington, Seattle, WA, 98195, USA
Miqin Zhang, Email: mzhang@u.washington.edu, Department of Materials Science and Engineering, University of Washington, Seattle, WA, 98195, USA; Department of Neurological Surgery, University of Washington, Seattle, WA, 98195, USA.
References
- 1.a) Huse JT, Holland EC. Nat. Rev. Cancer. 2010;10:319. doi: 10.1038/nrc2818. [DOI] [PubMed] [Google Scholar]; b) Hjelmeland AB, Lathia JD, Sathornsumetee S, Rich JN. Nat. Neurosci. 2011;14:1375. doi: 10.1038/nn.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lucio-Eterovic AK, Piao Y, de Groot JF. Clin. Cancer Res. 2009;15:4589. doi: 10.1158/1078-0432.CCR-09-0575. [DOI] [PubMed] [Google Scholar]
- 3.a) Veiseh O, Kievit FM, Ellenbogen RG, Zhang M. Adv Drug Deliv Rev. 2011;63:582. doi: 10.1016/j.addr.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lefranc F, Brotchi J, Kiss R. J. Clin. Oncol. 2005;23:2411. doi: 10.1200/JCO.2005.03.089. [DOI] [PubMed] [Google Scholar]
- 4.a) Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, Oh EY, Gaber MW, Finklestein D, Allen M, Frank A, Bayazitov IT, Zakharenko SS, Gajjar A, Davidoff A, Gilbertson RJ. Cancer Cell. 2007;11:69. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]; b) Burden-Gulley SM, Qutaish MQ, Sullivant KE, Lu H, Wang J, Craig SEL, Basilion JP, Wilson DL, Brady-Kalnay SM. Cancer Res. 2011;71:5932. doi: 10.1158/0008-5472.CAN-11-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Toole BP. Nat. Rev. Cancer. 2004;4:528. doi: 10.1038/nrc1391. [DOI] [PubMed] [Google Scholar]; b) Toole BP. Clin. Cancer Res. 2009;15:7462. doi: 10.1158/1078-0432.CCR-09-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giese A, Westphal M. Neurosurgery. 1996;39:235. doi: 10.1097/00006123-199608000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, Zhou B, Yan J, Xu B, Tai P, Li J, Peng S, Zhang M, Xia G. J Endocrinol. 2008;197:409. doi: 10.1677/JOE-07-0592. [DOI] [PubMed] [Google Scholar]
- 8.Johnson J, Nowicki MO, Lee CH, Chiocca EA, Viapiano MS, Lawler SE, Lannutti JJ. Tissue Eng Part C Methods. 2009;15:531. doi: 10.1089/ten.tec.2008.0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. Cell. 2011;144:646. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Ananthanarayanan B, Kim Y, Kumar S. Biomaterials. 2011;32:7913. doi: 10.1016/j.biomaterials.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paulus W, Roggendorf W, Schuppan D. Virchows Arch A Pathol Anat Histopathol. 1988;413:325. doi: 10.1007/BF00783025. [DOI] [PubMed] [Google Scholar]
- 12.a) Jin SG, Jeong YI, Jung S, Ryu HH, Jin YH, Kim IY. J. Korean. Neurosurg. Soc. 2009;46:472. doi: 10.3340/jkns.2009.46.5.472. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Coquerel B, Poyer F, Torossian F, Dulong V, Bellon G, Dubus I, Reber A, Vannier JP. Glia. 2009;57:1716. doi: 10.1002/glia.20884. [DOI] [PubMed] [Google Scholar]
- 13.a) Colen CB, Shen YM, Ghoddoussi F, Yu PY, Francis TB, Koch BJ, Monterey MD, Galloway MP, Sloan AE, Mathupala SP. Neoplasia. 2011;13:620. doi: 10.1593/neo.11134. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Oellers P, Schallenberg M, Stupp T, Charalambous P, Senner V, Paulus W, Thanos S. Nat. Protoc. 2009;4:923. doi: 10.1038/nprot.2009.62. [DOI] [PubMed] [Google Scholar]
- 14.Agudelo-Garcia PA, De Jesus JK, Williams SP, Nowicki MO, Chiocca EA, Liyanarachchi S, Li PK, Lannutti JJ, Johnson JK, Lawler SE, Viapiano MS. Neoplasia. 2011;13:831. doi: 10.1593/neo.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Bhattarai N, Li ZS, Gunn J, Leung M, Cooper A, Edmondson D, Veiseh O, Chen MH, Zhang Y, Ellenbogen RG, Zhang MQ. Adv. Mater. 2009;21:2792. [Google Scholar]; b) Cooper A, Jana S, Bhattarai N, Zhang MQ. J. Mater. Chem. 2010;20:8904. [Google Scholar]; c) Cooper A, Bhattarai N, Zhang M. Carbohydrate Polymers. 2011;85:149. [Google Scholar]
- 16.a) Formolo CA, Williams R, Gordish-Dressman H, MacDonald TJ, Lee NH, Hathout Y. Journal of proteome research. 2011;10:3149. doi: 10.1021/pr200210w. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) DeHauwer C, Camby I, Darro F, Decaestecker C, Gras T, Salmon I, Kiss R, VanHam P. Biochem Bioph Res Co. 1997;232:267. doi: 10.1006/bbrc.1997.6291. [DOI] [PubMed] [Google Scholar]; c) Burgoyne AM, Palomo JM, Phillips-Mason PJ, Burden-Gulley SM, Major DL, Zaremba A, Robinson S, Sloan AE, Vogelbaum MA, Miller RH, Brady-Kalnay SM. Neuro-Oncology. 2009;11:767. doi: 10.1215/15228517-2009-019. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Zhang MX, Herion TW, Timke C, Han N, Hauser K, Weber KJ, Peschke P, Wirkner U, Lahn M, Huber PE. Neoplasia. 2011;13:537. doi: 10.1593/neo.11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bettinger CJ, Langer R, Borenstein JT. Angew Chem Int Edit. 2009;48:5406. doi: 10.1002/anie.200805179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.a) Winkler F, Kienast Y, Fuhrmann M, Von Baumgarten L, Burgold S, Mitteregger G, Kretzschmar H, Herms J. Glia. 2009;57:1306. doi: 10.1002/glia.20850. [DOI] [PubMed] [Google Scholar]; b) Farin A, Suzuki SO, Weiker M, Goldman JE, Bruce JN, Canoll P. Glia. 2006;53:799. doi: 10.1002/glia.20334. [DOI] [PubMed] [Google Scholar]
- 19.a) Mikheeva S, Mikheev A, Petit A, Beyer R, Oxford R, Khorasani J-P, Maxwell L, Glackin C, Wakimoto H, Gonzalez-Herrero I, Sanchez-Garcia I, Silber J, Horner P, Rostomily R. Molecular Cancer. 2010;9:194. doi: 10.1186/1476-4598-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Cell. 2004;117:927. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 20.a) Myung J, Cho BK, Kim YS, Park SH. Neuropathology. 2010;30:224. doi: 10.1111/j.1440-1789.2009.01072.x. [DOI] [PubMed] [Google Scholar]; b) Han SP, Kim JH, Han ME, Sim HE, Kim KS, Yoon S, Baek SY, Kim BS, Oh SO. Cell Mol Neurobiol. 2011;31:489. doi: 10.1007/s10571-010-9643-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.a) Bruna A, Darken RS, Rojo F, Ocana A, Penuelas S, Arias A, Paris R, Tortosa A, Mora J, Baselga J, Seoane J. Cancer Cell. 2007;11:147. doi: 10.1016/j.ccr.2006.11.023. [DOI] [PubMed] [Google Scholar]; b) Rich JN. Front Biosci. 2003;8:e245. doi: 10.2741/992. [DOI] [PubMed] [Google Scholar]
- 22.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. Cell. 2008;133:704. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.a) Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, Lasorella A, Aldape K, Califano A, Iavarone A. Nature. 2010;463:318. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sherry MM, Reeves A, Wu JK, Cochran BH. Stem Cells. 2009;27:2383. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wang H, Lathia JD, Wu Q, Wang J, Li Z, Heddleston JM, Eyler CE, Elderbroom J, Gallagher J, Schuschu J, MacSwords J, Cao Y, McLendon RE, Wang XF, Hjelmeland AB, Rich JN. Stem Cells. 2009;27:2393. doi: 10.1002/stem.188. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Guryanova OA, Wu Q, Cheng L, Lathia JD, Huang Z, Yang J, MacSwords J, Eyler CE, McLendon RE, Heddleston JM, Shou W, Hambardzumyan D, Lee J, Hjelmeland AB, Sloan AE, Bredel M, Stark GR, Rich JN, Bao S. Cancer Cell. 2011;19:498. doi: 10.1016/j.ccr.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.a) Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Nature. 2006;444:756. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]; b) Chen MS, Woodward WA, Behbod F, Peddibhotla S, Alfaro MP, Buchholz TA, Rosen JM. J Cell Sci. 2007;120:468. doi: 10.1242/jcs.03348. [DOI] [PubMed] [Google Scholar]; c) Woodward WA, Chen MS, Behbod F, Alfaro MP, Buchholz TA, Rosen JM. Proc Natl Acad Sci U S A. 2007;104:618. doi: 10.1073/pnas.0606599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Brien CA, Kreso A, Jamieson CH. Clin Cancer Res. 2010;16:3113. doi: 10.1158/1078-0432.CCR-09-2824. [DOI] [PubMed] [Google Scholar]
- 26.a) Lanzetti L, Di Fiore PP. Traffic. 2008;9:2011. doi: 10.1111/j.1600-0854.2008.00816.x. [DOI] [PubMed] [Google Scholar]; b) Palamidessi A, Frittoli E, Garre M, Faretta M, Mione M, Testa I, Diaspro A, Lanzetti L, Scita G, Di Fiore PP. Cell. 2008;134:135. doi: 10.1016/j.cell.2008.05.034. [DOI] [PubMed] [Google Scholar]; c) Scita G, Di Fiore PP. Nature. 2010;463:464. doi: 10.1038/nature08910. [DOI] [PubMed] [Google Scholar]
- 27.a) Kosodo Y, Roper K, Haubensak W, Marzesco AM, Corbeil D, Huttner WB. Embo J. 2004;23:2314. doi: 10.1038/sj.emboj.7600223. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lathia JD, Hitomi M, Gallagher J, Gadani SP, Adkins J, Vasanji A, Liu L, Eyler CE, Heddleston JM, Wu Q, Minhas S, Soeda A, Hoeppner DJ, Ravin R, McKay RD, McLendon RE, Corbeil D, Chenn A, Hjelmeland AB, Park DM, Rich JN. Cell Death Dis. 2011;2:e200. doi: 10.1038/cddis.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elias MC, Tozer KR, Silber JR, Mikheeva S, Deng M, Morrison RS, Manning TC, Silbergeld DL, Glackin CA, Reh TA, Rostomily RC. Neoplasia. 2005;7:824. doi: 10.1593/neo.04352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strack RL, Strongin DE, Bhattacharyya D, Tao W, Berman A, Broxmeyer HE, Keenan RJ, Glick BS. Nat Methods. 2008;5:955. doi: 10.1038/nmeth.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]