Abstract
Tau pathologically aggregates in Alzheimer’s disease, and evidence suggests that reducing tau expression may be safe and beneficial for the prevention or treatment of this disease. We sought to examine the role of the 3’-untranslated region (3’-UTR) of human tau mRNA in regulating tau expression. Tau expresses two 3’-UTR isoforms, long and short, as a result of alternative polyadenylation. Using luciferase reporter constructs, we found that expression from these isoforms is differentially controlled in human neuroblastoma cell lines M17D and SH-SY5Y. Several microRNAs were computationally identified as candidates that might bind the long, but not short, tau 3’-UTR isoform. A hit from a screen of candidates, miR-34a, was subsequently shown to repress the expression of endogenous tau protein in M17D cells. Conversely, inhibition of endogenously expressed miR-34 family members leads to increased endogenous tau expression. Additionally, through an unbiased screen of fragments of the human tau 3’-UTR using a luciferase reporter assay, we identified several other regions in the long tau 3’-UTR isoform that contain regulatory cis-elements. Improved understanding of the regulation of tau expression by its 3’-UTR may ultimately lead to the development of novel therapeutic strategies for the treatment of Alzheimer’s disease and other tauopathies.
Keywords: 3’-Untranslated region, Alzheimer’s disease, Gene regulation, MAPT, Neurodegenerative disease
Introduction
The microtubule-associated protein tau promotes the assembly and stability of microtubules (Drubin and Kirschner, 1986; Weingarten, 1975). Tau is implicated in a number of neurodegenerative diseases, collectively known as tauopathies (Lee et al., 2001). In Alzheimer’s disease (AD)§, tau is a major component of intraneuronal protein aggregates known as neurofibrillary tangles (NFTs) (Kosik et al., 1986; Nukina and Ihara, 1986; Wood et al., 1986). While the presence of tau aggregates in AD and other tauopathies provides a correlative association between tau and disease (Lee et al., 2001), the strongest evidence for a causal relationship between tau and neurodegenerative disease comes from the presence of dominant mutations of the microtubule-associated protein tau (MAPT) gene in rare familial cases of frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17) (Hutton et al., 1998; Poorkaj et al., 1998; Spillantini et al., 1998).
In the amyloid cascade hypothesis of AD pathogenesis, altered tau protein and NFT formation are considered to be downstream consequences of amyloid β (Aβ) toxicity (Hardy and Higgins, 1992). Although alterations in tau are not considered the earliest event in AD pathogenesis, several studies have provided strong evidence that tau plays a key role in this process. Cultured hippocampal neurons from homozygous tau knockout mice failed to develop Aβ-induced cytotoxicity (Rapoport et al., 2002) and axonal transport defects (Vossel et al., 2010) seen in wild-type hippocampal neurons. Similarly, knock out of tau in transgenic mouse models of AD results in improved learning and memory, increased survival, and reduced excitotoxicity compared with littermates with wild-type tau (Ittner et al., 2010; Roberson et al., 2007). Taken together, these studies suggest that a reduction of tau expression may be an effective therapeutic strategy for treating AD. As a first step toward identifying novel therapeutic strategies to reduce tau levels, our laboratory undertook the present study to investigate the role of the human tau 3’-untranslated region (3’-UTR) in regulating tau expression.
In general, 3’-UTRs have a variety of functions in regulating gene expression, such as 3’-end processing, translational regulation, stability, and subcellular localization of the transcript (Barrett et al., 2012). These regulatory roles are mediated by the interaction of cis-elements in the 3’-UTR and trans-factors, which include proteins and microRNAs (miRNAs), in the cell (Chen et al., 2006). miRNAs are endogenous, approximately 22 nucleotide (nt) long RNAs that regulate gene expression via translation repression or mRNA cleavage (Bartel, 2004). An additional level of regulation results from alternative polyadenylation (APA), which can occur in transcripts that contain two or more polyadenylation signals (PASs). Transcripts that undergo APA express alternative 3’-UTR isoforms, which can be differentially regulated by either the inclusion or exclusion of regulatory cis-elements (Shi, 2012).
The human tau 3’-UTR, as well as that of rodents, contains two PASs in tandem and can undergo APA to produce transcripts of approximately 2 or 6 kb (Poorkaj et al., 2001). Profiling of the tau 3’-UTR isoform expression pattern by Northern blot has shown that the isoform expression pattern is tissue and cell type-specific (Goedert et al., 1988; Wang et al., 1993). In the frontal cortex of normal subjects, the long tau 3’-UTR is more highly expressed. However, in the frontal cortex of subjects with AD, levels of the short tau 3’-UTR isoform are apparently increased, such that the levels of the short and long isoforms are approximately equal (Goedert et al., 1988).
Despite these findings, the functional consequences of the APA of tau on its expression have not been investigated. A few studies on the rat or mouse tau 3’-UTR have identified a regulatory cis-element and trans-factors involved in the stability (Aranda-Abreu et al., 1999; Aronov et al., 1999), translational efficiency (Atlas et al., 2007), and axonal localization (Aronov et al., 2001; Behar et al., 1995; Larcher et al., 2004) of rodent tau mRNA. The 91-nt T-rich regulatory cis-element identified in these studies of rodent tau 3’-UTR is not well-conserved in humans (Poorkaj et al., 2001), making it difficult to infer the functional regulatory cis-elements and trans-factors for the human tau 3’-UTR.
In this study, we sought to investigate the role of the human tau 3’-UTR in regulating tau expression with the hope that a deeper understanding of these regulatory mechanisms may lead to the identification of novel therapeutic approaches to treat AD and other tauopathies. Here, we report that expression from the long and short tau 3’-UTR isoforms is differentially controlled, suggesting a role of APA in regulating tau expression. Furthermore, we show that miR-34a can bind the human tau 3’-UTR and reduce tau levels, whereas inhibition of endogenous miR-34 family members results in increased tau levels. Finally, we present the preliminary identification of additional regulatory cis-elements in the human tau 3’-UTR.
Experimental Procedures
3’-RACE
Rapid Amplification of cDNA 3’-Ends (3’-RACE) was performed on human frontal cortex total RNA (Agilent Technologies) with the FirstChoice RLM-RACE Kit (Life Technologies) using primers targeting the short and long human tau 3’-UTR isoforms (Supplemental Table S1). The 3’-RACE products were sequenced and aligned with human MAPT using ClustalW (Larkin et al., 2007).
Cloning and Site-Directed Mutagenesis
The wild-type human tau 3’-UTR sequence was amplified from genomic DNA derived from primary human neonatal dermal fibroblasts (ATCC) by PCR using KAPA HiFi HotStart ReadyMix (Kapa Biosystems). This sequence was inserted into the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega) between the XhoI and SalI restriction sites using the In-Fusion HD EcoDry Cloning Kit (Clontech). Additional tau 3’-UTR sequences were subcloned from the wild-type human tau 3’-UTR vector using the same method. Primers used for cloning are listed in Supplemental Table S2. Site-directed mutagenesis was performed using the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies). Primers used for site-directed mutagenesis are listed in Supplemental Table S3.
Cell Culture
All cells were cultured at 37 °C in a 5% CO2 atmosphere. Primary human neonatal dermal fibroblasts (ATCC) were cultured in Fibroblast Basal Medium supplemented with Fibroblast Growth Kit—Low Serum (ATCC). M17D human neuroblastoma cells (kind gift of Dennis Selkoe) and HEK 293 cells (ATCC) were cultured in DMEM (Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Sigma-Aldrich) and l-glutamine (Life Technologies). SH-SY5Y human neuroblastoma cells were cultured in 1:1 Eagle’s Minimal Essential Medium (EMEM): Ham’s F-12 Medium (Sigma-Aldrich) supplemented with 10% FBS (ATCC). SH-SY5Y cells were differentiated using a modified version of a previously published protocol (Encinas et al., 2000). Briefly, SH-SY5Y cells were plated on fibronectin-coated plates (BD-Biosciences) and treated with 10 µM retinoic acid (Sigma-Aldrich) in 1:1 EMEM:Ham’s F-12 Medium supplemented with 10% FBS for 7 days. The SH-SY5Y cells were then treated with 50 ng/mL brain-derived neurotrophic growth factor (BDNF) (Life Technologies) in 1:1 EMEM:Ham’s F-12 Medium (without FBS) for 3 days.
Prediction of miRNAs Binding the Human Tau 3’-UTR
microrna.org (Betel et al., 2007), PicTar (Krek et al., 2005), TargetScan (Lewis et al., 2005), and DIANA-microT (Maragkakis et al., 2009) were searched for miRNAs predicted to bind the human MAPT gene.
Transfections
M17D and HEK cells were transfected using Lipofectamine 2000 (Life Technologies), and differentiated SH-SY5Y cells were transfected using Lipofectamine LTX (Life Technologies) according to the manufacturer’s protocols. Plasmids were transfected at 1 µg per well of a 24-well plate. Co-transfections included 200 ng plasmid and 30 nM Pre-miR miRNA Precursor (Life Technologies) per well of a 24-well plate. Transfection of Pre-miR miRNA Precursors was performed at a concentration of 50 nM. Transfection of miRCURY LNA microRNA Inhibitors (Exiqon) was performed at a concentration of 20 nM per LNA. Sequences of these oligonucleotides are listed in Supplemental Table S4.
Luciferase Assay
Firefly and Renilla luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega), which uses luciferin as a substrate for firefly luciferase, and coelenterazine as a substrate for Renilla luciferase, according to the manufacturer’s protocol. Luminescence signal was measured using a Synergy H1 Hybrid Multi-mode Microplate Reader (BioTek).
RNA Isolation and Quantitative PCR
RNA was isolated using the Nucleospin RNA II kit (Macherey-Nagel). Containing DNA was removed from the RNA by DNase treatment using the TURBO DNA-free Kit (Life Technologies). The RNA was reverse transcribed to cDNA using Double Primed RNA to cDNA EcoDry Premix (Clontech). Quantitative PCR (qPCR) was performed using TaqMan chemistry (Life Technologies) using the absolute quantification method of analysis on the ViiA 7 Real-Time PCR System (Life Technologies) using the pmirGLO vector as a standard for the firefly luciferase gene luc2 and the Renilla luciferase gene hRluc and custom GeneArt Strings DNA Fragments (Life Technologies) as standards for tau and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Supplemental Table S5). Custom TaqMan probes (Life Technologies) were designed for the luc2 and hRluc genes in the pmirGLO vector. TaqMan Gene Expression Assays (Life Technologies) were used for tau and GAPDH. Additional information on the TaqMan probes is provided in Supplemental Table S6.
Western Blot
Cells were lysed in M-PER Mammalian Protein Extraction Reagent (Thermo Scientific) containing Halt Protease Inhibitor Cocktail (Thermo Scientific). The protein in the lysates was quantified using the Pierce BCA Protein Assay Kit (Thermo Scientific) according to the manufacture’s instructions. For each lysate, 3 µg total protein, NuPAGE Sample Reducing Agent (Life Technologies), and NuPAGE LDS Sample Buffer (Life Technologies) were combined and heated to 70 °C for 10 minutes (min). The samples were subjected to lithium dodecyl sulfate polyacrylamide gel electrophoresis (LDS-PAGE) on a NuPAGE Novex 4–12% Bis-Tris Gel (Life Technologies) in XT MES Buffer (Bio-Rad) at 200 V for 35 min. The proteins were transferred to an Immobilon-P membrane (Millipore) in Novex Tris-Glycine Buffer (Life Technologies) with 20% methanol at 180 mA for 75 min. The blots were blocked in 5% milk in tris-buffered saline (TBS) with 0.1% Tween-20 (TBST) and probed with primary antibodies (1:12,000 dilution) for total tau (Dako), sirtuin 1 (SIRT1) (Abcam), and GAPDH (Novus Biologicals) in 5% milk in TBST for 1 h at room temperature or overnight at 4 °C. Secondary antibodies conjugated to horseradish peroxidase (HRP) (GE Healthcare) were used as a dilution of 1:8,000 in 5% milk in TBST for 1 h at room temperature. The HRP activity was visualized using Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare Life Sciences), which uses luminol as a substrate for HRP. Blots were stripped using Restore Plus Western Blot Stripping Buffer (Thermo Scientific) according to the manufacturer’s instructions.
Quantification of western blots
Densitometry was carried out with ImageJ image processing software, using the gel analysis method (http://imagej.nih.gov/ij/)(Abramoff et al. 2004). Loading variability was controlled for tau and SIRT1 by normalizing to the GAPDH signal for the corresponding lane. All values were expressed as a percentage of the average value for the corresponding negative control condition. Analysis by t-test was carried out using GraphPad Prism.
Statistical Analysis
Data are presented as a mean of biological replicates, and variation is indicated as the SEM. Comparisons of more than two groups were conducted using a one-way ANOVA followed by a correction for multiple comparisons. Comparisons of two groups were conducted using an unpaired Student’s t-test.
Results
3’-RACE and cloning of human tau 3’-UTR isoform constructs
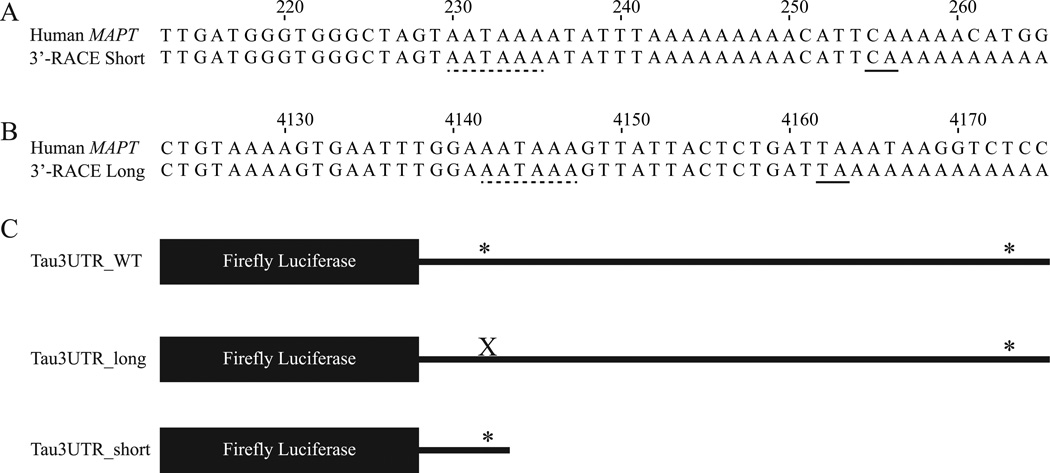
In order to define the 3’-ends of the human tau 3’-UTR isoforms, total RNA from human frontal cortex was subjected to 3’-RACE. The 3’-RACE products were sequenced and aligned with the human MAPT gene to identify the site where cleavage and polyadenylation occurs for each isoform. The short 3’-UTR isoform is 256 nt long beginning at the first nucleotide after the stop codon, has a canonical AATAAA PAS, and has a canonical CA cleavage site (Figure 1A). The long tau 3’-UTR isoform is 4163 nt in length, has a canonical AATAAA PAS, and has a non-canonical TA cleavage site (Figure 1B). These data were used to clone the wild-type and short tau 3’-UTR sequences into the pmirGLO luciferase reporter construct. A construct that expresses the long only tau 3’-UTR was made by mutating the proximal tau PAS (A233C) in the wild-type tau 3’-UTR constructs so that cleavage and polyadenylation does not occur at this site. A schematic of these constructs is shown in Figure 1C.
Figure 1.
Human tau 3’-UTR isoforms. A and B. ClustalW alignment of the 3’-UTR of the human MAPT gene with (A) the short 3’-RACE product and (B) the long 3’-RACE product. The numbering begins at the first nucleotide of the human tau 3’-UTR. The PAS is indicated by a dashed line, and the cleavage site is indicated by a solid line. C. Diagram of the luciferase reporter constructs containing WT, long, and short tau 3’-UTRs. * indicates a PAS. X represents mutation of the proximal PAS.
The expression of human tau 3’-UTR isoforms is differentially regulated
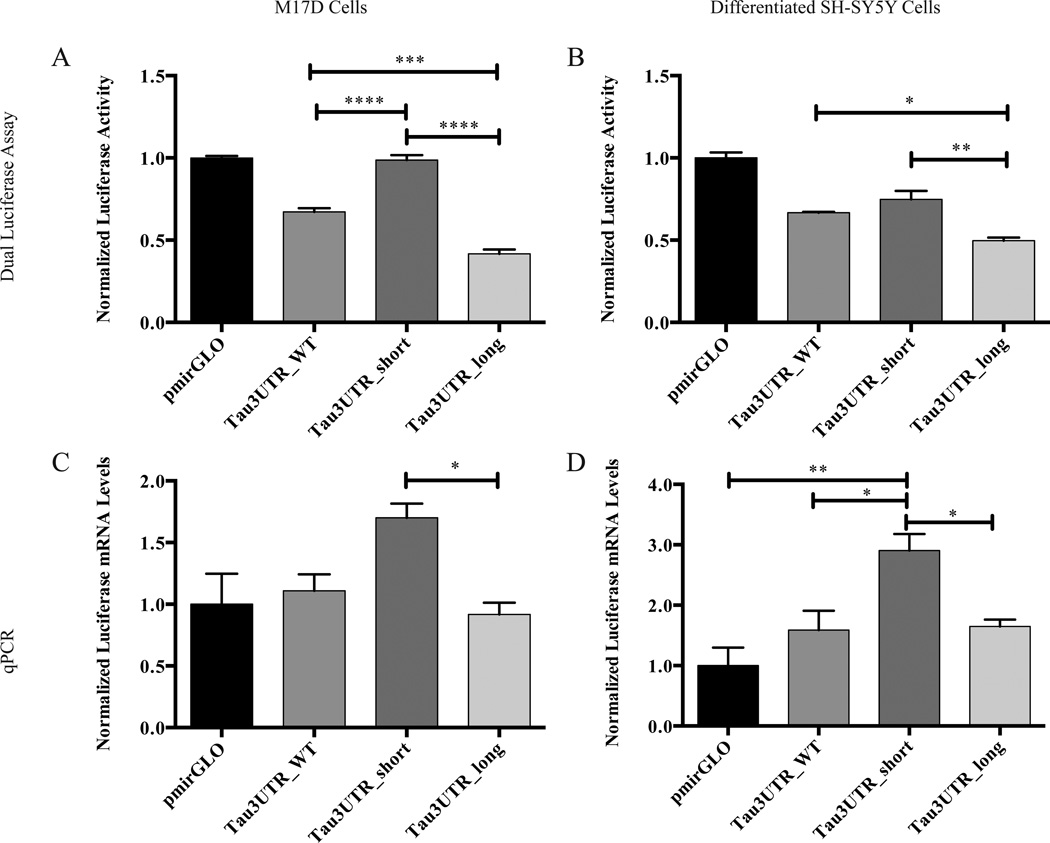
We used the luciferase reporter constructs to investigate the potential role of alternative polyadenylation in regulating tau expression. The tau 3’-UTR isoform constructs or empty pmirGLO vector were transfected into two human neuroblastoma cells lines, M17D and differentiated SH-SY5Y cells. In the pmirGLO-based constructs, firefly luciferase, which has the tau 3’-UTR isoforms cloned downstream of it, serves as the experimental reporter, whereas the Renilla luciferase serves as a normalization control for transfection efficiency. After 48 h, expression of the firefly and Renilla luciferase reporters was determined at the protein level by a dual luciferase assay and at the mRNA level by qPCR, which revealed that the expression of the short isoform construct is significantly higher than that of the long isoform construct in both cell lines at the level of both protein and mRNA (Figure 2). In general, the wild-type tau 3’-UTR reporter, which contains both PASs, had intermediate expression levels (Figure 2). Since M17D cells and differentiated SH-SY5Y cells gave similar results, we elected to perform most future experiments in M17D cells since they were transfected with much higher efficiency (data not shown).
Figure 2.
Expression of human tau 3’-UTR isoforms is differentially regulated. The indicated constructs were transfected into (A and C) M17D cells or (B and D) differentiated SH-SY5Y cells. After 48 h, luciferase expression was determined at the protein level by (A and B) dual luciferase assay or at the mRNA level by (C and D) qPCR. For each experiment, n = 3 biological replicates. Error bars represent SEM. Data analyzed by one-way ANOVA followed by Tukey test. *,**,***,****, adjusted p < 0.05, 0.01, 0.001, 0.0001, respectively. Note that in (A), comparisons of pmirGLO with Tau3UTR_WT and Tau3UTR_long reached statistical significance and in (B), all comparisons with pmirGLO reached statistical significance. These comparisons with pmirGLO are not indicated on the graph for the sake of clarity. No comparisons with pmirGLO are indicted in (C) with the vector control because no comparisons were significant with vector control.
Identification of miR-34 family members as regulators of tau expression
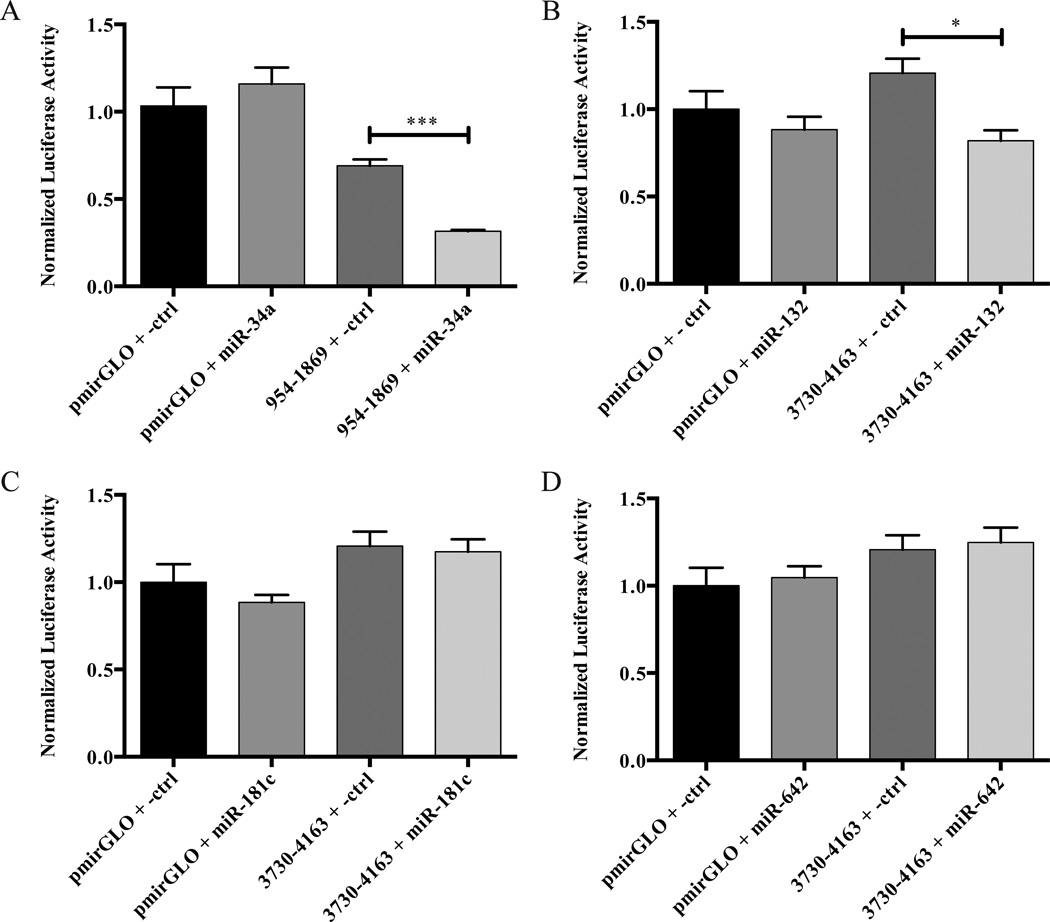
Since miRNAs often bind 3’-UTRs and regulate gene expression, we considered whether one or more miRNAs might contribute to the control of tau expression. As a first step in exploring this question, we searched microrna.org (Betel et al., 2007), PicTar (Krek et al., 2005), TargetScan (Lewis et al., 2005), and DIANA-microT (Maragkakis et al., 2009) for miRNAs predicted to bind the human tau 3’-UTR. We narrowed the list of candidate miRNAs by selecting those with conserved seed regions and/or previous reports of dysregulation in AD (Cogswell et al., 2008; Maes et al., 2009) (Table 1). In order to screen the four candidate microRNAs for functional effects on constructs expressing the tau 3’-UTR, we co-transfected HEK 293 cells with the Pre-miR precursor miRNA mimic corresponding to the candidate miRNA or a non-targeting negative control Pre-miR and a luciferase reporter vector containing a fragment of the human tau 3’-UTR predicted to contain the miRNA binding site. As a control for specificity, co-transfections of Pre-miRs and the empty pmirGLO vector were also performed. In this screen, a specific reduction in the tau 3’-UTR luciferase reporter activity was observed for Pre-miR-34a and Pre-miR-132; however, no effect was seen with Pre-miR-181c or Pre-miR642 (Figure 3).
Table 1.
Candidate miRNAs Predicted to Target the Human Tau 3’-UTR.
| miRNA | Prediction Program Identifying this miRNA | Location of Predicted Binding Site in 3’-UTR |
|---|---|---|
| hsa-miR-34a | microRNA.org, PicTar, TargetScan, DIANA-microT | 1172–1194 |
| hsa-miR-132 | microRNA.org, DIANA-microT | 4104–4121 |
| hsa-miR-181c | microRNA.org, DIANA-microT | 4056–4074 |
| hsa-miR-642 | DIANA-microT | 4019–4036 |
Figure 3.
Screening candidate miRNAs for effects on luciferase reporter activity from tau 3’-UTR construct expression. The indicated luciferase reporter constructs and Pre-miRs were co-transfected into HEK 293 cells. After 48 h, luciferase protein levels were determined by a dual luciferase assay. For each experiment, n = 3 biological replicates. The error bars indicate SEM. Data were analyzed by unpaired t-test. *,***, p < 0.05, 0.001, respectively. Luciferase assays performed on lysates from cells treated with (A) Pre-miR-34a, (B) Pre-miR-132, (C) Pre-miR-181c, and (D) Pre-miR-642.
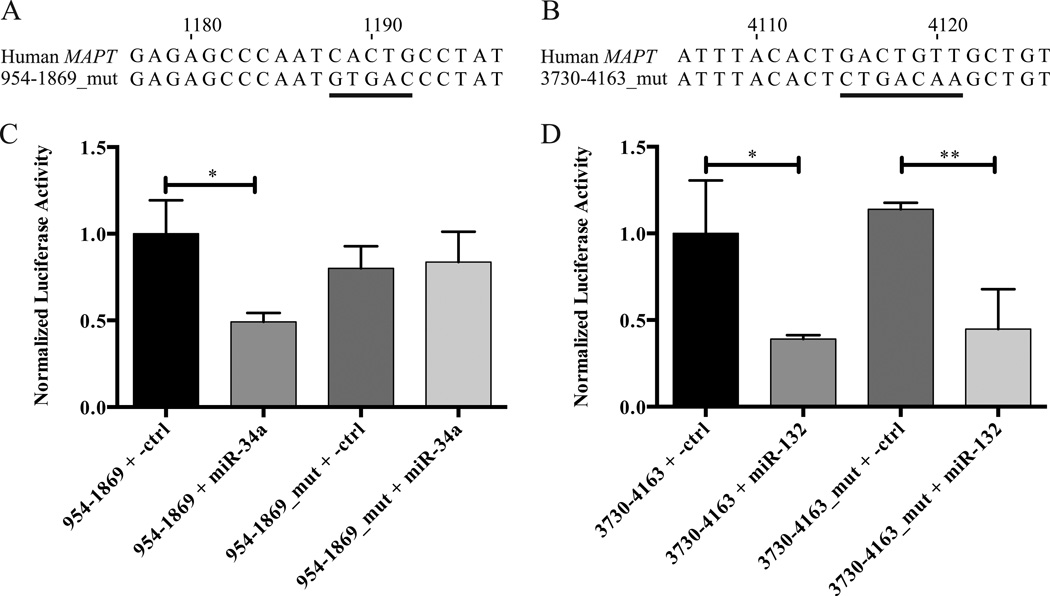
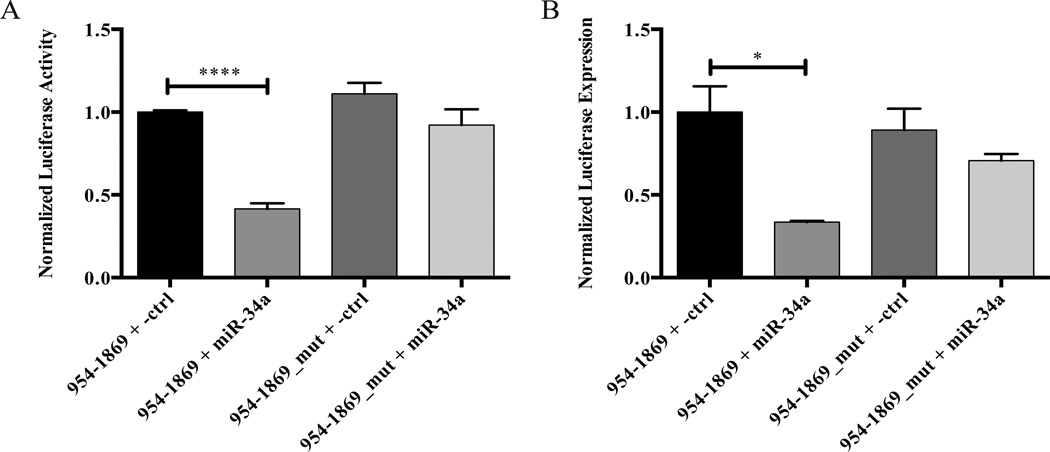
In a follow-up experiment, the seed regions of miR-34a and miR-132 were mutated in the luciferase reporter constructs by site-directed mutagenesis (Figures 4A & 4B). Co-transfection of Pre-miR-34a with the luciferase reporter construct containing the wild-type miR-34a binding site in HEK 293 cells resulted in lower normalized luciferase reporter expression compared with the non-targeting negative control Pre-miR (Figure 4C). In contrast, the miR-34a seed region mutant luciferase reporter vector was insensitive to Pre-miR-34a (Figure 4C), demonstrating that miR-34a does indeed interact specifically with this seed region. A similar experiment was performed with Pre-miR-132. In this case, Pre-miR-132 repressed the expression of both the wild-type and mutated miR-132 seed region tau 3’-UTR luciferase reporter constructs (Figure 4D), indicating that the observed effect was not due to binding of miR-132 to its predicted binding site in the human tau 3’-UTR. As the effect of miR-132 appears to be indirect, we decided to focus on exploring the role of miR-34a in regulating tau expression.
Figure 4.
Effect of mutating the miR-34a and miR-132 seed regions in tau 3’-UTR luciferase constructs. A and B. Alignment of the mutated (A) miR-34a or (B) miR-132 seed region tau 3’-UTR construct with the wild-type human MAPT sequence. The numbering begins at the first nucleotide of the human tau 3’-UTR. The mutated nucleotides of the seed regions are underlined. C and D. The indicated luciferase reporter constructs and Pre-miRs were cotransfected into HEK 293 cells. After 48 h, luciferase expression was determined by a dual luciferase assay. n = 3 biological replicates. Error bars indicate SEM. Data analyzed by unpaired t-test. *,**, adjusted p < 0.05 and 0.01, respectively.
In order to test the effect of miR-34a on the luciferase reporter constructs in a system more relevant to the nervous system, the co-transfections with Pre-miR-34a were repeated in M17D neuroblastoma cells. After 48 h, the cells were lysed and a dual luciferase assay was performed, which yielded the same result in M17D cells (Figure 5A) as was seen in HEK 293 cells (Figure 4C). In order to examine the effect of Pre-miR-34a on the luciferase reporters at the mRNA level, the co-transfections were repeated in M17D cells, and the expression levels of firefly and Renilla luciferase mRNAs were measured by qPCR. Again, the construct with wild-type miR-34a seed region was sensitive to Pre-miR-34a, whereas the miR-34a seed region mutant was not (Figure 5B). These results indicate that miR-34a can affect the expression of reporter constructs containing the miR-34a binding site at both the level of protein and mRNA.
Figure 5.
The Pre-miR-34a mediated repression of expression from a tau 3’-UTR luciferase reporter construct is miR-34a seed region-dependent in M17D cells. The indicated luciferase reporter constructs and Pre-miRs were co-transfected into M17D cells. After 48 h, luciferase expression was determined at the protein level by (A) dual luciferase assay or at the mRNA level by (B) qPCR. For each experiment, n = 3 biological replicates. Error bars indicate SEM. Data analyzed by unpaired t-test. *,****, adjusted p < 0.05 and 0.0001, respectively.
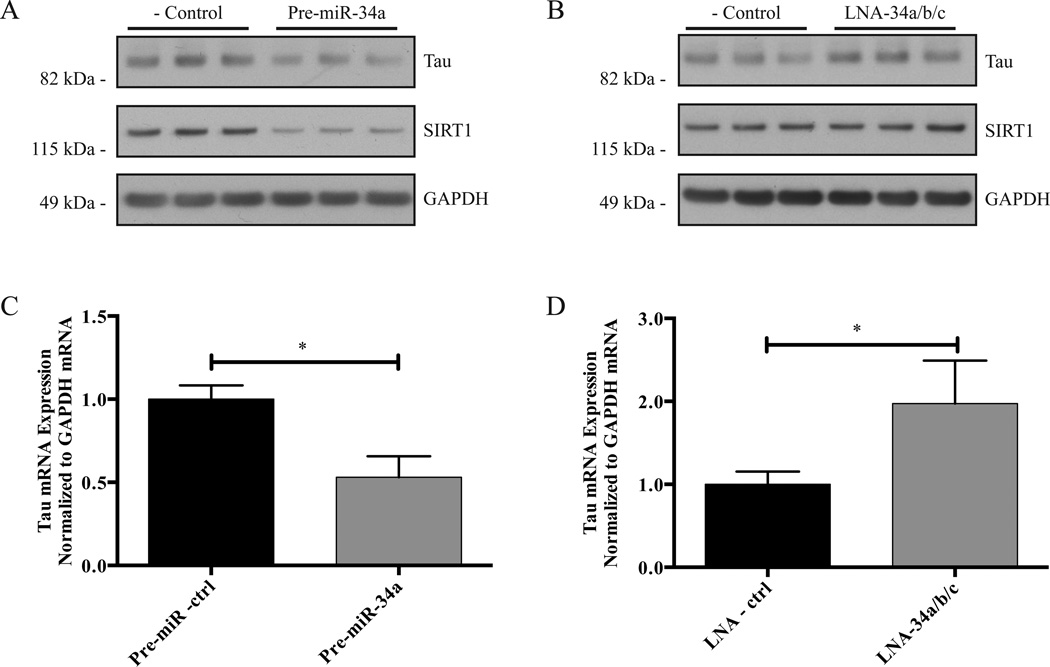
In order to investigate the ability of miR-34a to regulate the expression of endogenous tau, we transfected Pre-miR-34a or a non-targeting negative control Pre-miR into M17D cells. After 48 h, the cells were lysed, and a Western blot probing for total tau, SIRT1, and GAPDH was performed. SIRT1 was included as a positive control, as it is a known target of miR-34a (Yamakuchi et al., 2008), and GAPDH was included as a loading control. Cells transfected with Pre-miR-34a had reduced expression of tau and SIRT1 compared to cells transfected with the negative control Pre-miR (Figure 6A; quantification in Fig. S1), demonstrating that miR-34a can control endogenous tau protein expression. To determine the ability of endogenous miRNAs to regulate endogenous tau expression, we co-transfected locked nucleic acid (LNA) microRNA inhibitors of miR-34a, -34b, and -34c into M17D cells and performed the western blot analysis as described above. We used a combination of LNA inhibitors of all miR-34 family members to prevent possible compensation by other family members if an inhibitor of only one family member were used. The western blot analysis revealed that inhibition of endogenous miR-34 family members led to a moderate increase in endogenous tau protein expression (Figure 6B; quantification in Fig. S2). SIRT1 levels did not appear to be affected, suggesting that the role of endogenous miR-34 family members in regulating SIRT1 expression in this cell line may be marginal compared with other SIRT1-regulating trans-factors. We then examined the effect of Pre-miR-34a or LNA inhibition of miR-34 family members on the expression of endogenous total tau mRNA in M17D cells using qPCR, with GAPDH mRNA serving as a normalization control. In agreement with the western blot data, Pre-miR-34a reduced tau mRNA expression (Figure 6C), whereas inhibition of miR-34 family members led to an increase in tau mRNA expression (Figure 6D). Taken together, these data indicate that endogenous miR-34 family members regulate tau expression at both the protein and mRNA levels.
Figure 6.
miR-34 family members regulate the expression of endogenous tau. The indicated oligonucleotides were transfected into M17D cells. After 48 h, the cells were lysed. A and B. Western blot analysis was performed using primary antibodies for tau, SIRT1, and GAPDH. See Supplemental Figures S1 and S2 for quantification by densitometry. C and D. qPCR was performed using TaqMan probes for tau and GAPDH. For each experiment, n = 3 biological replicates. Error bars indicate SEM. Data analyzed by unpaired t-test. *, adjusted p < 0.05.
Unbiased search for additional regulatory cis-elements in the human tau 3’-UTR
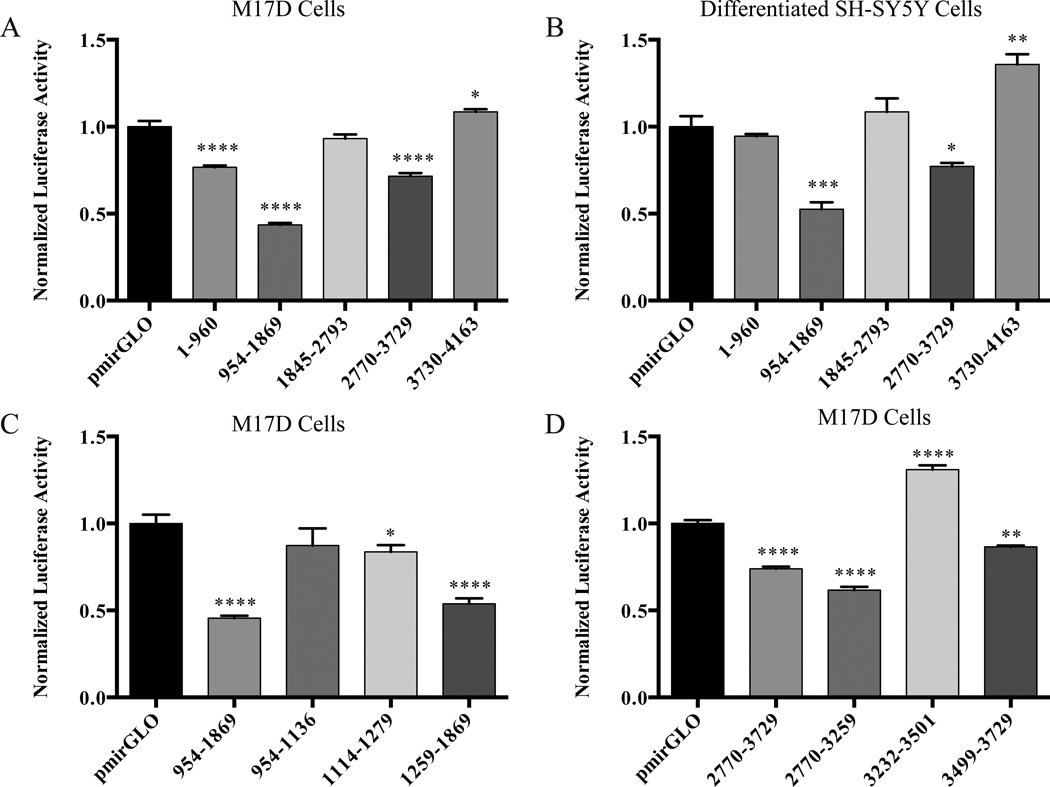
Recognizing that there may be additional regulatory cis-elements in the human tau 3’-UTR besides the miR-34 family binding site, we decided to take an unbiased approach to identify additional cis-elements by cloning sequential fragments of approximately 1,000 bp of the human tau 3’-UTR into the pmirGLO vector. We transfected these constructs, along with the empty pmirGLO vector into M17D cells and differentiated SH-SY5Y cells. After 48 h, a dual luciferase assay was performed. While several fragments had a normalized luciferase activity that was significantly different from the empty pmirGLO vector in M17D cells (Figure 7A), we chose to focus on fragments containing nucleotides 954–1869 and 2770–3729 in the human tau 3’-UTR since these fragments also consistently showed a similar pattern of decreased normalized luciferase activity compared to empty pmirGLO vector in differentiated SH-SY5Y cells (Figure 7B) and HEK 293 cells (data not shown). In order to further define the regulatory cis-elements in these fragments, smaller sub-fragments were cloned into the pmirGLO vector, which were tested in the same manner as the larger fragments. For the sub-fragments of the 954–1869 fragment, a relatively small, but statistically significant, difference in the normalized luciferase activity of the sub-fragment containing the miR-34a binding site (1114–1279) compared to empty pmirGLO vector was observed (Figure 7C). Additionally, a substantial and significant decrease in normalized luciferase activity was observed for sub-fragment 1259–1869 (Figure 7C). For the sub-fragments of the 2770–3729 fragment, the normalized luciferase activity of sub-fragments 2770–3259 and 3499–3729 was significantly decreased compared to empty pmirGLO vector, whereas the normalized luciferase activity of sub-fragment 3232–3501 was significantly increased (Figure 7D).
Figure 7.
Identification of regulatory cis-elements in the human tau 3’-UTR. The indicated constructs were transfected into the indicated cells, and after 48 h, a dual luciferase assay was performed. A. Analysis of fragments from the entire human tau 3’-UTR in M17D cells. B. Analysis of fragments from the entire human tau 3’-UTR in differentiated SH-SY5Y cells. C. Analysis of fragments within nucleotides 954 to 1869 of the tau 3’-UTR in M17D cells. D. Analysis of fragments within nucleotides 2770 to 3729 of the tau 3’-UTR in M17D cells. For each experiment, n = 3 biological replicates. The error bars indicate SEM. Data were analyzed by a one-way ANOVA followed by Dunnett’s test, compared with pmirGLO. *,**,***,****, adjusted p< 0.05, 0.01, 0.001, and 0.0001, respectively.
Discussion
Because of its role in neurodegenerative diseases, tau has been extensively investigated in the past few decades. However, the role of the tau 3’-UTR in regulating tau expression has been relatively understudied, especially in humans. Here, we present the first evidence that the expression of the two human tau 3’-UTR isoforms is differentially regulated, with effects at the level of both protein and mRNA. This indicates that APA is a mechanism by which tau expression can be modulated. Because APA is spatially and temporally regulated, it is an important determinant of gene expression profiles in different tissues and in different developmental stages (Shi, 2012). Additionally, changes in APA have been associated with disease. For example, global shifts toward the use of proximal PASs have been observed in cancer (Lin et al., 2012) and cardiac hypertrophy (Park et al., 2011). Since APA is an important mechanism of regulating gene expression, further investigations of the physiological and/or pathological contexts in which the APA of tau is involved are warranted.
In examining the regulatory cis-elements and trans-factors responsible for the differential regulation of the human tau 3’-UTR isoforms, we identified a miR-34a binding site in the long tau 3’-UTR isoform and demonstrated that miR-34a can inhibit the expression of endogenous tau. miR-34a is one of three members of the miR-34 family and is processed from a primary transcript encoded on human chromosome 1p36. The other two members of the miR-34 family, miR-34b and miR-34c are co-transcribed from the same locus on human chromosome 11q23 (Yang, 2011). Because they are transcribed from different loci, the miR-34 family members can have different expression profiles, with the expression of miR-34a being highest in the brain and the expression of miR-34b and miR-34c being highest in the lungs (Bommer et al., 2007). miR-34a has been found to regulate neuronal differentiation and neurite outgrowth (Agostini et al., 2011a; 2011b; Aranha et al., 2011), and miR-34a expression is upregulated in brain tissue (Cogswell et al., 2008) and blood mononuclear cells (Schipper et al., 2007) from patients with AD. Our findings that miR-34a can regulate the expression of tau, a key player in AD pathogenesis, together with the alteration of miR-34a expression in AD raise the questions if and how the changes in miR-34a expression relate to the pathophysiology of AD. One possibility is that the upregulation of miR-34a may act as a compensatory mechanism to decrease the expression of tau as tau protein accumulates and aggregates. This idea, however, is speculative, and miR-34a would presumably be only one of many factors conspiring to control overall tau expression.
During the preparation of this manuscript, Wu and colleagues published a report that exogenous miR-34c can bind the human tau 3’-UTR and repress tau expression in gastric cancer cells (Wu et al., 2013). While their study was focused on understanding mechanisms underlying paclitaxel resistance in gastric cancers and their miRNA of interest was miR-34c (Wu et al., 2013), their findings are consistent with our identification of miR-34a as a regulator of tau expression. As members of the same miRNA family, miR-34a and miR-34c have the same seed region, and they are predicted to bind to the same region of the tau 3’-UTR. Taken together, our finding that LNA inhibition of endogenous miR-34 family members leads to an increase in tau expression and the findings of Wu and colleagues indicate a role for miR-34 family members in regulating tau expression.
In addition to the miR-34 binding site, we have sought to identify other regulatory cis-elements in the human tau 3’-UTR. Our preliminary investigations have identified several regions containing cis-elements, most of which are repressive. In these experiments, the fragment containing the miR-34a binding site (1114–1279) had a small but significant reduction in expression compared with the pmirGLO vector. The relatively small magnitude of this effect may be due to the fact that the overexpression of the luciferase reporter makes the levels of the luciferase reporter mRNA relatively high compared with the expression levels of endogenous miR-34 family members. We plan to continue these studies in order to identify the minimal regulatory cis-elements present in the human tau 3’-UTR. Additionally, we plan to identify the trans-factors that bind these cis-elements and regulate tau expression. Ultimately, we hope the increased understanding of the regulation of tau expression by its 3’-UTR from this and future studies will lead to the identification of new therapeutic approaches for the prevention or treatment of AD and other tauopathies.
Supplementary Material
Acknowledgements
The authors would like to thank Rachel Moda for technical assistance with site-directed mutagenesis and the members of the Wolfe Lab for helpful discussions. J.D. and M.W. conceived of and designed the studies. J.D., C.K., and D.M. were responsible for data acquisition and analysis. J.D. drafted the article. All authors were responsible for interpretation of data, critical revision of the manuscript, and approving the final version for publication. This work was supported by a scholarship from the University of Southern Denmark (C.K.), the Lundbeck Foundation (B.F., C.K.), and NIH T32 grant GM007306 (J.D.).
Footnotes
ARRIVE Guidelines are not applicable. No animal experiments performed.
The abbreviations used are: 3’-RACE, Rapid Amplification of cDNA 3’-Ends; 3’-UTR, 3’-untranslated region; Aβ, amyloid β; AD, Alzheimer’s disease; APA, alternative polyadenylation; BDNF, brain-derived neurotrophic growth factor; DMEM, Dulbecco’s Modified Eagle Medium EMEM, Eagle’s Minimal Essential Medium; FBS, fetal bovine serum; FTDP-17, frontotemporal dementia with parkinsonism linked to chromosome 17; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HRP, horseradish peroxidase; LDS-PAGE, lithium dodecyl sulfate PAGE; LNA, locked nucleic acid; MAPT, microtubule-associated protein tau; min, minutes; miRNA, microRNA; NFTs, neurofibrillary tangles; nt, nucleotide; PAS, polyadenylation signal; qPCR, quantitative PCR; SIRT1, sirtuin 1; TBS, tris-buffered saline; TBST, TBS with 0.1% Tween-20; WT, wild-type.
The authors declare no conflict of interest.
References
- Abramoff MD, Magalhaes PJ, Ram SJ. Image Processing with Image. J. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Agostini M, Tucci P, Killick R, Candi E, Sayan BS, Rivetti di Val Cervo P, Nicotera P, McKeon F, Knight RA, Mak TW, et al. Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proceedings of the National Academy of Sciences U.S.A. 2011a;108:21093–21098. doi: 10.1073/pnas.1112061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M, Tucci P, Steinert JR, Shalom-Feuerstein R, Rouleau M, Aberdam D, Forsythe ID, Young KW, Ventura A, Concepcion CP, et al. microRNA-34a regulates neurite outgrowth, spinal morphology, and function. Proceedings of the National Academy of Sciences U.S.A. 2011b;108:21099–21104. doi: 10.1073/pnas.1112063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda-Abreu GE, Behar L, Chung S, Furneaux H, Ginzburg I. Embryonic lethal abnormal vision-like RNA-binding proteins regulate neurite outgrowth and tau expression in PC12 cells. Journal of Neuroscience. 1999;19:6907–6917. doi: 10.1523/JNEUROSCI.19-16-06907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranha MM, Santos DM, Solá S, Steer CJ, Rodrigues CMP. miR-34a Regulates Mouse Neural Stem Cell Differentiation. PLoS ONE. 2011;6:e21396. doi: 10.1371/journal.pone.0021396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronov S, Aranda G, Behar L, Ginzburg I. Axonal tau mRNA localization coincides with tau protein in living neuronal cells and depends on axonal targeting signal. Journal of Neuroscience. 2001;21:6577–6587. doi: 10.1523/JNEUROSCI.21-17-06577.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronov S, Marx R, Ginzburg I. Identification of 3'UTR region implicated in tau mRNA stabilization in neuronal cells. J Mol Neurosci. 1999;12:131–145. doi: 10.1007/BF02736927. [DOI] [PubMed] [Google Scholar]
- Atlas R, Behar L, Sapoznik S, Ginzburg I. Dynamic association with polysomes during P19 neuronal differentiation and an untranslated-region-dependent translation regulation of the tau mRNA by the tau mRNA-associated proteins IMP1, HuD, G3BP1JNeurosci. Res. 2007;85:173–183. doi: 10.1002/jnr.21099. [DOI] [PubMed] [Google Scholar]
- Barrett LW, Fletcher S, Wilton SD. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. CMLS, Cell. Mol. Life Sci. 2012;69:3613–3634. doi: 10.1007/s00018-012-0990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Behar L, Marx R, Sadot E, Barg J, Ginzburg I. cis-acting signals and transacting proteins are involved in tau mRNA targeting into neurites of differentiating neuronal cells. Int. J. Dev. Neurosci. 1995;13:113–127. doi: 10.1016/0736-5748(95)00001-w. [DOI] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Research. 2007;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, et al. p53-Mediated Activation of miRNA34 Candidate Tumor-Suppressor Genes. Current Biology. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- Chen J-M, Férec C, Cooper DN. A systematic analysis of disease-associated variants in the 3′ regulatory regions of human protein-coding genes I: general principles and overview. Hum Genet. 2006;120:1–21. doi: 10.1007/s00439-006-0180-7. [DOI] [PubMed] [Google Scholar]
- Cogswell JPJ, Ward JJ, Taylor IAI, Waters MM, Shi YY, Cannon BB, Kelnar KK, Kemppainen JJ, Brown DD, Chen CC, et al. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- Drubin DG, Kirschner MW. Tau protein function in living cells. J. Cell Biol. 1986;103:2739–2746. doi: 10.1083/jcb.103.6.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encinas MM, Iglesias MM, Liu YY, Wang HH, Muhaisen AA, Ceña VV, Gallego CC, Comella JXJ. Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factordependent, human neuron-like cells. J Neurochem. 2000;75:991–1003. doi: 10.1046/j.1471-4159.2000.0750991.x. [DOI] [PubMed] [Google Scholar]
- Goedert M, Wischik CM, Crowther RA, Walker JE, Klug A. Cloning and sequencing of the cDNA encoding a core protein of the paired helical filament of Alzheimer disease: identification as the microtubule-associated protein tau. Proc. Natl. Acad. Sci. U.S.A. 1988;85:4051–4055. doi: 10.1073/pnas.85.11.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy JA, Higgins GA. Alzheimer's disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, et al. Association of missense and 5'-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- Ittner LM, Ke YD, Delerue F, Bi M, Gladbach A, van Eersel J, Wölfing H, Chieng BC, Christie MJ, Napier IA, et al. Dendritic Function of Tau Mediates Amyloid β Toxicity in Alzheimer's Disease Mouse Models. Cell. 2010;142:387–397. doi: 10.1016/j.cell.2010.06.036. [DOI] [PubMed] [Google Scholar]
- Kosik KS, Joachim CL, Selkoe DJ. Microtubule-associated protein tau (tau) is a major antigenic component of paired helical filaments in Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 1986;83:4044–4048. doi: 10.1073/pnas.83.11.4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nature Genetics. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Larcher J-C, Gasmi L, Viranaïcken W, Eddé B, Bernard R, Ginzburg I, Denoulet P. Ilf3 and NF90 associate with the axonal targeting element of Tau mRNA. The FASEB Journal. 2004;18:1761–1763. doi: 10.1096/fj.04-1763fje. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lee VM, Goedert M, Trojanowski JQ. Neurodegenerative tauopathies. Annu. Rev. Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates that Thousands of Human Genes are MicroRNA Targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lin Y, Li Z, Ozsolak F, Kim SW, Arango-Argoty G, Liu TT, Tenenbaum SA, Bailey T, Monaghan AP, Milos PM, et al. An in-depth map of polyadenylation sites in cancer. Nucleic Acids Research. 2012;40:8460–8471. doi: 10.1093/nar/gks637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes OC, Chertkow HM, Wang E, Schipper HM. MicroRNA: Implications for Alzheimer Disease and other Human CNS Disorders. Curr. Genomics. 2009;10:154–168. doi: 10.2174/138920209788185252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Research. 2009;37:W273–W276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nukina N, Ihara Y. One of the antigenic determinants of paired helical filaments is related to tau protein. J. Biochem. 1986;99:1541–1544. doi: 10.1093/oxfordjournals.jbchem.a135625. [DOI] [PubMed] [Google Scholar]
- Park JY, Li W, Zheng D, Zhai P, Zhao Y, Matsuda T, Vatner SF, Sadoshima J, Tian B. Comparative Analysis of mRNA Isoform Expression in Cardiac Hypertrophy and Development Reveals Multiple Post-Transcriptional Regulatory Modules. PLoS ONE. 2011;6:e22391. doi: 10.1371/journal.pone.0022391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poorkaj P, Bird TD, Wijsman E, Nemens E, Garruto RM, Anderson L, Andreadis A, Wiederholt WC, Raskind M, Schellenberg GD. Tau is a candidate gene for chromosome 17 frontotemporal dementia. Ann Neurol. 1998;43:815–825. doi: 10.1002/ana.410430617. [DOI] [PubMed] [Google Scholar]
- Poorkaj P, Kas A, D'Souza I, Zhou Y, Pham Q, Stone M, Olson MV, Schellenberg GD. A genomic sequence analysis of the mouse and human microtubule-associated protein tau. Mammalian Genome. 2001;12:700–712. doi: 10.1007/s00335-001-2044-8. [DOI] [PubMed] [Google Scholar]
- Rapoport M, Dawson HN, Binder LI, Vitek MP, Ferreira A. Tau is essential to beta-amyloid-induced neurotoxicity. Proc. Natl. Acad. Sci. U.S.A. 2002;99:6364–6369. doi: 10.1073/pnas.092136199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson ED, Scearce-Levie K, Palop JJ, Yan F, Cheng IH, Wu T, Gerstein H, Yu GQ, Mucke L. Reducing Endogenous Tau Ameliorates Amyloid-Induced Deficits in an Alzheimer's Disease Mouse Model. Science. 2007;316:750–754. doi: 10.1126/science.1141736. [DOI] [PubMed] [Google Scholar]
- Schipper HM, Maes OC, Chertkow HM, Wang E. MicroRNA expression in Alzheimer blood mononuclear cells. Gene Regul Syst Bio. 2007;1:263–274. doi: 10.4137/grsb.s361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. Alternative polyadenylation: New insights from global analyses. RNA. 2012;18:2105–2117. doi: 10.1261/rna.035899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillantini MG, Murrell JR, Goedert M, Farlow MR, Klug A, Ghetti B. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc. Natl. Acad. Sci. U.S.A. 1998;95:7737–7741. doi: 10.1073/pnas.95.13.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vossel KA, Zhang K, Brodbeck J, Daub AC, Sharma P, Finkbeiner S, Cui B, Mucke L. Tau reduction prevents Abeta-induced defects in axonal transport. Science. 2010;330:198. doi: 10.1126/science.1194653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Loomis PA, Zinkowski RP, Binder LI. A novel tau transcript in cultured human neuroblastoma cells expressing nuclear tau. J. Cell Biol. 1993;121:257–267. doi: 10.1083/jcb.121.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarten MD. A Protein Factor Essential for Microtubule Assembly. Proceedings of the National Academy of Sciences U.S.A. 1975;72:1858–1862. doi: 10.1073/pnas.72.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JG, Mirra SS, Pollock NJ, Binder LI. Neurofibrillary tangles of Alzheimer disease share antigenic determinants with the axonal microtubule-associated protein tau (tau) Proc. Natl. Acad. Sci. U.S.A. 1986;83:4040–4043. doi: 10.1073/pnas.83.11.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Huang M, Lu M, Zhu W, Shu Y, Cao P, Liu P. Regulation of microtubule-associated protein tau (MAPT) by miR-34c-5p determines the chemosensitivity of gastric cancer to paclitaxel. Cancer Chemother Pharmacol. 2013 doi: 10.1007/s00280-013-2108-y. [DOI] [PubMed] [Google Scholar]
- Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proceedings of the National Academy of Sciences U.S.A. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang microRNA-34 family and treatment of cancers with mutant or wild-type p53 (Review) Int J Oncol. 2011;38 doi: 10.3892/ijo.2011.970. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.