Abstract
Murine regenerating islet-derived 3β (Reg3β) represents a homologue of human hepatocarcinoma-intestine-pancreas/pancreatic-associated protein and enhanced mouse susceptibility to the acetaminophen (APAP)-induced hepatotoxicity. Our objective was to determine if and how knockout of Reg3β (KO) affected the APAP (300 mg/kg, ip)-mediated protein nitration in mouse liver. The APAP injection produced greater levels of hepatic protein nitration in the KO than the wild-type mice. Their elevated protein nitration was alleviated by a prior injection of recombinant mouse Reg3β protein, and was associated with an accelerated depletion of the peroxynitrite (ONOO−) scavenger glutathione by an up-regulated hepatic glutathione peroxidase-1 (GPX1) activity. The enhanced GPX1 production in the KO mice was mediated by an 85% activity rise (p < 0.05) of selenocysteine lyase (Scly), a key enzyme to mobilize Se for selenoprotein biosynthesis. Knockout of Reg3β enhanced AP-1 protein and its binding activity to the gene promoter of Scly, up-regulating its gene transcription. However, knockout of Reg3β did not affect gene expression of other key factors for selenoprotein biosynthesis. In conclusion, our finding unveiled a new metabolic role of Reg3β in protein nitration and a new biosynthesis control of GPX1 by a completely “unrelated” regenerating protein Reg3β via transcriptional activation of Scly in coping with hepatic protein nitration. Linking selenoproteins to tissue regeneration will have profound implications in understanding mechanism for Se functions and physiological coordination of tissue regeneration with intracellular redox control.
Keywords: Reg3β, GPX1, Selenoproteins, Selenocysteine lyase, Protein nitration
Introduction
Regenerating islet-derived 3β (Reg3β) is the mouse homologue of human hepatocarcinoma-intestine-pancreas/pancreatic-associated protein (HIP/PAP). As a member of the Reg family involved in tissue regeneration [1,2], Reg3β plays key roles in protecting against liver injuries produced by oxidants and toxins [3]. Knockout of Reg3β rendered mouse sensitive to the Fas-induced hepatotoxicity [3], and overexpression of the protein protected against the analgesic and antipyretic acetaminophen (APAP) toxicity [4]. Metabolically, APAP and its reactive metabolite N-acetyl-p-benzoquinoneimine (NAPQI) may react with target proteins to form NAPQI-protein adducts, leading to liver injury [5, 6]. Meanwhile, protein nitration has been found to contribute to the APAP-induced hepatotoxicity. There was strong immune-histochemical staining of nitrated proteins in centrilobular necrotic areas of APAP-exposed animals [7]. The APAP-induced protein nitration was associated with mitochondrial dysfunction [8]. However, roles of Reg3b in the APAP-induced hepatic protein nitration and the underlying mechanism have not been studied. While Reg3β knockout (KO) mice seem to be an ideal model to answer that question, secondary changes associated with the genetic manipulation could confound target responses in these global knockouts [9]. Thus, additional experiments are needed to verify direct findings from the KO mice as actual primary responses.
Protein nitration derives from reactions between tyrosine residues of target proteins and peroxynitrite (ONOO−, PN) [10]. As a main post-translational modification, the production of nitrotyrosine may alter protein function or stability, and consequently contribute to pathogeneses of various chronic diseases [11–13]. Apparently, the net rate of nitrotyrosine production is controlled by the balance between the formation and scavenge of PN [10,14]. The formation of PN depends on reactions of nitric oxide (NO) and superoxide anion [10]. Whereas the former, one of the important mediators of APAP-induced hepatotoxicity [15], can be regulated by inducible NO synthase (iNOS) [16] through PI3K-Akt signaling pathway [17,18]. The latter can be generated by cytochrome P450 2E1 (CYP2E1) [19] and converted into H2O2 by superoxide dismutase (SOD) [20]. Meanwhile, glutathione (GSH) is one of the most effective scavengers of PN [14,21], and enhancing intracellular GSH protects against the APAP overdose [22]. In general, cellular GSH is controlled by activities of key enzymes related to GSH synthesis: γ-glutamylcysteine synthetase (γ-GCS) [23] and glutathione synthetase (GS) [23], GSH regeneration: glutathione reductase (GR) [24], and GSH consumption: glutathione S-transferase (GST) [25] and cellular glutathione peroxidase-1 (GPX1) [26].
Among the 25 selenium (Se)-dependent proteins identified in mammals [27], GPX1 is the first characterized [28] and the most abundant [29]. While metabolic functions of Se are presumably performed mainly by various selenoproteins [29,30], biosynthesis of these proteins is controlled by selenophosphate synthetases 1 and 2 (SPS1 and SPS2), selenocysteine insertion sequence (SECIS)-binding protein (SBP2), and Se recycling enzyme selenocysteine lyase (Scly) [31]. As a GSH-consuming enzyme, overexpression of GPX1 actually sensitized mice to the APAP toxicity [32] by accelerating depletion of intracellular GSH. Reciprocally, knockout of GPX1 protected primary hepatocytes against protein nitration induced by authentic PN [33]. Although both Reg3β and GPX1 exert strong impacts on the APAP hepatotoxicity [4,32], these two distinctly different family proteins have never been recognized to be functionally related. Likewise, neither Se nor selenoproteins have been found to be involved in tissue regeneration despite their broad roles in antioxidant defense [34], thyroid hormone [35], reproduction [36], immune function [34], anti-tumorigenesis [37,38], cardiovascular diseases [39], and energy metabolism [31].
The initial objective of the present study was to determine if and how the Reg3β knockout affected the APAP-mediated hepatic protein nitration. To follow through the ultimate mechanism for the observed genotype difference in this regard, we took stepwise approaches to elucidate effects and mechanisms of the Reg3β knockout on the formation and scavenging of PN, synthesis and consumption of GSH, and functional expression of GPX1 and Scly. To exclude possible confounding effects associated with the global knockout of Reg3β, we expressed the mouse recombinant Reg3β protein (rcReg3β) in a yeast system and used the purified rcReg3β to verify our in vitro and in vivo findings. It is most exciting that we revealed a novel up-regulation of hepatic GPX1 activity by the knockout of Reg3β via transcriptional activation of Scly, and provided the first link between tissue regeneration and selenoprotein synthesis.
Materials and methods
Animals and APAP treatment
Our animal protocol was approved by the Institutional Animal Care and Use Committee at Cornell University and conducted in accordance with the NIH guidelines for animal care. The KO mouse breeders were provided by Dr. Jamila Faivre at Institut National de la Sante et de la Recherche Medicale (INSERM). These mice were generated as previously described [3] using a C57BL/6J genetic background. Heterozygous breeders of Reg3β (+/−) were used to produce littermates of KO and wild-type (WT) mice for the planned experiments. Genotyping was performed by PCR analysis (Supplementary Fig. S1) using the primer pairs (Supplementary Table S1) as descried previously [3]. Mice were housed in plastic boxes located in an environmentally-controlled room (23 ± 3°C, 40–60% humidity, and 12/12 h dark-light cycle), and were given free access to water and a Se-adequate Torula yeast-based diet [40]. All experimental mice were male and 8-wk of age. The APAP preparation and injection (intraperitoneally or ip at 300 mg/kg) were the same as previously described [41]. Mice were fasted for 8 h before the injection and killed to collect blood and liver tissue at 5 h after the injection. The blood was centrifuged immediately to prepare fresh plasma for the determination of alanine aminotransferase (ALT) activity. Liver was frozen in liquid nitrogen and stored at −80 °C until further analysis. The pre-treatment of purified rcReg3β protein in the KO mice was also administrated by the ip injection (0, 1 and 6 mg/kg) at 6 h before the APAP treatment.
Recombinant mouse Reg3β cloning, expression, and purification
To amplify the mature protein sequence of the Reg3β gene (NM_011036.1), total RNA extracted from the liver of the WT mouse was used to obtain cDNA. The gene was cloned into the expression vector pPICZαA (Invitrogen, Carlsbad, CA), transformed into E. coli DH5α, and electroporated into Pichia pastoris X33 for methanol-induced protein expression (6 d). The expressed Reg3β protein was purified by gel filtration through a Sephadex G-25 column, and ion-exchange chromatography on a Q-Sepharose column (Bio-Rad Laboratories, Hercules, CA). The purified protein was measured using the bicinchoninic acid (BCA) protein assay kit (Pierce Chemical Co., Rockford, IL) and confirmed by SDS-PAGE and Western blot using rabbit Reg3β antibody (provided by Dr. Stephen Hunt, University College London, UK).
Enzyme activity, GSH, and NO analyses
Plasma ALT activity was assayed using a kit as described by the manufacturer’s instructions (Thermo Scientific, Waltham, MA). Total hepatic GSH was measured by the GSH recycling assay [42] and oxidized glutathione (GSSG) was assayed using the same procedure after 2-vinylpyridine and triethanolamine were added to supernatant to incubate for 1 h. As an indicator of NO production, plasma nitrate and nitrite concentrations were measured as previously described [33] to test if the impact of Reg3β knockout on hepatic protein nitration was related to NO formation. Nitrate was reduced to nitrite using nitrate reductase. The fluorescence was measured with excitation at 360 nm and emission at 450 nm. Hepatic SOD1 activity was measured using a water-soluble formazan dye kit (Dojindo Molecular Technologies, Gaithersburg, MD) according to the manufacturer’s instructions. One unit of SOD1 was defined as the activity needed to inhibit 50% 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium monosodium salt reduction. Hepatic CYP2E1 activity was assayed in microsomal fractions by the hydroxylation of aniline into p-aminophenol [43]. Hepatic γ-GCS activity was determined as previously described [23], and GS activity was determined by measuring the formation of ADP in reaction mixtures [23]. Hepatic GR activity was measured and defined as nmol of the oxidized GSH form reduced, using a molar extinction coefficient of 6.22 ×103 for NADPH [33]. Hepatic GST activity was determined and defined as nmol of S-2,4-dinitrophenylglutathione formed per min with 1-chloro-2,4-dinitrobenzene as substrate [33]. Hepatic GPX1 activity was determined by the coupled assay of NADPH oxidation using H2O2 as substrate and expressed as nmol of GSH oxidized per minute and mg of protein [33]. Hepatic Scly activity was measured by the release of hydrogen selenide that reacts with lead acetate, and amounts of the produced hydrogen selenide were calculated using an apparent molar coefficient for lead selenide of 2.35 ×104 [44]. For testing possible direct effects of the rcReg3β protein on Scly activity, the purified rcReg3β protein (0, 0.1, or 1.0 μg) was mixed with 100 μg of liver homogenate protein and incubated for 30 min at room temperature before the Scly activity assay.
In vitro analysis of protein nitration
Liver samples of WT and KO mice were homogenized in 50 mM potassium phosphate buffer, pH 7.8, 0.1% Triton X-100, 1.34 mM diethylenetriaminepentaacetic acid, and centrifuged at 14,000 g for 10 min at 4 °C to collect the supernatants. Thereafter, 130 μg of liver homogenate protein (20 μl) of WT and KO mice wase incubated with rcReg3β (0, 0.1, and 1.0 μg/20 μl) or GSH (0, 0.13, and 1.3 μg/20 μl) for 30 min at room temperature. Subsequently, 16.4 nmol of PN (Calbiochem, San Diego, CA) was mixed with the reaction mixture for 5 min at 37°C [41], and the reaction was terminated by putting the samples on ice. After the PN treatment, 50 μg of protein from the mixture was used for analysis of nitrotyrosine formation by Western blotting.
Western blot analysis
Protein concentrations of liver homogenates were determined using BCA protein assay reagent (Pierce). Liver homogenates (40 to 100 μg proteins/lane) was subjected to SDS-PAGE gels (12 % gel) and transferred to nitrocellulose membranes [33]. Membranes were incubated with appropriate primary antibodies (Supplementary Table S2) overnight at 4°C, and were washed and incubated with either anti-mouse (Pierce) or anti-rabbit secondary antibody (Bio-Rad) at room temperature for 1 h. Signals were detected with SuperSignal West Pico Kit (Pierce) and exposed to imaging films (Kodak, Rochester, NY) according to the manufacturer’s instructions. Densitometry of the target bands was analyzed using Image J (National Center for Biotechnology Information, NCBI).
Quantitative PCR (qPCR) analysis of gene expression
Total RNA was isolated with TRIzol Reagent (Invitrogen, Carlsbad, CA) from liver tissue. The RNA integrity and quantity were assessed by Agilent Bioanalyzer 2100 using RNA 6000 Labchip kit (Agilent Technologies, Amstelveen, The Netherlands). Reverse-transcription was performed using Superscript and oligo(dT) primer following the manufacturer’s protocol (Invitrogen). Real-time qPCR was conducted with SYBR Green Master Mix reagents (Applied Biosystems, Foster City, CA) and an ABI 7700 (Applied Biosystems). Primers used are shown in Supplementary Table S1. The normalized expression levels of genes were calculated as 2−ΔΔCt.
Transcriptional regulation of Scly
The proximal promoter region of Scly, defined as 2,000 bases upstream of the transcription start site, was retrieved from the NIH/NCBI Entrez Gene database (http://www.ncbi.nlm.nih.gov/gene). A widely-used transcription factor database, Transcription Element Search System (TESS) (http://www.cbil.upenn.edu/cgi-bin/tess/tess) was used to identify putative binding sites for transcription factor in the promoter region of Scly. Chromatin was isolated from liver of WT and KO mice, fragmented, and immune-precipitated using an Imprint® Chromatin Immunoprecipitation (ChIP) kit (Sigma, St. Louis, MO) according to the manufacturer’s directions. The immunoprecipitated DNA was quantified by PCR. Primer sequences are shown in Supplementary Table S1. Relative binding intensity was measured by Image J (NCBI).
Statistical analysis
Treatment effects or genotype differences were analyzed using one-way ANOVA, and mean differences were tested by post hoc Tukey’s multiple range tests using Minitab® 14 software (Minitab Inc., State College, PA). Data are presented as means ± SEM. Differences were considered to be significant at p < 0.05.
Results
Knockout of Reg3β aggravated the APAP-induced liver injury and protein nitration
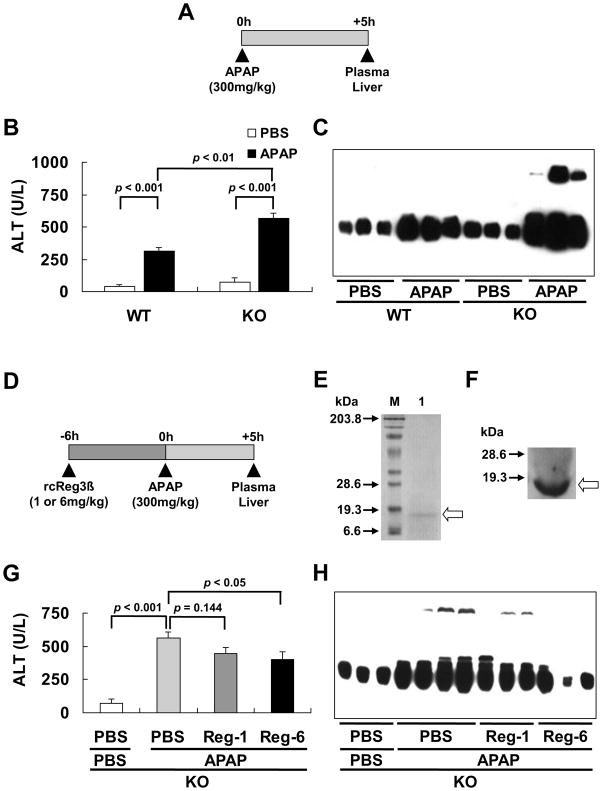
While the single ip injection of APAP (at 300 mg/kg for 5 h, Fig. 1A) caused liver injuries in both genotypes, plasma ALT activity was 1.8-fold greater (p < 0.01) in the KO than the WT mice after APAP treatment (Fig. 1B). Likewise, the APAP injection induced hepatic nitrotyrosine formation in both genotypes, but the intensity was much greater in the KO mice than in WT mice (Fig. 1C).
Fig. 1.
Aggravated hepatic protein nitration in the KO mice overdosed with APAP was rescued by a pre-treatment of rcReg3β protein. (A) Experimental design of the APAP treatment (ip injection, and PBS as a vehicle control). (B) The APAP-induced rise of plasma ALT activity, n = 4–6/group. (C) The APAP-induced hepatic nitrotyrosine formation analyzed by Western blot. (D) Experimental design of the pre-treatment (ip injection) with 1 (Reg-1) or 6 mg/kg (Reg-6) of rcReg3β protein (PBS as a vehicle control). (E) The purified rcReg3β (arrow) applied to a 12% SDS-PAGE and stained with Coomassie brilliant blue: M, sizes (kDa) of molecular weight markers; Lane 1, purified fraction of rcReg3β after ion exchange chromatography. (F) Western blot analysis of the purified rcReg3β (open arrow) using the rabbit Reg3β antibody. (G) Dose-dependent effect of the rcReg3β protein pre-treatment on the APAP-induced plasma ALT activity rises, n = 4-6/group. (H) Dose-dependent effect of the rcReg3β protein pre-treatment on the APAP-induced hepatic nitrotyrosine formation analyzed by Western blotting.
To clarify if the above-observed impact of Reg3β knockout on the APAP-induced liver injury and protein nitration was a primary response, we pre-treated the KO mice with an ip injection of 1 or 6 mg/kg of the purified rcReg3β protein at 6 h before the APAP administration (Fig. 1D). The purified protein was confirmed by SDS-PAGE (Fig. 1E) and Western blot (Fig. 1F). Compared with the vehicle (PBS)-treated control, the pre-treatment of rcReg3β resulted in a dose-dependent suppression on the APAP-induced plasma ALT activity rise (p < 0.05, Fig. 1G) and hepatic nitrotyrosine formation (Fig. 1H).
Knockout of Reg3β did not affect PN formation
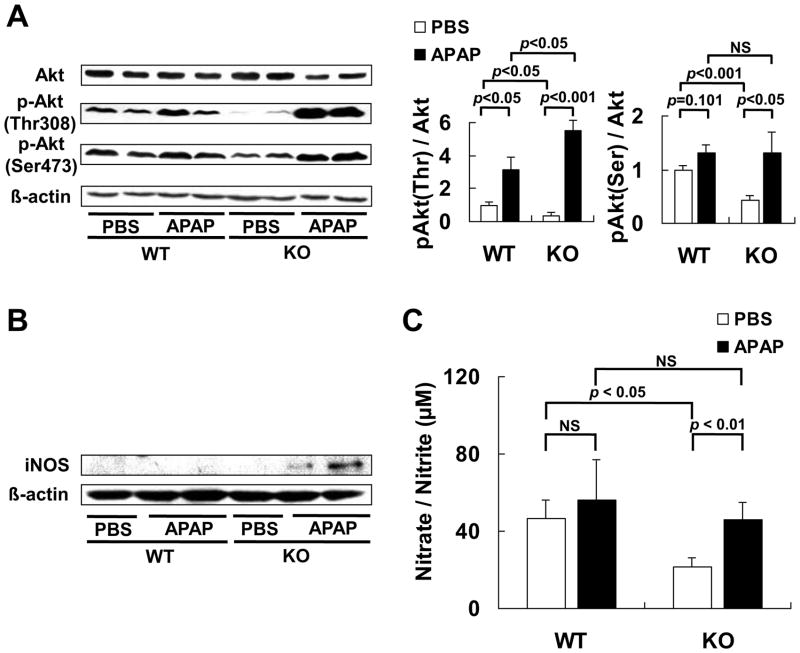
Because Reg3β was shown to stimulate hepatic Akt phosphorylation [4], and Akt signaling was involved in up-regulating iNOS for production of NO [17], one of factors for PN formation [10], we determined the effects of Reg3β knockout on hepatic Akt protein and phosphorylation in mice treated with or without APAP (Fig. 2A). Compared with the WT mice, knockout of Reg3β did not alter liver Akt protein, but suppressed the phosphorylation of Akt in the liver tissue, leading to 63 % (p < 0.05) and 57 % (p < 0.001) lower ratio of p-Akt (Thr308)/Akt and p-Akt (Ser473)/Akt, respectively, in the KO mice than the WT mice treated with PBS. Meanwhile, the APAP-induced Akt phosphorylation at Thr308 was much higher (p < 0.05) in KO mice than that in WT mice. And, the APAP treatment elevated Akt phosphorylation (p < 0.05) at Ser473 residue only in the KO mice. Hepatic iNOS protein band was detectable only in the APAP-treated KO mice (Fig. 2B). Plasma nitrate/nitrite concentrations were 53% lower (p < 0.05) in the PBS-treated KO mice than the WT mice, and were elevated 112 % (p < 0.01) by the APAP treatment only in the KO mice. This resulted in no significant difference in plasma NO between the two genotypes after the APAP injection (Fig. 2C).
Fig. 2.
Knockout of Reg3β did not alter plasma NO production after the APAP treatment. (A) Western blot analysis of hepatic Akt protein and Akt phosphorylation at Ser473 and Thr308 (left panel) and quantitative analysis of the gel data (right panel) normalized to the levels of β-actin, n = 5/group, NS, not significant. (B) Western blot analysis of hepatic iNOS protein. (C) Tissue NO formation measured by plasma nitrate and nitrite concentration, n = 5/group, NS, not significant.
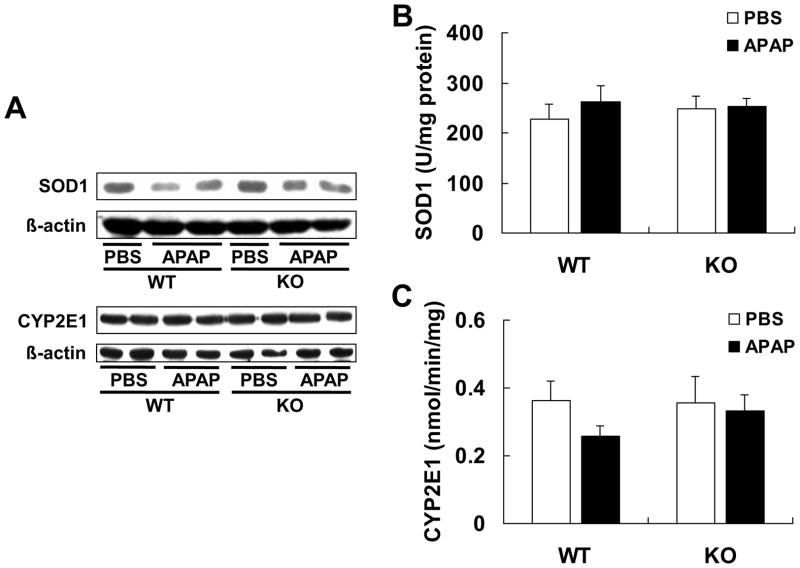
Because superoxide anion, another factor for PN formation, can be generated by CYP2E1 [19] and converted to H2O2 by SOD1 [20], we determined hepatic activities of CYP2E1 and SOD1 as indicators of PN formation and turnover [26,45]. There was no genotype difference in protein expressions of liver SOD1 and microsomal CYP2E1 (Fig. 3A) before or after the APAP treatment. The same was also true of the hepatic activity of SOD1 (Fig. 3B) and microsomal CYP2E1 (Fig. 3C).
Fig. 3.
Knockout of Reg3β did not affect hepatic SOD1 and CYP2E1 protein or activity. (A) Western blot analysis of hepatic SOD1 and microsomal CYP2E1. (B) Hepatic SOD1 activity, n = 5/group. (C) Microsomal CYP2E1 activity in the liver tissue, n = 4–5/group.
Enhanced protein nitration in the KO mice was involved in down-regulated PN scavenge
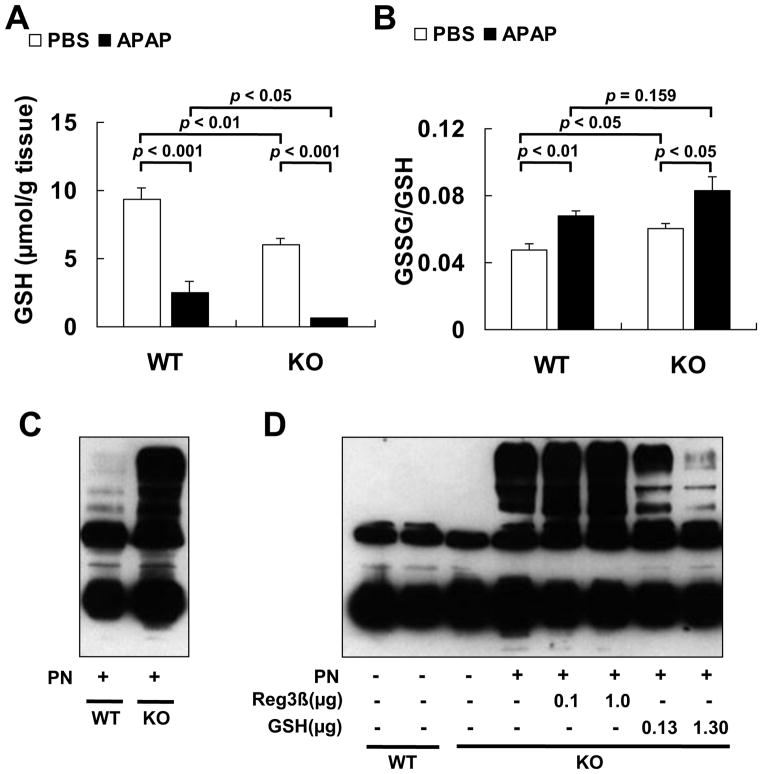
The baseline level of hepatic GSH, one of the most effective scavengers of PN [14,21], in KO mice was 36% lower (p < 0.01) than that in WT mice. The APAP treatment led to a greater loss (p < 0.05) of hepatic GSH in the KO mice than in the WT mice (Fig. 4A). The ratio of GSSG/GSH was higher in the KO mice than in the WT mice (Fig. 4B). To reveal if the genotype difference in the APAP-induced hepatic protein nitration was associated with the suppression of PN scavenge related to the changed hepatic GSH, we first incubated liver homogenates of both genotypes with the same amount of authentic PN and found a much stronger nitrotyrosine formation in the KO homogenate (Fig. 4C). Subsequently, we added 2 levels of the purified rcReg3β protein (0.1 or 1 μg) into the homogenate reaction mixture (130 μg) to determine whether the Reg3β protein functioned as a direct player or helper in scavenging PN. As shown in Fig. 4D, the purified rcReg3β protein exerted no impact on the PN-mediated protein nitration of liver homogenates of either genotype. In contrast, adding GSH at the level of 0.13 μg, comparable to the genotype baseline difference (3.34 μmol/g), or 10-fold of that level (1.30 μg) into the liver homogenate resulted in a dose-dependent suppression of the PN-mediated protein nitration.
Fig. 4.
Enhanced nitrotyrosine formation in the KO mice was associated with down-regulated PN scavenging. (A) Hepatic GSH concentration, n = 5/group. (B) Hepatic GSSG/GSH ratio, n = 5/ group. (C) Western blot analysis of nitrotyrosine formation in liver homogenates of the WT and KO mice induced by the same amount of extrinsic PN. (D) Western blot analysis of the pre-incubation effects of rcReg3β or GSH on the PN-mediated protein nitration in liver homogenates of KO mice. The rcReg3β protein or GSH was incubated with the reaction mixture for 30 min at room temperature before the PN addition.
Decreased GSH was associated with up-regulated GPX1 activity in the liver of KO mice
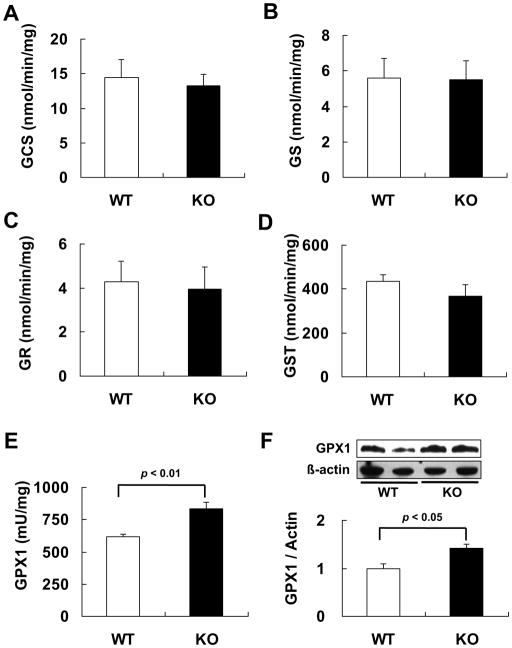
To reveal biochemical mechanisms for the lower baseline level of hepatic GSH in the KO mice, we measured activities of 5 GSH-related enzymes. Knockout of Reg3β did not alter activities of 2 rate-limiting enzymes of GSH synthesis, GCS and GS (Figs. 5A and B), or the key enzyme in GSH regeneration, GR (Fig. 5C). Meanwhile, activity of the GSH consuming enzyme GST was 15% lower in the KO mice than the WT mice, but the difference was not significant (Fig. 5D). In contrast, the KO mice had 34% greater hepatic GPX activity (p < 0.01, Fig. 5E) and 41% greater hepatic GPX1 protein (p < 0.05, Fig. 5F) than the WT mice, respectively.
Fig. 5.
Decreased GSH was associated with up-regulated GPX1 activity in the liver of KO mice. (A–D) Hepatic activities of GCS (A), GS (B), GR (C), and GST (D) in the WT and KO mice, n = 4–5/group. (E) Hepatic GPX1 activity in the WT and KO mice, n = 5/group. (F) Western blot analysis of hepatic GPX1 protein (top panel) and densitometric quantitation of the gel image normalized to the levels of β-actin, n = 5/group (bottom panel).
Elevated hepatic GPX1 in the KO mice was correlated with enhanced Scly function
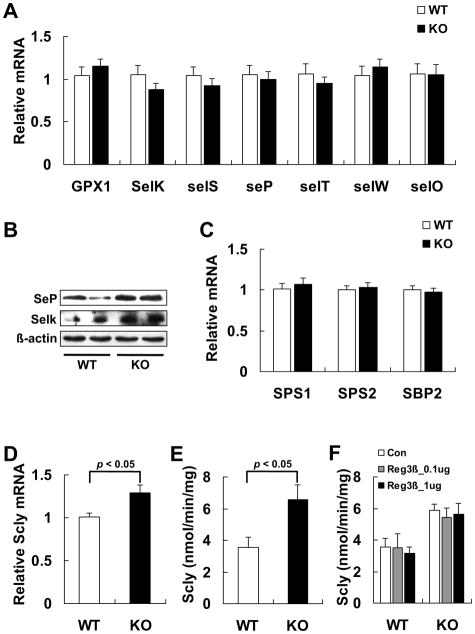
To reveal if the knockout of Reg3β specifically up-regulated hepatic GPX1 production, we determined hepatic protein levels of other selenoproteins (selenoprotein P, SeP and selenoprotein K, SelK). These proteins showed similar increases in the KO mice despite no changes in their mRNA levels (Figs. 6A and B). To explore if there was an overall up-regulation of selenoprotein biosynthesis in the KO mice, we determined gene expression of key factors for selenoprotein biosynthesis including SPS1, SPS2, SBP2, and Scly [31,46], and found a significant increase in only Scly mRNA level (Figs. 6C and D). Because Scly is a key enzyme for biosynthesis of selenoproteins, in particular GPX1 and SeP [29–31,47], we measured its activity and found an 85% increase in the KO mice compared with the WT mice (Fig. 6E). To exclude a possible direct inhibition of the Scly activity by Reg3β, we compared the enzyme activity in the liver homogenates of both genotypes before and after the addition of the purified rcReg3β in reaction mixture (Fig. 6F).
Fig. 6.
Elevated hepatic GPX1 in the KO mice was correlated with enhanced Scly function. (A) qPCR analysis of mRNA levels of hepatic selenoproteins in the WT and KO mice that were normalized to the levels of Gapdh, n = 9/group. (B) Western blot analysis of hepatic SeP and SelK. (C) qPCR analysis of hepatic Sps1, Sps2, and Sbp2 mRNA levels in the WT and KO mice that were normalized to the levels of Gapdh, n = 8–10/group. (D) qPCR analysis of hepatic Scly mRNA levels in the WT and KO mice that were normalized to the levels of Gapdh, n = 13–15/group. (E) Hepatic Scly activity in the WT and KO mice, n = 5/group. (F) Effect of pre-incubation of the rcReg3β protein (0, 0.1, or 1.0 μg) on Scly activity in the liver homogenates (100 μg proteins) of the WT and KO mice, n = 3–4/group. The pre-incubation was carried out for 30 min at room temperature before the Scly activity assay.
Knockout of Reg3β up-regulated transcription of Scly
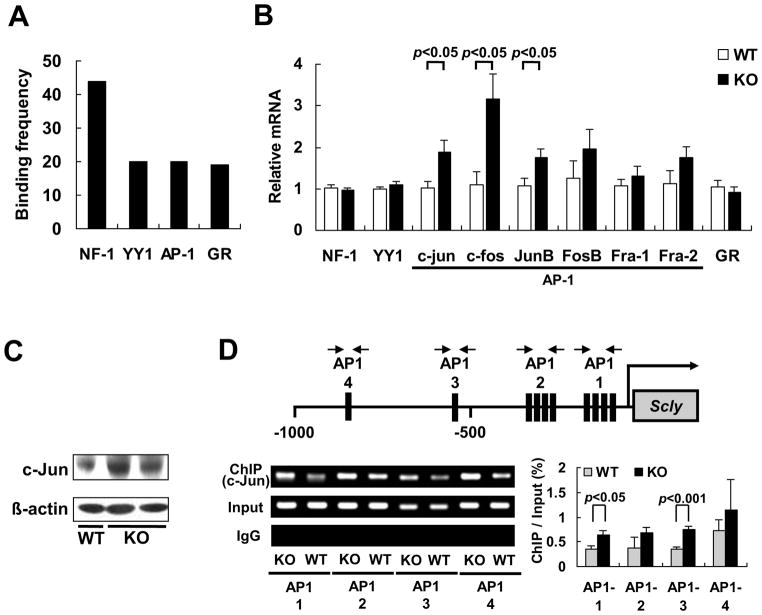
Because up-regulation of the Scly activity in the KO mice was accompanied with the rise in its mRNA level, we postulated an induced Scly transcription by the Reg3β knockout. To search for potential transcription factors for this proposed event, we analyzed the promoter region of Scly (2000 base pair upstream of the transcription start site) using TESS. The analysis identified 4 transcription factors, nuclear factor 1 (NF-1), Yin Yang 1 (YY1), activator protein 1 (AP-1) and glucocorticoid receptor (GR), with the highest frequencies of binding sites in the promoter region of Scly (Fig. 7A). Further qPCR analysis showed that gene expressions of only the AP-1 family members c-jun, c-fos, and JunB were significantly up-regulated by the knockout of Reg3β (p < 0.05, Fig. 7B). Consistently, the KO mice had greater amounts of hepatic c-Jun protein (Fig. 7C). Because AP-1 was suggested to bind the promoter region of rat Scly [48], we conducted ChIP assay to determine if c-Jun actually bound these sites of mouse Scly (Supplementary Table S3 and Fig. S2). In fact, c-Jun was able to bind all these putative sites and the binding was enhanced (p < 0.05) in 2 out of the 4 sites by the knockout of Reg3β (Fig. 7D).
Fig. 7.
Knockout of Reg3β up-regulated transcription of Scly. (A) Binding frequencies of the four most frequent transcriptional factors in the promoter region (2000 base pair upstream of the transcription start site) of Scly. (B) qPCR analysis of hepatic mRNA levels for NF-1, YY1, c-jun, c-fos, JunB, FosB, Fra-1, Fra-2, and GR in the WT and KO mice that were normalized to the levels of Gapdh, n = 4–5/group. (C) Western blot analysis of hepatic c-Jun protein. (D) The ChIP assay of c-Jun binding to the AP-1 sites in the Scly promoter. The top panel shows the schematic overview of the AP-1 sites in the Scly promoter (the black box represents the AP-1 binding sites for each primer sets). The bottom left panel is a representative DNA gel image to show genotype differences in the binding of c-Jun protein to different AP-1 sites in the Scly promoter. The bottom right panel is a quantitative analysis of the ChIP assays based on three independent experiments.
Discussion
The most direct finding of the present study is that knockout of Reg3β rendered mice susceptible to the APAP-induced protein nitration. Remarkably, pre-treating the KO mice with the purified rcReg3β protein before the APAP administration attenuated the induced hepatic protein nitration and plasma ALT rise. Previous studies have shown that overexpression of Reg3β protects against Fas- or APAP-induced liver failure and stimulates liver regeneration with early activation/deactivation of STAT3 [3,4], whereas knockout of Reg3β sensitizes liver to oxidative stress and delays liver regeneration with persistent TNF-a/IL6/STAT3 signaling [3]. Although the lack of appropriate reagent or functional assay for the Reg3β protein did not allow us to estimate the physiological level or the recovery of the protein after the ip injection in the liver, our study reveals a novel protection of Reg3β against hepatic nitrotyrosine formation mediated by the APAP overdose. While future research will be needed to assess if the injected doses of rcReg3β (1 or 6 mg/kg) are physiological or pharmacological, the effectiveness of the Reg3β pre-treatment in protecting mice against the APAP toxicity illustrates a new strategy to treat the APAP overdose [49]. It extends previous findings [4] on the role of Reg3β in APAP hepatotoxicity, and helps clarify PN and the PN-induced nitrotyrosine formation as a main mechanism for the APAP-induced hepatotoxicity [21].
A down-regulation of PN scavenger GSH in the liver of KO mice provides a strong explanation to their aggravated protein nitration after the APAP overdose compared with the WT mice. Three lines of evidences support this view. First, the KO mice did not have greater NO level than the WT mice following the APAP administration, despite the induced iNOS protein production, activation of Akt phosphorylation at Ser473, and greater level of Akt phosphorylation at Thr308. Intriguingly, the KO mice showed a lower baseline of NO production than the WT mice. Second, knockout of Reg3β did not affect protein production and activity of upstream PN-forming superoxide-related enzymes SOD1 or microsomal CYP2E1 [19,20,26,45]. Third, hepatic GSH was much lower in the KO mice than in the WT mice at the baseline, and was nearly diminished after the APAP administration. Furthermore, the addition of the same amount of PN to the liver homogenates produced remarkably more nitrotyrosine in the KO mice than in the WT mice. This probably resulted from the decreased PN scavenging. More convincingly, adding GSH to the liver homogenates of KO mice produced a dose-dependent suppression on the PN-induced protein nitration. It is noteworthy that Reg3β protein itself was not directly involved in the PN-mediated protein nitration because adding the protein at multiple doses into the liver homogenate of KO mice showed no effect on the reaction. This further supports a functional regulation of hepatic GSH by the knockout of Reg3β as the mechanism for the accelerated protein nitration in the APAP-overdosed KO mice.
Searching for biochemical mechanism for the attenuated hepatic GSH in the KO mice led us to the most significant discovery that knockout of Reg3β up-regulated selenoenzyme GPX1 via enhanced Scly function [31,47]. Among the five major GSH-synthesis, regeneration, or consuming enzymes assayed in the present study, GPX1 was the only one affected by the Reg3β knockout. The 34% elevation of hepatic GPX activity in the KO mice signified a strong reason for the aggravated APAP toxicity in these mice because GPX1 uses GSH as a substrate in its catalytic reactions of hydroperoxide removal [26] and thus competes against the need for GSH to directly scavenge PN in the early phase of APAP overdose [22]. Mirochnitchenko et al. [32] showed that overexpression of GPX1 sensitized mice to the APAP toxicity by accelerating hepatic GSH depletion. Reciprocally, our laboratory demonstrated a protective role of GPX1 knockout in primary hepatocytes against the PN-induced protein nitration and cell death by sparing cytosolic GSH from the voided enzymatic reaction [50]. Meanwhile, the increased hepatic GPX1 activity in the KO mice led us to observe a corresponding elevation of hepatic GPX1 protein. Most fascinatingly, we serendipitously discovered protein nitration of Scly in the APAP-treated KO mouse liver from a preliminary proteomic analysis. This enzyme breaks down selenocysteine into alanine and selenide that synthesize selenophosphate as a donor of Se for the biosynthesis of selenoproteins, and thus plays a key role in mobilizing Se from the decomposition of selenocysteine for selenoprotein synthesis [51]. A recent study has shown that knockout of Scly decreased SeP and SelS, along with GPX1 activity [31]. Because of the concurrent rise of GPX1 and Scly activity in the liver of KO mice, we postulated a possible overall up-regulation of selenoprotein biosynthesis in these mice mediated by elevated function of Scly [31,47]. Indeed, these mice had greater levels of hepatic SeP and SelK in addition to GPX1 than the WT mice, without changes in their mRNA levels. Thereby, we have herein revealed a novel in vivo regulation of selenoprotein biosynthesis, via Scly, by a previously unknown or completely-unrelated factor of the tissue regenerating protein Reg3β. Although mammalian selenoproteome and machinery of selenoprotein biosynthesis is well-characterized [27,51,52], current understanding of regulation of selenoprotein biosynthesis has been limited to only Se [53]. Evidently, our findings link this biosynthesis to tissue regeneration and offer the first evidence for the involvement of Se in a completely new metabolic process beyond its recognized roles in antioxidant defense [34], thyroid hormone metabolism [35], reproduction [36], immune function [34], anti-tumorigenesis [37,38], cardiovascular diseases [39], and energy metabolism [31]. Meanwhile, unveiling this link will provide a completely new clue to help explain yet resolved pathogenesis mechanisms of tissue degenerations in the classical Se/vitamin E deficiency diseases [54] and the anti-cancer function of Se [37,38]. Illustrating this link will also render Se or selenoproteins as a new regulator or modulator of tissue regeneration.
Substantial evidences generated from our mechanistic experiments indicate that knockout of Reg3β enhanced transcriptional factor AP-1 protein that activated transcription of Scly, leading to an up-regulation of Scly activity. First of all, adding the purified rcReg3β protein to the liver homogenates of KO or WT mice did not inhibit Scly activity. Thus, the activity rise in the KO mice was not caused by the Reg3β absence itself. Second, the activity rise in the KO mice was accompanied with consistent increases in the enzyme mRNA, suggesting a potential transcriptional up-regulation [55]. Notably, knockout of Reg3β affected gene expression of only Scly, but not SPS1, SPS2, or SBP2 that are also key players of selenoprotein biosynthesis [31,46]. Apparently, the relationship between Scly and Reg3β was specific. Third, bioinformatics analysis identified AP-1 as one of the top four transcription factors (NF-1, YY1, AP-1 and GR) with high frequencies of binding sites in the promoter region of Scly, and mRNA and(or) protein levels of AP-1 family c-jun and c-fos were actually greater in the KO mice than the WT mice. Fourth, the ChIP assay revealed four binding sites in the promoter of Scly for c-Jun protein and binding at two of these sites were up-regulated in the KO mice. Jafari et al. [48] previously reported that AP-1 binding sites in Scly promoter were related to the regulation of its transcription in the kidney of rats. Our study has confirmed AP-1 binding to the promoter region of Scly in the liver tissue of mice and offered a novel regulation for these induced binding activities by the Reg3β knockout. However, future research will be needed to identify other transcriptional factors that also bind the promoter region and their impact on transcription of Scly.
In summary, our study has elucidated a new function of Reg3β in the APAP-induced protein nitration via modulating tissue levels of PN scavenger GSH, which helps explain the previously-observed impacts of Reg3β knockout [3] or overexpression [3,4] on toxicities of APAP and other pro-oxidants. Most strikingly, our research has unveiled that Reg3β, a liver regenerating protein [3], served as a novel regulator of selenoprotein biosynthesis via the newly discovered pathway of AP-1→Scly→selenoproteins (GPX1, SeP, and SelK). Overall, our findings have not only identified new metabolic roles of Reg3β in the APAP-induced protein nitration, but also linked Se metabolism to tissue regeneration control. These results will have profound implications in understanding metabolism of Se, functions of selenoproteins, and physiological coordination of tissue regeneration and repairing.
Supplementary Material
Highlights.
Reg3β knockout aggravated APAP-induced hepatic protein nitration
Reg3β knockout accelerated GSH depletion via GPX1 up-regulation
Reg3β knockout enhanced GPX1 activity via Scly up-regulation
Reg3β knockout up-regulated AP-1 binding activity to the Scly promoter
Reg3β may link tissue regeneration to metabolism of selenoproteins
Acknowledgments
The KO mouse breeders were provided as a gift from Dr. Jamila Faivre at Institut National de la Sante et de la Recherche Medicale (INSERM). We thank Lvhui Sun, Xi Yan, and Carol A. Roneker for their technical help. This study was supported in part by National Institutes of Health Grant DK 53018 to X.G.L.
Abbreviations
- AKT
protein kinase B
- ALT
alanine aminotransferase
- AP-1
activator protein 1
- APAP
acetaminophen
- CYP2E1
cytochrome P450 2E1
- γ-GCS
γ-glutamylcysteine synthetase
- GPX
glutathione peroxidase
- GR
glucocorticoid receptor
- GR
glutathione reductase
- GS
glutathione synthetase
- GSH
glutathionnes
- GSSG
oxidized glutathione
- GST
glutathione S-transferase
- HIP/PAP
hepatocarcinoma-intestine-pancreas/pancreatic-associated protein
- iNOS
inducible NO synthase
- IP
intraperitoneal
- NF-1
nuclear factor 1
- NO
nitric oxide
- PN
peroxynitrite
- Reg3β
regenerating islet-derived 3β
- SBP
selenocysteine insertion sequence-binding protein
- Scly
selenocysteine lyase
- Se
selenium
- SECIS
selenocysteine insertion sequence
- SelK
selenoprotein K
- SeP
selenoprotein P
- SOD
superoxide dismutase
- SPS
selenophosphate synthetases
- YY1
Yin Yang 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nishimune H, Vasseur S, Wiese S, Birling MC, Holtmann B, Sendtner M, Iovanna JL, Henderson CE. Reg-2 is a motoneuron neurotrophic factor and a signalling intermediate in the CNTF survival pathway. Nat Cell Biol. 2000;2:906–914. doi: 10.1038/35046558. [DOI] [PubMed] [Google Scholar]
- 2.Malka D, Vasseur S, Bodeker H, Ortiz EM, Dusetti NJ, Verrando P, Dagorn JC, Iovanna JL. Tumor necrosis factor alpha triggers antiapoptotic mechanisms in rat pancreatic cells through pancreatitis-associated protein I activation. Gastroenterology. 2000;119:816–828. doi: 10.1053/gast.2000.16491. [DOI] [PubMed] [Google Scholar]
- 3.Lieu HT, Simon MT, Nguyen-Khoa T, Kebede M, Cortes A, Tebar L, Smith AJH, Bayne R, Hunt SP, Brechot C, Christa L. Reg2 inactivation increases sensitivity to Fas hepatotoxicity and delays liver regeneration post-hepatectomy in mice. Hepatology. 2006;44:1452–1464. doi: 10.1002/hep.21434. [DOI] [PubMed] [Google Scholar]
- 4.Lieu HT, Batteux F, Simon MT, Cortes A, Nicco C, Zavala F, Pauloin A, Tralhao JG, Soubrane O, Weill B, Brechot C, Christa L. HIP/PAP accelerates liver regeneration and protects against acetaminophen injury in mice. Hepatology. 2005;42:618–626. doi: 10.1002/hep.20845. [DOI] [PubMed] [Google Scholar]
- 5.Hart SG, Cartun RW, Wyand DS, Khairallah EA, Cohen SD. Immunohistochemical localization of acetaminophen in target tissues of the CD-1 mouse: correspondence of covalent binding with toxicity. Fundam Appl Toxicol. 1995;24:260–274. doi: 10.1006/faat.1995.1029. [DOI] [PubMed] [Google Scholar]
- 6.Roberts DW, Bucci TJ, Benson RW, Warbritton AR, McRae TA, Pumford NR, Hinson JA. Immunohistochemical localization and quantification of the 3-(cystein-S-yl)-acetaminophenproteinadductinacetamino-phen hepatotoxicity. Am J Pathol. 1991;138:359–371. [PMC free article] [PubMed] [Google Scholar]
- 7.Hinson JA, Pike SL, Pumford NR, Mayeux PR. Nitrotyrosine-protein adducts in hepatic centrilobular areas following toxic doses of acetaminophen in mice. Chem Res Toxicol. 1998;11:604–607. doi: 10.1021/tx9800349. [DOI] [PubMed] [Google Scholar]
- 8.Abdelmegeed MA, Jang S, Banerjee A, Hardwick JP, Song BJ. Robust protein nitration contributes to acetaminophen-induced mitochondrial dysfunction and acute liver injury. Free Radic Biol Med. 2013;60:211–222. doi: 10.1016/j.freeradbiomed.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petrenko AB, Yamakura T, Kohno T, Sakimura K, Baba H. Reduced immobilizing properties of isoflurane and nitrous oxide in mutant mice lacking the N-methyl-D-aspartate receptor GluR(epsilon)1 subunit are caused by the secondary effects of gene knockout. Anesth Analg. 2010;110:461–465. doi: 10.1213/ANE.0b013e3181c76e73. [DOI] [PubMed] [Google Scholar]
- 10.Abello N, Kerstjens HA, Postma DS, Bischoff R. Protein tyrosine nitration: selectivity, physicochemical and biological consequences, denitration, and proteomics methods for the identification of tyrosine-nitrated proteins. J Proteome Res. 2009;8:3222–3238. doi: 10.1021/pr900039c. [DOI] [PubMed] [Google Scholar]
- 11.Banks BA, Ischiropoulos H, McClelland M, Ballard PL, Ballard RA. Plasma 3-nitrotyrosine is elevated in premature infants who develop bronchopulmonary dysplasia. Pediatrics. 1998;101:870–874. doi: 10.1542/peds.101.5.870. [DOI] [PubMed] [Google Scholar]
- 12.Shishehbor MH, Aviles RJ, Brennan ML, Fu XM, Penn MS, Sprecher DL, Gokce N, Keaney JF, Vita JA, Hazen SL. Levels of nitrotyrosine, an inflammatory marker generated by nitric oxide derived oxidants, is associated with risk of coronary artery disease. J Am Coll Cardiol. 2003;41:303a–303a. [Google Scholar]
- 13.Ischiropoulos H. Biological selectivity and functional aspects of protein tyrosine nitration. Biochem Biophys Res Commun. 2003;305:776–783. doi: 10.1016/s0006-291x(03)00814-3. [DOI] [PubMed] [Google Scholar]
- 14.Bajt ML, Knight TR, Farhood A, Jaeschke H. Scavenging peroxynitrite with glutathione promotes regeneration and enhances survival during Acetaminophen-induced liver injury in mice. J Pharmacol Exp Ther. 2003;307:67–73. doi: 10.1124/jpet.103.052506. [DOI] [PubMed] [Google Scholar]
- 15.Gardner CR, Heck DE, Yang CS, Thomas PE, Zhang XJ, DeGeorge GL, Laskin JD, Laskin DL. Role of nitric oxide in acetaminophen-induced hepatotoxicity in the rat. Hepatology. 1998;27:748–754. doi: 10.1002/hep.510270316. [DOI] [PubMed] [Google Scholar]
- 16.Alexander B. The role of nitric oxide in hepatic metabolism. Nutrition. 1998;14:376–390. doi: 10.1016/s0899-9007(97)00492-9. [DOI] [PubMed] [Google Scholar]
- 17.Jo HY, Kim CK, Suh IB, Ryu SW, Ha KS, Kwon YG, Kim YM. Co-localization of inducible nitric oxide synthase and phosphorylated Akt in the lesional skins of patients with melasma. J Dermatol. 2009;36:10–16. doi: 10.1111/j.1346-8138.2008.00579.x. [DOI] [PubMed] [Google Scholar]
- 18.Tazi KA, Moreau R, Hervé P, Dauvergne A, Cazals-Hatem D, Bert F, Poirel O, Rabiller A, Lebrec D. Norfloxacin reduces aortic NO synthases and proinflammatory cytokine up-regulation in cirrhotic rats: role of Akt signaling. Gastroenterology. 2005;129:303–314. doi: 10.1053/j.gastro.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 19.Lu YK, Wang XD, Cederbaum AI. Lipopolysaccharide-induced liver injury in rats treated with the CYP2E1 inducer pyrazole. Am J Physiol Gastrointest Liver Physiol. 2005;289:G308–G319. doi: 10.1152/ajpgi.00054.2005. [DOI] [PubMed] [Google Scholar]
- 20.Beckman JS, Beckman TW, Chen J, Marshall PA, Freeman BA. Apparent Hydroxyl Radical Production by Peroxynitrite - Implications for Endothelial Injury from Nitric-Oxide and Superoxide. Proc Natl Acad Sci USA. 1990;87:1620–1624. doi: 10.1073/pnas.87.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knight TR, Ho YS, Farhood A, Jaeschke H. Peroxynitrite is a critical mediator of acetaminophen hepatotoxicity in murine livers: Protection by glutathione. J Pharmacol Exp Ther. 2002;303:468–475. doi: 10.1124/jpet.102.038968. [DOI] [PubMed] [Google Scholar]
- 22.Saito C, Zwingmann C, Jaeschke H. Novel Mechanisms of Protection Against Acetaminophen Hepatotoxicity in Mice by Glutathione and N-Acetylcysteine. Hepatology. 2010;51:246–254. doi: 10.1002/hep.23267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim SJ, Shin YH, Kim K, Park EH, Sa JH, Lim CJ. Regulation of the gene encoding glutathione synthetase from the fission yeast. J Biochem Mol Biol. 2003;36:326–331. doi: 10.5483/bmbrep.2003.36.3.326. [DOI] [PubMed] [Google Scholar]
- 24.Kanzok SM, Fechner A, Bauer H, Ulschmid JK, Muller HM, Botella-Munoz J, Schneuwly S, Schirmer RH, Becker K. Substitution of the thioredoxin system for glutathione reductase in Drosophila melanogaster. Science. 2001;291:643–646. doi: 10.1126/science.291.5504.643. [DOI] [PubMed] [Google Scholar]
- 25.Habig WH, Pabst MJ, Jakoby WB. Glutathione S-transferases The first enzymatic step in mercapturic acid formation. J Biol Chem. 1974;249:7130–7139. [PubMed] [Google Scholar]
- 26.Fukai T, Ushio-Fukai M. Superoxide dismutases: role in redox signaling, vascular function, and diseases. Antioxid Redox Signaling. 2011;15:1583–1606. doi: 10.1089/ars.2011.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 28.Rotruck JT, Pope AL, Ganther HE, Swanson AB, Hafeman DG, Hoekstra WG. Selenium - Biochemical Role as a Component of Glutathione Peroxidase. Science. 1973;179:588–590. doi: 10.1126/science.179.4073.588. [DOI] [PubMed] [Google Scholar]
- 29.Hill KE, Wu S, Motley AK, Stevenson TD, Winfrey VP, Capecchi MR, Atkins JF, Burk RF. Production of selenoprotein P (Sepp1) by hepatocytes is central to selenium homeostasis. J Biol Chem. 2012;287:40414–40424. doi: 10.1074/jbc.M112.421404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sunde RA. Regulation of glutathione peroxidase-1 expression. In: Hatfield DL, Berry MJ, Gladyshev VN, editors. Selenium: its molecular biology and role in human health. New York: Springer Science Media; 2006. pp. 149–160. [Google Scholar]
- 31.Seale LA, Hashimoto AC, Kurokawa S, Gilman CL, Seyedali A, Bellinger FP, Raman AV, Berry MJ. Disruption of the selenocysteine lyase-mediated selenium recycling pathway leads to metabolic syndrome in mice. Mol Cell Biol. 2012;32:4141–4154. doi: 10.1128/MCB.00293-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mirochnitchenko O, Weisbrot-Lefkowitz M, Reuhl K, Chen L, Yang C, Inouye M. Acetaminophen toxicity Opposite effects of two forms of glutathione peroxidase. J Biol Chem. 1999;274:10349–10355. doi: 10.1074/jbc.274.15.10349. [DOI] [PubMed] [Google Scholar]
- 33.Zhu JH, Lei XG. Double null of selenium-glutathione peroxidase-1 and copper, zinc-superoxide dismutase enhances resistance of mouse primary hepatocytes to acetaminophen toxicity. Exp Biol Med. 2006;231:545–552. doi: 10.1177/153537020623100508. [DOI] [PubMed] [Google Scholar]
- 34.Rayman MP. The importance of selenium to human health. Lancet. 2000;356:233–241. doi: 10.1016/S0140-6736(00)02490-9. [DOI] [PubMed] [Google Scholar]
- 35.Kohrle J. Selenium and the control of thyroid hormone metabolism. Thyroid. 2005;15:841–853. doi: 10.1089/thy.2005.15.841. [DOI] [PubMed] [Google Scholar]
- 36.Ursini F, Heim S, Kiess M, Maiorino M, Roveri A, Wissing J, Flohe L. Dual function of the selenoprotein PHGPx during sperm maturation. Science. 1999;285:1393–1396. doi: 10.1126/science.285.5432.1393. [DOI] [PubMed] [Google Scholar]
- 37.Irons R, Carlson BA, Hatfield DL, Davis CD. Both selenoproteins and low molecular weight selenocompounds reduce colon cancer risk in mice with genetically impaired selenoprotein expression. J Nutr. 2006;136:1311–1317. doi: 10.1093/jn/136.5.1311. [DOI] [PubMed] [Google Scholar]
- 38.Meplan C, Hughes DJ, Pardini B, Naccarati A, Soucek P, Vodickova L, Hlavata I, Vrana D, Vodicka P, Hesketh JE. Genetic variants in selenoprotein genes increase risk of colorectal cancer. Carcinogenesis. 2010;31:1074–1079. doi: 10.1093/carcin/bgq076. [DOI] [PubMed] [Google Scholar]
- 39.Steinbrenner H, Sies H. Protection against reactive oxygen species by selenoproteins. Biochim Biophys Acta. 2009;1790:1478–1485. doi: 10.1016/j.bbagen.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 40.Cheng WH, Valentine BA, Lei XG. High levels of dietary vitamin E do not replace cellular glutathione peroxidase in protecting mice from acute oxidative stress. J Nutr. 1999;129:1951–1957. doi: 10.1093/jn/129.11.1951. [DOI] [PubMed] [Google Scholar]
- 41.Zhu JH, Zhang XM, Roneker CA, McClung JP, Zhang S, Thannhauser TW, Ripoll DR, Sun Q, Lei XG. Role of copper, zinc-superoxide dismutase in catalyzing nitrotyrosine formation in murine liver. Free Radic Biol Med. 2008;45:611–618. doi: 10.1016/j.freeradbiomed.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson ME. Tissue glutathione. In: Greenwald RA, editor. CRC Handbook of Method for Oxygen Radical Research. Boca Raton: CRC press; 1985. pp. 317–320. [Google Scholar]
- 43.Imai Y, Ito A, Sato R. Evidence for biochemically different types of vesicles in the hepatic microsomal fraction. J Biochem. 1966;60:417–428. doi: 10.1093/oxfordjournals.jbchem.a128453. [DOI] [PubMed] [Google Scholar]
- 44.Deagen JT, Butler JA, Beilstein MA, Whanger PD. Effects of dietary selenite, selenocystine and selenomethionine on selenocysteine lyase and glutathione peroxidase activities and on selenium levels in rat tissues. J Nutr. 1987;117:91–98. doi: 10.1093/jn/117.1.91. [DOI] [PubMed] [Google Scholar]
- 45.Abdelmegeed MA, Moon KH, Chen C, Gonzalez FJ, Song BJ. Role of cytochrome P450 2E1 in protein nitration and ubiquitin-mediated degradation during acetaminophen toxicity. Biochem Pharmacol. 2010;79:57–66. doi: 10.1016/j.bcp.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allmang C, Wurth L, Krol A. The selenium to selenoprotein pathway in eukaryotes: More molecular partners than anticipated. Biochim Biophys Acta. 2009;1790:1415–1423. doi: 10.1016/j.bbagen.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 47.Kurokawa S, Takehashi M, Tanaka H, Mihara H, Kurihara T, Tanaka S, Hill K, Burk R, Esaki N. Mammalian Selenocysteine Lyase Is Involved in Selenoprotein Biosynthesis. J Nutr Sci Vitaminol. 2011;57:298–305. doi: 10.3177/jnsv.57.298. [DOI] [PubMed] [Google Scholar]
- 48.Jafari C, Panzer U, Steinmetz OM, Zahner G, Stahl RAK, Harendza S. Enhanced expression of selenocysteine lyase in acute glomerulonephritis and its regulation by AP-1. Cell Mol Biol Lett. 2006;11:424–437. doi: 10.2478/s11658-006-0035-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahmed MME, Wang T, Luo Y, Ye SL, Wu Q, Guo ZS, Roebuck BD, Sutter TR, Yang JY. Aldo-Keto Reductase-7A Protects Liver Cells and Tissues From Acetaminophen-Induced Oxidative Stress and Hepatotoxicity. Hepatology. 2011;54:1322–1332. doi: 10.1002/hep.24493. [DOI] [PubMed] [Google Scholar]
- 50.Fu YX, Sies H, Lei XG. Opposite roles of selenium-dependent glutathione peroxidase-1 in superoxide generator diquat- and peroxynitrite-induced apoptosis and signaling. J Biol Chem. 2001;276:43004–43009. doi: 10.1074/jbc.M106946200. [DOI] [PubMed] [Google Scholar]
- 51.Hoffmann PR, Berry MJ. Selenoprotein synthesis: A unique translational mechanism used by a diverse family of proteins. Thyroid. 2005;15:769–775. doi: 10.1089/thy.2005.15.769. [DOI] [PubMed] [Google Scholar]
- 52.Xu XM, Mix H, Carlson BA, Grabowski PJ, Gladyshev VN, Berry MJ, Hatfield DL. Evidence for direct roles of two additional factors, SECp43 and soluble liver antigen, in the selenoprotein synthesis machinery. J Biol Chem. 2005;280:41568–41575. doi: 10.1074/jbc.M506696200. [DOI] [PubMed] [Google Scholar]
- 53.Papp LV, Lu J, Holmgren A, Khanna KK. From selenium to selenoproteins: Synthesis, identity, and their role in human health. Antioxid Redox Signaling. 2007;9:775–806. doi: 10.1089/ars.2007.1528. [DOI] [PubMed] [Google Scholar]
- 54.Hill KE, Motley AK, Li X, May JM, Burk RF. Combined selenium and vitamin E deficiency causes fatal myopathy in guinea pigs. J Nutr. 2001;131:1798–1802. doi: 10.1093/jn/131.6.1798. [DOI] [PubMed] [Google Scholar]
- 55.Tiedge M, Lortz S, Drinkgern J, Lenzen S. Relation between antioxidant enzyme gene expression and antioxidative defense status of insulin-producing cells. Diabetes. 1997;46:1733–1742. doi: 10.2337/diab.46.11.1733. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.