Abstract
γD-crystallin is an abundant structural protein of the lens that is found in native and modified forms in cataractous aggregates. We establish that UV-B irradiation of γD-crystallin leads to structurally specific modifications and precipitation via two mechanisms: amorphous aggregates and amyloid fibers. UV-B radiation causes cleavage of the backbone, in large measure near the interdomain interface, where side chain oxidations are also concentrated. 2D IR spectroscopy and expressed protein ligation localize fiber formation exclusively to the C-terminal domain of γD-crystallin. The native β-sandwich domains are not retained upon precipitation by either mechanism. The similarity between the amyloid forming pathway when induced by either UV-B radiation or low pH suggests that it is the propensity for the C-terminal β-sandwich domain to form amyloid β-sheets that determines the misfolding pathway independent of the mechanism of denaturation.
Keywords: cataract, crystallin, protein aggregation, amyloid, ultraviolet, infrared spectroscopy, 2D IR spectroscopy, expressed protein ligation, isotope labeling, mass spectrometry
Cataracts are a common protein misfolding disease of the ocular lens, which affects approximately 50% of the population over the age of 65 (1, 2). This disease results from accumulated damage to lens crystallin proteins, which destabilizes their folds and causes them to aggregate, resulting in the blurring of vision (1). Currently, the only treatment for cataracts is invasive surgical extraction that is carried out in the advanced stages of the disease. As a result, there is much interest in understanding the cause of cataracts and the mechanism by which they form. However, this task is hindered by the difficulty of experimentally probing the structures of protein aggregates, which has also been an issue in the study of other diseases such as Alzheimer's disease, type II diabetes.
A complicating factor for cataracts is the likely diversity of destabilizing factors that induce the aggregation and which could result in complex mixtures of different structural states. A wide variety of processes such as oxidation, cross-linking, cleavage, and deamidation of crystallins have been identified as factors in their formation based on the distribution of modifications observed in proteins extracted from tissue (1, 3, 4). These modifications arise from environmental factors such as exposure to ultraviolet light (5-10) and oxidative stress (3, 4, 7, 8, 11-13). Because lens crystallins are not regenerated after development, damage to the proteins can accumulate over multiple years leading to the formation of age-onset cataracts (1, 2).
In order to understand the structures of lens crystallin aggregates and their mechanisms of formation, classes of crystallin proteins have been examined under a variety of denaturation conditions in vitro. Typically, proteins are denatured using heat, chemicals, or pH shifts (14-19). Although these conditions have been utilized extensively, the resulting crystallin molecular structures are still poorly understood. Two broad classes of aggregates have been observed: amorphous aggregates and amyloid aggregates. Amyloid aggregates result from a massive conformational shift in the protein upon precipitation (14, 15, 17, 19, 20). Recently, we applied segmental 13C labeling and ultrafast two-dimensional infrared (2D IR) spectroscopy to examine the acid-induced amyloid fiber structure of the abundant human structural lens protein γD-crystallin (15). By isotope labeling the domains individually, we were able to spectroscopically monitor the structures and dynamics associated with the C-terminal and N-terminal domains of γD-crystallin. We found that β-sheets of the amyloid fiber originate from residues in the protein's C-terminal domain, while its N-terminal domain becomes highly disordered (15). Using this information, along with spectral simulations as well as enzyme digests and mass spectrometry (21), we built and refined a structural model of the acid-induced fibers.
While acid-induced denaturation is a useful way to initiate crystallin aggregation, radiation induced aggregation may be more relevant the disease. Exposure to ultraviolet radiation in sunlight has been linked to the formation of age-related cataracts (1, 5, 7, 13). Human γD-crystallin absorbs ultraviolet light and has a large cross section in the UV-B range due to the presence of aromatic amino acids, particularly tryptophan and tyrosine (22). The four tryptophans have been shown to efficiently funnel UV-B excitation to thermal energy through a process involving rapid energy transfer and internal conversion thereby protecting the protein from UV initiated photochemistry (22-24). However, rare quenching events may lead to ultraviolet photodamage and it has recently been shown that the conversion of tryptophans in γD-crystallin to kynurenine (a known marker of cataract formation) destabilizes its fold (6). Here, we induce aggregation of human γD-crystallin by exposure to UV-B radiation. We show that this results in oxidative damage to side chains, as well as cross-linking and regiospecific scission of the polypeptide backbone. We demonstrate that the polypeptide backbone is cleaved photochemically, and use mass spectrometry to show that two regions of the protein, in the vicinity of specific tryptophan residues and the interdomain interface, are highly susceptible to peptide bond cleavage. Using thioflavin T binding, transmission electron microscopy, segmental isotope labeling, and 2D IR spectroscopy, we conclusively demonstrate that the aggregates formed by this process contain amyloid fibers, and that the fiber forming residues originate from the C-terminal domain. Using this information, we propose a mechanism for the initiation of aggregation by UV-B radiation. Since UV radiation is known to correlate with cataract formation in adults,(1, 5, 7, 13) our results suggest that amyloid fibers of γD-crystallin could form in the lens, via side chain damage, polypeptide crosslinking, or fragmentation, all of which can destabilize the protein enough to allow entry into an amyloid misfolding pathway.
Experimental Procedures
All chemicals were purchased from Sigma-Aldrich and used as received, unless otherwise noted. His-tagged human γD-crystallin, γD-crystallin (S84C), and its isolated domains were expressed in E. coli as described previously(15). Segmentally-labeled proteins were prepared by expressing one of the two domains in 13C labeled media and linking it to the other through expressed protein ligation (15). A bank of four 8 W medium pressure mercury UV-B lamps were used to irradiate samples at a distance of approximately 30 cm, with an irradiance of 35 W/m2. Samples, at approximately 5 μM concentration, were agitated gently using an orbital shaker during irradiation. Oxygen-free samples were prepared by lyophilizing the protein to dryness, purging with nitrogen, and redissolving the sample in deoxygenated buffer prepared using three freeze-pump-thaw cycles on a Schlenk line. A modified Fenton reagent composed of 0.1 mM (NH4)2Fe(SO4)2, 0.2 mM EDTA, 1 mM ascorbic acid, and 0.03% H2O2 was used to prepare samples damaged by hydroxyl radicals (25).
Samples for mass spectrometry were prepared by illuminating γD-crystallin for 6 hours, followed by digestion with bovine trypsin in the presence of 0.01% ProteaseMax surfactant (Promega). Digestions were performed at 42 °C for three hours, and the samples were acidified with 0.05% TFA. Samples were centrifuged to sediment degraded surfactant and insoluble materials. LC-MS/MS data was collected using a Thermo Scientific LTQ Orbitrap XL mass spectrometer. Collision-induced dissociation (CID) spectra of the tryptic peptides were used to generate sequence data. The data was analyzed as described previously (21) against the E. coli protein sequence database, containing 4,205 entries, with the sequence of the recombinant human γD-crystallin appended to it. Sequence matches were identified and semitryptic fragments and amino acid adducts were identified from the sequence data.
UV-vis samples were prepared by treating aggregated γD-crystallin samples with bovine trypsin (1:50 ratio), resulting in resolubilization. Spectra were measured in a 1 cm quartz cell (Starna). Undigested samples were prepared for SDS-PAGE by dissolving the samples in Laemmli sample buffer containing 20 mM 2-mercaptoethanol. SDS gels were analyzed using ImageJ software (available free of charge from http://rsbweb.nih.gov/ij/)
Transmission electron micrographs were taken at the University of Wisconsin Medical Sciences Electron Microscopy Facility, using a Philips CM 120 transmission electron microscope. Samples were negatively stained using methylamine tungstate for imaging. The size distributions of structures in the TEM images were determined via random selection and diameter measurement of 50 structures.
ThT binding assays were performed by removing aliquots of γD-crystallin at a series of time points during UV-B illumination, adding 2.5 equivalents of ThT, and incubating for 15 minutes. Fluorescence emission at 486 nm was measured after excitation of ThT at 430 nm. The measurements were performed for three samples prepared in parallel, and averaged.
2D IR spectra were collected on UV-B damaged γD-crystallin after 6 hours of illumination in deuterated buffers. Samples were then placed between CaF2 plates separated by a 56 μm Teflon spacer. Spectra were collected with perpendicular pump-probe polarization, and processed as described previously (15, 26).
Results
Aggregation of human γD-crystallin variants was induced by illuminating dilute samples (5-50 μM in 20 mM sodium phosphate, 100 mM NaCl, pH 7.0) with UV-B radiation from a bank of four unfiltered 8 W medium-pressure mercury vapor lamps (Spectroline, Inc.) The maximum output of the lamp was in the 275-325 nm range, overlapping with the UV absorption spectrum of human γD-crystallin (Figure 1A). The total power is 35 W/m2 at a distance of 30 cm. Under these conditions, the total energy incident on the sample in 1 hour is approximately equal to that from sea-level solar radiation over the course of 3 years assuming 1 hour per day of direct exposure (27). Although the power of the UV-B light bank is roughly 103 times that of sunlight on the earth's surface, multiphoton processes are still unlikely. Thus, these conditions are representative of exposure to UV-B solar radiation, albeit on a compressed timescale.
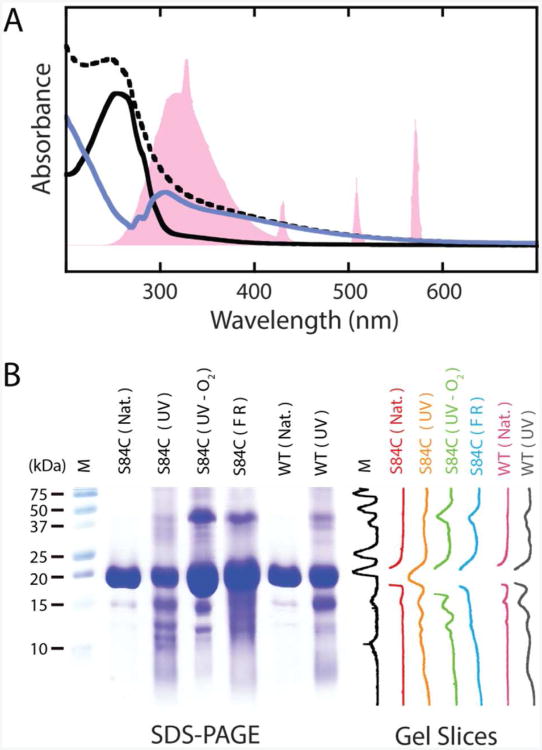
Figure 1.
UV-B photodamage products of human γD-crystallin. A. UV-vis spectra of γD-crystallin before and after UV-B illumination. The output spectrum of the mercury vapor lamp, normalized to the protein absorption, is shown in pink. The spectrum of the protein before illumination (solid, black) shows absorption below 300 nm consistent with the tyrosine and tryptophan content of the protein. After exposure to UV-B light, the spectrum (dashed, black) becomes broadened into the visible range. Subtraction of the pre-UV-B spectrum from the spectrum of the photodamaged protein yields a difference spectrum (solid, blue) with a bleach between 250 and 305 nm. The band structure of this bleach suggests a loss of tryptophan absorption. B. SDS-PAGE characterization of γD-crystallin degradation via UV-B photodamage and chemical methods. From left to right are molecular weight marker (M), undamaged γD-crystallin (S84C) (S84C (Nat.)), UV-B photodamaged γD-crystallin (S84C) (S84C (UV)), UV-B photodamaged γD-crystallin (S84C) in deoxygenated buffer (S84C (UV-O2)), γD-crystallin (S84C) exposed to Fenton's reagent (S84C (FR)), undamaged wild type γD-crystallin (WT (Nat.)), and UV-B photodamaged wild type γD-crystallin (WT (UV)), respectively. Vertical slices of the gel image are shown for comparison of each sample.
Exposure of γD-crystallin to UV-B light resulted in visible turbidity in the samples within 1 hour, and the continued production of precipitates for as long as we measured (10 hours). For the purposes of this paper, an exposure time of 6 hours is examined. The precipitation of γD-crystallin is accompanied by the formation of a pale yellow color. In order to solubilize the precipitates without chemically altering the side chains, we used bovine trypsin to digest the aggregates. Figure 1A shows the UV-vis spectra of otherwise identical samples with and without exposure to UV-B light. A clear extension of the spectrum into the visible is present in the UV-B exposed sample, accounting for the color change. This extension of the absorbance into the visible range is consistent with the photooxidative modification of aromatic side chains in proteins (28), and is similar to color changes that are sometimes observed in the lens with the formation of mature cataracts. Additionally, an apparent bleach occurs between 250 and 305 nm in the difference spectrum between the two samples, consistent with the loss of absorption from aromatic amino acids after UV-B exposure. The sequence of γD-crystallin contains 14 tyrosine and 4 tryptophan residues (29), both of which have been implicated in UV-B photodamage of proteins (9, 30-32). Tyrosine and tryptophan absorb between 250 and 300 nm. The bleach in the difference spectrum in Figure 1A has a band structure comparable to the vibronic bands of the 1Lb mode of tryptophan, indicating that tryptophan side chains are damaged (30, 33). From the magnitude of the bleach, we estimate that at least 75% of the tryptophans in the sample have been photodamaged within 6 hours.
In addition to damage to aromatic side chains, UV-B exposure also results in the cleavage of the γD-crystallin polypeptide backbone. SDS-PAGE analysis (Figure 1B) shows that insoluble high molecular weight aggregates, covalent cross-linked products, and discrete cleavage products are produced. Slices of the gel image show stain intensity profiles of each lane. Approximately 70% of the protein components of this sample exist in cross-linked and cleaved photoproducts. The linkages that produce the high-molecular weight products are not disulfide bonds because samples were reduced with 2-mercaptoethanol prior to electrophoresis. Thus, they must arise from other covalent linkages such as dityrosine adducts (28), which are a known product of the irradiation of proteins with UV-B light. Four major cleavage products are observed, with molecular weights estimated at 6.4, 10.5, 12.4, and 14.9 kDa. The small number of cleavage products indicates that the cleavage sites are sequence or structure dependent. Because the S84C mutation, which is required for our isotope labeling and 2D IR experiments, introduces an additional redox-active amino acid into the γD-crystallin sequence, we also performed this experiment on the wild type protein. A similar cleavage pattern is observed in the wild type protein (Figure 1) indicating that the UV-B induced cleavage is not dependent on the presence of an extra redox-active cysteine. We also performed the UV-damage experiment in deoxygenated buffer, and the resulting cleavage pattern observed by SDS-PAGE shows similarly sized fragments but with a lower yield, indicating that molecular oxygen, or reactive oxygen species, may be involved in the photodamage mechanism. To test whether the product distribution results from direct photoexcitation of the protein or its interaction with photochemically produced reactive oxygen species, we also used a modified Fenton reagent, which produces hydroxyl radicals in situ, to damage the protein (25). The highly specific UV-B cleavage pattern is not observed in the protein after damage by hydroxyl radicals; instead, a broad distribution of products is observed on the gel. Thus, we conclude that the mechanism of cleavage is initiated by UV-B photoexcitation of the protein itself, but is facilitated by molecular oxygen. This result is interesting in light of the fact that molecular oxygen is indeed present in the lens, but at smaller levels in the lens nucleus than in the outer portions of the lens (3).
To identify damaged side chains and determine the damage sites that lead to the UV-B induced fragments, we digested the UV-B damaged protein with bovine trypsin and analyzed the resulting cleavage products using LC-MS/MS. This method is similar to the one we used to identify the amyloid fiber core sequence of acid-induced γD-crystallin fibers in a previous publication (21). A sequence coverage of 94% was obtained from the analyzed tryptic fragments generated by this method. Representative MS/MS spectra of tryptic and semitryptic fragments are shown in Figure 2A-F. In the mixture of peptides generated by sequential UV-B photodamage and trypsin digest, we observe a variety of species generated from the same sequence. Figure 2A shows the CID spectrum and assignments of an unmodified peptide covering residues 117-139. In addition to this peptide, a variety of others corresponding to the same sequence region were also observed. For example, multiple oxidation products were identified in the 117-139 tryptic peptide, with Trp130 and His121 residues being oxidized (Figure 2B). The locations of oxidation are determined from +16 m/z shift in b-series ions compared to the control (Figure 2A). A number of residues have been shown to be similarly modified in natural cataracts (4, 7, 11). This damage is consistent with the loss of aromatic amino acid absorbances in the UV-vis difference spectrum shown in Figure 1A, and may also act to destabilize the protein fold resulting in aggregation.
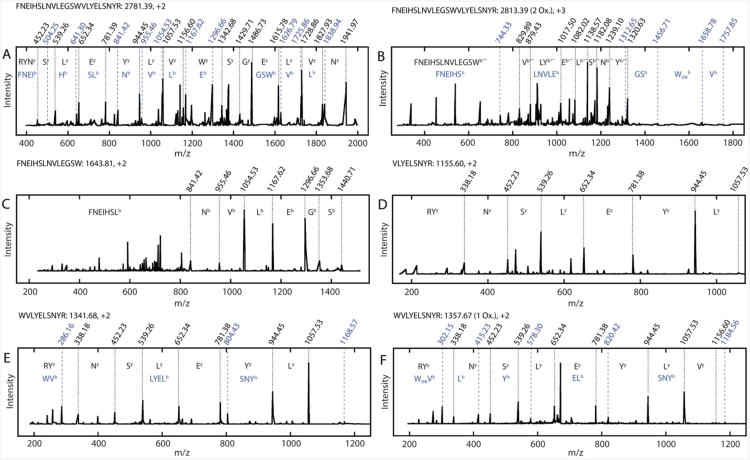
Figure 2.
Set of representative CID spectra of tryptic and semitryptic peptides used in the analysis of backbone cleavage and side chain oxidation. The most complete ion series identified (black) are indicated, as are ion series containing information revealing oxidation sites (blue) where appropriate. The m/z values for each assigned peak are indicated on each spectrum, as are the amino acids and/or peptides to which they correspond. Superscripts on peak assignments denote the ion series to which the peak belongs. Full ion data are shown in Tables S1-6. A. CID spectrum of unmodified (control) γD-crystallin tryptic peptide 117-139 prior to UV-B exposure. B. CID spectrum of tryptic peptide 117-139 after UV-B exposure, with oxidation at W130. C. CID spectrum of semitryptic peptide 117-139. D. CID spectrum of semitryptic peptide 131-139. E. CID spectrum of semitryptic peptide 130-139. F. CID spectrum of semitryptic peptide 130-139, with oxidation at W130.
In addition to oxidized products, smaller semitryptic peptides were observed. In these peptides, one end does not correspond to a tryptic site. It is unlikely that these result from residual chymotrypsin activity in the bovine trypsin we used for the cleavage reaction, because they do not all correspond to chymotryptic sites and the trypsin was treated with L-1-tosylamide-2-phenylethyl chloromethyl ketone to inhibit chymotryptic activity. Furthermore, these sites are not observed in tryptic digests of γD-crystallin that have not been treated with UV-B radiation (21). Thus, they most likely arise from cleavage of peptide bonds as a direct result of UV-B irradiation. Two such peptides are shown in Figure 2C-D: one containing residues 117-130 and another containing residues 131-139. These peptides arise from backbone cleavage between Trp130 and Val131. Together, they cover the entirety of the tryptic peptide 117-139. A variety of other semitryptic peptides and peptide pairs were observed for this region and others in γD-crystallin. Notably, many of the semitryptic peptides also contain internal oxidations, indicating that these modifications are closely linked. An example of such a peptide is the semitryptic peptide 130-139, described in Figure 2E,F. In Figure 2E, this peptide is identified without oxidative modification. Figure 2F shows a similar fragment pattern, but a number of the b-series ions are shifted by +16 m/z. Again, from the full ion assignment (Tables S5-6) we determine that the oxidation is on the tryptophan residue Trp130.
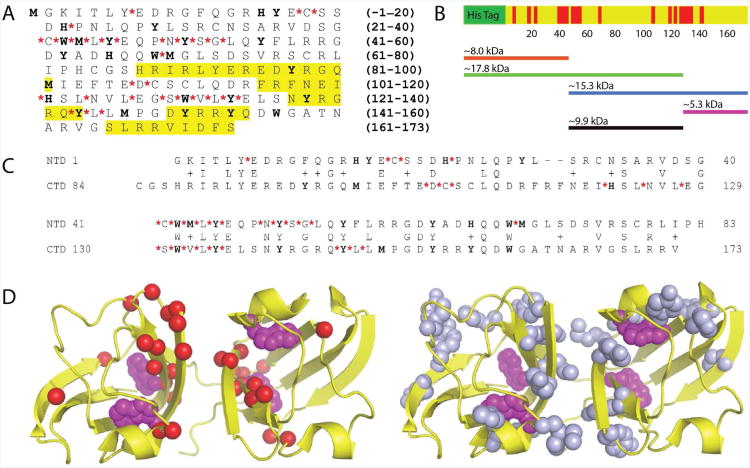
By mapping the sites of oxidized and non-tryptic cleavage to the primary structure of γD-crystallin (Figure 3A), we obtain a picture of the sequence context of both kinds of damage sites. There appears to be a close association between the positions of semitryptic sites and tryptophan residues in the sequence, while other redox-active amino acids (Met, Cys, Tyr, His) are not obviously correlated with the cleavage sites. Perhaps the most striking observation is that there are two stretches of sequences, containing about 15 amino acids, which are particularly susceptible to cleavage. The locations of these sites in the protein's primary structure, and the approximate lengths of expected cleavage fragments, are represented in schematic form in Figure 3B. Each domain contains one such sequence, centered around analogous tryptophans (W42 and W130). Cleavage of the γD-crystallin backbone within one or both of these would result in fragments with molecular masses of approximately 15.4 (17.8), 15.3, 9.9, 5.6 (8.), and 5.3 kDa (His-tagged molecular masses in parentheses), consistent with the SDS-PAGE results presented above. Unlike the backbone cleavage sites, which occur in clusters in the peptide primary structure, the oxidized amino acid side chains we identified appear to be distributed throughout the protein with no obvious sequence association. However, a sequence alignment of the N-terminal and C-terminal domains (Figure 3C) shows that the oxidized amino acids occur in similar regions of both domains, even when their identities are different.
Figure 3.
A. Sequence of human γD-crystallin with the locations non-tryptic cleavage sites (red asterisks) and side chain damage sites (bold) identified by LC-MS/MS. Sequences highlighted in yellow were previously identified as containing modifications by antibody reactivity. B. Distribution and density of non-tryptic cleavage sites (red) in the primary structure of γD-crystallin. C. Sequence alignment of the N-terminal domain and C-terminal domain of human γD-crystallin, with UV-B induced modifications identified as in (A). D. Left: Locations of non-tryptic cleavage sites (red spheres) in the native crystal structure of human γD-crystallin (PDB ID: 1HK0). Photoactive tryptophans are also shown (purple spheres). Right: Side chains oxidized by UV-B radiation (blue non-Trp and purple Trp) are shown as spheres.
Visualization of side chain oxidation and non-tryptic cleavage sites in the native crystal structure (29) of γD-crystallin (Figure 3D) reveals that they occur in β-hairpins located near the domain interface, and are quasi-symmetric about the interface. These interfacial β-hairpins are relatively occluded from solvent, and it is therefore unlikely that they form primary attack sites for radical species in solution (34, 35). Together with the comparison of photochemically generated and hydroxyl radical generated fragmentation in Figure 1B, this result suggests that the oxidation and fragmentation occur as a result of the absorption of UV-B light by γD-crystallin itself and not indirect processes arising from the radiolysis of solvent species. Furthermore, the close structural association between oxidation and non-tryptic cleavage sites suggests that they may arise from a common mechanism.
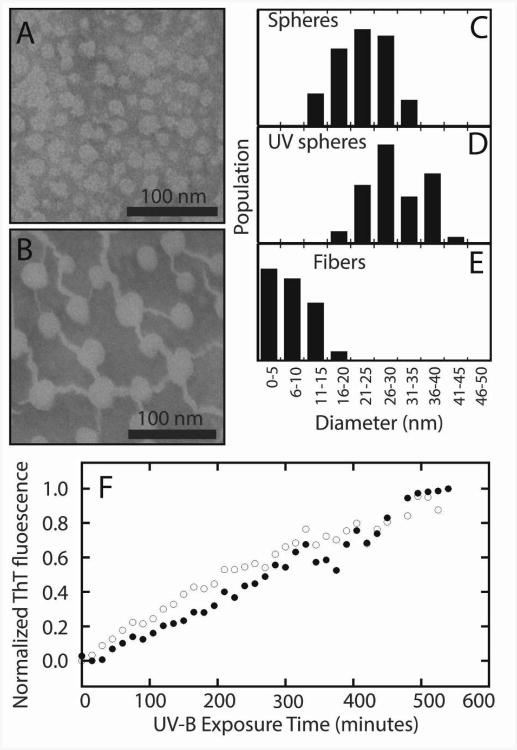
The above analysis provides no structural information on the aggregates, so we turn to transmission electron microscopy, Thioflavin-T (ThT) fluorescence, and two-dimensional infrared (2D IR) spectroscopy. Samples of aggregates taken prior to illumination, and after 6 hours of illumination, were negatively stained with methylamine tungstate. Representative TEM images are shown in Figure 4A-B, and the diameters of the observed structures are shown in Figure 4C-E as histograms. Prior to UV-B damage, spherical structures with a diameter of ∼20 nm are seen in γD-crystallin samples. Such structures were also observed in early acid-induced aggregates of γD-crystallin, prior to fiber formation (14). After irradiation for 6 hours, fibers are observed in addition to the spheres. The fibers have mean diameters of approximately ∼6 nm, similar to those observed in TEM images of acid-induced γD-crystallin amyloid fibers (21), but have a relatively wide distributions of diameters. These results are in striking contrast to previous studies on UV-induced aggregates of mammalian γ-crystallins, which did not show the presence of fibers in TEM images despite FTIR evidence for extended vibrational coupling in β-sheets (16).
Figure 4.
Aggregates of human γD-crystallin. TEM images of undamaged (A) and UV-B photodamaged (B) γD-crystallin (S84C) show the presence of spherical bodies in both samples, and the formation of fibers in the photodamaged sample after 6 hours of illumination. Analysis of the diameters of spheres and fibers in both samples (C-E) shows a slight increase in sphere sizes from ∼20 nm to ∼30 nm and the formation of fibers with a mean diameter of ∼6 nm. The aggregation of wild type (open circles) and S84C (closed circles) γD-crystallin is accompanied by an increase in ThT fluorescence over the course of 9 hours (F).
ThT is a fluorescent dye that acts as an indicator for amyloid-like β-sheets, based on an increase in fluorescence quantum yield at 486 nm after excitation at 430 nm (36). We tested for the presence of amyloid-like structures in UV-B induced γD-crystallin (S84C) and wild type γD-crystallin aggregates by removing aliquots over the course of a 10 hour illumination, and adding ThT in a 2.5:1 dye:protein ratio. The normalized fluorescence intensity change is plotted in Figure 4F. The aggregation kinetics show a slow rise in fluorescence over the course of 10 hours, after which the heterogeneity of the samples prevented the collection of ThT fluorescence data. ThT binds to amyloid β-sheets and thus the increase in fluorescence is evidence for the formation of amyloid fibers (36). The kinetics of the ThT fluorescence increase in S84C mutant and wild type samples are nearly identical, indicating that the mutation is not responsible for ThT binding. No clear lag phase is observed in either sample, indicating that nucleation events are not the rate limiting step in UV-B induced fiber formation. Moreover, the steady rise in ThT binding with exposure time suggests that only damaged proteins are contributing to amyloid formation; if damaged proteins were acting as seed to recruit the aggregation of undamaged proteins, then we would expect a sigmoidal dependence on fluorescence with exposure time. Thus, ThT binding provides evidence for amyloid fiber formation. However, ThT does not always accurately reflect structural changes in fibers (37, 38) and it is not known if ThT can also bind to the spherical morphologies observed in the TEM and which increase in size upon UV-B irradiation (Figure 4A-B). For a more structurally specific probe, we turn to 2D IR spectroscopy.
2D IR spectroscopy has recently emerged as a useful technique in structural analysis of proteins, especially in cases involving complex, heterogeneous, or disordered samples such as membrane proteins and protein aggregates (15, 21, 39-48). 2D IR spectroscopy is sensitive to protein secondary structure, vibrational coupling, and solvent exposure based on frequencies and cross-peaks. Unlike FTIR spectroscopy, 2D IR signals scale with the fourth power of the transition dipole moment, leading to better resolution of peaks in the spectra (49). Isotope labeling strategies, including segmental 13C labeling (15, 21), allow further resolution of signals from specific residues within the protein (26). Here, we use 2D IR spectroscopy and segmental 13C labeling to examine the secondary structure content of UV-B induced γD-crystallin aggregates and the domain origin of the fiber β-sheets.
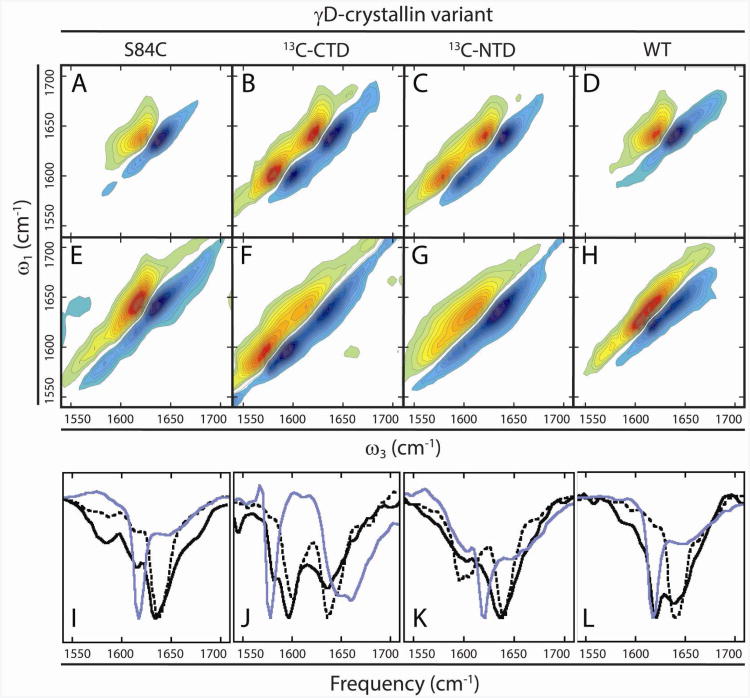
Figure 5A-D shows 2D IR spectra of unlabeled γD-crystallin, γD-crystallin (S84C) with a uniformly 13C labeled C-terminal domain (CTD), γD-crystallin (S84C) with a uniformly 13C labeled N-terminal domain (NTD), and unlabeled wild type γD-crystallin. The expressed protein ligation protocol used to isotope label the domains results in an S84C mutation, which was replicated in the unlabeled protein so that the unlabeled protein sequence does not differ from the labeled protein sequence. The S84C mutation has no effect on the protein stability, spectra or aggregation (15)Fig. 5I-L show slices through the diagonal of these 2D IR spectra (black, dashed line). The features of these spectra were discussed in detail in previous publications (15, 21) and have been calculated from molecular dynamics simulations (50). Briefly, the most intense signals along the diagonals in these spectra arise from the short, antiparallel β-sheets of the protein's β-sandwich motifs. In the segmentally labeled spectra (Figure 5B-C), the signal from the labeled domains is red-shifted by approximately 40 cm−1, consistent with replacement of 12C with 13C in an entire domain. Because vibrational couplings between residues within the domains are conserved (15), the lineshapes of the individual β-sandwich domains in the labeled proteins are similar to that of the unlabeled proteins. Thus, by isotope labeling, we can separately resolve the structure changes associated with residues in the CTD or NTD.
Figure 5.
2D IR spectra of γD-crystallin variants. In native, undamaged γD-crystallin (A-D), unlabeled β-sandwich domains appear at ∼1638 cm−1 along the diagonal, and labeled domains appear at ∼1598 cm−1. Segmentally labeled S84C proteins (B,C) have highly similar spectra with two major diagonal peaks. UV-B photodamaged samples of these proteins (E-H) reveal shifts in the spectra consistent with denaturation of the N-terminal domain and the formation of low-frequency amyloid β-sheet signals arising from the C-terminal domain. Diagonal slices (I-L) of native (dashed, black), UV-B photodamaged (solid, black), and acid-induced amyloid fibers (solid, blue) of each variant reveal the appearance of signals consistent with amyloid fiber formation in the UV-B photodamaged samples.
2D IR spectra of the three protein variants after 6 hours of UV-B exposure are shown in Figure 5E-H, along with slices in Figs. 5I-L (black, solid line). It is clear from these spectra that UV-B photodamage results in conformational changes in γD-crystallin based on changes in 2D frequencies and lineshapes compared to the spectra of the native proteins. The spectrum of UV-B aggregated unlabeled protein displays a new, narrow diagonal feature at approximately 1624 cm−1 that does not exist in the native protein spectra (Fig. 5E vs. 5A). We assign this feature to extensively coupled peptide bonds in an amyloid-like β-sheet conformation. Although both the native and amyloid states of γD-crystallin contain β-sheets, they appear at different frequencies due to the differences in β-sheet architecture, which in turn influences the vibrational coupling between peptide bonds. Because neighboring peptide bonds in adjacent β-strands have large negative vibrational coupling constants, large shifts to lower frequency can occur when multiple β-strands form a β-sheet, delocalizing vibrations. The magnitude of this effect is demonstrated by a survey of reported values in the literature for the frequencies of β-sheets in soluble proteins and amyloid fibers (Table 1). In soluble proteins such as native γD-crystallin, concanavalin A, and small β -hairpins, the β-sheet frequency appears between 1630 cm−1 and 1640 cm−1. Reported amide I frequencies for the β-sheets in well-ordered amyloid fibers, on the other hand, fall between 1617 cm−1 and 1620 cm−1(15, 41, 42, 47, 49, 51), much closer to the 1624 cm−1 reported here.
Table 1.
Representative IR frequencies of β-sheet amide I modes.
| Protein | β-sheet fold | Amide I frequency (12C) | Amide I frequency (13C) | References |
|---|---|---|---|---|
| γD-crystallin (UV-B induced aggregates) | Amyloid | 1624 cm−1 | 1584 cm−1 | This publication |
| γD-crystallin (native) | Greek key | 1638 cm−1 | 1598 cm−1 | (14, 15, 50) |
| γD-crystallin (acid induced fibers) | Amyloid | 1617 cm−1 | 1575 cm−1 | (14, 15) |
| hIAPP | Amyloid | 1617 cm−1 | N/A | (44, 47, 49) |
| Aβ | Amyloid | 1630 cm−1 | N/A | (42) |
| TrpZip | β-hairpin | 1635 cm−1 | N/A | (52) |
| Concanavalin A | Antiparallel β-sheet | 1635 cm−1 | N/A | (53) |
| Parallel β-dimer | Non-natural | 1640 cm−1 | N/A | (54) |
For a comparison of the UV-B induced aggregates to well-ordered amyloid fibers, we plot slices through previously published 2D IR spectra (15) of γD-crystallin that was induced to aggregate by acid denaturation (blue). Acid denaturation causes amyloid fibers to form (14, 15, 21), and the resulting peak lies at a similar frequency to that for UV-B induced aggregation. In addition to this amyloid β-sheet feature, both spectra show significant broadening of the amide I peak near 1640 cm−1, indicating a disordering of the non-amyloid components of the aggregates. Thus, the conformational changes in UV-B induced aggregates are not as severe as those in the acid-induced amyloid fibers, but it is clear that the UV-B aggregates share the main features of the acid induced amyloid fibrils, indicating broad similarities in their aggregate structures. UV-B induced aggregates of the unlabeled wild type protein (Figure 5H, L) show an amyloid peak in a similar position to that of the S84C mutant (Figure 5E,I), indicating again that the mutation itself does not cause amyloid fiber formation.
To study the structure of the UV-B aggregates of γD-crystallin in greater detail, we turn to the spectra of proteins with 13C labeled domains (15). The 2D IR spectra of the C-terminally labeled aggregates are shown in Figure 5F with diagonal slices in Figure 5J. These spectra allow us to assign the ordered fiber β-sheet signal to residues derived from the protein's two domains. If well-formed amyloid β-sheets are formed from unlabeled NTD residues, then a sharp peak will appear at 1624 cm−1 like in the wild type spectrum above. If the sharp peak is instead observed at 1624 - 40 = 1584 cm−1, then 13C labeled residues in the CTD are forming the amyloid sheets since isotope labeling causes a 40 cm−1 frequency shift. In Fig. 5F, a narrow feature is visible on the low-frequency side of the 13C labeled diagonal amide I peak, at 1584 cm−1. A comparison to our previously published acid-induced denaturation of γD-crystallin (15), using the same isotope labeling scheme, shows that the sharp amyloid features appear at similar frequencies. Thus, residues from the C-terminal domain are the source of well-ordered amyloid β-sheets. These amyloid features are small compared to the majority of the CTD features, which absorb at 1595 cm−1. 1595 cm−1 is about 10 cm−1 lower frequency than before UV-B damage (compare solid to dashed lines). The only reasonable way that a lower frequency can be created is by larger negative coupling constants – dehydration would produce a higher frequency shift and hydration caused by unfolding would create a peak no lower than 1610 cm−1 (55). This frequency range is most consistent with a β-sheet structure that is more strongly coupled than the β-sandwich domains in native γD-crystallin, but less ordered than fully formed amyloid fibers (See Table 1). Kinetics experiments following amyloid fiber formation have revealed a progressive shift to lower frequency as fibers form (15). Thus, we postulate that in addition to well-formed amyloid fibers, there are also β-sheet intermediates. In contrast, the 12C amide I signal of the NTD is centered at 1640 cm−1, which matches that of the native protein, but is significantly broadened compared to the native protein (the broadening is more apparent in the 2D IR spectra themselves, Figure 5F, which shows an elongation along the diagonal). Thus, the unlabeled N-terminal domain becomes disordered upon UV-B exposure.
No sharp peak is observed at 1624 cm−1 in Figure 5F, G, implying that the NTD does not contribute to the amyloid β-sheets, but since this region is spectrally congested, we also swapped our labeling scheme. Shown in Figure 5G is the 2D IR spectrum of γD-crystallin with a 13C labeled NTD. No narrow β-sheet feature is visible at 1584 cm−1, confirming that the NTD does not contribute to the fibers. As above, the NTD features are broadened, consistent with the conclusion that the NTD becomes disordered upon aggregation. This spectrum also emphasizes a broadening to the width of the CTD, in addition to the lower frequency. Thus, we conclude that fiber forming residues are exclusively derived from the C-terminal domain and that the β-sandwich motifs are not conserved upon UV-B exposure.
Discussion
Our results provide the most detailed analysis of the influence of UV-B radiation on lens crystallin structure to date. This information is important because exposure to ultraviolet radiation (including UV-B) from sunlight is a known cause of cataracts (1, 5, 7, 13). Based on SDS-PAGE, mass spectrometry, transmission electron microscopy, ThT binding, and 2D IR spectroscopy, we identify the location of side chain oxidation and polypeptide cleavage induced by UV-B radiation, describe the morphology of the resulting aggregates, and obtain unprecedented details of the molecular structures of proteins within the aggregates. Among the structures observed are amyloid fibers that form from residues in the C-terminal domain of γD-crystallin, a finding that is the first definitive proof that amyloid fibers can form under conditions relevant to cataract formation. This finding is particularly interesting because the presence of amyloid fibers in natural cataract material has not been conclusively established, possibly due to the structural complexity of the aggregates and the difficulty associated with obtaining detailed structural information about them.
In contrast to acid-induced amyloid fiber formation (21), the UV-B induced aggregation of γD-crystallin is accompanied by clear changes in the covalent structure of the protein, in the form of cross-linking, polypeptide cleavage, and side chain damage (Figure 1). However, these modifications are similar in nature to those observed in cataractous lenses where a variety of protein fragments and post-translational modifications have been characterized (1, 3, 4). From our LC-MS/MS results (Figure 2), it is clear that UV-B photodamage of γD-crystallin is structurally specific (Figure 3), with both oxidative damage and backbone cleavage occurring at sites near the interdomain interface.
The locations of these damage sites within the sequence and structure of γD-crystallin provide clues to the mechanism by which the protein is initially modified and destabilized to induce aggregation. First, backbone cleavage occurs largely in two specific stretches of sequence, between residues 40-53 and 120-134 where cleavage of nearly every polypeptide bond is observed. These sequences contain tryptophan residues W42 and W130, which have been shown to participate in photoprotective energy transfer mechanisms that are thought to prevent damage from UV radiation (22-24). Interestingly, they are a homologous pair that has much higher fluorescence quantum yields than W68 and W156 in single tryptophan mutants (22). This may also indicate that they are more susceptible to other excited state quenching processes including side chain oxidation and backbone cleavage. Because we showed that the specific polypeptide cleavage patterns are light-dependent (Figure 1B), we therefore invoke a model in which the failure of the hypothesized photoprotection mechanism of γD-crystallin results in distributed damage to the protein in regions near photochemically active tryptophan residues. In previous reports on the UV-B photoinduced aggregation of chicken egg white lysozyme, tryptophan and tyrosine photochemistry induced the reduction of native disulfide bonds, thereby inducing aggregation (56, 57). The close tertiary structural association of backbone cleavage and side chain oxidation sites in γD-crystallin suggests that the two kinds of damage arise from this initial tryptophan photochemistry, and the presence of oxidized residues in both tryptic and semitryptic peptides (Figure 2) indicates that both kinds of damage can occur in the same molecule.
The details of the chemical mechanism of UV-B photodamage, while currently unknown, are consistent with a radical transfer mechanism initiated by tryptophan photooxidation. In previous reports on other proteins, oxidative photodamage of aromatic amino acid side chains (Trp and Tyr) was shown to be capable of initiating peptide bond cleavage (35, 58). For example, the formation of N-formylkynurenine from tryptophan results in increased peptide bond hydrolysis (58, 59). Additionally, the formation of side chain radials via photooxidation can result in radical migration to the main chain (Cα) and subsequent peptide bond cleavage (35). Based on the broad distribution of side chain oxidation and main chain cleavage products within specific regions of sequence, as well as the dependence of backbone cleavage on UV-B light, we believe that such a radical based mechanism is the most plausible explanation for our results. This mechanism, initiated by the failure of the hypothesized energy transfer mechanism of photoprotection, would likely become more pronounced as the protein is destabilized by accumulated photodamage.The cleavage of the polypeptide backbone in γD-crystallin is an obvious mode of protein destabilization, since it compromises the integrity of the molecule that is required for folding. However, the presence of side chain oxidation in the region near the interdomain interface suggests another mode of protein destabilization in the fraction of protein that is not cleaved by UV-B radiation. The interface region has been shown to be particularly important in the stabilization of γD-crystallin (60), and modification of amino acid side chains at the interface are likely to facilitate unfolding of the protein. Because only local unfolding is required to nucleate fibers (61), even relatively small changes in stability due to oxidation can result in the entry of the protein into the amyloid aggregation pathway.
Although these results suggest mechanisms for aggregation initiation in vitro, they do not necessarily describe the aggregation of γD-crystallin in vivo despite the consistency of the types of modification we observe with lens proteomics results that identified a variety of oxidation and cleavage products in lens crystallins. Unfortunately, only a few studies have examined these modifications, and their role in cataract formation, in detail. One such study identified the locations of side chain modifications in three isoforms of a γD-crystallin fragment covering residues 87-173, or the entire C-terminal domain, using antibodies raised against six peptides contained within this region (62). While the epitopes in that study did not cover the entire C-terminal sequence (see regions highlighted in yellow in Figure 3A), large changes in reactivity to the antibodies were observed at residues 114-120, as well as 137-143, 149-154, and 165-173 suggesting side chain modifications in the natural fragments (62). Our MS/MS results, which cover a much larger fraction of the γD-crystallin sequence, also show side chain oxidations in this region (Figure 3A), drawing a link between natural modifications in cataracts and our in vitro results. In fact, our results show that the region of the C-terminal domain that is most susceptible to damage lies in a gap between antibody recognition sites in the previous study (62), indicating that damage in the lens may in fact be much more extensive. Despite the similarities in the locations of side chain oxidations to the natural γD-crystallin 87-173 fragment, our results suggest an alternative polypeptide cleavage pattern than that which was observed in the previous study on human lens material. However, it must be noted that characterization of a single, high abundance protein fragment does not preclude the presence of other fragments that exist at lower concentrations or that have such a large degree of modification that they are difficult to identify using immunoassays.
From TEM images, we learn that γD-crystallin samples contain aggregates with spherical morphology both before and after UV-B illumination, with a slight increase in diameter after illumination. However, the UV-B exposed samples also contain fibers that are not present in samples not exposed to UV-B light. These fibers are predominantly ∼5-10 nm across, within the range typical for amyloid fibers and similar to those produced by acid denaturation of γD-crystallin (14, 21). Perhaps the most striking aspect of these fibers is that they are closely associated with spheres, and appear to terminate at the spheres. On the other hand, spheres without fibers are also observed in the TEM images. This suggests that the fibers are growing from the spheres. Such a fiber formation mechanism is possible if two aggregation processes are at work: initial precipitation and subsequent fiber formation. It is known that single point mutations can cause crystallins to precipitate very quickly, which is thought to be the origin of congenital juvenile cataracts that form amorphous aggregates (1, 63-66). A similar change in solubility may be expected upon covalent modification by UV-B radiation. Therefore, we hypothesize that the initial precipitation into spherical aggregates occurs as a result of UV-B photodamage, and the spherical aggregates then reorganize into fibers. The ThT binding kinetics study (Figure 4F) supports this model. Typical amyloid aggregation is characterized by the presence of a lag phase due to the fact that fiber nucleation is the rate determining step in fiber formation (67). The UV-B induced aggregation, monitored by ThT fluorescence, is characterized by a steady rise in aggregate concentration with no discernible lag phase. Thus, it is likely that the initial aggregation of the protein is fast and therefore not rate limiting, and the steady rise in ThT fluorescence occurs as precipitated proteins reorganize into fibers and additional proteins precipitate as photodamage is continued.
2D IR spectroscopy, which is sensitive to protein secondary structure (15, 21, 39-48, 68), provides the molecular structure information required to link the observed UV-B photodamage pattern in γD-crystallin to the morphology of the aggregates. From the 2D IR spectra of unlabeled γD-crystallin (S84C and wild type, Figures 5E,H) we learn that the UV-B induced aggregates contain both a narrow β-sheet feature at ∼1624 cm−1 and a broadened feature at ∼1640 cm−1. The samples used to collect these spectra, like those shown in TEM images, contain both fibers and spheres that cannot currently be analyzed separately. However, it is clear from a comparison of these spectra to those of the corresponding native proteins (Figures 5A,D) that the native β-sandwich fold is not retained in the aggregates. This result is also important because it rules out domain swapping as the predominant mechanism for UV-B induced γD-crystallin aggregation. In γD-crystallin, the two domains are connected by a flexible loop, and make contact with each other via amino acid side chains at the interdomain interface (60). Mutation studies of this interface have shown that it contributes to protein stability (60), but the individual domains are also quite stable and the presence of both domains is not necessary for their proper folding (14, 69). The β-crystallins also contain two domains, but associate into domain-swapped oligomers (1, 2). Thus, one postulate for γD-crystallin aggregation is that damage destabilizes the interface leading to propagation of domain swapping that ultimately causes enough protein clusters to induce precipitation (70). Although this mechanism is not consistent with the amide I lineshapes in the 2D IR spectra of the final UV-B aggregates, the spectra are congested enough that domain swapped aggregates might constitute a minority population. Additionally, they may play a role in an early step in the aggregation mechanism that is not observed in our current data, as has been observed in molecular dynamics simulations (70). However, due to the degree of side chain damage observed at the interface, specific contacts are likely lost due to the same damage that destabilizes the protein, thus preventing domain swapping from occurring.
The appearance of the amide I signal at ∼1624 cm−1 in unlabeled UV-B photodamaged γD-crystallin allows us to examine the architecture of β-sheets in the resulting aggregates. β-sheets in proteins have infrared signatures that cover a relatively broad range of frequencies, from ∼1617 cm−1 to ∼1640 cm−1, depending on their size and organization (15, 42, 47, 49, 52-54, 71). From a survey of frequencies reported in the literature for both soluble and fibrillar proteins containing β-sheets (Table 1, and references therein), we know that β-sheets in amyloid fibers have frequencies near 1620 cm−1, while those in soluble proteins have frequencies greater than 1630 cm−1. In acid induced fibers of γD-crystallin, the β-sheet frequency is 1617 cm−1, and the native β-sandwich frequency is 1638 cm−1(14, 15). Thus, we conclude that the fiber components of the UV-B induced aggregates are indeed amyloid fibers. The frequency difference of ∼6-8 cm−1 between UV-B induced and acid induced γD-crystallin amyloid fibers reflects a decreased degree of vibrational coupling in the UV-B induced fibers (15, 49, 51, 68). It has been shown that the IR frequency of the β-sheet amide I feature depends on the number of residues in the β-sheet (49, 51). Early intermediates in the acid-induced pathway, which are presumably made up of small aggregates in the β-sheet conformation, also have higher β-sheet frequencies (15). During aggregation, these frequencies progressively shift to lower values as the fibers grow (15). However, the fibers we observe in the TEM images of UV-B induced aggregates are already longer than 10 nm and up to >100 nm along their axis (Figure 4B). Thus, it is unlikely that the amide I frequency difference between acid and UV-B induced fibers results from differences in fiber length. Instead, the higher β-sheet frequency in the UV-B fibers probably arises from structural disorder, which can also reduce vibrational coupling. Based on our results, this disorder may arise from either of two sources. First, the assembly of polypeptide fragments of different lengths may disrupt normal fiber packing. A more likely source of disorder, though, is oxidative damage to side chains, which can prevent efficient side chain packing between fiber β-sheets or recruit water into hydrophobic interfaces, thereby disrupting local secondary structure and vibrational coupling.
Using segmental 13C labeling, which resolves the two domains in 2D IR spectra (15, 21), we previously showed that in the acid-induced fibers of γD-crystallin, only the C-terminal domain forms β-sheets while the N-terminal domain becomes disordered (15). This result was surprising because both domains, which are highly similar, can form amyloid fibers in isolation (14). Careful consideration of the sequences of the domains, however, reveals that the C-terminal domain contains five additional carboxylic acids, which are protonated at low pH (≤ 3) and are a likely source of destabilization. This is in contrast to the UV-B induced photodamage in γD-crystallin, which is nearly symmetrical between the two domains (Figure 3C,D). Based on this, and the fact that backbone cleavage destabilizes and separates the domains, one might conclude that both the N-terminal and C-terminal domains are likely to form fibers in UV-B induced aggregates. However, our 2D IR spectra of segmentally 13C labeled γD-crystallin (Figure 5F,G) show that, like in the acid induced fibers, only residues from the C-terminal domain form amyloid fibers in UV-B induced aggregates. Thus, we conclude that the C-terminal domain has a higher intrinsic amyloidogenicity than the N-terminal domain, and forms fibers more readily regardless of the method of denaturation used. It should be noted that there are some conditions under which the N-terminal domain can in fact form fibers, as was demonstrated by previous experiments (14). Within the product mixture generated by UV-B photodamage, though, the amorphous aggregation pathway appears to be favored over the amyloid aggregation pathway for N-terminal domain sequences.
From these results, we learn that the destabilization of the C-terminal domain induces its entry into the amyloid misfolding pathway. However, a number of questions remain unresolved about the mechanism of fiber formation and the structures of the proteins in the fibers. The 2D IR spectra of segmentally labeled proteins show that some fraction of the C-terminal domain remains outside the fiber cores in a denatured conformation. While it is clear that only the C-terminal domain forms amyloid β-sheets, our data does not show which components of the complex photodamage product mixture form fibers, spheres, or soluble components. The fibers may contain full-length proteins destabilized by side chain oxidation, fragments containing some C-terminal residues, or both.
Conclusions
Despite the numerous unresolved structural and mechanistic details of UV-B induced γD-crystallin aggregation, the incorporation of only the C-terminal domain of γD-crystallin into UV-B induced amyloid fibers has important implications for the formation of cataracts in vivo. The γ-crystallins constitute ∼25% of lens proteins (72), and both γ-crystallins and other crystallin proteins have been found to form amyloid fibers under relatively mild denaturing conditions (17, 19, 20, 73). Because both backbone cleavage and side chain oxidation are observed in UV-B damaged γD-crystallin as well as in natural cataracts(1-13, 62, 74-76), it is reasonable to assume that the mechanisms we have identified here could also apply in the lens. However, the presence of amyloid fibers in natural cataracts has not been conclusively established, and their role in cataract formation remains a subject of debate. Only a few studies have provided evidence that the amyloid misfolding pathway may be physiologically relevant. For example, FTIR spectra of decapsulated cataract tissue exhibits an absorption at 1620 cm−1, which is especially pronounced in cataract tissue taken from patients that suffer from glaucoma (77). This amide I frequency indicative of the extended vibrational coupling in amyloid β-sheets (Table 1) (49, 51). Additionally, the presence of heavily modified C-terminal fragments of γD-crystallin, such as the 9 kDa fragment discussed above (62), suggests that highly amyloidogenic protein fragments are produced in the lens. This raises the question: if the propensity of crystallins is to form amyloid fibers in vitro, and amyloidogenic species are present in the lens, why is there so little evidence for amyloid species in cataract material? One suggestion has been that there is a competition between fast precipitation into unfolded structures, such as the ones reported here as spheres in the TEM images, and amyloid fiber formation (78). But in this report, our results suggest that disordered precipitates can reorganize into fibers, so presumably a similar mechanism could occur in vivo. It is also possible that protein crowding, or interactions with α-crystallins (which are chaperone proteins) prevent such fragments from forming fibers (79, 80); in vitro studies have shown that α-crystallins do indeed bind fibers formed by proteins such as α-synuclein (81) and modulate the amyloid aggregation of γ-crystallins (80). Finally, it is possible that fibers are indeed present in vivo, but standard techniques for studying cataract material cannot detect them due to the congestion of the samples with other structures that prevent resolution in TEM images or fiber-specific fluorescent dye binding. This latter point emphasizes the important role that 2D IR spectroscopy may play in the study of cataract formation. Due to its ability to resolve secondary structures in complex, heterogeneous samples, it is a unique and powerful tool for investigating protein aggregate architecture, both in vitro, and potentially, in samples extracted from tissue.
Supplementary Material
Acknowledgments
The authors would like to thank G. Sabat of the University of Wisconsin Biotechnology Mass Spectrometry Facility for help with LC-MS/MS data analysis and useful discussions, as well as R. Massey of the University of Wisconsin Medical Sciences Electron Microscopy Facility for his help in the collection of TEM images. This work was supported by the NIH NIDDK DK79895.
Footnotes
Supporting Information. Fragment ions used in the analysis of LC-MS/MS data are tabulated in Tables S1-6 in the Supporting Information. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Wang Y, King JA. Protein Misfolding Diseases. John Wiley & Sons, Inc; 2010. Cataract as a protein-aggregation disease; pp. 487–515. [Google Scholar]
- 2.Bloemendal H, de Jong W, Jaenicke R, Lubsen NH, Slingsby C, Tardieu A. Ageing and vision: structure, stability and function of lens crystallins. Prog Biophys Mol Biol. 2004;86:407–485. doi: 10.1016/j.pbiomolbio.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Truscott RJW. Age-related nuclear cataract—oxidation is the key. Experimental Eye Research. 2005;80:709–725. doi: 10.1016/j.exer.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Hanson SRA, Hasan A, Smith DL, Smith JB. The Major in vivo Modifications of the Human Water-insoluble Lens Crystallins are Disulfide Bonds, Deamidation, Methionine Oxidation and Backbone Cleavage. Experimental Eye Research. 2000;71:195–207. doi: 10.1006/exer.2000.0868. [DOI] [PubMed] [Google Scholar]
- 5.Varma SD, Hegde KR, Kovtun S. UV-B-induced damage to the lens in vitro: prevention by caffeine. Journal of ocular pharmacology and therapeutics: the official journal of the Association for Ocular Pharmacology and Therapeutics. 2008;24:439–444. doi: 10.1089/jop.2008.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia Z, Yang Z, Huynh T, King JA, Zhou R. UV-radiation Induced Disruption of Dry-Cavities in Human gammaD-crystallin Results in Decreased Stability and Faster Unfolding. Scientific reports. 2013;3:1560. doi: 10.1038/srep01560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies MJ, Truscott RJW. Photo-oxidation of proteins and its role in cataractogenesis. Journal of Photochemistry and Photobiology B: Biology. 2001;63:114–125. doi: 10.1016/s1011-1344(01)00208-1. [DOI] [PubMed] [Google Scholar]
- 8.Hott JL, Borkman RF. Analysis of photo-oxidized amino acids in tryptic peptides of calf lens γ-ii crystallin. Photochemistry and Photobiology. 1992;56:257–262. doi: 10.1111/j.1751-1097.1992.tb02155.x. [DOI] [PubMed] [Google Scholar]
- 9.Schauerte JA, Gafni A. Photodegradation of Tryptophan Residues and Attenuation of Molecular Chaperone Activity in α-Crystallin Are Correlated. Biochemical and Biophysical Research Communications. 1995;212:900–905. doi: 10.1006/bbrc.1995.2054. [DOI] [PubMed] [Google Scholar]
- 10.Wang Steven SS, Wen WS. Examining the influence of ultraviolet C irradiation on recombinant human γD-crystallin. Mol Vis. 2010;16:2777–2790. [PMC free article] [PubMed] [Google Scholar]
- 11.Hains PG, Truscott RJ. Post-translational modifications in the nuclear region of young, aged, and cataract human lenses. Journal of proteome research. 2007;6:3935–3943. doi: 10.1021/pr070138h. [DOI] [PubMed] [Google Scholar]
- 12.Kubiak K, Kowalska M, Nowak W. Molecular dynamics study of early events during photooxidation of eye lens protein γB-crystallin. Journal of Molecular Structure: THEOCHEM. 2003;630:315–325. [Google Scholar]
- 13.Balasubramanian D, Du X, Zigler JS. The reaction of singlet oxygen with proteins, with special reference to crystallins. Photochemistry and Photobiology. 1990;52:761–768. doi: 10.1111/j.1751-1097.1990.tb08679.x. [DOI] [PubMed] [Google Scholar]
- 14.Papanikolopoulou K, Mills-Henry I, Tho SL, Wang YT, Gross AAR, Kirschner DA, Decatur SM, King J. Formation of amyloid fibrils in vitro by human γD-crystallin and its isolated domains. Mol Vis. 2008;14:81–89. [PMC free article] [PubMed] [Google Scholar]
- 15.Moran SD, Woys AM, Buchanan LE, Bixby E, Decatur SM, Zanni MT. Two-dimensional IR spectroscopy and segmental 13C labeling reveals the domain structure of human gammaD-crystallin amyloid fibrils. Proc Natl Acad Sci U S A. 2012;109:3329–3334. doi: 10.1073/pnas.1117704109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fatima U, Sharma S, Guptasarma P. Structures of differently aggregated and precipitated forms of γB crystallin: an FTIR spectroscopic and EM study. Protein Pept Lett. 2010;17:1155–1162. doi: 10.2174/092986610791760414. [DOI] [PubMed] [Google Scholar]
- 17.Ecroyd H, Carver JA. Crystallin proteins and amyloid fibrils. Cell Mol Life Sci. 2009;66:62–81. doi: 10.1007/s00018-008-8327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosinski-Collins MS, King J. In vitro unfolding, refolding, and polymerization of human γD crystallin, a protein involved in cataract formation. Protein Sci. 2003;12:480–490. doi: 10.1110/ps.0225503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meehan S, Berry Y, Luisi B, Dobson CM, Carver JA, MacPhee CE. Amyloid fibril formation by lens crystallin proteins and its implications for cataract formation. J Biol Chem. 2004;279:3413–3419. doi: 10.1074/jbc.M308203200. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Petty S, Trojanowski A, Knee K, Goulet D, Mukerji I, King J. Formation of amyloid fibrils in vitro from partially unfolded intermediates of human γC-crystallin. Invest Ophthalmol Vis Sci. 2010;51:672–678. doi: 10.1167/iovs.09-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moran SD, Decatur SM, Zanni MT. Structural and Sequence Analysis of the Human gammaD-Crystallin Amyloid Fibril Core Using 2D IR Spectroscopy, Segmental (13)C Labeling, and Mass Spectrometry. J Am Chem Soc. 2012 doi: 10.1021/ja307898g. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Callis PR, King J. Mechanism of the Very Efficient Quenching of Tryptophan Fluorescence in Human γD- and γS-Crystallins: The γ-Crystallin Fold May Have Evolved To Protect Tryptophan Residues from Ultraviolet Photodamage†. Biochemistry. 2009;48:3708–3716. doi: 10.1021/bi802177g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Flaugh SL, Callis PR, King J. Mechanism of the highly efficient quenching of tryptophan fluorescence in human gammaD-crystallin. Biochemistry. 2006;45:11552–11563. doi: 10.1021/bi060988v. [DOI] [PubMed] [Google Scholar]
- 24.Xu J, Chen J, Toptygin D, Tcherkasskaya O, Callis P, King J, Brand L, Knutson JR. Femtosecond fluorescence spectra of tryptophan in human gamma-crystallin mutants: site-dependent ultrafast quenching. J Am Chem Soc. 2009;131:16751–16757. doi: 10.1021/ja904857t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu G, Chance MR. Hydroxyl Radical-Mediated Modification of Proteins as Probes for Structural Proteomics. Chem Rev. 2007;107:3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- 26.Middleton CT, Woys AM, Mukherjee SS, Zanni MT. Residue-specific structural kinetics of proteins through the union of isotope labeling, mid-IR pulse shaping, and coherent 2D IR spectroscopy. Methods. 2010;52:12–22. doi: 10.1016/j.ymeth.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bilbao J, de Miguel A. Estimation of UV-B irradiation from total global solar meteorological data in central Spain. Journal of Geophysical Research. 2010;115 [Google Scholar]
- 28.Dyer JM, Bringans SD, Bryson WG. Characterisation of photo-oxidation products within photoyellowed wool proteins: tryptophan and tyrosine derived chromophores. Photochemical & photobiological sciences: Official journal of the European Photochemistry Association and the European Society for Photobiology. 2006;5:698–706. doi: 10.1039/b603030k. [DOI] [PubMed] [Google Scholar]
- 29.Basak A, Bateman O, Slingsby C, Pande A, Asherie N, Ogun O, Benedek GB, Pande J. High-resolution X-ray Crystal Structures of Human γD Crystallin (1.25 Å) and the R58H Mutant (1.15 Å) Associated with Aculeiform Cataract. Journal of Molecular Biology. 2003;328:1137–1147. doi: 10.1016/s0022-2836(03)00375-9. [DOI] [PubMed] [Google Scholar]
- 30.Creed D. The photophysics and photochemistry of the near-uv absorbing amino acids–i. tryptophan and its simple derivatives. Photochemistry and Photobiology. 1984;39:537–562. [Google Scholar]
- 31.Kerwin BA, Remmele RL., Jr Protect from light: photodegradation and protein biologics. Journal of pharmaceutical sciences. 2007;96:1468–1479. doi: 10.1002/jps.20815. [DOI] [PubMed] [Google Scholar]
- 32.Vladimirov YA, Roshchupkin DI, Fesenko EE. Photochemical reactions in amino acid residues and inactivation of enzymes during U.V.-irradiation. A review. Photochemistry and Photobiology. 1970;11:227–246. doi: 10.1111/j.1751-1097.1970.tb05992.x. [DOI] [PubMed] [Google Scholar]
- 33.Callis PR. 1La and 1Lb transitions of tryptophan: Applications of theory and experimental observations to fluorescence of proteins. In: Ludwig Brand MLJ, editor. Methods in Enzymology. Academic Press; 1997. pp. 113–150. [DOI] [PubMed] [Google Scholar]
- 34.Cheal SM, Ng M, Barrios B, Miao Z, Kalani AK, Meares CF. Mapping protein-protein interactions by localized oxidation: consequences of the reach of hydroxyl radical. Biochemistry. 2009;48:4577–4586. doi: 10.1021/bi900273j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu G, Chance MR. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chemical reviews. 2007;107:3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- 36.Biancalana M, Koide S. Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochimica et biophysica acta. 2010;1804:1405–1412. doi: 10.1016/j.bbapap.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Middleton CT, Marek P, Cao P, Chiu CC, Singh S, Woys AM, de Pablo JJ, Raleigh DP, Zanni MT. Two-dimensional infrared spectroscopy reveals the complex behaviour of an amyloid fibril inhibitor. Nature chemistry. 2012;4:355–360. doi: 10.1038/nchem.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolfe LS, Calabrese MF, Nath A, Blaho DV, Miranker AD, Xiong Y. Protein-induced photophysical changes to the amyloid indicator dye thioflavin T. Proceedings of the National Academy of Sciences. 2010;107:16863–16868. doi: 10.1073/pnas.1002867107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woys AM, Mukherjee SS, Skoff DR, Moran SD, Zanni MT. A Strongly Absorbing Class of Non-Natural Labels for Probing Protein Electrostatics and Solvation with FTIR and 2D IR Spectroscopies. The journal of physical chemistry B. 2013 doi: 10.1021/jp402946c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dunkelberger EB, Buchanan LE, Marek P, Cao P, Raleigh DP, Zanni MT. Deamidation accelerates amyloid formation and alters amylin fiber structure. J Am Chem Soc. 2012;134:12658–12667. doi: 10.1021/ja3039486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Londergan CH, Wang J, Axelsen PH, Hochstrasser RM. Two-dimensional infrared spectroscopy displays signatures of structural ordering in peptide aggregates. Biophysical journal. 2006;90:4672–4685. doi: 10.1529/biophysj.105.075812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim YS, Liu L, Axelsen PH, Hochstrasser RM. 2D IR provides evidence for mobile water molecules in β-amyloid fibrils. Proceedings of the National Academy of Sciences. 2009;106:17751–17756. doi: 10.1073/pnas.0909888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Remorino A, Korendovych IV, Wu Y, DeGrado WF, Hochstrasser RM. Residue-Specific Vibrational Echoes Yield 3D Structures of a Transmembrane Helix Dimer. Science. 2011;332:1206–1209. doi: 10.1126/science.1202997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L, Middleton CT, Singh S, Reddy AS, Woys AM, Strasfeld DB, Marek P, Raleigh DP, de Pablo JJ, Zanni MT, Skinner JL. 2DIR spectroscopy of human amylin fibrils reflects stable beta-sheet structure. J Am Chem Soc. 2011;133:16062–16071. doi: 10.1021/ja204035k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manor J, Mukherjee P, Lin YS, Leonov H, Skinner JL, Zanni MT, Arkin IT. Gating mechanism of the influenza A M2 channel revealed by 1D and 2D IR spectroscopies. Structure. 2009;17:247–254. doi: 10.1016/j.str.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mukherjee P, Kass I, Arkin IT, Zanni MT. Picosecond dynamics of a membrane protein revealed by 2D IR. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3528–3533. doi: 10.1073/pnas.0508833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shim SH, Gupta R, Ling YL, Strasfeld DB, Raleigh DP, Zanni MT. Two-dimensional IR spectroscopy and isotope labeling defines the pathway of amyloid formation with residue-specific resolution. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6614–6619. doi: 10.1073/pnas.0805957106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woys AM, Lin YS, Reddy AS, Xiong W, de Pablo JJ, Skinner JL, Zanni MT. 2D IR line shapes probe ovispirin peptide conformation and depth in lipid bilayers. J Am Chem Soc. 2010;132:2832–2838. doi: 10.1021/ja9101776. [DOI] [PubMed] [Google Scholar]
- 49.Strasfeld DB, Ling YL, Gupta R, Raleigh DP, Zanni MT. Strategies for extracting structural information from 2D IR spectroscopy of amyloid: application to islet amyloid polypeptide. The journal of physical chemistry B. 2009;113:15679–15691. doi: 10.1021/jp9072203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lam AR, Moran SD, Preketes NK, Zhang TO, Zanni MT, Mukamel S. Study of the γD-Crystallin Protein Using Two-Dimensional Infrared (2D IR) Spectroscopy: Experiment and Simulation. Submitted. 2013 doi: 10.1021/jp405159v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hahn S, Kim SS, Lee C, Cho M. Characteristic two-dimensional IR spectroscopic features of antiparallel and parallel beta-sheet polypeptides: simulation studies. The Journal of chemical physics. 2005;123:084905. doi: 10.1063/1.1997151. [DOI] [PubMed] [Google Scholar]
- 52.Jones KC, Peng CS, Tokmakoff A. Folding of a heterogeneous β-hairpin peptide from temperature-jump 2D IR spectroscopy. Proceedings of the National Academy of Sciences. 2013;110:2828–2833. doi: 10.1073/pnas.1211968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheatum CM, Tokmakoff A, Knoester J. Signatures of beta-sheet secondary structures in linear and two-dimensional infrared spectroscopy. The Journal of chemical physics. 2004;120:8201–8215. doi: 10.1063/1.1689637. [DOI] [PubMed] [Google Scholar]
- 54.Woys AM, Almeida AM, Wang L, Chiu CC, McGovern M, de Pablo JJ, Skinner JL, Gellman SH, Zanni MT. Parallel β-Sheet Vibrational Couplings Revealed by 2D IR Spectroscopy of an Isotopically Labeled Macrocycle: Quantitative Benchmark for the Interpretation of Amyloid and Protein Infrared Spectra. Journal of the American Chemical Society. 2012;134:19118–19128. doi: 10.1021/ja3074962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manor J, Feldblum ES, Zanni MT, Arkin IT. Environment Polarity in Proteins Mapped Noninvasively by FTIR Spectroscopy. The journal of physical chemistry letters. 2012;3:939–944. doi: 10.1021/jz300150v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie JB, Cao Y, Pan H, Qin M, Yan ZQ, Xiong X, Wang W. Photoinduced fibrils formation of chicken egg white lysozyme under native conditions. Proteins: Structure, Function, and Bioinformatics. 2012;80:2501–2513. doi: 10.1002/prot.24132. [DOI] [PubMed] [Google Scholar]
- 57.Xie J, Qin M, Cao Y, Wang W. Mechanistic insight of photo-induced aggregation of chicken egg white lysozyme: The interplay between hydrophobic interactions and formation of intermolecular disulfide bonds. Proteins: Structure, Function, and Bioinformatics. 2011;79:2505–2516. doi: 10.1002/prot.23074. [DOI] [PubMed] [Google Scholar]
- 58.Previero A, Coletti Previero MA, Jollès P. Nonenzymatic cleavage of tryptophyl peptide bonds in peptides and proteins: I. Cleavage of C-tryptophyl peptide bonds in model peptides through their conversion into N′-formylkynurenine derivatives. Biochemical and Biophysical Research Communications. 1966;22:17–21. doi: 10.1016/0006-291x(66)90595-x. [DOI] [PubMed] [Google Scholar]
- 59.Sakiyama F. Selective Chemical Cleavage of the Peptide Bond at N'-Formylkynurenine in Ozone-Oxidized Hen Egg-White Lysozyme. Journal of Biochemistry. 1977;82:365–375. [PubMed] [Google Scholar]
- 60.Flaugh SL, Kosinski-Collins MS, King J. Interdomain side-chain interactions in human γD crystallin influencing folding and stability. Protein Science. 2005;14:2030–2043. doi: 10.1110/ps.051460505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brubaker WD, Freites JA, Golchert KJ, Shapiro RA, Morikis V, Tobias DJ, Martin RW. Separating instability from aggregation propensity in gammaS-crystallin variants. Biophysical journal. 2011;100:498–506. doi: 10.1016/j.bpj.2010.12.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Srivastava OP, Srivastava K. Characterization of Three Isoforms of a 9kDa γD-Crystallin Fragment Isolated From Human Lenses. Experimental Eye Research. 1996;62:593–604. doi: 10.1006/exer.1996.0070. [DOI] [PubMed] [Google Scholar]
- 63.Sandilands A, Hutcheson AM, Long HA, Prescott AR, Vrensen G, Löster J, Klopp N, Lutz RB, Graw J, Masaki S, Dobson CM, MacPhee CE, Quinlan RA. Altered aggregation properties of mutant γ-crystallins cause inherited cataract. The EMBO Journal. 2002;21:6005–6014. doi: 10.1093/emboj/cdf609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Santhiya ST, Shyam Manohar, Rawlley D, Vijayalakshmi P, Namperumalsamy P, Gopinath PM, Löster J, Graw J. Novel mutations in the γ-crystallin genes cause autosomal dominant congenital cataracts. Journal of Medical Genetics. 2002;39:352–358. doi: 10.1136/jmg.39.5.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stephan DA, Gillanders E, Vanderveen D, Freas-Lutz D, Wistow G, Baxevanis AD, Robbins CM, Van Auken A, Quesenberry MI, Bailey-Wilson J, Juo SHH, Trent JM, Smith L, Brownstein MJ. Progressive juvenile-onset punctate cataracts caused by mutation of the γD-crystallin gene. Proceedings of the National Academy of Sciences. 1999;96:1008–1012. doi: 10.1073/pnas.96.3.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Evans P, Wyatt K, Wistow GJ, Bateman OA, Wallace BA, Slingsby C. The P23T Cataract Mutation Causes Loss of Solubility of Folded γD-Crystallin. Journal of Molecular Biology. 2004;343:435–444. doi: 10.1016/j.jmb.2004.08.050. [DOI] [PubMed] [Google Scholar]
- 67.Marek P, Mukherjee S, Zanni MT, Raleigh DP. Residue-Specific, Real-Time Characterization of Lag-Phase Species and Fibril Growth During Amyloid Formation: A Combined Fluorescence and IR Study of p-Cyanophenylalanine Analogs of Islet Amyloid Polypeptide. Journal of Molecular Biology. 2010;400:878–888. doi: 10.1016/j.jmb.2010.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hamm P, Zanni MT. Concepts and Methods of 2D Infrared Spectroscopy. Cambridge University Press; New York: 2011. [Google Scholar]
- 69.Mills IA, Flaugh SL, Kosinski-Collins MS, King JA. Folding and stability of the isolated Greek key domains of the long-lived human lens proteins gammaD-crystallin and gammaS-crystallin. Protein science: a publication of the Protein Society. 2007;16:2427–2444. doi: 10.1110/ps.072970207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Das P, King JA, Zhou R. Aggregation of gamma-crystallins associated with human cataracts via domain swapping at the C-terminal beta-strands. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10514–10519. doi: 10.1073/pnas.1019152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Demirdöven N, Cheatum CM, Chung HS, Khalil M, Knoester J, Tokmakoff A. Two-Dimensional Infrared Spectroscopy of Antiparallel β-Sheet Secondary Structure. Journal of the American Chemical Society. 2004;126:7981–7990. doi: 10.1021/ja049811j. [DOI] [PubMed] [Google Scholar]
- 72.Su S, Liu P, Zhang H, Li Z, Song Z, Zhang L, Chen S. Proteomic Analysis of Human Age-related Nuclear Cataracts and Normal Lens Nuclei. Investigative Ophthalmology & Visual Science. 2011;52:4182–4191. doi: 10.1167/iovs.10-7094. [DOI] [PubMed] [Google Scholar]
- 73.Meehan S, Knowles TPJ, Baldwin AJ, Smith JF, Squires AM, Clements P, Treweek TM, Ecroyd H, Tartaglia GG, Vendruscolo M, MacPhee CE, Dobson CM, Carver JA. Characterisation of Amyloid Fibril Formation by Small Heat-shock Chaperone Proteins Human αA-, αB- and R120G αB-Crystallins. Journal of Molecular Biology. 2007;372:470–484. doi: 10.1016/j.jmb.2007.06.060. [DOI] [PubMed] [Google Scholar]
- 74.Srivastava OP, McEntire JE, Srivastava K. Identification of a 9 kDa γ-crystallin fragment in human lenses. Experimental Eye Research. 1992;54:893–901. doi: 10.1016/0014-4835(92)90153-j. [DOI] [PubMed] [Google Scholar]
- 75.Srivastava OP, Srivastava K, Silney C. Covalent Modification at the C-Terminal End of a 9 kDa γD-Crystallin Fragment in Human Lenses. Experimental Eye Research. 1994;58:595–604. doi: 10.1006/exer.1994.1054. [DOI] [PubMed] [Google Scholar]
- 76.Santhoshkumar P, Udupa P, Murugesan R, Sharma KK. Significance of Interactions of Low Molecular Weight Crystallin Fragments in Lens Aging and Cataract Formation. Journal of Biological Chemistry. 2008;283:8477–8485. doi: 10.1074/jbc.M705876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee S, Lin S, Li M, Liang R. Possible mechanism of exacerbating cataract formation in cataractous human lens capsules induced by systemic hypertension or glaucoma. Ophthalmic Res. 1997;29:83–90. doi: 10.1159/000268001. [DOI] [PubMed] [Google Scholar]
- 78.Kosinski-Collins MS, King J. In vitro unfolding, refolding, and polymerization of human γD crystallin, a protein involved in cataract formation. Protein Science. 2003;12:480–490. doi: 10.1110/ps.0225503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rekas A, Jankova L, Thorn DC, Cappai R, Carver JA. Monitoring the prevention of amyloid fibril formation by α-crystallin. FEBS Journal. 2007;274:6290–6304. doi: 10.1111/j.1742-4658.2007.06144.x. [DOI] [PubMed] [Google Scholar]
- 80.Acosta-Sampson L, King J. Partially folded aggregation intermediates of human gammaD-, gammaC-, and gammaS-crystallin are recognized and bound by human alphaB-crystallin chaperone. J Mol Biol. 2010;401:134–152. doi: 10.1016/j.jmb.2010.05.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shammas Sarah L, Waudby Christopher A, Wang S, Buell Alexander K, Knowles Tuomas PJ, Ecroyd H, Welland Mark E, Carver John A, Dobson Christopher M, Meehan S. Binding of the Molecular Chaperone αB-Crystallin to Aβ Amyloid Fibrils Inhibits Fibril Elongation. Biophysical journal. 2011;101:1681–1689. doi: 10.1016/j.bpj.2011.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.