Abstract
Transition metals, including manganese, are required for proper virulence and persistence of many pathogenic bacteria. In Streptococcus pneumoniae (Spn), manganese homeostasis is controlled by a high affinity Mn(II) uptake complex, PsaBCA, and a constitutively expressed efflux transporter, MntE. PsaBCA expression is transcriptionally regulated by the DtxR/MntR family metalloregulatory protein pneumococcal surface antigen repressor (PsaR) in Spn. Here, we present a comprehensive analysis of the metal and DNA-binding properties of PsaR. PsaR is a homodimer in the absence and presence of metals and binds two manganese or zinc per protomer (four per dimer) in two pairs of structurally distinct sites, termed site 1 and site 2. Site 1 is likely filled with Zn(II) in vivo (KZn1≥1013 M−1; KMn1≈108 M−1). The Zn(II)-site 1 complex adopts a pentacoordinate geometry by x-ray absorption spectroscopy containing a single cysteine, and appears analogous to the Cd(II) site observed in S. gordonii ScaR. Site 1 is necessary but not sufficient for full positive allosteric activation of DNA operator binding by metals as measured by ΔGc, the allosteric coupling free energy, since mutants in site 1 show intermediate ΔGc. Site 2 is the primary regulatory site and governs specificity for Mn(II) over Zn(II) in PsaR, with ΔGcZn,Mn>>ΔGcZn,Zn despite the fact that Zn(II) binds site 2 with 40-fold higher affinity relative to Mn(II), i.e., KZn2>KMn2. Mutational studies reveal that Asp7 in site 2 is a critical ligand for Mn(II)-dependent allosteric activation of DNA binding. These findings are discussed in the context of other well-studied DtxR/MntR Mn(II)/Fe(II) metallorepressors.
INTRODUCTION
Transition metals constitute an array of critical micronutrients for nonpathogenic and pathogenic bacteria alike and provide required structural or catalytic functions tied to optimal growth or pathogenesis.1,2 Although required under normal conditions, deprivation or excess of these metals can severely impair bacterial growth. As a result, bioavailable metal concentrations are actively maintained inside the cell to allow for optimal growth under a wide range of prevailing conditions. This process, termed metal ion homeostasis, is orchestrated via coordinated transcriptional control of the expression of both metal uptake and efflux systems.3,4
Manganese has emerged as a biologically significant micronutrient as a result of accumulating evidence of its role in modulating the oxidative stress response inside cells.5 Indeed, vertebrates employ a Ca2+-activated Mn(II) and Zn(II) chelating protein, calprotectin, which provides broad-range inhibition of microbial growth via extracellular sequestration of Mn(II) and/or Zn(II), in an effort to deprive microorganisms of this essential metal(s).6–9 In E. coli transcriptional activation of the oxidative stress sensor OxyR induces this iron-centric bacterium to import manganese and replace mononuclear Fe(II) in a number of enzymes with Mn(II); this protects these enzymes against H2O2 damage.10–12 Manipulation of intracellular manganese concentrations has also been connected to protecting other bacteria from oxidative damage through both enzyme-mediated, e.g., via Mn(II) superoxide dismutase, and enzyme-independent means.13–16 Streptococcus pneumoniae may well utilize similar mechanisms to combat both endogenous and exogenous oxidative stress encountered during infections.17 We have previously shown that S. pneumoniae concentrates manganese to a whole cell-associated concentration approaching that of zinc and greater than iron despite a concentration of manganese that is ≈50-fold lower than either metal in the growth medium.18 Additionally, it has been shown that extracellular zinc stress induces an ≈40% reduction in total cell-associated Mn(II) and a growth defect likely caused by competition at the Mn(II) uptake transporter protein PsaA; as a result, there is transcriptional upregulation of the PsaR regulon under these conditions. Addition of excess manganese restores both normal growth and manganese concentrations.18,19 These data tie bioavailable manganese directly to the viability of S. pneumoniae leading us to further explore how manganese levels are regulated in the cell.
Members of the MntR/DtxR repressor family are primarily responsible for mediating transcription control of iron or manganese uptake.20,21 The three most extensively studied members of this family are the homologous Fe(II)-sensors Corynebacterium diphtheriae DtxR and Mycobacterium tuberculosis IdeR22,23 and Mn(II)-sensor in Bacillus subtilis, MntR.24–31 Although similar in both primary structure and quaternary structure of the activated metal-bound forms (see Supplementary Fig. S1), DtxR and MntR induce transcriptional co-repression by binding their cognate metal ions in distinct ways. DtxR requires binding to a pair of ancillary sites near the C-terminal SH3-like domain to induce protein dimerization; further binding to a pair of regulatory sites allosterically activates DNA binding and promotes transcriptional repression.22,23 B. subtilis MntR also requires the binding of four metal ions for activation of DNA binding, but lacks the C-terminal SH3-like domain of DtxR and instead employs a binuclear Mn(II) cluster to activate DNA binding.32 The precise coordination structure of the binuclear cluster observed in crystallographic studies is dependent on temperature, giving rise to so-called A/B (only observed at 100 °C) and A/C Mn(II) clusters. The A/C cluster conformation is thought to be the physiologically relevant one, in which the Mn(II) ions are separated by 4.4 Å; the A/C conformer is also most consistent with the EPR studies in solution which shows a spin-coupling interaction consistent with a longer (>4 Å) distance between the two Mn(II) ions.28 Both Mn(II) ions must be bound to fully activate DNA binding, with the A-site thought to function as a “Mn(II) selectivity filter” that favors occupancy of the C-site with cognate Mn(II). Binding of non-cognate Zn(II), Co(II) and Fe(II), for example, only fills the A-site and none strongly activate DNA binding.33 Recent experiments reveal that the A-site Mn(II) ion in the A/C cluster can be replaced by a positively charged Lys ε-NH2 chain via mutagenesis (E11K) in MntR with the retention of full biological activity.33
These differences between DtxR and MntR suggest that this common core DtxR/MntR family scaffold may have evolved unique mechanisms of allosteric activation of DNA operator binding in related repressors in other bacteria as well.33 Consistent with this, the crystallographic structure of the dimeric Mn(II)-sensing repressor ScaR from Streptococcus gordonii34 solved to ≈2.7 Å resolution reveals either a Zn(II) or Cd(II) ion bound to a site positioned between the dimerization and FeoA domains that is distinct from that previously found for the ancillary sites in DtxR.34 The N-terminal winged helical DNA-binding domains in each protomer in this highly asymmetric dimer are splayed out relative to one another. This suggests that the primary activating site is not filled in this structure and as a result, likely corresponds to the inactive DNA-binding state. Metal binding studies showed between two and three Mn(II) ions may bind per protomer.34 The overall conclusion from this work is that ScaR and related Mn(II)-regulated repressors from other streptococci are superficially similar to DtxR and are capable of binding Mn(II) which in turn, activates DNA operator-promoter binding. The structural mechanism, however, remains incompletely understood.
In this work, we use the various crystallographic structures of B. subtilis MntR 24,25,32,33 and the Cd(II)-bound Streptococcus gordonii (Sgo) ScaR structure34 as well as our previous findings18 as a guide to elucidate the metal binding properties of Spn PsaR in solution. PsaR is 76% identical in sequence to Sgo ScaR. We present new insights into the mechanism of allosteric activation of psaBCA operator (PsaO) binding by PsaR induced by Mn(II) vs. Zn(II). We show that PsaR is a stable homodimer in all metallation states and harbors two pairs of transition metal sites (four per dimer) denoted sites 1 and 2. Metal binding studies show that site 1 is likely filled with Zn(II) in vivo with a KZn1 ≥ 1013 M−1; site 1 is required for full allosteric activation and may function more as a structural site that dampens conformational heterogeneity of the Spn PsaR homodimer. X-ray absorption spectroscopy reveals that this metal site is analogous to the Cd(II) site observed in the Sgo ScaR structure.34 The metal selectivity and DNA activation of Spn PsaR is dictated solely by site 2. Although site 2 binds non-cognate metal Zn(II) with a ≈40-fold higher affinity than Mn(II) (pH 8.0, 0.2 M NaCl), Mn(II) bound to site 2 to form PsaRZn,Mn is characterized by an allosteric coupling free energy, ΔGc, that is greater than twice that of Zn(II) bound to site 2 to form PsaRZn,Zn thus making PsaR a Mn(II)-specific repressor. Mutational studies of site 1 and 2 reveal that Asp7 in site 2 is a critical ligand for Mn(II)-dependent allosteric activation of DNA binding, a finding consistent with the analogous residue in this position functioning as a key driver of the Mn(II)-induced conformational switching in MntR24 and as an important metal specificity determinant [Mn(II) vs. Fe(II)] in B. subtilis MntR.25
MATERIALS AND METHODS
Chemicals and reagents
All water used in these experiments was Milli-Q deionized (>18 MΩ) and the buffers were obtained from Sigma. Indicator dyes mag-fura-2 (mf2) and quin-2 were obtained from Invitrogen and Sigma, respectively. Metal stocks were made with Ultra Pure Alfa Aesar metals. All other reagents were as indicated in the text. An Ätka 10 purifier (GE) was used for all chromatographic separations. All metal binding and DNA binding experiments were performed under aerobic conditions as described below.
Subcloning and introduction of missense mutations into the psaR gene
The psaR gene (locus tag SPD_1450) was amplified from S. pneumoniae D39 genomic DNA using cloning primers containing NdeI and BamHI restriction sites. The psaR insert was subcloned into the expression vector pET3a restricted with NdeI and BamHI to create pET-PsaR containing the wild-type psaR gene under transcriptional control of the T7 promoter. PsaR mutants were generated using the protocol supplied by QuikChange (Stratagene) using the pET-PsaR as the template and an appropriate mutagenic primer. The integrity of all expression plasmids was verified by double-stranded dideoxy sequencing. The plasmids were transformed into Rosetta™ BL21 (DE3) pLysS cells for protein expression.
Expression and purification of wild-type and mutant PsaRs
Competent cells were transformed with the desired pET-PsaR plasmid, plated onto an ampicillin and chloramphenicol plate (100 µg/mL and 37 µg/mL, respectively), and grown overnight at 37°C. Single colonies were used to inoculate 100 mL LB cultures containing both ampicillin and chloramphenicol, and were grown overnight at 37 °C with shaking. The overnight cultures were diluted into 1 L LB and grown at 37 °C with shaking. Overexpression of PsaR was accomplished by induction of 1 L of mid-log LB cultures with IPTG (Inalco) to a final concentration of 0.4 mM for 2.5 h at 37°C. The cells were harvested and resuspended in 25 mM MES, pH 6.0, 750 mM NaCl, 2 mM DTT, 5 mM EDTA. The resuspended cells were lysed by sonication and centrifuged at 6,750 × g for 20 min. A 10% polyethyleneimine (PEI) solution was added to the supernatant to a final concentration of 0.15% v/v and allowed to mix for 30 min at 4 °C to precipitate nucleic acids. The precipitate was removed by centrifugation for 20 min. Finally, a two-step ammonium sulfate precipitation protocol (%) was used to precipitate PsaRs from PEI and other contaminating proteins. The final 50% ammonium sulfate pellet was obtained by centrifugation for 15 min and resuspended in Buffer M (25 mM MES, pH 6.0, 2 mM DTT, 5 mM EDTA). The resuspended sample was dialyzed extensively into Buffer M containing 100 mM NaCl, with the supernatant loaded onto a cation exchange resin (SP-Sepharose, flow rate 2 mL/min, NaCl gradient 0.1 to 0.75 M over 90 min). PsaR-containing fractions were determined using SDS-PAGE, pooled, concentrated using Centrifugal filter units (Millipore) and applied onto a size exclusion column (Superdex 200 16/60 prep grade). All PsaRs migrated with an elution time consistent with that of a dimer. The PsaR-containing fractions were collected and dialyzed into 50 mM Tris, pH 8.0, 200 mM NaCl, 2 mM TCEP (with 10 g/L Chelex 100 resin), filtered, aliquoted into small aliquots and stored at −80 °C for further use. Wild-type and mutant PsaRs were subjected to mass spectrometry using ESI-MS to confirm the expected mass. Protein concentrations were calculated using a predicted extinction coefficient of 11,920 M−1 cm−1 at 280 nm (ProtParam).
Metal-binding stoichiometry and affinity determinations
Purified Apo-PsaR was diluted into chelexed metal binding buffer (50 mM Tris, pH 8.0, 100 mM NaCl). A fixed concentration of metal-free mag-fura-2 (mf2) or quin-2 was added to the solution to reach a final volume of 1 mL. Metal stocks of Mn(II) or Zn(II) in the same buffer were titrated into the protein-competitor solution and the spectra from 250 nm to 600 nm taken following an incubation of 2 min.. For mag-fura-2 experiments, the absorbance at 360 nm and 325 nm were monitored which report on the λmax for the metal-free and metal-bound states of the mag-fura-2, respectively. For quin-2 experiments, the decrease in the absorbance at 265 nm was monitored following an incubation time of 20 min after each addition. All metal binding experiments were acquired with a Hewlett-Packard model 8452A spectrophotometer at room temperature. The mf2 binding curves were fitted to an appropriate two-step or one-step competition binding model with a KZnmf2 = 5.0 × 107 M−1 35 or KMnmf2= 2.4 × 106 M−1 determined by direct titration (see Supplementary Fig. S2). Quin-2 binding curves were fitted to a one-step binding model with a KZnquin2 = 2.7 × 1011 M−1.36 A Ω value of 5 was used to account for the Tris-Zn(II) competition under these solution conditions;37 no correction was necessary for the Tris-Mn(II) competition (Ω≈1).38 All binding curves were carried out in triplicate and globally fitted to simple competition model using Dynafit (see Supplementary Fig. S3 for sample Dynafit script file).39
DNA binding assays
A MerMade 4 instrument was used to synthesize complementary oligonucleotides corresponding to the psaBCA operator-promoter region (5’–TTCAAAAATTAACTTGACTTAATTTTTTTT-Flu – 3’ and 5’–AAAAAAAATTAAGTCAAGTTAATTTTTGAA–3’, where Flu=fluorescein). The crude oligonucleotides were cleaved from the column using AMA (1:1 methylamine and ammonium hydroxide), ethanol precipitated, dried overnight and were resuspended in 750 µL 50 mM potassium phosphate, pH 6.0, 7 M urea. Purified oligonucleotides were obtained using a ResQ column (flow rate 1 mL/min, 0 to 1 M NaCl over 60 min). The concentration of the primers was determined by their predicted extinction coefficients of 308,300 and 286,500 M−1 cm−1 for the fluorescein-labeled and unlabeled strands, respectively. The ssDNAs were annealed with a slight excess of unlabeled primer in the presence of high salt (500 mM NaCl), heated at 65 °C for 10 min and allowed to cool in the dark for several hrs for optimal annealing to obtain PsaO DNA. Aliquots of the annealed PsaO duplex were diluted to approximately 1.5 µM and used as is.
Fluorescence anisotropy measurements were performed by using a Biotek Synergy H1 Hybrid Multi-mode microplate reader with λex of 487 nm. A typical experiment was done in triplicate in a 96-well format with 10 nM duplex DNA operator in 50 mM Tris, 200 mM NaCl, 2 mM TCEP, pH 8.0, at 25.0 °C unless noted otherwise, and various concentrations of PsaR. Metallated PsaRs were prepared by adding 0.95 mol protomer equiv of Zn(II) followed by no addition (PsaRZn,apo) or 1.05 mol equiv of the desired divalent metal, e.g., Mn(II) to create PsaRZn,Mn or Zn(II) to create PsaRZn,Zn. To ensure full occupancy of the second metal site when desired, an additional 10 µM of the indicated divalent metal was added to the anisotropy buffer. The resulting data were fit to a dissociable dimer-binding model that invokes the dimerization of PsaR (Kdimer), followed by dimer binding to the DNA (KDNA) using Dynafit.39 Kdimer was fixed to a values of 2.0 × 106 M−1 as estimated by an unconstrained fit to a titration of PsaRZn,Mn to DNA.18 Using this model with Kdimer constrained to this value for all PsaRs, the association constant, KDNA, was determined (see Supplementary Fig. S4 for a sample Dynafit script file). The Gibbs allosteric coupling free energy (ΔGc) of interest is operationally defined as ΔGc= −RT ln(KDNAZn,Mn/KDNAZn,apo). The errors associated with this value were determined as previously described.40
Multi-angle light scattering measurements
Wild-type PsaRs with different metallation states were prepared by dialyzing apo-PsaR into a MALS buffer (20 mM HEPES, pH 7.0, 200 mM NaCl, 1 mM TCEP), followed by addition of nothing (PsaRapo.,apo), 0.95 mol protomer equiv of Zn(II) (PsaRZn,apo), followed by 1.05 mol equiv of Zn(II) (PsaRZn,Zn) or Mn(II) (PsaRZn,Mn). MALS measurements were performed on a Wyatt Technology DAWN HELEOS II multi-angle light scattering (MALS) instrument coupled to a Wyatt Technology Optilab rEx refractive index (RI) detector. 100 µL of each sample (ranging from 150–160 µM protomer) was loaded onto a pre-equilibrated Superdex G75 10/300GL (GE) size exclusion column at a flow rate of 0.5 mL/min. SEC-MALS data were analyzed using Wyatt Technology ASTRA software to obtain molar mass.
X-ray absorption spectroscopy (XAS)
Wild-type PsaRZn,apo and PsaRZn,Mn were prepared by addition of 0.98 mol protomer equiv of Zn(CH3COO)2• 2H2O, followed no addition, or addition of 1.05 mol equiv of MnSO4, respectively, in 50 mM Tris pH 8.0, 200 mM NaBr and 1 mM TCEP. 620 µM PSAZn,apo and 560 µM PsaRZn,Mn samples were prepared by syringe injection into polycarbonate XAS holders wrapped in kapton tape and frozen rapidly in liquid nitrogen. XAS data were collected as previously described41 under dedicated ring conditions on beam line X3b at the National Synchrotron Light Source (NSLS), Brookhaven National Laboratories. The samples were loaded into an aluminum sample holder, which was cooled to ~50 K by using a He displex cryostat. Data were collected under ring conditions of 2.8 GeV and 120–300 mA using a sagitally focusing Si(111) double-crystal monochromator. Harmonic rejection was accomplished with a Ni-coated focusing mirror. X-ray fluorescence was collected using a 30-element fluorescence detector (Canberra). XANES were collected from ± 200 eV relative to the metal edge. The x-ray energy for Zn Kα-edge was internally calibrated to the first inflection point of the corresponding metal foil, Zn, 9660.7 eV. EXAFS was collected to 13.5 – 16 k above the edge energy (Eo), depending on the signal:noise at high values of k.
Data Reduction and Analysis
The XAS data reported are the average of 6–8 scans. XANES and EXAFS data were analyzed using EXAFS12342 and SixPack43, respectively. The SixPack fitting software builds on the ifeffit engine.44,45 For the EXAFS analysis, each data set was background-corrected and normalized. The data were converted to k-space using the
relationship, where me is the mass of the electron, ħ is Plank’s constant divided by 2π, and Eo is the threshold energy of the absorption edge. The threshold energies chosen for Zn was 9670 eV.41 A Fourier-transform of the data was produced using the data range k = 2 – 12.5 ¥−1, where the upper limit was determined by signal:noise. Scattering parameters for EXAFS fitting were generated using FEFF8 software package.44 The k3-weighted data were fit in r-space. The first coordination sphere was determined by setting the number of scattering atoms in each shell to integer values and systematically varying the combination of N/O- and S-donors (Supporting Information, Tables S1 and S2).
Multiple-scattering parameters for imidazole ligands bound to various metals were generated from crystallographic coordinates using the FEFF8 software package with previously published crystal structures as input.46 The best fits resulted in four prominent multiple-scattering features and paths of similar overall lengths were combined to make four imidazole scattering paths, matching these four prominent features as outlined by Costello et al.47,48 The four combined paths were used to fit the data by setting the number of imidazole ligands per metal ion to integral values while allowing R and σ2 to vary.47,48 To compare different models of the same data set, the nonlinear least squares fitting algorithm “ifeffit” (FEFF8) utilizes three goodness of fit parameters: χ2, reduced χ2, and the R-factor. These parameters are not to be confused with nomenclature where R is used for interatomic distance and χ for k-space EXAFS. The statistical parameter, χ2, that is minimized in a fit, is given by eq.1, where Nidp is the number of independent data points, Nε2 is the number of uncertainties to minimize, Re( ) and Im( ) denote the real and imaginary parts of the EXAFS fitting function and χ̃(Ri) is the Fourier-transformed data or model function:
| (1) |
Reduced χ2 = χ2/(Nidp – Nvarys) where Nvarys is the number of refining parameters and represents the degrees of freedom in the fit. Additionally ifeffit calculates the R-factor for each fit, which is directly proportional to χ2 and a measure of absolute misfit between the data and theory. The R-factor is given by eq. 2, and is scaled to the magnitude of the data making it proportional to χ2.
| (2) |
In comparing different models, the R-factor and reduced χ2 parameter were used to determine which model was the best fit for the data. The R-factor will generally improve with an increasing number of adjustable parameters, while reduced χ2 will go through a minimum and then increase, indicating that the model is over-fitting the data.49
RESULTS
Spn PsaR is a homodimer in solution independent of metallation state
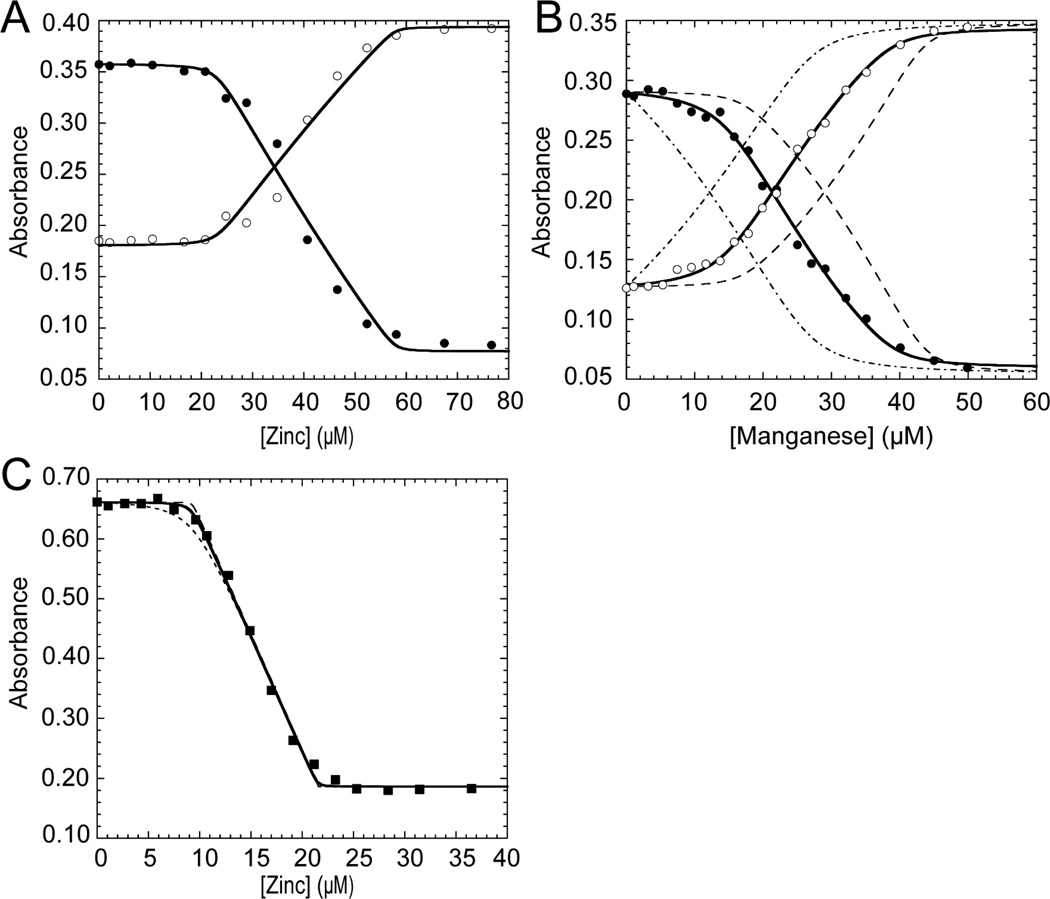
Wild-type PsaRapo,apo, PsaRZn,apo, PsaRZn,Zn and PsaRZn,Mn were subjected to size exclusion chromatography with the elution profile analyzed by multi-angle light scattering (MALS) in order to assess the homogeneity and quaternary structure of distinct metallation states of Spn PsaR (Fig. 1). Representative experiments are shown for PsaRapo,apo (Fig. 1A) and PsaRZn,apo (Fig. 1B) (≈160 µM protomer loaded onto the column, pH 7.0, 0.2 M NaCl) with the results shown for all wild-type PsaRs compiled in Table 1. Comparison of the elution profiles of PsaRapo,apo and PsaRZn,apo reveals a trailing tail on a major peak for PsaRapo,apo which is not observed in PsaRZn,apo. The apparent molecular weight of PsaRZn,apo is 48 kD from this experiment (Table 1). The addition of a second molar protomer equivalent (mol protomer equiv) of metal, as Zn(II) or Mn(II), reveals no change in the elution pattern or measured assembly state (Table 1). These data support the conclusion that all forms of PsaR are homodimeric under these conditions, and that the metal-free apo state may well be hydrodynamically somewhat larger and/or more structurally heterogeneous than metallated PsaRs, with Zn(II) binding to the high affinity site (see below) quenching this heterogeneity. No evidence for monomeric PsaR was obtained in these profiles.
Figure 1.
Representative multi-angle light scattering (MALS) traces of wild-type PsaRapo,apo (left) and PsaRZn,apo (right). Red trace, refractive index; blue trace, Raleigh ratio. The red peak at ≈40 min in each panel marks the included volume of the column and is not protein. Green trace, determined molar mass (kDa) across the main eluting species. Parameters for all metallated derivatives are complied in Table 1.
Table 1.
Average experimentally determined molecular weights for wild-type Spn PsaR loaded with various metals by MALS.
| PsaR Metallation Statea | Molecular Weight (D) |
|---|---|
| Monomer (reduced), expected | 24,852.6 |
| Dimer (reduced), expected | 49,705.2 |
| PsaRapo,apo | 50,000 (±4000) |
| PsaRZn,apo | 48,000 (±1000) |
| PsaRZn,Zn | 47,000 (±3000) |
| PsaRZn,Mn | 47,000 (±2000) |
Metallation state subscripts refer to metal loaded in the site 1 (high affinity), followed by site 2 (low affinity). Solution conditions: 20 mM HEPES, pH 7.0, 200 mM NaCl, 1 mM TCEP, ambient temperature. 150–160 µM protomer (100 µL) was used for each column run.
Mn(II) and Zn(II) bind to the PsaR dimer in each of two distinguishable steps
Following determination of the dominant assembly state of wild-type PsaR, we next measured metal binding affinities and stoichiometries for both cognate Mn(II) and non-cognate Zn(II). Chelator competition experiments were conducted with two different divalent metal ion competitors with vastly different affinities, mag-fura-2 (mf2) (KZn=5.0 × 107 M−1) and quin-2 (KZn=2.7 × 1011 M−1), and the data were fit to the appropriate one-site or two-site binding model. Representative experiments are shown for the titration of a solution of wild-type PsaR and mf2 with Zn(II) (Fig. 2A) or Mn(II) (Fig. 2B). Both titrations can be satisfactorily fit to a 2:1 metal:protomer binding model (or four metal ions per dimer), with no need to invoke cooperativity of metal binding within the homodimer (Table 2). Thus, binding to site 1 and site 2 on each protomer are therefore considered as independent events. For Zn(II) binding, the first mol protomer equiv of Zn(II) binds to PsaR since there is no change in the spectrum of the chelator; after that site is filled, Zn(II) binds to the second site in competition with mf2. Thus, only a lower limit of KZn1 ≥109 M−1 for site 1 can be obtained from this experiment; nonlinear least squares fitting reveals a KZn2 of 2.2 (±0.5) × 108 M−1 (Table 2). To further estimate KZn1, a quin-2 competition experiment was performed (Fig. 2C). Here, as with mf2 (Fig. 2A), quin-2 provides little competition to PsaR for Zn(II) binding to site 1; again, simulated curves (Fig. 2C) reveal that only a lower limit of KZn1≥ 1013 M−1 can be obtained from this experiment (Table 2).
Figure 2.
Representative isotherms obtained when a mixture of wild-type apo-PsaR and magfura-2 (mf2) (A,B; open symbols A325; filled symbols, A360) or quin-2 (C; filled symbols, A265) is titrated with the indicated metal salt. (A) Zn(II) titration into mag-fura-2 (12 µM) and PsaRapo,apo (23 µM monomer). (B) Mn(II) titration into a solution of mag-fura-2 (10 µM) and PsaR (17 µM monomer). (C) Zinc titration into a solution of quin-2 (12 µM) and PsaR (10 µM monomer). The continuous lines represent a nonlinear global least squares fit employing either a two-site (mag-fura-2) or single site (quin-2) competition model with the optimized binding parameters from multiple experiments compiled in Table 2. The broken lines in panels B and C represent simulated curves with each KMn (panel B) or KZn (panel C) 10-fold higher or 10-fold lower than the optimized value, for visual comparison with the fitted curves (Table 2).
Table 2.
Zn(II) and Mn(II) binding affinities for sites 1 and 2 for wild-type and mutant Spn PsaRs (pH 8.0, 0.2 M NaCl, 25 °C)a
| Spn PsaR | log KZn1 (M−1) | log KZn2 (M−1) | log KMn1 (M−1) | log KMn2 (M−1) |
|---|---|---|---|---|
| wild-type | ≥13b | 8.35 (±0.08) | 7.70 (±0.07) | 6.09 (±0.03) |
| D7A (site2) | ≥9c | 8.20 (±0.06) | 5.15 (±0.06) | ≤4d |
| E99A (site2) | ≥9c | 7.97 (±0.04) | 6.31 (±0.04) | ≤4 |
| H125A (site1) | n.d.e | n.d. | 7.4 (±0.1) | 6.61 (±0.04) |
| D160A (site1) | ≥9 | 7.88 (±0.06) | 7.7 (±0.1) | 6.62 (±0.04) |
| H76A | n.d. | n.d. | 7.67 (±0.08) | 5.73 (±0.05) |
| H76F | n.d. | n.d. | 8.0 (±0.2) | 6.39 (±0.05) |
Determined from mf2 competition experiments like those shown in Fig. 2 unless otherwise noted. The numbers in parentheses represent the standard error in log K from three or more independent experiments.
Determined from a quin-2 competition experiment and represents a lower limit.
A lower limit can only be determined from these mf2 competition experiments.40
Could not be detected in mf2 competition experiment and the number shown represents an upper limit.
n.d., not determined due to protein precipitation under these conditions.
The Mn(II) binding titrations of PsaR-mf2 (KMn=2.4 × 106 M−1) mixtures (Fig. 2B), like Zn(II), can also be fit to two sites per protomer (or four per dimer) that bind Mn(II) with distinct affinities. In contrast to Zn(II), fitting of replicate binding isotherms reveals that both sites effectively compete with mf2, with KMn1=5.0 (±0.2) × 107 M−1 and KMn2=1.2 (± 0.2) × 106 M−1 (Table 2). These results reveal that PsaR binds two mol protomer equiv of Mn(II) or Zn(II), with Zn(II) binding preferentially to both sites 1 and 2, a general finding consistent with predictions from the Irving-William series of metal-complex stabilities (KZn>>KMn), with this preference far greater at site 1. In fact, the disparity between KZn1 and KMn1 (Table 2) and the absolute magnitude of KZn1 suggests strongly that in vivo, site 1 is filled with zinc, which makes Spn PsaR a zinc metalloprotein. Interestingly, the putative regulatory site 2 also favors zinc over manganese by ≈40-fold; thus, in order for PsaR to function as a Mn(II)-specific repressor in the cell one would have to postulate that intracellular zinc concentrations do not rise under normal conditions to the point where Zn(II) would effectively compete with Mn(II) for occupancy of this site (see Discussion).
Structural identification of site 1 by x-ray absorption spectroscopy
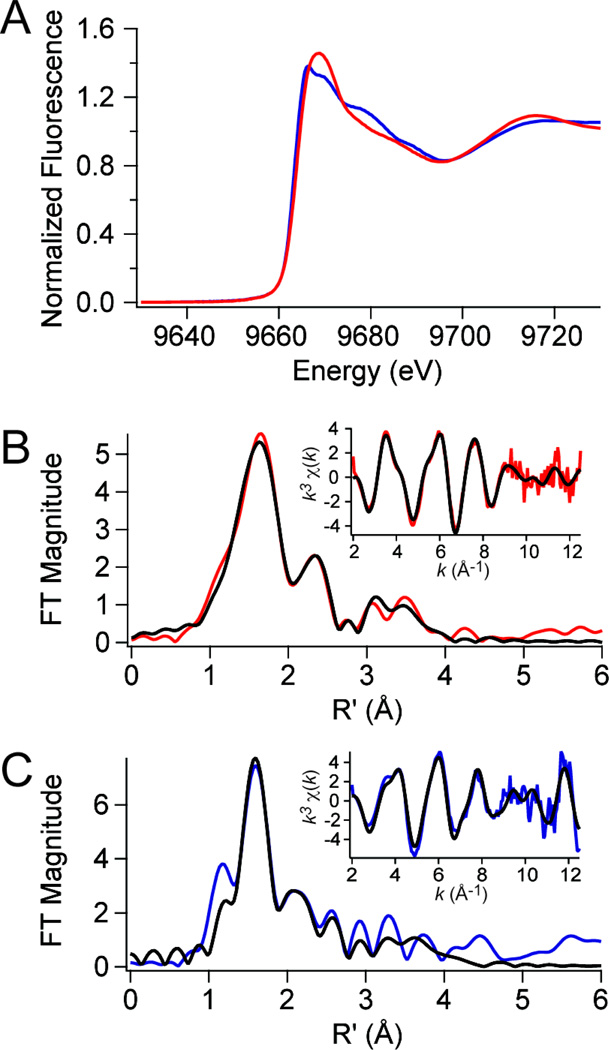
We next exploited the finding that site 1 could be preferentially loaded with Zn(II) to prepare wild-type PsaRZn,apo and PsaRZn,Mn samples for analysis by x-ray absorption spectroscopy. This experiment provides structural information on the first coordination sphere of the site 1 Zn(II) and allows a determination of the degree to which filling site 2 with the cognate Mn(II) influences the structure of this site. Note that Mn(II) is transparent in this experiment. The zinc x-ray absorption near-edge spectra (XANES) are shown for PsaRZn,apo (red trace) and PsaRZn,Mn (blue trace) in Fig. 3A, with the corresponding Fourier-transformed extended x-ray absorption fine structure (EXAFS) data and unfiltered k3-weighted EXAFS spectra shown for PsaRZn,apo (Fig. 3B) and PsaRZn,Mn (Fig. 3C) samples. The threshold energies chosen for Zn was 9670 eV.41 Edge normalization and background subtraction were performed using a Gaussian pre-edge function (between −200 and −50 eV with respect to Eo) and a second-order polynomial for the post-edge region (between 150 and 861 eV) followed by normalization of the edge jump to 1.0. The normalized fluorescence intensity of the near edge increases with increasing coordination number, and assumes a value of ≈1.3 for four-coordinate structures, and between 1.3 and 2.0 for five- and six-coordinate complexes, with six-coordinate complexes the most intense.50 For PsaRZn,apo, the normalized intensity of ≈1.45 and an edge energy is ~9663.9 eV is more consistent with a five-coordinate complex. However for PsaRZn,Mn, the normalized intensity of ≈1.38 and an edge energy is ~9663.3 eV is more consistent with a four-coordinate complex.
Figure 3.
Zn K-edge X-ray absorption spectroscopy of wild-type PsaRZn,apo (red traces) and PsaRZn,Mn (blue traces). (A) Zinc XANES spectra for each species. (B) Fourier-transformed EXAFS data (main panel) obtained for PsaRZn,apo (red trace) superimposed on best-fit coordination complex fits (black lines) as described by the parameters in Table 3. Inset, unfiltered k3-weighted EXAFS spectrum and fits for PsaRZn,apo. (C) Fourier-transformed EXAFS data (main panel) obtained for PsaRZn,Mn (blue trace) superimposed on best-fit coordination complex fits (black lines) (see Table 3). Inset, unfiltered k3-weighted EXAFS spectrum and fits for PsaRZn,Mn.
These conclusions are consistent with the EXAFS analysis (Figs. 3B–C; Table 3) and a nonlinear least squares analysis of the EXAFS data (black continuous curves, Fig. 3B–C). Fitted parameters from the best fit, as judged by minimizing %R and χ2, and optimizing meaningful σ2 values are compiled in Table 3 (the results of other fits are shown in Supplementary Tables S1 and S2). The best fits for PsaRZn,apo is a five-coordinate complex composed of four N/O donor ligands at 2.05 Å, with one from an imidazole (histidine) ligand (as evidenced by the spectral features at r=3–4 Å), and a single sulfur (S) scatterer at 2.31 Å from the Zn(II). The presence of a single S-donor suggests that the high affinity site 1 in PsaR is analogous to the Cd(II) site previously observed in Sgo ScaR formed by conserved residues Glu80, Cys123, His125 and Asp160, with one of the carboxylate ligands adopting bidentate coordination. In contrast, addition of Mn(II) to site 2 in PsaRZn,apo to form PsaRZn,Mn is best fit by a four-coordinate complex featuring one fewer N/O-donor, but otherwise likely involving the same donor residues. A five-coordinate fit, while also acceptable, returns a statistically somewhat poorer fit relative to the four-coordinate fit (Supplementary Table S2). These data taken collectively reveal Mn(II) binding to site 2 induces a detectable change in the first coordination sphere of the Zn(II) bound in site 1 (Table 3). The structural basis of this rearrangement is of course not known but could be accomplished by a change in bidentate to monodentate coordination by Glu80, which is bidentate in the Sgo ScaR Cd(II)-bound structure and is closest to the proposed regulatory site ligands (vide infra). In any case, these two metal sites in the PsaR dimer clearly communicate with one another and may be reporting on a more global conformational transition associated with binding of the cognate metal to site 2.
Table 3.
Zinc XANES and EXAFS analysis for wild-type PsaRZn,apo and wild-type PsaRZn,Mn.
| XANES analysis | EXAFS analysis | ||||||
|---|---|---|---|---|---|---|---|
| Metal ion | K-edge energy (eV) |
CNa | Shellb | r (Å) | σ2 (×10−3 Å2) | ΔEo (eV) | % R |
| PsaRZn,apo | |||||||
| Zn(II) | 9663.9 | 5 | 4 N/O (1 Im) | 2.05 (1)c | 8 (1) | −5 (1) | 1.14 |
| 1 S | 2.31 (1) | 6 (1) | |||||
| PsaRZn,Mn | |||||||
| Zn(II) | 9663.3 | 4 | 3 N/O (1 Im) | 2.03 (1) | 2 (1) | 3 (3) | 5.19 |
| 1 S | 2.37 (2) | 3 (2) | |||||
Coordination number.
Im, number of imidazole (histidine) ligands among N/O donors.
Numbers in parentheses are the estimated uncertainties in the corresponding parameters in the last significant figure given. These uncertainties are calculated by SixPack and reflect the change in the variable that will result in an increase in χ2 of 1.0.
Metal binding properties of PsaR mutants
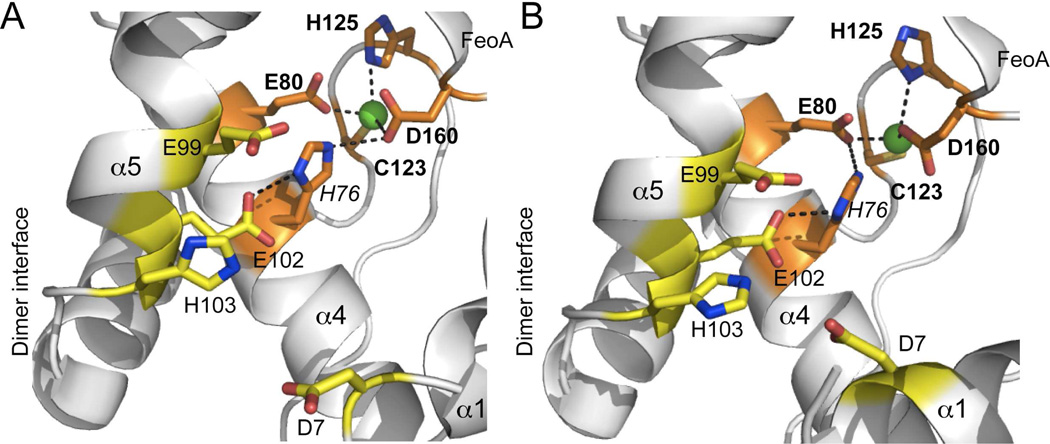
Having established the order of metal ion binding and structural identification of site 1 as the metal site positioned between the dimerization and FeoA domains, we next determined the metal [Zn(II) and Mn(II)] binding affinities and stoichiometries for two putative site 1 nonliganding mutants, H125A and D160A, two candidate site 2 mutants from a multiple sequence alignment of MntR, DtxR and PsaR, D7A and E99A (Supplementary Fig. S1), and two His76 mutants, H76F and H76A. His76 is not a predicted ligand to either metal in PsaR, but is a direct ligand to the ancillary metal ion in DtxR and to the A-site Mn(II) in MntR, and is absolutely conserved. In the ScaR structure, H76 appears to “bridge” site 1 (via Glu80 or D160) and candidate site 2 (Glu102) residues via hydrogen bonding, based on inspection of the two Cd(II) complexes in the Sgo ScaR dimer structure (Fig. 4). The prediction is that mutation of His76 would disrupt allosteric linkage between DNA and cognate regulatory site binding, with no affect on the metal binding affinities. We used the same chelator competition assays with mf2 to assess binding parameters for Zn(II) and Mn(II) exactly as described above for wild-type PsaR, with all results summarized in Table 2. Site 1, site 2 and His76 mutants are discussed in turn.
Figure 4.
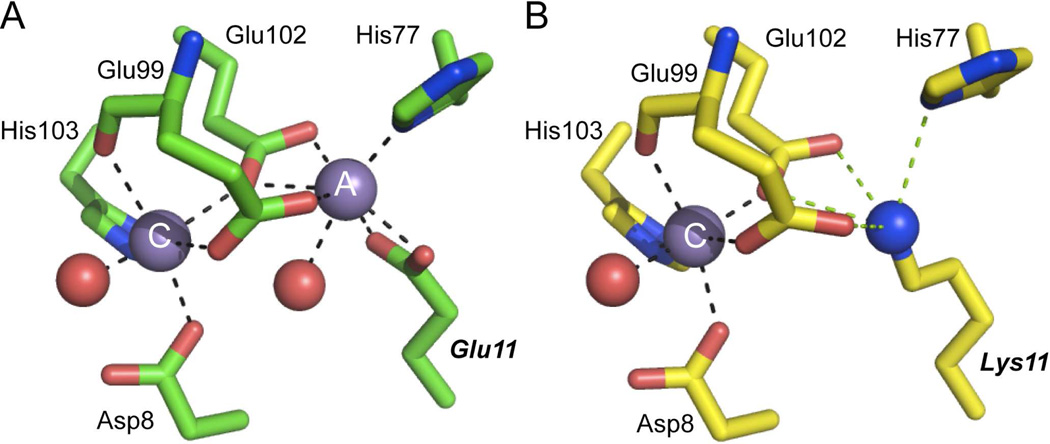
Close-up view of the Cd(II) site 1 in each of the two subunits (A,B) in Sgo ScaR (PDB code 3HRT)34 (ligating residues shaded orange; E80, C123, H125 and D160), emphasizing the physical location of H76 and immediately adjacent candidate regulatory site 2 ligands D7, E99, E102 and H103 (shaded yellow). Side-chains are shown in stick in elemental shading; Cd(II) ion, green sphere. Note that the homodimer is strongly asymmetric and this asymmetry extends to the metal site region which adopts distinct structures in each protomer. D7 makes much closer approach to the other candidate site 2 ligands in the α5 helix in the right (panel B) protomer relative to the left (panel A). Note also metal coordination bonds are unreasonably long in all cases, likely attributed to the modest resolution (2.7 Å) of the structure.
Both H125A and D160A PsaR site 1 mutants bind two mol protomer equiv of Mn(II) with KMn for both site 1 and site 2 within a factor of 2–3 of wild-type PsaR (Table 2). Although Zn(II) binding measurements for H125A PsaR could not be carried out due to extensive protein precipitation, the affinity of D160A PsaR for Zn(II) is within a factor of ≈2 of wild-type PsaR at the low affinity site 2 (KZn2=7.6 × 107 M−1), with only a lower limit obtained for the high affinity site 1 (KZn1≥109 M−1). Thus, to a first approximation, single amino acid substitutions in site 1 have only small effects on metal binding to either site, with the possible exception of Zn(II) binding to site 1 (Table 2).
In contrast, both D7A and E99A candidate site 2 mutants exhibit the shared characteristic of dramatically lowering the cognate Mn(II) affinity of both sites, with correspondingly little effect on non-cognate Zn(II) binding affinity. KZn1 remains too tight to measure in mf2 competition experiments, with KZn2≈108 M−1, essentially as described for wild-type PsaR (Table 2). For Mn(II), in contrast, only one binding site is competitive with mf2 (which we attribute to the high affinity site 1) but is characterized by KMn1 values which are 200-fold (D7A PsaR) and 20-fold (E99A PsaR) weaker than wild-type PsaR. The second Mn(II) binding event is not observable in either D7A or E99A PsaRs, i.e., KMn2≤104 M−1 under these conditions (Table 2).
Characterization of the metal binding properties of candidate allosteric mutants H76F and H76A PsaRs reveals that each binds two mol protomer equiv of Mn(II) (Table 2) with affinities largely indistinguishable, e.g., within a factor of 2–3 of wild-type PsaR. Zn(II) binding affinities could not be confidently measured under these conditions due to complications from precipitation of the high protein concentration required by this experiment. This did not preclude quantitative DNA binding experiments to extract ΔGc (Fig. 5), which are described below. In any case, these data suggest to a first approximation that substitution of His76 has only a small effect on metal binding affinities, much like the site 1 mutants (Table 2).
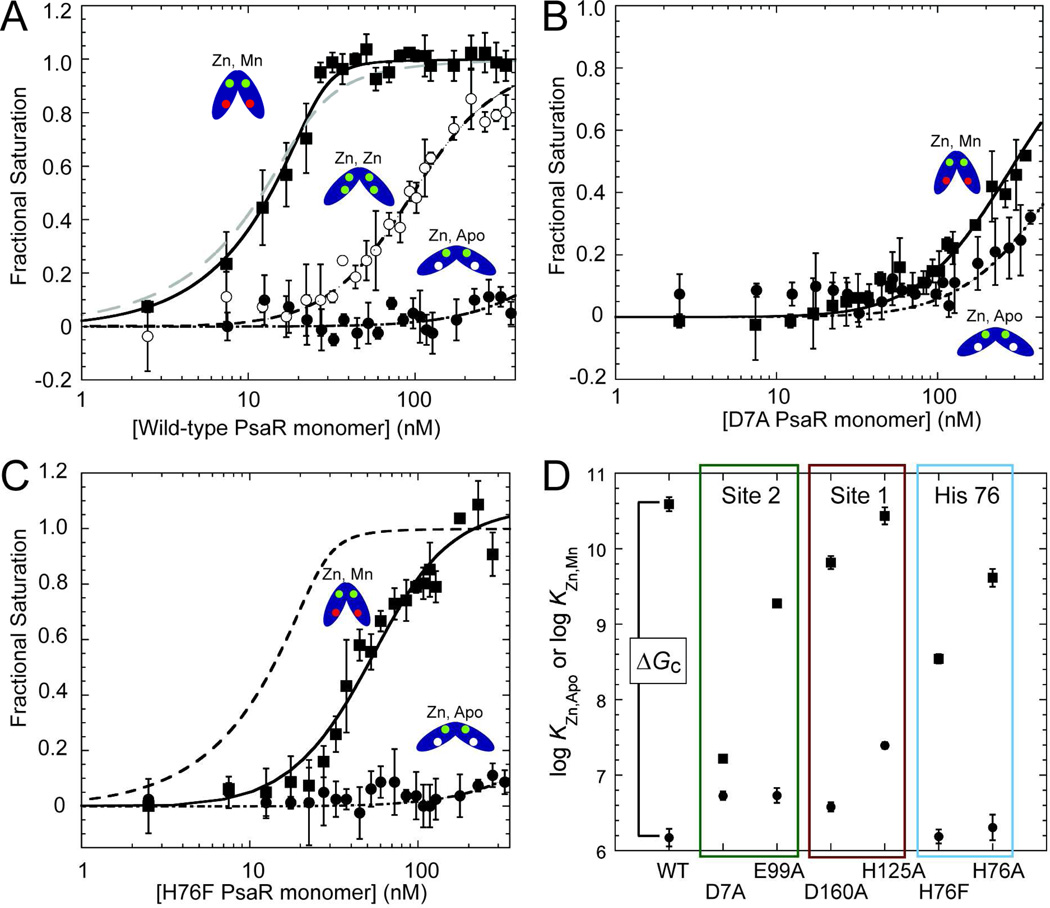
Figure 5.
DNA binding isotherms for various metallated states of wild-type PsaR (A), D7A PsaR (B) and H76F PsaR (C). In each case, data for PsaRZn,apo and PsaRZn,Zn and PsaRZn,Mn species are shown in filled circles, open circles and filled squares respectively. The smooth curves through each data set correspond to that derived from a dissociable dimer binding model (see Materials and Methods for details) with KDNAi vales complied in Table 4. The grey dashed line in panel (A) shows the fit for a nondissociable dimer model for PsaR binding (see Table 4). The dashed line in panel (C) shows the fit for wild-type PsaRZn,Mn for comparison only. (D) Summary of the DNA binding affinities of PsaRZn,apo (filled circles) and PsaRZn,Mn (fillled squares) for wild-type PsaR and indicated mutants with the vertical separation between these Ki values proportional to ΔGc from ΔGcMn = −RTln(KDNAZn,Mn/KDNAZn,apo) or for Zn(II), ΔGcZn = −RTln(KDNAZn,Zn/KDNAZn,apo). ΔGc is equivalent to a ΔΔG, or the difference in PsaO DNA binding free energy between the appropriate metallated form (PsaRZn,Mn or PsaRMn,Mn) and PsaRZn,apo.
DNA operator binding by various metallated wild-type and mutant PsaRs
Having determined the cognate Mn(II) and non-cognate Zn(II) binding affinities and stoichiometries for wild-type PsaR and various site 1, site 2 and H76 mutants, we next quantified the degree to which each metallated species is capable of metal-dependent [Mn(II) vs. Zn(II)] allosteric positive regulation of psaBCA operator (PsaO) binding. To do this, we carried out quantitative DNA binding assays by monitoring the anisotropy of the fluorescence of a 32-bp fluorescein-labeled PsaO-containing duplex (10 nM) in a 96-well microtiter plate format with each point in a binding curve measured in triplicate. The DNA binding affinities (KDNA) were determined for all four metallated states of wild-type PsaR (Fig. 5A; Table 4), i.e., PsaRapo.apo, PsaRZn,apo, PsaRZn,Zn, and PsaRZn,Mn, while for all other mutant proteins, only PsaRZn,apo and PsaRZn,Mn were measured (Fig. 5, Table 4). The allosteric coupling free energy, ΔGc, was then determined from these KDNA values as a measure of the degree to which cognate Mn(II) or non-cognate Zn(II) bound to site 2 allosterically activates DNA binding upon saturation of site 2 relative to an empty site 2 (with site 1 filled with Zn(II) in all cases, PsaRZn,apo) (Table 4).
Table 4.
DNA binding affinities of the indicated metallated derivatives of wild-type and mutant PsaRs as analyzed by a dimer linkage modela
| Spn PsaR | log KDNA (M−1) | Fractional change in KDNA from corresponding wild-type PsaR |
ΔGc (kcal mol−1) |
|---|---|---|---|
| wild-type PsaRZn,apo | 6.2 (±0.1) | 1 | — |
| wild-type PsaRZn,Zn | 8.02 (±0.03) | 0.0027b | −2.5 (±0.2)d |
| wild-type PsaRZn,Mn | 10.6 (±0.1) | 1 | −6.0 (±0.3)d |
| wild-type PsaRMn,Mn | 10.1 (±0.1) | 0.30 | −5.4 (±0.4)d |
| D7A PsaRZn,apo | 6.73 (±0.05) | 3.6c | — |
| D7A PsaRZn,Mn | 7.22 (±0.03) | 0.00042 | −0.7 (±0.1)e |
| E99A PsaRZn,apo | 6.7 (±0.1) | 3.6 | — |
| E99A PsaRZn,Mn | 9.27 (±0.05) | 0.048 | −3.5 (±0.2)e |
| H125A PsaRZn,apo | 7.39 (±0.04) | 16 | — |
| H125A PsaRZn,Mn | 10.4 (±0.1) | 0.71 | −4.1 (±0.2)e |
| D160A PsaRZn,apo | 6.58 (±0.06) | 2.5 | — |
| D160A PsaRZn,Mn | 9.8 (±0.1) | 0.17 | −4.4 (±0.2)e |
| H76A PsaRZn,apo | 6.3 (±0.2) | 1.4 | — |
| H76A PsaRZn,Mn | 9.6 (±0.1) | 0.11 | −4.5 (±0.4)e |
| H76F PsaRZn,apo | 6.2 (±0.1) | 1.0 | — |
| H76F PsaRZn,Mn | 8.54 (±0.06) | 0.0089 | −3.2 (±0.2)e |
Determined from experiments like those shown in Fig. 5 with the numbers in parentheses defined as the unweighted standard error of the fit from triplicate titrations.
The numbers in standard text are calculated from KDNA(mutant PsaRi,j)/KDNA(wild-type PsaRZn,apo), where i=j=Zn or Mn.
The numbers in italicized font are given by the ratio KDNA(mutant PsaRZn,apo)/KDNA(wild-type PsaRZn,apo).
ΔGc= −RTln(KDNA(wild-type PsaRi,j)/KDNA(wild-type PsaRZn,apo), where i=j=Zn or Mn.
ΔGc= −RTln(KDNA(mutant PsaRZn,Mn)/KDNA(mutant PsaRZn,apo).
Inspection of these binding isotherms for wild-type PsaR (Fig. 5A) shows clearly that apo PsaR, PsaRZn,apo, binds weakly under these conditions (pH 8.0, 0.2 M NaCl) as expected, and that non-cognate Zn(II), while capable of activating PsaO binding when bound to site 2 (PsaRZn,Zn), is far less effective than cognate Mn(II). Analysis using a dimer linkage model and assuming the same Kdimer in each species reveals a ≈380-fold decrease in PsaO affinity of the non-cognate PsaRZn,Zn complex vs., the cognate PsaRZn,Mn complex (Table 4), and does not give rise to significant activation until a protein concentration reaches a level where the Mn(II)-bound form saturates the DNA. The Gibbs coupling free energy, ΔGc, operationally defined as described in Table 4, is more than 2-fold greater for Mn(II) vs. Zn(II) bound to site 2, at −6.0 vs. −2.5 kcal mol−1 under these conditions. Interestingly, PsaRMn,Mn is slightly less capable than PsaRZn,Mn in driving allosteric activation of PsaO binding, although the difference is small, ΔGc=−5.3 kcal mol−1 (Table 4). Although this operationally defined value of ΔGc is obviously dependent upon the value measured for Kdimer,40 there is clearly some detectable linkage to the monomer-dimer equilibrium as a fit to a nondissociable dimer binding model for PsaRZn,Mn does not fit nearly as well (Fig. 5A, grey dashed line). This justifies use of this simple linkage model to analyze all subsequent binding isotherms.
Both site 1 mutations, H125A and D160A, significantly lower ΔGc to ≈4 kcal mol−1 or by about 35% relative to wild-type PsaR, and thus have a detectable influence on allosteric linkage (Table 4) in the absence of large changes in Mn(II) binding affinity (Table 2). The underlying origins are different however in the two mutants. For H125A PsaR, the apo form (PsaRZn,apo) binds the operator DNA ≈10-fold more tightly than wild-type PsaR, while the fully activated affinity of PsaRZn,Mn is indistinguishable from that of wild-type PsaR (Fig. 5D, Table 4). In contrast, in D160A PsaR, the apoprotein affinity is only 2-fold higher with a fully activating affinity of PsaRZn,Mn ≈8-fold lower than wild-type PsaR. The characteristics of these mutants reveal that mutations in metal site 1 disrupt allosteric switching, which itself is dependent strongly on occupancy of site 2 with cognate Mn(II), i.e, the structural integrity of both sites is required for full allosteric activation of PsaO binding by PsaR (Fig. 5D).
Like the two site 1 mutants, both site 2 mutants, D7A and E99A PsaRs, have similar DNA binding characteristics; however, the D7A mutant is far more functionally compromised than E99A PsaR. In fact, these findings generally track with the relative perturbations in metal site affinities for each mutant (vide supra, Table 2). Both mutants bind the PsaO with a slightly elevated affinity in the absence of activating metal (PsaRZn,apo) (Fig. 5D; Table 4); however, the D7A mutation essentially abrogates Mn(II)-dependent activation of PsaO binding with ΔGc for PsaRZn,Mn of −0.7 kcal mol−1 under these conditions (see also Fig. 5C). E99A PsaR is also severely functionally compromised, although the PsaRZn,Mn form binds ≈100-fold more strongly to the PsaO than D7A PsaR, giving a ΔGc for E99A PsaRZn,Mn of −3.2 kcal mol−1 or ≈50% that of wild-type PsaR. We conclude that the structural integrity of site 2 is critically tied to allosteric switching, with the candidate ligand derived from near the N-terminus, D7, playing a major role in this process.
Finally characterization of the predicted allosteric switching mutants H76A and H76F PsaRs reveals that H76 functions in some way in this process. Both PsaRZn,apo mutants bind the PsaO with wild-type-like low affinities (Table 4; Fig. 5C), but each is distinct in allosteric activation of the PsaRZn,Mn forms. Interesting, the H76F PsaR mutant (Fig, 5C) is far more functionally compromised relative to H76A PsaR, with an activation potential comparable to that of E99A PsaR (ΔGc=−3.2 kcal mol−1) with the H76A mutant most quantitatively similar to that of the site 1 mutant D160A PsaR (ΔGc=−4.5 kcal mol−1). The characterization of these two H76 mutants highlight the potential impact of the second coordination shell to either or both sites 1 (see Fig. 4) and 2 metal complexation in moving from PsaRZn,apo to the activated PsaRZn,Mn state.
DISCUSSION
In this work, we elucidate the quaternary structure, metal binding and DNA binding properties of Spn PsaR, a member of an emerging subclass of Mn(II)-sensing repressors in the DtxR/MntR metalloregulator family.4 PsaR is a stable homodimer at high concentration (≈160 µM) irrespective of metallation state, consistent with previous studies on both Bsu MntR and Sgo ScaR.29,34 We establish that the PsaR homodimer harbors two pairs of divalent metal binding sites (four per dimer) that are functionally non-equivalent and structurally distinct. The high affinity site 1 that straddles the dimerization and FeoA domains is likely filled by Zn(II) in the cell given KZn1≥1013 M−1>>KMn1, thus establishing that PsaR is a zinc metalloprotein. Occupancy of this site by Zn(II) or Mn(II) is necessary for full activation by Mn(II), but maintains PsaR in an inactive DNA-binding state. It is only when the lower affinity site 2 is bound by cognate Mn(II) that PsaRZn,Mn is maximally activated to bind to the PsaO (Fig. 5). Zn(II) binding to site 2 is weakly effective in allosteric activation, despite a higher binding affinity, and PsaRZn,Zn is characterized by an allosteric coupling free energy only 40% that of PsaRZn,Mn. The high affinity metal site 1 is unique to streptococcal clade of DtxR/MntR repressors. The two metal binding sites in PsaR clearly communicate with one another since mutations of ligating residues in site 1 significantly diminish ΔGc while mutations in the activating metal site 2 significantly alter the affinity of Mn(II), and perhaps Zn(II), for site 1. Furthermore, occupancy of cognate Mn(II) in site 2, detectably alters the first coordination sphere around the Zn(II) ion bound to site 1 (Fig. 3; Table 3), which may or may not be mediated by the “bridging” H76 and E80 in site 1 (Fig. 4).
One striking finding is that the Mn(II) binding affinity of the regulatory site 2 in PsaR measured here at pH 8.0, 0.2 M NaCl, KMn2=1.2 (±0.2) × 106 M−1 (Kd,Mn2=0.83 µM), is ≈10-fold larger that that directly measured for the two sites in MntR32,51 and for Sgo ScaR34 under similar conditions and the same pH. In contrast, the Zn(II) affinities of all three proteins are more similar than different, and in the range of 107–108 M−1. The set-point model for metal-sensing in the cell posits that the dissociation constant (Kd) of the regulator for its cognate metal establishes the intracellular bioavailability or “free” concentration, e.g., [Mn]free, in the cell.52 As a result, one could hypothesize that S. pneumoniae maintains a lower bioavailable pool of Mn(II) than does B. subtilis. Total cell-associated Mn(II) is on par with that Zn(II) in S. pneumoniae and significantly greater than that of Fe(II),18 but to our knowledge similar measurements have not yet been reported for B. subtilis. In any case, with relatively few annotated Mn(II)-specific metalloenzymes in S. pneumoniae, including a Mn(II) SodA53, SapH54, PhpP, PpaC and CpsB55, we hypothesize that much of the cell-associated Mn(II) may be bioavailable in low molecular weight complexes as found in other bacteria.14 This may well protect the pneumococcus from endogenous and exogenous oxidative damage and from immune system killing.19 Modest Zn(II) toxicity is known to reduce cell-associated Mn(II) levels by ≈40% which gives rise to strong upregulation of the PsaR regulon;18,19,56 this suggests the formal possibility that under these conditions, Zn(II) effectively competes with Mn(II) and drives PsaR off the DNA as a result of its lower ΔGc (Fig. 5, Table 4). For this to happen inside of the cell, [Zn]free would only have to rise into the ≈0.01 µM range under these conditions; this seems possible given the modest (≈107–108 M−1) affinity of the zinc efflux regulator SczA for Zn(II) (G. Campanello and D. Giedroc, unpublished results).52 Direct measurement of [Zn]free is required to substantiate the proposal that excess intracellular Zn(II) causes mis-regulation of PsaR function in the cell.
The general finding of two pairs of functionally nonequivalent metal sites is also found in the Fe(II)-sensor DtxR. Here, an ordered or step-wise binding to a higher affinity ancillary site 1 is required for complete activation of DNA binding, but metal specificity is achieved by cognate metal binding to a lower affinity site 2 which incorporates a ligand analogous to that the D7 (M8 in DtxR) from the DNA binding domain.22 DtxR contains a Met (M8) and Cys (C102; often substituted with D in crystallographic studies) at positions corresponding to D7 and E99 in PsaR studied here; M8 is a clear metal specificity determinant when one compares DtxR and MntR using both structural and biological approaches.25,57 The molecular details of allosteric activation by Fe(II) or other divalent metal ions bound to site 2 have been the subject of much discussion since the original structure of the Ni(II)-activated C102D DtxR bound to the tox operator-promoter was reported,58 and may well involve a coil-to-helix transition in the α1 helix harboring M8 (D7 in PsaR; see Fig. 4).58 In MntR, Mn(II)-binding strongly stabilizes the α1 helix (which harbors two metal ligands to each of the A- and C-sites of the binuclear Mn(II)-cluster; see Fig. 6A) against H–D exchange relative to the apo-state59 and limits the conformational disorder of the DNA-binding domain relative to the dimerization domain.24 In any case, what seems clear is that direct coordination by M8 in DtxR/IdeR and D7 in MntR by the activating metal drives a global “caliper-like” closure of the DNA-binding domains in each subunit relative to one another (Supplementary Fig. S1), which brings them into alignment to bind to successive major grooves in the DNA operator.24,33,58,60,61 A globally similar mechanism almost certainly characterizes PsaR given the essentiality of D7 to allosteric activation of PsaO binding (Table 4; Fig. 5) and the finding that each empty site 2 of the “apo” structure of Sgo ScaR adopts a distinct structure with D7 making closer approach to the bound metal site 1 (Fig. 4). Also of interest is the early finding that the C-terminal SH3 domain of DtxR (not found in MntR) makes direct contact with site 1 metal ligands and the winged helical domain in the metal-activated DNA-bound form of DtxR60 thus potentially quantitatively influencing allosteric activation in that system. It is unclear what role the FeoA domain in PsaR might play in this process except to say that mutagenesis of metal site 1 ligand D160, found in the loop between the dimerization and FeoA domains, reduces the allosteric coupling free energy by ≈35% (Table 4).
Figure 6.
Comparison of the Mn(II)-bound chelate regions of wild-type MntR (2F5D)32 (A) and E11K MntR (4HX4)33 (B). The A and C Mn(II) ions are indicated in each panel and all residues are shown in stick representation. Coordination bonds are represented by black dashed lines, with potential electrostatic interactions deriving the εNH2 group of Lys11in E11K PsaR shown as green dashed lines in panel B. All residues are conserved in PsaR with identical residue numbers except that His77 is His76 in PsaR (see text for details). E11K MntR33 (B) is an excellent model for the Mn(II)-bound in site 2 in PsaR (this work).
The C-site Mn(II) complex in MntR is likely an excellent model for the activating site 2 in PsaR, since D8, E99, E102 and H103 in MntR precisely correspond to D7, E99, E102 and H103 in PsaR (Fig. 6). The coordination geometry of the C-site can be described as distorted octahedral, which involved monodentate coordination by each of these four side chains, in addition to the backbone carbonyl oxygen atom of E99 and a water molecule (Fig. 6A). Recent experiments reveal that the A-site metal cation can be functionally replaced by the ε-NH3+ group of Lys11 in a E11K mutant; further, substitution of the only non-bridging ligand to the A-site, H77, with Ala, still results in specific Mn(II)-dependent allosteric activation upon occupancy of the C-site (Fig. 6B). Strikingly, Sgo ScaR and Spn PsaR and closely related streptococcal Mn(II) sensors harbor a Lys residue in this position while the Fe(II) sensors DtxR and IdeR harbor an Arg here (Supplementary Fig. S1C); this would be predicted to destroy the A-site-like metal binding in PsaR, which is dependent on E99 and E102 bridging the A- and C-sites, with His76 completing the A-site coordination complex (Fig. 6A).33 These studies provide strong support for the conclusion that Spn PsaR is simply a C-site single Mn(II) variant of Bsu MntR (Fig. 6B).33 The strongly functionally compromised D7A and E99A PsaRs are fully consistent with this conclusion, with D7 Mn(II)-coordination absolutely required for the Mn(II)-dependent activation. Finally, it is interesting to note that H77 in MntR corresponds precisely to His76 in PsaR, and the analogous H76A and H76F PsaRs are functionally compromised, without donation of the metal-ligand bond to either metal, although to differing degrees (Fig. 5; Table 4). The location of the H76 side chain between site 1 and site 2 in PsaR suggests that it may function as an important part of a network that effectively transitions the PsaR homodimer to a high-affinity DNA binding state when Mn(II) is loaded into site 2 (Fig. 4).
Why is Zn(II) not a potent allosteric activator in PsaR? There is significant support for the hypothesis that distinct coordination environments of cognate vs. non-cognate metal complexes in metal sensor proteins of widely disparate structures is a key determinant of the functional selectivity in the cell.25,35,40,62–68 This may not, however, be the case for all metallosensors36,51,69 and in these cases, the influence of cellular metal bioavailability is likely a major factor that controls metal sensing so that illegitimate cross-talk between metal homeostasis systems can be avoided.2,51,57,70,71 This hypothesis predicts that Mn(II) would adopt a coordination geometry that is distinct from that of Zn(II). We do not yet know the coordination chemistries around either the Mn(II) or Zn(II)-bound regulatory site 2; however, it is the case that substitution of D7 (and E99) with Ala has a substantial ≥100-fold decrease in the Mn(II)-binding affinity, while the Zn(II)-binding affinity of site 2 is only minimally, if at all, affected (Table 2). This suggests the possibility that Zn(II) does not stably recruit D7 into a lower coordination number complex, making a “caliper-like” closure of the DNA-binding domains far less energetically favorable thus reducing ΔGc for Zn(II) over cognate Mn(II). It is the case that the Zn(II) affinity of site 2 in PsaR is quite modest (≈108 M−1) which might imply a smaller set of protein-derived ligands to this non-cognate metal. In Bsu MntR, the structure of Zn(II)-bound MntR reveals that the α1 helix is unwound and Asp7 is not coordinated to what would be the C-site Zn(II), since Zn(II) is bound only to the A-site in wild-type MntR (see Fig. 6A).32 The coordination structure of the Mn(II)-sensing complex is PsaR is not yet known, but likely adopts a six-coordinate C-site-like complex exactly like found in the binuclear wild-type and mononuclear E11K MntRs (Fig. 6A).32,33 EPR studies should allow us to determine this structure, with the mixed metal PsaRZn,Mn species an ideal target for these studies.72 High resolution crystallographic studies will be required to elucidate further the structural basis of allosteric regulation of PsaR and in particular, the key roles played by Asp7 and the “bridging” residue His76 is driving this allosteric switch, as well as its ability to distinguish Mn(II) from Fe(II) and thereby regulate cross-talk between these two metals in the cell.51,73 These future studies will have implications for understanding of how Mn(II) homeostasis impacts microbial physiology and virulence of this important human pathogen.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Crisjoe Joseph, Heidi Hu and Carolyn Carr from the Maroney laboratory for XAS data collection, Dr. Todd Stone for the training on and use of the DAWN HELEOS II and OptiLab rEX instruments at the Indiana University Physical Biochemistry Instrumentation Facility (PBIF) and Dr. Lichun Li, Indiana University, for training and use of the Biotek Synergy H1 Hybrid Multi-Mode Microplate reader.
Funding
Funding was provided by the National Institutes of Health grants GM042569 (D.P.G.), GM097225 (D.P.G.) and GM069696 (M.J.M.) and a fellowship awarded by the Graduate Training Program in Quantitative and Chemical Biology (QCB TP) at Indiana University (J.P.L.). The National Synchrotron Light Source at Brookhaven National Laboratory is supported by the U.S. Department of Energy, Division of Materials Sciences and Division of Chemical Sciences. Beamline X3B at NSLS is supported by the NIH. This publication was made possible by the Center for Synchrotron Biosciences grant, P30-EB-009998, from the National Institute of Biomedical Imaging and Bioengineering (NIBIB).
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting information
Supplementary Figures S1–S4 and Supplementary Tables S1 and S2. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal ions in biological catalysis: from enzyme databases to general principles. J Biol Inorg Chem. 2008;13:1205–1218. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- 2.Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460:823–830. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- 3.Klein JS, Lewinson O. Bacterial ATP-driven transporters of transition metals: physiological roles, mechanisms of action, and roles in bacterial virulence. Metallomics. 2011;3:1098–1108. doi: 10.1039/c1mt00073j. [DOI] [PubMed] [Google Scholar]
- 4.Ma Z, Jacobsen FE, Giedroc DP. Coordination Chemistry of Bacterial Metal Transport and Sensing. Chem Rev. 2009;109:4644–4681. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol. 2013;11:443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, Caprioli RM, Nacken W, Chazin WJ, Skaar EP. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 7.Kehl-Fie TE, Chitayat S, Hood MI, Damo S, Restrepo N, Garcia C, Munro KA, Chazin WJ, Skaar EP. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe. 2011;10:158–164. doi: 10.1016/j.chom.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kehl-Fie TE, Zhang Y, Moore JL, Farrand AJ, Hood MI, Rathi S, Chazin WJ, Caprioli RM, Skaar EP. MntABC and MntH contribute to systemic Staphylococcus aureus infection by competing with calprotectin for nutrient manganese. Infect Immun. 2013;81:3395–3405. doi: 10.1128/IAI.00420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Damo SM, Kehl-Fie TE, Sugitani N, Holt ME, Rathi S, Murphy WJ, Zhang Y, Betz C, Hench L, Fritz G, Skaar EP, Chazin WJ. Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc Natl Acad Sci U S A. 2013;110:3841–3846. doi: 10.1073/pnas.1220341110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anjem A, Varghese S, Imlay JA. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol Microbiol. 2009;72:844–858. doi: 10.1111/j.1365-2958.2009.06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sobota JM, Imlay JA. Iron enzyme ribulose-5-phosphate 3-epimerase in Escherichia coli is rapidly damaged by hydrogen peroxide but can be protected by manganese. Proc Natl Acad Sci U S A. 2011;108:5402–5407. doi: 10.1073/pnas.1100410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anjem A, Imlay JA. Mononuclear Iron Enzymes Are Primary Targets of Hydrogen Peroxide Stress. J Biol Chem. 2012;287:15544–15556. doi: 10.1074/jbc.M111.330365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Archibald FS, Fridovich I. Manganese, superoxide dismutase, and oxygen tolerance in some lactic acid bacteria. J Bacteriol. 1981;146:928–936. doi: 10.1128/jb.146.3.928-936.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daly MJ, Gaidamakova EK, Matrosova VY, Vasilenko A, Zhai M, Venkateswaran A, Hess M, Omelchenko MV, Kostandarithes HM, Makarova KS, Wackett LP, Fredrickson JK, Ghosal D. Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science. 2004;306:1025–1028. doi: 10.1126/science.1103185. [DOI] [PubMed] [Google Scholar]
- 15.Daly MJ, Gaidamakova EK, Matrosova VY, Vasilenko A, Zhai M, Leapman RD, Lai B, Ravel B, Li SM, Kemner KM, Fredrickson JK. Protein oxidation implicated as the primary determinant of bacterial radioresistance. PLoS Biol. 2007;5:e92. doi: 10.1371/journal.pbio.0050092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barnese K, Gralla EB, Cabelli DE, Valentine JS. Manganous phosphate acts as a superoxide dismutase. J Am Chem Soc. 2008;130:4604–4606. doi: 10.1021/ja710162n. [DOI] [PubMed] [Google Scholar]
- 17.Yesilkaya H, Andisi VF, Andrew PW, Bijlsma JJE. Streptococcus pneumoniae and reactive oxygen species: an unusual approach to living with radicals. Trends Microbiol. 2013;21:187–195. doi: 10.1016/j.tim.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Jacobsen FE, Kazmierczak KM, Lisher JP, Winkler ME, Giedroc DP. Interplay between manganese and zinc homeostasis in the human pathogen Streptococcus pneumoniae. Metallomics. 2011;3:38–41. doi: 10.1039/c0mt00050g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDevitt CA, Ogunniyi AD, Valkov E, Lawrence MC, Kobe B, McEwan AG, Paton JC. A molecular mechanism for bacterial susceptibility to zinc. PLoS Path. 2011;7:e1002357. doi: 10.1371/journal.ppat.1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papp-Wallace KM, Maguire ME. Manganese transport and the role of manganese in virulence. Ann Rev Microbiol. 2006;60:187–209. doi: 10.1146/annurev.micro.60.080805.142149. [DOI] [PubMed] [Google Scholar]
- 21.Lee JW, Helmann JD. Functional specialization within the Fur family of metalloregulators. Biometals. 2007;20:485–499. doi: 10.1007/s10534-006-9070-7. [DOI] [PubMed] [Google Scholar]
- 22.D'Aquino JA, Tetenbaum-Novatt J, White A, Berkovitch F, Ringe D. Mechanism of metal ion activation of the diphtheria toxin repressor DtxR. Proc Natl Acad Sci U S A. 2005;102:18408–18413. doi: 10.1073/pnas.0500908102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding X, Zeng H, Schiering N, Ringe D, Murphy JR. Identification of the primary metal ion-activation sites of the diphtheria tox repressor by X-ray crystallography and site-directed mutational analysis. Nat Struct Biol. 1996;3:382–387. doi: 10.1038/nsb0496-382. [DOI] [PubMed] [Google Scholar]
- 24.DeWitt MA, Kliegman JI, Helmann JD, Brennan RG, Farrens DL, Glasfeld A. The conformations of the manganese transport regulator of Bacillus subtilis in its metal-free state. J Mol Biol. 2007;365:1257–1265. doi: 10.1016/j.jmb.2006.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glasfeld A, Guedon E, Helmann JD, Brennan RG. Structure of the manganese-bound manganese transport regulator of Bacillus subtilis. Nat Struct Biol. 2003;10:652–657. doi: 10.1038/nsb951. [DOI] [PubMed] [Google Scholar]
- 26.Golynskiy M, Li S, Woods VL, Jr, Cohen SM. Conformational studies of the manganese transport regulator (MntR) from Bacillus subtilis using deuterium exchange mass spectrometry. J Biol Inorg Chem. 2007;12:699–709. doi: 10.1007/s00775-007-0216-z. [DOI] [PubMed] [Google Scholar]
- 27.Golynskiy MV, Davis TC, Helmann JD, Cohen SM. Metal-induced structural organization and stabilization of the metalloregulatory protein MntR. Biochemistry. 2005;44:3380–3389. doi: 10.1021/bi0480741. [DOI] [PubMed] [Google Scholar]
- 28.Golynskiy MV, Gunderson WA, Hendrich MP, Cohen SM. Metal Binding Studies and EPR Spectroscopy of the Manganese Transport Regulator MntR. Biochemistry. 2006;45:15359–15372. doi: 10.1021/bi0607406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lieser SA, Davis TC, Helmann JD, Cohen SM. DNA-binding and oligomerization studies of the manganese(II) metalloregulatory protein MntR from Bacillus subtilis. Biochemistry. 2003;42:12634–12642. doi: 10.1021/bi0350248. [DOI] [PubMed] [Google Scholar]
- 30.Moore CM, Gaballa A, Hui M, Ye RW, Helmann JD. Genetic and physiological responses of Bacillus subtilis to metal ion stress. Mol Microbiol. 2005;57:27–40. doi: 10.1111/j.1365-2958.2005.04642.x. [DOI] [PubMed] [Google Scholar]
- 31.Que Q, Helmann JD. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol Microbiol. 2000;35:1454–1468. doi: 10.1046/j.1365-2958.2000.01811.x. [DOI] [PubMed] [Google Scholar]
- 32.Kliegman JI, Griner SL, Helmann JD, Brennan RG, Glasfeld A. Structural basis for the metal-selective activation of the manganese transport regulator of Bacillus subtilis. Biochemistry. 2006;45:3493–3505. doi: 10.1021/bi0524215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGuire AM, Cuthbert BJ, Ma Z, Grauer-Gray KD, Brunjes Brophy M, Spear KA, Soonsanga S, Kliegman JI, Griner SL, Helmann JD, Glasfeld A. Roles of the A and C sites in the manganese-specific activation of MntR. Biochemistry. 2013;52:701–713. doi: 10.1021/bi301550t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stoll KE, Draper WE, Kliegman JI, Golynskiy MV, Brew-Appiah RA, Phillips RK, Brown HK, Breyer WA, Jakubovics NS, Jenkinson HF, Brennan RG, Cohen SM, Glasfeld A. Characterization and structure of the manganese-responsive transcriptional regulator ScaR. Biochemistry. 2009;48:10308–10320. doi: 10.1021/bi900980g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pennella MA, Shokes JE, Cosper NJ, Scott RA, Giedroc DP. Structural elements of metal selectivity in metal sensor proteins. Proc Natl Acad Sci U S A. 2003;100:3713–3718. doi: 10.1073/pnas.0636943100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reyes-Caballero H, Guerra AJ, Jacobsen FE, Kazmierczak KM, Cowart D, Koppolu UM, Scott RA, Winkler ME, Giedroc DP. The metalloregulatory zinc site in Streptococcus pneumoniae AdcR, a zinc-activated MarR family repressor. J Mol Biol. 2010;403:197–216. doi: 10.1016/j.jmb.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grossoehme NE, Giedroc DP. Illuminating allostery in metal sensing transcriptional regulators. Methods Mol Biol. 2012;875:165–192. doi: 10.1007/978-1-61779-806-1_8. [DOI] [PubMed] [Google Scholar]
- 38.Hanlon DP, Watt DS, Westhead EW. The interaction of divalent metal ions with tris buffer in dilute solution. Anal Biochem. 1966;16:225–233. doi: 10.1016/0003-2697(66)90150-3. [DOI] [PubMed] [Google Scholar]
- 39.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 40.Campanello GC, Ma Z, Grossoehme NE, Guerra AJ, Ward BP, DiMarchi RD, Ye Y, Dann Iii CE, Giedroc DP. Allosteric Inhibition of a Zinc-Sensing Transcriptional Repressor: Insights into the Arsenic Repressor (ArsR) Family. J Mol Biol. 2013;425:1143–1157. doi: 10.1016/j.jmb.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leitch S, Bradley MJ, Rowe JL, Chivers PT, Maroney MJ. Nickel-specific response in the transcriptional regulator, Escherichia coli NikR. J Am Chem Soc. 2007;129:5085–5095. doi: 10.1021/ja068505y. [DOI] [PubMed] [Google Scholar]
- 42.Padden KM, Krebs JF, MacBeth CE, Scarrow RC, Borovik AS. Immobilized metal complexes in porous organic hosts: development of a material for the selective and reversible binding of nitric oxide. J Am Chem Soc. 2001;123:1072–1079. doi: 10.1021/ja003282b. [DOI] [PubMed] [Google Scholar]
- 43.Webb SM. SIXpack: A graphical user interface for XAS analysis using IFEFFIT. Physica Scripta. 2005;T115:1011–1014. [Google Scholar]
- 44.Ankudinov AL, Ravel B, Rehr JJ, Conradson SD. Real-space multiple-scattering calculation and interpretation of x-ray-absorption near-edge structure. Phys Rev B. 1998;58:7565–7576. [Google Scholar]
- 45.Zabinsky SI, Rehr JJ, Ankudinov A, Albers RC, Eller MJ. Multiple-scattering calculations of x-ray-absorption spectra. Phys Rev B Cond Matt. 1995;52:2995–3009. doi: 10.1103/physrevb.52.2995. [DOI] [PubMed] [Google Scholar]
- 46.Gasper R, Scrima A, Wittinghofer A. Structural insights into HypB, a GTP-binding protein that regulates metal binding. J Biol Chem. 2006;281:27492–27502. doi: 10.1074/jbc.M600809200. [DOI] [PubMed] [Google Scholar]
- 47.Costello AL, Sharma NP, Yang KW, Crowder MW, Tierney DL. X-ray absorption spectroscopy of the zinc-binding sites in the class B2 metallo-beta-lactamase ImiS from Aeromonas veronii bv. sobria. Biochemistry. 2006;45:13650–13658. doi: 10.1021/bi061547e. [DOI] [PubMed] [Google Scholar]
- 48.Costello A, Periyannan G, Yang KW, Crowder MW, Tierney DL. Site-selective binding of Zn(II) to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia. J Biol Inorg Chem. 2006;11:351–358. doi: 10.1007/s00775-006-0083-z. [DOI] [PubMed] [Google Scholar]
- 49.Herbst RW, Guce A, Bryngelson PA, Higgins KA, Ryan KC, Cabelli DE, Garman SC, Maroney MJ. Role of conserved tyrosine residues in NiSOD catalysis: a case of convergent evolution. Biochemistry. 2009;48:3354–3369. doi: 10.1021/bi802029t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacquamet L, Aberdam D, Adrait A, Hazemann JL, Latour JM, Michaud-Soret I. X-ray absorption spectroscopy of a new zinc site in the fur protein from Escherichia coli. Biochemistry. 1998;37:2564–2571. doi: 10.1021/bi9721344. [DOI] [PubMed] [Google Scholar]
- 51.Ma Z, Faulkner MJ, Helmann JD. Origins of specificity and cross-talk in metal ion sensing by Bacillus subtilis Fur. Mol Microbiol. 2012;86:1144–1155. doi: 10.1111/mmi.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reyes-Caballero H, Campanello GC, Giedroc DP. Metalloregulatory proteins: metal selectivity and allosteric switching. Biophys Chem. 2011;156:103–114. doi: 10.1016/j.bpc.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yesilkaya H, Kadioglu A, Gingles N, Alexander JE, Mitchell TJ, Andrew PW. Role of manganese-containing superoxide dismutase in oxidative stress and virulence of Streptococcus pneumoniae. Infect Immun. 2000;68:2819–2826. doi: 10.1128/iai.68.5.2819-2826.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang Y-L, Zhang J-W, Yu W-L, Cheng W, Zhang C-C, Frolet C, Di Guilmi A-M, Vernet T, Zhou C-Z, Chen Y. Structural and Enzymatic Characterization of the Streptococcal ATP/Diadenosine Polyphosphate and Phosphodiester Hydrolase Spr1479/SapH. J Biol Chem. 2011;286:35906–35914. doi: 10.1074/jbc.M111.228585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morona JK, Morona R, Miller DC, Paton JC. Streptococcus pneumoniae capsule biosynthesis protein CpsB is a novel manganese-dependent phosphotyrosine-protein phosphatase. J Bacteriol. 2002;184:577–583. doi: 10.1128/JB.184.2.577-583.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kloosterman TG, Witwicki RM, van der Kooi-Pol MM, Bijlsma JJ, Kuipers OP. Opposite effects of Mn2+ and Zn2+ on PsaR-mediated expression of the virulence genes pcpA, prtA, and psaBCA of Streptococcus pneumoniae. J Bacteriol. 2008;190:5382–5393. doi: 10.1128/JB.00307-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guedon E, Helmann JD. Origins of metal ion selectivity in the DtxR/MntR family of metalloregulators. Mol Microbiol. 2003;48:495–506. doi: 10.1046/j.1365-2958.2003.03445.x. [DOI] [PubMed] [Google Scholar]
- 58.White A, Ding X, vanderSpek JC, Murphy JR, Ringe D. Structure of the metal-ion-activated diphtheria toxin repressor/tox operator complex. Nature. 1998;394:502–506. doi: 10.1038/28893. [DOI] [PubMed] [Google Scholar]
- 59.Golynskiy M, Li S, Woods VL, Jr, Cohen SM. Conformational studies of the manganese transport regulator (MntR) from Bacillus subtilis using deuterium exchange mass spectrometry. J. Biol. Inorg. Chem. 2007;12:699. doi: 10.1007/s00775-007-0216-z. [DOI] [PubMed] [Google Scholar]
- 60.Pohl E, Holmes RK, Hol WG. Crystal structure of a cobalt-activated diphtheria toxin repressor-DNA complex reveals a metal-binding SH3-like domain. J Mol Biol. 1999;292:653–667. doi: 10.1006/jmbi.1999.3073. [DOI] [PubMed] [Google Scholar]
- 61.Pohl E, Holmes RK, Hol WG. Crystal structure of the iron-dependent regulator (IdeR) from Mycobacterium tuberculosis shows both metal binding sites fully occupied. J Mol Biol. 1999;285:1145–1156. doi: 10.1006/jmbi.1998.2339. [DOI] [PubMed] [Google Scholar]
- 62.Changela A, Chen K, Xue Y, Holschen J, Outten CE, O'Halloran TV, Mondragon A. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science. 2003;301:1383–1387. doi: 10.1126/science.1085950. [DOI] [PubMed] [Google Scholar]
- 63.Pennella MA, Arunkumar AI, Giedroc DP. Individual metal ligands play distinct functional roles in the zinc sensor Staphylococcus aureus CzrA. J Mol Biol. 2006;356:1124–1136. doi: 10.1016/j.jmb.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 64.Leitch S, Bradley MJ, Rowe JL, Chivers PT, Maroney MJ. Nickel-Specific Response in the Transcriptional Regulator, Escherichia coli NikR. J Am Chem Soc. 2007;129:5085–5095. doi: 10.1021/ja068505y. [DOI] [PubMed] [Google Scholar]
- 65.Ma Z, Cowart DM, Ward BP, Arnold RJ, DiMarchi RD, Zhang L, George GN, Scott RA, Giedroc DP. Unnatural amino acid substitution as a probe of the allosteric coupling pathway in a mycobacterial Cu(I) sensor. J Am Chem Soc. 2009;131:18044–18045. doi: 10.1021/ja908372b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ma Z, Cowart DM, Scott RA, Giedroc DP. Molecular insights into the metal selectivity of the copper(I)-sensing repressor CsoR from Bacillus subtilis. Biochemistry. 2009;48:3325–3334. doi: 10.1021/bi900115w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee CW, Chakravorty DK, Chang FM, Reyes-Caballero H, Ye Y, Merz KM, Jr, Giedroc DP. Solution structure of Mycobacterium tuberculosis NmtR in the apo state: insights into Ni(II)-mediated allostery. Biochemistry. 2012;51:2619–2629. doi: 10.1021/bi3001402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trepreau J, Girard E, Maillard AP, de Rosny E, Petit-Haertlein I, Kahn R, Coves J. Structural basis for metal sensing by CnrX. J Mol Biol. 2011;408:766–779. doi: 10.1016/j.jmb.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 69.Foster AW, Patterson CJ, Pernil R, Hess CR, Robinson NJ. Cytosolic Ni(II) sensor in cyanobacterium: nickel detection follows nickel affinity across four families of metal sensors. J Biol Chem. 2012;287:12142–12151. doi: 10.1074/jbc.M111.338301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Waldron KJ, Robinson NJ. How do bacterial cells ensure that metalloproteins get the correct metal? Nat Rev Microbiol. 2009;7:25–35. doi: 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
- 71.Ma Z, Lee JW, Helmann JD. Identification of altered function alleles that affect Bacillus subtilis PerR metal ion selectivity. Nucl Acids Res. 2011;39:5036–5044. doi: 10.1093/nar/gkr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hayden JA, Brophy MB, Cunden LS, Nolan EM. High-affinity manganese coordination by human calprotectin is calcium-dependent and requires the histidine-rich site formed at the dimer interface. J Am Chem Soc. 2013;135:775–787. doi: 10.1021/ja3096416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Puri S, Hohle TH, O'Brian MR. Control of bacterial iron homeostasis by manganese. Proc Natl Acad Sci U S A. 2010;107:10691–10695. doi: 10.1073/pnas.1002342107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.