Abstract
Glucokinase (GCK) is responsible for maintaining glucose homeostasis in the human body. Dysfunction or misregulation of GCK causes hyperinsulinemia, hypertriglyceridemia and type 2 diabetes. In the liver GCK is regulated by interaction with the glucokinase regulatory protein (GKRP), a 68-kDa polypeptide that functions as a competitive inhibitor of glucose binding to GCK. Formation of the mammalian GCK-GKRP complex is stimulated by fructose 6-phosphate and antagonized by fructose 1-phosphate. Here we report the crystal structure of the mammalian GCK-GKRP complex in the presence of fructose 6-phosphate at a resolution of 3.50 Å The interaction interface, which totals 2060 Å2 of buried surface area, is characterized by a small number of polar contacts and substantial hydrophobic interactions. The structure of the complex reveals the molecular basis of disease states associated with impaired regulation of GCK by GKRP. It also offers insight into the modulation of complex stability by sugar phosphates. The atomic description of the mammalian GCK-GKRP complex provides a framework for the development of novel diabetes therapeutic agents that disrupt this critical macromolecular regulatory unit.
Keywords: Glucokinase, Glucokinase Regulatory Protein, Type 2 Diabetes
Glucokinase (GCK) functions as the body’s principal glucose sensor and plays a central role in glucose homeostasis.1, 2 GCK catalyzes the rate-limiting step of glucose metabolism in the pancreas, where it governs the rate of insulin secretion from β-cells. It also displays a high flux control coefficient upon glycogen synthesis in the liver.3 In both tissues GCK is regulated by a unique cooperative kinetic response to glucose characterized by a K0.5 value that approximates physiological plasma glucose levels.4 GCK cooperativity results from slow, glucose-mediated order-disorder transitions within the enzyme’s intrinsically disordered small domain.5, 6 In the liver GCK is regulated by the glucokinase regulatory protein (GKRP), a 68-kDa polypeptide that functions as a competitive inhibitor of glucose binding to GCK.7, 8 Under low glucose concentrations GCK associates with GKRP and the inactive complex is recruited to the hepatocyte nucleus.9, 10 When glucose levels rise, the GCK-GKRP complex dissociates and GCK returns to the cytosol to participate in glycogen metabolism. In mammals, formation of the complex is promoted by fructose 6-phosphate and antagonized by fructose 1-phosphate.
The GCK-GKRP system is essential for proper glucose sensing and regulation in humans. Inactivating mutations in the gck locus, of which more than 600 have been identified, are linked to maturity onset diabetes of the young type 2 (MODY-II) and permanent neonatal diabetes mellitus (PNDM).11 Conversely, gain-of-function gck mutations result in persistent hyperinsulinemic hypoglycemia of infancy (PHHI).12 Genome-wide association studies have identified polymorphisms in the gckr locus strongly associated with hyperlipidemia and altered triglyceride concentrations.13, 14 GKRP knockout mice display decreased hepatic glucokinase expression and impaired glucose clearance.15 Due to the importance of GCK and GKRP in glucose homeostasis, significant efforts have been directed toward targeting this system for anti-diabetic therapy. Allosteric activators of GCK that stimulate enzyme activity in vivo have been actively pursued as potential drugs for type 2 diabetes mellitus.16, 17 More recently, targeted disruption of the GCK-GKRP complex has emerged as a promising avenue to increase glucose metabolism in a hepatocyte specific manner.18 A major limitation in targeting the GCK-GKRP assembly with anti-diabetic agents, however, is the lack of a description of the complex at atomic resolution. Here we present the first X-ray crystallographic structure of the mammalian GCK-GKRP complex. Our data provide a framework for understanding the molecular details of complex assembly and for reprogramming the GCK-GKRP regulatory system with small-molecule therapeutics.
Experimental Procedures
Protein Production and Purification
Recombinant, N-terminal hexahistidine tagged pancreatic human glucokinase was produced in BL21(DE3) cells and purified using a combination of immobilized metal affinity and size-exclusion chromatography, as previously described.19 Recombinant, C-terminal hexahistidine tagged rat GKRP was produced from pET22(b) in BL21(DE3) cells grown at 37°C until OD600 nm reached 0.5, at which time the temperature was reduced to 16°C and IPTG was added to a final concentration of 0.5 mM. Following 40 hours of incubation at 16°C, cells were harvested by centrifugation and resuspended in GKRP loading buffer (50 mM Tris-HCl pH 7.6, 25 mM imidazole, 1 mM dithiothreitol and 10% w/v glycerol). Cells were lysed by sonication, clarified by centrifugation at 35,000g for 30 min at 4°C and the supernatant was loaded onto a 5 mL HisTrap FF affinity column (GE Lifesciences) previously equilibrated in GKRP loading buffer. The column was washed with 100 mL of GKRP loading buffer and protein was eluted with 40 mL of GKRP loading buffer containing 0.25 M imidazole. GKRP was dialyzed overnight at 4°C in a 14,000 MWCO dialysis bag against 2 L of GKRP SEC buffer (50 mM potassium phosphate, pH 7.6 containing 1 mM dithiothreitol). Following dialysis, GKRP was concentrated to a final volume of ~0.5 mL with an Amicon 30,000 MWCO spin concentrator, and the protein was injected onto a Superdex 200 HR 10/30 column (24 mL), previously equilibrated in GKRP SEC buffer, at a flow rate of 0.25 mL/min. Pure GKRP eluted as a single peak centered at 13.5 mL (Figure S1).
To form the complex, aliquots of size-exclusion purified GCK and GKRP were mixed at a 1:1.1 stoichiometry. Fructose 6-phosphate (3 mM) was added to this mixture and the sample was incubated 10 min at 4°C to promote complex formation. The complex was concentrated to a final volume of 0.5 mL using an Amicon 30,000 MWCO spin concentrator and the sample was immediately injected at a flow rate of 0.25 mL/min onto a Superdex 200 HR 10/30 column (24 mL) previously equilibrated in GCK/GKRP SEC buffer (10 mM HEPES pH 7.1, 25 mM KCl, 3 mM dithiothreitol and 3 mM fructose 6-phosphate). Pure GCK-GKRP-fructose 6-phosphate ternary complex eluted at 11.9 mL (Figure S1). Fractions displaying the highest A280 nm values were pooled and concentrated to 10 mg/mL using an Amicon 30,000 MWCO spin concentrator. Samples were used immediately in crystallization trials.
Crystallization
Suitable crystallization conditions were identified using commercial sparse matrix screening conducted at the NCCR Structural Biology facility at the University of Zurich. Initial hits were optimized by grid screens. Crystals were obtained at 20°C using the sitting drop vapor diffusion method by mixing 200 nL of GCK-GKRP-fructose 6-phosphate complex solution with 100 nL of reservoir solution containing 0.1 M HEPES, pH 7.1 and 8% w/v PEG 6000. Crystals typically appeared within 12 hours of setup and reached final dimensions within 3 days (Figure S1). After adding 200 nL of mother liquor supplemented with 35% w/v glycerol and 3 mM fructose 6-phosphate to the crystallization drop, crystals were flash frozen in liquid N2.
Structure Determination and Refinement
Diffraction data were collected at 100 K on the PX-I beamline at the Swiss Light Source (Villigen, CH). Data were processed with XDS20 and scaled with SADABS21. The structure was solved with Phaser22 using a combined search with three different ensembles: (1) the isolated large domain of GCK from the open structure (1V4T), (2) the isolated small domain of GCK from the open structure (1V4T), and (3) the “capless” GKRP structure (4BB9). In the search model, GCK was separated into its component domains to omit bias concerning the relative orientation of these domains with respect to one another in the search model. A representative electron density map generated by molecular replacement using only the large domain from the open GCK (1V4T) and the “capless” GKRP structure (4BB9) as a search model is shown in Figure S2. The electron density clearly indicates that the small domain of GCK adopts a conformation similar to the open form. Use of the intact, closed structure for GCK (3F9M) as a search model produced a model of poor quality (low log likelihood gain score and six packing clashes in Phaser, which did not occur in the combined search with the three ensembles as described above).
Initial automatic and manual rebuilding was carried out with PHENIX Autobuild25 and Coot26. Initial refinement was carried out with Refmac27 within the CCP4 suite28, using Prosmart29 to generate external restraints based on reference structures for closed GCK (3F9M) and GKRP (4BB9). For refinement, we chose the closed GCK structure since it was determined to higher resolution and displays improved geometrical characteristics. It is important to recognize that the use of ProSMART based restraints introduces bias into the model. To minimize this bias, we chose reference models with high quality statistics and high degrees of sequence identity with our target (100% for GCK and 89% for GKRP). Additionally, we generated reference model restraints only for the main chain to minimize bias for the position of the side chains. We inspected the fit of the model to the electron density thoroughly to ensure that the reference model restraints did not distort our model, especially in the regions of structural difference. The map sharpening utility in Refmac29 was crucial for building the N-terminus and cap domain in GKRP and residues 180–205 in GCK. This program was also used to prepare the omit map for Figures 1B. Subsequent refinement was carried out with PHENIX25, using structures of closed GCK (3F9M) and GKRP (4BB9) as reference models. The model was improved by iterative rounds of refinement and manual rebuilding. The N- and C-termini are disordered for both proteins, as are residues 151–180 in GCK and three surface loops (residues 282–285, 368–392 and 428–429) in GKRP. Distribution of thermal displacement parameters is shown in Figure S4. UNIPROT accession numbers for GCK and GKRP used in this study are P35557 and Q07071, respectively. Data collection statistics and refinement details are shown in Table S1. Resolution estimation was performed by using an I/sigma(I) criterion of 2 (3.50 Å). Additionally, the split half correlation CC(1/2) criterion was used to avoid discarding highly important data.30 Thus, data up to 3.40 Å were used for refinement. Data statistics and refinement in Table S1 are based on this data cut-off. A model of fructose 6-phosphate was fit into the electron density in a similar location to the position of fructose 1-phosphate in the isolated GKRP structure (4BB924). At the sugar-binding site, two sites with clear electron density above 1.5 σ are observed (Figure 1B). The phosphate group was placed into the site with the more pronounced electron density, in a similar position as the phosphate moiety of fructose 1-phosphate in the isolated GKRP structure (Figure S5). The remaining electron density was more consistent with the furanose form of fructose 6-phosphate, rather than the linear keto form.31 The electron density did not allow a clear positioning of the furanose ring, thus an orientation similar to the fructose 1-phosphate position was picked. This procedure is unavoidably biased in favor of the high-resolution fructose 1-phosphate structure; however the high degree of structural similarity between the reference (fructose 1-phosphate) and model (fructose 6-phosphate) ligands provides a reasonable rationale for this decision.
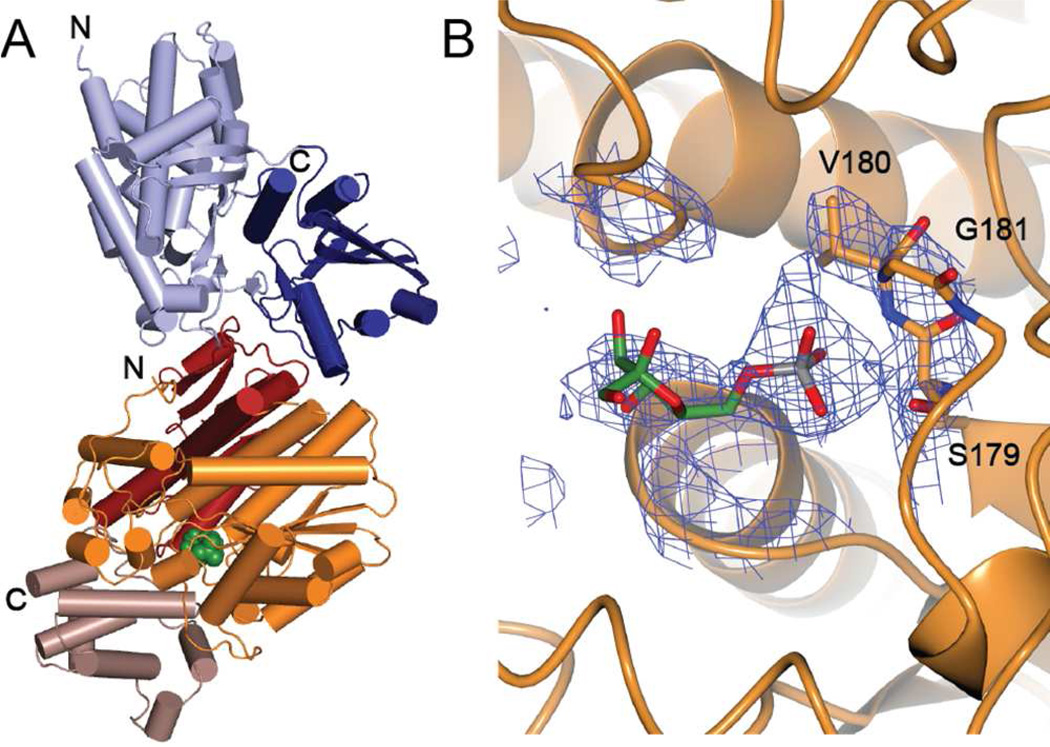
Figure 1.
Structure of the mammalian GCK-GKRP-fructose 6-phosphate complex. (A) Cartoon representation of the complex with GCK depicted in light blue (large domain) and dark blue (small domain), and GKRP depicted in orange (sugar isomerase domain 1), red (sugar isomerase domain 2) and brown (cap). Fructose 6-phosphate (green, space-filling) binds where the cap meets the sugar isomerase domains. (B) Fructose 6-phosphate binding site with carbon, oxygen, nitrogen and phosphorus atoms shown in green, red, blue and grey, respectively. Electron density is contoured at 1.5 σ, clipped at 5 Å around the ligand and calculated without the ligand (omit map). Residues that contact the phosphoryl moiety are labeled.
Model validation was carried out with the Molprobity server32, which indicated that 95.2% residues are in the most favored regions of the Ramachandran plot and 4.6% are in the additionally allowed regions, with only two outliers (0.2 %); Molprobity score: 1.57 (this structure is in the 100th percentile for N=638, 3.400Å ± 0.25 Å). The programs PyMOL (DeLano Scientific LLC) and CCP4MG28 were employed for figure preparation.
Determination of Interaction Interfaces
Interaction interfaces were determined with the PDB PISA server.33 The main interface is located within the asymmetric unit and the interface area is calculated as 2060 Å2; it involves 32 residues of GCK and 34 residues of GKRP (for details see text). For the second interaction interface present in the crystal structure, the main interactions are found between the N-terminus (residues 7–8, 17–20) and residues 63–67 of GKRP and residues 103–109 and 452–460 of a GCK molecule related by crystallographic symmetry (x−1/2, −y+1/2, −z+1). In the present structure, residues 7–27 of GKRP adopt a new conformation compared to the isolated GKRP structure, where it contacts the cap domain.24 Since the cap domain adopts a different conformation in the GCK-GKRP complex (now the cap is ajar), the N-terminus of GKRP has to reorient (Figure S6). Therefore it is very likely that the present orientation of the N-terminus of GKRP found in the GCK-GKRP complex structure is due to crystallization contacts. The second interface as determined by the PISA server involves 23 residues of GCK and 16 of GKRP with a total buried area of 1210 Å2, which is ~50% of the area of the main interface. The interface is composed of only one hydrogen bond (GCK Met107 carbonyl O with amide NH of GKRP Gln8) and one salt bridge (GCK His105 with GKRP Glu18) and several hydrophobic contacts.
Results
Structure of the Mammalian GCK-GKRP Complex
To understand the basis for GKRP-mediated inhibition of GCK, we determined the crystal structure of the complex between rat liver GKRP and human pancreatic GCK in the presence of fructose 6-phosphate (Figure 1). We chose rat GKRP, which is 89% identical to human GKRP, because this protein behaves similarly to the human protein and can be produced in large quantities from recombinant sources, unlike human GKRP. The biochemical properties of rat GKRP have been thoroughly characterized and sugar phosphates modulate the inhibitory action of rat GKRP, as they do for human GKRP. Human pancreatic GCK, which differs from the liver isozyme only by 15 N-terminal residues uninvolved in GKRP association8, was selected because it proved amenable to crystallization. Using immobilized metal affinity and size-exclusion chromatography we successfully prepared highly purified samples of isolated rat GKRP and human GCK. Incubation of these proteins in the presence of fructose 6-phosphate resulted in the formation of a stable complex with an estimated molecular mass of 120 kDa (Figure S1). Following purification of the complex via size-exclusion chromatography, the GCK-GKRP complex was crystallized and the structure was determined to a resolution of 3.50 Å Crystallization conditions, data collection statistics and refinement details are provided in supporting information. The final model contains residues 12–461 of GCK and 7–605 of GKRP. N- and C-termini of both proteins are disordered, as are GCK residues 151–180 and three GKRP surface loops containing residues 282–285, 368–392 and 428–429, respectively. Although the structure of the complex was determined to modest resolution, the model displays excellent geometrical characteristics (Ramachandran plot, Molprobity score), compared with other structures at similar resolution. High-resolution crystal structures of each isolated protein are available23, 24 and inclusion of restraints based on these reference models ensured the quality of the final model of the GCK-GKRP complex. The structure has been deposited with the RCSB PDB with accession code 4LC9.
The structure of the mammalian GCK-GKRP complex is consistent with previous crystallographic studies of each isolated protein.23, 24 GCK adopts a prototypic hexokinase fold consisting of a large and a small domain separated by a variable opening angle. GKRP contains three domains – two sugar isomerase folds (SIS1 and SIS2) formed from flavodoxin nucleotide binding motifs, and a C-terminal alpha helical cap domain. The structure of the complex confirms that GCK and GKRP form a heterodimeric assembly with a 1:1 stoichiometry. The structure also reveals a single fructose 6-phosphate binding site located within GKRP that overlaps with the binding site for fructose 1-phosphate.24, 34 Specifically, fructose 6-phosphate binds in a pocket near the interface of the two sugar isomerase domains, the entrance to which is partially excluded from solvent by the alpha helical cap domain (Figure 1A). The electron density map supports binding of fructose 6-phosphate in the furanose form rather than in the less populated, linear keto conformation (Figure 1B).31, 35 The furanose form is also supported by the 1.47 Å structure of human GKRP in complex with fructose 1-phosphate, which unambiguously shows the carbohydrate bound in the ring-closed conformation.24 The possibility remains that the electron density observed in the sugar phosphate binding site represents a species other than fructose 6-phosphate. A logical alternate ligand would be inorganic phosphate, which was included in the first step of the purification procedure of GKRP. This appears unlikely, however, because fructose 6-phosphate is expected to bind more tightly to GKRP compared to phosphate. In addition, phosphate was excluded, but fructose 6-phosphate was included, in the size exclusion buffer used in the second step of complex purification.
Mechanism of Competitive Inhibition of GCK by GKRP
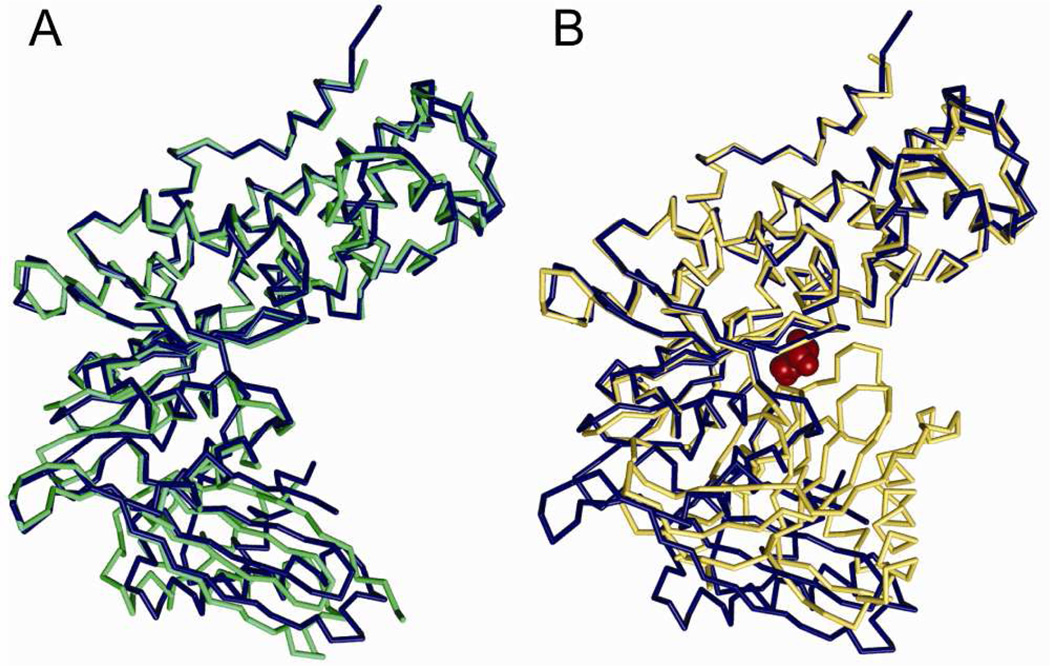
The structure of the complex provides an explanation for the observation that GKRP acts as a competitive inhibitor of glucose binding to GCK. When bound to GKRP, GCK adopts a conformation closely resembling one previously observed in the crystal structure of the unliganded enzyme.23 In this conformation, termed the “super-open” state, the large and small domains are separated by a large opening angle and the binding site for glucose is absent. In the GCK-GKRP complex, the opening angle is slightly smaller than previously observed for unliganded GCK (Figure 2A). This small degree of closure is reflected in a ~10° movement of the central β-sheet and the α2-α3 helices of the small domain toward the large domain. By contrast, glucose association with GCK produces a much larger conformational change involving a 99° closure of the cleft between the large and small domains (Figure 2B).23
Figure 2.
Structure of GCK. (A) GKRP acts as a competitive inhibitor of glucose by binding to the “super-open” conformation of GCK (blue), which differs from the unliganded GCK structure (green, PDB 1V4T) by a subtle ~10° closure of the opening angle between the large and small domains. (B) Comparison of GCK as found in the complex (blue) with the “closed” conformation of GCK (yellow, PDB 1V4S) in the presence of glucose (red).
The GCK active site loop comprised of residues 151–180 is disordered in the crystal structure of unliganded GCK.23 This loop undergoes major structural reconfiguration upon glucose binding, adopting a β-hairpin structure. Dynamic reorganization of this structural element is linked to the allosteric regulation and kinetic cooperativity of GCK.6 Interestingly, in the structure of the GCK-GKRP complex, the 151–180 loop remains disordered. The retention of a dynamic, solvent accessible active site loop following association with GKRP likely facilitates glucose-mediated disruption of the inhibitory complex. This feature is required for GCK activation and relocalization to the hepatocyte cytosol upon glucose stimulation.
GCK-GKRP Interaction Interface
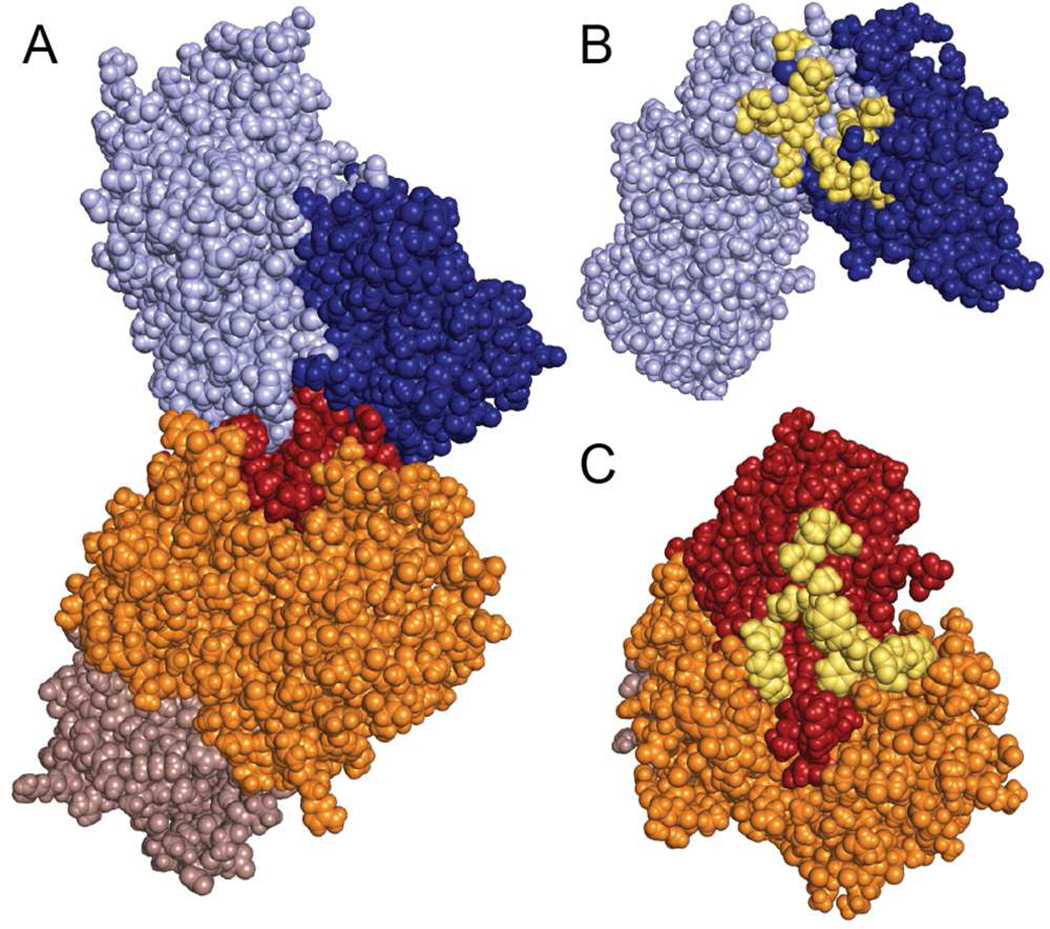
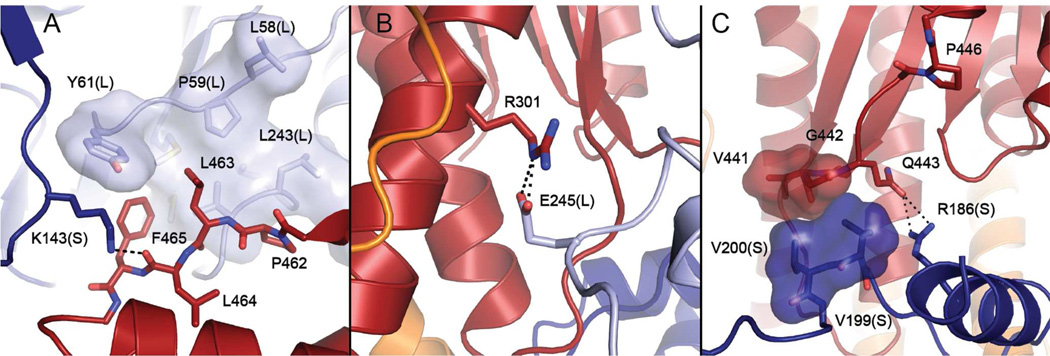
The interaction between GCK and GKRP reveals a large buried surface area, characterized by a limited number of polar and coulombic contacts (Figure S3). The protein-protein interface totals 2060 Å2 of buried surface area and includes a substantial number by hydrophobic contacts (Figure 3), consistent with the results of prior calorimetric studies.36 The magnitude of the buried surface area observed in the GCK-GKRP complex structure is similar to that observed in the structures of other protein-protein recognition sites.37 The structure confirms previous investigations identifying residues 141–144, located in the GCK small domain, as important for GKRP binding. Site-directed mutagenesis of residues 141–144 produces up to a 250-fold reduction in affinity of GCK for GKRP.38 In the complex, residues 141–144 are located on a loop that contacts GKRP. The side chain of one loop residue, Lys143, forms a hydrogen bond with the backbone carbonyl of GKRP residue Leu464 (Figure 4A). This interaction is reinforced by hydrophobic contacts involving a cleft on GCK, comprised of Leu47, Leu58, Pro59, Tyr61, Met238, Leu243, Val244 and Met251 from the large domain, which forms van der Waals contacts with a protruding loop of GKRP consisting of residues 462–470. Adjacent to this hydrophobic contact is a coulombic interaction between the side chain of Glu245 from GCK and Arg301 from GKRP (Figure 4B). A second potential coulombic interaction is nearby, involving Asp247 from GCK and Arg297 from GKRP. A smaller hydrophobic contact interface exists between Val199 and Val200 of GCK and Val441 and Gly442 from GKRP (Figure 4C). Notably, a conserved segment of GKRP spanning Leu181–Gly188, which was previously identified as an important GCK recognition element, is distant from the interface.39
Figure 3.
GCK-GKRP interaction interface. (A) Space-filling representation of the complex with GCK depicted in light blue (large domain) and dark blue (small domain), and GKRP depicted in orange (SIS1), red (SIS2) and brown (cap). (B) Surface representation of GCK and (C) GKRP, following a 120 degree rotation of each protein, with interacting residues highlighted in yellow.
Figure 4.
GCK-GKRP contact regions. (A) A hydrophobic cleft on the GCK large domain (light blue, L) consisting of Leu47, Leu58, Pro59, Tyr61, Met238, Leu243, and Val244 interacts with GKRP loop residues Pro462-Phe465 (red). These contacts are reinforced by a hydrogen bond between Lys143 from the GCK small domain (dark blue, S) and the backbone carbonyl group of Leu464. (B) The interaction interface includes a coulombic interaction between Glu245 from GCK and Arg301 from GKRP. (C) A hydrophobic contact between GCK small domain residues Val199 and Val200 and GKRP residues Val441 and Gly442 is reinforced by an adjacent hydrogen bond between Arg186 of GCK and Gln443 of GKRP. Located nearby is Pro446, substitution of which impairs GKRP-mediated regulation of GCK and is linked to hypertriglyceridemia.
Discussion
Implications of the GCK-GKRP Complex Structure for Human Disease
The structure of the mammalian GCK-GKRP complex provides insight into the molecular basis of metabolic disorders associated with impaired GCK regulation by GKRP. Genome-wide association studies reveal a link between single nucleotide polymorphisms in the gckr locus and the development of disease states associated with glucose homeostatic dysfunction.13, 14 The most common clinically observed GKRP variant is Pro446Leu, which is strongly correlated with elevated triglyceride levels and an increased risk of cardiovascular disease.40 Biochemical characterization of the Pro446Leu variant reveals reduced GCK inhibition and decreased GKRP-mediated nuclear sequestration of the enzyme.41, 42 The GCK-GKRP complex structure shows that Pro446 is located at the C-terminal end of a loop that interacts with GCK (Figure 4C). In particular, the side chain of Gln443, located within this loop, forms a putative hydrogen bond with the side chain of Arg186 of GCK. In addition, GKRP loop residues Val441 and Gly442 form hydrophobic interactions with GCK residues Val199 and Val200 (vide supra). Based on this data, we postulate that substitution of Pro446 alters the structure of the loop, negatively impacting this contact site and resulting in decreased GKRP inhibitory action.
The other disease-associated GKRP mutations, which are rare in human populations40, are distributed throughout the GKRP structure. These substitutions do not appear at the GCK-GKRP interface, nor are they located in the sugar phosphate binding site. This observation suggests a more complex mode of action for these mutations. GKRP is also modified at Ser481 via phosphorylation by AMP-activated protein kinase, which impairs GKRP-mediated inhibition and translocation of GCK.43 The structure demonstrates that Ser481 is buried in the interior of GKRP, at a location more than 12 Å from the GCK interface. This information suggests that phosphorylation of Ser481 does not impact GKRP-mediated regulation of GCK by directly disrupting the protein-protein interface. It also raises questions about the physiological role of this post-translational modification.
Several hyperactive pancreatic GCK variants linked to PHHI are located near the GKRP interface. For example, single amino acid substitutions of Ser64, Thr65 and Gly68 produce an activated GCK that displays a reduced glucose K0.5 value.11 These three residues are located in close proximity to Leu58, Pro59 and Tyr61, which form part of the hydrophobic cleft of GCK that contacts the 462–470 loop from GKRP (Figure 4A). Similarly, the Met197Val and Met197Ile mutations have been shown to stimulate GCK activity by increasing the enzyme’s sensitivity to glucose.44, 45 Met197 is located immediately upstream of Val199 and Val200, two residues that form a second hydrophobic patch on GCK that interacts with GKRP (Figure 4C). Although the phenotypic consequences associated with substitutions of Ser64, Thr65, Gly68 and Met197 have been attributed to alterations in the kinetic response of the pancreatic isozyme, substitutions of these residues in the liver isozyme may also impact hepatic GCK regulation by reducing affinity toward GKRP.
Modulation of Complex Stability by Sugar Phosphates
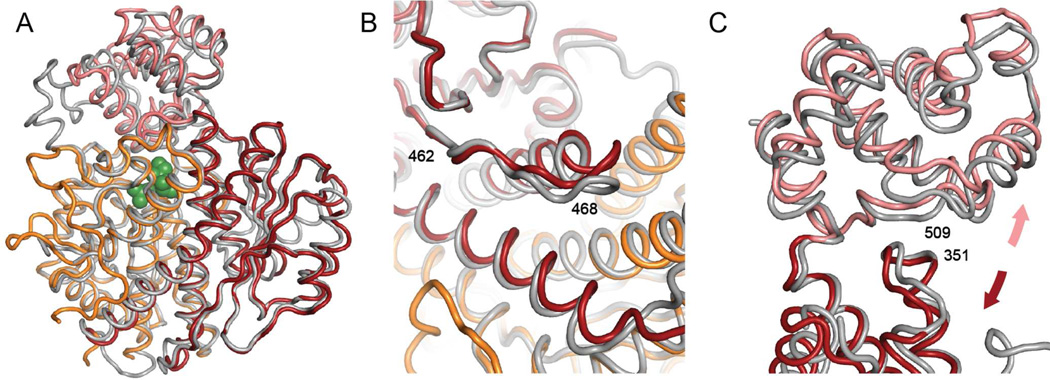
The structure of the mammalian GCK-GKRP assembly provides insight into the mechanism of complex regulation by sugar phosphates. Unexpectedly, the GKRP sugar phosphate binding site is more than 30 Å away from the GCK interaction interface (Figure 1A). Rat GKRP displays moderate affinity for human GCK in the absence of a bound carbohydrate (KD = 1 µM).36 This affinity is enhanced 20-fold in the presence of saturating concentrations of fructose 6-phosphate, but is reduced 7-fold upon association with fructose 1-phosphate. The differential impact of these two carbohydrates on complex stability equates to a modest 3 kcal/mol of binding energy. To understand this difference, we compared the structure of the complex determined in the presence of fructose 6-phosphate with the recently published structure of the “inactive” GKRP-fructose 1-phosphate binary complex.24 Alignment of the two structures using GKRP residues 45–415, which constitute the body of GKRP and include both sugar isomerase domains, produces an RMSD of 0.72 Å(Figure 5A). Based on this alignment, several structural characteristics appear to depend upon the identity of the bound ligand. In the presence of fructose 1-phosphate (or inorganic phosphate) the cap domain of GKRP adopts a closed structure, with the carbohydrate buried near the sugar isomerase domain interface.24 In the fructose 6-phosphate-GCK-GKRP ternary complex the cap is ajar, moving away from the sugar isomerase domains by ~10°. In addition to this change in cap position, we observe a notable change to the interaction interface involving a restructuring of GKRP loop residues 462–470 (Figure 5B). This loop makes several hydrophobic contacts with the hinge region of GCK (Figure 4A). Thus, alterations in the structure and/or position of the GKRP 462–470 loop, which appear to be coupled to changes in the cap domain, facilitate the modulation of complex stability by sugar phosphates.
Figure 5.
Sugar phosphate modulation of complex stability. (A) Comparison of the inactive GKRP-fructose 1-phosphate structure (grey, PDB 4BB9) with the structure of GKRP in complex with GCK and fructose 6-phosphate. Cap is colored pink, SIS1 is orange, and SIS2 is red. Structural alignment was performed using residues 45–415. (B) Repositioning of the GKRP 462–470 loop between the fructose 1-phosphate structure (grey) and the fructose 6-phosphate structure (orange and red). (C) In the presence of fructose 6-phosphate, the cap domain (pink) and SIS2 domain (red) move apart, breaking a backbone hydrogen bond between Arg509 (cap) and His351 (SIS2).
To analyze the impact of sugar phosphate binding upon GKRP structure in more detail, we compared the contact surface between the cap domain and SIS2 when different ligands are bound. When GKRP binds fructose 1-phosphate, the interaction between the cap and SIS2 domains results in the burial of 930 Å2 of surface area. When fructose 6-phosphate replaces fructose 1-phosphate, the buried surface area decreases by 160 Å2 (Figure 5C). This change involves the disruption of a backbone hydrogen bond between the amide nitrogen of Arg509 from the cap domain and the carbonyl oxygen of SIS2 domain residue His351. As a result, both regions become more solvent exposed, likely causing the interaction between the cap and the SIS2 domain to weaken.
Our observations provide a model for the regulation of GCK-GKRP complex stability by sugar phosphates involving a subtle reorganization of the GKRP scaffold. We postulate that in the presence of fructose 6-phosphate, which promotes association with GCK, the interaction between the cap and SIS2 domains of GKRP is destabilized. This allows repositioning of residues 462–470 from the SIS2 domain into a conformation that is more favorable for interaction with GCK. In the presence of the antagonist fructose 1-phosphate, the SIS2 and cap domains move toward one another, stabilizing a conformation of SIS2 that is less favorable for GCK interaction. This structural alteration involves a reorganization of the relative positions of the two sugar isomerase domains with respect to one another. Our postulate differs from the results of MD simulations using models of the mammalian GCK-GKRP complex extrapolated from the crystal structure of the amphibian complex, which shares limited sequence identity with the human proteins and is insensitive to modulation by sugar phosphates.46 These studies suggested that contacts between the side chains of Glu348, His351 and bound ligand are responsible for mediating the differential impact of different sugar phosphate upon complex stability. Based on our structural data, the modest difference in affinity produced by fructose phosphate esters results from subtle alterations in the interaction between the cap and SIS2 domains. The sensitivity of the complex to small changes in GKRP structure may also provide a rationale for understanding the variety of phenotypically mild disorders linked to GKRP and GCK polymorphisms.11, 40
It is important to note that the resolution of the final model introduces some uncertainty into the nature of the conformational reorganizations in response to ligand binding. The thermal displacement parameters of the final structural model are lowest near the interaction interface, providing confidence in the loop reorganizations depicted in Figure 5B. The thermal displacement parameters in the cap domain are larger, indicating more uncertainty in the nature and magnitude of the structural changes near the SIS2-cap domain interface (Figure 5C). Ongoing work to improve the resolution of the complex, as well as to determine the complex structure in the presence of other, more potent effector molecules, is expected to clarify this issue.
Conclusion
The importance of the GCK-GKRP complex in maintaining glucose homeostasis in the liver provides impetus for targeting this complex in therapeutic development for diseases including type 2 diabetes. Indeed, a strategy aimed at disrupting the GCK-GKRP complex with small-molecules has recently emerged as an attractive potential method to increase GCK activity in a liver-specific manner.18 The determination of the atomic structure of the mammalian GCK-GKRP complex formed in the presence of the agonist fructose 6-phosphate is an important step in that direction. In principle, any small molecule capable of disrupting the GCK-GKRP interaction, without decreasing glucokinase activity, could be useful in stimulating glucose metabolism in the hepatocyte. Of particular note are the two hydrophobic patches located at the GCK-GKRP interface, which could be attractive targets for future drug design efforts. In addition, one could envision developing allosteric effectors that disrupt the complex by associating at or near the sugar phosphate binding site. Elucidating the structural basis for the mode of action of such compounds should provide important insights into the mechanism by which these putative therapeutic agents could function to combat diseases of glucose homeostasis.
Supplementary Material
Acknowledgments
We thank Professor Donald Hilvert for helpful comments, encouragement and support. We also thank Beat Blattmann and the staff at the NCCR Structural Biology facility at the University of Zurich, the staff at the X-ray facility of the Laboratory of Organic Chemistry, ETH Zürich and the beamline staff at the Swiss Light Source, Villigen for assistance.
Funding: This work was supported by the ETH Zürich, a Marie Curie Postdoctoral Fellowship within the FP7-PEOPLE Program (IEF-2011-299400) to TB and a grant from the National Institute of Diabetes and Digestive and Kidney Diseases (DK081358) to BGM.
Abbreviations
- GCK
glucokinase
- GKRP
glucokinase regulatory protein
- MODY
maturity onset diabetes of the young
- PNDM
permanent neonatal diabetes mellitus
- PHHI
persistent hypoglycemic hyperinsulinemia
- gck
glucokinase gene
- gckr
glucokinase regulatory protein gene
- MWCO
molecular weight cutoff
- SIS
sugar isomerase
- RMSD
root mean square deviation
Footnotes
Supporting Information
Supporting Information Available: (1) Data statistics and refinement details; (2) Preparation and crystallization of the GCK-GKRP-fructose 6-phosphate complex; (3) Electron density map after molecular replacement with a partial search model; (4) Electrostatic depiction of the GCK-GKRP interface; (5) Distribution of thermal displacement parameters in the GCK-GKRP-fructose 6-phosphate complex; (6) Overlay of fructose 1-phosphate and fructose 6-phosphate binding sites; (7) Orientation of the GKRP N-terminus. This material is available free of charge via the Internet at http://pubs.acs.org
Related Content
The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4LC9)
The authors declare no competing financial interests
References
- 1.Bell GI, Polonsky KS. Diabetes mellitus and genetically programmed defects in β-cell function. Nature. 2001;414:788–791. doi: 10.1038/414788a. [DOI] [PubMed] [Google Scholar]
- 2.Vionnet N, Stoffel M, Takeda J, Yasuda K, Bell GI, Zouali H, Lesage S, Velho G, Iris F, Passa P, Froguel P, Cohen D. Nonsense mutations in the glucokinase gene causes early onset non-insulin-dependent diabetes mellitus. Nature. 1992;356:721–722. doi: 10.1038/356721a0. [DOI] [PubMed] [Google Scholar]
- 3.Agius L. Glucokinase and molecular aspects of liver glycogen metabolism. Biochem. J. 2008;414:1–18. doi: 10.1042/BJ20080595. [DOI] [PubMed] [Google Scholar]
- 4.Larion M, Miller BG. Homotropic allosteric regulation in monomeric mammalian glucokinase. Arch. Biochem. Biophys. 2012;519:103–111. doi: 10.1016/j.abb.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin SX, Neet KE. Demonstration of a slow conformational change in liver glucokinase by fluorescence spectroscopy. J. Biol. Chem. 1990;265:9670–9675. [PubMed] [Google Scholar]
- 6.Larion M, Salinas RK, Bruschweiler-Li L, Miller BG, Brüchweiler R. Order-disorder transitions govern allostery and kinetic cooperativity in monomeric human glucokinase. PLoS Biol. 2012;10:e1001452. doi: 10.1371/journal.pbio.1001452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Schaftingen E. A protein from rat liver confers to glucokinase the property of being antagonistically regulated by fructose 6-phosphate and fructose 1-phosphate. Eur. J. Biochem. 1989;179:179–184. doi: 10.1111/j.1432-1033.1989.tb14538.x. [DOI] [PubMed] [Google Scholar]
- 8.Van Schaftingen E, Veiga-da-Cunha M, Niculescu L. The regulatory protein of glucokinase. Biochem. Soc. Trans. 1996;25:136–140. doi: 10.1042/bst0250136. [DOI] [PubMed] [Google Scholar]
- 9.Agius L, Peak M, Newgard CB, Gomez-Foix AM, Guinovart JJ. Evidence for a role of glucose-induced translocation of glucokinase in the control of hepatic glycogen synthesis. J. Biol. Chem. 1996;271:30479–30486. doi: 10.1074/jbc.271.48.30479. [DOI] [PubMed] [Google Scholar]
- 10.de la Iglesia N, Veigha-da-Cunha M, Van Schaftingen E, Guinovart JJ, Ferrer JC. Glucokinase regulatory protein is essential for the proper subcellular localization of liver glucokinase. FEBS Lett. 1999;456:332–338. doi: 10.1016/s0014-5793(99)00971-0. [DOI] [PubMed] [Google Scholar]
- 11.Osbak KK, Colclough K, Saint-Martin C, Beer NL, Bellane-Chantelot C, Ellard S, Gloyn AL. Update on mutations in glucokinase (GCK), which cause maturity-onset diabetes of the young, permanent neonatal diabetes, and hyperinsulinemic hypoglycemia. Hum. Mutat. 2009;30:1512–1526. doi: 10.1002/humu.21110. [DOI] [PubMed] [Google Scholar]
- 12.Glaser B, Kesavan P, Heyman M, Davis E, Cuesta A, Buchs A, Stanley CA, Thornton PS, Permutt MA, Matschinsky FM, Herold KC. Familial hyperinsulinism caused by an activating glucokinase mutation. N. Engl. J. Med. 1998;338:226–230. doi: 10.1056/NEJM199801223380404. [DOI] [PubMed] [Google Scholar]
- 13.Saxena R, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 14.Johansen CT, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat. Genet. 2010;42:684–687. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimsby J, Coffey JW, Dvorozniak MT, Magram J, Li G, Matschinsky FM, Shiota C, Kaur S, Magnuson MA, Grippo JF. Characterization of glucokinase regulatory protein-deficient mice. J. Biol. Chem. 2000;275:7826–7831. doi: 10.1074/jbc.275.11.7826. [DOI] [PubMed] [Google Scholar]
- 16.Grimsby J, et al. Allosteric activators of glucokinase: potential role in diabetes therapy. Science. 2003;301:370–373. doi: 10.1126/science.1084073. [DOI] [PubMed] [Google Scholar]
- 17.Matschinsky FM. Assessing the potential of glucokinase activators in diabetes therapy. Nat. Rev. Drug Discov. 2009;8:399–416. doi: 10.1038/nrd2850. [DOI] [PubMed] [Google Scholar]
- 18.Pfefferkorn JA. Strategies for the design of hepatoselective glucokinase activators to treat type 2 diabetes. Expert Opin. Drug Discov. 2013;8:319–330. doi: 10.1517/17460441.2013.748744. [DOI] [PubMed] [Google Scholar]
- 19.Larion M, Miller BG. 23-residue C-terminal α-helix governs kinetic cooperativity in monomeric human glucokinase. Biochemistry. 2009;48:6157–6165. doi: 10.1021/bi9007534. [DOI] [PubMed] [Google Scholar]
- 20.Kabsch W. XDS. Acta Crystallogr. D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheldrick GM. SADABS. University of Göttingen. 2010 [Google Scholar]
- 22.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Reed RJ. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamata K, Mitsuya M, Nishimura T, Eiki JI, Nagata Y. Structural basis for allosteric regulation of the monomeric allosteric enzyme human glucokinase. Structure. 2004;12:429–438. doi: 10.1016/j.str.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Pautsch A, Stadler N, Lohle A, Rist W, Berg A, Glocker L, Nar H, Reinert D, Lenter M, Heckel A, Schnapp G, Kauschke SG. Crystal structure of glucokinase regulatory protein. Biochemistry. 2013;52:3523–3531. doi: 10.1021/bi4000782. [DOI] [PubMed] [Google Scholar]
- 25.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Section D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. Section D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Section D. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr. Section D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicholls R, Long F, Murshudov GN. Low-resolution refinement tools in REFMAC5. Acta Crystallogr. Section D. 2012;68:404–417. doi: 10.1107/S090744491105606X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Schaftingen E, Vandercammen A, Detheux M, Davies DR. The regulatory protein of liver glucokinase. Advan. Enzyme Regul. 1992;32:133–148. doi: 10.1016/0065-2571(92)90013-p. [DOI] [PubMed] [Google Scholar]
- 32.Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Section D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 34.Veigha-da-Cunha M, Van Schaftingen E. Identification of fructose 6-phosphate and fructose 1-phosphate binding residues in the regulatory protein of glucokinase. J. Biol. Chem. 2000;277:8466–8473. doi: 10.1074/jbc.M105984200. [DOI] [PubMed] [Google Scholar]
- 35.Detheux M, Vandercammen A, Van Schaftingen E. Effectors of the regulatory protein acting on liver glucokinase: A kinetic investigation. Eur. J. Biochem. 1991;200:553–561. doi: 10.1111/j.1432-1033.1991.tb16218.x. [DOI] [PubMed] [Google Scholar]
- 36.Anderka O, Boyken J, Aschenbach U, Batzer A, Boscheinen O, Schmoll D. Biophysical characterization of the interaction between hepatic glucokinase and its regulatory protein: impact of physiological and pharmacological effectors. J. Biol. Chem. 2008;283:31333–31340. doi: 10.1074/jbc.M805434200. [DOI] [PubMed] [Google Scholar]
- 37.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 38.Veigha-da-Cunha M, Courtois S, Michel A, Gosselain E, Van Schaftingen E. Amino acid conservation in animal glucokinase. Identification of residues implicated in the interaction with the regulatory protein. J. Biol. Chem. 1996;271:6292–6297. doi: 10.1074/jbc.271.11.6292. [DOI] [PubMed] [Google Scholar]
- 39.Baltrusch S, Lenzen S, Okar DA, Lange AJ, Tiedge M. Characterization of glucokinase-binding protein epitopes by a phage-displayed peptide library. Identification of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase as a novel interaction partner. Diabetes. 2001;45:1670–1677. doi: 10.1074/jbc.M105470200. [DOI] [PubMed] [Google Scholar]
- 40.Rees MG, Ng D, Ruppert S, Turner C, Beer NL, Swift AJ, Morken MA, Below JE, Blech I, Mullikin JC, McCarthy MI, Biesecker LG, Gloyn AL, Collins FS NISC Comparative Sequencing Program. Correlation of rare coding variants in the gene encoding human glucokinase regulatory protein with phenotypic, cellular and kinetic outcomes. J. Clin. Inves. 2012;122:205–217. doi: 10.1172/JCI46425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beer NL, Tribble ND, McCulloch LJ, Roos C, Johnson PRV, Orho-Melander M, Gloyn AL. The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum. Mol. Genet. 2009;18:4081–4804. doi: 10.1093/hmg/ddp357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rees MG, Wincovitch S, Schultz J, Waterstradt R, Beer NL, Baltrusch S, Collins FS, Gloyn AL. Cellular characterization of the GCKR P446L variant associated with type 2 diabetes risk. Diabetologia. 2011;55:114–122. doi: 10.1007/s00125-011-2348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukhtar MH, Payne VA, Arden C, Harbottle A, Khan S, Lange AJ, Agius L. Inhibition of glucokinase translocation by AMP-activated protein kinase is associated with phosphorylation of both GKRP and 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008;29:R766–R774. doi: 10.1152/ajpregu.00593.2007. [DOI] [PubMed] [Google Scholar]
- 44.Pal P, Miller BG. Activating mutations in the human glucokinase gene revealed by genetic selection. Biochemistry. 2008;48:814–816. doi: 10.1021/bi802142q. [DOI] [PubMed] [Google Scholar]
- 45.Sayed S, Langdon DR, Odili S, Chen P, Buettger C, Schiffman AB, Suchi M, Taub R, Grimsby J, Matschinsky FM, Stanley CA. Extremes of clinical and enzymatic phenotypes in children with hyperinsulinism caused by glucokinase activating mutations. Diabetes. 2009;58:1419–1427. doi: 10.2337/db08-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi J, Seo M, Kyeong H, Kim E, Kim H. Molecular basis of the role of glucokinase regulatory protein as the allosteric switch for glucokinase. Proc. Natl. Acad. Sci. USA. 2013;110:10171–10176. doi: 10.1073/pnas.1300457110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.