Abstract
Adeno-associated virus (AAV) is a key candidate in the development of gene therapy. In this report, we used surface plasmon resonance spectroscopy to study the interaction between AAV and heparin and other glycosaminoglycans. Surface plasmon resonance results revealed that heparin binds to AAV with extremely high affinity. Solution competition studies shows that AAV binding to heparin is chain length dependent. AAV prefers to bind full chain heparin. All sulfo groups (especially N-sulfo and 6-O-sulfo groups) on heparin are important for the AAV- heparin interaction. Higher levels of sulfo group substitution in GAGs enhance their binding affinities. Atomic force microscopy was also performed to image AAV-2 complexed with heparin.
Keywords: Surface Plasmon Resonance, Adeno-associated virus, glycosaminoglycans, heparin
Adeno-asssociated virus (AAV) is a small DNA-containing human parvovirus that is a leading candidate vector for in vivo human gene therapy (1). Gene therapy involves the delivery of genes into cells to treat or prevent a disease, supplying a replacement for a faulty or missing gene, or sensitizing cells to (cancer) pro-drugs (2). Development of gene therapy currently centers mostly on basic science, but early clinical successes include demonstrated long-term efficacy in experimental treatments for retinopathies (3, 4) and hemophilia B (5). Viruses are the usual choice for gene delivery as they are naturally evolved for this purpose. The above examples show that among several viruses that have been engineered for gene therapy, AAV is increasingly prominent, considered a relatively safe vector for moderately sized genes < 5 kb. AAV is much less immunogenic than adenoviruses (no relation), and less oncogenic than retroviruses, because they transduce cells episomally, or integrate with partial site specificity (1, 6–9). A lack of understanding of the tropism and cell specificity of AAV has impeded its application in gene therapy.
In vivo, multiple factors determine AAV tissue tropism, including its traversal of the endothelium, immune neutralization, and blood clearance (10–13). Interactions with host cell molecules, starting with the primary receptor, are important both in vivo and in vitro. Cell entry for AAV starts with attachment to extracellular glycoconjugates followed by binding to one or more membrane-anchored co-receptors (or secondary receptors) prior to endosomal entry (14–20). Viral-glycan interactions, involving glycoproteins, glycolipids or proteoglycans (PGs), figure prominently in the cell entry of many viruses (21). However, the importance of these viral-glycan interactions varies, particularly with respect to their role in conferring cell and tissue specificity (22–26). Multiple lines of evidence implicate an important role of glycans in AAV attachment, including the affect of mutations at the glycan binding site on both in vitro cell specificity and in vivo tissue tropism (11, 27–32). AAV serotypes have different preferences for glycans in their attachment. AAV-2 and AAV-3 are thought to use heparan sulfate (HS) PGs as the primary receptor in cell attachment (33, 34). These serotypes, as well as AAV-6 to lesser extent, bind to heparin, a commonly used HSPG analog (35, 36). In vivo, AAV-6 has a preference for glycans terminated with sialic acid, as do AAV-1, -4 and -5 (37, 38), whereas AAV-9 binds to a galactose-terminated glycan (39, 40).
Heparin/HS belongs to the glycosaminoglycan (GAG) family of anionic, and often highly sulfated, complex polydisperse linear polysaccharides. An increasing number of GAG-binding proteins have been reported in the literature (41). A number of diverse patho-physiological processes are mediated through the interaction between heparin/HS and proteins including: blood coagulation, cell growth and differentiation, host defense and viral infection, lipid transport and metabolism, cell-to-cell and cell-to-matrix signaling, inflammation and cancer (41–44). Generally, an understanding of heparin/HS-virus particles interactions at the molecular level is of fundamental importance to biology and will aid in the development of highly specific glycan-based therapeutic agents (41, 45). For AAV, it is hoped that an improved understanding of GAG interactions will eventually allow more exquisite and rational modulation of cell attachment, transduction and tissue tropism, beyond the gross changes demonstrated with chimeric transducing vectors (30, 35, 36).
Heparin has been used as a model GAG in the study of HS-protein interaction, since it mimics the interactions of proteins with the HS present on cell surfaces and in the extracellular matrix (21). The goal of this study is to analyze molecular interactions of heparin/GAGs with Adeno-associated virus (AAV). Three different variants were used: natural human serotypes AAV-2 and AAV-6, and a chimeric gene-shuffled recombinant, AAV-DJ, which has been developed as a hepatotropic gene therapy vector. A BIAcore 3000 system was used to measure the strength of AAV-glycan interactions by surface plasmon resonance (SPR). Atomic force microscopy was also performed to image the complexes from AAV-2- heparin interaction.
EXPERIMENTAL PROCEDURES
Materials
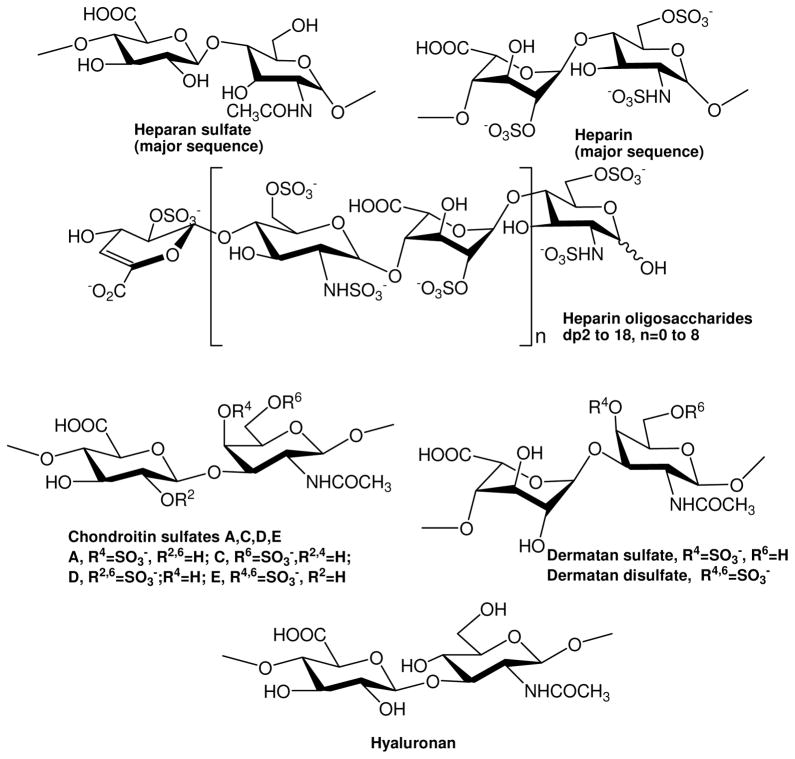
The GAGs used were porcine intestinal heparin (16 kDa), low molecular weight (LMW) heparin (4.8 kDa) and porcine intestinal heparan sulfate (12kDa) (Celsus Laboratories, Cincinnati, OH); chondroitin sulfate A (20 kDa) from porcine rib cartilage (Sigma, St. Louis, MO), dermatan sulfate (also known as chondroitin sulfate B, 30 kDa, from porcine intestine, Sigma), dermatan disulfate (4,6 disulfo DS, 33 kDa, Celsus) prepared through the regioselective chemical 6-O-sulfonation of dermatan sulfate(46), chondroitin sulfate C (20 kDa, from shark cartilage, Sigma), chondroitin sulfate D (20 kDa, from whale cartilage, Seikagaku, Tokyo, Japan), chondroitin sulfate E (20 kDa from squid cartilage, Seikagaku), and hyaluronic acid sodium salt (100 kDa, from Streptococus zooepidemicus, Sigma). Fully desulfated heparin (14 kDa), N-desulfated heparin (14 kDa) and 2-O-desulfated IdoA heparin (13 kDa) were all prepared based on Yates et al. (47). 6-O-desulfated heparin (13 kDa) was a generous gift from Dr. Lianchun Wang of the Complex Carbohydrate Research Center, University of Georgia. Heparin oligosaccharides included disaccharide (degree of polymerization (dp)2) tetrasaccharide (dp4), hexasaccharide (dp6), octasaccharide (dp8), decasaccharide (dp10), tetradecasaccharide (dp14), hexadecasaccharide (dp16) and octadecasaccharide (dp18) and were prepared from controlled partial heparin lyase 1 treatment of bovine lung heparin (Sigma) followed by size fractionation. Chemical structures of these GAGs and heparin oligosaccharides are showed in Figure 1. Sucrose octasulfate (SOS) was from Toronto Biochemicals (Toronto, Canada). Sensor SA chips were from BIAcore AB (AB, Uppsala, Sweden). SPR measurements were performed on a BIAcore 3000 operated using BIAcore 3000 control and BIAevaluation software (version 4.0.1).
Figure 1.
Chemical structures of heparin, heparin-derived oligosaccharides and GAGs.
Atomic force microscopy (AFM) was operated on an MFP-3D AFM (Asylum Research, Santa Barbara CA) using Asylum Research 6.22 software. AFM scans were performed with 2 nm (±1 nm) Super Sharp Silicon-NCLR (Non-Contact Long-cantilever Reflex-Coating) AFM cantilevers (Nano and More, Soquel, CA) on highest-grade (V1) mica sheets (Ted Pella, Redding, CA). Milli-Q water was degassed and re-filtered using 0.22 μm cellulose acetate membranes (Millipore, Billerica, MA).
AAV Expression and Purification
Production of AAV-2 and AAV-DJ Virus-like particles (VLPs) in SF9 cells followed methods developed for AAV-2 (48) that used the Bac-to-Bac Baculovirus Expression Vector System (Invitrogen), and were adapted for AAV-DJ as previously described (49). VLPs are devoid of infectious viral DNA, but the outer protein capsids appear structurally and functionally identical to wild-type virus. These empty capsids were purified by three rounds of CsCl density gradient ultracentrifugation, followed by heparin-affinity chromatography, eluting with a NaCl gradient. AAV-6 was prepared in HeLa cells as infectious virions and purified by three rounds of CsCl gradient ultracentrifugation, all using protocols that have been described previously (50). All samples were dialyzed into phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 and 1.8 mM KH2PO4, pH 7.4).
Preparation of the heparin biochip
The biotinylated heparin was prepared by reaction of sulfo-N-hydroxysuccinimide long-chain biotin (Piece, Rockford, IL) with the free amino groups of unsubstituted glucosamine residues in the polysaccharide chain, following a published procedure (51). The biotinylated heparin was immobilized to a streptavidin (SA) chip based on the manufacturer’s protocol. In brief, a 20-μL solution of the heparin-biotin conjugate (0.1 mg/mL) in HBS-EP running buffer was injected over flow cell 2 (FC2) of the SA chip, at a flow rate of 10 μL/min. The successful immobilization of heparin was confirmed by the observation of a ~50 resonance unit (RU) increase in the sensor chip. The control flow cell (FC1) was prepared by 1 min injection with saturated biotin.
Measurement of the Interaction between Heparin and AAV using the BIAcore SPR
The protein samples were diluted in HBS-EP buffer (0.01 M HEPES, 0.15 M NaCl, 3 mM EDTA, 0.005% surfactant P20, pH 7.4,). Different dilutions of protein samples were injected at a flow rate of 30 μL/min. At the end of the sample injection, the same buffer was flowed over the sensor surface to facilitate dissociation. After a 3 min dissociation time, the sensor surface was regenerated with 30 μL of 0.5% SDS, then with 2 M NaCl to get fully regenerated surface. The response was monitored as a function of time (sensorgram) at 25 °C.
Inhibition of Heparin-binding by Oligosaccharides
AAV-2 at 8.3 pM (equivalent to capsid protein at 0.5 nM) was mixed with 1000 nM of heparin oligosaccharides, including disaccharide (dp2) tetrasaccharide (dp4), hexasaccharide (dp6), octasaccharide (dp8), decasaccharide (dp10), tetradecasaccharide (dp14), hexadecasaccharide (dp16) and octadecasaccharide (dp18), all in HBS-EP buffer. Samples were injected over the heparin chip at a flow rate of 30 μL/min. After each run, dissociation and regeneration were performed as described above. For each set of competition experiments on SPR, a control experiment (target without ligand) was performed to make sure the surface was completely regenerated and that the results obtained between runs were comparable.
Inhibition of Heparin-binding by GAGs and Chemically Modified Heparin
AAV-2 at 8.3 pM was pre-mixed with 100 nM of GAG or chemical modified heparin. Otherwise, SPR was performed as described above.
Atomic Force Microscopy (AFM)
AAV-2 was combined with 100 nM GAG and incubated at 22°C for 10 min. A 20 μL aliquot was spotted on a sheet of immobilized mica and allowed to set for 1 h before washing with Milli-Q water. The sample was dried for 6 h. Raster images were obtained using 10 μm squares and a representative area was re-scanned at higher resolution with 2 μm squares. Using Asylum Research 6.22 software, particle heights were obtained for the 10 μm squares in representative sections.
RESULTS
Kinetics of the AAV-heparin Interactions
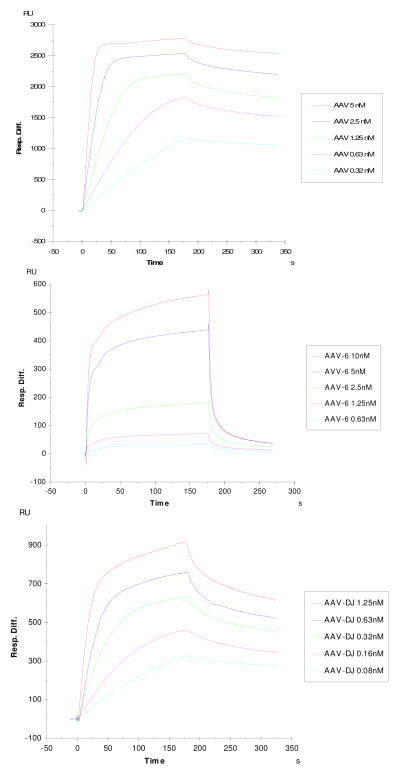
The binding to immobilized heparin was measured for the three virions: AAV-2, AAV-6 and AAV-DJ, using SPR. The SPR binding sensorgrams of the AAV-2/heparin, AAV-6/heparin and AAV-DJ/heparin interactions are displayed in Figure 2 showing different binding profiles. The sensorgrams were fit globally to obtain apparent on (ka) and off (kb) rates for the binding equilibrium (Table 1), using the BiaEvaluation software and assuming a 1:1 Langmuir model.
Figure 2.
Top: SPR sensorgrams of the AAV2-heparin interaction. Concentrations of AAV (from top to bottom): 5, 2.5, 1.25, 0.63 and 0.32 nM, respectively.
Middle: SPR sensorgrams of the AAV6-heparin interaction. Concentrations of AAV (from top to bottom): 10, 5, 2.5, 1.25, and 0.63, respectively.
Bottom: SPR sensorgrams of the AAV DJ-heparin interaction. Concentrations of AAV (from top to bottom): 1.25, 0.63, 0.32, 0.16 and 0.08 nM, respectively.
Table 1.
Summary of kinetic data of AAV- heparin interactions
| Interaction | ka (1/MS) | kd (1/S) | KD (M) |
|---|---|---|---|
| AAV-2/Heparin | 1.2 × 107 | 1.2 × 10−3 | 1.0 × 10−10 |
| AAV-6/Heparin | 1.61 × 106 | 0.0197 | 1.22 × 10−8 |
| AAV-DJ/Heparin | 4.3 ×107 | 4.6 × 10−3 | 1.1 × 10−10 |
Polymer Length
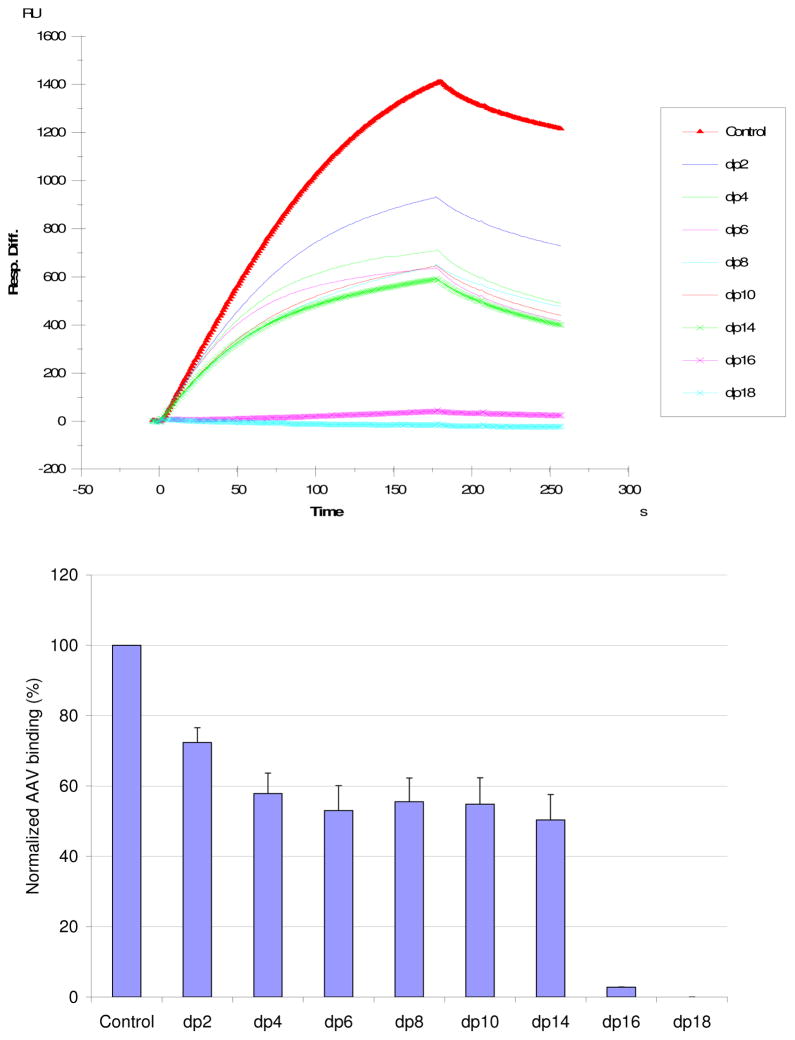
Relative avidity, determined through SPR measurements of AAV2 -heparin binding inhibited by different oligosaccharides, was used to establish the dependence of binding on saccharide chain length. These heparin-derived oligosaccharides ranged from disaccharides to octadecasaccharides (dp2 to dp18). Competitive inhibition increases in two steps, once near a chain length of 4 saccharide units, and the other at 16 saccharide units (Fig. 3). This can be rationalized if the binding sites on the side of each spike are about 5 saccharides in length, and if a chain length of 16 saccharide units is the minimum for a polysaccharide to bind to two neighboring sites on spikes related by one of the viral 3-fold axes of symmetry (52).
Figure 3.
Top: Sensorgrams for the AAV2-heparin interaction inhibited by solution oligosaccharides of different chain-length. The AAV2 concentration was 0.5 nM, and concentrations of heparin oligosaccharides were 1000 nM). Bottom: Bar graphs (errors from triplicate experiments) showing relative AAV2 binding in the presence of competing heparin oligosaccharides.
Specificity for Different GAGs
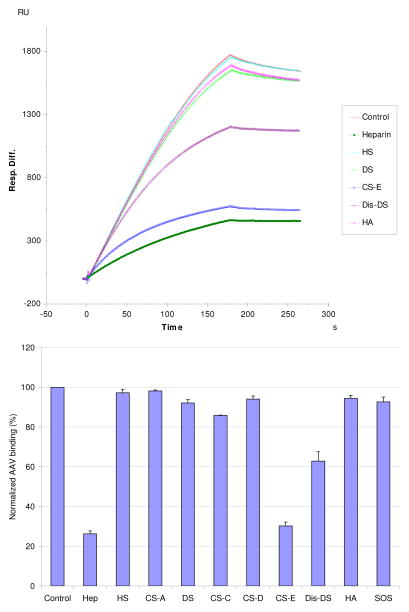
The SPR competition assay was also utilized to determine the binding preference of AAV-2 to various GAGs (Fig. 1). SPR competition sensorgrams and inhibition levels are displayed in Figure 4. The results showed that CSE provided the strongest inhibitory effect (70–80%) on AAV-2 binding to immobilized heparin. DiS-DS also inhibited binding by ~40%. Other GAGs and SOS (as a non-GAG polyanion control) had little effect. The data suggest that the binding interactions are dependent on GAG structure and highly influenced by the level of GAG sulfation.
Figure 4.
Top: Sensorgrams for the AAV2-heparin interaction inhibited by GAGs of various types. The AAV2 concentration was 0.5 nM, and concentrations of GAGs were 100 nM). Bottom:
Bar graphs (errors from triplicate experiments) showing relative AAV2 binding in the presence of different competing GAGs.
Chemically Modified Heparins
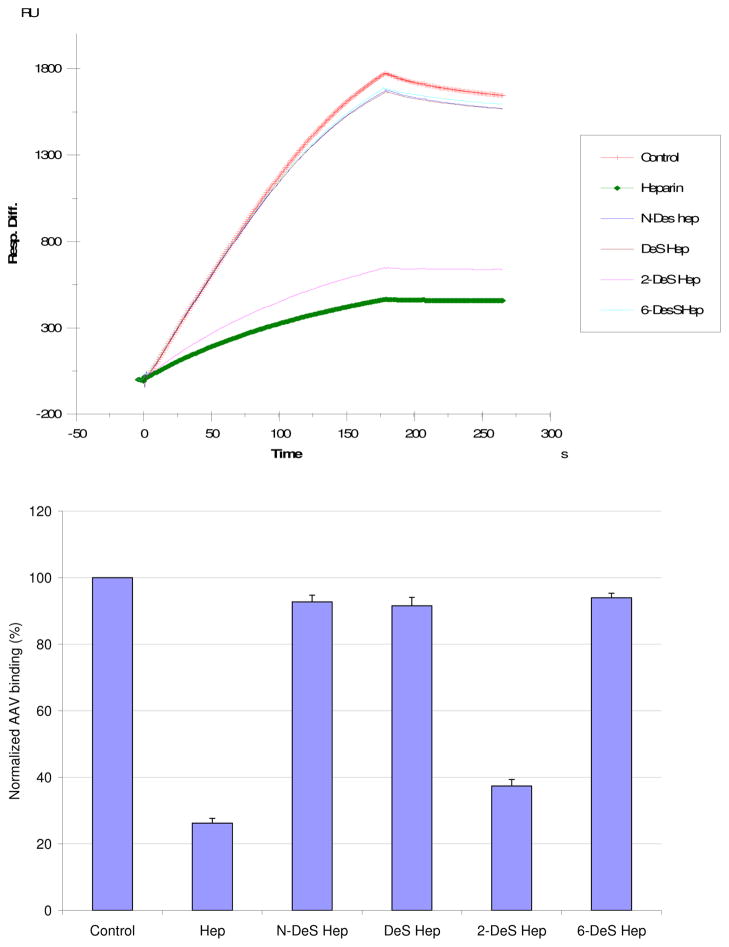
SPR competition sensorgrams and inhibition profiles with chemical modified heparins are displayed in Figure 5 for AAV-2. Inhibitory activity is lost upon all types of desulfation (fully desulfated heparin, N-desulfated heparin, 2-O-desulfated heparin and 6-O-desulfated heparin). However, with 2-O-desulfated heparin, the loss of inhibition is much less with AAV-2. This implies that N-sulfo groups and 6-O-sulfo groups are more critical than 2-O-sulfo groups to the AAV-2-heparin binding interaction.
Figure 5.
Top: Sensorgrams for the AAV2-heparin interaction inhibited by heparins that have been chemically desulfonated in various ways. The AAV2 concentration was 0.5 nM, and concentrations of chemical modified heparin were 100 nM). Bottom: Bar graphs (errors from triplicate experiments) showing relative AAV2 binding in the presence of GAGs with different sulfonation patterns.
AFM Analysis
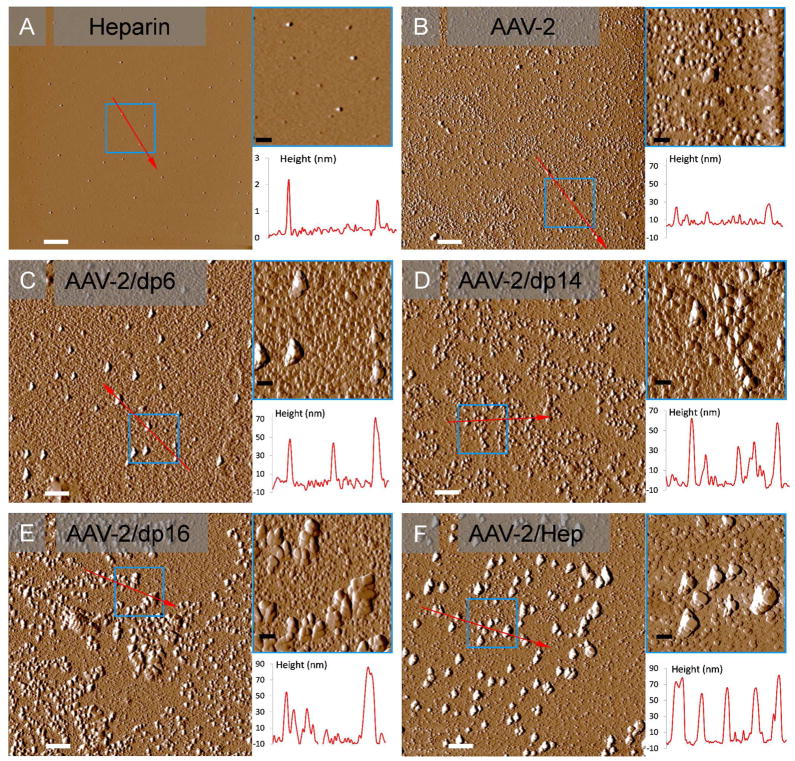
In the absence of heparin oligosaccharides, AFM shows a field of approximately round particles of AAV-2 with heights in the range of 15 – 30 nm (Figure 6). This is consistent with particle heights reported earlier by Chen and the observation that empty particles were measured with somewhat reduced diameters due to deformation under the ATM tip (59, 60). In the presence of heparin oligosaccharides, the average particle size increases with chain-length, indicating aggregation (Table 2 and Figure 6). Even with dp6 some larger particles are observed, suggesting a limited ability to cross-link. There is a marked increase both in average size and frequency of larger aggregates in dp14, dp16 and full-length heparin. The large particles of 60 – 90 nm heightseen with dp16 oligosaccharides and dominant with heparin correspond to several particle diameters and indicate an increasing ability for multiple virions to be bound along the GAG chain and for each particle to host interactions with multiple GAGs, presumably at binding sites related by icosahedral symmetry.
Figure 6.
AFM images of AAV-2 and complexes with different heparin oligosaccharides. (A) heparin alone (control). (B) AAV-2 particles alone (control). (C) AAV-2/heparin dp 6 complexes. (D) AAV-2/heparin dp 14 complexes. (E) AAV-2/heparin dp 16 complexes. (F) AAV-2/heparin (full chain) complexes. White and black scale bars represent 1 μm and 250 nm, respectively. All panels show 10 μm images. The upper insets are expansions (5X) of the areas depicted in small blue squares, and the lower plots are particle height distributions along the red arrows.
Table 2.
Description of particle heights observed with AFM on AAV-2/heparin oligosaccharide complexes
| Combination | Predominant particle size range | Minor morphologies |
|---|---|---|
| AAV-2 only | 15 – 30 nm particles | |
| AAV-2/dp6 | 15 – 30 nm particles | 30 – 80 nm particles |
| AAV-2/dp14 | 15 – 30 nm particles | 30 – 80 nm particles |
| AAV-2/dp16 | 15 – 60 nm particles | 60 – 90 nm particles |
| AAV-2/heparin | 60 – 90 nm particles | 15 – 30 nm particles |
Molecular Modeling
An extended chain model of β-D-GlcA-(1,4′)-α-D-GlcNAc-(1,4′))8 was generated using the GLYCAM Web-tool (http://www.glycam.com) (53). This was fit into the difference density obtained by subtracting the cryo-EM reconstruction of AAV-2 from that of the AAV-2/heparin complex at 8.3 Å resolution (52). Coot (54) was used for computer-aided manual fitting of the model into the difference density contoured at 6 σ. Stereochemical distortions, introduced during modeling, were corrected through molecular mechanics energy minimization using NAMD (55). Close contacts with the virus were resolved and the fit to density improved using Coot, before final energy minimization with NAMD, and checking of stereochemical parameters. Figure 7 was generated using PyMol (56).
Figure 7.
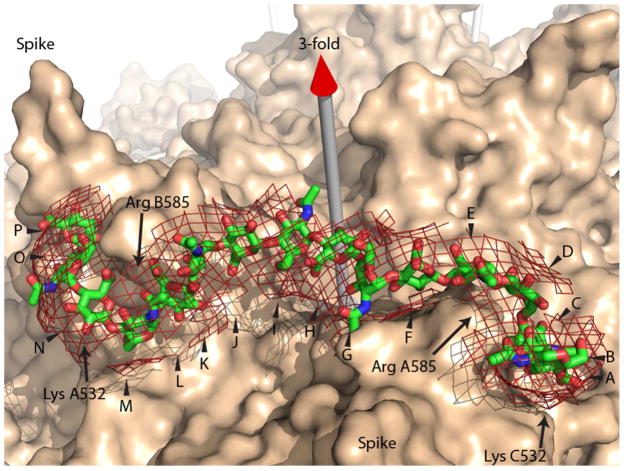
Binding of oligosaccharides to AAV-2. A 16-residue glycosaminoglycan (GAG) stick model has been fit approximately into the heparin difference density from a cryo-EM study at 8 Å resolution (52) which is shown as a net at a 6 σ contour level on top of the van der Waals surface of the AAV-2 crystal structure (57). The GAG oligomer is modeled bridging between 3-fold symmetry-related binding sites on the sides of adjacent viral spikes. A chain length of at least 13 is needed to make contact with the five positively charged residues implicated at each of two binding sites. Density extending over 16 residues (lettered A through P) suggests additional interactions and the possibility that the stronger binding of a 16-mer in Figure 5 might correspond to an increase in avidity with optimization of two-site binding. Note that a single asymmetric oligosaccharide chain passing through two symmetry-related binding sites must have somewhat different interactions, suggesting that the binding sites have evolved some adaptability.
DISCUSSION
Through SPR, we now have fully quantitative measures of the avidity of AAV for heparin. Prior quantitation used the proxy of NaCl concentration needed to elute AAV from a heparin affinity column for rank ordering (29, 35, 36). Heparin-affinity elution had shown that, of natural serotypes, AAV-2 binds strongest, and AAV-6 the weakest of those measurable. SPR is consistent with this, and now shows a 100-fold difference in avidity (Table 1). AAV-DJ is a chimeric combination of AAV-2, -8 and -9 created by random gene shuffling, followed by selection for liver cell transduction and escape from human polyclonal neutralizing sera (11). There are several potential causes for the improved in vivo (mouse) hepatotropism, of which altered attachment has been considered as a possibility. SPR, like heparin affinity (49) and cell-binding (11), indicates weakened binding relative to AAV-2. Quantitatively, the SPR-derived 10% difference in apparent KD (Table 1) is consistent with the structures of AAV-2 and AAV-DJ being very similar in the heparin-binding region, and differing in calculated electrostatic potential only on the periphery of the binding site (49, 57). A 10% difference in binding avidity is unlikely to account, through cell attachment alone, for > 10-fold differences in in vitro transduction efficiencies and in vivo tissue tropism (11). Suspicion then naturally falls on the region of the structure where there are greatest differences, the external loop that constitutes variable region 1. Given that this is the center of the AAV-2 epitope for neutralizing monoclonal antibody A20 (58), it is likely that structural differences resulted here from AAV-DJ’s stringent selection by escape from neutralizing sera (11). It is possible that changed tissue tropism in vivo results from altered immune clearance, sequestration or other effects. As for changes to transduction in vitro, quantitatively similar dissociation constants draws attention away from initial attachment to speculation that selection of neutralization escape variants may have serendipitously altered co-receptor binding, if neutralizing epitopes are well-represented at currently uncharacterized co-receptor binding sites.
The dependence of AAV2-GAG interactions on chain-length can be interpreted in in light of the cryo-electron microscopy (EM) structure of AAV-2 complexed with a 17 kDa fragment of heparin (Figure 7) (52). The heparin in the cryo-EM visualization lacks molecular detail, because of the modest 8 Å experimental resolution, and because we see the average of heterogeneous sequences that are bound to the thousands of particles imaged. The virus structure in the complex is better defined, because the 3 Å resolution crystal structure (57) can be docked as a rigid body by superimposing the icosahedral symmetry. The maximal heparin density, implying tightest binding, is adjacent to Arg585 and Arg588. Strong density extends about the length of a pentasaccharide to cover other positively-charged residues implicated by mutagenesis in heparin-binding: Arg484, Arg487 and Lys532 (31, 59), as well as Lys527. With short oligosaccharides, Figure 5B shows progressively stronger binding, until presumably all the main interactions of a single site on AAV are contributing. The next big change comes between dp14 and dp16, which might correspond to the chain length required for a change in avidity with the contribution of two symmetry-related sites on the virus. Density bridging between adjacent sites in the EM reconstruction is about half the maximum height, implying looser association, but still experimentally significant at 6 σ. Modeling a canonical glycosaminoglycan through the path of this density shows that a minimum chain length of 13 is required to make contacts with two symmetry-related copies of Lys532 (Figure 7). Intriguingly, density continues away from the 3-fold in each direction such that a 16-mer can be fit into the EM density. This indicates continuing GAG-virus interactions beyond Lys532. It may be that the stronger binding of dp16 evident in Figure 5B and the length of the density seen in the EM reconstruction are both consequences of two-site attachment becoming optimal with a chain length of at least 15. Some caution is warranted, because structures much better than 8 Å resolution would be needed to estimate the strength of atomic interactions at the ends of the 16-mer. Nonetheless, with indications both from the EM and the current binding studies, evidence is building that AAV might get wrapped by GAGs binding at two or more sites – with an interesting implication. If at one site the polysaccharide is bound running towards the 3-fold axis, at the adjacent binding site it is running away. Thus, a single polysaccharide chain cannot match the viral symmetry at adjacent sites, and therefore the site must be tolerant of binding in either direction.
The inhibitory effects of soluble GAGs on the AAV-immobilized heparin interaction were greatest for heparin with 2.8 mol of sulfo group per disaccharide repeating unit, followed by CS-E and Dis-DS with 1.5–2 mol sulfo group per disaccharide, then HS, DS, CS-A, and CS-C with ≤1 mol sulfo group per disaccharide. Combining the structural requirements in the modified heparin and chondroitin/dermatan family of the GAGs suggests an AAV binding motif that prefers disaccharide units with two sulfo groups on a hexosamine residue linked to a uronic acid having a carboxyl group. On heparin these two charged groups per disaccharide repeating unit are N-sulfo and 6-O-sulfo groups and on the chondroitin/dermatan family are the 4-O-sulfo and 6-O-sulfo groups found on CS-E or on Dis-DS. These results are similar to the GAG sulfation preferences of other proteins, such as Shh, Ihog, FGF1, FGF2, and CrataBL (60–62).
Prior to the current work, nothing was known about the internal sequence preferences within the GAGs to which AAVs attach. While much remains for a full characterization of any of the serotypes, the inhibition patterns for heparins desulfated at specific locations begins to offer guiding principles. The presence of sulfo groups at the N- and 6-O- positions is more critical than at 2-O- position. Such information might support the first step in an interative process through which one might home in on heparin oligomers with stronger binding and inhibitory activity, and thereby advance our understanding of AAV attachment by combining structural and virological studies.
Acknowledgments
Funding This work was supported by grants from the National Institutes of Health in the form of GM-38060 to R.J.L. and GM66875 to (M.S.C.) and a Pilot Project Grant from the OHSU Center for Spatial Systems Biomedicine (OCSSB) to M.S.C. TFL was supported by the Interactions at Microbe-Host Interface training grant from the National Institutes of Health (T32AI007472).
ABBREVIATIONS
- GAG
glycosaminoglycan
- AAV
adeno-associated virus
- SPR
surface plasmon resonance
- AFM
atomic force microscopy
- LMWH
low molecular weight heparin
- HS
heparan sulfate
- PG
proteoglycan
- CS-A
chondroitin sulfate A
- DS
dermatan sulfate
- Dis-DS
dermatan disulfate
- CS-C
chondroitin sulfate C
- CS-D
chondroitin sulfate D
- CS-E
chondroitin sulfate E
- SOS
sucrose octasulfate
- SA
streptavidin
- dp
degree of polymerization
- HA
hyaluronan
References
- 1.Muzyczka N, Berns KI. Parvoviridae: The Viruses and Their Replication. In: Fields BN, Knipe DM, Howley PM, editors. Virology. 4. Lippincott Williams & Wilkins; Philadelphia: 2001. pp. 2327–2360. [Google Scholar]
- 2.Schambach A, Maetzig T, Baum C. Retroviral vectors for Cell and Gene Therapy. In: Templeton NS, editor. Gene and cell therapy: therapeutic mechanisms and strategies. CRC Press; Boca Raton: 2008. pp. 3–15. [Google Scholar]
- 3.Simonelli F, Maguire AM, Testa F, Pierce EA, Mingozzi F, Bennicelli JL, Rossi S, Marshall K, Banfi S, Surace EM, Sun J, Redmond TM, Zhu X, Shindler KS, Ying GS, Ziviello C, Acerra C, Wright JF, McDonnell JW, High KA, Bennett J, Auricchio A. Gene therapy for Leber’s congenital amaurosis is safe and effective through 1.5 years after vector administration. Mol Ther. 2010;18:643–650. doi: 10.1038/mt.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cideciyan AV, Hauswirth WW, Aleman TS, Kaushal S, Schwartz SB, Boye SL, Windsor EAM, Conlon TJ, Sumaroka A, Roman AJ, Byrne BJ, Jacobson SG. Vision 1 Year after Gene Therapy for Leber’s Congenital Amaurosis. New England Journal of Medicine. 2009;361:725–727. doi: 10.1056/NEJMc0903652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nathwani AC, Tuddenham EG, Rangarajan S, Rosales C, McIntosh J, Linch DC, Chowdary P, Riddell A, Pie AJ, Harrington C, O’Beirne J, Smith K, Pasi J, Glader B, Rustagi P, Ng CY, Kay MA, Zhou J, Spence Y, Morton CL, Allay J, Coleman J, Sleep S, Cunningham JM, Srivastava D, Basner-Tschakarjan E, Mingozzi F, High KA, Gray JT, Reiss UM, Nienhuis AW, Davidoff AM. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samulski RJ, Zhu X, Xiao X, Brook JD, Housman DE, Epstein N, Hunter LA. Targeted Integration of Adenoassociated virus (AAV) into Human Cheomosome-19. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dyall J, Szabo P, Berns KI. Adeno-associated virus (AAV) site-specific integration: formation of AAV-AAVS1 junctions in an in vitro system. Proc Natl Acad Sci U S A. 1999;96:12849–12854. doi: 10.1073/pnas.96.22.12849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huttner NA, Girod A, Schnittger S, Schoch C, Hallek M, Buning H. Analysis of site-specific transgene integration following cotransduction with recombinant adeno-associated virus and a rep encodingplasmid. J Gene Med. 2003;5:120–129. doi: 10.1002/jgm.324. [DOI] [PubMed] [Google Scholar]
- 9.Carter BJ, Burstein H, Peluso RW. Adeno-associated Virus and AAV Vectors for Gene delivery. In: Templeton NS, editor. Gene and cell therapy: therapeutic mechanisms and strategies. CRC Press; Boca Raton: 2008. pp. 115–156. [Google Scholar]
- 10.Gao G, Lu Y, Calcedo R, Grant RL, Bell P, Wang L, Figueredo J, Lock M, Wilson JM. Biology of AAV serotype vectors in liver-directed gene transfer to nonhuman primates. Mol Ther. 2006;13:77–87. doi: 10.1016/j.ymthe.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 11.Grimm D, Lee JS, Wang L, Desai T, Akache B, Storm TA, Kay MA. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J Virol. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotchey NM, Adachi K, Zahid M, Inagaki K, Charan R, Parker RS, Nakai H. A potential role of distinctively delayed blood clearance of recombinant adeno-associated virus serotype 9 in robust cardiac transduction. Mol Ther. 2011;19:1079–1089. doi: 10.1038/mt.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halbert CL, Standaert TA, Wilson CB, Miller AD. Successful readministration of adeno-associated virus vectors to the mouse lung requires transient immunosuppression during the initial exposure. J Virol. 1998;72:9795–9805. doi: 10.1128/jvi.72.12.9795-9805.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asokan A, Hamra JB, Govindasamy L, Agbandje-McKenna M, Samulski RJ. Adeno-associated virus type 2 contains an integrin alpha5beta1 binding domain essential for viral cell entry. J Virol. 2006;80:8961–8969. doi: 10.1128/JVI.00843-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kashiwakura Y, Tamayose K, Iwabuchi K, Hirai Y, Shimada T, Matsumoto K, Nakamura T, Watanabe M, Oshimi K, Daida H. Hepatocyte growth factor receptor is a coreceptor for adeno-associated virus type 2 infection. J Virol. 2005;79:609–614. doi: 10.1128/JVI.79.1.609-614.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ling C, Lu Y, Kalsi JK, Jayandharan GR, Li B, Ma W, Cheng B, Gee SW, McGoogan KE, Govindasamy L, Zhong L, Agbandje-McKenna M, Srivastava A. Human hepatocyte growth factor receptor is a cellular coreceptor for adeno-associated virus serotype 3. Hum Gene Ther. 2010;21:1741–1747. doi: 10.1089/hum.2010.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pasquale GD, Davidson BL, Stein CS, Martins I, Scudiero D, Monks A, Chiorini JA. Identification of PDGFR as a receptor for AAV-5 transduction. Nat Med. 2003;9:1306–1312. doi: 10.1038/nm929. [DOI] [PubMed] [Google Scholar]
- 18.Qing K, Mah C, Hansen J, Zhou S, Dwarki V, Srivastava A. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat Med. 1999;5:71–77. doi: 10.1038/4758. [DOI] [PubMed] [Google Scholar]
- 19.Akache B, Grimm D, Pandey K, Yant SR, Xu H, Kay MA. The 37/67-kilodalton laminin receptor is a receptor for adeno-associated virus serotypes 8, 2, 3, and 9. J Virol. 2006;80:9831–9836. doi: 10.1128/JVI.00878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weller ML, Amornphimoltham P, Schmidt M, Gutkind S, Chiorini JA. American Society for Virology. FASEB; Vancouver: 2009. Epidermal Growth Factor Receptor is a Receptor for Adeno-Associated Virus Serotype 6; p. 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of glycobiology. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 2009. [PubMed] [Google Scholar]
- 22.Patel M, Yanagishita M, Roderiquez G, Bou-Habib DC, Oravecz T, Hascall VC, Norcross MA. Cell-surface heparan sulfate proteoglycan mediates HIV-1 infection of T-cell lines. AIDS Res Hum Retroviruses. 1993;9:167–174. doi: 10.1089/aid.1993.9.167. [DOI] [PubMed] [Google Scholar]
- 23.Saphire AC, Bobardt MD, Gallay PA. Host cyclophilin A mediates HIV-1 attachment to target cells via heparans. EMBO J. 1999;18:6771–6785. doi: 10.1093/emboj/18.23.6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson T, Ellard FM, Ghazaleh RA, Brookes SM, Blakemore WE, Corteyn AH, Stuart DI, Newman JW, King AM. Efficient infection of cells in culture by type O foot-and-mouth disease virus requires binding to cell surface heparan sulfate. J Virol. 1996;70:5282–5287. doi: 10.1128/jvi.70.8.5282-5287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang W, Heil M, Kuhn RJ, Baker TS. Heparin binding sites on Ross River virus revealed by electron cryo-microscopy. Virology. 2005;332:511–518. doi: 10.1016/j.virol.2004.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gillet L, Colaco S, Stevenson PG. The Murid Herpesvirus-4 gL regulates an entry-associated conformation change in gH. PLoS One. 2008;3:e2811. doi: 10.1371/journal.pone.0002811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Work LM, Buning H, Hunt E, Nicklin SA, Denby L, Britton N, Leike K, Odenthal M, Drebber U, Hallek M, Baker AH. Vascular bed-targeted in vivo gene delivery using tropism-modified adeno-associated viruses. Mol Ther. 2006;13:683–693. doi: 10.1016/j.ymthe.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 28.Perabo L, Goldnau D, White K, Endell J, Boucas J, Humme S, Work LM, Janicki H, Hallek M, Baker AH, Buning H. Heparan sulfate proteoglycan binding properties of adeno-associated virus retargeting mutants and consequences for their in vivo tropism. J Virol. 2006;80:7265–7269. doi: 10.1128/JVI.00076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Z, Asokan A, Grieger JC, Govindasamy L, Agbandje-McKenna M, Samulski RJ. Single Amino Acid Changes Can Influence Titer, Heparin Binding, and Tissue Tropism in Different Adeno-Associated Virus (AAV) Serotypes. J Virol. 2006;80:11393–11397. doi: 10.1128/JVI.01288-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asokan A, Conway JC, Phillips JL, Li C, Hegge J, Sinnott R, Yadav S, DiPrimio N, Nam HJ, Agbandje-McKenna M, McPhee S, Wolff J, Samulski RJ. Reengineering a receptor footprint of adeno-associated virus enables selective and systemic gene transfer to muscle. Nat Biotechnol. 2010;28:79–82. doi: 10.1038/nbt.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kern A, Schmidt K, Leder C, Muller OJ, Wobus CE, Bettinger K, Von der Lieth CW, King JA, Kleinschmidt JA. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J Virol. 2003;77:11072–11081. doi: 10.1128/JVI.77.20.11072-11081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller OJ, Leuchs B, Pleger ST, Grimm D, Franz WM, Katus HA, Kleinschmidt JA. Improved cardiac gene transfer by transcriptional and transductional targeting of adeno-associated viral vectors. Cardiovasc Res. 2006;70:70–78. doi: 10.1016/j.cardiores.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 33.Handa A, Muramatsu S, Qiu J, Mizukami H, Brown KE. Adeno-associated virus (AAV)-3-based vectors transduce haematopoietic cells not susceptible to transduction with AAV-2-based vectors. J Gen Virol. 2000;81:2077–2084. doi: 10.1099/0022-1317-81-8-2077. [DOI] [PubMed] [Google Scholar]
- 34.Summerford C, Samulski RJ. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lerch TF, Chapman MS. Identification of the heparin binding site on adeno-associated virus serotype 3B (AAV-3B) Virology. 2012;423:6–13. doi: 10.1016/j.virol.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie Q, Lerch TF, Meyer NL, Chapman MS. Structure-function analysis of receptor-binding in adeno-associated virus serotype 6 (AAV-6) Virology. 2011;420:10–19. doi: 10.1016/j.virol.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu Z, Miller E, Agbandje-McKenna M, Samulski RJ. Alpha2,3 and alpha2,6 N-linked sialic acids facilitate efficient binding and transduction by adeno-associated virus types 1 and 6. J Virol. 2006;80:9093–9103. doi: 10.1128/JVI.00895-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaludov N, Brown KE, Walters RW, Zabner J, Chiorini JA. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J Virol. 2001;75:6884–6893. doi: 10.1128/JVI.75.15.6884-6893.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen S, Bryant KD, Sun J, Brown SM, Troupes A, Pulicherla N, Asokan A. Glycan binding avidity determines the systemic fate of adeno-associated virus type 9. J Virol. 2012;86:10408–10417. doi: 10.1128/JVI.01155-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bell CL, Gurda BL, Van Vliet K, Agbandje-McKenna M, Wilson JM. Identification of the galactose binding domain of the AAV9 capsid. J Virol. 2012 doi: 10.1128/JVI.00448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Capila I, Linhardt RJ. Heparin-protein interactions. Angew Chem Int Ed Engl. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 42.Hacker U, Nybakken K, Perrimon N. Heparan sulphate proteoglycans: the sweet side of development. Nat Rev Mol Cell Biol. 2005;6:530–541. doi: 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- 43.Parish CR. The role of heparan sulphate in inflammation. Nat Rev Immunol. 2006;6:633–643. doi: 10.1038/nri1918. [DOI] [PubMed] [Google Scholar]
- 44.Powell AK, Yates EA, Fernig DG, Turnbull JE. Interactions of heparin/heparan sulfate with proteins: appraisal of structural factors and experimental approaches. Glycobiology. 2004;14:17R–30R. doi: 10.1093/glycob/cwh051. [DOI] [PubMed] [Google Scholar]
- 45.Sasisekharan R, Raman R, Prabhakar V. Glycomics approach to structure-function relationships of glycosaminoglycans. Annu Rev Biomed Eng. 2006;8:181–231. doi: 10.1146/annurev.bioeng.8.061505.095745. [DOI] [PubMed] [Google Scholar]
- 46.Brister SJ, Buchanan MR, Griffin CC, Van Gorp CL, Linhardt RJ. Dermatan disulfate, an inhibitor of thrombin and complement activation. #5,922,690. US Patent. 1999
- 47.Yates EA, Santini F, Guerrini M, Naggi A, Torri G, Casu B. 1H and 13C NMR spectral assignments of the major sequences of twelve systematically modified heparin derivatives. Carbohydr Res. 1996;294:15–27. doi: 10.1016/s0008-6215(96)90611-4. [DOI] [PubMed] [Google Scholar]
- 48.Urabe M, Ding C, Kotin RM. Insect cells as a factory to produce adeno-associated virus type 2 vectors. Hum Gene Ther. 2002;13:1935–1943. doi: 10.1089/10430340260355347. [DOI] [PubMed] [Google Scholar]
- 49.Lerch Thomas F, O’Donnell Jason K, Meyer Nancy L, Xie Q, Taylor Kenneth A, Stagg Scott M, Chapman Michael S. Structure of AAV-DJ, a Retargeted Gene Therapy Vector: Cryo-Electron Microscopy at 4.5 Å Resolution. Structure. 2012;20:1310–1320. doi: 10.1016/j.str.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie Q, Ongley HM, Hare J, Chapman MS. Crystallization and preliminary X-ray structural studies of adeno-associated virus serotype 6. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64:1074–1078. doi: 10.1107/S1744309108033290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hernaiz M, Liu J, Rosenberg RD, Linhardt RJ. Enzymatic modification of heparan sulfate on a biochip promotes its interaction with antithrombin III. Biochem Biophys Res Commun. 2000;276:292–297. doi: 10.1006/bbrc.2000.3453. [DOI] [PubMed] [Google Scholar]
- 52.O’Donnell J, Taylor KA, Chapman MS. Adeno-associated virus-2 and its primary cellular receptor - Cryo-EM structure of a heparin complex. Virology. 2009;385:434–443. doi: 10.1016/j.virol.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kirschner KN, Yongye AB, Tschampel SM, Gonzalez-Outeirino J, Daniels CR, Foley BL, Woods RJ. GLYCAM06: a generalizable biomolecular force field. Carbohydrates. J Comput Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
- 57.Xie Q, Bu W, Bhatia S, Hare J, Somasundaram T, Azzi A, Chapman MS. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc Natl Acad Sci U S A. 2002;99:10405–10410. doi: 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCraw DM, O’Donnell JK, Taylor KA, Stagg SM, Chapman MS. Structure of adeno-associated virus-2 in complex with neutralizing monoclonal antibody A20. Virology. 2012;431:40–49. doi: 10.1016/j.virol.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Opie SR, Warrington KH, Jr, Agbandje-McKenna M, Zolotukhin S, Muzyczka N. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J Virol. 2003;77:6995–7006. doi: 10.1128/JVI.77.12.6995-7006.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang F, McLellan JS, Ayala AM, Leahy DJ, Linhardt RJ. Kinetic and structural studies on interactions between heparin or heparan sulfate and proteins of the hedgehog signaling pathway. Biochemistry. 2007;46:3933–3941. doi: 10.1021/bi6025424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang F, Zhang Z, Lin X, Beenken A, Eliseenkova AV, Mohammadi M, Linhardt RJ. Compositional analysis of heparin/heparan sulfate interacting with fibroblast growth factor fibroblast growth factor receptor complexes. Biochemistry. 2009;48:8379–8386. doi: 10.1021/bi9006379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang F, Walcott B, Zhou D, Gustchina A, Lasanajak Y, Smith DF, Ferreira RS, Correia MT, Paiva PM, Bovin NV, Wlodawer A, Oliva ML, Linhardt RJ. Structural Studies of the Interaction of Crataeva tapia Bark Protein with Heparin and Other Glycosaminoglycans. Biochemistry. 2013 doi: 10.1021/bi400077b. [DOI] [PMC free article] [PubMed] [Google Scholar]