Abstract
Background
The diterpene cyclase ent-copalyl diphosphate synthase (CPS) catalyzes the first committed step in the biosynthesis of gibberellins. The previously reported 2.25 Å resolution crystal structure of CPS complexed with (S)-15-aza-14,15-dihydrogeranylgeranyl thiolodiphosphate (1) established the αβγ domain architecture, but ambiguities regarding substrate analog binding remained.
Method
Use of crystallization additives yielded CPS crystals diffracting to 1.55 Å resolution. Additionally, active site residues that hydrogen bond with D379, either directly or through hydrogen bonded water molecules, were probed by mutagenesis.
Results
This work clarifies structure-function relationships that were ambiguous in the lower resolution structure. Well-defined positions for the diphosphate group and tertiary ammonium cation of 1 as well as extensive solvent structure, are observed.
Conclusions
Two channels involving hydrogen bonded solvent and protein residues lead to the active site, forming hydrogen bonded "proton wires" that link general acid D379 with bulk solvent. These proton wires may facilitate proton transfer with the general acid during catalysis. Activity measurements made with mutant enzymes indicate that N425, which donates a hydrogen bond directly to D379, and T421, which hydrogen bonds with D379 through an intervening solvent molecule, help orient D379 for catalysis. Residues involved in hydrogen bonds with the proton wire, R340 and D503, are also important. Finally, conserved residue E211, which is located near the diphosphate group of 1 is proposed to be a ligand to Mg2+ required for optimal catalytic activity.
General Significance
This work establishes structure-function relationships for class II terpenoid cyclases.
Keywords: protein crystallography, terpene cyclase, enzyme mechanism, gibberellin biosynthesis
1. Introduction
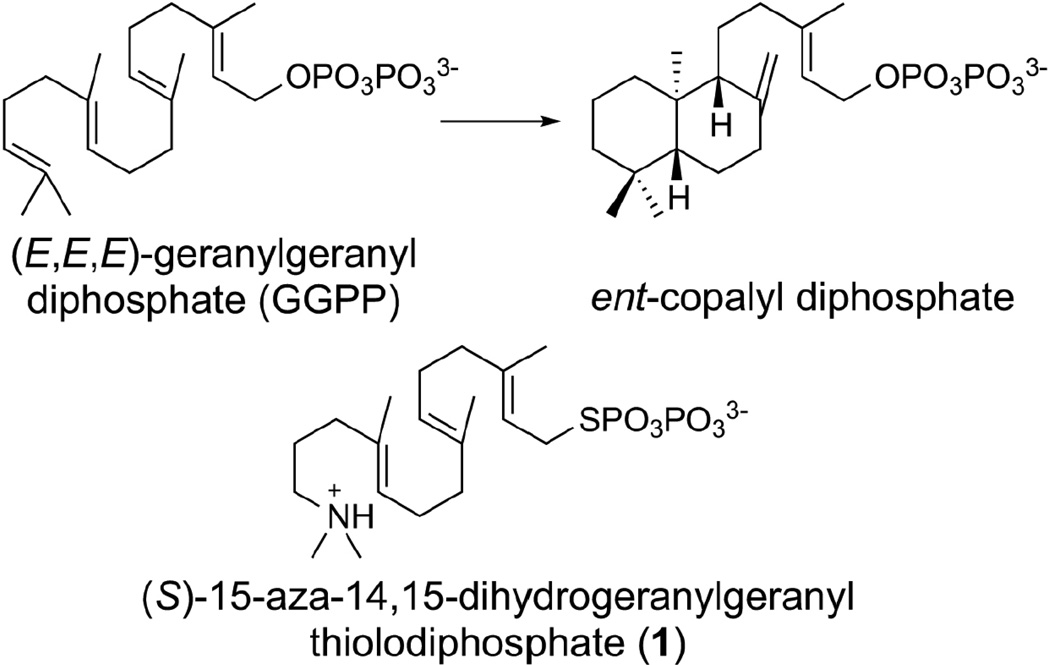
The diterpene cyclase ent-copalyl diphosphate synthase (CPS) from Arabidopsis thaliana is a class II terpenoid cyclase that initiates the cyclization of the substrate, (E,E,E)-geranylgeranyl diphosphate (GGPP), by protonation of the C14,C15 double bond to form a tertiary carbocation, thereby triggering a cascade of carbon-carbon bond forming reactions that ultimately yield the bicyclic product ent-copalyl diphosphate (Figure 1) [1–3]. This comprises the first committed step in the biosynthesis of gibberellins, ubiquitous phytohormones that function in plant growth and development [4–6]. The active site of CPS plays a critical role as a template in this reaction, since the highly flexible C20-isoprenoid substrate must be fixed in a specific conformation to direct carbon-carbon bond formation with structural and stereochemical precision. While Mg2+ is required for optimal catalytic activity, this metal ion does not participate directly in the chemistry of catalysis. Instead, Mg2+ is believed to anchor the position of the substrate diphosphate group to help chaperone the substrate binding conformation [2].
Figure 1.
Cyclization reaction catalyzed by ent-copalyl diphosphate synthase and the unreactive substrate analogue (S)-15-aza-14,15-dihydrogeranylgeranyl thiolodiphosphate (1).
The general acid responsible for substrate protonation is the "middle" aspartic acid residue in the DXDD amino acid sequence motif that characterizes a class II terpenoid cyclase [7–10]. This motif has diverged to VXDC in the related oxidosqualene cyclases [11–13], in which the aspartic acid protonates the oxirane ring of the substrate to generate the initial carbocation. Neither general acid motif is to be confused with the "DDXXD" motif in which the first and sometimes the last aspartate residue coordinate to metal ions in the active site of a class I terpenoid cyclase [8, 14]. While carbon-carbon bond forming chemistry similarly proceeds through multiple carbocation intermediates in class I cyclase reactions, initial carbocation formation is achieved by the metal-dependent ionization of the substrate diphosphate group to generate an allylic carbocation. Thus, class I and class II diterpene cyclases utilize the same substrate, GGPP, but they initiate carbocation formation at opposite ends of this substrate. The evolution of alternative cyclase classes reflects the evolution of alternative regiochemical strategies for carbocation generation in a common substrate.
Four crystal structures of class II terpenoid cyclases have been reported to date: the C30-triterpene cyclases squalene-hopene cyclase and oxidosqualene cyclase at 2.0 Å and 2.1 Å resolution, respectively, and the C20-diterpene cyclases abietadiene synthase (a bifunctional class I and class II cyclase) and CPS at 2.3 Å and 2.25 Å resolution, respectively [15–19]. Each of these cyclases exhibits the characteristic αβγ domain architecture predicted in an elegant bioinformatics study [20]. Common structure-function relationships include the DXDD (or VXDC) general acid motif positioned at the "bottom" of a hydrophobic active site cleft at the interface of the β and γ domains. This cleft is complementary to the nonpolar portion of the isoprenoid substrate; additional polar channels leading to the general acid motif could be implicated in proton transfer during catalysis. Given that the protonation of a carbon-carbon double bond (C=C) typically requires a strong acid such as H3O+ (pKa = −1.7), it is curious as to how the aspartic acid general acid in the active site of a class II terpenoid cyclase can provide a sufficiently acidic proton to efficiently protonate a C=C bond. The environment of the aspartic acid presumably modulates its pKa from ~4 measured for the free aspartic acid side chain to a value comparable to that measured for H3O+ to ensure an appropriately strong acid function in the active site.
Here, we report the 1.55 Å-resolution crystal structure of CPS complexed with (S)-15-aza-14,15-dihydrogeranylgeranyl thiolodiphosphate (1) (Figure 1), which mimics the tertiary carbocation formed upon protonation of the C14,C15 double bond of substrate GGPP. The structure of this complex prepared in the presence of Mg2+ ion clarifies various aspects of the previously-reported structure at 2.25 Å-resolution, including a well-defined position for the diphosphate group and tertiary ammonium cation of 1. Additionally, the 1.55 Å-resolution structure provides an unparalleled view of solvent structure in the active site of a class II terpenoid cyclase. Intriguingly, hydrogen bonded water molecules appear to form "proton wires" that link general acid D379 with bulk solvent through secondary channels distinct from the main entrance to the active site cavity. These proton wires define possible trajectories for regenerating the active form of the general acid during catalysis. The results of site-directed mutagenesis support the proposed roles for active site residues in substrate binding and catalysis.
2. Materials and Methods
2.1. Crystallization and structure refinement
The crystallization of CPS has been previously described [19]. Briefly, a 2-µL drop of protein solution [5 mg/mL CPS, 25 mM 3-morpholino-2-hydroxypropanesulfonic acid (pH 6.8), 2.0 mM 1, 2.0 mM MgCl2, 10% glycerol, 300 mM NaCl, 1.0 mM dithiothreitol (DTT) (incubated at 4 °C for 6 hrs and centrifuged to clear the solution)] was added to a 1.6-µL drop of precipitant solution [100 mM sodium citrate (pH 5.4), 30% polyethylene glycol 400, 200 mM KH2PO4]. Subsequently, 0.4-µL drop of 40% (v/v) 1,4-butanediol was added to this mixture and equilibrated against a 100-µL reservoir of precipitant solution. Rectangular prism-shaped plates appeared in 2–3 weeks and grew to maximal dimensions of 50 µm × 100 µm × 200 µm. Crystals were flash-cooled after transfer to a cryoprotectant solution consisting of the mother liquor augmented with 10% glycerol.
Crystals of the CPS-1 complex diffracted X-rays to 1.55 Å-resolution at the National Synchrotron Light Source (NSLS), Brookhaven National Laboratory, beamline X29A, using incident radiation with λ = 0.9795 Å. All diffraction data were processed with HKL2000 [21]. Crystals of the CPS-1 complex belonged to space group P212121 with unit cell parameters a = 130.2 Å, b = 51.4 Å, c = 114.3 Å. Data collection and reduction statistics are recorded in Table 1. The atomic coordinates from the previously reported for the CPS-1 complex determined at 2.25 Å-resolution (PDB ID: 3PYA) [19] were used as the starting point for refinement with PHENIX [22]. Manual model rebuilding was performed with COOT [23]. In the final model of the CPS-1 complex, 678 of 727 residues are present; disordered segments excluded from the final model include N-terminal residues M84-S89 (M84 is the N-terminus of the construct), the C-terminal hexahistidine tag and one of its associated linker residues (S804-H810), and surface loops I617-G638, G687-E689, and R730-E740. The refined structure also includes one S-hydroxy-L-cysteine residue at position 411, presumably the result of radiation damage to C411. Refinement statistics are recorded in Table 1. Ramachandran plot statistics were calculated with PROCHECK [24] and simulated-annealing omit maps were calculated with CNS [25]. Protein structure figures were prepared with the graphics program PyMol (http://www.pymol.org). Atomic coordinates of the 1.55 Å-resolution structure of the CPS-1 complex have been deposited in the Protein Data Bank (www.rcsb.org) with accession code 4LIX.
Table 1.
Data Collection and Refinement Statistics for the CPS-1 Complex
| A. Data Collection | |
| Incident wavelength (Å) | 0.9795 |
| Resolution range (Å) | 50.0-1.55 |
| No. of reflections (total/unique) | 644,615 / 110,705 |
| Completenessa (%) | 98.6 (96.9) |
| Redundancya | 5.8 (5.4) |
| I/σ | 24.4 (2.6) |
| Rmergeb | 0.061 (0.734) |
| B. Refinement | |
| Rwork/Rfreec | 0.169 / 0.196 |
| Protein atomsd | 5,647 |
| Solvent atomsd | 673 |
| Ligand atomsd | 40 |
| R.m.s. deviations | |
| Bonds (Å) | 0.015 |
| Angles (°) | 1.6 |
| Dihedral angles (°) | 21.8 |
| Improper dihedral angles (°) | 1.6 |
| Average B factors (Å2) | |
| Main chain | 22 |
| Side chain | 28 |
| Ligand | 44 |
| Solvent | 37 |
| Ramachandran plot | |
| Allowed (%) | 93.5 |
| Additionally allowed (%) | 6.3 |
| Generously allowed (%) | 0.2 |
| Disallowed (%) | 0 |
Number in parentheses refer to the outer shell of data.
Rmerge = Σ|I-〈I〉|/ΣI, where I is the observed intensity and 〈I〉I is the average intensity calculated from replicate data.
Rwork = Σ||Fo|-|Fc||/Σ|Fo| for reflections contained in the working set, and Rfree = Σ||Fo|-|Fc||/Σ|Fo| for reflections contained in the test set held aside during refinement (1% of the total number of reflections). |Fo| and |Fc| are the observed and calculated structure factor amplitudes, respectively.
Per asymmetric unit.
2.2. Mutagenesis
The following forward and reverse primer sets were used for site directed mutagenesis of selected residues by using the QuickChange protocol (Agilent, USA): CPS_T421A_F, CAATCAAACCAAGCAGTAGCCGGTATGTTCAACCTATAC; CPS_T421A_R, GTATAGGTTGAACATACCGGCTACTGCTTGGTTTGATTG; CPS_T421S_FN, GTGGGGCAATCAAACCAAGCAGTATCCGGTATGTTCAACCTATACCGGGC; CPS_T421S_RN, GCCCGGTATAGGTTGAACATACCGGATACTGCTTGGTTTGATTGCCCCAC; CPS_F424A_FN, CAATCAAACCAAGCAGTAACCGGTATGGCCAACCTATACCGGGCATCACAATTG; CPS_F424A_RN, CAATTGTGATGCCCGGTATAGGTTGGCCATACCGGTTACTGCTTGGTTTGATTG; CPS_N425A_FN, CCAAGCAGTAACCGGTATGTTCGCCCTATACCGGGCATCACAATTGG; CPS_N425A_RN, CCAATTGTGATGCCCGGTATAGGGCGAACATACCGGTTACTGCTTGG; CPS_N425H_FN, CCAAGCAGTAACCGGTATGTTCCACCTATACCGGGCATCACAATTGG; CPS_N425H_RN, CCAATTGTGATGCCCGGTATAGGTGGAACATACCGGTTACTGCTTGG; CPS_E211A_FN, GATGAGCATATGCCAATCGGATTCGCAGTAGCATTCCCATCGTTGCTTGAG; CPS_E211A_RN, CTCAAGCAACGATGGGAATGCTACTGCGAATCCGATTGGCATATGCTCATC; CPS_R340A_FN, CACATATGGATAGTGGATCGGTTACAAGCTTTAGGGATATCGAGATACTTTGAAG; CPS_R340A_RN, CTTCAAAGTATCTCGATATCCCTAAAGCTTGTAACCGATCCACTATCCATATGTG; CPS_D503A_FN, GATCAATATGGTGGAGAAAACGCCGTTTGGATTGGCAAGACTCTTTATAG; CPS_D503A_RN, CTATAAAGAGTCTTGCCAATCCAAACGGCGTTTTCTCCACCATATTGATC.
2.3. Enzyme Purification and Assay
The various CPS constructs described here were expressed from pET22bCV (a custom-made variant of pET22b) as 6xHis tagged fusion proteins, for ease of purification, in the C41 OverExpress strain of Escherichia coli (Lucigen), much as previously described [19,26]. Briefly, the recombinant E. coli were grown in 1 L NZY media to 0.6 A600 at 37 °C, then shifted to 16 °C for an hour prior to induction with 0.5 mM IPTG, followed by overnight fermentation at 16 °C. Cells were harvested by centrifugation for (5,000 g × 20 min. at 4°C). Cells were resuspended in 40 mL lysis buffer (50 mM Bis-Tris, pH 6.8, 150 mM KCl, 10 mM MgCl2, 1 mM DTT, 10% glycerol) and lysed by passing twice through an Avestin Emulsiflex-C5 homogenizer at 15,000 psi, with the resulting lysate clarified by centrifugation (15,000 g × 20 min. at 4°C). The tagged enzymes were purified over Ni-NTA His-bind resin on a Bio-Rad Logic LP system, using a running buffer of 50 mM NaPO4, 150 mM NaCl, and 10 mM MgCl2, with 10 mM (buffer A) or 250 mM (buffer B) imidazole. Cell lysate was passed over a 5 mL column equilibrated with buffer A; this was then washed with 30 mL of buffer A, and the tagged CPS eluted with a 0–100% gradient of buffer B over 20 mL. Fractions were checked by SDS-PAGE, with those containing >90% pure CPS pooled (≥5 mL total) and dialyzed overnight against 1 L of storage buffer (10 mM Bis-Tris, pH 6.8, 150 mM KCl, 10 mM MgCl2, 1 mM DTT, 10% glycerol). Enzymatic kinetic assays were conducted as previously described [3]. Briefly, assays were performed at 30°C in 1 mL of assay buffer (50 mM HEPES, pH 7.75, 100 mM KCl, 0.1 mM MgCl2, 10% glycerol). Reactions were terminated by the addition of 110 µL of 20 mM N-ethylmaleimide in 0.5 M glycine (pH 11), and incubation at 75°C for 5 min. Excess N-ethylmaleimide was deactivated by the addition of 20 µL 1M DTT and incubation for 15 min at room temperature. The samples were then neutralized with 60 µL of 1M HCl. ZnCl2 and MgCl2 were added to final concentrations of 10 mM and 100 mM (12 µL) before the addition of 4 units of bovine alkaline phosphatase (Invitrogen) to dephosphorylate both substrate and products. The samples were either incubated at room temperature, overnight, or at 37°C for 3 h. The resulting alcohols (geranylgeraniol and copalol) were extracted thrice with 1mL hexanes; extractions were pooled, dried under N2, and resuspended in 50 µL hexanes for GC analysis. The production of CPP was verified by GC-MS, with subsequent quantification of the relative catalytic turnover via GC-FID, as previously described [3]. For the reported kinetics analyses, enzyme concentration and incubation time were varied to achieve 2–8% turnover for GGPP (Isoprenoid Co). To correct for handling errors, catalytic rates were derived from the observed fractional turnover (i.e. the ratio of product to the sum of product and substrate), along with the known starting concentration of GGPP. Kinetic parameters were calculated by fitting the resulting reaction rate data to the substrate inhibition equation for wild-type, E211A, R340A, T421S, T421A, N425A and D503A CPS enzymes, or the Michaelis-Menten equation for N425H CPS (KaleidaGraph 4.0; Synergy Software, Reading, PA, USA). For all curve fits, R2 ≥ 0.90, except N425H where R2 = 0.8.
3. Results and Discussion
3.1. Structure of the CPS-1 complex at 1.55 Å-resolution
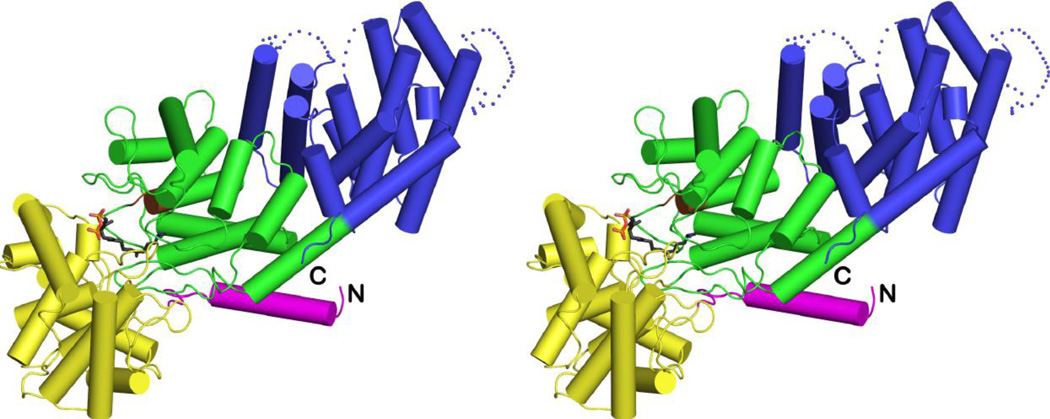
The overall fold of CPS in the 1.55 Å-resolution structure of the CPS-1 complex is essentially identical to that observed in the previously-reported 2.25 Å-resolution structure [19]. The enzyme adopts αβγ domain architecture with the active site located at the interface of the β and γ domains (Figure 2). However, there are important differences at the atomic level that now clarify structure-mechanism relationships.
Figure 2.
Overall structure of the CPS-1 complex color-coded as follows: α domain = blue; β domain = green; γ domain = yellow; N-terminal helix = magenta; DXDD general acid motif = brown. The N- and C-termini are indicated by "N" and "C", respectively. The γ domain is inserted between the N-terminal helix and the second helix of the β domain. Inhibitor 1 is shown as a stick figure bound in the active site at the interface of the β and γ domains.
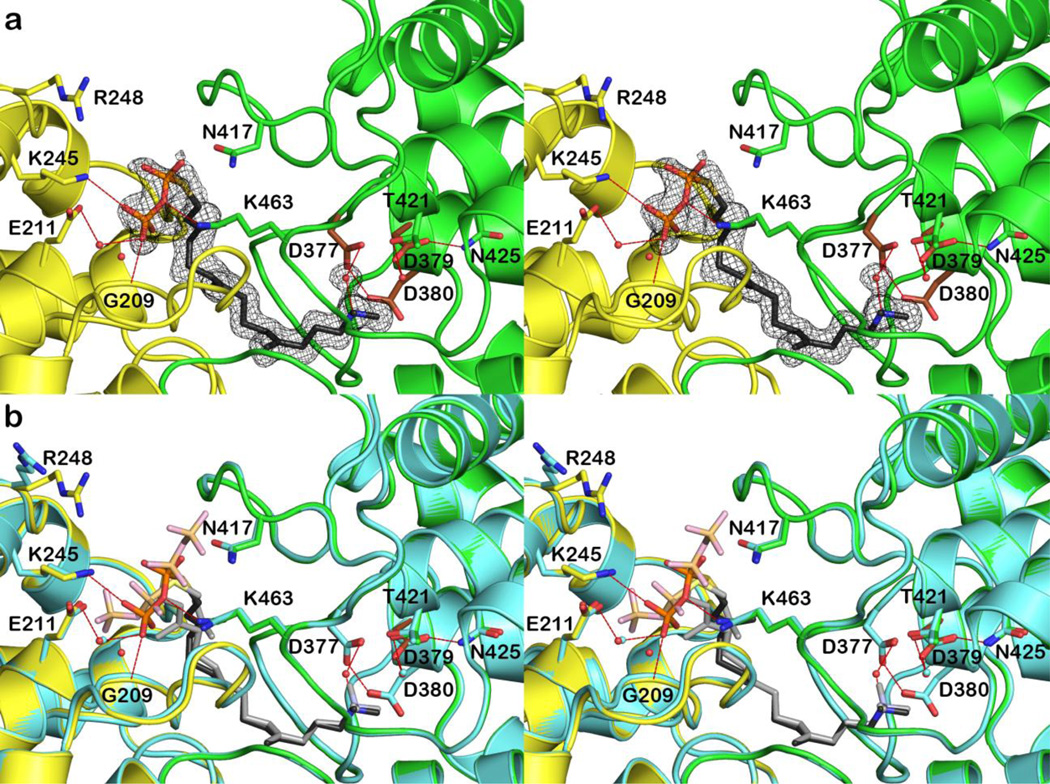
The key difference between the previously reported 2.25 Å-resolution structure of CPS [19] and the 1.55 Å-resolution structure reported herein is the presence of 1.0 mM MgCl2 in the mother liquor of the crystal. Although this magnesium salt was added with the intention of locating the catalytically-important Mg2+ binding site [2], no bound Mg2+ ions are interpretable in the electron density map, even at the higher resolution of the current crystal structure determination. The lack of Mg2+ binding is attributed to the relatively low pH of the crystal structure determination (the pH of the crystallization drop was measured to be approximately 5.4 [19]), which would weaken metal-ligand coordination interactions. We were unable to prepare crystals at higher pH values. However, the presence of Mg2+ in the crystallization buffer apparently affects the conformation of the diphosphate group of 1, which was previously disordered between two positions in the absence of Mg2+ and is now ordered in a single position in the presence of Mg2+ (Figure 3a).
Figure 3.
(a) Simulated annealing omit map (contoured at 4.0σ) of the CPS-1 complex at 1.55 Å resolution. Atoms are color-coded as follows: C = green (β domain), brown (DXDD general acid motif in the β domain), or yellow (γ domain), N = blue, O = red, P = orange, S = yellow; selected water molecules are shown as small red spheres, and hydrogen bond interactions are indicated by dash red lines. (b) Superposition of the 1.55 Å-resolution structure of the CPS-1 complex color-coded as in (a) with the 2.25 Å-resolution structure (all atoms cyan).
Since 1 binds with an extended conformation, as previously noted in the 2.25 Å-resolution structure of the CPS-1 complex [19], it cannot mimic the overall conformation or intermolecular interactions of GGPP required for cyclization to ent-copalyl diphosphate. However, the binding conformation of 1 may reflect the penultimate precatalytic substrate binding conformation, such that the substrate diphosphate group is not likely to shift too far away from the conserved basic segment K241-R248 upon assuming a cyclization-competent precatalytic conformation. The diphosphate group of 1 accepts direct hydrogen bonds from conserved residues K245 and K463, and the backbone NH group of G209. Since electron density defining the side chains of K245 and K463 is very weak, hydrogen bonds with these residues may not be strong. The carboxamide group of N417 is close to the diphosphate group of 1, but with a separation of 3.5 Å this interaction is too long to be considered a hydrogen bond. The diphosphate group of 1 also accepts a hydrogen bond from a water molecule that also donates a hydrogen bond to E211 (Figures 3a and 3b). A second water molecule is 3.5 Å away from the diphosphate group of 1, but this interaction is too long to be considered a hydrogen bond. The bound conformation of 1 may reflect an incompletely-desolvated substrate binding mode that precedes formation of the productive precatalytic enzyme-substrate complex. We hypothesize that E211 could serve as a ligand to the catalytically-obligatory Mg2+ ion, which could then be coordinated by the substrate diphosphate group once it is completely desolvated.
A second difference evident in the higher resolution structure of the CPS-1 complex is a single conformation for the tertiary ammonium group that mimics the initial carbocation generated by protonation of the C14-C15 bond of GGPP. In the 2.25 Å-resolution structure, the electron density map indicated two alternative conformations of this group [19], whereas the 1.55 Å-resolution electron density map definitively shows a single conformation of this group and a hydrogen bonded water molecule (Figure 3a). The ammonium nitrogen atom (which would correspond to the C15 atom of substrate GGPP) makes no other hydrogen bond interactions and is 4.2 Å from general acid D379. The C14 atom of 1, which corresponds to the C14 atom of GGPP that would be protonated by the general acid, is 5.7 Å from D379. A productive substrate binding conformation would enable direct proton transfer from D379 to the substrate C14 atom. While it might be tempting to speculate that the water molecule hydrogen bonded to the ammonium nitrogen atom of 1 could be involved in the substrate protonation step, this water molecule does not hydrogen bond with general acid D379.
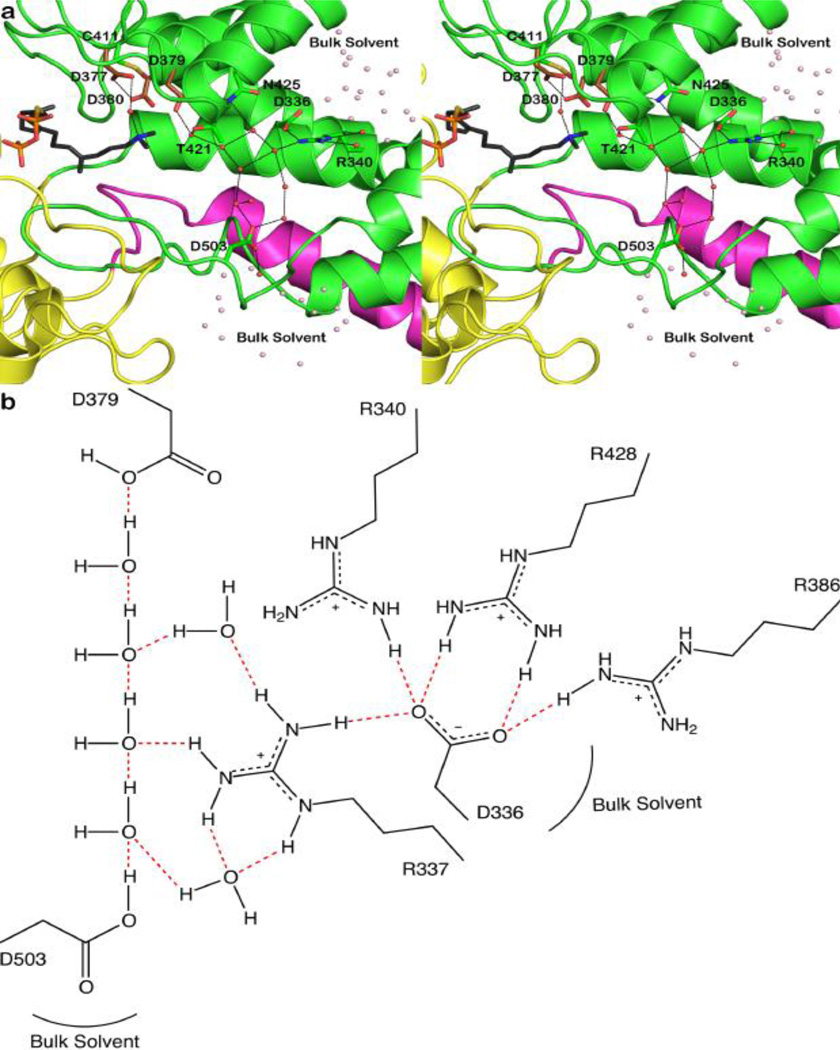
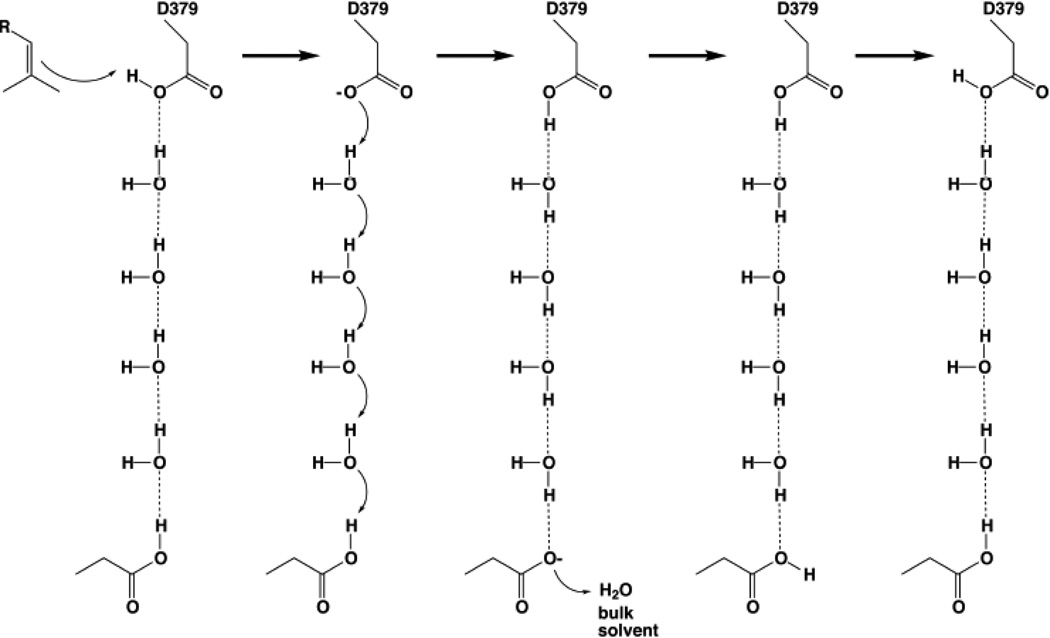
The final difference of note between the 2.25 Å-resolution structure and the new 1.55 Åresolution structure of the CPS-1 complex is a more accurate and extensive view of solvent structure: 263 water molecules are observed in the lower resolution structure [19], whereas 673 water molecules are observed in the higher resolution structure (Table 1). Extensive solvent structure is observed at the interface of the α and β domains and in the active site at the interface of the β and γ domains. Additionally, two hydrogen-bonded solvent channels lead away from general acid D379 (Figure 4). These channels are distinct from the main active site cleft, i.e., the substrate entry and product egress route, and may influence proton transfer to and from D379 during catalysis. The hydrogen bond geometry around D379 suggests that this residue serves its general acid function through the more acidic [27] anti-oriented proton; the resulting carboxylate conjugate base form of D379 can subsequently be reprotonated through the more basic syn geometry by Grotthuss diffusion through one of the hydrogen bonded solvent channels, as illustrated in Figure 5.
Figure 4.
(a) Cut-away view of the CPS active site showing solvent channels leading to general acid D379. One channel leads to D503 and the solvent-exposed surface of the β domain (green) near the interface with the γ domain (yellow); the other channel leads to D336-R340, which are exposed to bulk solvent at the interface of the α (blue) and β domains. These channels are distinct from the main active site cleft, which opens to the left in this view. (b) Molecular scheme showing hydrogen bond interactions linking general acid D379 with bulk solvent through the two polar solvent channels shown in (a). For clarity, R337, R386, and R428 are omitted from the view in (a) but are shown here to emphasize the highly polar nature of the hydrogen bond networks with D379.
Figure 5.
General acid D379 could be reprotonated by Grotthuss diffusion of a proton across a 4-solvent "proton wire" in a polar channel leading to another aspartic acid serving as a proton shuttle to bulk solvent, such as D503 or D336; in order for D336 to serve in this capacity, its hydrogen bond with R340 must be displaced by a solvent molecule.
3.2. Catalytic activity of CPS mutants
In order to probe the function of residues that could influence the general acid function of D379, we prepared site-specific mutants of CPS in which adjacent residues T421 and N425 were substituted by alternative amino acids. Additionally, we prepared exploratory mutants in which selected polar residues lining the solvent channels, D336 and R340, were substituted. Finally, we studied a mutant of the proposed Mg2+ ligand, E211. Steady-state kinetic parameters measured from these mutants are recorded in Table 2.
Table 2.
Steady-State Kinetic Parameters for CPS Mutants
| kcat (s−1) | KM (µM) | kcat/KM (M−1s−1) | |
|---|---|---|---|
| Wild-type | 0.9 ± 0.2 | 3 ± 1 | 3 × 105 |
| T421S | 0.3 ± 0.1 | 3 ± 2 | 1 × 105 |
| T421A | 0.006 ± 0.002 | 6 ± 3 | 1 × 103 |
| N425H | 0.012 ± 0.001 | 0.8 ± 0.4 | 2 × 104 |
| N425A | 0.071 ± 0.007 | 1.8 ± 0.5 | 4 × 104 |
| E211A | 0.0019 ± 0.0003 | 1.7 ± 0.6 | 1.1 × 103 |
| D503A | 0.13 ± 0.02 | 2.9 ± 0.7 | 4.5 × 104 |
| R340A | 0.0011 ± 0.0001 | 2.0 ± 0.5 | 5.5 × 102 |
n.m., not measurable.
The side chain hydroxyl group of T421 hydrogen bonds with a water molecule, which in turn donates a syn-oriented hydrogen bond to the side chain Oδ2 atom of D379 (Figure 4). The T421A mutant exhibits a 163-fold reduction in kcat (KM is increased only 2-fold), suggesting that this residue and its hydrogen bonded water molecule play an important role in catalysis. That kcat is reduced by less than 3-fold and KM is essentially invariant in T421S CPS further supports an important role for the γ-hydroxyl group of residue 421. Possibly, the water-bridged hydrogen bond between T421 and D379 is important for orienting the carboxylic acid side chain of D379 for its general acid function.
The side chain carboxamide group of N425 donates a syn-oriented hydrogen bond directly to the side chain Oδ1 atom of D379 (Figure 4), so it is likely that this residue similarly plays an important role in orienting the general acid catalyst. In accord with this expectation, the catalytic activity of N425A CPS exhibits a 13-fold reduction in kcat. The corresponding residue in squalene-hopene cyclase is a histidine [15, 16], and the substitution of a histidine residue in N425H CPS exhibits a 75-fold reduction in kcat.
The substitution of selected polar residues lining solvent channels leading to general acid D379 influences kcat, while KM remains relatively invariant. Although D336A CPS is not sufficiently stable to allow for kinetic measurements, D503A CPS exhibits a 7-fold reduction in kcat; this residue, through hydrogen bonding, connects the active site solvent network directly to the surface of the β domain. In contrast, R340A CPS exhibits an 850-fold reduction in kcat; this residue hydrogen bonds with D336 and blocks the solvent channel leading to the interface between the α and β domains. However, since R340 hydrogen bonds with solvent in the channel as well as bulk solvent, R340 and/or its hydrogen bond partner D336 could mediate proton transfer to D379. While the structural consequences of these mutations must be fully evaluated in future X-ray crystal structure determinations to confirm that no unanticipated structural changes are triggered by the mutations, our working hypothesis is that the solvent channel leading to the interface between the α and β domains is important for catalytic function. This channel corresponds to that first identified in the X-ray crystal structure determination of oxidosqualene cyclase [17], which exhibits βγ domain architecture.
Finally, deletion of the putative Mg2+ ligand in E211A CPS results in a nearly 500-fold reduction in kcat but only a slight reduction in KM. In the high resolution structure, a water molecule bridges the diphosphate group of 1 and the E211 side chain. While this confirms the catalytic importance of E211, the kinetic results suggest that the postulated coordination of the substrate diphosphate group to Mg2+ may be more important for stabilizing the cyclization-competent conformation in the transition state [28].
3.3. General acid function of D379
It is unusual to find general acid catalysis function within a cluster of carboxylic acid-containing amino acids in a protein structure, such as the DXDD motif of a class II terpenoid cyclase. More typically, a carboxylate cluster is implicated in binding metal ions [29], as is the case for the DDXXD motif that characterizes a class I terpenoid cyclase. While the middle aspartic acid in the DXDD motif is protonated and serves as the general acid in catalysis by class II terpenoid cyclases, the first and third carboxylic acids in this motif are equally interesting in that they are sufficiently close to require a hydrogen bond between them. In the CPS-1 complex, D380 donates an anti-oriented hydrogen bond to D378, which receives the hydrogen bond with syn-orientation. Based on this hydrogen bond interaction and the presumed protonation state of D379, the carboxylic acid cluster in the DXDD motif of a class II terpenoid cyclase bears a net charge of −1. Following substrate protonation by D379, the carboxylic acid cluster bears a net charge of −2. It is proposed that this additional negative charge provides electrostatic stabilization for carbocation intermediates in the cyclization cascade [11].
As noted upon the first crystal structure determination of a class II terpenoid cyclase, squalene-hopene cyclase [15], the general acid aspartic acid is located such that the more acidic anti-oriented proton is oriented toward the substrate [11]. This is also the case for CPS (Figure 3) [19] and abietadiene synthase [18]. Interestingly, this is not the case for oxidosqualene cyclase [17], perhaps due to the fact that the moiety being protonated is not an isoprenoid C=C bond, but instead an epoxide ring; the epoxide is more reactive and may not require as powerful a Brønsted acid for efficient protonation. Additionally, oxidosqualene cyclase does not have a residue comparable to N425 of CPS or H451 of squalene-hopene cyclase to orient the general acid aspartic acid side chain. Instead, these residues appear as A536 in oxidosqualene cyclase.
Since the anti-oriented conformer of a carboxylic acid is less stable than the syn-oriented conformer by 6–8 kcal/mol [30–32], the enzyme active site must provide an environment that stabilizes the anti-oriented conformer of D379 so that it is poised for general acid catalysis. The hydrogen bond network involving D379, N425, and T421 ensures that only the anti-oriented position of the side chain Oδ2 atom of D379 is oriented toward the substrate binding site. Mutagenesis results suggest that N425 helps to orient D379 for catalysis; T421 plays a similar role, and may also orient the bridging solvent molecule to enable proton transfer with D379 to regenerate the active form of the general acid at the conclusion of the cyclization cascade.
Finally, regeneration of the anti-oriented general acid D379 requires reprotonation after each catalytic turnover, and this may occur through either one or both of the solvent channels identified in the structure of the CPS-1 complex (Figure 5). Proton transfer across hydrogen bonded solvent networks in such channels through Grotthuss diffusion is more efficient than the hydrodynamic diffusion of H3O+ [33] and may account for mutagenesis results with our exploratory site-specific mutants D503A and R340A (Table 2). Future studies will probe structure-function relationships in these active site solvent channels in greater depth.
Highlights.
The 1.55 Å-resolution structure of ent-copalyl diphosphate synthase is described.
The enzyme is complexed with an unreactive substrate analogue.
Ambiguities in the prior 2.25 Å-resolution structure of this complex are clarified.
Mutagenesis of active site residues provides insight on general acid catalysis.
Acknowledgments
We thank the National Synchrotron Light Source at Brookhaven National Laboratory for access to beamline X29A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This work was supported by US National Institutes of Health Grant GM56838 to D.W.C. and GM076324 to R.J.P.
References
- 1.Sun T-P, Kamiya Y. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell. 1994;6:1509–1518. doi: 10.1105/tpc.6.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prisic S, Peters RJ. Synergistic substrate inhibition of ent-copalyl diphosphate synthase: a potential feed-forward inhibition mechanism limiting gibberellin metabolism. Plant Physiol. 2007;144:445–454. doi: 10.1104/pp.106.095208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prisic S, Xu J, Coates RM, Peters RJ. Probing the role of the DXDD motif in class II diterpene cyclases. ChemBioChem. 2007;8:869–874. doi: 10.1002/cbic.200700045. [DOI] [PubMed] [Google Scholar]
- 4.Fleet CM, Sun T-P. A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 2005;8:77–85. doi: 10.1016/j.pbi.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi S. Gibberellin metabolism and its regulation. Ann. Rev. Plant Biol. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- 6.Hedden P, Thomas SG. Gibberellin biosynthesis and its regulation. Biochem. J. 2012;444:11–25. doi: 10.1042/BJ20120245. [DOI] [PubMed] [Google Scholar]
- 7.Wendt KU, Schulz GE. Isoprenoid biosynthesis: manifold chemistry catalyzed by similar enzymes. Structure. 1998;6:127–133. doi: 10.1016/s0969-2126(98)00015-x. [DOI] [PubMed] [Google Scholar]
- 8.Christianson DW. Structural biology and chemistry of the terpenoid cyclases. Chem. Rev. 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- 9.Siedenburg G, Jendrossek D. Squalene-hopene cyclases. Appl. Environ. Microbiol. 2011;77:3905–3915. doi: 10.1128/AEM.00300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao Y, Honzatko RB, Peters RJ. Terpenoid synthase structures: a so far incomplete view of complex catalysis. Nat. Prod. Rep. 2012;29:1153–1175. doi: 10.1039/c2np20059g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wendt KU, Schulz GE, Corey EJ, Liu DR. Enzyme mechanisms for polycyclic triterpene formation. Angew. Chem. Int. Ed. 2000;39:2812–2833. [PubMed] [Google Scholar]
- 12.Wendt KU. Enzyme mechanisms for triterpene cyclization: new pieces of the puzzle. Angew. Chem. Int. Ed. 2005;44:3966–3971. doi: 10.1002/anie.200500804. [DOI] [PubMed] [Google Scholar]
- 13.Abe I. Enzymatic synthesis of cyclic triterpenes. Nat. Prod. Rep. 2007;24:1311–1331. doi: 10.1039/b616857b. [DOI] [PubMed] [Google Scholar]
- 14.Aaron JA, Christianson DW. Trinuclear metal clusters in catalysis by terpenoid synthases. Pure Appl. Chem. 2010;82:1585–1597. doi: 10.1351/PAC-CON-09-09-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wendt KU, Poralla K, Schulz GE. Structure and function of a squalene cyclase. Science. 1997;277:1811–1815. doi: 10.1126/science.277.5333.1811. [DOI] [PubMed] [Google Scholar]
- 16.Wendt KU, Lenhart A, Schulz GE. The structure of the membrane protein squalene-hopene cyclase at 2.0 Å resolution. J. Mol. Biol. 1999;286:175–187. doi: 10.1006/jmbi.1998.2470. [DOI] [PubMed] [Google Scholar]
- 17.Thoma R, Schulz-Gasch T, D'Arcy B, Benz J, Aebi J, Dehmlow H, Hennig M, Stihle M, Ruf A. Insight into steroid scaffold formation from the structure of human oxidosqualene cyclase. Nature. 2004;432:118–122. doi: 10.1038/nature02993. [DOI] [PubMed] [Google Scholar]
- 18.Zhou K, Gao Y, Hoy JA, Mann FM, Honzatko RB, Peters RJ. Insights into diterpene cyclization from structure of bifunctional abietadiene synthase from Abies grandis . J. Biol. Chem. 2012;287:6840–6850. doi: 10.1074/jbc.M111.337592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Köksal M, Hu H, Coates RM, Peters RJ, Christianson DW. Structure and mechanism of the diterpene cyclase ent-copalyl diphosphate synthase. Nat. Chem. Biol. 2011;7:431–433. doi: 10.1038/nchembio.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao R, Zhang Y, Mann FM, Huang C, Mukkamala D, Hudock MP, Mead ME, Prisic S, Wang K, Lin FY, Chang TK, Peters RJ, Oldfield E. Diterpene cyclases and the nature of the isoprene fold. Proteins: Struct. Funct. Bioinf. 2010;78:2417–2432. doi: 10.1002/prot.22751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 25.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 26.Zhou K, Peters RJ. Investigating the conservation pattern of a putative second terpene synthase divalent metal binding motif in plants. Phytochemistry. 2009;70:366–369. doi: 10.1016/j.phytochem.2008.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gandour RD. On the importance of orientation in general base catalysis by carboxylate. Bioorg. Chem. 1981;10:169–176. [Google Scholar]
- 28.Peters RB, Croteau RB. Abietadiene synthase catalysis: conserved residues involved in protonation-initiated cyclization of geranylgeranyl diphosphate to (+)-copalyl diphosphate. Biochemistry. 2002;41:1836–1842. doi: 10.1021/bi011879d. [DOI] [PubMed] [Google Scholar]
- 29.Needham JV, Chen TY, Falke JJ. Novel ion specificity of a carboxylate cluster Mg(II) binding site: strong charge selectivity and weak size selectivity. Biochemistry. 1993;32:3363–3367. doi: 10.1021/bi00064a020. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Houk KN. Theoretical elucidation of the origin of the anomalously high acidity of Meldrum's acid. J. Am. Chem. Soc. 1988;110:1870–1872. [Google Scholar]
- 31.Wiberg KB, Laidig KE. Barriers to rotation adjacent to double bonds. 3. The carbon-oxygen barrier in formic acid, methyl formate, acetic acid, and methyl acetate. The origin of ester and amide resonance. J. Am. Chem. Soc. 1987;109:5935–5943. [Google Scholar]
- 32.Wiberg KB, Laidig KE. Acidity of (Z)- and (E)-methyl acetates: relationship to Meldrum's acid. J. Am. Chem. Soc. 1988;110:1872–1874. [Google Scholar]
- 33.Knight C, Voth GA. The curious case of the hydrated proton. Acc. Chem. Res. 2012;45:101–109. doi: 10.1021/ar200140h. [DOI] [PubMed] [Google Scholar]