Summary
Microbial products can be recognized by pattern recognition receptors (PRRs) expressed by immune and parenchymal cells and drive innate immunity that can in turn shape adaptive immune responses to microbial and transplant antigens. In transplanted patients, the signals and their downstream inflammatory cytokines elicited in response to infections can modulate ongoing alloimmune responses and modify the fate of transplanted organs. In recent years, it has become apparent that microbial signals can be generated not only by active pathogenic infections, but also by commensal microbiota thus opening a new field of research into the interplay between the microbiota and the immune system in homeostasis and disease. The wide use of antibiotics and immunosuppressive drugs in transplanted patients can have dramatic consequences on the microbiota that can, in turn, shape immune responses and perhaps alloresponses, whereas the ongoing immune responses can in turn affect the commensal or pathogenic microorganisms in a feed-forward circle. Here, we discuss known and hypothesized mechanisms for how infections or microbiota-derived signals may affect local or systemic alloimmunity and briefly review data on downstream effects of antibiotics and vaccinations.
Keywords: Infections, transplantation, rejection, microbiota, tolerance
Introduction
Transplant patients are at higher risk for infections than the general population; these infections in turn can threaten the survival of the transplanted organs, both directly via microbial-mediated damage and indirectly by activating alloimmune responses. Several factors contribute to the increased infectious risk, including greater exposure to pathogens prior to transplantation because of the underlying disease (cystic fibrosis, diabetes, virally-mediated cirrhosis, chronic catheterizations for example), possible transmission of pathogens from the donor at the time of transplantation, infections in the peri-transplant period, and increased susceptibility to infections by the recipient because of pharmacological immunosuppression following transplantation (1). In addition, transplant patients receive anti-microbial agents as prophylactic or therapeutic drugs and these can cause dysbiosis in the recipient microbiota that in turn may have immune consequences or favor growth of pathogenic organisms. Here we review how microbes can influence T cell alloimmunity.
Infections and alloimmunity
Infections prior to transplantation
It is increasingly documented that antigen recognition by T cells can be promiscuous with a single TCR capable of binding different peptide/MHC combinations with different affinities (2, 3). A recent report demonstrates that adult humans harbor memory CD4+ T cells reacting with viruses to which they have never been exposed (4). These T cells are thought to have acquired a memory phenotype from cross-reactivity to environmental antigens encountered in their hosts’ lifetime, inasmuch the T cell populations specific for the same viruses are all naïve in neonates (4). Several TCRs cross-reacting between viral peptides/self-MHC and allo-MHC/donor self-peptide have been reported in both mice and humans (5). Thus, infections occurring prior to transplantation have the potential to generate memory T cells that can bind to both viral antigens and alloantigens expressed in a subsequently transplanted organ. This is significant because memory T cells, by virtue of increased cell numbers, alterations in signaling and epigenetic changes, are more difficult to immunosuppress than naïve T cells and can constitute a barrier to transplantation tolerance in mice (6). In humans, the presence of IFN-γ-producing anti-donor T cells has been correlated with increased risk of post-transplant rejection episodes (7), and virus-specific lymphocytes have been shown to contribute significantly to the alloresponse in certain responder-stimulator HLA combinations (8). In animal models, sequential exposure to different viruses generated a population of alloreactive memory CD8+ T cells (9) while LCMV-reactive memory CD8+ T cells could drive skin allograft rejection following adoptive transfer into T cell-deficient recipients (10). In addition to these experiments with viruses, exposure to the parasite Leishmania major also generated heterologous immunity to alloantigen that could prevent tolerance induction to subsequent skin grafts (11) and at least conceptually, microbes that produce superantigens that can stimulate whole families of T cells expressing a particular TCRVβ chain may be able to generate heterologous alloreactivity in an antigen-independent manner.
Infections after transplantation
Following transplantation, infections can theoretically influence ongoing alloimmunity of both naïve and memory alloreactive T cells by different means, independently of TCR cross-reactivity. First, microbial molecules can activate pattern-recognition receptors (PRRs) expressed on many cell types including donor and recipient hematopoietic, endothelial and epithelial cells, resulting in production of inflammatory cytokines. While antigen-specific immune responses against microbial antigens are enhanced by the PRR signals, inflammatory cytokines secreted during an infection have been shown to reduce the threshold for activation (i.e. the amount of antigen needed) of other CD8+ T cells (12) and as such may be able to potentiate ongoing alloresponses. In addition, a given antigen-presenting cell (APC) can present distinct antigens on different MHC molecules such that PRR ligands from a pathogen could serve as adjuvants for APCs presenting alloantigens, resulting in a stronger alloresponse. Finally, cytokines elicited during a response to a worm infection in draining lymph nodes have been shown to permeate the whole lymph node thus determining the polarization of non-worm-reactive T cells (13) such that an ongoing infection may dictate the particular differentiation of an alloreactive T cell that is encountering alloantigen. Thus, one could imagine the phenotype of a differentiating alloreactive T cell to mimic that of the antimicrobial ones, with viruses and intracellular bacteria promoting Th1 differentiation, extracellular bacteria and some fungi driving Th17 differentiation and parasites facilitating the Th2 pathway. These effector T cells can have distinct pathological consequences to allografts, as the specific complement of cytokines and chemokines released by each T cell phenotype can cause the differential recruitment and activation of macrophages, neutrophils, or eosinophils (14, 15).
In support of these models, it has been shown that exposure to TLR ligands at the time of transplantation can prevent the ability of immunosuppressive regimens to induce long-term graft acceptance in mice (16–18), correlating with enhanced anti-donor responses, and in some models, dependent on enhanced Th1 or Th17 differentiation (19, 20). Similarly, peri-transplant infection with Listeria monocytogenes or Staphylococcus aureus could also prevent costimulation-blockade-mediated long-term graft acceptance in mice, dependent on signaling by Type I IFN or IL-6, respectively (21, 22). In a mouse model of kidney transplantation, mouse polyoma virus infection was also shown to enhance anti-donor immunity (23). Moreover, graft nephropathy did not correlate with viral load suggesting that the mechanism for renal injury is not direct viral cytopathology but more likely the interplay between the infection and the alloimmune response (24). In patients, infections have also been associated with episodes of acute rejection. While evidence is stronger for infections within the allograft (25), there are data suggesting that infections distant from the transplanted organ, and thus independently from direct microbial damage, can spur acute and chronic rejection (25).
Infections after tolerance induction
Although it is relatively easy to prevent the induction of transplantation tolerance in animal models with TLR agonists, viral, bacterial and parasitic infections at the time of transplantation, once tolerance has been achieved it is quite challenging to abrogate it in mouse models. Indeed, even combinations of TLR agonists at half-lethal doses have failed to break established tolerance to cardiac allografts (unpublished results). However, our group has reported that Listeria monocytogenes infection that results in systemic production of type I IFN and IL-6 could drive cardiac allograft rejection long after stable tolerance induction by costimulation blockade therapy, and these innate cytokines were both necessary and sufficient for the abrogation of established tolerance (26).
In patients, transplantation tolerance is a rare event for renal transplant recipients. In those patients whose state of tolerance is revealed following the stopping of their immunosuppression for various reasons, it was recently reported that this state of tolerance was not permanent. Long-term follow up of 27 patients who met the criteria for operational tolerance revealed that 8 rejected after a mean duration of 10 ± 5 years of drug-free stable graft function (27). Notably, those who lost tolerance had a higher incidence of bacterial (50 versus 10.5%) or viral (25 versus 5%) infections during the drug-free period compared to those who maintained their tolerance. While the numbers of patients in this study were small, these observations nonetheless are consistent with the notion that infections may promote transplant loss in operationally tolerant recipients.
Vaccinations and alloimmunity
Vaccinations have been widely used in transplantation to prevent infections in immunocompromized hosts. Influenza vaccination has been shown to be safe and generally well tolerated in both adult and pediatric transplant recipients, but responses to the vaccine depend on various factors including time from transplantation and specific immunosuppression regimen (28). The protection reported following trivalent inactivated influenza vaccination (29, 30) has not always been observed after vaccination against other viral illnesses (31, 32), perhaps because in contrast to influenza, people lack preexisting memory responses to these other viruses. Even with influenza vaccination, the response in transplanted patients is often blunted (33, 34).
Following vaccination of transplant recipients on immunosuppression, it is controversial whether vaccine-induced memory T and B cells may promote transplant rejection by mechanisms described above or in contrast protect the graft (and the patient) by preventing a more serious infection. Indeed, a recent report (35) suggested that adjuvanted inactivated H1N1 influenza vaccine was correlated with increased incidence of acute cellular rejection diagnosed with routine post-transplant endomyocardial biopsies in heart transplant patients. While the study population was small (N=15), the rejection rate was 6 of 15 in the vaccinated and 1 of 45 in the control non-vaccinated group, with odds ratios following multivariate analysis of predictors of acute cellular rejection being 26.5 (p<0.006). Conversely, in a recent study of 51,730 Medicare primary patients, influenza vaccination was actually associated with a lower risk for allograft loss and death compared to those who were not vaccinated (36). Thus, clinical observations such as these underscore the need to carefully dissect the effect of vaccinations/adjuvants that confer protection against infections but may also have unanticipated effects on alloreactivity and transplantation outcomes in the clinic.
Microbiota and alloimmune responses
Microbiota and the immune system
The microbiota plays an important role in preventing colonization by pathogenic microbes, drawing energy from otherwise inaccessible food components, synthesizing vitamin K and regulating host fat storage. But commensal bacteria can also shape local and distal immune responses and may determine susceptibility to autoimmunity (37) and may even be responsible for hormone-dependent gender-based autoimmune susceptibility (38). Some mechanisms by which the microbiota may affect immune cells are beginning to emerge. Microbial signals have been shown recently to promote dendritic cell migration from tissue to lymph nodes for T cell priming (39) and to poise APCs to rapidly produce type I IFN following both systemic and local viral infections (40, 41). The model suggests that intestinal microbial products translocate out of the gut to bind distant PRRs on APCs and induce permissive epigenetic modifications of anti-viral genes. Thus acute viral infections can drive a rapid production of IFN-I in the presence of normal microbiota to promote viral clearance. In contrast, in antibiotic-treated or germ-free mice, this response is impaired resulting in poorly controlled viral replication and death (41). Thus commensal flora enables a lower threshold for activation of APCs at sites distant from the intestine, and this may explain why intestinal microbiota appears to be important not only in local but also systemic immune responses and theoretically could affect both innate and subsequent adaptive immune responses to transplanted organs.
The microbiota has also been shown to modulate T cell responses, either through direct or indirect mechanisms. For instance, intestinal segmented filamentous bacterium (SFB) is known to confer resistance to the intestinal pathogen Citrobacter rodentium (42) but also to drive the development of rheumatoid arthritis (43), in both settings seemingly via its capacity to facilitate Th17 differentiation. Thus by affecting distant T cell response profiles, it is conceivable that the intestinal microbiota may affect alloimmunity not only to intestinal transplants but also to all other types of transplanted organs or tissues. In addition, the microbiota may also contribute to generation of memory alloreactive T cells. Indeed, during a gastrointestinal infection, Hand and colleagues were able to detect the onset of immune responses not only to the pathogen but also to gut intestinal commensals resulting in commensal-reactive T-cell memory (44). As discussed above, cross-reactivity of TCRs could mean that a pool of memory cells that react to alloantigens may in fact be the consequence of anti-commensal T cell memory.
Importantly, the presence of intestinal microbiota increases the number of colonic mucosa FoxP3+ regulatory T cells (Tregs) after weaning when the microbial diversity widens (45) and some of these colonic Tregs recognize microbial antigens (46, 47). In particular indigenous Clostridium species (48), and Bacteroides fragilis (49) have been shown to promote colonic iTreg differentiation. iTregs are important to prevent mucosal effector T cell responses at environmental interfaces (50) and maintain local T cell homeostasis, but have also been identified in accepted transplanted organs in mice (51) and demonstrated to play a role in the maintenance of transplantation tolerance, at least of skin grafts (52). Thus, the modulation of iTreg homeostasis by the microbiota may have a role in graft acceptance.
Though most studies have focused on intestinal microbiota, a recent report indicates that local skin but not gut microbiota facilitates protective immunity against the cutaneous pathogen Leishmania major, by augmenting IL-1α signaling and amplifying immune responses according to the local inflammatory milieu (53). By analogy, it is conceivable that the local microbiota may also influence alloimmune responses to colonized transplanted organs such as skin, lung and intestine that are observed to be more prone to rejection than sterile organs, suggesting a hypothesis that local microbiota may function as an adjuvant to enhance alloimmune responses. While seemingly intuitive in the context of our current understanding of how innate immune responses are stimulated and how they in turn license adaptive immunity, this hypothesis remains to be proven clinically or even experimentally.
Finally, not only do microbes affect immune responses, but particular immune signals can also profoundly alter microbial properties. For instance, IFN-γ production by immune cells can induce expression of virulence factors of Pseudomonas aeruginosa that can disrupt intestinal epithelium cell function (54), with further downstream consequences on immune and possibly alloimmune responses.
Microbiota and transplantation
Little is known yet about the effect of the microbiota in alloimmune responses. Descriptive investigations in patients have identified a correlation between different graft outcomes and particular changes in the gut or allograft microbiota; however, whether these are a cause or consequence of the immune response to the graft and/or are due to alterations in clinical therapeutics has yet to be delineated in most of these studies. A possible causal role was best illustrated in a bone marrow transplantation model (BMT) where a cross-talk between the microbiota and immune cells could be identified with inflammation changing the commensal profile and microbiota, which in turn further exacerbated inflammation associated with graft versus host disease (GVHD) (55). Indeed, a reduction in overall diversity with expansion of Lactobacillales and loss of Clostridiales was associated with GVHD in a mouse model of BMT and selectively decreasing or reintroducing Lactobacillales aggravated or protected against GVHD, respectively. Remarkably, a similar loss in bacterial diversity accompanied by increases in Lactobacillales and decreases in Clostridiales, was observed in BMT patients undergoing GVHD (55). Another recent study reported that GVHD targeted the Paneth cells that secrete antimicrobial peptides, α-defensins (56). Thus increased GVHD severity correlated with reductions in the diversity of the intestinal microbiota while allowing the expansion of rare Escherichia coli. Furthermore treatment with polymyxin B inhibited outgrowth of E. coli and ameliorated GVHD. These observations reveal a mechanism responsible for the shift in gut flora during GVHD, which may in turn affect systemic immunity.
In solid organ transplantation, historical experiments from the ‘60s examining the fate of minor mismatched (CBA into C3H) skin grafts in germ-free mice did not report prolonged survival compared with conventional animals. However, some delays were observed if recipients were thymectomized neonatally prompting the authors to conclude that “in the absence of bacterial contamination, the cellular mechanism for the rejection of foreign skin grafts is defective in thymectomized recipients” (57). This modest delay in skin graft rejection and the technical challenges in performing solid organ transplantations under germ-free conditions stymied further investigations into the role of the microbiota in transplantation. However, in recent years, the roles of innate immunity and the microbiota have become more fully appreciated and new approaches to analyze the microbiota have become available, allowing for a re-examination of the connection between the microbiota and acute rejection.
Acute rejection of intestinal transplants in patients was reported to be associated with a reduced proportion of Firmicutes and an increased proportion of Proteobacteria, 2 bacterial phyla present in the intestinal lumen (58). Lung transplant recipients have been shown to develop diminished bacterial diversity in the lung following transplantation, while displaying increased bacterial load in their bronchoalveolar lavage (59). In a study investigating the change in microbiota after transplantation and the risk for developing bronchiolitis obliterans syndrome (BOS), the reestablishment of dominant populations present in patients’ lungs before transplantation, notably Pseudomonas aeruginosa in individuals with cystic fibrosis, was negatively correlated with BOS (60). In contrast, de novo acquisition of microbial populations often belonging to the same genera as the original dominant bacteria may have increased the risk of BOS (60). As an explanation, the authors reasoned that long-term colonizers of the cystic fibrotic lung were selected for persistence, exhibited decreased invasiveness and elicited reduced host immune responses, whereas acute infection, even with related species, would result in a more vigorous immune response. Furthermore, they observed that P. aeuroginosa in the microbiota protected against colonization by Aspergillus fumigatus, whose colonization was recently associated with development of BOS (61). These studies are just beginning to unravel what is likely to be a complex interplay between the microbiota and local and/or systemic immune responses, including alloreactivity.
Antibiotic use and transplantation
With new tools for high throughput sequencing of bacterial genes, investigators have been able to assess the impact of antibiotic use on the intestinal microbiota. For instance, a single dose of Clindamycin has been shown to reduce bacterial microbial diversity of the intestine by 90% for over 4 weeks, rendering mice extremely susceptible to infection with Clostridium difficile spores with subsequent colitis and lethality (62). In turn, duodenal infusion with microbiota from healthy people was recently shown to cure refractory C. difficile infection in 12/13 patients (63).
Intriguingly, certain viruses can exploit intestinal commensal bacteria for replication and transmission such that antibiotic treatment has been shown to result in reduced enteral infection by polio virus (64) and diminished transmission of an MMTV murine retrovirus by pregnant mothers to their progeny (65). Conversely, antibiotic-treated mice exhibited impaired innate and adaptive antiviral immune responses resulting in delayed viral clearance after exposure to systemic LCMV or mucosal influenza virus (40). Whether antibiotic treatment will reduce infectivity or on the contrary diminish immunity against viruses that affect transplanted patients is not known.
Thus, caution should be exercised in antibiotic usage, as it may result in complex alterations in microbiota that may potentially enhance or reduce alloreactivity as well as immunity against bacterial and viral pathogens. On the other hand, antibiotics are probably essential to eliminate pathogens whose inflammatory signature, such as type I IFN and IL-6 observed after Listeria monocytogenes in mice (27), can precipitate graft rejection.
Conclusion
A tight interplay is emerging between immune responses to microbial antigens, be it pathogenic or commensals, and T cell alloimmunity with microbial immunity playing a role in shaping the TCR repertoire of potentially cross-reactive memory T cells, determining the quality of alloresponses, and possibly eroding or breaking transplantation tolerance when it can be achieved. Table I and Figure I illustrate some of these points. The use of antibiotics can dramatically affect this interplay and alter not only the fate of infections and their direct effects on alloimmunity, but also perhaps the cross-talk between the microbiota and local and distant alloimmunity. Similarly, vaccinations may prevent infections but in some cases may generate immunity detrimental to the allografts. Future research that focuses on the molecular pathways by which infections and microbial signals can sculpt alloimmunity may help identify therapeutic targets to differentially modulate microbial and alloimmune responses. For instance, agents that block TLRs and are being tested in pre-clinical and clinical trials to reduce pathology in autoimmune or septic patients [see review in (66)] may also diminish the pro-rejection effects driven by infection-dependent production of inflammatory factors. Similarly, selective and transient blockade of inflammatory mediators such as type I IFN and IL-6 that can trigger rejection of established allografts (26) may prevent infections from augmenting anti-donor immunity. Such treatments would probably need to be used judiciously since inhibition of PRRs or inflammatory cytokines can increase susceptibility to infections and be ultimately detrimental. Finally, as more is known about the commensal microbiota and its effects on local and distal immune responses, it may become possible to alter this microbiota to improve systemic Tregs to T effector ratios and thus dampen alloimmunity.
Table I.
Putative interplay between microbes, the alloimmune response and the allograft fate.
| Type of Microbe | Immune Consequence | Postulated Effect on Allograft |
|---|---|---|
| Resident commensals | Induced local Tregs | Prevents local and/or distal rejection ? OR reduces protective anti-microbial immunity resulting in infections promoting alloreactivity? |
| Resident commensals | Induced local T effectors | Promotes local and/or distal rejection? OR promotes protective anti-microbial immunity to prevent infections from driving rejection? |
| Resident commensals | Bacterial products traveling systemically poise APC to present antigen (alloantigen?) | Promotes distal rejection? OR promotes protective anti-microbial immunity to prevent infections from driving rejection? |
| Acute intestinal infection | Breach in barrier integrity can result in development of T and B cell memory to commensal antigens | Potential cross reactivity of these memory lymphocytes with alloantigens and resistance to immunosuppression => promotes rejection? |
| Acute infection | PRR activation and inflammatory cytokines can enhance alloantigen presentation and determine T cell differentiation | Promotes local (?) and distal rejection |
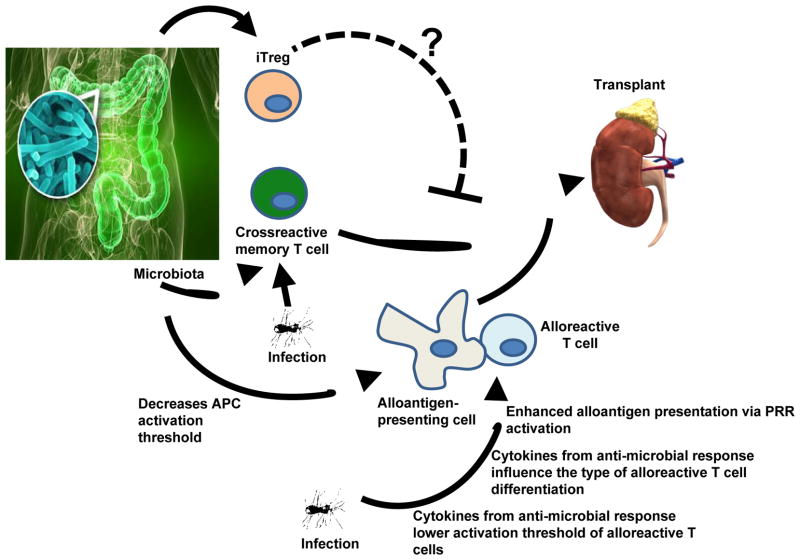
Figure 1. Interplay between microbes and the alloimmune response.
Infections prior to transplantation as well as inadvertent immunity to commensal microbiota may generate cross-reacting alloreactive memory T cells. Microbiota-derived microbial products transported systemically may also lower the activation threshold of APCs that may be presenting alloantigen. Infections at the time of or after transplantation may enhance the alloresponse directly by providing ligands to PRRs on APCs thus boosting alloantigen presentation, or indirectly via the cytokines produced during the anti-microbial response that can lower the activation threshold of an ongoing alloresponse or can dictate the phenotype of a differentiating alloreactive T cell. In addition to these effects that would be detrimental to an allograft, microbes may also help prevent alloimmunity. For instance, intestinal microbiota can generate iTregs and it is theoretically conceivable that these play a role in dampening the alloimmune response.
Acknowledgments
MLA and ASC wrote the review. MLA and ASC are funded by NIH/NIAID PO1 AI097113
Abbreviations
- APC
antigen-presenting cell
- BMT
bone marrow transplantation
- BOS
bronchiolitis obliterans syndrome
- GVHD
graft versus host disease
- PRR
pattern recognition receptors
Footnotes
These authors do not have conflicts of interest to disclose.
References
- 1.Fishman JA, Issa NC. Infection in organ transplantation: risk factors and evolving patterns of infection. Infect Dis Clin North Am. 2010;24(2):273–283. doi: 10.1016/j.idc.2010.01.005. 210.1016/j.idc.2010.1001.1005. [DOI] [PubMed] [Google Scholar]
- 2.Gras S, Kjer-Nielsen L, Chen Z, Rossjohn J, McCluskey J. The structural bases of direct T-cell allorecognition: implications for T-cell-mediated transplant rejection. Immunol Cell Biol. 2011;89(3):388–395. doi: 10.1038/icb.2010.150. Epub 2011 Feb 2018. [DOI] [PubMed] [Google Scholar]
- 3.Felix NJ, Allen PM. Specificity of T-cell alloreactivity. Nat Rev Immunol. 2007;7(12):942–953. doi: 10.1038/nri2200. [DOI] [PubMed] [Google Scholar]
- 4.Su LF, Kidd BA, Han A, Kotzin JJ, Davis MM. Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults. Immunity. 2013;38(2):373–383. doi: 10.1016/j.immuni.2012.10.021. 310.1016/j.immuni.2012.1010.1021. Epub 2013 Feb 1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith C, Miles JJ, Khanna R. Advances in direct T-cell alloreactivity: function, avidity, biophysics and structure. Am J Transplant. 2012;12(1):15–26. doi: 10.1111/j.1600-6143.2011.03863.x. 10.1111/j.1600-6143.2011.03863.x. Epub 02011 Dec 03867. [DOI] [PubMed] [Google Scholar]
- 6.Taylor DK, Neujahr D, Turka LA. Heterologous immunity and homeostatic proliferation as barriers to tolerance. Curr Opin Immunol. 2004;16(5):558–564. doi: 10.1016/j.coi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Heeger PS, et al. Pretransplant frequency of donor-specific, IFN-gamma-producing lymphocytes is a manifestation of immunologic memory and correlates with the risk of posttransplant rejection episodes. J Immunol. 1999;163(4):2267–2275. [PubMed] [Google Scholar]
- 8.Macedo C, et al. Contribution of naive and memory T-cell populations to the human alloimmune response. Am J Transplant. 2009;9(9):2057–2066. doi: 10.1111/j.1600-6143.2009.02742.x. Epub 2009 Jul 2016. [DOI] [PubMed] [Google Scholar]
- 9.Adams AB, et al. Heterologous immunity provides a potent barrier to transplantation tolerance. J Clin Invest. 2003;111(12):1887–1895. doi: 10.1172/JCI17477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brehm MA, et al. Allografts stimulate cross-reactive virus-specific memory CD8 T cells with private specificity. Am J Transplant. 2010;10(8):1738–1748. doi: 10.1111/j.1600-6143.2010.03161.x. 1710.1111/j.1600-6143.2010.03161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pantenburg B, Heinzel F, Das L, Heeger PS, Valujskikh A. T cells primed by Leishmania major infection cross-react with alloantigens and alter the course of allograft rejection. J Immunol. 2002;169(7):3686–3693. doi: 10.4049/jimmunol.169.7.3686. [DOI] [PubMed] [Google Scholar]
- 12.Richer MJ, Nolz JC, Harty JT. Pathogen-specific inflammatory milieux tune the antigen sensitivity of CD8(+) T cells by enhancing T cell receptor signaling. Immunity. 2013;38(1):140–152. doi: 10.1016/j.immuni.2012.09.017. 110.1016/j.immuni.2012.1009.1017. Epub 2012 Dec 1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perona-Wright G, Mohrs K, Mohrs M. Sustained signaling by canonical helper T cell cytokines throughout the reactive lymph node. Nat Immunol. 2010;11(6):520–526. doi: 10.1038/ni.1866. 510.1038/ni.1866. Epub 2010 Apr 1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu J, Paul WE. Heterogeneity and plasticity of T helper cells. Cell Res. 2010;20(1):4–12. doi: 10.1038/cr.2009.138. 10.1038/cr.2009.1138. Epub 2009 Dec 1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alegre ML, Florquin S, Goldman M. Cellular mechanisms underlying acute graft rejection: time for reassessment. Curr Opin Immunol. 2007;19(5):563–568. doi: 10.1016/j.coi.2007.07.019. Epub 2007 Aug 2027. [DOI] [PubMed] [Google Scholar]
- 16.Thornley TB, et al. TLR Agonists Abrogate Costimulation Blockade-Induced Prolongation of Skin Allografts. J Immunol. 2006;176(3):1561–1570. doi: 10.4049/jimmunol.176.3.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thornley TB, et al. Type 1 IFN mediates cross-talk between innate and adaptive immunity that abrogates transplantation tolerance. J Immunol. 2007;179(10):6620–6629. doi: 10.4049/jimmunol.179.10.6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, et al. TLR engagement prevents transplantation tolerance. Am J Transplant. 2006;6(10):2282–2291. doi: 10.1111/j.1600-6143.2006.01489.x. [DOI] [PubMed] [Google Scholar]
- 19.Porrett PM, et al. Mechanisms underlying blockade of allograft acceptance by TLR ligands. J Immunol. 2008;181(3):1692–1699. doi: 10.4049/jimmunol.181.3.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L, et al. TLR signals promote IL-6/IL-17-dependent transplant rejection. J Immunol. 2009;182(10):6217–6225. doi: 10.4049/jimmunol.0803842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang T, et al. Prevention of allograft tolerance by bacterial infection with Listeria monocytogenes. J Immunol. 2008;180(9):5991–5999. doi: 10.4049/jimmunol.180.9.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmed EB, Wang T, Daniels M, Alegre ML, Chong AS. IL-6 induced by Staphylococcus aureus infection prevents the induction of skin allograft acceptance in mice. Am J Transplant. 2011;11(5):936–946. doi: 10.1111/j.1600-6143.2011.03476.x. 910.1111/j.1600-6143.2011.03476.x. Epub 02011 Mar 03430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han Lee ED, et al. A mouse model for polyomavirus-associated nephropathy of kidney transplants. Am J Transplant. 2006;6(5 Pt 1):913–922. doi: 10.1111/j.1600-6143.2006.01265.x. [DOI] [PubMed] [Google Scholar]
- 24.Albrecht JA, et al. Adaptive immunity rather than viral cytopathology mediates polyomavirus-associated nephropathy in mice. Am J Transplant. 2012;12(6):1419–1428. doi: 10.1111/j.1600-6143.2012.04005.x. 1410.1111/j.1600-6143.2012.04005.x. Epub 02012 Mar 04015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmed EB, Daniels M, Alegre ML, Chong AS. Bacterial infections, alloimmunity, and transplantation tolerance. Transplant Rev. 2011;25(1):27–35. doi: 10.1016/j.trre.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang T, et al. Infection with the intracellular bacterium, Listeria monocytogenes, overrides established tolerance in a mouse cardiac allograft model. Am J Transplant. 2010;10(7):1524–1533. doi: 10.1111/j.1600-6143.2010.03066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brouard S, et al. The natural history of clinical operational tolerance after kidney transplantation through twenty-seven cases. Am J Transplant. 2012;12(12):3296–3307. doi: 10.1111/j.1600-6143.2012.04249.x. 3210.1111/j.1600-6143.2012.04249.x. Epub 02012 Sep 04213. [DOI] [PubMed] [Google Scholar]
- 28.Kumar D, et al. Influenza vaccination in the organ transplant recipient: review and summary recommendations. Am J Transplant. 2011;11(10):2020–2030. doi: 10.1111/j.1600-6143.2011.03753.x. 2010.1111/j.1600-6143.2011.03753.x. [DOI] [PubMed] [Google Scholar]
- 29.Dopp JM, et al. Effect of annual influenza immunization on antibody response in lung transplant patients. Prog Transplant. 2009;19(2):153–159. doi: 10.1177/152692480901900209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beck CR, et al. Influenza vaccination for immunocompromised patients: systematic review and meta-analysis from a public health policy perspective. PLoS One. 2011;6(12):e29249. doi: 10.1371/journal.pone.0029249. 29210.21371/journal.pone.0029249. Epub 0022011 Dec 0029222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eckerle I, Rosenberger KD, Zwahlen M, Junghanss T. Serologic vaccination response after solid organ transplantation: a systematic review. PLoS One. 2013;8(2):e56974. doi: 10.1371/journal.pone.0056974. 56910.51371/journal.pone.0056974. Epub 0052013 Feb 0056922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Danziger-Isakov L, Kumar D the ASTIDCoP . Vaccination in Solid Organ Transplantation. American Journal of Transplantation. 2013;13(s4):311–317. doi: 10.1111/ajt.12122. [DOI] [PubMed] [Google Scholar]
- 33.Mazzone PJ, et al. The humoral immune response to influenza vaccination in lung transplant patients. Eur Respir J. 2001;18(6):971–976. doi: 10.1183/09031936.01.00215201. [DOI] [PubMed] [Google Scholar]
- 34.Cordero E, Manuel O. Influenza vaccination in solid-organ transplant recipients. Curr Opin Organ Transplant. 2012;17(6):601–608. doi: 10.1097/MOT.0b013e3283592622. 610.1097/MOT.1090b1013e3283592622. [DOI] [PubMed] [Google Scholar]
- 35.Schaffer SA, Husain S, Delgado DH, Kavanaugh L, Ross HJ. Impact of adjuvanted H1N1 vaccine on cell-mediated rejection in heart transplant recipients. Am J Transplant. 2011;11(12):2751–2754. doi: 10.1111/j.1600-6143.2011.03743.x. 2710.1111/j.1600-6143.2011.03743.x. Epub 02011 Sep 03711. [DOI] [PubMed] [Google Scholar]
- 36.Hurst FP, Lee JJ, Jindal RM, Agodoa LY, Abbott KC. Outcomes associated with influenza vaccination in the first year after kidney transplantation. Clin J Am Soc Nephrol. 2011;6(5):1192–1197. doi: 10.2215/CJN.05430610. 1110.2215/CJN.05430610. Epub 05432011 Apr 05430621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chervonsky AV. Influence of microbial environment on autoimmunity. Nat Immunol. 2010;11(1):28–35. doi: 10.1038/ni.1801. Epub 2009 Dec 2017. [DOI] [PubMed] [Google Scholar]
- 38.Markle JG, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339(6123):1084–1088. doi: 10.1126/science.1233521. 1010.1126/science.1233521. Epub 1232013 Jan 1233517. [DOI] [PubMed] [Google Scholar]
- 39.Ichinohe T, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A. 2011;108(13):5354–5359. doi: 10.1073/pnas.1019378108. 5310.1073/pnas.1019378108. Epub 1019372011 Mar 1019378114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abt MC, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37(1):158–170. doi: 10.1016/j.immuni.2012.04.011. 110.1016/j.immuni.2012.1004.1011. Epub 2012 Jun 1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ganal SC, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37(1):171–186. doi: 10.1016/j.immuni.2012.05.020. 110.1016/j.immuni.2012.1005.1020. Epub 2012 Jun 1028. [DOI] [PubMed] [Google Scholar]
- 42.Ivanov, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. 410.1016/j.cell.2009.1009.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu HJ, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32(6):815–827. doi: 10.1016/j.immuni.2010.06.001. 810.1016/j.immuni.2010.1006.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hand TW, et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science. 2012;337(6101):1553–1556. doi: 10.1126/science.1220961. Epub 2012 Aug 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geuking MB, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34(5):794–806. doi: 10.1016/j.immuni.2011.03.021. Epub 2011 May 2019. [DOI] [PubMed] [Google Scholar]
- 46.Lathrop SK, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478(7368):250–254. doi: 10.1038/nature10434. 210.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cebula A, et al. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature. 2013;497(7448):258–262. doi: 10.1038/nature12079. 210.1038/nature12079. Epub 12013 Apr 12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–341. doi: 10.1126/science.1198469. Epub 2010 Dec 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Round JL, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332(6032):974–977. doi: 10.1126/science.1206095. Epub 2011 Apr 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Josefowicz SZ, et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482(7385):395–399. doi: 10.1038/nature10772. 310.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fan Z, et al. In vivo tracking of ‘color-coded’ effector, natural and induced regulatory T cells in the allograft response. Nat Med. 2010;16(6):718–722. doi: 10.1038/nm.2155. Epub 2010 May 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kendal AR, et al. Sustained suppression by Foxp3+ regulatory T cells is vital for infectious transplantation tolerance. J Exp Med. 2011;208(10):2043–2053. doi: 10.1084/jem.20110767. Epub 2011 Aug 2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Naik S, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337(6098):1115–1119. doi: 10.1126/science.1225152. 1110.1126/science.1225152. Epub 1222012 Jul 1225126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu L, et al. Recognition of host immune activation by Pseudomonas aeruginosa. Science. 2005;309(5735):774–777. doi: 10.1126/science.1112422. [DOI] [PubMed] [Google Scholar]
- 55.Jenq RR, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J Exp Med. 2012;209(5):903–911. doi: 10.1084/jem.20112408. 910.1084/jem.20112408. Epub 20112012 Apr 20112430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eriguchi Y, et al. Graft-versus-host disease disrupts intestinal microbial ecology by inhibiting Paneth cell production of alpha-defensins. Blood. 2012;120(1):223–231. doi: 10.1182/blood-2011-12-401166. 210.1182/blood-2011-1112-401166. Epub 402012 Apr 401124. [DOI] [PubMed] [Google Scholar]
- 57.Miller JF, Dukor P, Grant G, Sinclair NR, Sacquet E. The immunological responsiveness of germ-free mice thymectomized at birth. I. Antibody production and skin homograft rejection. Clin Exp Immunol. 1967;2(5):531–542. [PMC free article] [PubMed] [Google Scholar]
- 58.Oh PL, et al. Characterization of the ileal microbiota in rejecting and nonrejecting recipients of small bowel transplants. Am J Transplant. 2012;12(3):753–762. doi: 10.1111/j.1600-6143.2011.03860.x. 710.1111/j.1600-6143.2011.03860.x. Epub 02011 Dec 03867. [DOI] [PubMed] [Google Scholar]
- 59.Charlson ES, et al. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med. 2012;186(6):536–545. doi: 10.1164/rccm.201204-0693OC. 510.1164/rccm.201204-200693OC. Epub 202012 Jul 201212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Willner DL, et al. Reestablishment of Recipient-associated Microbiota in the Lung Allograft Is Linked to Reduced Risk of Bronchiolitis Obliterans Syndrome. Am J Respir Crit Care Med. 2013;187(6):640–647. doi: 10.1164/rccm.201209-1680OC. 610.1164/rccm.201209-201680OC. Epub 202013 Jan 201217. [DOI] [PubMed] [Google Scholar]
- 61.Weigt SS, et al. Colonization with small conidia Aspergillus species is associated with bronchiolitis obliterans syndrome: a two-center validation study. Am J Transplant. 2013;13(4):919–927. doi: 10.1111/ajt.12131. 910.1111/ajt.12131. Epub 12013 Feb 12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buffie CG, et al. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun. 2012;80(1):62–73. doi: 10.1128/IAI.05496-11. 10.1128/IAI.05496-05411. Epub 02011 Oct 05417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Nood E, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368(5):407–415. doi: 10.1056/NEJMoa1205037. 410.1056/NEJMoa1205037. Epub 1202013 Jan 1205016. [DOI] [PubMed] [Google Scholar]
- 64.Kuss SK, et al. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334(6053):249–252. doi: 10.1126/science.1211057. 210.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kane M, et al. Successful transmission of a retrovirus depends on the commensal microbiota. Science. 2011;334(6053):245–249. doi: 10.1126/science.1210718. 210.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chong AS, Alegre ML. The impact of infection and tissue damage in solid-organ transplantation. Nat Rev Immunol. 2012;12(6):459–471. doi: 10.1038/nri3215. 410.1038/nri3215. [DOI] [PMC free article] [PubMed] [Google Scholar]