Abstract
Natural protein assemblies have many sophisticated architectures and functions, creating nanoscale storage containers, motors and pumps1–3. Inspired by these systems, protein monomers have been engineered to self-assemble into supramolecular architectures4 including symmetrical5,6, metal-templated7,8 and cage-like structures8–10. The complexity of protein machines, however, has made it difficult to create assemblies with both defined structures and controllable functions. Here we report protein assemblies that have been engineered to function as light-controlled nanocontainers. We show that an adenosine-5′-triphosphate (ATP)-driven group II chaperonin11,12, which resembles a barrel with a builtin lid, can be reprogrammed to open and close on illumination with different frequencies of light. By engineering photoswitchable azobenzene-based molecules into the structure, light-triggered changes in interatomic distances in the azobenzene moiety are able to drive large-scale conformational changes of the protein assembly. The different states of the assembly can be visualized with single particle cryo-electron microscopy, and the nanocages can be used to capture and release non-native cargos. Similar strategies switching atomic distances with light could be used to build other controllable nanoscale machines.
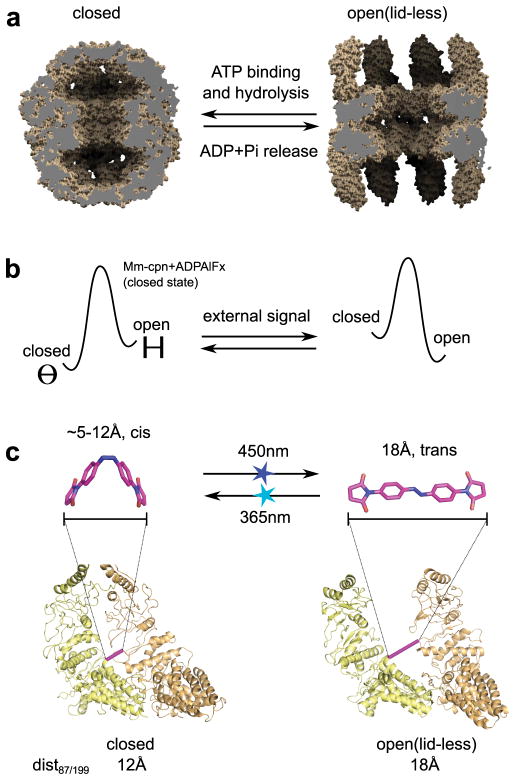
Group II chaperonins are protein assemblies comprising two eight-membered rings stacked back-to-back, each enclosing a central chamber of approximately ~150,000 Å3. The chamber is capped by a built-in lid that, in the natural system, opens and closes to transiently encapsulate client substrate proteins in a cycle driven by ATP binding and hydrolysis12. Structures of the group II homo-oligomeric chaperonin from Methanococcus maripaludis, Mm-cpn11, in the open and closed states illustrate the major conformational change (Fig. 1a) that occurs through an ATP-driven reorientation of the monomers12,13 via an incompletely understood mechanism.
Fig. 1.
Design strategy to control the shape transition of Mm-cpn with light by reversibly switching interatomic distances in the assembly. a Models derived from cryo-EM data of the atomic structure of Mm-cpn in the closed state (pdb code 3IYE9) and the open state of a truncated Mm-cpn construct missing the built-in lid (residues 241–267 replaced by a 4 residue linker, pdb code 3IYF9) illustrating the shape change of Mm-cpn, which in the natural system is dependent on ATP binding, hydrolysis and ADP/phosphate (Pi) release. b Design strategy for external control of the conformation of Mm-cpn by selective perturbation of the equilibrium between the closed (Θ symbol) and open (H symbol) states. c Molecular structure of the crosslinker ABDM in the 2 isomerization states cis and trans (upper panels, models created with the software Avogadro) and crosslinking site derived from the computational screen (lower panels). Shown are backbone models of two neighbouring Mm-cpn subunits in the closed and open states (as indicated). Bars connect the Cα atoms of the crosslinked residues in the mutant ΔC/K87C/S199C (pink) and dist87199 is the expected distance for the side chain sulphur atoms of an engineered cysteine at both positions. Light pulses that trigger the ABDM cis-trans (450nm) or the trans-cis isomerization (365nm) are indicated by blue or cyan stars, respectively.
To control these nano-containers externally, we devised a mechanism that could switch between the open and closed states of Mm-cpn with light. Light inputs, which offer exquisite spatial and temporal resolution, have been used to control peptide folding/unfolding transitions14, enzyme activity15 and ion channel activation16, and to modulate substrate release in the group I chaperonin GroEL17. However, it has been challenging to use similar approaches to trigger defined conformational states of protein machines. Our design strategy tests a simple premise based on the assumption of an equilibrium between the open and closed states of Mm-cpn: If one could reversibly switch between two interatomic distances in the assembly so that one distance was only compatible with the open and the second distance was only compatible with the closed state, switching should lead to selective stabilization of one of the two states and hence reversible interconversion between them (Fig. 1b).
To control interatomic distances in the assembly, we chose the thiol-reactive photo-switchable small molecule azobenzene-dimaleimide18 (ABDM, Fig. 1c), which crosslinks cysteine residues. Azobenzenes reversibly change their end-to-end distance through cis-trans isomerization upon illumination with either near UV or visible light (Fig. 1c), leading to a distance change from 18Å in the trans state to a 5–12Å distribution in the cis state19. We used two main criteria for designing cysteine mutations in Mm-cpn to create the attachment sites for the ABDM crosslinker: First, the distance of the side chain sulphur atoms of the engineered cysteines in the open and closed states of Mm-cpn must match the end-to-end distance of ABDM in its trans and cis state, respectively. Second, the side chain sulphur atoms of the engineered cysteines must be solvent accessible for cross-linking. We performed a computational screen guided by structural information from high-resolution cryo-electron microscopy data12 and selected three pairs of attachment sites (Methods, Supplementary Table S1). Each of these designed pairs crosslinks two neighbouring monomers in the same ring.
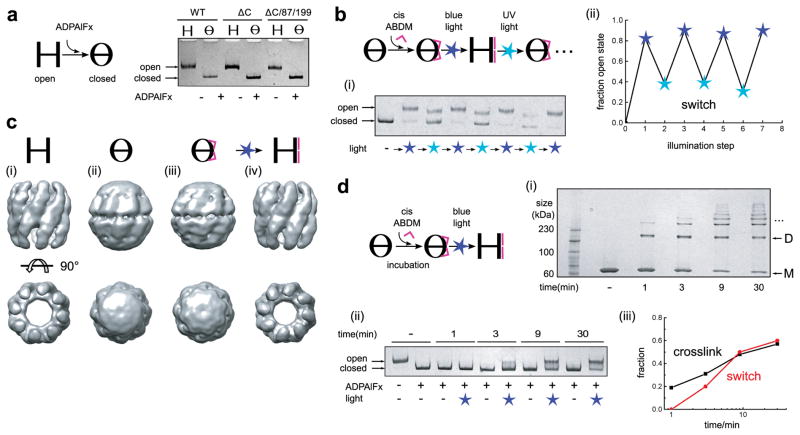
To ensure site-specific crosslinking of ABDM, we first removed all native cysteines in Mm-cpn to create the mutant C140S-C237S-C286A-C359T-C393A-C470S-C484S (termed ΔC). We verified that ΔC and the top ranked mutant introducing a new cysteine pair, ΔC/K87C/S199C (Fig. 1c), maintained the ability to adopt the open and closed states. Using a well-established native gel assay that uses AlFX (a phosphate analogue which binds to hydrolysed ATP after phosphate release and forms the transition state mimic ADP-AlFx) to bias Mm-cpn towards the closed conformation12,20,21, we observed clear distinct bands for both the open (nucleotide-free) and closed (ADP-AlFx complexed) states of wild-type, ΔC and ΔC/K87C/S199C Mm-cpn (Fig. 2a).
Fig. 2.
xMm-cpn switches reversibly between the closed and open conformations when illuminated with blue and near UV light, respectively. a Non-denaturing PAGE gel of Mm-cpn in the open and the ADP-AlFX-complexed closed conformation of wild type Mm-cpn (WT), the cysteine free construct (ΔC) and the double cysteine mutant ΔC/K87C/S199C (ΔC/87/199). b Crosslinking of ΔC/K87C/S199C Mm-cpn with cis-ABDM (pink ∧) to generate xMm-cpn, and switching between the open and closed state of xMm-cpn by alternating blue and UV light illumination: (i) Non-denaturing PAGE gel. (ii) Quantification of the fraction of open state present in the sample after each illumination step using either blue (450nm, blue star) or UV (365nm, cyan star) light from data shown in (i). c 15–19Å single particle cryo-electron microscopy reconstructions: (i) ΔC/K87C/S199C (open). (ii) ADP-AlFX complexed ΔC/K87C/S199C (closed). (iii) xMm-cpn (unlit). (iv) xMm-cpn (blue light illuminated). d Blue light-induced closed→open transition for xMm-cpn at different ABDM crosslinking ratios: (i) Formation of covalently linked Mm-cpn multimers after subunit crosslinking by ABDM, analysed on a SDS-PAGE gel at indicated crosslinking durations (M, monomer; D, dimer; …, higher oligomers). (ii) Blue light-induced closed→open transition of xMm-cpn at different ABDM crosslinking ratios (as shown in (i)) analysed on a non-denaturing PAGE gel. (iii) Comparison of the efficiency of blue light-induced opening (expressed as fraction of open state, red line) and crosslinking (as fraction of crosslinks to possible crosslinking sites, black line) as a function of ABDM incubation time. Shown is a quantification of the data in (i) and (ii).
We then created a crosslinked Mm-cpn variant (xMm-cpn) by incubating the closed state of ΔC/K87C/S199C with cis-ABDM (Fig. 2b). Consistent with our predictions, blue-light illumination opened the assembly and UV light closed it again, as visualized by the expected shifted bands on the native gel (Fig. 2b,i). In contrast, ABDM-incubated single cysteine mutants (ΔC/K87C and ΔC/S199C) showed no structural response to illumination, indicating that crosslinking is necessary for light-induced cavity opening (Supplementary Fig. S1). For xMm-cpn, blue light populated the open state efficiently, while conversion to the closed state with UV light was incomplete. A weaker band similar to the open state remained under UV illumination (Fig. 2b,i). We hypothesize that this smaller population resembles open xMm-cpn with a mixture of ABDM in the trans and cis states, as UV light is unable to shift the isomer equilibrium completely to the cis state18,22 (Supplementary Fig. S2). Remarkably, however, switching between predominantly open and closed states was reversible over multiple cycles (Fig. 2b,ii).
To validate the interpretation of the light-induced shape changes of xMm-cpn suggested by the native gel assays, we used single particle cryo-electron microscopy. The cryo-electron microscopy maps of non-crosslinked ΔC/K87C/S199C in the apo (Fig. 2c,i, open) and ADP-AlFX complexed form (Fig. 2c,ii, closed) showed the expected large-scale conformational changes, as previously seen for wild-type Mm-cpn12 (Supplementary Fig. S3). Unlit xMm-cpn (Fig. 2c,iii) showed an essentially identical conformation to the closed state, while subsequent blue light illumination switched xMm-cpn to the open structure (Fig. 2c,iv). These results confirm that the conformational states of the light-controlled designed xMm-cpn closely resemble the corresponding states of the naturally occurring protein machine.
To quantify the dependency of successful photoswitching on the stoichiometry of ABDM crosslinking, we generated a set of xMm-cpn samples with increasing crosslinking ratio by varying the ABDM incubation time. This increase was visualized by the appearance of higher-molecular weight species (dimers, trimers, etc.) when assayed via sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 2d,i). We then analysed the conformational state of the protein complex on a native gel before and after illumination with blue light. While non-illuminated (dark) samples remained in the closed state, illuminated samples showed a mixture of open and closed states (Fig. 2d,ii), which shifted to larger populations of open state with increasing crosslinking ratio (Fig. 2d,iii), consistent with our initial assumption of an equilibrium between the open and closed state. Furthermore, the two other designed candidates derived from our computational analysis (Supplementary Fig. S4), with cross-linking sites at sequence positions 132/202 (Supplementary Fig. S5) and 129/204 (Supplementary Fig. S6), showed a similar dependency of the fraction of light-induced open state on the crosslinking ratio (Supplementary Fig. S7a), even though these mutants achieved different maximum levels of crosslinking and resulting switching efficiency (Supplementary Figs. S5, S6). A simple model assuming a constant energy perturbation per crosslink (Supplementary Methods, Fig. S7b) is able to explain the experimental data for all three mutants by a requirement of approximately eight (out of 16 possible) crosslinks per Mm-cpn molecule to switch the equilibrium (Supplementary Fig. S7c–g). This finding is consistent with the expectation of allosteric coupling of the orientations of the subunits in the complex and can be compared with recent reports that in the related hetero-oligomeric group II chaperonin TRiC only eight of the sixteen subunits per chaperonin bind ATP under physiological conditions21,23.
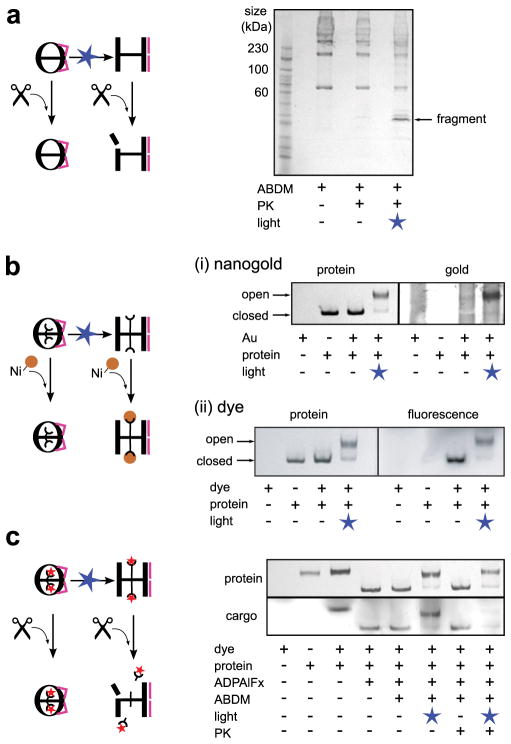
Next we investigated the physical properties of the designed light-gated nanocage. The level of spontaneous cavity opening of xMm-cpn without illumination (e.g. caused by thermal relaxation of cis ABDM to its more stable trans state) was low. Even after 4.5 hours of incubation at 37°C, the unlit xMm-cpn remained in the closed conformation, but preserved the ability to switch to the open state in response to blue light illumination (Supplementary Fig. S8). We also tested stability against protease digestion. Wild type Mm-cpn is protected against digestion in the closed but not the open state20. This behaviour is also seen for xMm-cpn, which is protected in the closed state but becomes accessible to digestion after blue light-induced cage opening, resulting in low molecular weight fragments on a SDS-PAGE gel (Fig. 3a).
Fig. 3.
xMm-cpn is a protease-resistant nanocage and can be used for light-triggered release of non-native cargos. a Unlit closed xMm-cpn is protected against proteinase K (PK, scissors symbol). A SDS-PAGE gel of unlit and blue light-illuminated xMm-cpn after PK incubation shows the appearance of smaller molecular weight fragments only after light-induced cage opening. b Blue light-illumination renders the C-terminal 6His-tag (half circle) of xMm-cpn accessible for binding of Ni-NTA coupled probes (yellow sphere): Unlit and blue light illuminated xMm-cpn incubated with either (i) Ni-NTA-nanogold (Au) or (ii) fluorescent Ni-NTA-Atto647N (dye) analysed on non-denaturing PAGE gels. The protein is visualized by coomassie staining (left panels) while the Ni-NTA-compounds are visualized (right panels) using either gold specific stain (Ni-NTA-nanogold) or via their fluorescence (Ni-NTA-Atto647N). Note the co-localization of Ni-NTA-Atto647N with both open and closed states, whereas Ni-NTA-nanogold is only co-localized to the open state. c Ni-NTA-Atto647N (red star) bound to the C-terminal 6His-tag of xMm-cpn in open and closed states stays bound to xMm-cpn in the presence of PK in the closed state, but is cleaved off from the cage after blue light-illumination (disappearance of the cargo band in rightmost lane) as analysed on a non-denaturing PAGE gel. The cargo (Ni-NTA-Atto647N) is visualized by its fluorescence (bottom gel panel).
The group I chaperonin GroEL (which is missing a built-in lid but can be assembled into tubular structures) has been shown to be able to host and release artificial cargos controlled by the ATP concentration in different environments24,25. To probe the accessibility of the Mm-cpn cavity for small molecules, we took advantage of the C-terminal 6His-tag of xMm-cpn that is located inside the cavity12,13. A dye specifically binding 6His-tags (Ni-NTA-Atto647N, volume~625 Å3) was able to bind to xMm-cpn in both the closed and the light-induced open state (Fig. 3b,ii). However, a 6His-tag binding gold particle (18 Å Ni-NTA-Nanogold with a size of 30 Å including coating, volume ~14,000 Å3) only bound to the open state (Fig. 3b,i). This suggested that the small dye was able to diffuse into closed form of the cage, possibly through lateral fenestrae, while the gold particle is too big to enter the closed cage and can only access the tag when it is exposed in the open state.
Finally, we loaded xMm-cpn with Ni-NTA-Atto647N, which stayed bound to closed xMm-cpn and was stable in the presence of protease. Subsequent blue-light illumination opened the nanocage and led to cargo release in the presence of the protease (Fig. 3c). Taken together, these results demonstrate the use of xMm-cpn to capture, shield and release a molecular cargo controlled by light.
Given the size and complexity of the Mm-cpn protein assembly, the magnitude of the structural rearrangements during the open→closed transition and its unknown mechanism, our design strategy was remarkably successful. The energy landscape of Mm-cpn is likely biased towards two dominant states (open and closed) mediated via strong coupling of the orientation of the subunits in the protein complex. Protein assemblies that are optimized by evolution to act as robust conformational switches, such as the one targeted here, are valuable targets for structure guided protein design. For these cooperative systems, designed precise perturbations to the energy landscape will mainly affect the population of the predominant functional states instead of creating new conformations (Fig. 1b) as demonstrated here. As many biological molecular machines are thought to function similarly via coupled conformational equilibria, our design strategy that highlights and exploits the cooperativity of the Mm-cpn assembly should be generally applicable to these systems.
Our prototype caging experiments furthermore suggest that light-gated highly stable assemblies like xMm-cpn may have interesting applications as protein-based cargo containers and shuttles. With further improvements, our engineered light-triggered protein nano-container is a stepping-stone towards the many applications of protein materials and machines switchable with the high temporal and spatial precision of light.
Materials and Methods
Structure-based design
To design attachment sites for ABDM, we used atomic structures derived from high-resolution cryo-electron microscopy data12 with 4.3Å resolution for the closed state and 8.0Å resolution for the open state of Mm-cpn (pdb codes 3IYE, 3IYF). We calculated the expected distances between sulphur atoms for every possible pair of cysteines mutations in neighbouring monomers as well as the expected solvent accessible surface area of the sulphur using the software Pymol (Schroedinger, LCC). The data set was then screened for residue pairs with an expected sulphur distance of 5–14Å in the closed and 16.6–19.5Å in the open state and a minimum expected solvent accessible surface area for the sulphur atoms of 8Å2 (10% of the maximum solvent accessible surface area of the sulphur atom in a deprotonated cysteine residue). As the moderate resolution of the cryo-electron microscopy data for the open state does not allow an exact determination of the atomic coordinates, we repeated the analysis with another atomic model derived from the same data set12 that had been refined with the program Rosetta26. When comparing both data sets, we found 9 residue pairs satisfying our matching criteria in both structure sets (Supplementary Table S1). This list was visually inspected for candidates for which there is enough unoccupied space between the attachment sites to accommodate ABDM, leaving 3 candidate residue pairs for crosslinking (87/199, 132/202, 129/204), which were tested experimentally (a fourth promising double cysteine mutant derived from the computational screen, ΔC/M77C/E378C, turned out to be unstable and not suitable for in vitro experiments).
Plasmids
A pET21a+ vector containing the Mm-cpn gene was a gift from J. Frydman. All Mm-cpn constructs were produced by site-directed mutagenesis. To purify the proteins, we attached a tag with the sequence DPNSSSVDKLAAALEHHHHHH to the C-terminus of the protein sequence.
Protein purification
Mm-cpn was overexpressed in Escherichia coli Rosetta (DE3) pLysS cells (EMD Biosciences). Cultures were grown at 37 °C to OD600 of 1, induced by the addition of 0.1mM isopropyl β-D-1-thiogalactopyranoside (IPTG), switched to 16 °C, grown over night and harvested by centrifugation. The cells were lysed by sonication in buffer A (50mM KCl, 5mM MgCl2, 20mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 10% glycerol, pH 7.5) supplemented with 0.2mM phenylmethanesulfonyl fluoride (PMSF) and Dnase I. The protein was purified via affinity chromatography using a Ni-NTA resin (Quiagen) and subsequent polishing steps using Heparin 16/10 FF (GE Healthcare; elution with a 0–1M NaCl gradient) and Superose 6 300/10 GL columns (GE Healthcare; running buffer: 20mM HEPES pH 7.4, 50mM KCl, 5mM MgCl2). After purification the protein sample was supplemented with 10% glycerol, aliquotted, shock frozen in liquid nitrogen and stored at −80 °C.
Native gel assay
Mm-cpn wild type, the cysteine-free construct (ΔC) and the double cysteine mutant ΔC/K87C/S199C were incubated in buffer A at a concentration of 0.25μM. Mm-cpn was biased towards the closed state via complexation with AlFX (a phosphate analogue which binds to hydrolysed ATP after phosphate release) by adding 1mM ATP, 6mM Al(NO3)3, and 25mM NaF to the solution (buffer A+, pH 7.0) and incubating the samples for 20 min at 43 °C20. Samples were analysed on a 4% non-denaturing PAGE gel and imaged on an AlphaImager HP system (proteinsimple). To additionally confirm photoswitching was independent of ATP hydrolysis, we also observed light-induced opening under conditions where no ATP was present in the buffer (i.e. addition of ADP instead of ATP, Fig. S9).
ABDM crosslinking and photoswitching
ABDM was a gift from GA Woolley. ABDM was dissolved in DMF to a concentration of 1.2mM and stored at −20 °C. Prior to crosslinking, ABDM was illuminated for 1.5 min using a 1W LED emitting at 367nm (Advancemart, full width at half maximum: 20nm). UV illumination resulted in an accumulation of ~75% cis isomer in the solution, which was estimated by analysing the absorption spectrum of the sample18. ABDM was added to a 0.25μM solution of ADP-AlFX-complexed Mm-cpn mutant ΔC/K87C/S199C in buffer A+ (closed state, prepared as described in the previous paragraph) at a ratio of 1μl ABDM solution per 50μl protein solution. The crosslinking reaction was quenched after 40min incubation time by adding DTT to a concentration of 2mM. To switch azobenzene between the cis and the trans isomerization states, xMm-cpn samples were illuminated either for 20s with a blue 1W LED (sparkfun, emission maximum 452nm, full width at half maximum: 24nm) or for 90s with the UV LED. The photoswitching efficiency of xMm-cpn, defined as the fraction of light-induced open state, was determined by comparing the intensities of the open and closed state bands on a 4% non-denaturating PAGE gel using the ImageJ software package27. The crosslinking ratio was determined by analysing the samples on a 4–20% sodium dodecyl sulphate (SDS) -PAGE gel (Lonza), and calculating the sum of the relative intensities of the multimer bands (band intensity divided by the sum of the band intensities for all multimers) weighted by their ratio of crosslinks to subunits. Proteinase K (New England Biolabs) was added to a final concentration of 20 μg/ml to xMm-cpn samples and samples were incubated for 10min at room temperature before quenching the reaction by addition of 2mM PMSF. Ni-NTA-Atto647N (Sigma-Aldrich) or Ni-NTA-nanogold solution (goldiblot kit, Nanoprobes) were added to xMm-cpn and incubated for 30min at room temperature. For selective visualization of the cargo, the gold particles were stained using the goldiblot kit (Nanoprobes) and the fluorescent dye Atto647N was detected using a Typhoon 9400 imager (GE Healthcare).
Cryo-electron microscopy specimen preparation and imaging
ADP-AlFX complexation, ABDM crosslinking and blue-light illumination were performed as described in the previous sections. PEG8000 at 1% final concentration was added to improve particle dispersion in xMm-cpn after illumination. Octyl glucoside (OG) was subsequently added to a concentration of 0.1 % to increase the yield of side views of apo ΔC/K87C/S199C Mm-cpn and illuminated xMm-cpn (Supplementary Table S2)12. The resulting samples were applied to 200-mesh R1.2/1.3 holey-carbon grids (Quantifoil Micro Tools GmbH) that were acetone-cleaned and glow-discharged. Grids were blotted and immediately vitrified by plunging into liquid ethane using a Vitrobot (FEI Inc, Hillsboro, OR). The frozen, hydrated specimen grid was transferred to a Gatan 626 cryo-specimen holder. Images were collected on a JEM2010F electron microscope (JEOL, 200 kV) equipped with a field emission gun and a Gatan Ultrascan 4K × 4K CCD camera (Gatan US4000). The detector magnification was 69Kx (1.81Å/pixel) or 55Kx (2.17Å/pixel) under a dose of 20 electrons/Å2. Images were captured across an approximate defocus range of 1.5–3.5 μm, and archived in the EMEN database28. Imaging conditions of respective data set are described in Supplementary Table S2.
3D Cryo-electron microscopy map reconstruction
Image processing and 3D map reconstruction were performed with the single particle reconstruction modules of EMAN2 by imposing D8 symmetry29. Evaluation of conformational homogeneity in each data set was carried out using the EMAN2 multiple-model refinement strategy that can be used to sort different particle sub-populations from mixed conformations if present30. Open and closed state maps of mutant Mm-cpn after a 50Å Gaussian low pass-filter were used as two initial reference models for particle orientation parameter determination. In the xMm-cpn (blue light-illuminated) data set, ~85% of the particles were classified into the open conformation. In the xMm-cpn (ADP-AlFX, unlit) data set, the two sub-population particles converged to the closed conformation. These conformation distributions observed in the particle image classification are consistent with the fractions of unlit closed and light-induced open states determined by the native gel analysis (Fig. 2b,ii). Subsequently, the 85% of particles in the xMm-cpn (blue light illuminated) set classified into the open conformation and the entire set of the xMm-cpn (ADP-AlFX, unlit) particle images were subjected to a single-model refinement. Two independent maps for each data set were computed to estimate the resolution of the final map with the combined data set, according to the gold-standard protocol31 using the 0.143 Fourier shell correlation criterion (Supplementary Fig. S10)32. Each of the two independent reconstructions was carried out with an initial model that was a blob-like map (Supplementary Fig. S11) generated by applying a 100Å Gaussian low-pass filter to the closed state Mm-cpn map (electron microscope database (EMD) accession number 5137) and randomizing the phases beyond 40Å. The program e2refine_evenodd.py (EMAN2) was used for refinement of particle orientation parameters. Upon convergence, the two half-maps were combined to generate the final 3D reconstruction. The final maps for each specimen were reconstructed from a total number of particle images of 7389 (apo ΔC/K87C/S199C), 7759 (ADP-AlFX complexed ΔC/K87C/S199C), 8428 (unlit xMm-cpn) and 3718 (blue light-illuminated xMm-cpn). Map visualization was facilitated by UCSF Chimera33.
Supplementary Material
Acknowledgments
We thank Judith Frydman and Nick Douglas for discussions and advice throughout this work and for sharing the Mm-cpn plasmid; Andrew Woolley for discussions and a gift of ABDM; Steven J. Ludtke for advice on image processing; and James Fraser for comments on the manuscript. This research was supported by grants from the Program for Breakthrough Biomedical Research and the Sandler Family Foundation to T.K., the National Institute of Health (PN2EY016525 and P41GM103832 to W.C.), the National Science Foundation (NSF CBET-1134127 to T.K.) and a Deutsche Forschungsgemeinschaft postdoctoral fellowship (HO 4429/2-1 to D.H.).
Footnotes
Author contributions
T.K. and D.H. designed the research, D.H. performed the computational design, biochemical experiments and model building, S-H.R. performed the cryo-electron microscopy experiments, S-H.R. and W.C. analysed the cryo-electron microscopy data. All authors wrote the manuscript.
Supplementary information accompanies this paper at www.nature.com/naturenanotechnology.
Reprints and permission information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Tanaka S, et al. Atomic-level models of the bacterial carboxysome shell. Science. 2008;319:1083–1086. doi: 10.1126/science.1151458. [DOI] [PubMed] [Google Scholar]
- 2.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 3.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nature Rev Mol Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 4.Yeates TO, Padilla JE. Designing supramolecular protein assemblies. Curr Opin Struct Biol. 2002;12:464–470. doi: 10.1016/s0959-440x(02)00350-0. [DOI] [PubMed] [Google Scholar]
- 5.King NP, et al. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science. 2012;336:1171–1174. doi: 10.1126/science.1219364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gradisar H, et al. Design of a single-chain polypeptide tetrahedron assembled from coiled-coil segments. Nature Chem Biol. 2013;9:362–366. doi: 10.1038/nchembio.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brodin JD, et al. Metal-directed, chemically tunable assembly of one-, two- and three-dimensional crystalline protein arrays. Nature Chem. 2012;4:375–382. doi: 10.1038/nchem.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huard DJ, Kane KM, Tezcan FA. Re-engineering protein interfaces yields copper-inducible ferritin cage assembly. Nature Chem Biol. 2013;9:169–176. doi: 10.1038/nchembio.1163. [DOI] [PubMed] [Google Scholar]
- 9.Fletcher JM, et al. Self-Assembling Cages from Coiled-Coil Peptide Modules. Science. 2013 doi: 10.1126/science.1233936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai YT, Cascio D, Yeates TO. Structure of a 16-nm cage designed by using protein oligomers. Science. 2012;336:1129. doi: 10.1126/science.1219351. [DOI] [PubMed] [Google Scholar]
- 11.Kusmierczyk AR, Martin J. Nucleotide-dependent protein folding in the type II chaperonin from the mesophilic archaeon Methanococcus maripaludis. Biochem J. 2003;371:669–673. doi: 10.1042/BJ20030230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, et al. Mechanism of folding chamber closure in a group II chaperonin. Nature. 2010;463:379–383. doi: 10.1038/nature08701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pereira JH, et al. Crystal structures of a group II chaperonin reveal the open and closed states associated with the protein folding cycle. J Biol Chem. 2010;285:27958–27966. doi: 10.1074/jbc.M110.125344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumita JR, Smart OS, Woolley GA. Photo-control of helix content in a short peptide. Proc Natl Acad Sci USA. 2000;97:3803–3808. doi: 10.1073/pnas.97.8.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schierling B, et al. Controlling the enzymatic activity of a restriction enzyme by light. Proc Natl Acad Sci USA. 2010;107:1361–1366. doi: 10.1073/pnas.0909444107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banghart M, Borges K, Isacoff E, Trauner D, Kramer RH. Light-activated ion channels for remote control of neuronal firing. Nature Neurosci. 2004;7:1381–1386. doi: 10.1038/nn1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muramatsu S, Kinbara K, Taguchi H, Ishii N, Aida T. Semibiological molecular machine with an implemented “AND” logic gate for regulation of protein folding. J Am Chem Soc. 2006;128:3764–3769. doi: 10.1021/ja057604t. [DOI] [PubMed] [Google Scholar]
- 18.Umeki N, et al. Incorporation of an azobenzene derivative into the energy transducing site of skeletal muscle myosin results in photo-induced conformational changes. J Biochem. 2004;136:839–846. doi: 10.1093/jb/mvh194. [DOI] [PubMed] [Google Scholar]
- 19.Beharry AA, et al. Quantitative analysis of the effects of photoswitchable distance constraints on the structure of a globular protein. Biochemistry. 2012;51:6421–6431. doi: 10.1021/bi300685a. [DOI] [PubMed] [Google Scholar]
- 20.Douglas NR, et al. Dual action of ATP hydrolysis couples lid closure to substrate release into the group II chaperonin chamber. Cell. 2011;144:240–252. doi: 10.1016/j.cell.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Y, et al. Sensing cooperativity in ATP hydrolysis for single multisubunit enzymes in solution. Proc Natl Acad Sci USA. 2011;108:16962–16967. doi: 10.1073/pnas.1112244108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Behrendt R, et al. Photomodulation of the Conformation of Cyclic Peptides with Azobenzene Moieties in the Peptide Backbone. Angew Chem Int Ed Engl. 1999;38:2771–2774. doi: 10.1002/(sici)1521-3773(19990917)38:18<2771::aid-anie2771>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 23.Reissmann S, et al. A gradient of ATP affinities generates an asymmetric power stroke driving the chaperonin TRIC/CCT folding cycle. Cell Rep. 2012;2:866–877. doi: 10.1016/j.celrep.2012.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishii D, et al. Chaperonin-mediated stabilization and ATP-triggered release of semiconductor nanoparticles. Nature. 2003;423:628–632. doi: 10.1038/nature01663. [DOI] [PubMed] [Google Scholar]
- 25.Biswas S, et al. Biomolecular robotics for chemomechanically driven guest delivery fuelled by intracellular ATP. Nature Chem. 2013;5:613–620. doi: 10.1038/nchem.1681. [DOI] [PubMed] [Google Scholar]
- 26.Dimaio F, Zhang J, Chiu W, Baker D. Cryo-EM model validation using independent map reconstructions. Protein Sci. 2013;22:865–868. doi: 10.1002/pro.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature Meth. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ludtke SJ, Nason L, Tu H, Peng L, Chiu W. Object oriented database and electronic notebook for transmission electron microscopy. Microscopy and Microanalysis. 2003;9:556–565. doi: 10.1017/S1431927603030575. [DOI] [PubMed] [Google Scholar]
- 29.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Chen DH, Song JL, Chuang DT, Chiu W, Ludtke SJ. An expanded conformation of single-ring GroEL-GroES complex encapsulates an 86 kDa substrate. Structure. 2006;14:1711–1722. doi: 10.1016/j.str.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 31.Scheres SHW, Chen S. Prevention of overfitting in cryo-EM structure determination. Nature Meth. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 33.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comp Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.