Abstract
Eukaryotic glutaminyl-tRNA synthetase (GlnRS) contains an appended N-terminal domain (NTD) whose precise function is unknown. Although GlnRS structures from two prokaryotic species are known, no eukaryotic GlnRS structure has been reported. Here we present the first crystallographic structure of yeast GlnRS, finding that the structure of the C-terminal domain is highly similar to Escherichia coli GlnRS but that 214 residues, including the NTD, are crystallographically disordered. We present a model of the full-length enzyme in solution, using the structures of the C-terminal domain, and the isolated NTD, with small-angle X-ray scattering data of the full-length molecule. We proceed to model the enzyme bound to tRNA, using the crystallographic structures of GatCAB and GlnRS–tRNA complex from bacteria. We contrast the tRNA-bound model with the tRNA-free solution state and perform molecular dynamics on the full-length GlnRS–tRNA complex, which suggests that tRNA binding involves the motion of a conserved hinge in the NTD.
Keywords: eukaryotic glutaminyl-tRNA synthetase, structure, C-terminal domain, small-angle X-ray scattering, molecular dynamics
Introduction
Aminoacyl-tRNA synthetases are required in all three domains of life for covalently attaching amino acids to their cognate tRNA molecules for use in protein synthesis.1 These enzymes are separated into two structural classes,2,3 class I and class II, which operate via a two-step reaction. First, after binding ATP and the amino acid, an aminoacyl-adenylate is formed, followed by esterification of the amino acid moiety to either the 2′ OH (class I) or 3′ OH (class II) of tRNA. In addition, eukaryotic tRNA synthetases often contain additional domains appended to their N-terminal or C-terminal ends that are not found in their prokaryotic homologs.4 These domains are known to have various roles, including nucleic acid binding, protein–protein interactions, and hydrolytic editing mechanisms1; however, many of these domains are of unknown function. While structures of several eukaryotic tRNA synthetases have been determined,5 virtually all of these lack the appended domain. The structures of some appended domains have been determined in isolation, but few models of full-length eukaryotic tRNA synthetases exist.
While in most cases one synthetase exists for each amino acid, an exception occurs for glutamine and asparagine.6 In eukaryotes and some bacteria, the traditional pathway of aminoacylation exists for glutamine, in which glutaminyl-tRNA synthetase (GlnRS) binds to tRNAgln, glutamine, and ATP and first forms a glutaminyl-adenylate molecule that is then covalently attached to the 3′-end of tRNAgln with the release of AMP. A different pathway exists in most bacteria and all archaea, where a non-discriminating glutamyl-tRNA synthetase (GluRS) attaches glutamic acid to both tRNAglu and tRNAgln. The misacylated Glu-tRNAgln is then converted to Gln-tRNAgln by the GatCAB amidotransferase enzyme in bacteria and some archaea or by the GatDE amidotransferases in other archaea. GatCAB performs a similar function for asparaginyl-tRNAasn. Since this indirect pathway for aminoacylation exists in most prokaryotes, GlnRS is primarily a eukaryotic enzyme, and its presence in a small number of bacteria is believed to have occurred through a horizontal gene transfer event.7 Nonetheless, there are significant differences between the prokaryotic and eukaryotic GlnRS enzymes, as described below.
Structural data for two prokaryotic GlnRS species exist,8,9 yet no structure has been reported for any full-length eukaryotic GlnRS. Saccharomyces cerevisiae GlnRS (Gln4) is an 809-residue protein that contains a 215-residue domain appended to its N-terminus. This domain is nearly ubiquitous among eukaryotic GlnRS species but absent in prokaryotic homologs.10 Recently we described the structure of the N-terminal domain (NTD) of Gln4, revealing that it has an extraordinary structural resemblance to the region of the B subunit of the GatCAB amidotransferase (GatB)11 that binds to tRNAgln.12 Although deletion of the NTD distinctly affects catalytic activity, growth in yeast, and tRNA binding,12 the manner in which tRNA binding occurs is still unknown.
Here we present the first crystallographic structure of the C-terminal domain (CTD) of Gln4 from crystals of the full-length protein. Based on this structure, the structure of the isolated NTD,12 and small-angle X-ray scattering (SAXS) data of the full-length enzyme, we present a model of the full-length enzyme in solution. We extend this to model the full-length enzyme bound to tRNAgln from the crystallographic structures and homology with known transamidosome and GlnRS–tRNA complex structures11,13 yielding new insights into the structural rearrangements occurring in eukaryotic GlnRS–tRNAgln complex formation.
Results and Discussion
Overall structure
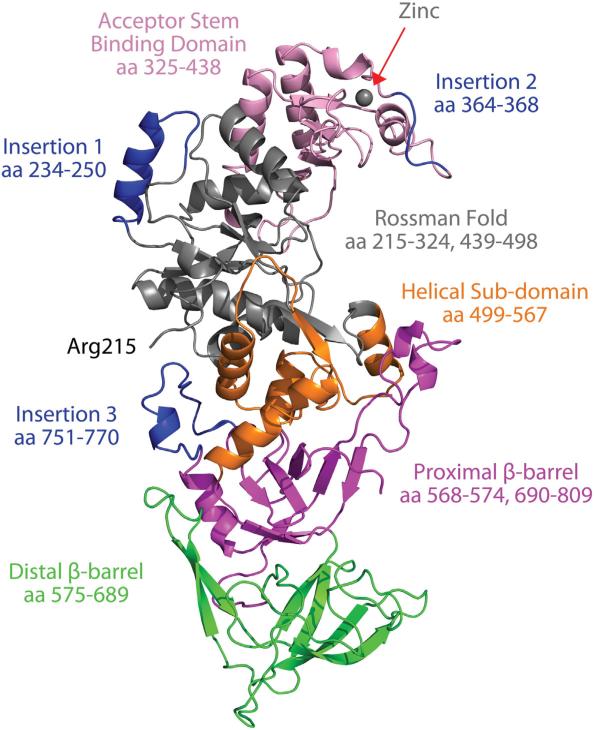
We solved the crystal structure of Gln4 to 2.15 Å resolution [Protein Data Bank (PDB) ID: 4H3S] by single-wavelength anomalous dispersion with a zinc anomalous signal identified from an initial excitation scan, coupled with molecular replacement using the crystal structure of the Escherichia coli GlnRS as a template (Fig. 1 and Table 1). Of the 809 amino acids in the primary sequence of Gln4, ~30% were not resolved in the electron density. The unresolved regions are residues 1–187 (which correspond to the NTD), residues 188–214 (which encode a nonconserved 26-residue region that is predicted to be unstructured and links the NTD and CTD; Fig. S1), and residues 672–678 (which encode a disordered loop within the CTD). The presence of the NTD in purified protein and in the crystals was confirmed by expressing Gln4 with a His6 tag on its N-terminus, followed by purification using Ni2+ affinity chromatography, crystallization, and antibody detection with anti-His antibody on dissolved crystals. Residues 1–214 were still unresolved in structural data from these crystals.
Fig. 1.
X-ray crystal structure of Gln4. Cartoon representation of the Gln4 structure is shown color-coded according to domains and insertions relative to E. coli GlnRS. Residues 1–214 are missing in the electron density. Domains are labeled with corresponding amino acid numbers.
Table 1.
Crystallographic data collection parameters and statistics
| Beamline | SSRL BL 11−1 |
|---|---|
| Wavelength (Å) | 1.169 |
| Space group | P3121 |
| Unit cell parameters | |
| a, b, c (Å) | 176.61, 176.61, 72.19 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution limits (outer shell) (Å) | 52.49–2.15 (2.227–2.15) |
| No. of unique reflections | 70,276 (6963) |
| Completeness (%) | 99.86 (99.84) |
| Rsim (%) | 0.068 (0.348) |
| 〈I/σ(/)〉 | 23.26 (2.98) |
| Reflections | |
| Working | 70,276 (6963) |
| Test (%) | 5 |
| Atoms | 10,566 |
| Protein | 5031 |
| Water | 447 |
| Ligands | 75 |
| R factor | 0.160 (0.222) |
| R free | 0.175 (0.242) |
| RMSD | |
| Bonds (Å) | 0.003 |
| Angles (°) | 0.73 |
| Average B-factor | |
| Overall | 48.10 |
| Macromolecules | 47.40 |
| Solvent (water) | 54.20 |
| Ramachandran analysis (%) | |
| Most favored regions | 99 |
| Outliers | 0.17 |
| Generously allowed regions | 0 |
| Disallowed regions | 0 |
| Clash score | 5.27 |
| PDB code | 4H3S |
Since the NTD of Gln4 is known to be structured12 and is present in the full-length protein in the crystal, but no electron density is observed for this region, we conclude that the NTD is crystallographically disordered (i.e., the position and/or orientation of the NTD varies between unit cells). It seems likely that the NTD is located in large solvent channels approximately 145–175 Å wide observed along the z-axis (Fig. S2). Consistent with this, we note that the channels can comfortably accommodate the structure of the NTD,12 which has dimensions of ~25 Å × 25 Å × 75 Å and a macromolecular volume of ~25,000 Å3. Moreover, the first residue resolved in the electron density, Arg215, is located adjacent to the solvent channels, and the preceding residues, 188–214, are predicted to be unstructured (Fig. S1). It is therefore likely that this region is the cause of the crystallographic disorder of the NTD.
Based on the considerations presented above, using the naming convention previously described for E. coli GlnRS,14 the overall structure of Gln4 consists of a folded NTD (residues 1–187); a positively charged 26-residue region linking the NTD and CTD (residues 188–214); a catalytic domain consisting of a Rossmann fold domain (residues 215–324 and 439–498), an acceptor stem binding domain (residues 325–438), and a helical subdomain (residues 499–567); and an anticodon binding domain consisting of proximal (residues 568–574 and 690–809) and distal (residues 575–689) β-barrels (Fig. 1). The structure of the CTD of Gln4 (residues 215–809) is highly similar to E. coli GlnRS (PDB ID: 1NYL; RMSD = 1.86 Å15) but has multiple insertions relative to the bacterial homolog. The Gln4 catalytic domain (residues 215–567) is highly conserved in sequence and structure, showing an E value of 1 × 10–101 relative to the E. coli GlnRS catalytic domain using PSI-BLAST and an RMSD of only 1.03 Å. The Gln4 anticodon binding domain (residues 568–809) shares moderate sequence homology but high structural conservation showing an E value of 5 × 10–32 and an RMSD of 1.13 Å. The structures of the Gln4 CTD and a second bacterial homolog, Deinococcus radiodurans GlnRS (PDB ID: 2HZ7),8 are also similar with an RMSD of 1.62 Å.
A detailed comparison of the Gln4 CTD structure with the unliganded (PDB ID: 1NYL)15 and tRNA-bound (PDB ID: 1QTQ)13E. coli GlnRS structures revealed both similarities and differences in the individual residues previously found to be important substrate recognition elements in E. coli GlnRS.16,17 The ATP-binding pocket formed by the HIGH and MSK (LSK in Gln4) is similar to that found in E. coli GlnRS, which are signature sequences conserved across class I aminoacyl-tRNA synthetases.1 The replacement of Met268 in E. coli GlnRS with Leu496 in Gln4 does not appear to significantly change the structure of this pocket. In E. coli GlnRS, residues Arg133, Arg192, Lys194, Thr68, and Asn69 were shown to be important in forming the hairpin loop of the tRNAgln CCA tail that is necessary to bring the 2′ hydroxyl of A76 into proximity with the glutamine adenylate molecule for glutamine transfer to tRNA.16 In Gln4, these residues (Arg357, Arg421, Leu422, Thr293, and Asn294, respectively) are conserved in position and orientation and therefore are likely to cause the CCA tail to form a similar hairpin loop. In the anticodon binding region of E. coli GlnRS, Arg341 interacts with anticodon base U35, resulting in distant, interdomain communication to the active site necessary for glutamine recognition.15 The corresponding residue in Gln4, Arg568, is in the same orientation and therefore could lead to similar interactions with U35. In E. coli GlnRS, Lys401 and Arg402 interact with G36, which is the discriminating base of the anticodon, and are crucial for recognition of cognate tRNAgln and discrimination against noncognate tRNAglu. In Gln4, while Arg402 is conserved as Arg631, Lys401 is replaced by Phe630. While both lysine and phenylalanine are large residues, it is unknown if the loss of the positively charged side chain results in different interactions with G36 in Gln4.
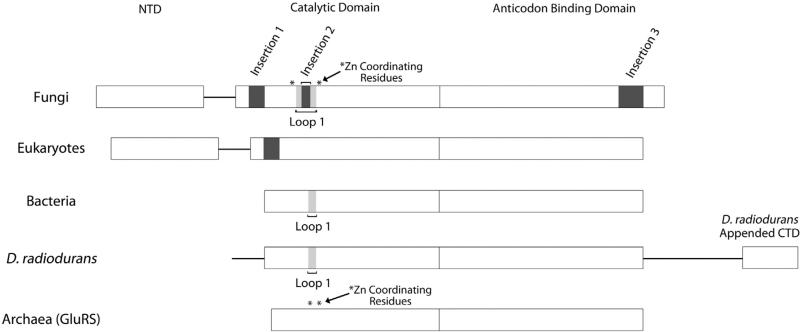
Multiple sequence analysis among eukaryotic GlnRS species revealed three insertions relative to E. coli GlnRS (Figs. 1 and 2). Insertion 1, located at residues 234–250 of Gln4, is present in all eukaryotes, including those species lacking an NTD, although its length and sequence are not conserved among different kingdoms. Comparison with the E. coli GlnRS structure reveals that this insertion is part of the Rossmann fold domain in the CTD. Insertion 2 is located at residues 364–368 and is situated in a zinc binding motif consisting of Cys346-X-Cys348-X24-Cys372-X-His374 (Fig. S3). The zinc binding motif and Insertion 2, which are always found together, are present in fungal GlnRS species but are absent in other eukaryotes. Insertion 3 is a loop located at residues 751–770 and is also only conserved among fungi and absent in other eukaryotes.
Fig. 2.
Domain architecture of GlnRS. Domains for GlnRS are shown for fungi, eukaryotes (other than fungi), bacteria, the bacterium D. radiodurans, and archaeal GluRS. Major domains are labeled NTD, catalytic domain, anticodon binding domain, and the unique CTD appended to the D. radiodurans GlnRS. Insertion 1, Insertion 2, Insertion 3, and Loop 1 are labeled along with the zinc coordinating residues of the zinc binding motif, which is present only in fungal GlnRS and archaeal GluRS.
GlnRS selects both the correct amino acid substrate, glutamine, and the cognate tRNAgln molecule. To gain insight into the tRNAgln discrimination mechanism in Gln4, we performed further sequence analysis incorporating the archaeal non-discriminating GluRS, an evolutionary precursor of GlnRS that has the ability to recognize both tRNAgln and tRNAglu.18 This revealed that Insertion 2 is part of a larger nine-residue loop, Loop 1, which is absent in archaeal GluRS (Fig. 2). In E. coli GlnRS, Loop 1 is only four residues long and has been suggested to provide the ability to discriminate against the G1:C72 base pair in tRNAglu and instead only recognizes tRNAgln by disrupting the weak U1:A72 base pair.18 Our results suggest that Loop 1 may play a different role in fungi and other eukaryotes than it does in bacteria. Although Loop 1 is present in Gln4, it differs in amino acid sequence compared to E. coli GlnRS and, with Insertion 2, is five residues longer. Additionally, while the first base pair of tRNAgln is U1:A72 in E. coli, the first base pair of tRNAgln is G1:C72 in fungi and other eukaryotes. It is therefore unlikely that the extended Loop 1 seen in Gln4 functions similarly to E. coli GlnRS Loop 1 by disrupting the first base pair, since if the extended Loop 1 in Gln4 were to disrupt the strong G1:C72 base pair in tRNAgln it might also disrupt the weak U1:A72 base pair in tRNAglu and fail to discriminate between them. While our observations are based on length, it is unknown whether sequence-specific contacts in Loop 1 contribute tRNA discrimination ability to Gln4. However, in all eukaryotes other than fungi, Loop 1 is entirely absent (Fig. 2) and this region of eukaryotic GlnRS more closely resembles archaeal GluRS, suggesting that Loop 1 is not likely to be a critical tRNA discriminatory motif in eukaryotes with the possible exception of fungi.
Motion of NTD in solution is limited in the absence of tRNA
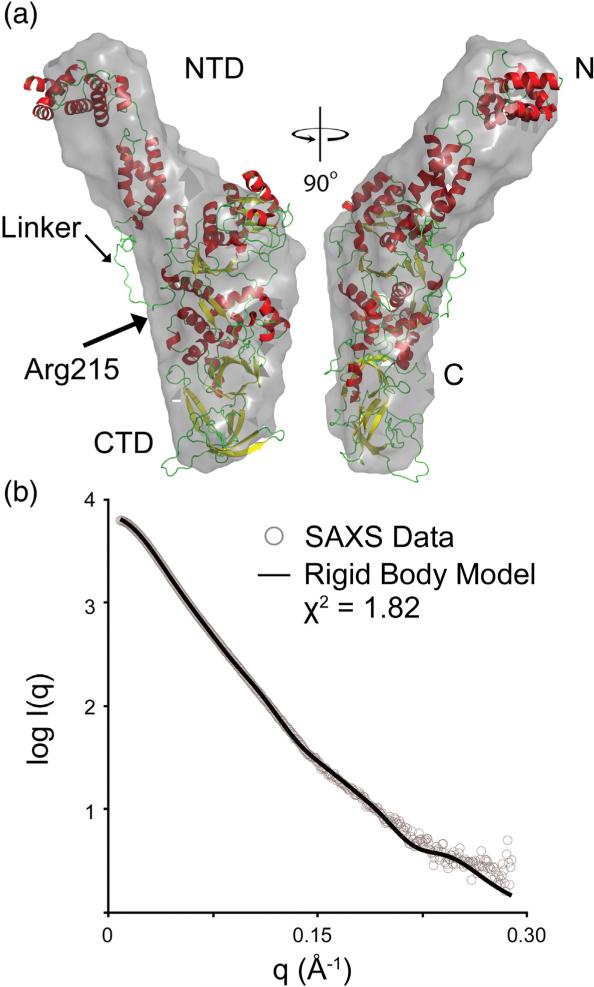
We collected SAXS data on full-length Gln4 (Fig. S4) because SAXS is a solution-based technique that is sensitive to the overall shape and size of a molecule and can also provide insight into protein dynamics.19,20 Analysis of the pair distribution function (the distribution of interatomic distances) yields a maximum particle dimension of ~147 Å and a radius of gyration (Rg) of 43.51 Å, in close agreement with that estimated by Guinier analysis,21 43.32 Å. This is significantly larger than the maximum dimension and Rg of the CTD from the crystal structure (108 Å and 32.3 Å, respectively). The increased values are consistent with the presence of an appended NTD. The SAXS data of full-length Gln4 suggest an elongated particle shape due to the large maximum dimension compared to the Rg and the left-skewed pair distribution function (Fig. S4d).
Since the crystallographic data showed that the NTD was disordered, we used the SAXS data to probe the motion of the NTD in solution. Information about protein flexibility can be obtained from a Porod–Debye analysis of SAXS data, where the decay in intensity is studied as a function of angle and reported as the Porod exponent.22 In cases of globular, rigid molecules, the Porod exponent will approach 4, whereas for flexible, random chains, it will approach 2. A Porod–Debye analysis of full-length Gln4 resulted in a Porod exponent of 3.4, suggesting a well-folded protein with mild flexibility, consistent with the crystallographic data. The total volume occupied by the protein can be measured from the SAXS data and is known as the Porod volume.23 The Porod volume of full-length Gln4 is ~153,000 Å3, corresponding to an estimated molecular mass of 92.6 kDa,22 similar to the molecular mass estimated from the forward scattering [I(0)] of 89.1 kDa and the expected molecular mass of 93.1 kDa (these data also confirm that the full-length protein is intact). This demonstrates that the volume occupied by the NTD is approximately limited to the size of the domain and does not occupy a larger region of conformational space.
Ten ab initio envelope reconstructions of full-length Gln4 were created and averaged, exhibiting an average normalized spatial discrepancy (NSD) of 0.633 ± 0.022. NSD is a measure of the variance among a set of multiple SAXS envelopes. NSD values less than 1 reflect uniqueness and low flexibility,24 whereas values greater than 1 reflect an inability to describe the protein using one unique shape. To generate a rigid-body model, we created 10,000 random conformations of full-length Gln4 by treating the NTD and CTD crystal structures as independent rigid bodies and allowing the linker (residues 188–214) to be flexible. The model whose calculated scattering best fit the experimental SAXS data was selected from this pool. This model is shown in Fig. 3 superposed onto the ab initio envelope. The rigid-body model fits well to the experimental scattering data with Χ2 = 1.82. The location of the NTD of Gln4 appears to be similar to the appended domain modeled in the hybrid structure of GlnRS from D. radiodurans.8D. radiodurans is unique because it is the only bacteria known to contain a GlnRS with an appended domain. However, the appended domain of D. radiodurans GlnRS is located on the C-terminus, downstream of the core enzyme (Fig. 2), and contains no sequence homology to the appended domain of eukaryotic GlnRS, which is located on the N-terminus, upstream of the core enzyme. The similar spatial location of both appended domains, despite being extensions at different termini of the core enzyme, is made possible by the much longer 80-residue linker in D. radiodurans GlnRS, compared to the 26-residue linker in Gln4. Additionally, the appended domain of D. radiodurans GlnRS was shown to cross-react with an antibody that recognizes the GatB subunit of the GatCAB amidotransferase used in the indirect pathway of Gln-tRNAgln formation used in most prokaryotes.8 Our previous study reported a high degree of structural similarity between the Gln4 NTD and the GatB subdomain,12 suggesting that the appended domains of GlnRS from eukaryotes and D. radiodurans perform similar functions and are likely an instance of convergent evolution.
Fig. 3.
SAXS rigid-body model of full-length Gln4. (a) Two orientations rotated 90° relative to each other are shown in cartoon representation and colored according to secondary structure and superposed onto the ab initio envelope shown in gray. (b) The calculated scattering of the rigid-body model (black continuous line) fit to the experimental SAXS data (gray circles).
Ab initio reconstructions and rigid-body modeling do not take into account the dynamic information present in SAXS data but represent an average of all conformations present in solution. Ensemble modeling can overcome this by representing a protein structure as an ensemble of multiple conformations. Using the ensemble optimization method24 with multiple conformations of the NTD did improve the overall fit to the scattering data (Χ2 = 1.75); however, the improvement was only marginal. The Rg distribution of conformers present in the ensemble was similar to that seen for simulated data of the rigid-body model alone, suggesting little flexibility. The limited flexibility from the Porod–Debye analysis, the low NSD for multiple ab initio envelope reconstructions, and the marginal improvement from an ensemble model compared to the rigid-body model demonstrate that the mobility of the NTD in solution is limited.
A model of full-length Gln4 bound to tRNAgln suggests a substantial conformational change of the NTD
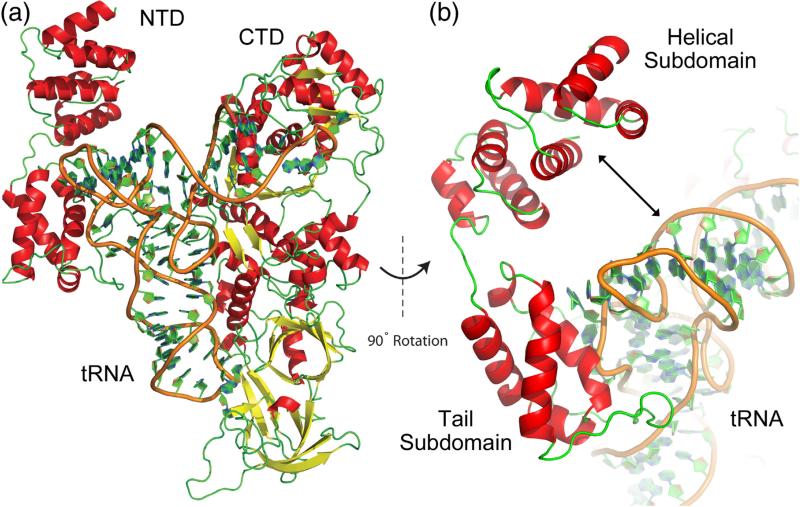
We used the structure of E. coli GlnRS bound to tRNA13 and the structure of Thermotoga maritima GatCAB bound to tRNA11 to obtain a model of full-length Gln4 bound to tRNA. First, the structure of E. coli GlnRS bound to tRNAgln (PDB ID: 1QTQ) was superposed onto the Gln4 CTD (residues 215–809). The structure of the tRNA molecule was then extracted, providing an initial model of tRNAgln complexed with the Gln4 CTD. Since the nucleotide sequence of E. coli tRNAgln differs from yeast tRNAgln, the tRNA molecule was computationally mutated to match that of yeast tRNAgln and a geometric minimization was performed using ModeRNA.25 Our model contains similar contacts between the tRNAgln anticodon and the Gln4 anticodon binding domain that were shown to be important identity elements in E. coli GlnRS9,13 due to the high degree of structural similarity between E. coli GlnRS and the Gln4 CTD.
To correctly place the NTD in the model, we utilized known structural homology of the NTD with subunit B of the GatCAB amidotransferase (GatB) from T. maritima (PDB ID: 3AL0), which was solved in complex with tRNAgln. The tail subdomain (residues 119–187) of the Gln4 NTD was superposed to the tail subdomain of GatB due to the high level of structural homology between these two regions.12 Finally, MODELLER26 was used for de novo modeling of the predicted flexible linker (residues 188–214) (Fig. 4).
Fig. 4.
Homology model of full-length Gln4 bound to tRNAgln. (a) Full-length Gln4 shown bound to tRNAgln. (b) Enlarged and rotated model showing gap between NTD helical subdomain and tRNA molecule.
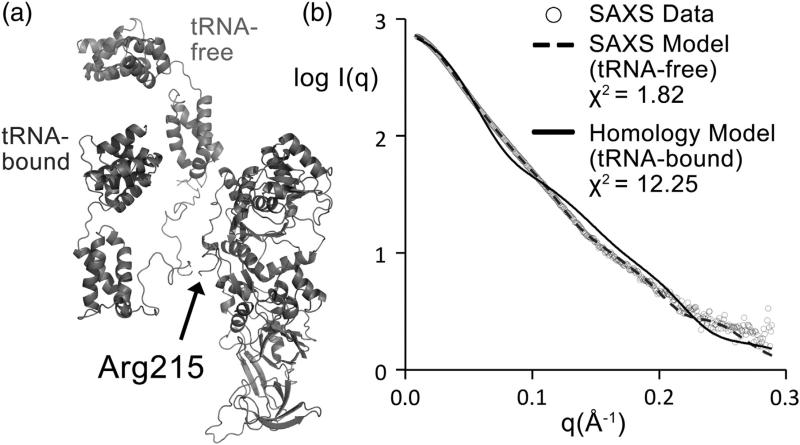
Our model of full-length Gln4 bound to tRNAgln shows a significant change in the NTD position when compared to the tRNA-free, SAXS-derived conformation (Fig. 5a). The model shows an ~160° rotation and an ~40-Å translation of the NTD with respect to the solution conformation according to an analysis by DynDom.27 To determine if this new conformation of the NTD exists in solution, we simulated the SAXS profile from our model of Gln4 bound to tRNA, after removing the tRNA molecule, and compared it to the experimental SAXS data. Fitting the simulated scattering of only the protein portion of the model of the protein–tRNA complex to the experimental SAXS data resulted in a poor fit, yielding Χ2 = 12.25 compared to 1.82 for the rigid-body model created using the experimental SAXS data (Fig. 5b). The limited flexibility of the NTD, coupled with the poor fit of the simulated scattering of the protein portion of the model bound to tRNAgln, suggests that, without tRNA bound, this conformation does not exist in solution.
Fig. 5.
Comparison of NTD position before and after tRNA binding. (a) Gln4 is shown in cartoon representation. The NTD position prior to tRNA binding determined by SAXS rigid-body modeling is shown in light gray. The NTD position upon binding tRNA predicted by homology modeling is shown in dark gray. The CTD is also shown in dark gray. (b) The simulated SAXS profile of the rigid-body model (broken line) and the homology model (continuous line; calculated without tRNA in the model) are shown fitted to the experimental SAXS data (circles). The better visual fit and lower Χ2 value of the tRNA-free model shows that the SAXS data are able to clearly distinguish between which model exists in solution.
Conformational change in NTD subdomains is predicted upon interaction with tRNAgln
In the compiled model of Gln4 bound to tRNA, the NTD subdomains appear to be in an “open” conformation resulting in the helical subdomain (residues 1–110) being too distant from the tRNA molecule to form stable contacts (Fig. 4b). This conformation more closely resembles that observed in the Staphylococcus aureus GatCAB structure solved without tRNA bound12 than the conformation seen in tRNA-bound T. maritima GatCAB. Thus, we considered that the molecule might undergo a conformational change upon tRNA binding. To probe the dynamics of full-length Gln4 in complex with tRNAgln, we carried out 70 ns of molecular dynamics simulation. The model stabilized within approximately 10 ns of simulation time and remained relatively unchanged for the remaining 60 ns (Fig. S5), showing that the simulation time captured the relevant dynamics.
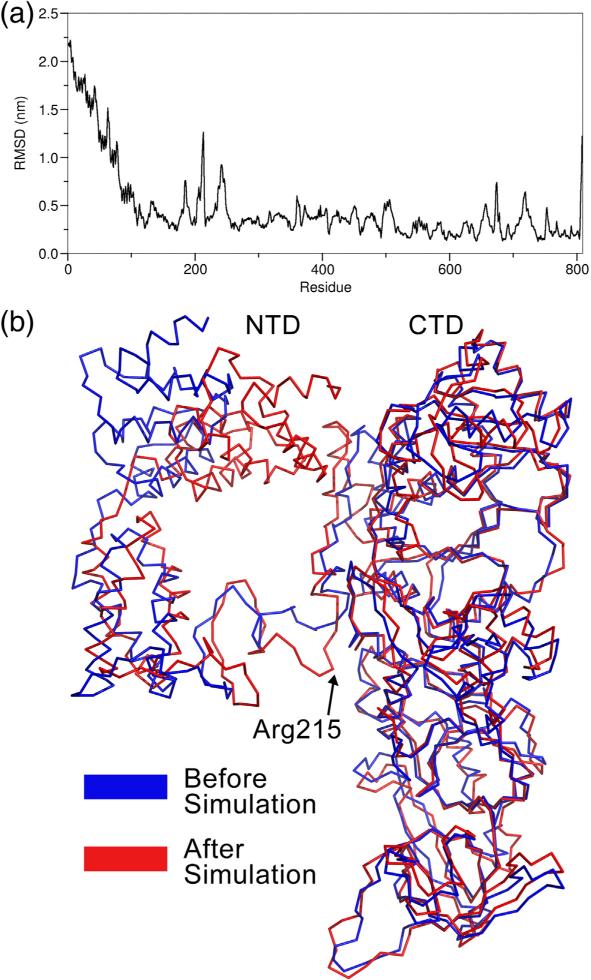
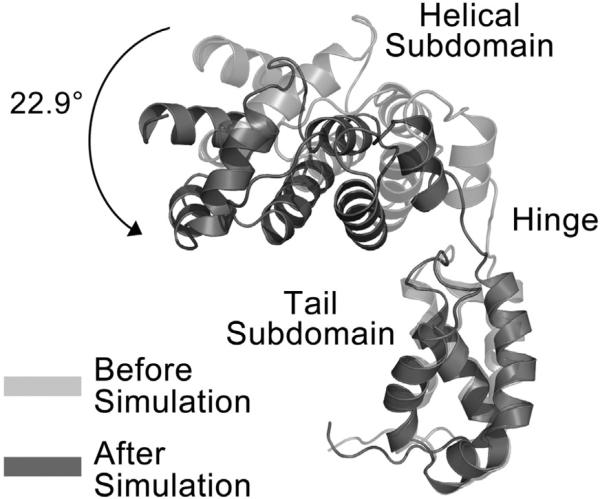
The resulting molecular dynamics trajectory (Figs. 6 and 7 and Fig. S5) shows that the helical subdomain of the NTD rotates about a conserved hinge by 22.9°, forming electrostatic interactions between the positively charged side chains of lysine residues 19, 20, and 26, which have been previously implicated in tRNA binding integrity,28 and the negatively charged phosphate backbone of tRNA. These electrostatic interactions along the inner face of the NTD were speculated12 to be responsible for the nonspecific RNA binding activity.29 This new conformation of the NTD is in agreement with the tRNA-bound conformation seen in the T. maritima GatB structure. The hinge motion in our model is strongly supported by our previous study demonstrating that mutations in conserved residues in the hinge region, connecting the helical and tail domains of the NTD, significantly reduce tRNA binding.12 In addition, our modeling predicts that residues K29, K63, G64, T65, and D66 of the helical subdomain of the NTD make several contacts with the CTD, which includes residues P238 and M241 of Insertion 1. Since Insertion 1 was shown above to be exclusive to eukaryotes, we speculate that this insertion may provide a means of communication between the NTD and CTD.
Fig. 6.
NTD undergoes conformational change after binding to tRNA. (a) Plot of backbone RMS fluctuation as a function of residue. The RMS has been calculated as the deviation from the starting structure. (b) Structure of Gln4 before (blue) and after (red) molecular dynamics simulation. The tRNA molecule has been removed for clarity.
Fig. 7.
Comparison of NTD structure before and after molecular dynamics simulation. The position of the NTD before the simulation is shown in light gray and that after the simulation is shown in dark gray. The black continuous arrow shows the degree and direction of angular motion calculated by DynDom.
The molecular dynamics simulation shows a significant distortion of the tRNA molecule as a result of the NTD binding to tRNA. At the start of the simulation, base U73 is pointed away from the inner core of the enzyme, making no specific contacts with the protein. Approximately 3 ns into the simulation, upon distortion of the tRNA molecule, this base flips inwards toward the enzyme, resulting in contacts with Leu463. This conformational change in the tRNA, caused by binding of the NTD, results in these previously unknown interactions between Leu463 and U73. Our previous study reported that the presence of the NTD across eukaryotes is strongly correlated with the existence of base U73 of the acceptor stem, immediately preceding the CCA tail.12 Furthermore, it was shown in E. coli GlnRS that any alterations to base 73 led to substantial increases in KM for glutamine.17 Our molecular dynamics simulation showing these concerted conformational changes, combined with the phylogenetic correlation of the NTD and base U73 and the increase in KM for glutamine when mutating base 73 in E. coli GlnRS, may explain the result that deletion of the NTD in Gln4 leads to increased KM for glutamine.12
Previously it was shown that GatB provides tRNA discrimination ability to GatCAB through the tail domain.30 This discrimination occurs through an interaction with U20B of tRNAglu that is absent in tRNAgln in prokaryotes. Analysis of tRNAgln and tRNAglu sequences in yeast reveals that base U20B is present in both tRNA sequences, suggesting that this base does not participate in tRNA discrimination interactions with Gln4. Our model of Gln4 bound to tRNAgln reveals no evidence of significant interactions between tRNAgln and the NTD that would otherwise be negated by replacement with tRNAglu. This is consistent with previous studies demonstrating nonspecific RNA binding activity of the NTD.28,29 Therefore, in contrast to the tRNA discrimination properties of GatB, our model provides no evidence that the Gln4 NTD makes any direct contacts with tRNAgln that would confer any additional capacity for discrimination against tRNAglu, suggesting that discrimination occurs via specific contacts with the CTD.
Conclusion
The structure of the CTD of Gln4 presented here was shown to be highly similar to the E. coli GlnRS structure. However, multiple insertions relative to E. coli GlnRS revealed insights into one structural motif common to all eukaryotes and two motifs specific to fungi. Since most eukaryotic GlnRS species lack the bacterial unpairing loop that is proposed to play a role in tRNA discrimination, then another mechanism must be employed to discriminate between tRNAglu and tRNAgln in eukaryotic GlnRS species. Therefore, in eukaryotes, there may be a compensating mechanism to discriminate between G1:C72 of tRNAgln from U1:A72 of tRNAglu or, as in the case of non-discriminating archaeal GluRS,18 the first base pair does not play a significant role in tRNA discrimination.
Our structure-based models of the first full-length eukaryotic GlnRS with and without tRNAgln bound suggest that CTD binding to tRNA results in a large conformational reorientation of the NTD allowing for interactions between the NTD and the tRNA. Given the distinct increases in KM and KD for tRNAgln following deletion of the NTD,10,12 the solution model of full-length Gln4 presented here suggests that the NTD plays a direct role in tRNA binding. Our molecular dynamics simulation revealed that the helical and tail subdomains of the NTD undergo a hinge motion after binding to tRNA, allowing for tighter binding between the NTD and tRNA. Our structural results and modeling suggest the intriguing possibility that the NTD communicates with the CTD through Insertion 1, which is found in all eukaryotes. The absence of such an interaction may explain the loss of the NTD in bacterial GlnRS evolution. In addition, since the NTD and the active site of Gln4 are too distant to interact directly and since deletion of the NTD also increases KM for glutamine and ATP, it seems plausible that the effects on glutamine and ATP are due to the concerted conformational changes in Gln4 that occur upon tRNA binding as was observed in E. coli GlnRS.10,12,13,15 Further experimental studies will be required to confirm these proposed interactions in eukaryotic GlnRS.
Materials and Methods
Protein expression and purification
To obtain highly purified Gln4 protein and its derivatives, we cloned open reading frames into the previously described LIC vectors BG2483 or BG2663 under PGAL1 control31 as previously described12 and expressed them in yeast strain BCY123.32 To purify full-length Gln4 from a Gln4-pt-tagged construct (QB721B), we performed large-scale growth, affinity purification on IgG Sepharose, removal of GST-3C protease, concentration of samples, and sizing on Superdex HiLoad 16/60 (GE Healthcare 17–1069, 10 mm × 300 mm bed dimension) as previously described.33 This purified Gln4 construct was used for crystallographic and structural analyses.
To ensure that full-length polypeptide was purified for crystallography, we added an N-terminal Met-Ala-His6 tag at the N-terminus of GLN4, during PCR amplification of the GLN4 gene using QB832ADFP and QB832ADRP primers (Supplementary Table 1). Full-length GLN4 was amplified with QB1012ADFP and QB832ADRP.
For crystallography of the full-length His6-Gln4 protein, the His6-Gln4-pt was purified from strain QB832AD (21.6 L at 6.5 OD/L) on 24 mL IgG Sepharose and eluted with 64 mL 3C cleavage buffer.33 After elution from IgG Sepharose and removal of GST-3C protease but prior to sizing, the sample containing the full-length Gln4 protein with the N-terminal His6 tag (QB832AD) was diluted with an equal volume of buffer T [25 mM Hepes (pH 7.5), 850 mM NaCl, 10% (v/v) glycerol, 2 mM β-mercaptoethanol (BME), and 2 mM phenylmethylsulfonyl fluoride], mixed with 3 mL prewashed Talon resin [washed in Wash buffer, 20 mM Hepes (pH 7.5), 0.5 M NaCl, 5% (v/v) glycerol, and 2 mM BME], incubated for 1 h at 4 °C, followed by centrifugation at 2000 rpm for 2 min and removal of supernatant, after which the Talon resin was washed once for 10 min with 40 mL Wash buffer containing 0.5 M NaCl [20 mM Hepes (pH 7.5), 0.5 M NaCl, 5% (v/v) glycerol, and 2 mM BME], centrifuged at 2000 rpm for 2 min, followed by two more washes of the resin with Wash buffer containing 1 M NaCl, then a wash with Wash buffer containing 0.5 M NaCl, followed by a wash with Wash buffer containing 0.5 M NaCl and 10 mM imidazole. The protein was eluted from the Talon resin with four sequential washes of the resin with Wash buffer that contains 0.5 M NaCl containing 250 mM imidazole, each of which was mixed for 10 min prior to the low-speed spin. Three fractions (elutions 1 and 2 as well as the 10 mM imidazole wash) were combined and diluted with 75 mL of no-salt buffer to bring the NaCl to 0.2 M, followed by concentration to 5 mL using an Amicon (Millipore UFC901024), and loaded onto a Superdex 200 sizing column as previously described.33
Crystallization
Initial crystallization conditions were identified using a high-throughput microbatch-under-oil method with a 1536-condition, incomplete-factorial-based screen.34 Conditions that produced crystals appearing suitable for optimization were prioritized according to their ease of cryoprotection35 and optimized using a drop volume ratio versus temperature technique.36 Crystals of full-length Gln4 were prepared for diffraction by combining 3.5 μL of protein solution [13.9 mg/mL protein in 200 mM NaCl, 5% (v/v) glycerol, 2 mM DTT, 0.025% (w/v) NaN3, and 20 mM Hepes buffer (pH 7.5)] with 2.0 μL of precipitant solution [50 mM NH4Br, 50 mM KC2H3O2, 100 mM Hepes (pH 7.5), and 20%(w/v) polyethylene glycol 20,000], incubated at 14 °C. The optimized 16:9 protein-to-precipitant ratio and 14 °C temperature were determined from a drop volume ratio versus temperature technique. Crystals appeared after 4 weeks.36
Single crystal data collection and structure solution
Crystals of the full-length protein were harvested and cryoprotected, then shipped to Stanford Synchrotron Radiation Lightsource (SSRL). Single crystal X-ray data were collected remotely on beamline 11-1.37 An initial excitation scan revealed the presence of zinc. Data were collected to 2.15 Å, integrated with XDS,38–40 and reduced with Scala,41 part of the CCP4 package.42 Initial molecular replacement with PHENIX43 using an ~40% sequence homology E. coli Gln-tRNA synthetase (PDB ID: 1GTS)14 failed, but a combined molecular replacement/single-wavelength anomalous dispersion approach with the zinc signal was successful. An iterative process of PHENIX refinement and manual model building through Coot44 was employed with validation using MolProbity.45 The coordinates were deposited as PDB ID: 4H3S. Data collection, processing, and refinement statistics are given in Table 1. The N-terminal region, residues 1–214, was unresolved in the electron density map. SDS-PAGE gels indicated that it was present in the crystals. A similar procedure was followed to determine the structure of the His tag purified protein. Western blot analysis again confirmed that the NTD residues were present in the crystals. PyMOL46 was used to generate symmetry mates, analyze solvent channels and crystallographic packing, and produce images.
Bioinformatics
The DISOPRED2 prediction of protein disorder server was used to predict disordered residues.32 Primary sequences were taken from the UniProt database.47 Structures of GlnRS from E. coli (PDB ID: 1NYL) and D. radiodurans (PDB ID: 2HZ7) and GluRS from Methanobacterium thermoautotrophicus (PDB ID: 3AII) were taken from the PDB.5,8,15,18 Sequence data and structural data were combined for use in the PROMALS3D multiple sequence and structure alignment server,48 and the resulting alignment was used to identify structural motifs discussed in the text. tRNA sequences were taken from the tRNAdb 2009 online database.49 BLAST (Basic Local Alignment Search Tool) was used to calculate expectation values (E values) for comparing sequences.50,51
Small-angle X-ray scattering
Small-angle X-ray solution scattering data were collected on beamline 4-2 of the SSRL.52 Data were collected from the full-length protein at a wavelength of 1.3 Å for eight consecutive 2-s exposures collected at five different concentrations ranging from 1.0 to 7.7 mg/mL. Data were collected from the flow-through buffer of the final purification column [200 mM NaCl, 5% (v/v) glycerol, 2 mM DTT, 0.025% (w/v) NaN3, and 20 mM Hepes buffer (pH 7.5)] and subtracted from the total protein solution scattering. The data were integrated with SasTool52 and then examined with PRIMUS.53 Analysis of eight consecutive time frames ensured that no radiation damage took place over the course of the experiment. The SAXS data for different protein concentrations were investigated for aggregation using Guinier plots.21 Radius of gyration estimates were derived by the Guinier approximation with qRg < 1.3 using the radius of gyration function of PRIMUS where q = 4πsinθ/λ. GNOM54 was used to compute the pair distribution function, P(r), and to determine the maximum particle dimension, Dmax. No evidence of concentration dependence was seen when comparing all five concentrations. Chicken egg white lysozyme was used as a protein standard to estimate the molecular weight from I(0) extrapolated from the scattering curve using GNOM in reciprocal space. Porod–Debye analysis and Porod volume estimations were performed using a pre-release version of the software SCÅTTER22 and confirmed with PRIMUS. Ten ab initio shape reconstructions were generated by DAMMIF55 and averaged with DAMAVER.56 Ensemble modeling was carried out with the ensemble optimization method24 using default parameters. Fifty identical runs were performed using 20 conformers in the ensemble selected from among 10,000 random conformations of full-length Gln4, while treating the NTD and CTD crystal structures as independent rigid bodies and allowing the linker (residues 188–214) to be flexible. Rigid-body modeling was performed similarly using only one conformer in the ensemble, which selected the best single conformation from among all 10,000 random conformations.
Molecular dynamics
The initial model used for molecular dynamics simulation was generated as described in the text. All structural alignments were performed using the “fit” function of PyMOL using carbon α atoms. Molecular dynamics simulations were performed in GROMACS with the AMBER99SB force field.57,58 The initial model was solvated using a cubic SPC/E water model59 and neutralized with ions prior to minimization via steepest descents. Distance restraints were added to keep the zinc ion in place. The model was then equilibrated under an isothermal–isochoric ensemble for 100 ps at 300 K followed by equilibration under an isothermal–isobaric ensemble for 100 ps to ensure stability prior to the simulation. Simulations were then performed at the Center for Computational Resources60 on 512 processors. Total simulation time was 70 ns. The DynDom Protein Domain Motion Analysis Web server27 was used to compare models. RMSD from the starting structure was calculated for each frame of the trajectory after fitting to the starting structure. RMS fluctuation was calculated as the deviation from the starting structure after fitting to the starting structure.
Supplementary Material
Acknowledgements
We would like to thank the staff at SSRL beamline 4-2, Drs. Thomas Weiss, Tsutomu Matsui, and Lester Carter. We gratefully thank Dr. Robert Rambo at the Advanced Light Source for access to a pre-release version of SCÅTTER and for helpful discussions. We would also like to acknowledge Dr. Alan Grossfield at the University of Rochester for helpful discussions regarding molecular dynamics protocols and analysis and Cynthia Cornelius at the University at Buffalo's Center for Computational Resources for technical assistance and advice. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program. Financial support was provided by the Defense Threat Reduction Agency (Grant HDTRA1-10-C-0057 to E.H.S.) and National Institutes of Health (R01 GM100494).
Dedication. We would like to dedicate this article in memory of the late Dr. Hiro Tsuruta who provided valuable guidance and instruction for the collection and analysis of small-angle X-ray scattering data.
Abbreviations used
- NTD
N-terminal domain
- CTD
C-terminal domain
- SAXS
small-angle X-ray scattering
- PDB
Protein Data Bank
- NSD
normalized spatial discrepancy
- BME
β-mercaptoethanol
- SSRL
Stanford Synchrotron Radiation Lightsource
Footnotes
Accession numbers
The coordinates and the structure factors have been deposited in the PDB with accession number 4H3S.
Supplementary Data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2013.03.043
References
- 1.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Cusack S, Berthet-Colominas C, Hartlein M, Nassar N, Leberman R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 Å. Nature. 1990;347:249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- 3.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 4.Guo M, Yang XL, Schimmel P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat. Rev., Mol. Cell Biol. 2010;11:668–674. doi: 10.1038/nrm2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr, Brice MD, Rodgers JR, et al. The Protein Data Bank: a computer-based archival file for macromolecular structures. Eur. J. Biochem. 1977;80:319–324. doi: 10.1111/j.1432-1033.1977.tb11885.x. [DOI] [PubMed] [Google Scholar]
- 6.Curnow AW, Hong K, Yuan R, Kim S, Martins O, Winkler W, et al. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc. Natl Acad. Sci. USA. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamour V, Quevillon S, Diriong S, N'Guyen VC, Lipinski M, Mirande M. Evolution of the GlxtRNA synthetase family: the glutaminyl enzyme as a case of horizontal gene transfer. Proc. Natl Acad. Sci. USA. 1994;91:8670–8674. doi: 10.1073/pnas.91.18.8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deniziak M, Sauter C, Becker HD, Paulus CA, Giege R, Kern D. Deinococcus glutaminyltRNA synthetase is a chimer between proteins from an ancient and the modern pathways of aminoacyl-tRNA formation. Nucleic Acids Res. 2007;35:1421–1431. doi: 10.1093/nar/gkl1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rould MA, Perona JJ, Söll D, Steitz TA. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 Å resolution. Science. 1989;246:1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- 10.Ludmerer SW, Wright DJ, Schimmel P. Purification of glutamine tRNA synthetase from Saccharomyces cerevisiae. A monomeric aminoacyl-tRNA synthetase with a large and dispensable NH2-terminal domain. J. Biol. Chem. 1993;268:5519–5523. [PubMed] [Google Scholar]
- 11.Ito T, Yokoyama S. Two enzymes bound to one transfer RNA assume alternative conformations for consecutive reactions. Nature. 2010;467:612–616. doi: 10.1038/nature09411. [DOI] [PubMed] [Google Scholar]
- 12.Grant TD, Snell EH, Luft JR, Quartley E, Corretore S, Wolfley JR, et al. Structural conservation of an ancient tRNA sensor in eukaryotic glutaminyl-tRNA synthetase. Nucleic Acids Res. 2012;40:3723–3731. doi: 10.1093/nar/gkr1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rath VL, Silvian LF, Beijer B, Sproat BS, Steitz TA. How glutaminyl-tRNA synthetase selects glutamine. Structure. 1998;6:439–449. doi: 10.1016/s0969-2126(98)00046-x. [DOI] [PubMed] [Google Scholar]
- 14.Perona JJ, Rould MA, Steitz TA. Structural basis for transfer RNA aminoacylation by Escherichia coli glutaminyl-tRNA synthetase. Biochemistry. 1993;32:8758–8771. doi: 10.1021/bi00085a006. [DOI] [PubMed] [Google Scholar]
- 15.Sherlin LD, Perona JJ. tRNA-dependent active site assembly in a class I aminoacyl-tRNA synthetase. Structure. 2003;11:591–603. doi: 10.1016/s0969-2126(03)00074-1. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez-Hernandez A, Perona JJ. Heat maps for intramolecular communication in an RNP enzyme encoding glutamine. Structure. 2011;19:386–396. doi: 10.1016/j.str.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 17.Ibba M, Hong KW, Sherman JM, Sever S, Söll D. Interactions between tRNA identity nucleotides and their recognition sites in glutaminyltRNA synthetase determine the cognate amino acid affinity of the enzyme. Proc. Natl Acad. Sci. USA. 1996;93:6953–6958. doi: 10.1073/pnas.93.14.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nureki O, O'Donoghue P, Watanabe N, Ohmori A, Oshikane H, Araiso Y, et al. Structure of an archaeal non-discriminating glutamyl-tRNA synthetase: a missing link in the evolution of Gln-tRNAGln formation. Nucleic Acids Res. 2010;38:7286–7297. doi: 10.1093/nar/gkq605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 20.Grant TD, Luft JR, Wolfley JR, Tsuruta H, Martel A, Montelione GT, Snell EH. Small angle X-ray scattering as a complementary tool for high-throughput structural studies. Biopolymers. 2011;95:517–530. doi: 10.1002/bip.21630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guinier A, Foumet F. Small Angle Scattering of X-rays. Wiley Interscience; New York, NY: 1955. [Google Scholar]
- 22.Rambo RP, Tainer JA. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers. 2011;95:559–571. doi: 10.1002/bip.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porod G. Die röntgenkleinwinkelstreuung von dichtgepackten kolloiden systemen. Colloid Polym. Sci. 1951;124:83–114. [Google Scholar]
- 24.Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using small-angle X-ray scattering. J. Am. Chem. Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 25.Rother M, Milanowska K, Puton T, Jeleniewicz J, Rother K, Bujnicki JM. ModeRNA server: an online tool for modeling RNA 3D structures. Bioinformatics. 2011;27:2441–2442. doi: 10.1093/bioinformatics/btr400. [DOI] [PubMed] [Google Scholar]
- 26.Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 27.Hayward S, Berendsen HJ. Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins. 1998;30:144–154. [PubMed] [Google Scholar]
- 28.Wang CC, Morales AJ, Schimmel P. Functional redundancy in the nonspecific RNA binding domain of a class I tRNA synthetase. J. Biol. Chem. 2000;275:17180–17186. doi: 10.1074/jbc.M001057200. [DOI] [PubMed] [Google Scholar]
- 29.Wang CC, Schimmel P. Species barrier to RNA recognition overcome with nonspecific RNA binding domains. J. Biol. Chem. 1999;274:16508–16512. doi: 10.1074/jbc.274.23.16508. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura A, Sheppard K, Yamane J, Yao M, Söll D, Tanaka I. Two distinct regions in Staphylococcus aureus GatCAB guarantee accurate tRNA recognition. Nucleic Acids Res. 2010;38:672–682. doi: 10.1093/nar/gkp955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malkowski MG, Quartley E, Friedman AE, Babulski J, Kon Y, Wolfley J, et al. Blocking S-adenosylmethionine synthesis in yeast allows selenomethionine incorporation and multiwavelength anomalous dispersion phasing. Proc. Natl Acad. Sci. USA. 2007;104:6678–6683. doi: 10.1073/pnas.0610337104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macbeth MR, Lingam AT, Bass BL. Evidence for auto-inhibition by the N terminus of hADAR2 and activation by dsRNA binding. RNA. 2004;10:1563–1571. doi: 10.1261/rna.7920904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quartley E, Alexandrov A, Mikucki M, Buckner FS, Hol WG, DeTitta GT, et al. Heterologous expression of L. major proteins in S. cerevisiae: a test of solubility, purity, and gene recoding. J. Struct. Funct. Genomics. 2009;10:233–247. doi: 10.1007/s10969-009-9068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luft JR, Collins RJ, Fehrman NA, Lauricella AM, Veatch CK, DeTitta GT. A deliberate approach to screening for initial crystallization conditions of biological macromolecules. J. Struct. Biol. 2003;142:170–179. doi: 10.1016/s1047-8477(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 35.Kempkes R, Stofko E, Lam K, Snell EH. Glycerol concentrations required for the successful vitrification of cocktail conditions in a high-throughput crystallization screen. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2008;64:287–301. doi: 10.1107/S0907444907067613. [DOI] [PubMed] [Google Scholar]
- 36.Luft JR, Wolfley JR, Said MI, Nagel RM, Lauricella AM, Smith JL, et al. Efficient optimization of crystallization conditions by manipulation of drop volume ratio and temperature. Protein Sci. 2007;16:715–722. doi: 10.1110/ps.062699707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soltis SM, Cohen AE, Deacon A, Eriksson T, Gonzalez A, McPhillips S, et al. New paradigm for macromolecular crystallography experiments at SSRL: automated crystal screening and remote data collection. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2008;64:1210–1221. doi: 10.1107/S0907444908030564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kabsch W. Automatic indexing of rotation diffraction patterns. J. Appl. Crystallogr. 1998;21:67–72. [Google Scholar]
- 39.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kabsch W. XDS. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evans P. Scaling and assessment of data quality. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 42.Collaborative Computational Project, N The CCP4 suite: programs for protein crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1. 2010:3r1. [Google Scholar]
- 47.Magrane M, Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011 doi: 10.1093/database/bar009. http://dx.doi.org/10.1093/database/bar009. [DOI] [PMC free article] [PubMed]
- 48.Pei J, Tang M, Grishin NV. PROMALS3D web server for accurate multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:W30–W34. doi: 10.1093/nar/gkn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 51.Karlin S, Altschul SF. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc. Natl Acad. Sci. USA. 1990;87:2264–2268. doi: 10.1073/pnas.87.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smolsky IL, Liu P, Niebuhr M, Ito K, Weiss TM, Tsuruta H. Biological small-angle X-ray scattering facility at the Stanford Synchrotron Radiation Laboratory. J. Appl. Crystallogr. 2007;40:S453–S458. [Google Scholar]
- 53.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 54.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 55.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 58.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins: Struct., Funct., Bioinf. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berendsen HJC, Grigera JR, Straatsma TP. The missing term in effective pair potentials. J. Phys. Chem. 1987;91:6269–6271. [Google Scholar]
- 60.Green ML, Miller R. Grid Computing in Buffalo New York. Ann. Eur. Acad. Sci. 2003:191–218. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.