Abstract
Polyploidy, the possession of more than 2 complete genomes, is a major force in plant evolution known to affect the genetic and genomic constitution and the phenotype of an organism, which will have consequences for its ecology and geography as well as for lineage diversification and speciation. In this review, we discuss phylogenetic patterns in the incidence of polyploidy including possible underlying causes, the role of polyploidy for diversification, the effects of polyploidy on geographical and ecological patterns, and putative underlying mechanisms as well as chromosome evolution and evolution of repetitive DNA following polyploidization. Spurred by technological advances, a lot has been learned about these aspects both in model and increasingly also in nonmodel species. Despite this enormous progress, long-standing questions about polyploidy still cannot be unambiguously answered, due to frequently idiosyncratic outcomes and insufficient integration of different organizational levels (from genes to ecology), but likely this will change in the near future. See also the sister article focusing on animals by Choleva and Janko in this themed issue.
Keywords: Chromosome evolution, Diversification, Ecogeography, Polyploidy, Repetitive DNA
Polyploidy, initially considered ‘a hindrance to the evolutionary success of higher plants’ [Stebbins, 1971], is now recognized as a major force in plant evolution, affecting diversification and speciation [Otto and Whitton, 2000; Adams and Wendel, 2005; Soltis et al., 2009a; Madlung, 2013]. This is also due to genomic data, which show that all angiosperms, regardless of current genome size and chromosome number, have been affected by whole genome duplications (WGD), most of them repeatedly [Wendel, 2000; Seoighe, 2003; Comai, 2005; Jiao et al., 2011; McGrath and Lynch, 2012]. Comparisons of polyploid model systems including synthetic polyploids (e.g. wheat [Feldman and Levy, 2009]; Brassica/Brassicaceae [Gaeta and Pires, 2010]; Spartina/Poaceae [Ainouche et al., 2012]; Senecio/Asteraceae [Hegarty et al., 2012]; tobacco [Kovarik et al., 2012]; Tragopogon/Asteraceae [Soltis et al., 2012]; cotton [Wendel et al., 2012]) with the older established polyploids [Xiong et al., 2011; Kovarik et al., 2012; Wendel et al., 2012] reveal that the evolutionary consequences of polyploidization are manifold. Such studies are now greatly facilitated by the use of next-generation sequencing [Egan et al., 2012] readily applicable also in nonmodel systems [Zimmer and Wen, 2012]. Extensive and partly directed genetic, epigenetic, transcriptomic, and genomic changes often occur immediately after polyploidization [Liu and Wendel, 2003; Osborn et al., 2003; Levy and Feldman, 2004; Adams and Wendel, 2005; Cifuentes et al., 2010; Gaeta and Pires, 2010; Parisod et al., 2010a, b] and may lead to long-term genetic, physiological and morphological differentiation, eventually resulting in novel phenotypes, the raw material for evolution. Some of the consequences of polyploidy shall be discussed in this review. After briefly introducing types of polyploids, we will discuss phylogenetic patterns in the incidence of polyploidy including possible underlying causes, the role of polyploidy for diversification (speciation and extinction), the effects of polyploidy on geographical and ecological patterns, and chromosome evolution and evolution of repetitive DNA following polyploidization. For other aspects of polyploid evolution in plants, such as genic redundancy and gene evolution or the process of diploidization, and for aspects of polyploid evolution in animals, the reader is referred to other contributions to this special issue.
Two basic types of polyploidy, auto- and allopolyploidy, are usually distinguished [Kihara and Ono, 1926]. Autopolyploidy refers to multiplication of genetically (nearly) identical chromosome sets within a single (sub)species [Ramsey and Schemske, 1998, 2002], whereas allopolyploidy entails the multiplication of chromosome sets accompanying hybridization between 2 genetically different (sub)species [Stebbins, 1971; Husband et al., 2013]. Furthermore, autopolyploids are usually characterized by polysomic inheritance and multivalent formation in meiosis, whereas allopolyploids show disomic inheritance and bivalent formation [Stebbins, 1971; Ramsey and Schemske, 2002; Comai, 2005; Le Comber et al., 2010; Parisod et al., 2010b; Zielinski and Mittelsten Scheid, 2012], although exceptions to these inheritance patterns do exist (e.g. in segmental allopolyploids [Stebbins, 1971] or in autopolyploids with regular bivalent formation [Weiss and Maluszynska, 2000]). The majority of well-analyzed polyploid systems are allopolyploids [te Beest et al., 2012], but with the availability of flow cytometry, which allows the incidence of autopolyploidy in natural populations to be quantitatively analyzed [e.g. Suda et al., 2007; Sonnleitner et al., 2010; Laport et al., 2012; Suda and Herben, 2013], autopolyploids have received more attention in the last years [Soltis et al., 2007; Parisod et al., 2010b].
A different classification system for polyploids is based on their evolutionary age, recognizing (in order of increasing age) neo-, meso- and paleopolyploids [Ehrendorfer, 1980; Ramsey and Schemske, 1998, 2002; Comai, 2005]. Their precise circumscription, however, differs depending on the defining characters used (e.g. chromosome pairing [Ramsey and Schemske, 2002]; cytological differentiation [Schubert and Lysak, 2011]).
Polyploidy Incidence and Evolutionary Potential for Diversification
Polyploidy is widespread in the plant kingdom, but its frequency and the maximum ploidies vary considerably among and within different taxonomic groups [Husband et al., 2013]. Polyploidy is absent in (or at least has played a minor role for diversification and evolution of) liverworts, hornworts and gymnosperms, while it has been very important in green algae, lycopods, ferns, and angiosperms [Barker, 2013; Husband et al., 2013; Murray, 2013; Weiss-Schneeweiss and Schneeweiss, 2013]. Even in groups, where polyploidy is rampant, it is unevenly distributed throughout different levels. For instance, within angiosperms, polyploidy is more frequent in monocots than in dicots [Otto and Whitton, 2000] and within monocots, polyploidy is more frequent in Poaceae than in Orchidaceae [Goldblatt, 1980]. Similar differences can also be observed at the interspecific level: a number of genera encompass species that participate in producing autopolyploids, allopolyploids or both, whereas related species are known from the diploid levels only [e.g. Kim et al., 2008; Brassac et al., 2012; Weiss-Schneeweiss et al., 2012].
This high variation in the frequency of polyploidy suggests different propensities for polyploidization and/or polyploid establishment in different lineages [Husband et al., 2013]. Underlying factors include rate of unreduced gamete formation [Bretagnolle and Thompson, 1995; Ramsey and Schemske, 1998, 2002; Ramsey, 2007] and features promoting polyploid formation, establishment and spreading, such as life and growth form (polyploids tend to be perennial and more often show vegetative propagation [Stebbins, 1971; Grant, 1981]), breeding system (e.g. apomixis will promote polyploid establishment and expansion [Levin, 2002]) or ecological pre-adaptations (e.g. better capabilities to invade harsher environments due to physiological changes and/or increased genetic buffering [Brochmann et al., 2004; Mable, 2004]). For allopolyploids, additional factors are the propensity for hybrid formation (e.g. wide geographic distributions or floral and inflorescence features enhancing cross-pollination such as attractiveness or longevity [Grant, 1981; Kim et al., 2008]) as well as parental divergence (i.e. higher chance of allopolyploid formation in cases of high parental divergence [Darlington, 1937; Chapman and Burke, 2007; Paun et al., 2009, 2011]), the latter probably reflecting higher rates of unreduced gamete formation in hybrids [Ramsey and Schemske, 1998], reduced number of meiotic abnormalities [Chapman and Burke, 2007], and/or increased transgressive segregation, adding to the hybrids’ evolutionary potential [Stelkens and Seehausen, 2009]. The incidence of polyploidy correlates negatively with certain genomic features, such as genome size and chromosome numbers [Leitch and Leitch, 2008; Wood et al., 2009; Husband et al., 2013].
The role of polyploidy for speciation and diversification of angiosperms is still controversial. Early on, Stebbins [1950] considered polyploids as evolutionary dead ends because the additional genomes not only masked deleterious, but also beneficial mutations and because new gene combinations were rarely formed due to increased self-fertilization, both factors reducing the rate of adaptive evolution. In contrast, more recent authors emphasize the significant evolutionary potential of polyploids highlighting several advantages of polyploids, including the increased number of alleles allowing masking of deleterious recessive mutations, often stable heterosis allowing transgressive performance, and the potential of duplicated genes to acquire new or slightly varied functions [Soltis and Soltis, 1993; Wendel, 2000; Adams and Wendel, 2005; Madlung, 2013].
A favorable role of polyploidy for the diversification of angiosperms is suggested by several lines of evidence: (1) the majority of polyploid taxa are of multiple and spatially and/or temporally recurrent origin potentially increasing the polyploid’s genomic diversity [Soltis and Soltis, 1999; Soltis et al., 2009b, 2012; Hegarty et al., 2012; Weiss-Schneeweiss et al., 2012]; (2) species richness and the frequency of polyploids are positively related, the greatest species richness being found in genera with 50–75% polyploid species [Petit and Thompson, 1999; Otto and Whitton, 2000; Vamosi and Dickinson, 2006]; (3) fossil evidence indicates polyploidy in the majority of angiosperms [Masterson, 1994]; and (4) all angiosperms show traces of at least one ancient WGD event [Jiao et al., 2011]. Accordingly, a dramatic increase in species richness in several clades of angiosperms after an ancient WGD has been interpreted as indicative for a higher diversification in polyploids [Soltis et al., 2009a], reduced risks of extinction due to functional redundancy, mutational robustness and increased rates of evolution and adaptation in polyploids [Crow and Wagner, 2006] being one possible mechanism. An increase in polyploids is also predicted by the nonadaptive null model of the polyploidy ratchet [Meyers and Levin, 2006], which is based on the assumption that polyploidy is largely irreversible [DeWet, 1968; Stebbins, 1980; Grant, 1981].
In contrast, Wood et al. [2009] found no correlation of polyploidization rates with species richness. Instead, despite ubiquitous polyploidy in ferns and angiosperms only 31 and 15%, respectively, of all speciation events were associated with polyploidization. Similarly, Mayrose et al. [2011] inferred that polyploidization events were overrepresented at the tips of a phylogenetic tree and that net diversification rates of polyploids were significantly less than the rates of diploids due to reduced or, if accounting for the reduced probability of polyploid speciation in an already polyploid lineage, unchanged speciation rates in established polyploids and due to increased extinction rates in neopolyploids. Taken together, this suggests that the observed high frequency of polyploids is a consequence of their high formation rate rather than the diversification of polyploids themselves [Arrigo and Barker, 2012]. Even if polyploidy often will be an evolutionary dead end, some of these events can result in very successful lineages, as is evident from the paleopolyploid ancestor of extant angiosperms. This may be because (1) polyploids originated at specific times favorable for polyploid diversification (due to e.g. ample availability of novel niches), as has been suggested for several clades of angiosperms at the Cretaceous-Tertiary mass extinction event [Fawcett et al., 2009], (2) WGD conferred increased plant complexity via retention of duplicated genes underlying transcriptional regulatory networks [DeBodt et al., 2005], or (3) WGD can increase the evolutionary potential over a long period of time [Fawcett et al., 2013]. Further research will be necessary to establish the precise role of polyploidy for lineage diversification over different temporal scales and its underlying mechanisms.
Ecogeographical Consequences of Polyploidy
Polyploidy is considered an important factor shaping the geographical range of a species. Initial evidence for a link between ploidy and distribution came from surveys of relative frequencies of polyploids in different floras or taxonomic groups. These revealed that the frequency of polyploids increases with increasing latitude [compilations in Löve and Löve, 1949–1951; Reese, 1958; Grant, 1981], even within the Arctic [Brochmann et al., 2004], increasing altitude [compilation in Hanelt, 1966; Löve and Löve, 1967] or other forms of increasing environmental extremeness, such as salinity [Tischler, 1937]. Of these mentioned, only the high incidence of polyploidy at high altitudes appears to hold [Husband et al., 2013]. A variety of factors have been suggested to explain such trends, including high genetic and biochemical diversity in polyploids [Otto and Whitton, 2000; Brochmann et al., 2004], better colonizing capabilities of polyploids especially into deglaciated areas [Stebbins, 1984, 1985; Brochmann et al., 2004], the age of a flora [Reese, 1958; Ehrendorfer, 1980], or conditions brought about by Milankovitch climatic oscillations. These conditions, because of lack of species specialized to the newly emerged environmental conditions, provide new expansion possibilities allowing direct competition with progenitors to be avoided, and promoting secondary contact zones [Dynesius and Jansson, 2000]. Testing these hypotheses is impeded by uncertainty concerning the general validity of a latitudinal trend because of geographically uneven sampling (subtropical and tropical regions remain understudied), high inconsistency in the geographic position of diploids and polyploids within a certain taxonomic group, and the failure to account for confounding effects of different biogeographic and phylogenetic histories [Husband et al., 2013]. These obstacles are not insurmountable: flow cytometric ploidy determination, calibrated by karyologically verified chromosome numbers, allows ploidy data to be gathered for many samples (preferably from the region under consideration [Ehrendorfer, 1980]) in a relatively short period of time [Suda et al., 2007] and phylogenetic history can be readily accommodated by using phylogenetic comparative techniques [Harvey and Pagel, 1991; Pagel, 1999]. Clearly, a broad comparison of a large number of diploid and polyploid species is needed to comprehensively test the long-standing hypothesis of a latitudinal ploidy gradient.
A corollary of the hypothesis that polyploids have greater genetic and biochemical diversity and thus greater ecological tolerance is that polyploids are expected to have larger geographic ranges and/or occur in more habitats than diploids [Levin, 2002]. Such relationships are indeed observed in specific taxonomic groups: e.g. broader ecological niches in hexaploids compared to diploids in the Claytonia perfoliata complex (Portulacaceae) [McIntyre, 2012b], the wide distribution of tetraploid Biscutella laevigata (Brassicaceae) compared to small distribution areas of diploids associated with Pleistocene glacial refugia [Tremetsberger et al., 2002; Parisod and Besnard, 2007], or significantly larger ranges in polyploid species of Clarkia (Onagraceae) compared to diploid ones [Lowry and Lester, 2006]. Survey data on percentages of polyploids in widespread European genera [Stebbins and Dawe, 1987], on the range of ecological requirements of Pyrenean plants [Petit and Thompson, 1999], and on extent and mean geographic and ecological ranges in North American plants [Martin and Husband, 2009] provide, however, no support for this hypothesis. Further studies employing large surveys while taking phylogenetic relationships into account will be necessary to ascertain that polyploidy does not translate into larger ranges and/or greater diversity of occupied habitats.
An indirect association between range size and polyploidy arises via invasiveness and rarity. Comparative evidence [Pandit, 2006; Pandit et al., 2011] shows that the incidence of polyploidy is positively correlated with invasiveness (invasive species tend to be widespread in their native ranges [Pyšek et al., 2009; Hovick et al., 2012]), but negatively correlated with rarity (rare species tend to be narrowly distributed [Gaston, 1996]). In general, polyploidy is considered an important factor in species invasion success via, e.g. pre-adaptation to novel conditions or higher adaptive potential due to increased genetic diversity [te Beest et al., 2012].
Even if there is no consistent trend in geographic patterns of polyploids, there is no doubt that polyploids usually are geographically and/or ecologically differentiated from their diploid ancestors. Ecogeographic isolation (i.e. the combined role of ecology and geography as a reproductive barrier) is recognized as an important factor in plant speciation in general [Sobel et al., 2010] and is amply documented in diploid-polyploid systems [e.g. Felber-Girard et al., 1996; Halverson et al., 2008; Duchoslav et al., 2010; Sonnleitner et al., 2010; Glennon et al., 2012; Kolář et al., 2012; McIntyre, 2012a, b], also on a local scale (fig. 1a) [Baack, 2004; Schönswetter et al., 2007; Hülber et al., 2009]. The pervasiveness of ecogeographic divergence is in line with modeling results [Fowler and Levin, 1984; Rodriguez, 1996; Oswald and Nuismer, 2011] showing that niche differentiation is an important way of escaping the minority cytotype exclusion [Levin, 1975], i.e. the exclusion of newly emerging and initially rare polyploids from a diploid population due to (unsuccessful) pollination (triploid block [Köhler et al., 2010]) with pollen from the much more common diploids [Suda and Herben, 2013]. Factors potentially reinforcing niche differentiation are reproductive assurance (via increased selfing) [Rodriguez, 1996; Rausch and Morgan, 2005], known for angiosperms [Mable, 2004; Barringer, 2007], or via asexuality, including apomixis [Hörandl, 2006; Kao, 2008] and vegetative propagation [Gustafsson, 1948, Otto and Whitton, 2000, but see Baldwin and Husband, 2013] or by higher assortative mating [Husband, 2000; Oswald and Nuismer, 2011], which may be achieved by local pollen and seed dispersal [Baack, 2005]. Although the minority cytotype exclusion principle has been developed for autopolyploids and segmental allopolyploids [Levin, 1975], fitness disadvantages due to e.g. stigma clogging [Lloyd and Webb, 1986; Webb and Lloyd, 1986] can also be expected for allopolyploids if in minority. Furthermore, niche differentiation is an important way of competition avoidance both for autopolyploids and allopolyploids, as it is for homoploid species.
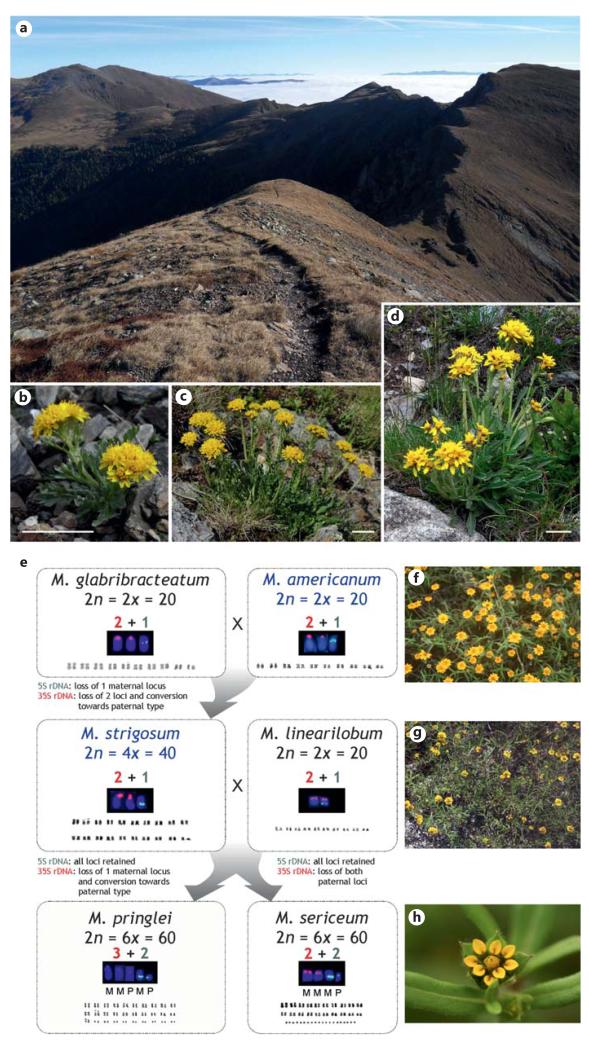
Fig. 1.
a In the easternmost Alps, the 3 main cytotypes of Senecio carniolicus (Asteraceae) occur in mixed populations, where they separate into different microhabitats: diploids (b) grow in open, rocky patches (foreground), tetraploids (c) are found on mostly north-exposed slopes with longer snow-cover (left side of the ridge), and hexaploids (d) occur in dense grass swards (right side of the ridge). The 3 cytotypes also differ morphologically with respect to overall size (scale bars in b–d approximately 5 cm), dimensions of the synflorescence and the flowering head, indumentum and leaf shape. e In Melampodium sect. Melampodium (Asteraceae), sequential allopolyploidization involved (maternal parent indicated in blue) diploid M. glabribracteatum and likely M. americanum (f), giving rise to allotetraploid M. strigosum, which in turn together with diploid M. linearilobum (g) resulted in the allohexaploids M. pringlei and M. sericeum (h). Whereas no gross karyotypic change is evident during allopolyploid evolution (karyotypes are given for each species), evolution of rDNA loci is characterized by loci loss and conversion (chromosomes carrying rDNA loci are shown for each species, 5S and 35S rDNA indicated by green and red fluorescence signals, respectively), which differ in the independently originated allohexaploids despite their identical parentage. (Photo credits: a M. Winkler, b– d M. Sonnleitner, f– h T.F. Stuessy and E. Ortiz.)
Valuable insights into the ecological factors (habitat features) that putatively underlie cytotype divergence can be gained from correlations of cytotype distribution with ecological (habitat) features. For instance, in Senecio carniolicus (Asteraceae) diploids occur in more open and usually also more exposed habitats, whereas polyploids are found in more dense vegetation (fig. 1a) [Hülber et al., 2009; Sonnleitner et al., 2010]; in Aster amellus (Asteraceae), sites of diploid and hexaploid populations differ significantly in vegetation and soil properties [Raabova et al., 2008]; compared to diploid Centaurea maculosa (Asteraceae) tetraploid cytotypes show a shift towards drier climates, which is even more pronounced in the introduced North American range [Treier et al., 2009]; the distribution of diploid and tetraploid races of Brachypodium distachyon (Poaceae) is geographically structured and associated with an aridity gradient [Manzaneda et al., 2012].
Establishing a causal link between ploidy and ecogeographic differentiation remains, however, challenging. First, little is known whether this divergence is due to nonadaptive processes (dispersal limitation, phylogeographic structure due to e.g. different Pleistocene refugia) or due to adaptive processes. For instance, current distribution patterns of tetra- and octoploid Cardamine torrentis s. l. (Brassicaceae) were suggested to be caused by historical factors, mainly Late Pleistocene climatic oscillations [Marhold et al., 2010]. Testing this or a similar hypothesis requires reciprocal transplant experiments. If nonadaptive processes were responsible, then cytotypes are expected to perform at least as well in the other cytotype’s habitat as in their own. Consequently, the lack of observing any advantage in the parental habitat, as seen in Ranunculus adoneus (Ranunculaceae) [Baack and Stanton, 2005], is indicative of nonadaptive divergence. In other cases, it has been shown that ecogeographic differentiation is due to local adaptation [e.g. Flégrová and Krahulec, 1999; Raabova et al., 2008; Ramsey, 2011]. Clearly, considerably more data will be necessary to address the relative importance of nonadaptive versus adaptive cytotype divergence.
Second, despite an early interest in the underlying mechanisms of ecogeographic differentiation of polyploids from diploids [e.g. Hagerup, 1932, 1933], effects of polyploidy on physiological and morphological traits determining the ecology of plants remain poorly understood [Soltis et al., 2010]. Numerous physiological traits have been found to differ between polyploids and diploids, including growth rates [Garbutt and Bazzaz, 1983], secondary metabolism [McArthur and Sanderson, 1999], cold tolerance [Liu et al., 2011], water relations [Maherali et al., 2009], or stress tolerance [Schlaepfer et al., 2010; Coate et al., 2013]. The adaptive potential of such differences has been recently shown in B. distachyon (Poaceae), where the higher water-use efficiency of tetraploids likely is co-responsible for its occurrence in more arid regions compared to the diploid cytotype [Manzaneda et al., 2012], and in Atriplex canescens (Chenopodiaceae), where higher ploidy cytotypes have greater xylem cavitation resistance than diploids, allowing them to grow on more drought-prone soils [Hao et al., 2013]. Polyploidization has an immediate effect on plant morphology via usually enlarged cell sizes [Müntzing, 1936], which via changes in cellular architecture, regulatory functions and replication times [Bennett and Leitch, 2005; Comai, 2005] may also affect metabolism and growth rates [Cavalier-Smith, 1978]. In allopolyploids, heterosis conferred by, for example, epigenetic modifications of circadian clock genes [Ni et al., 2009] can contribute to increased morphological vigor [Chen, 2010]. Morphologically, polyploids (at least in the first phase after their origin) often are larger in habit (fig. 1b–d) and have larger seeds [Garbutt and Bazzaz, 1983; Bretagnolle and Lumaret, 1995; Bretagnolle et al., 1995], which may lead to increased competiveness [Maceira et al., 1993; Schlaepfer et al., 2010; but see Burton and Husband, 2000; Hülber et al., 2011]. Furthermore, they usually have larger flowers (fig. 1b–d), which may increase pollinator visitation frequencies [Kennedy et al., 2006] or alter pollinator spectra [Segraves and Thompson, 1999], and they have more flowers per inflorescence [Vamosi et al., 2007], which may increase selfing rates via geitonogamy [Harder and Barrett, 1995]. Because of their hybrid origin, allopolyploids show earlier and more pronounced changes in gene expression than autopolyploids [Church and Spaulding, 2009; Hegarty et al., 2012], which may translate into more pronounced variation in physiological and morphological traits in allopolyploids [e.g. Oates et al., 2012]. Evidently, more studies, which can profit from data on trait evolution following polyploidy obtained from crop and non-crop model species [e.g. Pires et al., 2004b; Bansal et al., 2012; Miller et al., 2012], will be necessary to fully understand the role of traits and their interaction for cytotype divergence.
Finally, the commonly used approach of comparing diploids with naturally occurring polyploids fails to distinguish between differences caused by polyploidy per se from those evolved since the polyploid formation. Although recently (i.e. in historical times) evolved allopolyploids, such as North American Tragopogon species (Asteraceae; Soltis et al. [2004]), Spartina anglica (Poaceae; Ainouche et al. [2004]) or S. cambrensis (Asteraceae; Abbott and Lowe [2004]), constitute valuable model systems, especially size-related differences between cytotypes appear to decrease rapidly over a few generations [Otto and Whitton, 2000], rendering their assessment even in those systems difficult. An alternative approach is to use first generation polyploids (selected polyploid offspring of diploid parents or induced polyploids [Ramsey, 2007; Wu et al., 2012]) and compare these to both the diploid ancestors and established polyploid lineages (of usually unknown age). Using this approach, Ramsey [2011] found that newly generated hexaploids of Achillea borealis (Asteraceae) possessed a 70% survival advantage in habitats of established hexaploids (Mediterranean type habitats, such as sand dunes or oak woodlands) compared to the parental tetraploids (found in more mesic habitats). In Chamerion angustifolium (Onagraceae), newly generated polyploids had physiological tolerances to water stress comparable to the diploids [Maherali et al., 2009], higher evolvability of flowering time in response to selective pressure [Martin and Husband, 2012], lower pollen siring success than both diploids and established tetraploids [Baldwin and Husband, 2011], and low intrinsic cost of selfing [Husband et al., 2008]. Data from this powerful approach are still too scarce to allow generalizations, and it remains to be tested, to what extent changes in traits potentially conferring ecogeographic divergence are due to the polyploidization event versus subsequent changes due to selection.
Repetitive DNA and Chromosomal Evolution
In contrast to genomes of higher animals [Kejnovsky et al., 2009], plant genomes contain large proportions of repetitive DNA [Schmidt and Heslop-Harrison, 1998; Leitch and Leitch, 2008; Heslop-Harrison and Schwarzacher, 2011]. This genome fraction in plants consists of tandem repeats (rDNA, satellite DNA) and of dispersed repeats, mainly transposable elements (mostly LTR retro-elements comprising Ty1-copia and Ty3-gypsy elements [Kumar and Bennetzen, 1999; Bennetzen, 2002; Bento et al., this issue]). Evolution of transposable elements in most of the polyploid genomes is specific for the type of transposable element and very dynamic [Baumel et al., 2002; Petit et al., 2007, 2010; Parisod et al., 2012], with the possible exception of cotton [Wendel et al., 2012]. In the pre-genomic era, methodological constraints have restricted analysis of repetitive DNA in polyploids mostly to the highly conserved and ubiquitous rDNA [Maluszynska et al., 1998] or to a few available or newly isolated tandemly repeated DNA types [Zhao et al., 1998; Lim et al., 2000b, 2008; Pires et al., 2004a; Chester et al., 2010; Petit et al., 2010; Kolano et al., 2011]. This has changed with the availability of next-generation sequencing techniques allowing in depth characterization of repetitive DNA [e.g. Macas et al., 2007, 2011; Wicker et al., 2009; Hribová et al., 2010] also in polyploids (allopolyploid tobacco [Renny-Byfield et al., 2011; Buggs et al., 2012; Parisod et al., 2012], cotton [Wendel et al., 2012] and Tragopogon/Asteraceae [Soltis et al., 2012] as well as the likely autotetraploid [Schneeweiss et al., 2004] Orobanche gracilis [Piednoël et al., 2012]).
Polyploids typically show genome rearrangements involving rDNA repeat and/or locus silencing (nucleolar dominance [Preuss and Pikaard, 2007]) or loss [Vaughan et al., 1993; Lim et al., 2000a, b, 2007a, b; Weiss-Schneeweiss et al., 2007, 2012; Kotseruba et al., 2010; Kolano et al., 2011; Książczyk et al., 2011], interlocus recombination and complete or near-complete repeat replacement [Lim et al., 2000a; Kovarik et al., 2005; Matyasek et al., 2007; Mlinarec et al., 2012; Weiss-Schneeweiss et al., 2012], or redistribution and/or increase in loci number [Adams et al., 2000; Weiss-Schneeweiss et al., 2007]. The underlying mechanisms include physical locus loss, interchromosomal translocations [Weiss and Maluszynska, 2000], or differential and repeated amplification of minor and reduction of major loci of rDNA [Childs et al., 1981; Dubcovsky and Dvorak, 1995]. For 35S rDNA, locus loss as well as repeat type conversion and homogenization have been commonly reported especially in older allopolyploids [Clarkson et al., 2005; Weiss-Schneeweiss et al., 2012]. The evolution of 35S rDNA can be directional, loci of one parent always being lost or converted (fig. 1e) [Vaughan et al., 1993; Lim et al., 2000a; Jang et al., 2011; Renny-Byfield et al., 2011; Kovarik et al., 2012; Weiss-Schneeweiss et al., 2012]. The direction of conversion and homogenization of 35S rDNA can be biased towards the maternal [Weiss-Schneeweiss et al., 2012] or the paternal parent [Lim et al., 2000a, b; Matyasek et al., 2003; Renny-Byfield et al., 2011] and may differ in independently originated allopolyploids of identical parentage (fig. 1e) [Weiss-Schneeweiss et al., 2012]. In contrast to tobacco, in North American Tragopogon, containing the allopolyploids T. mirus (T. dubius × T. porrifolius) and T. miscellus (T. dubius × T. pratensis) that formed repeatedly following the introduction of 3 diploids to the United States ~80 years ago [Soltis et al., 2004], both rDNA loci number and distribution remain additive in the polyploids [Pires et al., 2004a]. Still, concerted evolution of 35S rDNA in Tragopogon allopolyploids is ongoing and incomplete [Kovarik et al., 2005], with transcription favoring the underrepresented T. dubius copies [Matyasek et al., 2007; Malinska et al., 2010]. In contrast to 35S rDNA, there is no evidence for interlocus homogenization of 5S rDNA in any of the polyploid systems analyzed (tobacco, Fulnecek et al. [2002]; cotton, Cronn et al. [1996]; Melampodium/Asteraceae (fig. 1e–h), Weiss-Schneeweiss et al. [2012]). 5S rDNA loci may, however, change in number, most frequently being lost [Clarkson et al., 2005; Weiss-Schneeweiss et al., 2012]. Extent and rate of rDNA loci loss roughly correlates with a polyploid’s age: in younger allopolyploids and in autopolyploids, the number of loci often equals the sum of those of their progenitors [Clarkson et al., 2005; Chester et al., 2012; Rebernig et al., 2012], whereas in older allopolyploids maternal and/or paternal loci are often lost [Clarkson et al., 2005; Weiss-Schneeweiss et al., 2007, 2012; Kolano et al., 2011].
Transposable elements are important components of genome size evolution, participating not only in genome size increase, but also in genome downsizing, which seems to be a general trend in polyploids [Leitch and Bennett, 2004]. Its extent is group specific [Weiss-Schneeweiss et al., 2007], but clearly depends on the age of the polyploid, with additive genome size compared to parental genomes in young polyploids [Rebernig et al., 2012; Soltis et al., 2012] and reaching significant levels of reduction in old polyploids, where genome downsizing might also be accompanied by chromosome number reductions [Clarkson et al., 2005; Schönswetter et al., 2007; Mandakova et al., 2010; Doyle, 2012]. In Nicotiana tabacum, an intersectional hybrid formed within the last 200,000 years from relatives of extant diploids N. silvestris (genome S) and N. tomentosiformis (genome T [Kenton et al., 1993; Kovarik et al., 2012]), established and newly synthesized polyploids show congruent patterns of retroelement dynamics (mobility and loss) and of changes in copy number of various repetitive DNAs, suggesting that retrotransposon amplification and genome restructuring in tobacco occurs rapidly and is directional [Skalická et al., 2005; Renny-Byfield et al., 2012; Kovarik et al., 2012]. Selected families of copia LTR-retrotransposons diversified subsequent to allopolyploidization, with retroelement losses being more frequent from the paternal T-genome and novel insertions targeting preferentially the maternal S-genome [Petit et al., 2007, 2010]. However, spreading of genome-specific transposons to the other parental genome has also been found in cotton [Zhao et al., 1998]. Genome downsizing in tobacco [Leitch et al., 2008] can be attributed to extensive losses across most repeat types (NicCL3; A1/A2 repeats; NsEPRV, Ty3/gypsy LTR elements, 35S rDNA) preferentially from the paternal T-genome [Renny-Byfield et al., 2011, 2012]. The directed preferential loss of repeats from one paternal genome may provide support for the nuclear-cytoplasmic interaction hypothesis whereby the paternal genome may be in disadvantage due to incompatibility in interactions with maternally inherited cytoplasm [Leitch et al., 2006].
Chromosomal rearrangements are often associated with rearrangements of the repetitive DNA fraction or result from extensive homeologous chromosome recombination [Gaeta et al., 2007] and may lead to considerable chromosome number reductions [Lysak et al., 2006; Mandakova et al., 2010; Weiss-Schneeweiss and Schneeweiss, 2013]. Changes in gross genome structure encompass numerous balanced (conversions, inversions, translocations, fusions, or fissions) and unbalanced (duplications, deletions or aneuploidy) chromosomal rearrangements [Lysak and Schubert, 2013] and have been addressed only in a few polyploid systems [Lim et al., 2000a, b, 2007a, b, 2008; Weiss-Schneeweiss et al., 2007, 2012; Mandakova et al., 2010; Książczyk et al., 2011; Renny-Byfield et al., 2011; Chester et al., 2012]. In tobacco, early cytogenetic analyses revealed several intergenomic translocations [Kenton et al., 1993; Moscone et al., 1996; Lim et al., 2004], some of which were fixed and some of which were also detected in synthetic tobacco [Skalická et al., 2005]. Intergenomic translocations are also known from the allopolyploid Avena/Poaceae [Jellen et al., 1994]. An extensive survey in T. miscellus revealed common intergenomic translocations, likely the result of homoeologous recombination, with some of the translocations being close to fixation [Chester et al., 2012]. Although most of the individuals analyzed were 2n = 24 and despite earlier observations of preferential bivalent pairing in meiosis [Ownbey, 1950], very few individuals had additive karyotypes (i.e. all parental chromosomes present in 2 copies [Lim et al., 2008; Chester et al., 2012]). This compensatory (reciprocal) aneuploidy, which may result from high homology between the 2 parental genomes and subsequent (bivalent) homeologous chromosome pairing, will influence disomic versus polysomic inheritance of some genes [Chester et al., 2012]. This deviation from gene balance may also have consequences, for example, in gene expression or phenotypic traits [Birchler and Veitia, 2012]. Compensatory aneuploidy is known from other synthetic or spontaneous neo-allopolyploids (e.g. synthetic B. napus [Xiong et al., 2011] or wheat [Mestiri et al., 2010]). A similar system occurs also in inter-cytotype polyploids of Prospero autumnale (Hyacinthaceae). Allopolyploids of the diploid cytotypes with 2n = 12 and 2n = 14, where 2n = 12 arose after a fusion of 2 chromosomes of the 2n = 14 cytotype [Vaughan et al., 1997], produce – due to preferential bivalent formation between homologous or homeologous chromosomes alike – F1 progeny with balanced chromosome numbers varying from 2n = 24 to 2n = 28, but most common 2n = 26 [Jang et al., 2011; Weiss-Schneeweiss et al., unpubl. data].
Chromosomal evolution of polyploids is usually driven by amplification and removal of repetitive DNA and by inflicted gross chromosomal changes, most commonly translocations, inversions, aneuploidy, or dysploidy [Ma and Gustafson, 2005]. Such changes not only affect chromosome numbers, karyotype structure or genome size, but are very important for restoring full fertility via proper chromosome pairing [Feldman and Levy, 2009]. This diploidization has 2 components: while genetic diploidization restores disomic inheritance, cytological diploidization manifests itself in regular bivalent pairing, despite homeology or redundant homology [Feldman and Levy, 2009]. As the time required for achievement of full bivalent pairing depends greatly on the degree of chromosome homeology, autopolyploids are expected to be much more prone to multivalent formation than allopolyploids. This might have consequences for establishment probabilities of newly formed polyploids, which may be higher in allopolyploids than in autopolyploids.
Conclusions
Although a lot has been learned about polyploidy both in model and, spurred by technological advances, also in nonmodel species, many long-standing questions about polyploidy concerning, for instance, its role in lineage diversification or its ecological consequences can still not be unambiguously answered. This is due to both still insufficient knowledge on many aspects of polyploidy (especially concerning ecology, physiology and evolution [Soltis et al., 2010]) and insufficient integration of different organizational levels (e.g. whether transcriptomic and genomic changes lead to greater adaptation [Madlung, 2013]). Although many of the evolutionary consequences of polyploidy reviewed herein apply to both autopolyploids and allopolyploids, their precise evolutionary trajectories are expected to differ, given that allopolyploids combine effects of genome doubling (polyploidy) with those of genome merger (hybridization). Integrative approaches combining phylogenetic, molecular and ecophysiological tools across autopolyploid and allopolyploid model systems have great potential to help us better understand the evolutionary and ecological role of polyploidy.
Acknowledgements
We thank the editors of this special issue, Dr. D.K. Lamatsch and Dr. M. Stöck, for inviting us to write this review. Our research in polyploidy is supported by the Austrian Science Fund (FWF grants P21440 and P25131 to H.W.-S. and P20736 to G.M.S.).
References
- Abbott RJ, Lowe AJ. Origins, establishment and evolution of new polyploid species: Senecio cambrensis and S. eboracensis in the British Isles. Biol J Linn Soc Lond. 2004;82:467–474. [Google Scholar]
- Adams SP, Leitch IJ, Bennett MD, Chase MW, Leitch AR. Ribosomal DNA evolution and phylogeny in Aloe (Asphodelaceae) Am J Bot. 2000;87:1578–1583. [PubMed] [Google Scholar]
- Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr Opin Plant Biol. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Ainouche ML, Baumel A, Salmon A. Spartina anglica C.E. Hubbard: a natural model system for analysing early evolutionary changes that affect allopolyploid genomes. Biol J Linn Soc Lond. 2004;82:475–484. [Google Scholar]
- Ainouche M, Chelaifa H, Ferreira J, Bellot S, Ainouche A, Salmon A. Polyploid evolution in Spartina: dealing with highly redundant hybrid genomes. In: Soltis PS, Soltis DE, editors. Polyploidy and Genome Evolution. Springer; Heidelberg: 2012. pp. 225–244. [Google Scholar]
- Arrigo N, Barker MS. Rarely successful polyploids and their legacy in plant genomes. Curr Opin Plant Biol. 2012;15:140–146. doi: 10.1016/j.pbi.2012.03.010. [DOI] [PubMed] [Google Scholar]
- Baack EJ. Cytotype segregation on regional and microgeographic scales in snow buttercups (Ranunculus adoneus: Ranunculaceae) Am J Bot. 2004;91:1783–1788. doi: 10.3732/ajb.91.11.1783. [DOI] [PubMed] [Google Scholar]
- Baack EJ. To succeed globally, disperse locally: effects of local pollen and seed dispersal on tetraploid establishment. Heredity. 2005;94:538–546. doi: 10.1038/sj.hdy.6800656. [DOI] [PubMed] [Google Scholar]
- Baack EJ, Stanton ML. Ecological factors influencing tetraploid speciation in snow buttercups (Ranunculus adoneus): niche differentiation and tetraploid establishment. Evolution. 2005;59:1936–1944. [PubMed] [Google Scholar]
- Baldwin SJ, Husband BC. Genome duplication and the evolution of conspecific pollen precedence. Proc R Soc Lond B Biol Sci. 2011;278:2011–2017. doi: 10.1098/rspb.2010.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin SJ, Husband BC. The association between polyploidy and clonal reproduction in diploid and tetraploid Chamerion angustifolium. Mol Ecol. 2013;22:1806–1819. doi: 10.1111/mec.12217. [DOI] [PubMed] [Google Scholar]
- Bansal P, Banga S, Banga SS. Heterosis as investigated in terms of polyploidy and genetic diversity using designed Brassica juncea amphiploid and its progenitor diploid species. PLoS One. 2012;7:e29607. doi: 10.1371/journal.pone.0029607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MS. Karyotype and genome evolution in pteridophytes. In: Leitch IJ, Greilhuber J, Doležel J, Wendel JF, editors. Plant Genome Diversity 2: Physical Structure, Behaviour and Evolution of Plant Genomes. Springer; Wien: 2013. pp. 245–253. [Google Scholar]
- Barringer BC. Polyploidy and self-fertilization in flowering plants. Am J Bot. 2007;94:1527–1533. doi: 10.3732/ajb.94.9.1527. [DOI] [PubMed] [Google Scholar]
- Baumel A, Ainouche M, Kalendar R, Schulman AH. Retrotransposons and genomic stability in populations of the young allopolyploid species Spartina anglica C.E. Hubbard (Poaceae) Mol Biol Evol. 2002;19:1218–1227. doi: 10.1093/oxfordjournals.molbev.a004182. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ. Genome size evolution in plants. In: Gregory TR, editor. The Evolution of the Genome. Elsevier Academic Press; Amsterdam: 2005. pp. 89–162. [Google Scholar]
- Bennetzen JL. Mechanisms and rates of genome expansion and contraction in flowering plants. Genetica. 2002;115:29–36. doi: 10.1023/a:1016015913350. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA. Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci USA. 2012;109:14746–14753. doi: 10.1073/pnas.1207726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brassac J, Jakob SS, Blattner FR. Progenitor-derivative relationships of Hordeum polyploids (Poaceae, Triticeae) inferred from sequences of TOPO6, a nuclear low-copy gene region. PLoS One. 2012;7:e33808. doi: 10.1371/journal.pone.0033808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretagnolle F, Lumaret R. Bilateral polyploidization in Dactylis glomerata L. subsp. lusitanica : occurrence, morphological and genetic characteristics of first polyploids. Euphytica. 1995;84:197–207. [Google Scholar]
- Bretagnolle F, Thompson JD. Gametes with the somatic chromosome number: mechanisms of their formation and role in the evolution of autopolyploid plants. New Phytol. 1995;129:1–22. doi: 10.1111/j.1469-8137.1995.tb03005.x. [DOI] [PubMed] [Google Scholar]
- Bretagnolle F, Thompson JD, Lumaret R. The influence of seed size variation on seed germination and seedling vigour in diploid and tetraploid Dactylis glomerata. Ann Bot. 1995;76:607–615. [Google Scholar]
- Brochmann C, Brysting AK, Alsos IG, Borgen L, Grundt HH, et al. Polyploidy in arctic plants. Biol J Linn Soc Lond. 2004;82:521–536. [Google Scholar]
- Buggs RJ, Renny-Byfield S, Chester M, Jordon-Thaden IE, Viccini LF, et al. Next-generation sequencing and genome evolution in allopolyploids. Am J Bot. 2012;99:372–382. doi: 10.3732/ajb.1100395. [DOI] [PubMed] [Google Scholar]
- Burton TL, Husband BC. Fitness differences among diploids, tetraploids, and their triploid progeny in Chamerion angustifolium : mechanisms of inviability and implications for polyploid evolution. Evolution. 2000;54:1182–1191. doi: 10.1111/j.0014-3820.2000.tb00553.x. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Nuclear volume control by nucleoskeletal DNA, selection for cell volume and cell growth rate, and the solution of the DNA C-value paradox. J Cell Sci. 1978;34:247–278. doi: 10.1242/jcs.34.1.247. [DOI] [PubMed] [Google Scholar]
- Chapman MA, Burke JM. Genetic divergence and hybrid speciation. Evolution. 2007;61:1773–1780. doi: 10.1111/j.1558-5646.2007.00134.x. [DOI] [PubMed] [Google Scholar]
- Chen ZJ. Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 2010;15:57–71. doi: 10.1016/j.tplants.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester M, Leitch AR, Soltis PS, Soltis DE. Review of the application of modern cytogenetic methods (FISH/GISH) to the study of reticulation (polyploidy/hybridisation) Genes. 2010;1:166–192. doi: 10.3390/genes1010166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester M, Gallagher JP, Symonds VV, Cruz da Silva AV, Mavrodiev EV, et al. Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae) Proc Natl Acad Sci USA. 2012;109:1176–1181. doi: 10.1073/pnas.1112041109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs G, Maxon R, Kedes LC. Orphons: dispersed genetic elements derived from tandem repetitive genes of eukaryotes. Cell. 1981;23:651–663. doi: 10.1016/0092-8674(81)90428-1. [DOI] [PubMed] [Google Scholar]
- Church SA, Spaulding EJ. Gene expression in a wild autopolyploid sunflower series. J Hered. 2009;100:491–495. doi: 10.1093/jhered/esp008. [DOI] [PubMed] [Google Scholar]
- Cifuentes M, Grandont L, Moore G, Chèvre AM, Jenczewski E. Genetic regulation of meiosis in polyploid species: new insights into an old question. New Phytol. 2010;186:29–36. doi: 10.1111/j.1469-8137.2009.03084.x. [DOI] [PubMed] [Google Scholar]
- Clarkson JJ, Lim KY, Kovarik A, Chase MW, Knapp S, Leitch AR. Long-term genome diploidization in allopolyploid Nicotiana section Repandae (Solanaceae) New Phytol. 2005;168:241–252. doi: 10.1111/j.1469-8137.2005.01480.x. [DOI] [PubMed] [Google Scholar]
- Coate JE, Powell AF, Owens TG, Doyle JJ. Transgressive physiological and transcriptomic responses to light stress in allopolyploid Glycine dolichocarpa (Leguminosae) Heredity. 2013;110:160–170. doi: 10.1038/hdy.2012.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. The advantages and disadvantages of being polyploid. Nat Rev Genet. 2005;6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- Cronn RC, Zhao XP, Paterson AH, Wendel JF. Polymorphism and concerted evolution in a tandemly repeated gene family: 5S ribosomal DNA in diploid and allopolyploid cottons. J Mol Evol. 1996;42:685–705. doi: 10.1007/BF02338802. [DOI] [PubMed] [Google Scholar]
- Crow KD, Wagner GP. What is the role of genome duplication in the evolution of complexity and diversity? Mol Biol Evol. 2006;23:887–892. doi: 10.1093/molbev/msj083. [DOI] [PubMed] [Google Scholar]
- Darlington CD. Recent Advances in Cytology. 2nd ed. P. Blakiston’s Son and Co.; Philadelphia: 1937. [Google Scholar]
- De Bodt S, Maere S, van de Peer Y. Genome duplication and the origin of angiosperms. Trends Ecol Evol. 2005;20:591–597. doi: 10.1016/j.tree.2005.07.008. [DOI] [PubMed] [Google Scholar]
- DeWet JMJ. Diploid-tetraploid-haploid cycles and the origin of variability in Dichanthium agamospecies. Evolution. 1968;22:394–397. doi: 10.1111/j.1558-5646.1968.tb05906.x. [DOI] [PubMed] [Google Scholar]
- Doyle JJ. Polyploidy in legumes. In: Soltis PS, Soltis DE, editors. Polyploidy and Genome Evolution. Springer; Heidelberg: 2012. pp. 147–180. [Google Scholar]
- Dubcovsky J, Dvorak J. Ribosomal RNA multi-gene loci: nomads of the Triticeae genomes. Genetics. 1995;140:1367–1377. doi: 10.1093/genetics/140.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchoslav M, Šafářová L, Krahulec F. Complex distribution patterns, ecology and coexistence of ploidy levels of Allium oleraceum (Alliaceae) in the Czech Republic. Ann Bot. 2010;105:719–735. doi: 10.1093/aob/mcq035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynesius M, Jansson R. Evolutionary consequences of changes in species’ geographical distributions driven by Milankovitch climate oscillations. Proc Natl Acad Sci USA. 2000;97:9115–9120. doi: 10.1073/pnas.97.16.9115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan AN, Schlueter J, Spooner DM. Applications of next-generation sequencing in plant biology. Am J Bot. 2012;99:175–185. doi: 10.3732/ajb.1200020. [DOI] [PubMed] [Google Scholar]
- Ehrendorfer F. Polyploidy and distribution. In: Lewis WH, editor. Polyploidy: Biological Relevance. Plenum Press; New York: 1980. pp. 45–60. [Google Scholar]
- Fawcett JA, Maere S, van de Peer Y. Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc Natl Acad Sci USA. 2009;106:5737–5742. doi: 10.1073/pnas.0900906106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett JA, van de Peer Y, Maere S. Significance and biological consequences of polyploidization in land plant evolution. In: Leitch IJ, Greilhuber J, Doležel J, Wendel JF, editors. Plant Genome Diversity 2: Physical Structure, Behaviour and Evolution of Plant Genomes. Springer; Wien: 2013. pp. 277–293. [Google Scholar]
- Felber-Girard M, Felber F, Buttler A. Habitat differentiation in a narrow hybrid zone between diploid and tetraploid Anthoxanthum alpinum. New Phytol. 1996;133:531–540. [Google Scholar]
- Feldman M, Levy AA. Genome evolution in allopolyploid wheat – a revolutionary reprogramming followed by gradual changes. J Genet Genomics. 2009;36:511–518. doi: 10.1016/S1673-8527(08)60142-3. [DOI] [PubMed] [Google Scholar]
- Flégrová M, Krahulec F. Anthoxanthum odoratum and A. alpinum : life history parameters at two different altitudes. Folia Geobot. 1999;34:19–31. [Google Scholar]
- Fowler NL, Levin DA. Ecological constraints on the establishment of a novel polyploid in competition with its diploid progenitor. Am Nat. 1984;124:701–711. [Google Scholar]
- Fulnecek J, Lim KY, Leitch AR, Kovarík A, Matyásek R. Evolution and structure of 5S rDNA loci in allotetraploid Nicotiana tabacum and its putative parental species. Heredity. 2002;88:19–25. doi: 10.1038/sj.hdy.6800001. [DOI] [PubMed] [Google Scholar]
- Gaeta RT, Pires JC. Homoeologous recombination in allopolyploids: the polyploid ratchet. New Phytol. 2010;186:18–28. doi: 10.1111/j.1469-8137.2009.03089.x. [DOI] [PubMed] [Google Scholar]
- Gaeta RT, Pires JC, Iniguez-Luy F, Leon E, Osborn TC. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell. 2007;19:3403–3417. doi: 10.1105/tpc.107.054346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbutt K, Bazzaz FA. Leaf demography, flower production and biomass of diploid and tetraploid populations of Phlox drummondii Hook. on a soil moisture gradient. New Phytol. 1983;93:129–141. [Google Scholar]
- Gaston KJ. The multiple forms of the interspecific abundance-distribution relationship. Oikos. 1996;76:211–220. [Google Scholar]
- Glennon KL, Rissler LJ, Church SA. Ecogeographic isolation: a reproductive barrier between species and between cytotypes in Houstonia (Rubiaceae) Evol Ecol. 2012;26:909–926. [Google Scholar]
- Goldblatt P. Polyploidy in angiosperms: monocotyledons. In: Lewis WH, editor. Polyploidy: Biological Relevance. Plenum Press; New York: 1980. pp. 219–239. [DOI] [PubMed] [Google Scholar]
- Grant V. Plant Speciation. 2nd ed. Columbia Univ Press; New York: 1981. [Google Scholar]
- Gustafsson A. Polyploidy, life-form, and vegetative reproduction. Hereditas. 1948;34:1–22. [Google Scholar]
- Hagerup O. Über Polyploidie in Beziehung zu Klima, Ökologie und Phylogenie. Hereditas. 1932;16:19–40. [Google Scholar]
- Hagerup O. Studies on polyploid ecotypes in Vaccinium uliginosum L. Hereditas. 1933;18:122–128. [Google Scholar]
- Halverson K, Heard SB, Nason JD, Stireman JO., 3rd Origins, distribution, and local co-occurrence of polyploid cytotypes in Solidago altissima (Asteraceae) Am J Bot. 2008;95:50–58. doi: 10.3732/ajb.95.1.50. [DOI] [PubMed] [Google Scholar]
- Hanelt P. Polyploidie-Frequenz und geographische Verbreitung bei Höheren Pflanzen. Biol Rundschau. 1966;4:183–196. [Google Scholar]
- Hao GY, Lucero ME, Sanderson SC, Zacharias EH, Holbrook NM. Polyploidy enhances the occupation of heterogeneous environments through hydraulic related trade-offs in Atriplex canescens (Chenopodiaceae) New Phytol. 2013;197:970–978. doi: 10.1111/nph.12051. [DOI] [PubMed] [Google Scholar]
- Harder LD, Barrett SCH. Mating cost of large floral displays in hermaphrodite plants. Nature. 1995;373:512–515. [Google Scholar]
- Harvey PH, Pagel MD. The Comparative Method in Evolutionary Biology. Oxford Univ Press; Oxford: 1991. [Google Scholar]
- Hegarty MJ, Abbott RJ, Hiscock SJ. Allopolyploid speciation in action: the origins and evolution of Senecio cambrensis. In: Soltis PS, Soltis DE, editors. Polyploidy and Genome Evolution. Springer; Heidelberg: 2012. pp. 245–270. [Google Scholar]
- Heslop-Harrison JS, Schwarzacher T. Organization of the plant genome in chromosomes. Plant J. 2011;66:18–33. doi: 10.1111/j.1365-313X.2011.04544.x. [DOI] [PubMed] [Google Scholar]
- Hörandl E. The complex causality of geographical parthenogenesis. New Phytol. 2006;171:525–538. doi: 10.1111/j.1469-8137.2006.01769.x. [DOI] [PubMed] [Google Scholar]
- Hovick SM, Peterson CJ, Carson WP. Predicting invasiveness and range size in wetland plants using biological traits: a multivariate experimental approach. J Ecol. 2012;100:1373–1382. [Google Scholar]
- Hribová E, Neumann P, Matsumoto T, Roux N, Macas J, Dolezel J. Repetitive part of the banana (Musa acuminata) genome investigated by low-depth 454 sequencing. BMC Plant Biol. 2010;10:204. doi: 10.1186/1471-2229-10-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülber K, Sonnleitner M, Flatscher R, Berger A, Dobrovsky R, et al. Ecological segregation drives fine-scale cytotype distribution of Senecio carniolicus in the Eastern Alps. Preslia. 2009;81:309–319. [PMC free article] [PubMed] [Google Scholar]
- Hülber K, Berger A, Gilli C, Hofbauer M, Patek M, Schneeweiss GM. No evidence for a role of competitive capabilities of adults in causing habitat segregation of diploid and hexaploid Senecio carniolicus (Asteracaeae) Alp Bot. 2011;121:123–127. doi: 10.1007/s00035-011-0091-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husband BC. Constraints on polyploid evolution: a test of the minority cytotype exclusion principle. Proc R Soc Lond B Biol Sci. 2000;267:217–223. doi: 10.1098/rspb.2000.0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husband BC, Ozimec B, Martin SL, Pollock L. Mating consequences of polyploid evolution in flowering plants: current trends and insights from synthetic polyploids. Int J Plant Sci. 2008;169:195–206. [Google Scholar]
- Husband BC, Baldwin SJ, Suda J. The incidence of polyploidy in natural plant populations: major patterns and evolutionary processes. In: Leitch IJ, Greilhuber J, Doležel J, Wendel JF, editors. Plant Genome Diversity 2: Physical Structure, Behaviour and Evolution of Plant Genomes. Springer; Wien: 2013. pp. 255–276. [Google Scholar]
- Jang TS, Emadzade K, Temsch E, Macas J, Leitch A, et al. Karyotype analysis of diploid and polyploidy species of the Prospero (Hyacinthaceae). Abstracts 43rd Symp of The Korean Society of Plant Taxonomists; Seoul. 2011. p. 401. [Google Scholar]
- Jellen EN, Gill BS, Cox TS. Genomic in situ hybridization differentiates between A/D- and C-genome chromatin and detects intergenomic translocations in polyploid oat species (genus Avena) Genome. 1994;37:613–618. doi: 10.1139/g94-087. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chander-bali AS, Landherr L, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- Kao RH. Origins and widespread distribution of co-existing polyploids in Arnica cordifolia (Asteraceae) Ann Bot. 2008;101:145–152. doi: 10.1093/aob/mcm271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kejnovsky E, Leitch IJ, Leitch AR. Contrasting evolutionary dynamics between angiosperm and mammalian genomes. Trends Ecol Evol. 2009;24:572–582. doi: 10.1016/j.tree.2009.04.010. [DOI] [PubMed] [Google Scholar]
- Kennedy B, Sabara HA, Haydon D, Husband BC. Pollinator-mediated assortative mating in mixed ploidy populations of Chamerion angustifolium (Onagraceae) Oecologia. 2006;150:398–408. doi: 10.1007/s00442-006-0536-7. [DOI] [PubMed] [Google Scholar]
- Kenton A, Parokonny AS, Gleba YY, Bennett MD. Characterization of the Nicotiana tabacum L. genome by molecular cytogenetics. Mol Gen Genet. 1993;240:159–169. doi: 10.1007/BF00277053. [DOI] [PubMed] [Google Scholar]
- Kihara H, Ono T. Chromosomenzahlen und systematische Gruppierung der Rumex-Arten. Zeitschr Zellf Mikrosk Anat. 1926;4:475–481. [Google Scholar]
- Kim ST, Sultan SE, Donoghue MJ. Allopolyploid speciation in Persicaria (Polygonaceae): insights from a low-copy nuclear region. Proc Natl Acad Sci USA. 2008;105:12370–12375. doi: 10.1073/pnas.0805141105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Mittelsten Scheid O, Erilova A. The impact of the triploid block on the origin and evolution of polyploid plants. Trends Genet. 2010;26:142–148. doi: 10.1016/j.tig.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Kolano B, Gardunia BW, Michalska M, Bonifacio A, Fairbanks D, et al. Chromosomal localization of two novel repetitive sequences isolated from the Chenopodium quinoa Willd. genome. Genome. 2011;54:710–717. doi: 10.1139/g11-035. [DOI] [PubMed] [Google Scholar]
- Kolář F, Fér T, Štech M, Trávníček P, Dušková E, et al. Bringing together evolution on serpentine and polyploidy: spatiotemporal history of the diploid-tetraploid complex of Knautia arvensis (Dipsacaceae) PLoS One. 2012;7:e39988. doi: 10.1371/journal.pone.0039988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotseruba V, Pistrick K, Blattner FR, Kumke K, Weiss O, et al. The evolution of the hexaploid grass Zingeria kochii (Mez) Tzvel. (2n = 12) was accompanied by complex hybridization and uniparental loss of ribosomal DNA. Mol Phylogenet Evol. 2010;56:146–155. doi: 10.1016/j.ympev.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Kovarik A, Pires JC, Leitch AR, Lim KY, Sherwood AM, et al. Rapid concerted evolution of nuclear ribosomal DNA in two Tragopogon allopolyploids of recent and recurrent origin. Genetics. 2005;169:931–944. doi: 10.1534/genetics.104.032839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovarik A, Renny-Byfield S, Grandbastien MA, Leitch A. Evolutionary implications of genome and karyotype restructuring in Nicotiana tabacum L. In: Soltis PS, Soltis DE, editors. Polyploidy and Genome Evolution. Springer; Heidelberg: 2012. pp. 209–224. [Google Scholar]
- Książczyk T, Kovarik A, Eber F, Huteau V, Khaitova L, et al. Immediate unidirectional epigenetic reprogramming of NORs occurs independently of rDNA rearrangements in synthetic and natural forms of a polyploid species Brassica napus. Chromosoma. 2011;120:557–571. doi: 10.1007/s00412-011-0331-z. [DOI] [PubMed] [Google Scholar]
- Kumar A, Bennetzen JL. Plant retrotransposons. Annu Rev Genet. 1999;33:479–532. doi: 10.1146/annurev.genet.33.1.479. [DOI] [PubMed] [Google Scholar]
- Laport RG, Minckley RL, Ramsey J. Phylogeny and cytogeography of the North American creosote bush (Larrea tridentata , Zygophyllaceae) Syst Bot. 2012;37:153–164. [Google Scholar]
- Le Comber SC, Ainouche ML, Kovarik A, Leitch AR. Making a functional diploid: from polysomic to disomic inheritance. New Phytol. 2010;186:113–122. doi: 10.1111/j.1469-8137.2009.03117.x. [DOI] [PubMed] [Google Scholar]
- Leitch AR, Leitch IJ. Genome plasticity and the diversity of polyploid plants. Science. 2008;320:481–483. doi: 10.1126/science.1153585. [DOI] [PubMed] [Google Scholar]
- Leitch AR, Lim KY, Skalická K, Kovarik A. Nuclear cytoplasmic interaction hypothesis and the role of translocations in Nicotiana allopolyploids. In: Cigna AAD, Yerevan M, editors. Radiation Risk Estimates in Normal and Emergency Situations: NATO Security through Science Series B: Physics and Biophysics (ARMENIA) Springer; Dordrecht: 2006. pp. 319–326. [Google Scholar]
- Leitch IJ, Bennett MD. Genome downsizing in polyploid plants. Biol J Linn Soc Lond. 2004;82:651–663. [Google Scholar]
- Leitch IJ, Hanson L, Lim KY, Kovarik A, Chase MW, et al. The ups and downs of genome size evolution in polyploid species of Nicotiana (Solanaceae) Ann Bot. 2008;101:805–814. doi: 10.1093/aob/mcm326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. Minority cytotype exclusion in local plant populations. Taxon. 1975;24:35–43. [Google Scholar]
- Levin D. The Role of Chromosomal Change in Plant Evolution. Oxford Univ Press; Oxford: 2002. [Google Scholar]
- Levy AA, Feldman M. Genetic and epigenetic reprogramming of the wheat genome upon allopolyploidization. Biol J Linn Soc Lond. 2004;82:607–613. [Google Scholar]
- Lim KY, Kovarik A, Matyasek R, Bezdĕk M, Lichtenstein CP, Leitch AR. Gene conversion of ribosomal DNA in Nicotiana tabacum is associated with undermethylated, decondensed and probably active gene units. Chromosoma. 2000a;109:161–172. doi: 10.1007/s004120050424. [DOI] [PubMed] [Google Scholar]
- Lim KY, Matyásek R, Lichtenstein CP, Leitch AR. Molecular cytogenetic analyses and phylogenetic studies in the Nicotiana section Tomentosae. Chromosoma. 2000b;109:245–258. doi: 10.1007/s004120000074. [DOI] [PubMed] [Google Scholar]
- Lim KY, Matyasek R, Kovarik A, Leitch A. Genome evolution in allotetraploid Nicotiana. Biol J Linn Soc Lond. 2004;82:599–606. [Google Scholar]
- Lim KY, Kovarik A, Matyasek R, Chase MW, Clarkson JJ, et al. Sequence of events leading to near-complete genome turnover in allopolyploid Nicotiana within five million years. New Phytol. 2007a;175:756–763. doi: 10.1111/j.1469-8137.2007.02121.x. [DOI] [PubMed] [Google Scholar]
- Lim KY, Matyasek R, Kovarik A, Leitch AR. Parental origin and genome evolution in the allopolyploid Iris versicolor. Ann Bot. 2007b;100:219–224. doi: 10.1093/aob/mcm116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KY, Soltis DE, Soltis PS, Tate J, Matyasek R, et al. Rapid chromosome evolution in recently formed polyploids in Tragopogon (Asteraceae) PLoS One. 2008;3:e3353. doi: 10.1371/journal.pone.0003353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Wendel JF. Epigenetic phenomena and the evolution of plant allopolyploids. Mol Phylogenet Evol. 2003;29:365–379. doi: 10.1016/s1055-7903(03)00213-6. [DOI] [PubMed] [Google Scholar]
- Liu SY, Chen SM, Chen Y, Guan ZY, Yin DM, Chen FD. In vitro induced tetraploid of Dendranthema nankingense (Nakai) Tzvel. shows an improved level of abiotic stress tolerance. Sci Horticult. 2011;127:411–419. [Google Scholar]
- Llyod DG, Webb CJ. The avoidance of interference between the presentation of pollen and stigmas in angiosperms I. Dichogamy. New Zeal J Bot. 1986;24:135–162. [Google Scholar]
- Löve Á , Löve D. The geobotanical significance of polyploidy. I. Polyploidy and latitude. Portugaliae Acta Biol Ser A. 1949–1951;273:352. [Google Scholar]
- Löve Á , Löve D. Polyploidy and altitude: Mt. Washington. Biol Zentralbl Suppl Vol. 1967:307–312. [Google Scholar]
- Lowry E, Lester SE. The biogeography of plant reproduction: potential determinants of species’ range sizes. J Biogeo. 2006;33:1975–1982. [Google Scholar]
- Lysak MA, Schubert I. Mechanisms of chromo-some rearrangements. In: Leitch IJ, Greilhuber J, Doležel J, Wendel JF, editors. Plant Genome Diversity 2: Physical Structure, Behaviour and Evolution of Plant Genomes. Springer; Wien: 2013. pp. 137–147. [Google Scholar]
- Lysak MA, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc Natl Acad Sci USA. 2006;103:5224–5229. doi: 10.1073/pnas.0510791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XF, Gustafson JP. Genome evolution of allopolyploids: a process of cytological and genetic diploidization. Cytogenet Genome Res. 2005;109:236–249. doi: 10.1159/000082406. [DOI] [PubMed] [Google Scholar]
- Mable BK. Why polyploidy is rarer in animals than in plants: myths and mechanisms. Biol J Linn Soc Lond. 2004;82:453–466. [Google Scholar]
- Macas J, Neumann P, Navrátilová A. Repetitive DNA in the pea (Pisum sativum L.) genome: comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genomics. 2007;8:427. doi: 10.1186/1471-2164-8-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macas J, Kejnovský E, Neumann P, Novak P, Koblížková A, Vyskot B. Next generation sequencing-based analysis of repetitive DNA in the model dioceous plant Silene latifolia. PLoS One. 2011;6:e27335. doi: 10.1371/journal.pone.0027335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maceira NO, Jacquard P, Lumaret R. Competition between diploid and derivative autotetraploid Dactylis glomerata L. from Galicia. Implications for the establishment of novel polyploid populations. New Phytol. 1993;124:321–328. doi: 10.1111/j.1469-8137.1993.tb03822.x. [DOI] [PubMed] [Google Scholar]
- Madlung A. Polyploidy and its effect on evolutionary success: old questions revisited with new tools. Heredity. 2013;110:99–104. doi: 10.1038/hdy.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali H, Walden AE, Husband BC. Genome duplication and the evolution of physiological responses to water stress. New Phytol. 2009;184:721–731. doi: 10.1111/j.1469-8137.2009.02997.x. [DOI] [PubMed] [Google Scholar]
- Malinska H, Tate JA, Matyasek R, Leitch AR, Soltis DE, et al. Similar patterns of rDNA evolution in synthetic and recently formed natural populations of Tragopogon (Asteraceae) allotetraploids. BMC Evol Biol. 2010;10:291. doi: 10.1186/1471-2148-10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maluszynska J, Hasterok R, Weiss H. rRNA genes: their distribution and activity in plants. In: Maluszynska J, editor. Plant Cytogenetics. Silesian Univ Press; Katowice: 1998. pp. 75–95. [Google Scholar]
- Mandakova T, Joly S, Krzywinski M, Mummen-hoff K, Lysak MA. Fast diploidization in close mesopolyploid relatives of Arabidopsis. Plant Cell. 2010;22:2277–2290. doi: 10.1105/tpc.110.074526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzaneda AJ, Rey PJ, Bastida JM, Weiss-Lehman C, Raskin E, Mitchell-Olds T. Environmental aridity is associated with cytotype segregation and polyploidy occurrence in Brachypodium distachyon (Poaceae) New Phytol. 2012;193:797–805. doi: 10.1111/j.1469-8137.2011.03988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhold K, Kudoh H, Pak JH, Watanabe K, Španiel S, Lihová J. Cytotype diversity and genome size variation in eastern Asian polyploid Cardamine (Brassicaceae) species. Ann Bot. 2010;105:249–264. doi: 10.1093/aob/mcp282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SL, Husband BC. Influence of phylogeny and ploidy on species ranges of North American angiosperms. J Ecol. 2009;97:913–922. [Google Scholar]
- Martin SL, Husband BC. Whole genome duplication affects evolvability of flowering time in an autotetraploid plant. PLoS One. 2012;7:e44784. doi: 10.1371/journal.pone.0044784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterson J. Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science. 1994;264:421–424. doi: 10.1126/science.264.5157.421. [DOI] [PubMed] [Google Scholar]
- Matyasek R, Lim KY, Kovarik A, Leitch AR. Ribosomal DNA evolution and gene conversion in Nicotiana rustica. Heredity. 2003;91:268–275. doi: 10.1038/sj.hdy.6800333. [DOI] [PubMed] [Google Scholar]
- Matyasek R, Tate JA, Lim YK, Srubarova H, Koh J, et al. Concerted evolution of rDNA in recently formed Tragopogon allotetraploids is typically associated with an inverse correlation between gene copy number and expression. Genetics. 2007;176:2509–2519. doi: 10.1534/genetics.107.072751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrose I, Zhan SH, Rothfels CJ, Magnuson-Ford K, Barker MS, et al. Recently formed polyploid plants diversify at lower rates. Science. 2011;333:1257. doi: 10.1126/science.1207205. [DOI] [PubMed] [Google Scholar]
- McArthur ED, Sanderson SC. Cytogeography and chromosome evolution of subgenus Tridentatae of Artemisia (Asteraceae) Am J Bot. 1999;86:1754–1775. [PubMed] [Google Scholar]
- McGrath CL, Lynch M. Evolutionary significance of whole-genome duplication. In: Soltis PS, Soltis DE, editors. Polyploidy and Genome Evolution. Springer; Heidelberg: 2012. pp. 1–20. [Google Scholar]
- McIntyre P. Cytogeography and genome size variation in the Claytonia perfoliata (Portulacaceae) polyploid complex. Ann Bot. 2012a;110:1195–1203. doi: 10.1093/aob/mcs187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre P. Polyploidy associated with altered and broader ecological niches in the Claytonia perfoliata (Portulacaceae) species complex. Am J Bot. 2012b;99:655–662. doi: 10.3732/ajb.1100466. [DOI] [PubMed] [Google Scholar]
- Mestiri I, Chague V, Tanguy AM, Huneau C, Huteau V, et al. Newly synthesized wheat allohexaploids display progenitor-dependent meiotic stability and aneuploidy but structural genomic additivity. New Phytol. 2010;186:86–101. doi: 10.1111/j.1469-8137.2010.03186.x. [DOI] [PubMed] [Google Scholar]
- Meyers LA, Levin DA. On the abundance of polyploids in flowering plants. Evolution. 2006;60:1198–1206. [PubMed] [Google Scholar]
- Miller M, Zhang CQ, Chen ZJ. Ploidy and hybridity effects on growth vigor and gene expression in Arabidopsis thaliana hybrids and their parents. G3 (Bethesda) 2012;2:505–513. doi: 10.1534/g3.112.002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlinarec J, Satovic Z, Malenica N, Ivancic-Bace I, Besendorfer V. Evolution of the tetraploid Anemone multifida (2n = 32) and hexaploid A. baldensis (2n = 48) (Ranunculaceae) was accompanied by rDNA loci loss and intergenomic translocation: evidence for their common genome origin. Ann Bot. 2012;110:703–712. doi: 10.1093/aob/mcs128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscone EA, Matzke MA, Matzke AJ. The use of combined FISH/GISH in conjunction with DAPI counterstaining to identify chromosomes containing transgene inserts in amphidiploid tobacco. Chromosoma. 1996;105:231–236. [PubMed] [Google Scholar]
- Müntzing A. The evolutionary significance of autopolyploidy. Hereditas. 1936;21:263–378. [Google Scholar]
- Murray BG. Karyotype variation and evolution in gymnosperms. In: Leitch IJ, Greilhuber J, Doležel J, Wendel JF, editors. Plant Genome Diversity 2: Physical Structure, Behaviour and Evolution of Plant Genomes. Springer; Wien: 2013. pp. 231–243. [Google Scholar]
- Ni Z, Kim ED, Ha M, Lackey E, Liu J, et al. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 2009;457:327–331. doi: 10.1038/nature07523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates KM, Ranney TG, Touchell DH. Influence of induced polyploidy on fertility and morphology of Rudbeckia species and hybrids. HortScience. 2012;47:1217–1221. [Google Scholar]
- Osborn TC, Pires JC, Birchler JA, Auger DL, Chen ZJ, et al. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 2003;19:141–147. doi: 10.1016/s0168-9525(03)00015-5. [DOI] [PubMed] [Google Scholar]
- Oswald BP, Nuismer SL. A unified model of autopolyploid establishment and evolution. Am Nat. 2011;178:687–700. doi: 10.1086/662673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP, Whitton J. Polyploid incidence and evolution. Annu Rev Genet. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- Ownbey M. Natural hybridization and amphiploidy in the genus Tragopogon. Am J Bot. 1950;37:487–499. [Google Scholar]
- Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- Pandit MK. Continuing the search for pattern among rare plants: are diploid species more likely to be rare? Evol Ecol Res. 2006;8:543–552. [Google Scholar]
- Pandit MK, Pocock MJO, Kunin WE. Ploidy influences rarity and invasiveness in plants. J Ecol. 2011;99:1108–1115. [Google Scholar]
- Parisod C, Besnard G. Glacial in situ survival in the Western Alps and polytopic autopolyploidy in Biscutella laevigata L. (Brassicaceae) Mol Ecol. 2007;16:2755–2767. doi: 10.1111/j.1365-294X.2007.03315.x. [DOI] [PubMed] [Google Scholar]
- Parisod C, Alix C, Just J, Petit M, Sarilar V, et al. Impact of transposable elements in organization and functioning of allopolyploid genomes. New Phytol. 2010a;186:37–45. doi: 10.1111/j.1469-8137.2009.03096.x. [DOI] [PubMed] [Google Scholar]
- Parisod C, Holderegger R, Brochmann C. Evolutionary consequences of autopolyploidy. New Phytol. 2010b;186:5–17. doi: 10.1111/j.1469-8137.2009.03142.x. [DOI] [PubMed] [Google Scholar]
- Parisod C, Mihri C, Clarkson J, Lim KY, Chase MW, et al. Differential impact of transposable elements on long-term genome diploidization in the allopolyploid Nicotiana section Repandae. PLoS One. 2012;7:e50352. doi: 10.1371/journal.pone.0050352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paun O, Fay MF, Forest F, Chase MW. Hybrid speciation in angiosperms: parental divergence drives ploidy. New Phytol. 2009;182:507–518. doi: 10.1111/j.1469-8137.2009.02767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paun O, Forest F, Fay MF, Chase MW. Parental divergence and hybrid speciation in angiosperms revisited. Taxon. 2011;60:1241–1244. [PMC free article] [PubMed] [Google Scholar]
- Petit C, Thompson JD. Species diversity and ecological range in relation to ploidy level in the flora of the Pyrenees. Evol Ecol. 1999;13:45–66. [Google Scholar]
- Petit M, Lim KY, Julio E, Poncet C, Dorlhac de Borne F, et al. Differential impact of retrotransposon populations on the genome of allotetraploid tobacco (N. tabacum) Mol Genet Genomics. 2007;278:1–15. doi: 10.1007/s00438-007-0226-0. [DOI] [PubMed] [Google Scholar]
- Petit M, Guidat C, Daniel J, Denis E, Motoriol E, et al. Mobilization of retrotransposons in synthetic allotetraploid tobacco. New Phytol. 2010;186:135–147. doi: 10.1111/j.1469-8137.2009.03140.x. [DOI] [PubMed] [Google Scholar]
- Piednoël M, Aberer AJ, Schneeweiss GM, Macas J, Novak P, et al. Next-generation sequencing reveals the impact of repetitive DNA across phylogenetically closely related genomes of Orobanchaceae. Mol Biol Evol. 2012;29:3601–3611. doi: 10.1093/molbev/mss168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires JC, Lim KY, Kovarik A, Matyasek R, Boyd A, et al. Molecular cytogenetic analysis of recently evolved Tragopogon (Asteraceae) allopolyploids reveal a karyotype that is additive of the diploid progenitors. Am J Bot. 2004a;91:1022–1035. doi: 10.3732/ajb.91.7.1022. [DOI] [PubMed] [Google Scholar]
- Pires JC, Zhao J, Schranz EM, Leon EJ, Quijada PA, et al. Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae) Biol J Linn Soc Lond. 2004b;82:675–688. [Google Scholar]
- Preuss S, Pikaard CS. rRNA gene silencing and nucleolar dominance: insights into a chromosome-scale epigenetic on/off switch. Biochim Biophys Acta. 2007;1769:383–392. doi: 10.1016/j.bbaexp.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyšek P, Jarosik V, Pergl J, Randall R, Chytry M, et al. The global invasion success of Central European plants is related to distribution characteristics in their native range and species traits. Divers Distrib. 2009;15:891–903. [Google Scholar]
- Raabova J, Fischer M, Münzbergerová Z. Niche differentiation between diploid and hexaploid Aster amellus. Oecologia. 2008;158:463–472. doi: 10.1007/s00442-008-1156-1. [DOI] [PubMed] [Google Scholar]
- Ramsey J. Unreduced gametes and neopolyploids in natural populations of Achillea borealis (Asteraceae) Heredity. 2007;98:143–150. doi: 10.1038/sj.hdy.6800912. [DOI] [PubMed] [Google Scholar]
- Ramsey J. Polyploidy and ecological adaptation in wild yarrow. Proc Natl Acad Sci USA. 2011;108:7096–7101. doi: 10.1073/pnas.1016631108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J, Schemske DW. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol Syst. 1998;29:467–501. [Google Scholar]
- Ramsey J, Schemske DW. Neopolyploidy in flowering plants. Annu Rev Ecol Syst. 2002;33:589–639. [Google Scholar]
- Rausch JH, Morgan MT. The effect of self-fertilization, inbreeding depression, and population size on autopolyploid establishment. Evolution. 2005;59:1867–1875. [PubMed] [Google Scholar]
- Rebernig CA, Weiss-Schneeweiss H, Blöch C, Turner B, Stuessy TF, et al. The evolutionary history of the white-rayed species of Melampodium (Asteraceae) involved multiple cycles of hybridization and polyploidization. Am J Bot. 2012;99:1043–1057. doi: 10.3732/ajb.1100539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese G. Polyploidie und Verbreitung. Z Botanik. 1958;46:339–354. [Google Scholar]
- Renny-Byfield S, Chester M, Kovarik A, Le Comber SC, Grandbastien MA, et al. Next generation sequencing reveals genome downsizing in allotetraploid Nicotiana tabacum, predominantly through the elimination of paternally derived repetitive DNAs. Mol Biol Evol. 2011;28:2843–2854. doi: 10.1093/molbev/msr112. [DOI] [PubMed] [Google Scholar]
- Renny-Byfield S, Kovarik A, Chester M, Nichols RA, Macas J, et al. Independent, rapid and targeted loss of highly repetitive DNA in natural and synthetic allopolyploids of Nicotiana tabacum. PLoS One. 2012;7:e36963. doi: 10.1371/journal.pone.0036963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez DJ. A model for the establishment of polyploidy in plants. Am Nat. 1996;147:33–46. [Google Scholar]
- Schlaepfer DR, Edwards P, Billeter R. Why only tetraploid Solidago gigantea (Asteraceae) became invasive: a common garden comparison of ploidy levels. Oecologia. 2010;163:661–673. doi: 10.1007/s00442-010-1595-3. [DOI] [PubMed] [Google Scholar]
- Schmidt T, Heslop-Harrison JS. Genomes, genes and junk: the large-scale organization of plant chromosomes. Trends Plant Sci. 1998;3:195–199. [Google Scholar]
- Schneeweiss GM, Palomeque T, Colwell AE, Weiss-Schneeweiss H. Chromosome numbers and karyotype evolution in holoparasitic Orobanche (Orobanchaceae) and related genera. Am J Bot. 2004;91:439–448. doi: 10.3732/ajb.91.3.439. [DOI] [PubMed] [Google Scholar]
- Schönswetter P, Lachmayer M, Lettner C, Prehsler D, Rechnitzer S, et al. Sympatric diploid and hexaploid cytotypes of Senecio carniolicus (Asteraceae) in the Eastern Alps are separated along an altitudinal gradient. J Plant Res. 2007;120:721–725. doi: 10.1007/s10265-007-0108-x. [DOI] [PubMed] [Google Scholar]
- Schubert I, Lysak MA. Interpretation of karyo-type evolution should consider chromosome structural constraints. Trends Genet. 2011;27:207–216. doi: 10.1016/j.tig.2011.03.004. [DOI] [PubMed] [Google Scholar]
- Segraves KA, Thompson JN. Plant polyploidy and pollination: floral traits and insect visits to diploid and tetraploid Heuchera grossulariifolia. Evolution. 1999;53:1114–1127. doi: 10.1111/j.1558-5646.1999.tb04526.x. [DOI] [PubMed] [Google Scholar]
- Seoighe C. Turning the clock back on ancient genome duplication. Curr Opin Genet Dev. 2003;13:636–643. doi: 10.1016/j.gde.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Skalická K, Lim KY, Matyasek R, Matzke M, Leitch AR, Kovarik A. Preferential elimination of repeated DNA sequences from the paternal, Nicotiana tomentosiformis genome donor of a synthetic, allotetraploid tobacco. New Phytol. 2005;166:291–303. doi: 10.1111/j.1469-8137.2004.01297.x. [DOI] [PubMed] [Google Scholar]
- Sobel JM, Chen GF, Watt LR, Schemske DW. The biology of speciation. Evolution. 2010;64:295–315. doi: 10.1111/j.1558-5646.2009.00877.x. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS. Molecular data and the dynamic nature of polyploidy. Crit Rev Plant Sci. 1993;12:243–273. [Google Scholar]
- Soltis DE, Soltis PS. Polyploidy: origins of species and genome evolution. Trends Ecol Evol. 1999;14:348–352. doi: 10.1016/s0169-5347(99)01638-9. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Pires JC, Kovarik A, Tate JA, Mavrodiev E. Recent and recurrent polyploidy in Tragopogon (Asteraceae): cytogenetic, genomic and genetic comparisons. Biol J Linn Soc Lond. 2004;82:485–501. [Google Scholar]
- Soltis DE, Soltis PS, Schemske DW, Hancock JF, Thompson JN, et al. Autopolyploidy in angiosperms: have we grossly underestimated the number of species? Taxon. 2007;56:13–30. [Google Scholar]
- Soltis DE, Albert VA, Leebens-Mack J, Bell CD, Paterson AH, et al. Polyploidy and angiosperm diversification. Am J Bot. 2009a;96:336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Buggs RJ, Barbazuk WB, Schnable PS, Soltis PS. On the origins of species: does evolution repeat itself in polyploid populations of independent origin? Cold Spring Harb Symp Quant Biol. 2009b;74:215–223. doi: 10.1101/sqb.2009.74.007. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Buggs RJA, Doyle JJ, Soltis PS. What we still don’t know about polyploidy. Taxon. 2010;59:1387–1403. [Google Scholar]
- Soltis DE, Buggs RJA, Barbazuk WB, Chamala S, Chester M, et al. The early stages of polyploidy: rapid and repeated evolution in Tragopogon. In: Soltis PS, Soltis DE, editors. Polyploidy and Genome Evolution. Springer; Heidelberg: 2012. pp. 271–292. [Google Scholar]
- Sonnleitner M, Flatscher R, Escobar García P, Rauchová J, Suda J, et al. Distribution and habitat segregation on different spatial scales among diploid, tetraploid and hexaploid cytotypes of Senecio carniolicus (Asteraceae) in the Eastern Alps. Ann Bot. 2010;106:967–977. doi: 10.1093/aob/mcq192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL. Variation and Evolution in Plants. Columbia Univ Press; New York: 1950. [Google Scholar]
- Stebbins GL. Chromosomal Evolution in Higher Plants. Edward Arnold; London: 1971. [Google Scholar]
- Stebbins GL. Polyploidy: future prospects. In: Lewis WH, editor. Polyploidy: Biological Relevance. Plenum Press; New York: 1980. pp. 495–520. [Google Scholar]
- Stebbins GL. Polyploidy and the distribution of the arctic-alpine flora: new evidence and a new approach. Bot Helv. 1984;94:1–13. [Google Scholar]
- Stebbins GL. Polyploidy, hybridization, and the invasion of new habitats. Ann Miss Bot Gard. 1985;72:824–832. [Google Scholar]
- Stebbins GL, Dawe JC. Polyploidy and distribution in the European flora: a reappraisal. Bot Jahrb Syst. 1987;108:343–354. [Google Scholar]
- Stelkens R, Seehausen O. Genetic distance between species predicts novel trait expression in their hybrids. Evolution. 2009;63:884–897. doi: 10.1111/j.1558-5646.2008.00599.x. [DOI] [PubMed] [Google Scholar]
- Suda J, Herben T. Ploidy frequencies in plants with ploidy heterogeneity: fitting a general gametic model to empirical population data. Proc Biol Sci. 2013;280:20122387. doi: 10.1098/rspb.2012.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda J, Kron P, Husband BC, Trávníček P. Flow cytometry and ploidy: applications in plant systematics, ecology and evolutionary biology. In: Doležel J, Greilhuber J, Suda J, editors. Flow Cytometry with Plant Cells. Analysis of Genes, Chromosomes and Genomes. Wiley-VCH; Weinheim: 2007. pp. 103–130. [Google Scholar]
- te Beest M, Le Roux JJ, Richardson DM, Brysting AK, Suda J, et al. The more the better? The role of polyploidy in facilitating plant invasions. Ann Bot. 2012;109:19–45. doi: 10.1093/aob/mcr277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler T. Die Halligenflora der Nordsee im Lichte cytologischer Forschung. Cytologia. 1937;8:162–170. [Google Scholar]
- Treier UA, Broennimann O, Normand S, Guisan A, Schaffner U, et al. Shift in cytotype frequency and niche space in the invasive plant Centaurea maculosa. Ecology. 2009;90:1366–1377. doi: 10.1890/08-0420.1. [DOI] [PubMed] [Google Scholar]
- Tremetsberger K, König C, Samuel R, Pinsker W, Stuessy TF. Infraspecific genetic variation in Biscutella laevigata (Brassicaceae): new focus on Irene Manton’s hypothesis. Plant Syst Evol. 2002;233:163–181. [Google Scholar]
- Vamosi JC, Dickinson TA. Polyploidy and diversification: a phylogenetic investigation in Rosaceae. Int J Plant Sci. 2006;167:349–358. [Google Scholar]
- Vamosi JC, Goring SJ, Kennedy BF, Mayberry RJ, Moray CM, et al. Pollination, floral display, and the ecological correlates of polyploidy. Funct Ecosyst Commun. 2007;1:1–9. [Google Scholar]
- Vaughan HE, Jamilena M, Ruiz Rejón C, Parker JS, Garrido-Ramos MA. Loss of nucleolar-organizer regions during polyploid evolution in Scilla autumnalis. Heredity. 1993;71:574–580. [Google Scholar]
- Vaughan HE, Taylor S, Parker JS. The ten cytological races of the Scilla autumnalis species complex. Heredity. 1997;79:371–379. [Google Scholar]
- Webb CJ, Lloyd DG. The avoidance of interference between the presentation of pollen and stigmas in angiosperms II. Herkogamy. New Zeal J Bot. 1986;24:163–178. [Google Scholar]
- Weiss H, Maluszynska J. Chromosomal rearrangement in autotetraploid plants of Arabidopsis thaliana. Hereditas. 2000;133:255–261. doi: 10.1111/j.1601-5223.2000.00255.x. [DOI] [PubMed] [Google Scholar]
- Weiss-Schneeweiss H, Schneeweiss GM. Karyotype diversity and evolutionary trends in angiosperms. In: Leitch IJ, Greilhuber J, Doležel J, Wendel JF, editors. Plant Genome Diversity 2: Physical Structure, Behaviour and Evolution of Plant Genomes. Springer; Wien: 2013. pp. 209–230. [Google Scholar]
- Weiss-Schneeweiss H, Schneeweiss GM, Stuessy TF, Mabuchi T, Park JM, et al. Chromosomal stasis in diploids contrasts with genome restructuring in auto- and allopolyploid taxa of Hepatica (Ranunculaceae) New Phytol. 2007;174:669–682. doi: 10.1111/j.1469-8137.2007.02019.x. [DOI] [PubMed] [Google Scholar]
- Weiss-Schneeweiss H, Blöch C, Turner B, Villaseñor JL, Stuessy TF, Schneeweiss GM. The promiscuous and the chaste: frequent allopolyploid speciation and its genomic consequences in American daisies (Melampodium sect. Melampodium; Asteraceae) Evolution. 2012;66:211–228. doi: 10.1111/j.1558-5646.2011.01424.x. [DOI] [PubMed] [Google Scholar]
- Wendel JF. Genome evolution in polyploids. Plant Mol Biol. 2000;42:225–249. [PubMed] [Google Scholar]
- Wendel JF, Flagel LE, Adams KL. Jeans, genes, and genomes: cotton as a model for studying polyploidy. In: Soltis PS, Soltis DE, editors. Polyploidy and Genome Evolution. Springer; Heidelberg: 2012. pp. 181–207. [Google Scholar]
- Wicker T, Taudien S, Houben A, Keller B, Graner A, et al. A whole-genome snapshot of 454 sequences exposes the composition of the barley genome and provides evidence for parallel evolution of genome size in wheat and barley. Plant J. 2009;59:712–722. doi: 10.1111/j.1365-313X.2009.03911.x. [DOI] [PubMed] [Google Scholar]
- Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH. The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci USA. 2009;106:13875–13879. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JH, Ferguson AR, Murray BG, Jia Y, Datson PM, Zhang J. Induced polyploidy dramatically increases the size and alters the shape of fruit in Actinidia chinensis. Ann Bot. 2012;109:169–179. doi: 10.1093/aob/mcr256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Gaeta RT, Pires JC. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc Natl Acad Sci USA. 2011;108:7908–7913. doi: 10.1073/pnas.1014138108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XP, Si Y, Hanson RE, Crane CF, Price HJ, et al. Dispersed repetitive DNA has colonized new genomes since polyploid formation in cotton. Genome Res. 1998;8:479–492. doi: 10.1101/gr.8.5.479. [DOI] [PubMed] [Google Scholar]
- Zielinski ML, Mittelsten Scheid O. Meiosis in polyploid plants. In: Soltis PS, Soltis DE, editors. Polyploidy and Genome Evolution. Springer; Heidelberg: 2012. pp. 33–55. [Google Scholar]
- Zimmer EA, Wen J. Using nuclear gene data for plant phylogenetics: progress and prospects. Mol Phylogenet Evol. 2012;65:774–785. doi: 10.1016/j.ympev.2012.07.015. [DOI] [PubMed] [Google Scholar]