Abstract
Background:
Processing of microRNAs (miRNAs) is a highly controlled process. Deregulation of miRNA expression was observed in several types of cancer but changes in the miRNA-processing enzymes have not been analysed until today. In this study, we analysed Argonaute2 (AGO2, EIF2C2), as one main factor of the miRNA processing ensemble, in the context of cancer development, especially in melanoma.
Methods:
We determined the AGO2 expression level in melanoma, as well as in other cancers, with biochemical approaches (qRT–PCR, western blot and immunofluorescence studies) and analysed the cell behaviour in migration assays.
Results:
Specifically in melanoma, we revealed a strong reduction of AGO2 expression compared with primary melanocytes. The reduction of AGO2 expression was only found on protein level, whereas the mRNA level stayed unchanged hinting to post-transcriptional regulation. We could show that re-expression of AGO2 in melanoma leads to a strong improvement of regulatory effects due to increased functionality of small-interfering RNAs and short hairpin RNAs.
Conclusion:
We identified melanoma-specific downregulation of AGO2 and corresponding reduced RNAi efficiency. These findings will help to understand the molecular basis of malignant melanoma and can potentially lead to an improvement of therapeutic strategies.
Keywords: malignant melanoma, AGO2, RNAi, miRNA, siRNA, shRNA, cJun
Malignant melanoma is an aggressive tumour derived from melanocytes. Several details are known about the processes leading to the development of malignant melanoma, but the detailed molecular causes of the disease remain largely unknown. One class of molecules recently being shown to have a role in melanoma development are microRNAs (miRNAs). MiRNAs are small non-coding RNA molecules that regulate gene expression on the post-transcriptional level (Lee et al, 1993; Krol and Loedige, 2010). It is known that miRNAs are first transcribed as primary transcripts (pri-miRNAs), which are further processed in the nucleus to reach the pre-miRNA hairpin intermediate. After the pre-miRNA transfer to the cytoplasm, mature miRNAs are formed (Lee et al, 2002; Bartel, 2009).
However, many enzymes are involved in the processing cascade and each of them is a critical factor for miRNA production. The RNA-binding protein DiGeorge Syndrome Critical Region 8 (DGCR8; also known as Pasha in invertebrates) binds to Drosha, an RNase III enzyme, to process pri-miRNAs to pre-miRNAs. The nucleocytoplasmic transporter Exportin-5 transfers the pre-miRNA into the cytoplasm, where a second RNase III enzyme Dicer processes the pre-miRNA to a double stranded miRNA:miRNA* intermediate. Finally, miRNAs are incorporated into RISC, where they directly bind to a member of the Argonaute (AGO) protein family (Peters and Meister, 2007; Hutvagner and Simard, 2008). This family in humans consists of four Argonauts (AGO1-4), but only AGO2 exhibits endonuclease activity (Hauptmann et al, 2013). MiRNAs guide the RISC to complementary target sites on mRNAs and modulate mRNA stability by inducing degradation of the mRNA (Huntzinger et al, 2010) or controlling translation (Meister and Tuschl, 2004; Huntzinger et al, 2010; Janga and Vallabhaneni, 2011; Vasudevan, 2012). Recent research has not only revealed the strong impact of miRNAs on almost every regulatory pathway in eukaryotic cells but has also clearly demonstrated their involvement in the tumorigenesis of a variety of human cancers (Calin and Croce, 2006).
Studies from our own group as well as from several other groups revealed that the expression of many miRNAs is deregulated in malignant melanoma cells, which is undoubtedly linked to important processes affecting melanoma formation and progression (reviewed by (Mueller and Bosserhoff, 2009; Voller et al, 2013; Kunz, 2013)). However, the deregulation of miRNAs in cancer development based on deregulated miRNA-processing proteins is not well understood and in the spotlight of current research. For example, upregulation of Dicer in cutaneous melanoma is associated with tumorigenesis (Ma et al, 2011; Pellegrino et al, 2013). In contrast to Dicer, the role of AGO2 as the key RISC protein in melanoma is still unknown. In this study, we first analysed the expression status and localisation of AGO2 in malignant melanoma and second in comparison with other ‘non-melanoma' tumours. In addition, we investigate whether AGO2 modulation is responsible for small-interfering RNA (siRNA) and miRNA functionality because of the fact that most miRNAs are upregulated in melanoma (Mueller et al, 2009) compared with the rather downregulated miRNA expression pattern in other tumours (Wang et al, 2012; Zhao et al, 2012). Some miRNAs are already known to have an important role by targeting specific genes in malignant melanoma (Chen et al, 2010; Deng et al, 2011; Xu et al, 2012), so we were interested in determining whether AGO2 is the responsible key factor in modulation of the miRNA pattern in melanoma. This finding could lead to new insights into miRNA processing and could also result in consequences for miRNA-based therapies in melanoma.
Materials and Methods
Cell culture and tissue samples
Melanoma cell lines Mel Ei, Mel Wei, Mel Ho, Mel Juso, Mel Im, Mel Ju, A375, HMB2, HTZ19d, SkMel3, SkMel28 and NHEMs were described previously (Jacob et al, 1998; Braig and Bosserhoff, 2013). The cell lines Mel Juso and A375 were derived from primary cutaneous melanomas; Mel Im, Mel Ju, HTZ19d, SkMel3, SkMel28 and HMB2 were derived from metastases of malignant melanomas. Cells were maintained in DMEM supplemented with penicillin (400 units per ml), streptomycine (50 μg ml−1), L-glutamine (300 μg ml−1), 10% FCS (Sigma-Aldrich, Steinheim, Germany) and split at a 1 : 5 ratio every 3 days. Normal human epidermal melanocytes (NHEMs; PromoCell, Heidelberg, Germany) were derived from neonatal skin. Isolation and cultivation of NHEMs were described previously (Muller and Bosserhoff, 2008). Cells were used between passages 2–6 and not later than 2 days after trypsinisation. Tissue samples of snap-frozen melanoma primary tumours (tumour bank: TB62, TB69, TB71, TB93, TB97, TB104 and TB126) and melanoma metastases (tumour bank: TB21, TB31, TB35, TB43, TB90, TB95 and TB331) with clear-cut pathological classification were obtained from the institute's tissue collection (Institute of Pathology, University of Regensburg, Germany). Sampling and handling of patient material was carried out in accordance with the ethical principles of the Declaration of Helsinki.
RNA isolation and reverse transcription
Total cellular RNA was isolated from cultured cells and tissues using the E.Z.N.A. Total RNA Kit I (Omega Bio-Tek, VWR, Darmstadt, Germany) according to the manufacturer's instructions. Complementary DNAs were generated by a reverse transcriptase reaction (500 ng of total RNA) using the Super Script II Reverse Transcriptase Kit (Invitrogen, Groningen, The Netherlands).
Analysis of gene expression by quantitative PCR
Quantitative real time-PCR (qRT–PCR) was performed on a Lightcycler (Roche, Mannheim, Germany). cDNA template (500 ng), 0.5 μl (20 μM) of forward and reverse primers and 10 μl of Sybr Premix Ex Taq (TaKaRa, Shiga, Japan) in a total volume of 20 μl were applied to the following PCR program: 30 s 95 °C (initial denaturation); 20 °C s−1 temperature transition rate up to 95 °C for 15 s, 10 s annealing, 20 s 72 °C, 10 s acquisition mode single, repeated for 45 times (amplification). Annealing and melting temperatures were optimised for each primer set (Table 1). β-Actin was used for normalisation.
Table 1. Oligonucleotide sequences and qRT–PCR conditions.
| Gene | Primer sequences (fwd/rev) | Ta (°C) | Tm (°C) |
|---|---|---|---|
|
β-Actin |
5′-CTA CGT CGC CCT GGA CTT CGA GC-3′
5′-GAT GGA GCC GCC GAT CCA CAC GG-3′ |
60–68 |
80 |
|
AGO2 |
5′-GTC TCT GAA GGC CAG TTC CA-3′
5′-ATA GAG GCC TCA CGG ATG G-3′ |
60 |
78 |
|
cJun |
5′-TTC CTC CCG TCC GAG AGC GG-3′
5′-TCG GCG TGG TGG TGA TGT GC-3′ |
60 |
74 |
| NRAS | 5′-ATG AGG ACA GGC GAA GGC T-3′ 5′-TGA GTC CCA TCA TCA CTG CTG-3′ | 60 | 66 |
Transfecting cells with pAGO2 or siRNA
In all, 2 × 104 cells were seeded each well in six-well plates and transfected with 0.5–2 μg of plasmids pAGO2 or pIRES by using Lipofectamine LTX (Invitrogen) or with 2–30 pmol siRNA by using Lipofectamine RNAiMAX reagent (Invitrogen).
Protein analysis in vitro (western blotting)
In all, 3 × 106 cells were resuspended in 200 μl radio-immune precipitation assay (RIPA) buffer (Roche) and lysed for 15 min at 4 °C. Insoluble fragments were removed by centrifugation at 13 000 r.p.m. for 10 min and the supernatant was stored at −20 °C. In total, 40 μg of RIPA complete cell lysates were loaded per lane and separated on SDS–PAGE gels (Invitrogen) and subsequently blotted onto a PVDF membrane. After blocking for 1 h with 3% BSA/PBS in case of anti-c-Jun and 5% non-fat dry milk/TBS-T in case of anti-AGO2, the membrane was incubated for 16 h with one of the following antibodies: anti-AGO2 (11A9) (1 in 50 dilution, (Rudel et al, 2008), anti-c-Jun (1 in 1000 dilution; Upstate, Merck, Darmstadt, Germany), anti-β-actin (1 in 5000 dilution; Sigma-Aldrich) and anti-tubulin (YL1/2) (1 in 500 dilution; Merck Millipore, Darmstadt, Germany). After three washing steps with TBS-T, the membrane was incubated for 1 h with an alkaline phosphate-coupled secondary anti-mouse (1 in 3000 dilution in TBS-T (Chemicon, Hofheim, Germany)), anti-rabbit (1 in 3000 dilution in TBS-T(Chemicon)) or anti-rat (1 in 5000 dilution in TBS-T (Sigma-Aldrich, St Louis, MO, USA) IgG antibody and then washed again for three times in TBS-T. Finally, immunoreactions were visualised by NBT/BCIP (Sigma-Aldrich) staining.
AGO2 co-immunoprecipitation
Protein-G-sepharose beads (GE Healthcare, Munich, Germany) were rinsed four times with ice cold PBS (used in all subsequent washing steps), and incubated with monoclonal anti-AGO2 over night at 4 °C. Beads were then washed four times with PBS. Mel Ju cells (3 × 106) were lysed in 100 μl RIPA buffer (Roche) and incubated for 15 min at 4 °C. Insoluble fragments were removed by centrifugation at 13 000 r.p.m. for 15 min and the supernatant was stored at –20 °C. One hundred microgram protein lysate were diluted in PBS to a total volume of 300 μl. The lysate was incubated with Protein-G-sepharose beads at 4 °C for 3 h. After co-IP, the beads were washed four times with ice cold PBS and finally loaded with 4 × loading dye on SDS–PAGE gels (Invitrogen).
Mass spectrometry of AGO2
Samples were separated by SDS–PAGE on NuPAGE Novex 4–12% Bis-Tris gels using the MOPS-buffer system. After silver staining (Blum et al, 1987) bands of interest were excised and washed according to Shevchenko et al (Shevchenko et al, 1996) ith slight modifications. In brief, gel slizes were washed twice alternately with 50 μl 50 mM NH4HCO3 (washing buffer A) and 25 mM NH4HCO3 in 50% acetonitrile (washing buffer B). For reduction of disulphide bridges, the samples were incubated for 30 min in 10 mM DTT in washing buffer A at 56 °C. Carbamidomethylation of cysteines was performed by incubation with 5 mM iodoacetamide in washing buffer A for 30 min. Subsequently, the gel slizes were washed twice alternately with washing buffer A and B again and dried in a vacuum centrifuge. The gel pieces were rehydrated using 6 μl of trypsin solution (10 ng μl−1 in 50 mM ammoniumbicarbonate) and incubated at 37 °C overnight for in-gel-digestion. Resulting peptides were eluted with 20 μl of 5% formic acid and subjected to nano-LC-MS/MS-analysis on an Ultimate 3000 nano-HPLC-system (Dionex GmbH, Idstein, Germany) using a 1-h binary gradient directly coupled to a QTOF mass spectrometer (QStar XL, Applied Biosystems, Darmstadt, Germany) as described previously (Thomas et al, 2013). The tandem-MS-spectra were searched against the Uniprot-database using the Mascot Distiller (version 2.4.2.0) and the Mascot algorithm (version 2.2; Matrix Science Ltd, London, UK) using the following adjustments:
trypsin as protease
max. one missed cleavage site
carbamidomethylation of cysteines as fixed modification
oxidation of methionine, pGlu for N-terminal Gln as variable modifications
100 p.p.m. tolerance for MS- and 0.2 Da-tolerance for MS/MS-signals
only doubly and triply charged ions.
Only proteins with at least two different identified peptides with significantly scored spectra, which passed manual verification were considered.
Immunofluorescence for AGO2
Immunofluorescence assays were performed as described previously. Briefly, cells were seeded in chamber slides, washed with PBS, fixed with ice cold acetone for 10 min at −20 °C, permeabilised using 0.1% TritonX-100 for 5 min, washed again and blocked for 1 h with 1% bovine serumalbumin/PBS. Subsequently, cells were incubated with anti-AGO2 antibody (200 μl Hybridoma) overnight at 4 °C. After washing, cells were incubated with the secondary antibody (1 : 50, fluoresceinisothiocyanate-conjugated anti-rat immunoglobulin) for 2 h, followed by rinsing with PBS and mounting with Vectashield Hard SetMounting Medium with DAPI H-1500 (Vector Laboratories, Burlingame, CA, USA). Images were collected by immunofluorescence microscopy using an Axio Imager Zeiss Z1 fluorescence microscope (Axiovision Rel. 4.6.3, Carl Zeiss AG, Oberkochen, Germany).
Migration assay
Assays were performed using Boyden Chambers containing polycarbonate filters with 8-μm pore size (Costar, Bodenheim, Germany), essentially as described previously (Rothhammer et al, 2005). Filters were coated with gelatin (5 mg l−1). The lower compartment was filled with fibroblast-conditioned medium, used as a chemo-attractant. Melanoma cells were harvested by trypsinisation for 2 min, resuspended in DMEM without FCS at a density of 3 × 104 cells per ml and placed in the upper compartment of the chamber. After incubation at 37 °C for 4 h, the filters were collected and the cells adhering to the lower surface were fixed, stained and counted. Experiments were carried out in triplicates and were repeated three times.
Software
For image analysis the freeware ImageJ v1.33 downloaded from the NIH website (http://rsb.info.nih.gov/ij) and for statistical analysis the GraphPad Prism were used.
Statistical analysis
In the bar graphs, results are expressed as mean±s.d. (range) or per cent. Comparison between groups was made using the Student's unpaired t-test. The one-way or two-way analyses of variance were used for comparisons of more than two groups. A P-value <0.05 was considered as statistically significant (NS, not significant, *P<0.05, **P<0.01, ***P<0.001). All calculations were performed using the GraphPad Prism Software (GraphPad Software, Inc., San Diego, USA).
Results
siRNA effectivity in malignant melanoma
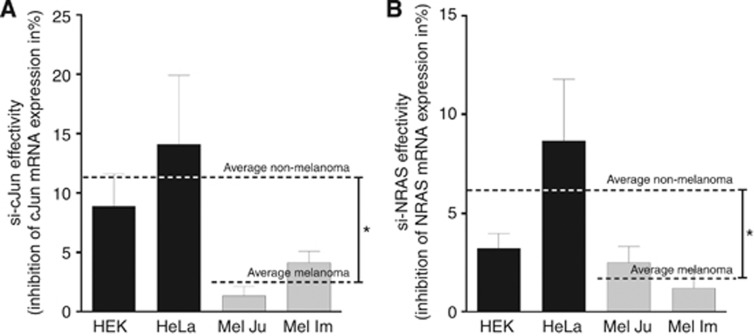
Recent data suggested that gene-targeting by siRNAs lead to minor effects in melanoma compared with effects in cell lines of other kinds of cancer. Further, higher amounts of siRNA were needed to achieve comparable effects. We, therefore, first aimed to quantitatively compare the capability of siRNAs for the gene knockdown in different cell lines. We analysed two siRNAs against cJun (Figure 1A) and NRAS (Figure 1B) in the melanoma cell lines Mel Ju and Mel Im and two ‘non-melanoma' cell lines CaCo2 (epithelial colorectal adenocarcinoma) and HeLa (cervical cancer). Figure 1 displays the reduction of gene expression per pmol transfected siRNA. In melanoma cells, both siRNAs achieved a significantly lower reduction of gene expression compared with ‘non-melanoma' cells.
Figure 1.
SiRNA effectivity in melanoma cell lines compared with ‘non-melanoma' cell lines. Inhibition of (A) cJun and (B) NRAS after 24 h treatment with si-cJun or si-NRAS in ‘non-melanoma' cell lines HEK and HeLa and melanoma cell lines Mel Im and Mel Ju. The average values of both groups are represented as dotted lines and pointed significant reductions of siRNA effectivities in melanoma cell lines. Statistical analysis was performed using the t-test, comparing ‘non-melanoma' and melanoma cell lines. *P<0.05.
AGO2 reduction in melanoma
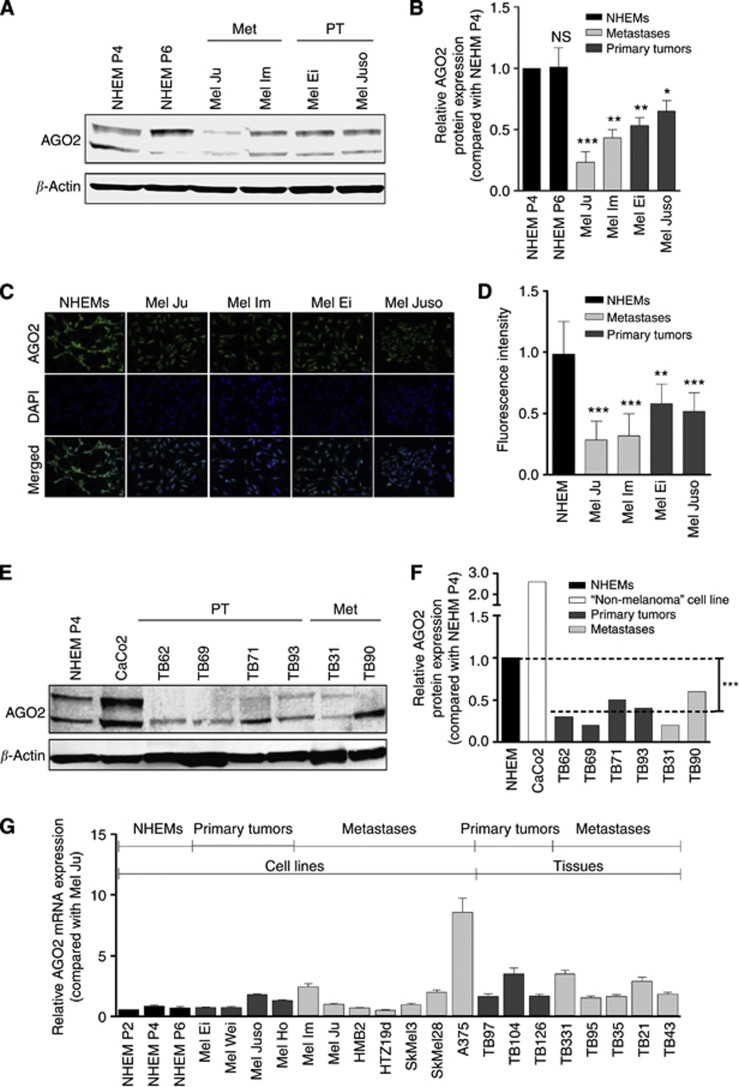
Little is known about alterations in the miRNA-processing machinery in melanoma. Some melanoma array studies determined that AGO2 stays unchanged compared with melanocytes but using cDNA arrays, post-transcriptional modifications are not detectable. Thus, we wanted to take a closer look into the expression of AGO2 as the main factor in miRNA function. We determined the AGO2 protein expression in melanoma cell lines derived from both primary tumours and metastases in comparison with normal human epidermal melanocytes (NHEM). The western blot analyses demonstrated a strong reduction of AGO2 in melanoma (Figure 2A). The AGO2 western blot displays two AGO2 bands, a lower band, which corresponds to the predicted AGO2 size of 99 kDa (Rudel et al, 2008) and an upper band that probably owns a post-translational modification. In Figure 2B, the quantification of AGO2 protein levels by analysing three different and independent analyses is presented, confirming the significant reduction of AGO2 in all melanoma cell lines. The reduction is stronger in both metastatic cell lines (Mel Ju, Mel Im) compared with primary tumour cell lines (Mel Ei, Mel Juso).
Figure 2.
AGO2 expression in melanoma cell lines and tissue samples. (A) AGO2 western blot and corresponding (B) AGO2 western blot quantifications of melanoma cell lines samples from both primary melanoma (Mel Ei and Mel Juso) and melanoma metastasis (Mel Ju and Mel Im) showed strong reduced AGO2 expression compared with normal human epidermal melanocytes (NHEM) in passage 4 and 6. β-Actin was used as loading control. We determined and quantified the total protein amount of both AGO2 bands. *P<0.05, **P<0.01, ***P<0.001. (C) Immunofluorescence staining for the detection of AGO2 deregulation in malignant melanoma compared with melanocytes. (D) AGO2 immunofluorescence quantification of 10 cells per cell line revealed strong reduction of AGO2. (E) AGO2 western blot of four melanoma primary tumour tissue samples and two melanoma metastases tissue samples compared with NHEMs in passage 4 and CaCo2 (human epithelial colorectal adenocarcinoma cells) demonstrate strong AGO2 reduction in melanoma tissues. β-Actin was used as loading control. (F) Western blot quantifications of AGO2 expression in melanoma tissue samples point strong AGO2 reductions compared with NHEMs and CaCo2. Comparison between the average melanoma tissue sample values (shown as dotted lines) and NHEMs revealed significant AGO2 reduction in melanoma tissues. (G) AGO2 qRT–PCR displays no AGO2 mRNA reduction in melanoma primary tumours and metastases in cell lines and tissue samples compared with NHEMs.
We further characterised the AGO2 protein level in melanoma cell lines by AGO2 immunfluorescence staining. The AGO2 expression is strongly reduced in all analysed melanoma cell lines compared with NHEMs (Figure 2C). To quantify these results, we measured the immunfluorescence intensity of 10 single cells of each cell line. The collected data are summarised in Figure 2D and display the strong and significant reduction of AGO2 in melanoma.
To characterise the AGO2 expression in tissue samples, we analysed tissue extract (RIPA) samples of primary tumours and metastases of melanoma and determined the AGO2 level by western blot (Figure 2E). The data as well as the corresponding western blot quantification (Figure 2F) revealed significantly reduced AGO2 expression in all tissue samples from both primary tumours and metastases in comparison with NHEMs and CaCo2.
Finally, we measured the AGO2 mRNA expression in NHEMs, cell lines and primary tumour samples of melanoma (Figure 2G). The melanoma cell lines compared with NHEMs displays equal AGO2 levels, only A375 showed a slightly higher expression rate. Also all tumour tissue samples (TB) indicated an AGO2 mRNA expression rate, which is close to the expression level of NHEMs. Significant differences between primary tumour tissues and metastases samples could not be observed.
AGO2 expression in melanoma and other ‘non-melanoma' tumours
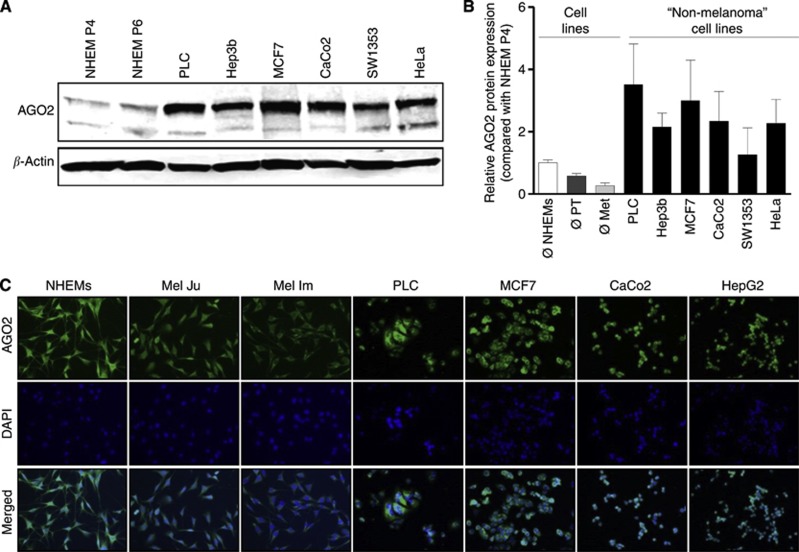
To determine whether loss of AGO2 is melanoma-specific or a general regulation in cancer cells, we analysed the AGO2 protein expression in six different tumours in comparison with NHEMs. The AGO2 expression in ‘non-melanoma' tumours (PLC (human hepatocellular carcinoma cell line), Hep3b (hepatocellular carcinoma cell line), MCF7 (Michigan Cancer Foundation—7, breast cancer cell line), CaCo2 (epithelial colorectal adenocarcinoma cell line), SW1353 (human bone chondrosarcoma, fibroblast-like cell line) and HeLa (human epithelial carcinoma cell line)) was higher than in NHEMs (Figure 3A). The AGO2 western blot quantification displayed that the AGO2 protein expression in all tumours was higher than in NHEMs and therefore also higher as in melanoma primary tumours or metastasis cell lines (Figure 3B).
Figure 3.
AGO2 expression in melanoma and ‘non-melanoma' tumours. (A) AGO2 western blots of NHEMs in passage 4 and 6 compared with six ‘non-melanoma' tumour cell lines indicate AGO2 reduction in NHEMs. (B) AGO2 western blot quantification display a high AGO2 expression in ‘non-melanoma' tumours in contrast to the lower amount of AGO2 in NHEMs and also to the strong reduced AGO2 expression in melanoma cell lines (PT=melanoma primary tumours, Met=melanoma metastases). (C) AGO2 immunofluorescence staining revealed strong AGO2 reduction in malignant melanoma compared with NHEMs and ‘non-melanoma' tumor cell lines.
To confirm the western blot results, we performed AGO2 immunofluorescence analyses (Figure 3C) and observed an equal and strong staining of NHEMs and all ‘non-melanoma' tumour cell lines (PLC, HeLa, SW1353 and HepG2). In contrast to that the Mel Ju and Mel Im AGO2 staining intensity is strongly reduced.
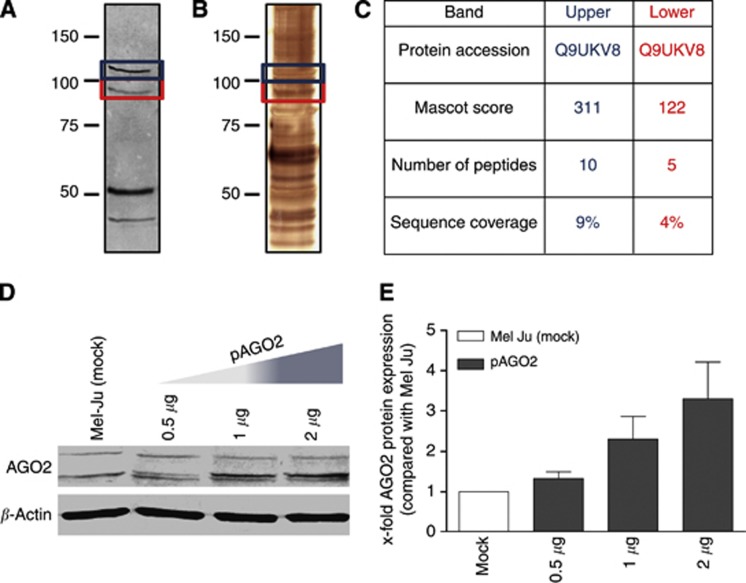
Analysis of the AGO2 protein
The performed western blot analyses of AGO2 display a specific and conserved band pattern consisting of two bands. To ensure that both bands are AGO2 and not a second target protein of the AGO2 antibody, we immunoprecipitated AGO2 from Mel Ju RIPA and analysed the double band by mass spectrometry. Figure 4A shows the AGO2 western blot of the AGO2 immunoprecipitation sample. Both AGO2 bands, which were further analysed, are highlighted in red and blue boxes. We identified the AGO2 bands likewise after silver staining of SDS gel analyses (red and blue boxes in Figure 4B) and characterised the containing proteins of both bands after excision in mass spectrometry. The result of the mass spectrometry analyses is present in Figure 4C and both bands contained AGO2.
Figure 4.
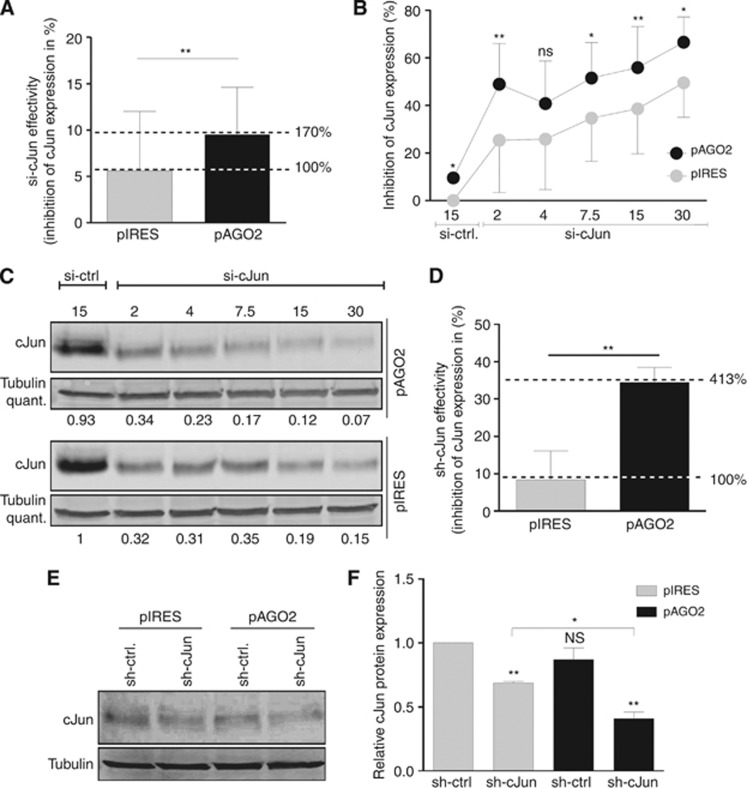
AGO2 analysis and re-expression in melanoma. (A) AGO2 western blot analysis and (B) silver staining of a AGO2 co-immunoprecipitation sample from Mel Ju cells. The blue box marked the upper (∼115 kDa) AGO2 band and the red box the lower (∼99 kDa) AGO2 band. (C) Mass spectrometry analysis of AGO2 co-immunoprecipitation demonstrates that both bands contain AGO2. (D) AGO2 western blot analysis and (E) corresponding AGO2 western blot quantifications of Mel-Ju cells after pAGO2 or pIRES (ctrl) transfections with increasing plasmid concentrations (0.5, 1 and 2 μg). The full colour version of this figure is available at British Journal of Cancer online.
To analyse the cellular consequences of AGO2 reduction in melanoma, we re-expressed AGO2 in the metastases cell line Mel Ju by using the plasmid pIRES-AGO2 (pAGO2), leading to expression of AGO2 protein with additional N-terminal HA- and Flag-tags (Meister et al, 2004). As can be seen in Figure 4D, AGO2 expression increased depending on the amount of transfected pAGO2 expression plasmid. The re-expressed AGO2 differed in size (plus 5.1 kDa) to the lower AGO2 band, based on the HA and Flag-tag modifications. The empty expression plasmid pIRES was used as a control and showed no modulation of AGO2 expression. We quantified the AGO2 western blot results to determine the amount of required pAGO2 plasmid concentrations to achieve the AGO2 expression level of NHEMs in Mel Ju cells. Figure 2B illustrates that the AGO2 level of Mel Ju cells is ∼25% compared with NHEMs. We obtained the AGO2 level of NHEMs in Mel Ju cells by transfection 2 μg pAGO2 expression plasmid.
Modulation of siRNA activity by AGO2 re-expression in melanoma
To analyse whether there is an influence of miRNA effectivity based on the melanoma-specific AGO2 reduction, we determined the efficiency of miRNA processing in the context of AGO2 modulation. We, therefore, used the transcription factor cJun as a model protein to analyse consequences of AGO2 reduction in melanoma cells for miRNA-regulated genes. cJun is strongly overexpressed in melanoma, which is a consequence of the reduced expression rate of miR-125b. The miR-125b targets cJun directly in its coding sequence (Kappelmann et al, 2013). Both are perfect attributes for the usage of cJun as a target protein to analyse the relevance of AGO2 in the miRNA pathway.
We treated Mel Ju cells with siRNA against cJun (si-cJun) or with a random siRNA (si-ctrl.) as negative control and determined the siRNA effectivity by analysing the cJun inhibition per pmol si-cJun (Figure 5A). We observed an increase in siRNA activity of 70% after AGO2 re-expression compared with mock-treated Mel Ju cells.
Figure 5.
si-cJun and sh-cJun activity in melanoma after AGO2 re-expression. (A) cJun-qRT–PCR analysis of Mel-Ju cells after 2 μg pAGO2 or 2 μg pIRES (mock) transfection and si-cJun treatment (cJun inhibition per pmol si-cJun). cJun inhibition increases after AGO2 re-expression up to 70% compared with mock. (B) cJun qRT–PCR (*P<0.05, **P<0.01) and (C) cJun western blot analyses demonstrate the increase of cJun inhibition in Mel Ju cells after pAGO2 (2 μg) or pIRES (2 μg) transfection and dose-dependent si-Jun (2, 4, 7.5, 15 and 30 pmol si-cJun and 15 pmol si-ctrl.) treatment. Tubulin was used as loading control. (D) cJun qRT–PCR analysis of Mel Ju cells treated with shRNA against cJun (2 μg pRS-cJun) or a control sh-RNA (2 μg pRS-ctrl.) demonstrate strong increased cJun inhibition after AGO2 re-expression (2 μg pAGO2) compared with mock (2 μg pIRES) transfected Mel Ju cells. (E) cJun western blot and (F) cJun western blot quantifications display the increase of cJun inhibition caused by sh-cJun (2 μg pRS-cJun) treatment in contrast to control cells (2 μg pRS-ctrl.) after AGO2 re-expression (2 μg pAGO2) compared with mock (2 μg pIRES) transfected cells. Tubulin was used as loading control.
Figure 5B illustrates the inhibition of cJun in cells treated with various si-cJun concentrations after pAGO2 and pIRES (mock) transfection (2 μg plasmid). The graphs display a strong increase of si-cJun effectivity after AGO2 re-expression. This effect can be found for all si-cJun concentrations (2 pmol, 4 pmol, 7.5 pmol, 15 pmol and 30 pmol) and is significant, with the exception of 4 pmol si-cJun. The low but reproducible differences of AGO2 re-expression for the cJun expression in si-ctrl. treated Mel Ju cells is, in our opinion, based on the AGO2 modulation of endogenous siRNA, including miR-125b.
To ensure that the AGO2 re-expression did not affect the siRNA transfection efficiency, we determined the amount of transfected fluorescence labelled siRNA (si-Glo) after AGO2 transfection in Mel Ju cells. Supplementary Figure 1S illustrates that AGO2 did not modulate the siRNA transfection efficiency.
Further, we wanted to confirm the increase in siRNA effectivity for the protein expression of the siRNA target proteins by re-expression of AGO2. Therefore, we determined and quantified the amount of cJun in western blot analyses after AGO2 re-expression and si-cJun treatment (Figure 5C). We observed a decrease of cJun corresponding to the increasing amount of si-cJun treatment in pAGO2 and pIRES (mock) transfected Mel Ju cells and also strong differences of AGO2 re-expressing cells compared with mock.
Next, we were interested to determine whether AGO2 reduction in melanoma affects only mature siRNAs or also miRNAs during their processing. Diederichs and Haber (2007) were able to show that AGO2 is also a miRNA-processing enzyme, which processes the pre-miRNA to an additional processing intermediate. This intermediate miRNA is termed ‘AGO2-cleaved precursor miRNA' or ‘ac-pre-miRNA'. Therefore, we used short hairpin (sh) RNA against cJun, which is processed by AGO2 and analysed the cJun inhibition in Mel Ju cells in relation to the amount of AGO2. Figure 5D demonstrates the significant effect of AGO2 re-expression in melanoma for the processing of miRNAs. The cJun mRNA degradation by sh-cJun is more than fourfold higher after AGO2 re-expression compared with mock (pIRES) transfection. This strong effect is also visible in the cJun western blot (Figure 5E). The sh-cJun is not strongly efficient in melanoma cells, only one-third reduction was observed compared with the control (sh-ctrl) in mock transfected cells. However, after re-expression of AGO2 the cJun inhibition doubles from 30 per cent inhibition to 60 per cent after short hairpin RNA (shRNA) treatment (Figure 5F).
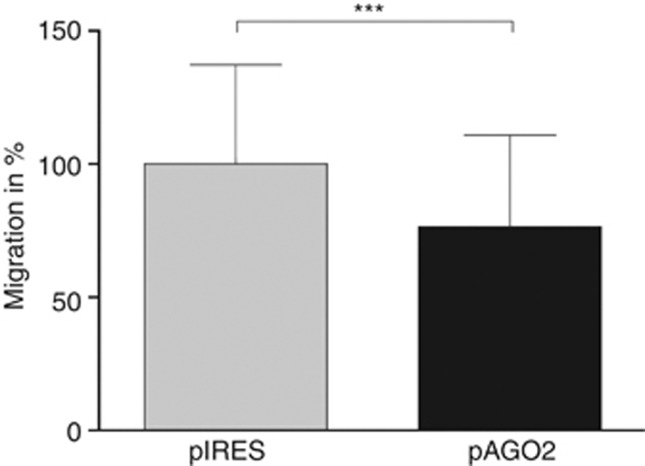
Modulation of specific siRNA or miRNA concentrations can lead to phenotypically variation of melanoma cells, for example changes in invasion (Luo et al, 2013), migration (Dynoodt et al, 2013) or proliferation (Nemlich et al, 2013). To analyse if the re-expression of AGO2 results in altered cell behaviour, we compared the migration potential of Mel Ju cells transfected with 2 μg pAGO2 with mock (2 μg pIRES) transfected cells. Figure 6 demonstrates that the migration potential of Mel Ju cells decrease about 20% after AGO2 re-expression.
Figure 6.
Melanoma migration potential after AGO2 re-expression. Boyden chamber analysis of Mel-Ju cells after 2 μg pAGO2 or 2 μg pIRES (mock) transfection to determine the migratory potential. The AGO2 re-expression reduces the migration behaviour ∼20%, ***P<0.001.
Discussion
Malignant melanoma is an aggressive tumour with a tendency to early metastasis and strong resistance to current therapeutic approaches (Russo et al, 2009; Shoo and Kashani-Sabet, 2009; Jemal et al, 2011). Aberrant expression patterns of certain miRNAs were shown to contribute to the initiation and progression of melanoma acting as so called oncomirs (Esquela-Kerscher and Slack, 2006; Mueller and Bosserhoff, 2009).
MiRNAs are known to control cell proliferation, differentiation and metabolism through their specific gene regulatory network with estimations that at least one-third of human protein-coding genes are targets of miRNAs. Further, it is known that one single miRNA can target and regulate a multitude of genes (Philippidou et al, 2010; Xu et al, 2012).
Several array studies analysed the miRNA expression pattern in melanoma in comparison with melanocytes (Mueller et al, 2009; Caramuta et al, 2010; Philippidou et al, 2010; Nazarov et al, 2013) and identified a general induction of miRNA expression with more miRNAs up- than downregulated in melanoma. Interestingly, this was not observed in other types of cancer hinting to melanoma specificity. It was speculated that regulation of genes by miRNAs could be in part differently regulated compared with other tumours. Our finding that AGO2 is strongly downregulated in melanoma suggests the RISC complex as a regulatory unit. Reduction in AGO2 would lead to preferential regulation by the miRNAs with strongest expression resulting from the competition for AGO2. Therefore, miRNAs, which are not upregulated, reduce their efficient probability to regulate their corresponding targets. This effect of shifted miRNA effectiveness is directly corresponding to the amount of miRNA and intensified the shortage of downregulated miRNAs. For example, Integrin β3 is part of the receptor for fibronectin, vitronectin and other matrix proteins and is strongly upregulated in melanoma. Overexpression of this Integrin β3 is a direct consequence of downregulated mir-let-7a and is associated with aggressive tumour growth, invasion and metastasis (Albelda et al, 1990; Muller and Bosserhoff, 2008). We speculate that the reduced amount of AGO2 is not able to load the remaining miR-let-7a efficiently and therefore extent the melanoma progression.
The competence of miRNAs to regulate multiple pathways at various checkpoints makes them in theory useful as therapeutics. Two possible strategies are practical to modulate the level of a specific miRNA (reviewed by Garzon et al (2010)). The first one is, in the case of downregulated miRNAs, to re-express the miRNA as so called pre-miRs. Second, which is more attractive in melanoma because of rather upregulated miRNAs, to express antagomiRs (miRNA-inhibitors) for inhibition of unwanted miRNAs. However, the functionality of all miRNAs depends on their availability to get processed or loaded by AGO2. We could show that AGO2 modulation directly results in altered phenotypically behaviour of melanoma. Therefore, it is necessary for the progress of therapeutically miRNA treatment in melanoma patients to understand the importance of the AGO2 reduction.
The Ago subfamily of human AGO proteins consists of four members (AGO1, AGO2, AGO3 and AGO4) and all are able to form miRNPs but they differ in their mechanism of action. Only the AGO2 and AGO3 enzyme own the ribonuclease active site in the AGO PIWI domain catalyses the cleavage of target mRNA. But in contrast to AGO2, AGO3-associated cleavage activity could not be observed (Meister et al, 2004). The specialised functions of AGO1, AGO3 and AGO4 remain unclear so far (Modzelewski et al, 2012). Therefore, ongoing analysis of the cause of reduced AGO2 expression in melanoma cells could lead to insights into fundamental process of RNase III free processing of miRNAs.
Possible explanation for the reduced amounts of AGO2 in melanoma could be the recent finding that the miRNA-processing enzymes DICER and AGO2 are targets for degradation by selective autophagy (Gibbings et al, 2012) or a fast degradation by the ubiquitin degradation pathway (Rybak et al, 2009).
Taken together, the results in this work demonstrate that the siRNA, shRNA and miRNA activity depends on the amount of AGO2. After AGO2 re-expression in melanoma cells, the functionality of siRNA and miRNA increases dramatically. This finding adds new information on the role of deregulated miRNAs in malignant melanoma. Furthermore, this study is important for a possible therapeutical potential of miRNAs for melanoma patients or general clinical research.
Acknowledgments
We would like to thank Corinna Ott and Lena Honold (Institute of Pathology, University of Regensburg, Regensburg, Germany) for critically reading the manuscript. Melanie Kappelmann (Institute of Pathology, University of Regensburg, Regensburg, Germany) for her patient support of the shRNA assays. Alexander Riechers (Institute of Pathology, University of Regensburg, Regensburg, Germany) for his grateful assistance in si-Glow measurements. Elisabeth Kremmer (Helmholtz Center Munich) for anti-AGO2 antibody production. This work was supported by the DFG (Grants to AB and GM, SFB960) and the Bavarian Ministry for education and science (BioSysNet). GM is supported by Grants from the Bavarian Ministry for education and science (BayGene), the European Union (ERC starting Grant ‘sRNAs', FP7 project ‘ONCOMIRs') and the Bundesministerium für Bildung und Forschung (BMBF, NGFN+).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Albelda SM, Mette SA, Elder DE, Stewart R, Damjanovich L, Herlyn M, Buck CA. Integrin distribution in malignant melanoma: association of the beta 3 subunit with tumour progression. Cancer Res. 1990;50:6757–6764. [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum H, Beier H, Gross H. Improved silver staining of plant proteins, RNA and DNA inpolyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- Braig S, Bosserhoff AK. Death inducer-obliterator 1 (Dido1) is a BMP target gene and promotes BMP-induced. Oncogene. 2013;32:837–848. doi: 10.1038/onc.2012.115. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Caramuta S, Egyhazi S, Rodolfo M, Witten D, Hansson J, Larsson C, Lui WO. MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. J Invest Dermatol. 2010;130:2062–2070. doi: 10.1038/jid.2010.63. [DOI] [PubMed] [Google Scholar]
- Chen J, Feilotter HE, Pare GC, Zhang X, Pemberton JG, Garady C, Lai D, Yang X, Tron VA. MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Am J Pathol. 2010;176:2520–2529. doi: 10.2353/ajpath.2010.091061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Deng H, Bi F, Liu J, Bemis LT, Norris D, Wang XJ, Zhang Q. MicroRNA-137 targets carboxyl-terminal binding protein 1 in melanoma cell lines. Int J Biol Sci. 2011;7:133–137. doi: 10.7150/ijbs.7.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Dynoodt P, Speeckaert R, De Wever O, Chevolet I, Brochez L, Lambert J, Van Gele M. miR-145 overexpression suppresses the migration and invasion of metastatic melanoma cells. Int J Oncol. 2013;42:1443–1451. doi: 10.3892/ijo.2013.1823. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings D, Mostowy S, Jay F, Schwab Y, Cossart P, Voinnet O. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol. 2012;14:1314–1321. doi: 10.1038/ncb2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauptmann J, Dueck A, Harlander S, Pfaff J, Merkl R, Meister G. Turning catalytically inactive human Argonaute proteins into active slicer enzymes. Nat Struct Mol Biol. 2013;20:814–817. doi: 10.1038/nsmb.2577. [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Braun JE, Heimstadt S, Zekri L, Izaurralde E. Two PABPC1-binding sites in GW182 proteins promote miRNA-mediated gene silencing. EMBO J. 2010;29:4146–4160. doi: 10.1038/emboj.2010.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Jacob K, Wach F, Holzapfel U, Hein R, Lengyel E, Buettner R, Bosserhoff AK. In vitro modulation of human melanoma cell invasion and proliferation by all-trans-retinoic acid. Melanoma Res. 1998;8:211–219. doi: 10.1097/00008390-199806000-00003. [DOI] [PubMed] [Google Scholar]
- Janga SC, Vallabhaneni S. MicroRNAs as post-transcriptional machines and their interplay with cellular networks. AdvExp Med Biol. 2011;722:59–74. doi: 10.1007/978-1-4614-0332-6_4. [DOI] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Kappelmann M, Kuphal S, Meister G, Vardimon L, Bosserhoff AK. MicroRNA miR-125b controls melanoma progression by direct regulation of c-Jun protein expression. Oncogene. 2013;32:2984–2991. doi: 10.1038/onc.2012.307. [DOI] [PubMed] [Google Scholar]
- Krol J, Loedige I. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Kunz M. MicroRNAs in melanoma biology. AdvExp Med Biol. 2013;774:103–120. doi: 10.1007/978-94-007-5590-1_6. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Tetteh PW, Merz PR, Dickes E, Abukiwan A, Hotz-Wagenblatt A, Holland-Cunz S, Sinnberg T, Schittek B, Schadendorf D, Diederichs S, Eichmuller SB. miR-137 inhibits the invasion of melanoma cells through downregulation of multiple oncogenic target genes. J Invest Dermatol. 2013;133:768–775. doi: 10.1038/jid.2012.357. [DOI] [PubMed] [Google Scholar]
- Ma Z, Swede H, Cassarino D, Fleming E, Fire A, Dadras SS. Up-regulated Dicer expression in patients with cutaneous melanoma. PLoS One. 2011;6:e20494. doi: 10.1371/journal.pone.0020494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Modzelewski AJ, Holmes RJ, Hilz S, Grimson A, Cohen PE. AGO4 regulates entry into meiosis and influences silencing of sex chromosomes in the male mouse germline. Dev Cell. 2012;23:251–264. doi: 10.1016/j.devcel.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller DW, Bosserhoff AK. Role of miRNAs in the progression of malignant melanoma. Br J Cancer. 2009;101:551–556. doi: 10.1038/sj.bjc.6605204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller DW, Rehli M, Bosserhoff AK. miRNA expression profiling in melanocytes and melanoma cell lines reveals miRNAs associated with formation and progression of malignant melanoma. J Invest Dermatol. 2009;129:1740–1751. doi: 10.1038/jid.2008.452. [DOI] [PubMed] [Google Scholar]
- Muller DW, Bosserhoff AK. Integrin beta 3 expression is regulated by let-7a miRNA in malignant melanoma. Oncogene. 2008;27:6698–6706. doi: 10.1038/onc.2008.282. [DOI] [PubMed] [Google Scholar]
- Nazarov PV, Reinsbach SE, Muller A, Nicot N, Philippidou D, Vallar L, Kreis S. Interplay of microRNAs, transcription factors and target genes: linking dynamic expression changes to function. Nucleic Acids Res. 2013;41:2817–2831. doi: 10.1093/nar/gks1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemlich Y, Greenberg E, Ortenberg R, Besser MJ, Barshack I, Jacob-Hirsch J, Jacoby E, Eyal E, Rivkin L, Prieto VG, Chakravarti N, Duncan LM, Kallenberg DM, Galun E, Bennett DC, Amariglio N, Bar-Eli M, Schachter J, Rechavi G, Markel G. MicroRNA-mediated loss of ADAR1 in metastatic melanoma promotes tumour growth. J Clin Invest. 2013;123:2703–2718. doi: 10.1172/JCI62980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino L, Jacob J, Roca-Alonso L, Krell J, Castellano L, Frampton AE. Altered expression of the miRNA processing endoribonuclease Dicer has prognostic significance in human cancers. Expert Rev Anticancer Ther. 2013;13:21–27. doi: 10.1586/era.12.150. [DOI] [PubMed] [Google Scholar]
- Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Philippidou D, Schmitt M, Moser D, Margue C, Nazarov PV, Muller A, Vallar L, Nashan D, Behrmann I, Kreis S. Signatures of microRNAs and selected microRNA target genes in human melanoma. Cancer Res. 2010;70:4163–4173. doi: 10.1158/0008-5472.CAN-09-4512. [DOI] [PubMed] [Google Scholar]
- Rothhammer T, Poser I, Soncin F, Bataille F, Moser M, Bosserhoff AK. Bone morphogenic proteins are overexpressed in malignant melanoma and promote cell invasion and migration. Cancer Res. 2005;65:448–456. [PubMed] [Google Scholar]
- Rudel S, Weinmann L, Kremmer E, Meister G. A multifunctional human Argonaute2-specific monoclonal antibody. RNA. 2008;14:1244–1253. doi: 10.1261/rna.973808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo AE, Torrisi E, Bevelacqua Y, Perrotta R, Libra M, McCubrey JA, Spandidos DA, Stivala F, Malaponte G. Melanoma: molecular pathogenesis and emerging target therapies (Review) Int J Oncol. 2009;34:1481–1489. doi: 10.3892/ijo_00000277. [DOI] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Hadian K, Smirnova L, Wulczyn EA, Michel G, Nitsch R, Krappmann D, Wulczyn FG. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat Cell Biol. 2009;11:1411–1420. doi: 10.1038/ncb1987. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Shoo BA, Kashani-Sabet M. Melanoma arising in African-, Asian-, Latino- and Native-American populations. SeminCutan Med Surg. 2009;28:96–102. doi: 10.1016/j.sder.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Thomas A, Klein MS, Stevens AP, Reinders Y, Hellerbrand C, Dettmer K, Gronwald W, Oefner PJ, Reinders J. Changes in the hepatic mitochondrial and membrane proteome in mice fed a. J Proteomics. 2013;80C:107–122. doi: 10.1016/j.jprot.2012.12.027. [DOI] [PubMed] [Google Scholar]
- Vasudevan S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip Rev RNA. 2012;3:311–330. doi: 10.1002/wrna.121. [DOI] [PubMed] [Google Scholar]
- Voller D, Ott C, Bosserhoff A. MicroRNAs in malignant melanoma. ClinBiochem. 2013;46:909–917. doi: 10.1016/j.clinbiochem.2013.01.008. [DOI] [PubMed] [Google Scholar]
- Wang W, Zhao LJ, Tan YX, Ren H, Qi ZT. Identification of deregulated miRNAs and their targets in hepatitis B virus-associated hepatocellular carcinoma. World J Gastroenterol. 2012;18:5442–5453. doi: 10.3748/wjg.v18.i38.5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Brenn T, Brown ER, Doherty V, Melton DW. Differential expression of microRNAs during melanoma progression: miR-200c, miR-205 and miR-211 are downregulated in melanoma and act as tumour suppressors. Br J Cancer. 2012;106:553–561. doi: 10.1038/bjc.2011.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Sun Y, Hou Y, Peng Q, Wang L, Luo H, Tang X, Zeng Z, Liu M. MiRNA expression analysis of cancer-associated fibroblasts and normal fibroblasts in breast cancer. Int J Biochem Cell Biol. 2012;44:2051–2059. doi: 10.1016/j.biocel.2012.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.