Abstract
Background:
The multidrug resistance and distant metastasis of cholangiocarcinoma result in high postoperative recurrence and low long-term survival rates. It has been demonstrated that the ectopic expression of miR-200 suppresses the multidrug resistance and metastasis of cancer. However, the expression and function of miR-200 in cholangiocarcinoma has not yet been described.
Methods:
In this study, we identified dysregulated microRNAs (miRNAs, miR) in cholangiocarcinoma tissue by microarray analysis, and subsequent real-time PCR and northern blot analyses validated the expression of candidate miR. We performed functional analyses and investigated the relationship between miR-200b/c expression and the properties of cholangiocarcinoma cells. A dual luciferase assay was applied to examine the effect of miRNAs on the 3′-UTR of target genes, and we demonstrated the function of the target gene by siRNA transfection identifying the downstream pathway via western blotting.
Results:
We found significantly downregulated expression of four miR-200 family members (miR-200a/b/c/429) and then confirmed that ectopic miR-200b/200c inhibits the migration and invasion of cholangiocarcinoma cells both in vitro and in vivo. We found that miR-200b/c influenced the tumourigenesis of cholangiocarcinoma cells including their tumour-initiating capacity, sphere formation, and drug resistance. We further found that miR-200b/c regulated migration and invasion capacities by directly targeting rho-kinase 2 and regulated tumorigenic properties by directly targeting SUZ12 (a subunit of a polycomb repressor complex).
Conclusion:
Our study shows that miR-200b/c has a critical role in the regulation of the tumorigenic and metastatic capacity of cholangiocarcinoma and reveals the probable underlying mechanisms.
Keywords: cholangiocarcinoma, microRNA, tumourigenesis, metastasis
Cholangiocarcinoma is generally acknowledged as being difficult to diagnose and treat. Despite improvements in treatment, the survival of patients remains poor (Khan et al, 2008), which is attributed to early extensive local tumour invasion and distant metastasis, as well as multichemoresistance (Khan et al, 2005; Blechacz and Gores, 2008). The development of cholangiocarcinoma is a multistep process, with an accumulation of genetic and epigenetic alterations (Evert et al, 2013; Timmer et al, 2013). Therefore, a better understanding of the molecular mechanisms of the multidrug resistance, distant metastasis, and tumorigenic properties of cholangiocarcinoma is likely to lead to novel therapeutic strategies for cholangiocarcinoma (Visvader and Lindeman, 2008).
MicroRNAs (miRNAs) are a well-known class of small RNAs that inhibit gene expression at the post-transcriptional level by directly targeting regions of sequence complementarity in the 3′-untranslated regions (3′-UTRs) of mRNAs (He and Hannon, 2004). The miR-200 family, including the miR-200a/141 and miR-200b/c/429 clusters, is involved in neurogenesis (Karres et al, 2007), the regulation of embryonic and adult stem cells, and cancer stem cells (CSCs) (Shimono et al, 2009; Wellner et al, 2009; Samavarchi-Tehrani et al, 2010), chemosensitivity, and apoptosis (Cochrane et al, 2010; Schickel et al, 2010). The miR-200 family also influences the epithelial-to-mesenchymal transition (EMT) through the Zeb–E-cadherin axis (Burk et al, 2008; Korpal et al, 2008; Park et al, 2008). However, recent studies have yielded conflicting results in terms of the tumour suppressor function of this family (Dykxhoorn et al, 2009), it remains unclear whether the metastasis suppressor functions of the miR-200 family are mediated entirely or only partially through the Zeb-E-cadherin axis (Korpal et al, 2011). Indeed, although the potential of other cancer-related functions of the miR-200 family warrants further research, few studies on the relationship between the expression levels of miRNAs and cholangiocarcinoma cells have been published. Therefore, we conducted miRNA profiling in cholangiocarcinoma tissues and found a differential expression of miRNAs, including four of the five miR-200 family members, which have been shown to have an important role in regulating the properties of metastasis and invasion. Nonetheless, the function of miR-200 in cholangiocarcinoma tumourigenesis has not been clearly elucidated.
Here, we demonstrate that miR-200b/c regulates the metastatic capacity and tumorigenic properties of cholangiocarcinoma, including tumour-initiating capacity, sphere formation, and drug resistance. We also show that miR-200b/c inhibits invasion and migration by directly targeting other genes such as rho-kinase2 (ROCK2). Moreover, miR-200b/c influences cholangiocarcinoma tumourigenesis by silencing SUZ12 (a subunit of a polycomb repressor complex). These results provide insight into the molecular mechanisms of cholangiocarcinoma regulation and may suggest a novel therapeutic strategy for cholangiocarcinoma treatment.
Materials and methods
Tumours
Human cholangiocarcinoma tissues were obtained from 14 patients with postoperative pathological diagnosed perihilar or distal biliary cholangiocarcinoma from September 2010 to May 2011 at the Department of Biliary-Pancreatic Surgery, Affiliated Tongji Hospital (Hubei, China) (Supplementary Table 1). Human sample collection procedures were approved by the China Ethical Review Committee.
MiRNA microarrays
Total RNA was extracted from three cholangiocarcinoma tissues and their corresponding normal bile ducts using a mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) according to the manufacturer's instructions. An Agilent Human miRNA Microarray Kit (V2) (Agilent Inc, Santa Clara, CA, USA) (Sanger database v.10.1) was used for analysis. RNA labelling and hybridisation on Agilent miRNA microarray chips were performed with an miRNA Labelling Reagent and Hybridization Kit (Agilent Technologies). Microarray data analyses including GO analysis, Pathway Analysis, Path-net, Signal-net, and MicroRNA-gene network were performed (Genminix Company, Shanghai, China). The microarray data were deposited in NCBI's Gene Expression Omnibus (GEO) public database (http://www.ncbi.nlm.nih.gov/geo/; Gene Expression Omnibus accession number, GSE47764).
Cell culture and transfections
The TFK-1 cell line was purchased from DSMZ by Professor Shengquan Zou of the Affiliated Tongji Hospital. The QBC939 cell line was provided by Professor Shu-guang Wang of The Third Military Medical University, Chongqing, China. All cell lines were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS), L-glutamine, and 1% penicillin/streptomycin at 37 °C in a 5% CO2 incubator. Cells were seeded in 6-well plates and transfected with miR-200b/c mimics/antagomir or negative control (NC) (Ambion) at a final concentration of 50 nM using siPORT NeoFX Transfection Agent (Ambion). Small interfering RNA and the NC were purchased from RiboBio (Guangzhou, China), The sequences of siRNA could be found in Supplementary Table 4. MiRNA-200b/c overexpression and knockdown lentiviruses, sh-SUZ12 and sh-ROCK2, were constructed by and purchased from Genechem (Shanghai, China). All transfections were carried out according to the manufacturer's instructions.
RNA extraction and RT-qPCR
Total RNA was isolated as described above. Real-time polymerase chain reaction was carried out using 50 ng or 500 ng of total RNA using a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA, USA) or PrimeScript Reverse Transcription Kit (TAKARA, Dalian, China) for miRNA and mRNA analyses, respectively. Real-time fluorescence quantitative polymerase chain reaction was performed using TaqMan MicroRNA Assays (Applied Biosystems) or SYBR Premix Ex Taq (TAKARA) for miRNA and mRNA analyses, respectively. All values were normalised to human RNU6B or β-actin endogenous controls, respectively, and run in triplicate. The sequences of primers could be found in Supplementary Table 4. Amplification and detection were performed using a CFX96 Real-Time PCR System (Bio-Rad, Hercules, CA, USA).
Northern blot
MiRNA northern blot assays was performed using the MiRNA Northern Blot Assay Kit (Signosis, Sunnyvale, CA, USA) according to the manufacturer's protocol and as described in Yu et al (2007). Total RNAs were extracted as described above. High-sensitive miRNA probe used was follows: miR-200b: 5′-TCATCATTACCAGGCAGTATTA-3′ miR-200c: 5′-TCCATCATTACCCGGCAGTATTA-3′ RNU6B: 5′-AGTATATGTGCTGCCGAAGCGAGCAC-3′. The image was acquired using a FluorChem FC2 imager (Alpha Innotech Corp, San Leandro, CA, USA). Expression of SUZ12/ROCK2 mRNA was assessed using DIG luminescent detection kit (Roche, Shanghai, China) as described (Almontashiri et al, 2013).
Wound healing assay
Cells (1 × 106 per well) were cultured in 6-well plates overnight, and then transfected with miR-200b/c mimics or antagomir and the respective controls. After 24 h of transfection, the cell layer was scratched with a sterile plastic tip, washed with PBS two times, and then cultured for 24 h with medium containing 1% FBS. Images of the plates were taken under a microscope.
Invasion assay
Transwell inserts (24-well insert, 8 μm pore size; Corning Inc., Corning, NY, USA) were used to determine the invasion capacity in vitro. Inserts were precoated with extracellular matrix gel (BD Biosciences, Bedford, MA, USA). After transfection, cells were serum starved in serum-free medium overnight, and then resuspended in medium containing 0.1% bovine serum albumin and placed in the upper chambers in triplicate. Lower chambers were filled with 10% FBS as the attractant, and the cells were incubated for 48 h. Then, the cells were removed from the upper surface of the membrane, and cells on the lower surface were fixed and stained with 0.4% paraformaldehyde and 0.1% crystal violet, respectively. Then, the stained cells were counted under a light microscope.
Liver metastasis model of cholangiocarcinoma cells in BALB/c nude mice
Female BALB/c nude mice, 4–6 weeks old, were purchased from the Animal Laboratory Unit of Peking Union Medical College (Beijing, China). The mice were maintained under standard conditions and reared according to the institutional guidelines for animal care. Lentivirus-transfected cells (1 × 105) were suspended in serum-free medium and then injected into the involucrum of the spleen. The mice underwent a lienectomy at 20 min after injection. The weight of the mice was measured once a week.
Xenograft experiments
A total of 1 × 105 transfected cells were subcutaneously injected into the right armpit of BALB/c nude mice. The weight of the mice and the diameter of tumours were measured every week.
Drug resistance experiments
A total of 2 × 105 transfected cells were first seeded into one well of a 24-well plate in the normal medium and cultured overnight. Then, the cells were treated with 5-fluorouracil at the indicated concentrations. At 24, 48, and 72 h, the survival rate was detected using a WST Cell Counting Kit-8 (Dojindo, Kumamoto, Japan), and IC50 values were calculated following the manufacturer's instructions.
Sphere culture and differentiation
Cells (1000 cells per ml) were cultured in suspension in serum-free DMEM-F12 (Hyclone, Logan, UT, USA), supplemented with B27 (1 : 50; Invitrogen, Carlsbad, CA, USA), 20 ng ml−1 epidermal growth factor (PeproTech EC, London, UK), 100 ng ml−1 basic fibroblast growth factor (PeproTech EC), and 100 ng ml−1 leukaemia inhibitory factor (Chemicon, Billerica, MA, USA) as described elsewhere (Hermann et al, 2007). Differentiation experiments were carried out as described (Yu et al, 2007).
Flow cytometry
Transfected cells were dissociated, washed two times with BD PharMingen Stain Buffer (BD Bioscience), and resuspended. Antibodies were added to the cells followed by incubation for 30 min at 4 °C. The antibodies used were CD24-FITC, CD44-APC, and CD133-PE (BD Bioscience) each at 10 μl per 1 × 106 cells. Flow cytometry was performed using a FACSAria (BD Immunocytometry Systems, Franklin Lakes, NJ, USA).
Luciferase reporter assay
The 3′-UTRs of eight putative SUZ12/ROCK2 genes were selected according to the genomic fragment of TargetScan containing the predicted miR-200b/c/429 binding sites. The mutant SUZ12/ROCK2 3′-UTR construct was designed to mutate three intermittent nucleotides complementary to the miR-200b/c/429 seed region. The TFK-1 cells were cultured in 24-well plates and transfected with 400 ng of either wild-type and one mutant pMIR/SUZ12 or three mutant pMIR/ROCK2 plasmids carrying firefly luciferase, together with 100 ng pRL-TK vector (Promega, Madison, WI, USA) carrying renilla luciferase. Transfection was performed using Lipofectamine 2000 (Invitrogen). Relative luciferase activity was calculated at 48 h post-transfection by a Dual Luciferase Reporter Assay (Promega).
Statistical analysis
Data are presented as the mean±standard error. Statistically significant differences were determined by the Student's t-test. Statistical analyses were performed by SPSS Software (version 19.0) (SPSS Inc, Chicago, IL, USA). A two-tailed value of P<0.05 was considered to indicate a statistically significant result.
Results
Differential expression of miRNAs in cholangiocarcinoma patients, and validation of microarray results by RT-qPCR
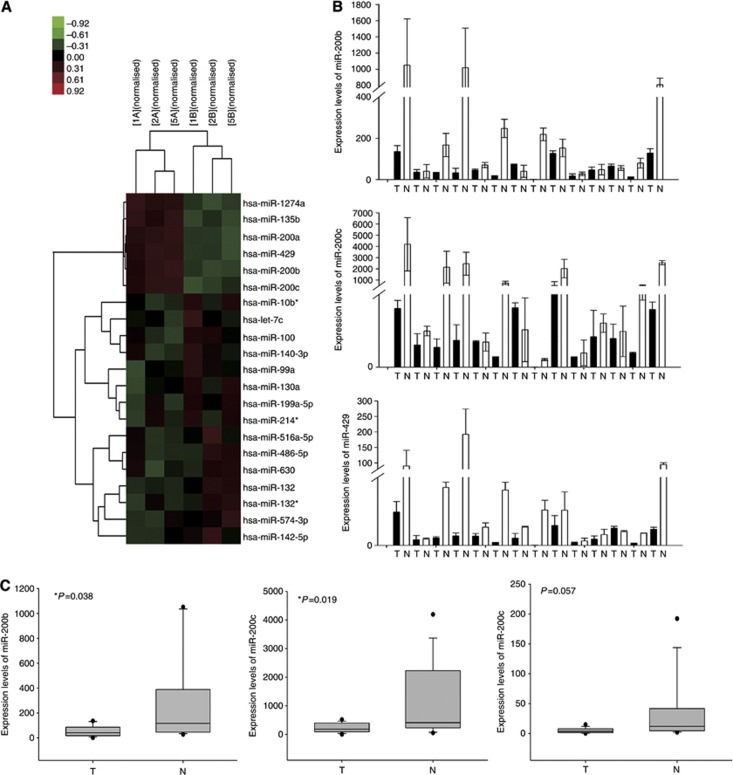
To identify miRNAs that are differentially expressed in cholangiocarcinoma, we compared the expression of miRNAs in three cholangiocarcinoma samples and the corresponding normal bile duct tissues using an Agilent miRNA Microarray. We found that 15 miRNAs were overexpressed and six miRNAs were underexpressed in the cholangiocarcinoma tumour group compared with that in the controls (Figure 1A). The 15 overexpressed miRNAs in the cholangiocarcinoma group were 2.25- to 42 222-fold overexpressed compared with the control group, and the six underexpressed miRNAs were 6.39- to 4032-fold underexpressed compared with the controls (Supplementary Table 2). Next, we predicted the possible targets of this differential expression and identified 2844 target genes. These target genes were then analysed for biological process and molecular function using information available in the Gene Ontology (GO) database. As shown in Supplementary Figure 1B and Supplementary Table 5, there were 78 upregulated GO terms and 82 downregulated terms. To determine whether the abnormal expression of these genes was due to dysregulated signalling pathways, we performed pathway analyses and found 20 upregulated pathways and 30 downregulated pathways in relation to the dysregulated genes (Supplementary Figure 1C and Supplementary Table 6). Using the significant pathway terms, a network of pathways and genes was generated using the KEGG database (Supplementary Figure 2A and B). Lastly, we identified the crucial target genes regulated by the differentially expressed miRNAs, including miR-200b and miR-200c (Supplementary Figure 1A). Three miRNAs, miR-200b, miR-200c, and miR-429, were selected for validation of the microarray results by RT-qPCR and northern blotting. The data analysis showed that miR-200b, miR-200c, and miR-429 were underexpressed in the cholangiocarcinoma group compared with the controls (Figure 1B and Supplementary Figure 7A). The data showed that the reduced expression of miR-200b and miR-200c was significant (Figure 1C, P=0.038 and P=0.019, respectively), whereas that of miR-429 was not (Figure 1C, P=0.057). The RT–PCR and northern results were consistent with those of the microarray analysis. Therefore, miR-200b and miR-200c were selected for further study.
Figure 1.
Differential expression of miRNAs in three cholangiocarcinoma samples and matching normal bile duct tissues as determined by an Agilent miRNA microarray. (A) Heatmap of miRNA profiling. A line indicates a microRNA gene, and a column indicates a sample. Red reflects a high expression value, whereas green reflects a low expression value. (B) Validation of differential miR-200b/c/429 expression in 14 cholangiocarcinoma and matching normal bile duct tissues by RT-qPCR. The expression levels of miR-200b, miR-200c, and miR-429 are shown. (C) Collected data from 14 pairs of cholangiocarcinoma patient tissues show the expression of miR-200b/c/429 compared with that in their normal bile duct tissue *P<0.05. Abbreviations: N=Normal bile duct tissue; T=cholangiocarcinoma (tumour group).
Ectopic miR-200b/c inhibits the migration and invasion of cholangiocarcinoma cells
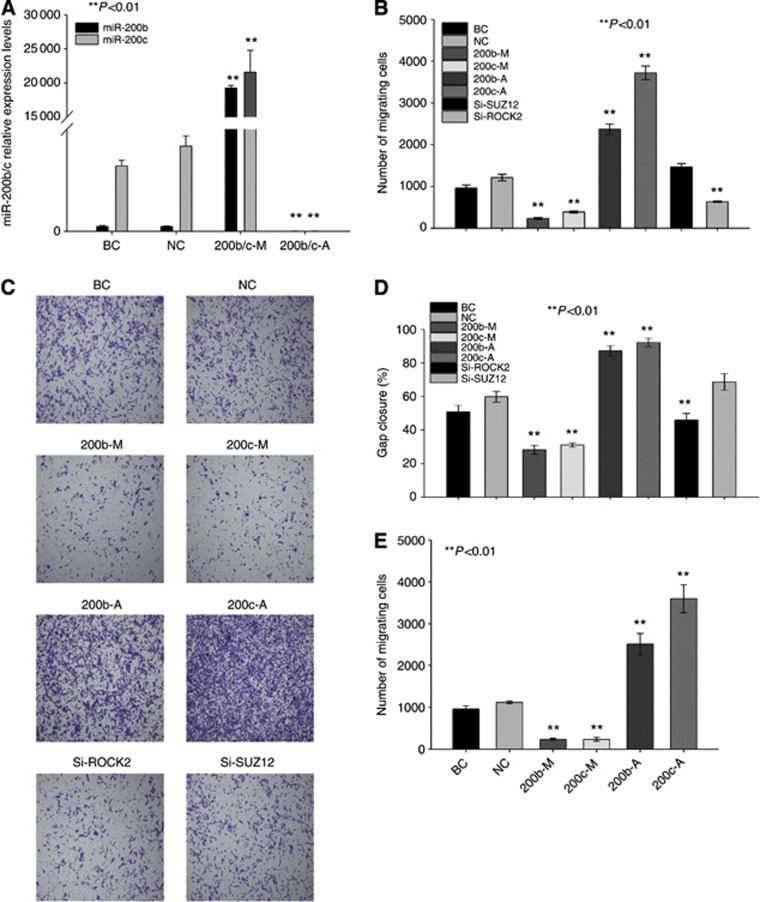
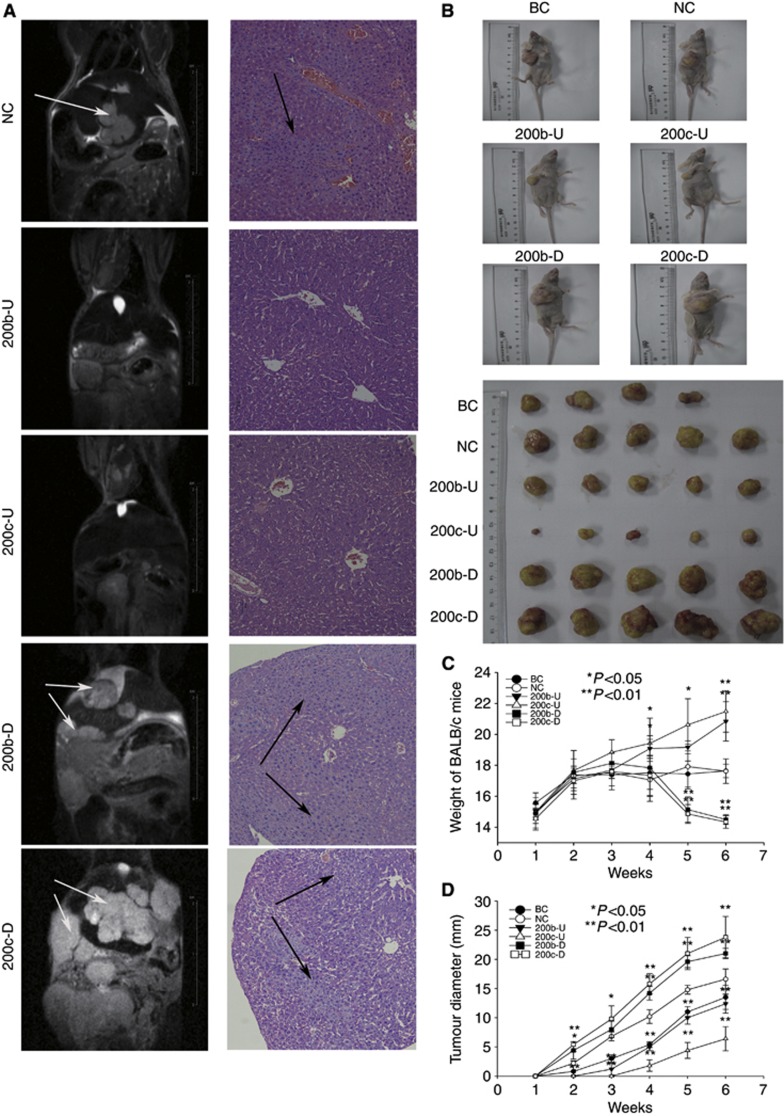
We determined whether miR-200b/c can alter the migration and invasion capacities of cholangiocarcinoma cells. As expected, the transfection of miR-200b/c mimics and antagomir into TFK-1 cells resulted in a substantial increase (by mimics) and decrease (by antagomir) of miR-200b/c expression, respectively, compared with that in the NC-transfected cells (Figure 2A, P<0.001). An invasion assay showed that the overexpression of miR-200b/c resulted in a reduction in the invasion rate of TFK-1 cells compared with that of the control (Figures 2C and B, 5.04±0.11- and 3.12±0.03-fold, respectively, P<0.001). The TFK-1 cells transfected with the miR-200b/c mimics also slowly closed wound gaps compared with the control (Figure 2D and Supplementary Figure 4, 2.13±0.24- and 1.92±0.03-fold, respectively, P<0.001). Similar results were observed using QBC939 cells (Figure 2E and Supplementary Figure 3A and B). In contrast, the knockdown of miR-200b/c promoted the invasion of the cholangiocarcinoma cells (Figure 2C): miR-200b/c knockdown in TFK-1 cells resulted in significant gap closure in the wound healing assay (Figure 2D, 0.69±0.01- and 0.65±0.03-fold, respectively, P<0.001) and a increase in cell invasion compared with that of the antagomir-NC group (Figure 2B, 1.96±0.10- and 3.07±0.19-fold, respectively, P<0.001). Moreover, we confirmed an effect of miR-200b/c on cholangiocarcinoma's metastatic capacity in vivo. We transfected TFK-1 cells with a lentivirus-based expression vector for either the overexpression or downregulation miR-200b/c, and then established a liver metastasis model of cholangiocarcinoma using these transfected cells. The results showed that the upregulation of miR-200b/c inhibited the distant metastasis of TFK-1 cells, whereas the knockdown of miR-200b/c promoted this metastatic process (Figure 3A). Because of the low resolution of MRI, we could not scan all the liver tumour metastases. Thus, we prepared serial sections of whole liver for haematoxylin and eosin (HE) staining, and observed the sections under a light microscope. According to the results, more micrometastases were detected in the miR-200b/c-knockdown group, and miR-200b/c upregulation reduced the formation of micrometastases compared with the NC groups (Figure 3A). Interestingly, miR-200c might have more of an influence on distant metastasis than miR-200b. In conclusion, these data show that miR-200b/c is an important participant in the regulation of the migration and invasion potentials of cholangiocarcinoma cells.
Figure 2.
Ectopic miR-200b/c expression inhibits the invasion and migration of cholangiocarcinoma cells. The effects of miR-200b/c mimics and an antagomir are shown in (A). (C) Upregulated miR-200b/c expression inhibited the invasion capacity of TFK-1 cells, whereas downregulated miR-200b/c expression promoted the capacity. The number of invasive cells is shown in (B). Wound-healing assay of transfected TFK-1 cells: the proportion of gap closure in the wound healing assay is shown in (D). All the data were compared with those from the NC group. MiR-200b/c upregulation inhibited the invasion capacity of QBC939 cells, whereas downregulated miR-200b/c promoted this capacity. The number of invasive cells is shown in (E). Abbreviations: BC=blank control group; NC=negative control group; 200b/c-M=200b/c-mimics group; 200b/c-A=200b/c-antagomir group.
Figure 3.
Ectopic miR-200b/c expression inhibits the migration and distant metastasis capacities of TFK-1 cells. (A) Lentivirus-transfected TFK-1 cells were used to construct a liver metastasis model of cholangiocarcinoma cancer cells. Images of the MRI scans are shown on the left line. Whole-liver serial sectioning for HE staining of the liver metastasis BALB/c-nude mice model. The micrometastases are shown on the right line. (B) Different tumour-initiating capacity of transfected TFK-1 cells. All the BALB/c-nude mice were killed at 6 weeks after injection. The weight data are shown in (C). The tumour diameters are shown in (D). All of the data were compared with those from the NC group. Abbreviations: BC=blank control group; NC=negative control group; 200b/c-U=200b/c-upregulated lentivirus group; 200b/c-D=200b/c downregulated lentivirus group.
MiR-200b/c expression is associated with 5-fluorouracil chemoresistance in cholangiocarcinoma cells
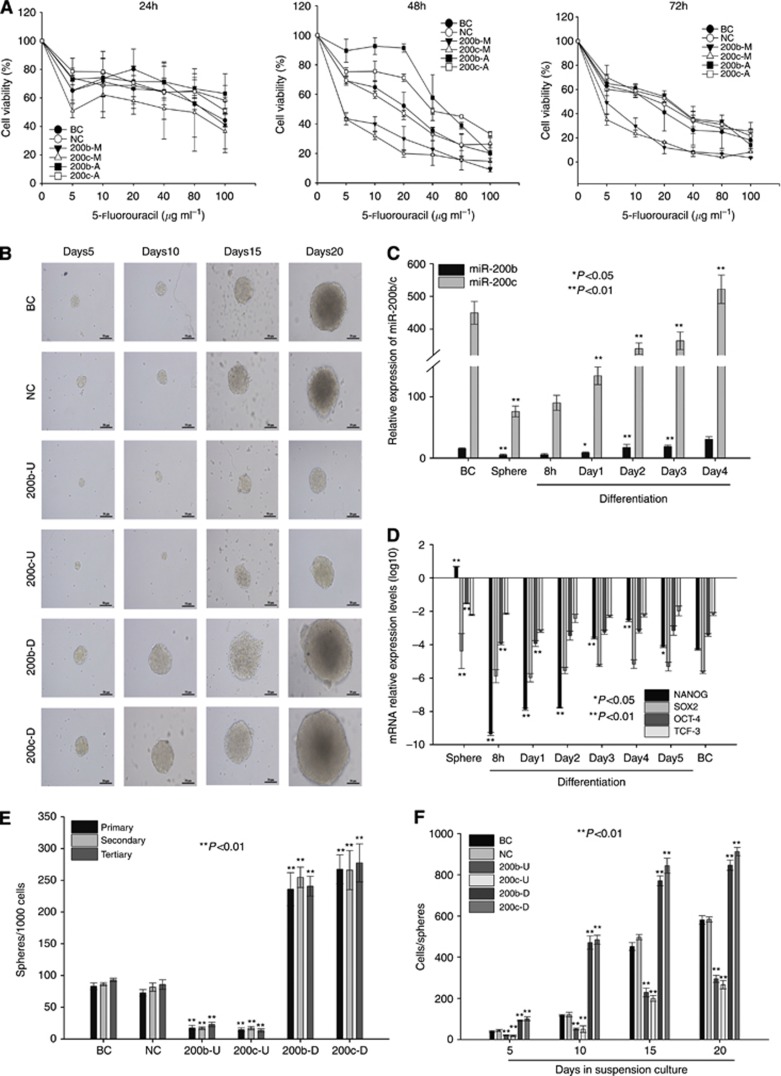
To determine whether miR-200b/c expression is also associated with chemoresistance in cholangiocarcinoma, we measured the expression of miR-200b/c in TFK-1 cells pretreated with 5-fluorouracil, which is widely used for cholangiocarcinoma treatment. The expression level of miR-200b/c in the 5-fluorouracil-pretreated TFK-1 cells was lower than that in the untreated controls (Supplementary Figure 3C). We then transfected TFK-1 cells as described above and found that pretreatment with miR-200b/c mimics promoted the cytotoxic effects of 5-fluorouracil on TFK-1 cells compared with the 5-fluorouracil+NC (Figure 4A). The IC50 values for the pre-miR-200b mimics+5-fluorouracil and pre-miR-200c mimics+5-fluorouracil groups at various time points are shown in Supplementary Table 3, with the IC50 value for the miR-200b/c mimics+5-fluorouracil group being significantly lower than that of the NC group. Conversely, the knockdown of miR-200b/c by an antagomir aggravated the chemoresistance against 5-fluorouracil. As shown in Supplementary Table 3, the IC50 values for the pre-miR-200b antagomir+5-fluorouracil and pre-miR-200c antagomir+5-fluorouracil groups at various time points were significantly higher than those of the NC+5-fluorouracil. These data illustrate that miR-200b/c significantly enhances the cytotoxic effect of 5-fluorouracil on cholangiocarcinoma cells.
Figure 4.
(A) 5-Fluorouracil chemoresistance. The proportion of cell viability is shown at 24, 48, and 72 h. (B) Different transfected TFK-1 cells were used for sphere formation and are shown at various time points (5, 10, 15, and 20 days). The expression of miR-200b and miR-200c (C) was detected during sphere formation and differentiation culture. The expression of NANOG, SOX2, TCF3, and OCT-4 mRNA during sphere formation and differentiation culture is shown in (D). The cell per sphere values (F) indicate the dimensions of the spheres. Sphere per 1000 cells (E) shows the capacity for sphere formation. All the data were compared with the data from the NC group.
Ectopic miR-200b/c expression in cholangiocarcinoma cells influences their self-renewal in vitro and tumorigenic potential in vivo, and CD133 expression can be influenced by exogenous miR-200b/c expression
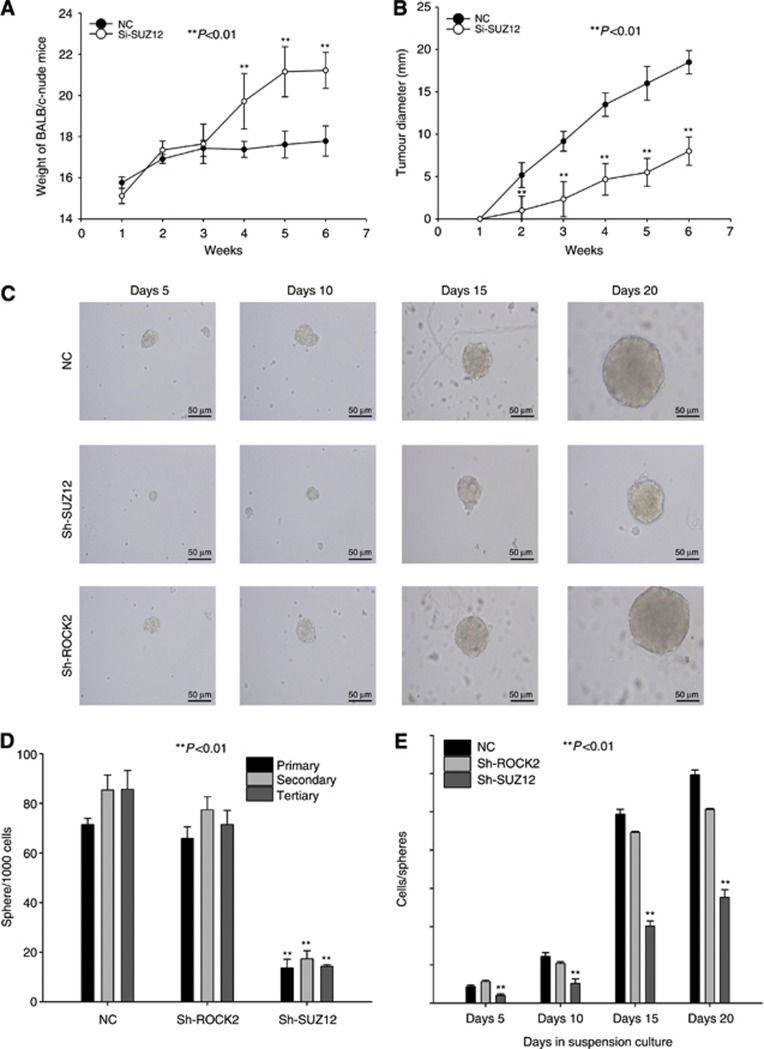
The relationship between miR-200b/c and metastasis/chemoresistance had been demonstrated, and it is well-known that there are multiple steps in tumourigenesis. To demonstrate whether miR-200b/c expression affects other aspects of cholangiocarcinoma tumorigenicity in vivo and in vitro, we transfected TFK-1 cells with the lentivirus-based expression vector, as described above. To investigate the effects of miR-200b/c on tumour formation, we next subcutaneously injected TFK-1 cells transfected with the control, miR-200b/c overexpression, and miR-200b/c knockdown vectors into the right armpit of BALB/c nude mice. Strikingly, even after 3 weeks, injection of 1 × 105 miR-200b/c-knockdown cells resulted in large tumours, whereas the miR-200b/c-overexpressing cells formed smaller tumours in five out of five mice compared with the control groups (Figure 3B, P<0.001). The tumour size was also dramatically altered, as shown in Figure 3D, the miR-200b/c-overexpressing cells formed tumours at a much slower rate than the NC group. The miR-200b/c-knockdown cells showed the opposite trend (P<0.001). The weight of the mice was also statistically significance from week 4 (Figure 3C, P<0.01). Figure 3B shows the different tumour-initiating capacities of the transfected cholangiocarcinoma cells. All these results confirm that the tumour-initiating capacity of TFK-1 cells is associated with low miR-200b/c expression, in support of our future research objectives.
To investigate whether miR-200b/c is important for influencing the tumorigenic properties of cholangiocarcinoma cells, we transfected TFK-1 cells with the lentiviral vectors described above and cultured the transfected cells in suspension under serum-free conditions to generate spheres. The number of spheres reflects the self-renewal capacity of the cells in vitro, whereas the number of cells per spheres indicates the self-renewal capacity of each sphere-generating cell (Yu et al, 2007). Over three passages, the miR-200b/c-overexpressing cells formed fewer spheres than the cells in the NC group (Figure 4E, 17.33±4.04, 16.67±2.08, and 22.67±3.51 for each passage of miR-200b-overexpressing cells, 14.33±3.05, 17.0±2.65, and 13.33±2.52 for each passage of miR-200c-overexpressing cells, and 72.67±5.13, 81.67±6.5, and 85.67±7.5 for each passage of NC cells, P<0.001). In contrast, the miR-200b/c-knockdown cells formed more spheres than the NC group over three passages (Figure 4E, 236±26, 254.7±16, and 240±15.6 for each passage of miR-200b-knockdown cells, and 267±22.7, 266±30, and 277.3±29.96 for each passage of miR-200c-knockdown cells, P<0.001). As shown in Figures 4B and F, after 20 days of culture, the miR-200b/c-knockdown cells formed larger spheres than the controls cells (847±24.6 and 912.67±20.31 vs 583.67±12.4, respectively, P<0.001), whereas the miR-200b/c-overexpressing cells formed smaller spheres than the controls cells (294.67±17.04 and 266.33±20.6, respectively, P<0.001). To imitate the differentiation of cancer cells, the dissociated spheres were plated on collagen IV in serum under differentiating conditions. After 6 days under differentiating conditions, the expression levels of differentiation markers OCT-4, NANOG, and SOX2 were decreased (Figure 4D), whereas the levels of miR-200b/c decreased after sphere formation and increased during differentiation (Figure 4C). Taken together with the slower growth rate and size of the miR-200b/c-overexpressing spheres and the inability to passage the cells as spheres, these data suggest that miR-200b/c-overexpressing spheres may have undergone the first step toward losing their self-renewal capacity. Conversely, the miR-200b/c-knockdown spheres appeared to have a greater capacity for sphere formation, indicating a higher self-renewal capacity.
Self-renewal capacity is an important characteristic of tumourigenesis and is also a potential stem-like property of cancer cells. Our previous studies showed that CD24, CD44, and CD133 are probable stem cell surface markers of cholangiocarcinoma (Wang et al, 2011a, 2011b). As our above results confirmed that miR-200b/c expression might be associated with the self-renewal capacity of cholangiocarcinoma, miR-200b/c may have a potential effect on stem-like cholangiocarcinoma cells. Therefore, we validated these results using flow cytometry by incubating the transfected TFK-1 cells, as described above, with antibodies against CD24, CD44, and CD133. As shown in Figures 5A and B, the proportion of CD133+ cells among the miR-200b/c-overexpressing cells was lower than that in the NC (29.72±0.15%, 16.55±0.11% vs 72.77±1.89%). However, the proportion of CD133+ cells among the miR-200b/c-knockdown cells was high (89.47±1.81 and 87.66±1.19%). Furthermore, the proportion of CD24+ and CD44+ cells was not related to miR-200b/c expression (Supplementary Figure 6B). Collectively, these data show that CD133 is a potential CSC marker of cholangiocarcinoma (Wang et al, 2011a), which can be inhibited by miR-200b/c.
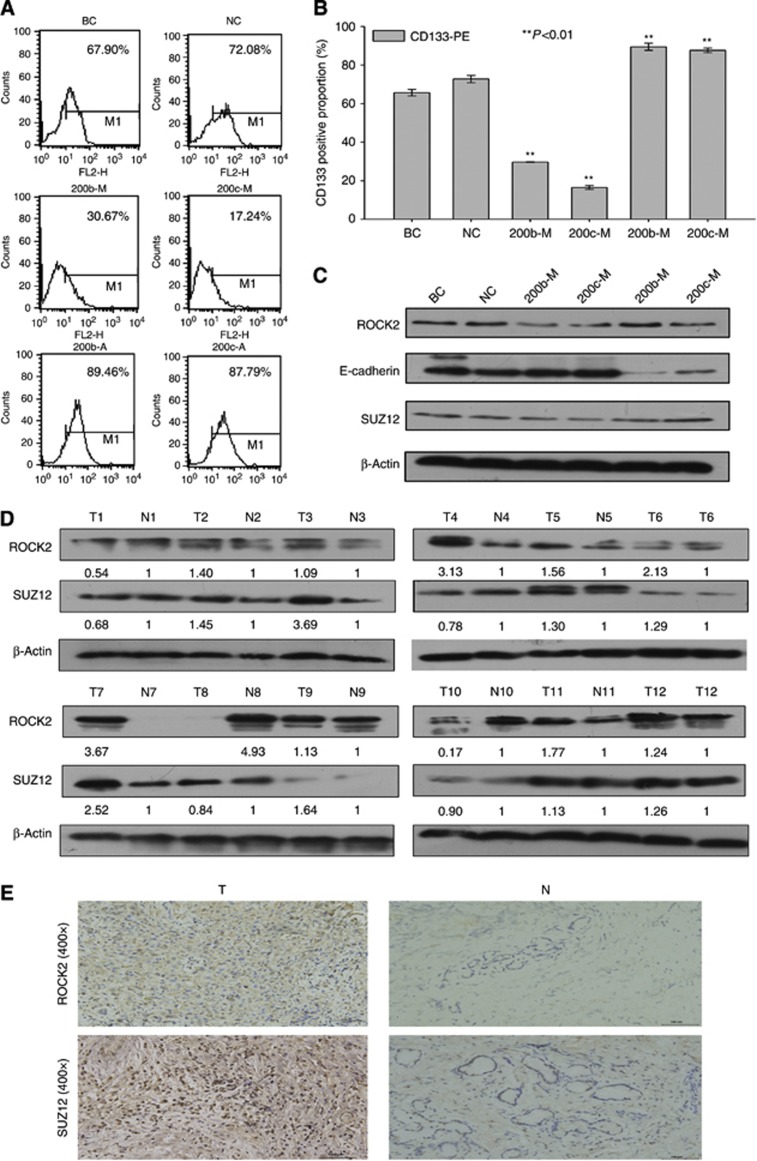
Figure 5.
(A) Proportion of CD133+ cells among transfected TFK-1 cells. At 48 h after transfection with miR-200b/c mimics/antagomir, the TFK-1 cells were resuspended, and the expression of CD133 was detected on the cell membrane. All the proportions are relative to the blank control and are shown in (B). (C) Protein expression of SUZ12, ROCK2, and E-cadherin. β-Actin was used as the control. All the data were compared with the data from the NC group. (D) Protein expression of SUZ12 and ROCK2 in 12 cholangiocarcinoma and matching normal bile duct tissues. (E) Immunohistochemistry of normal bile duct and cholangiocarcinoma tissues. The left and right images are the cholangiocarcinoma and normal bile duct tissues, respectively.
SUZ12 and ROCK2 are direct targets of miR-200b/c
According to our results, miR-200b/c affects migration, invasion, chemoresistance, tumour growth, and self-renewal capacities. Previous studies have shown that SUZ12 is a crucial CSC gene and a direct target of miR-200b in breast cancer cells. ROCK2 is an important downstream gene of the Rho/ROCK2 pathway, participating in the regulation of cytoskeletal signalling events and is crucial for cell motility (Supplementary Figures 2A and B). A TargetScan analysis (TargetScan Human 6.0) revealed that SUZ12 contains a classical and evolutionarily conserved miR-200b/c binding site in its 3′-UTR; ROCK2 contains one conserved and two poorly conserved miR-200b/c binding sites (Supplementary Figure 5). The ectopic expression of miR-200b/c in TFK-1 cells resulted in a reduction in SUZ12 and ROCK2 expression, whereas miR-200b/c inhibition led to increased expression (Figure 5C). However, their mRNA expression levels were unaffected by transfection (Supplementary Figure 7D). To further confirm whether SUZ12 and ROCK2 are direct targets of miR-200b/c, we detected the expression of the SUZ12 and ROCK2 proteins in 12 cholangiocarcinoma samples and the corresponding normal bile duct tissues using western blotting and immunohistochemistry. The results showed that both SUZ12 and ROCK2 were overexpressed in the cholangiocarcinoma tissues: 8 of 12 and 9 of 12 tumour tissues showed increased SUZ12 and ROCK2 levels, respectively, compared with the matched normal bile duct tissues (Figure 5D). Immunohistochemistry also showed that the protein expression levels of SUZ12 and ROCK2 were higher in the tumour tissues than in the normal bile duct tissues (Figure 5E, Supplementary Figures 3D and E, P=0.018 and <0.001, respectively). Regardless, there was little significant difference in the mRNA expression levels of SUZ12/ROCK2 between the cholangiocarcinoma and normal bile duct groups (Supplementary Figure 7B), suggesting that miR-200b/c might silence these two target only in a post-transcriptional manner. To further confirm our results, the miR-200b/c binding sites of SUZ12 and ROCK2 were each cloned into a luciferase reporter. When the SUZ12 wild-type sequence, but not a mutant sequence, was transfected into TFK-1 cells, we found that the ectopic expression of miR-200c resulted in a decrease in luciferase activity (SUZ12 has been reported to be direct target of miR-200b; Iliopoulos et al, 2010). Similar data were observed for ROCK2, showing that the forced expression of miR-200b/c in TFK-1 cells results in a decrease in luciferase activity using conserved wild-type binding sites compared with a mutant sequence (Figures 6A and B) and also indicating that the conserved predicted binding sites of miR-200b/c in the 3′-UTR of ROCK2 were functional. In conclusion, the above data demonstrate that SUZ12 and ROCK2 are direct target genes of miR-200b/c.
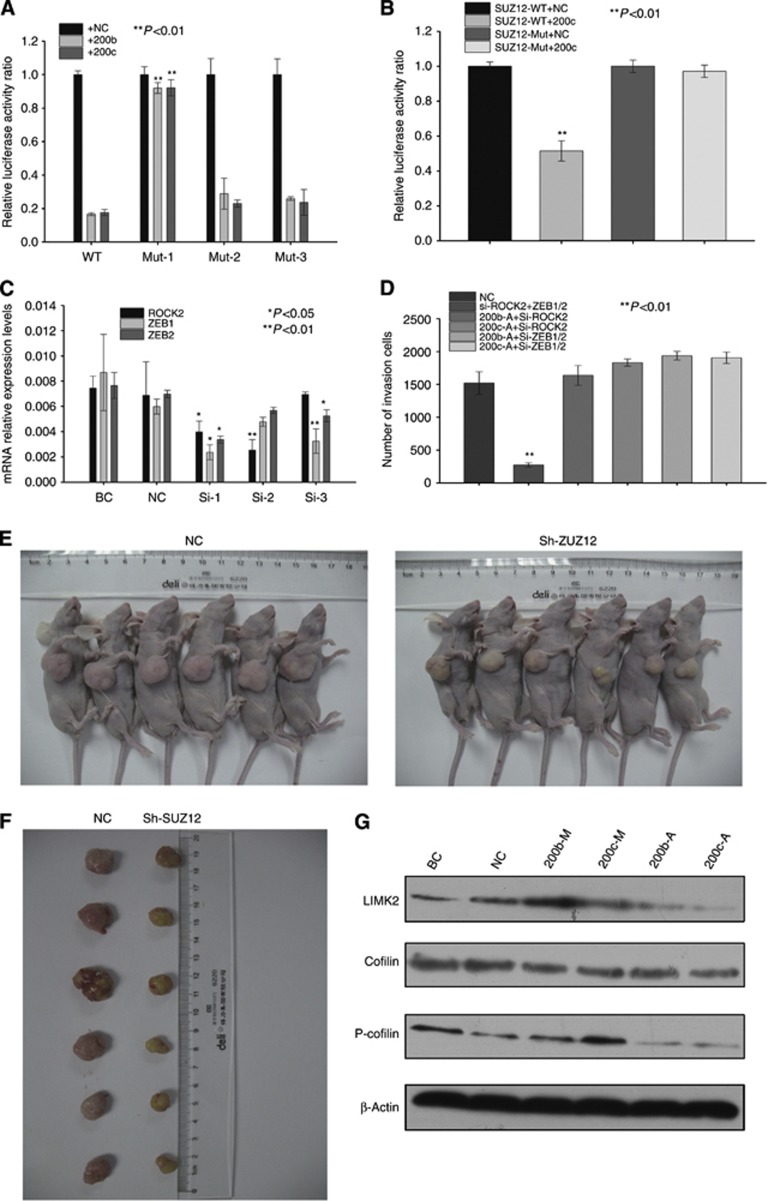
Figure 6.
Luciferase reporter assays of SUZ12 and ROCK2 expression are shown in (A) and (B). The mRNA expression of ROCK2 and ZEB1/2 after transfection of siRNA is shown in (C). Effect on the invasion capacity of TFK-1 cells after transfection with si-ROCK2+miR-200b/c antagomir, si-ZEB1/2+miR-200b/c antagomir and si-ROCK2+si-ZEB1/2. The number of invasive cells is shown in (D). (E) and (F) Lentivirus-based sh-SUZ12-transfected TFK-1 cells were used for tumour initiation. The BALB/c-nude mice were killed at 6 weeks after injection. All the data were compared with the NC group. (G) Downstream gene expression of ROCK2 after transfection with miR-200b/c mimics and antagomir.
SUZ12 and ROCK2 are potentially involved in the miR-200b/c-regulated properties of cholangiocarcinoma
Previous studies have concluded that miR-200-Zeb1/2-E-cadherin is correlated with the migration and invasion capacities of cancer cells. To determine whether SUZ12 and ROCK2 are related to these capacities, we transfected TFK-1 cells with SUZ12 and ROCK2 siRNAs to knockdown their expression and then detected the invasion capacity using a Transwell assay. The knockdown of SUZ12 did not affect the invasion capacity of TFK-1 cells: the number of invasive cells had no statistical significance compared to that of the control. In contrast, ROCK2 knockdown reduced the number of invasive cells compared with the controls (Figures 2B and C). Furthermore, we transfected TFK-1 cells with ROCK2 and ZEB1/2 siRNAs (Figure 6C), to confirm their involvement in the migration and invasion capacities of cancer cells. However, the silencing of neither ROCK2 nor ZEB1/2 in the miR-200b/c-knockdown cells could recover the invasive capacity compared with that of the controls. In addition, the silencing of both ROCK2 and ZEB1/2 resulted in the lowest number of invasive cells (Supplementary Figures 6A and D). Collectively, the results showed that the effect of miR-200b/c on the invasion capacities of cholangiocarcinoma cells is mediated by both the Zeb1/2-E-cadherin axis and also the ROCK2 pathway, whereas SUZ12 is not associated with the migration and invasion capacities of cholangiocarcinoma cells. Furthermore, we transfected TFK-1 cells with miR-200b/c mimics/antagomir and detected selected downstream genes of the ROCK2 pathway. As shown in Figures 5C and 6G, the overexpression of miR-200b/c decreased the protein expression level of ROCK2 and then increased the expression level of LIMK2. Increased LIMK2 could phosphorylate Cofilin and thus might reduce the invasion capacity of cholangiocarcinoma cells.
To confirm the mechanism regulating the effect of SUZ12 on the tumourigenesis properties of cholangiocarcinoma, we transfected TFK-1 cells with a lentivirus-based shRNA vector to silence the SUZ12 and ROCK2 genes (Supplementary Figure 6C) and we repeated the sphere formation and tumour-initiating assays. The SUZ12 shRNA group formed smaller spheres than the NC (Figure 7C), and the spheres of the control group increased in size and cell number more quickly than those of the SUZ12 shRNA group (Figure 7D and E). However, the sphere formation capacity of the sh-ROCK2 shRNA group showed no statistical significance compared with the NC group (Figure 7C–E). These changes were also associated with the self-renewal capacity, and the blank vector group formed tumours much more rapidly than the SUZ12-shRNA group (Figures 6E, F and 7A, B). However, the chemoresistance capacities of the SUZ12 shRNA group were not significant in comparison to the NC group. Collectively, the results indicate that SUZ12 is an important gene that is associated with some cholangiocarcinoma properties, such as sphere formation and tumour initiation, and is regulated by miR-200b/c.
Figure 7.
The weight data for the BALB/c-nude mice from the sh-SUZ12 tumour-initiating assay are shown in (A). The data for the tumour diameters are shown in (B). Lentivirus-based si-SUZ12-transfected TFK-1 cells were used for the sphere culture and differentiation assay (C). The cell per sphere values (D) and sphere per 1000 cell (E) values were determined.
Discussion
The initial studies on cholangiocarcinoma focused on its multichemoresistance and distant metastasis. To investigate the possible mechanism of the malignant biological behaviour of cholangiocarcinoma and to identify crucial regulators, we established that there was a dysregulated miRNA profile in cholangiocarcinoma tissues and found four miR-200 family members among the top six down-regulated miRNAs. Although the miR-200 family members have been demonstrated to be cancer suppressors in many forms of cancer, the function of miR-200 in cholangiocarcinoma cells has not been elucidated to date. Here, we investigated the dysregulation of miR-200b/c as a possible cause of the invasion and metastasis capacities and tumorigenic properties of cholangiocarcinoma, and we clarify a novel downstream mechanism. Overall, our results suggest a novel therapeutic strategy for cholangiocarcinoma treatment.
In comparison to normal controls, miR-200 family members are downregulated in many human cancers, including breast cancer (Teng et al, 2013), pancreatic tumours (Li et al, 2010), and hepatic cancer (Kim et al, 2011; Zhang et al, 2013). Most researchers have demonstrated that the expression of miR-200 is negatively correlated with the invasion and metastasis capacities of cancer (Hur et al, 2012; Soubani et al, 2012; Guo et al, 2013). Although the existence of the miR-200-ZEB1/2-E-cadherin axis during the EMT process was confirmed in initial studies (Teng et al, 2013), several recent studies have reported conflicting roles of miR-200 in metastatic progression. These contradictory findings may in part be due to the different source organisation and models (Korpal et al, 2011); indeed, there are different expression levels and mechanisms in different tissues. Thus, in our study, we demonstrated that miR-200b/c is downregulated in cholangiocarcinoma and that the knockdown of miR-200b/c promotes the invasion and metastasis capacities of cholangiocarcinoma cells. Similarly, we demonstrated that inhibiting the expression of ROCK2 reduces the invasion of cholangiocarcinoma cells (although the inhibition of SUZ12 had no effect). Both the overexpression of miR-200b/c and inhibition of ROCK2 can reduce the invasion of cholangiocarcinoma cells, yet ROCK2-knockdown cholangiocarcinoma cells may retain a stronger invasion capacity. The ROCK2-knockdown cholangiocarcinoma cells exhibited enhanced invasion and metastasis capacities in comparison to the miR-200b/c-overexpression cells, indicating that the influence of miR-200 on cholangiocarcinoma invasion extends beyond the regulation of ROCK2 and most likely involves other targets. We found that the combined suppression of ZEB1/2 and ROCK2 resulted in a similar inhibition effect as that of miR-200b/c overexpression. Therefore, both ZEB1/2 and ROCK2 are crucial regulatory proteins of the invasion capacity of cholangiocarcinoma cells and are silenced by miR-200b/c.
In addition to the invasion and metastasis capacities, many researchers have demonstrated that the expression of miR-200 is associated with the stemness of cancer cells. Indeed, our experiments examining sphere formation, drug resistance, and tumour-initiating capacity addressed CSC-like properties, suggesting a possible link between miR-200b/c and cholangiocarcinoma stem-like cells. Previous studies have shown that stemness-related genes such as those of the ZEB family, which have been demonstrated to be direct targets of miR-200, consist of transcriptional repressors, including ZEB1 and ZEB2. The functions of this protein family are extensive and include development, the activation of the transforming growth factor (TGF)-β signalling pathway in EMT, and stem cell pathways (Vandewalle et al, 2009; Sanchez-Tillo et al, 2011; Gheldof et al, 2012). According to the miR-200-ZEB dual loop theory, not only does the miR-200 family directly target ZEB1/2 but the overexpression of ZEB1/2 also reduces miR-200 levels. Based on the above findings, there may be two pathways by which the miR-200 family influences the stemness of cancer: (1) the suppression of ZEB1/2, thereby influencing the stemness of CSCs via downstream proteins, or (2) the direct targeting of certain stemness-related proteins, thereby directly influencing CSC properties. There are many studies supporting the hypotheses of pathway 1, but less experimental evidence supports pathway 2. Remarkably, our results show that the protein expression level of ZEB1 in TFK-1 cells is deficient, suggesting that the proportion of ZEB family related CSC-like cells in the TFK-1 cell line may be very low. Our integrated analysis of miR-200 and stemness shows that miR-200 regulates a very large number of target mRNA sequences. Thus, it appears likely that the effect of miR-200 on cholangiocarcinoma tumorigenic or stem cell-like properties is not mediated solely through the miR-200-ZEB axis and that there are some target mRNAs directly regulated by miR-200. The miR-200 family members may influence the stemness of cancer via direct suppression. Our results establish that miR-200b/c influences the drug resistance, tumour-initiating capacity, sphere formation, and CD133 expression level, a potential CSC marker, of cholangiocarcinoma cells. SUZ12 has been reported to be a direct target of miR-200b in breast CSCs, and the direct targeting of SUZ12 by miR-200c was demonstrated in our study. The direct knockdown of SUZ12 by shRNA achieved a similar suppressive effect on the sphere formation and tumour-initiating capacities of cholangiocarcinoma as that of miR-200b/c. This result confirms that SUZ12 is important in tumourigenesis and that related genes may be directly regulated by miR-200b/c. Based on the enrichment of low-differentiation cholangiocarcinoma cells by sphere formation and drug resistance, we considered that the proportion of cholangiocarcinoma stem-like cells had been increased, and we next validated the miR-200b/c expression levels of the parental and differentiated spheres and drug-resistant cells. Interestingly, the expression of miR-200b/c was decreased during low-differentiation cell enrichment and increased upon differentiation, indicating that there may be negative feedback between miR-200b/c and cholangiocarcinoma stem-like cells.
In conclusion, our findings demonstrate that miR-200b/c inhibits the invasion and metastasis capacities, and tumorigenic capacity (or the so-called CSC-like properties), including drug resistance, tumour-initiating capacity, sphere formation, and CD133 expression level, of cholangiocarcinoma cells. Such a suppression of these properties can be imitated by directly repressing ROCK2 and SUZ12, two direct targets of miR-200b/c. Thus, our observations provide very strong experimental results regarding the involvement of miR-200 in the invasion and metastasis capacities of cholangiocarcinoma, suggesting a novel therapeutic strategy for cholangiocarcinoma treatment and proposing a potential surface marker for cholangiocarcinoma stem-like cells.
Acknowledgments
This study was funded by The National Natural Science Foundation of China (nos. 81071775 and 81272659) to RQ, no. 81101621 to MW, no. 81160311 to JJ, no. 81172064 to MS, no. 81001068 to RT, no. 81272425 to XW, and no. 81101870 to Jun Hu; National ‘Eleventh Five-Year' Scientific and Technological Support Projects (no. 2006BAI02A13-402) to RQ; Key Projects of Science Foundation of Hubei Province (no. 2011CDA030) to RQ; and Research Fund of Young Scholars for the Doctoral Program of Higher Education of China (no. 20110142120014) to MW.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Almontashiri NA, Fan M, Cheng BL, Chen HH, Roberts R, Stewart AF. Interferon-gamma activates expression of p15 and p16 regardless of 9p21.3 coronary artery disease risk genotype. J Am Coll Cardiol. 2013;61 (2:143–147. doi: 10.1016/j.jacc.2012.08.1020. [DOI] [PubMed] [Google Scholar]
- Blechacz B, Gores GJ. Cholangiocarcinoma: advances in pathogenesis, diagnosis, and treatment. Hepatology. 2008;48 (1:308–321. doi: 10.1002/hep.22310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9 (6:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane DR, Howe EN, Spoelstra NS, Richer JK. Loss of miR-200c: a marker of aggressiveness and chemoresistance in female reproductive cancers. J Oncol. 2010;2010:821717. doi: 10.1155/2010/821717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykxhoorn DM, Wu Y, Xie H, Yu F, Lal A, Petrocca F, Martinvalet D, Song E, Lim B, Lieberman J. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS One. 2009;4 (9:e7181. doi: 10.1371/journal.pone.0007181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evert M, Dombrowski F, Biao F, Ribback S, Chen X, Calvisi DF.2013On the role of Notch1 and adult hepatocytes in murine intrahepatic cholangiocarcinoma development Hepatologye-pub ahead of print 23 March 2013doi: 10.1002/hep.26411 [DOI] [PubMed]
- Gheldof A, Hulpiau P, van Roy F, De Craene B, Berx G. Evolutionary functional analysis and molecular regulation of the ZEB transcription factors. Cell Mol Life Sci. 2012;69 (15:2527–2541. doi: 10.1007/s00018-012-0935-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Chen C, Shi M, Wang F, Chen X, Diao D, Hu M, Yu M, Qian L, Guo N.2013Stat3-coordinated Lin-28-let-7-HMGA2 and miR-200-ZEB1 circuits initiate and maintain oncostatin M-driven epithelial-mesenchymal transition Oncogenee-pub ahead of print 14 January 2013doi: 10.1038/onc.2012.573 [DOI] [PubMed]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5 (7:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1 (3:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Hur K, Toiyama Y, Takahashi M, Balaguer F, Nagasaka T, Koike J, Hemmi H, Koi M, Boland CR, Goel A. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2012;62 (9:1315–1326. doi: 10.1136/gutjnl-2011-301846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, Struhl K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol cell. 2010;39 (5:761–772. doi: 10.1016/j.molcel.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 2007;131 (1:136–145. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- Khan SA, Thomas HC, Davidson BR, Taylor-Robinson SD. Cholangiocarcinoma. Lancet. 2005;366 (9493:1303–1314. doi: 10.1016/S0140-6736(05)67530-7. [DOI] [PubMed] [Google Scholar]
- Khan SA, Toledano MB, Taylor-Robinson SD. Epidemiology, risk factors, and pathogenesis of cholangiocarcinoma. Off J Int Hepato Pancreato Biliary Assoc. 2008;10 (2:77–82. doi: 10.1080/13651820801992641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Veronese A, Pichiorri F, Lee TJ, Jeon YJ, Volinia S, Pineau P, Marchio A, Palatini J, Suh SS, Alder H, Liu CG, Dejean A, Croce CM. p53 regulates epithelial–mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J Exp Med. 2011;208 (5:875–883. doi: 10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celia-Terrassa T, Mercatali L, Khan Z, Goodarzi H, Hua Y, Wei Y, Hu G, Garcia BA, Ragoussis J, Amadori D, Harris AL, Kang Y. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat Med. 2011;17 (9:1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283 (22:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Omura N, Hong SM, Vincent A, Walter K, Griffith M, Borges M, Goggins M. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res. 2010;70 (13:5226–5237. doi: 10.1158/0008-5472.CAN-09-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22 (7:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7 (1:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Sanchez-Tillo E, Siles L, de Barrios O, Cuatrecasas M, Vaquero EC, Castells A, Postigo A. Expanding roles of ZEB factors in tumorigenesis and tumor progression. Am J Cancer Res. 2011;1 (7:897–912. [PMC free article] [PubMed] [Google Scholar]
- Schickel R, Park SM, Murmann AE, Peter ME. miR-200c regulates induction of apoptosis through CD95 by targeting FAP-1. Mol Cell. 2010;38 (6:908–915. doi: 10.1016/j.molcel.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, Dirbas FM, Somlo G, Pera RA, Lao K, Clarke MF. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138 (3:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubani O, Ali AS, Logna F, Ali S, Philip PA, Sarkar FH. Re-expression of miR-200 by novel approaches regulates the expression of PTEN and MT1-MMP in pancreatic cancer. Carcinogenesis. 2012;33 (8:1563–1571. doi: 10.1093/carcin/bgs189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y, Mei Y, Hawthorn L, Cowell JK.2013WASF3 regulates miR-200 inactivation by ZEB1 through suppression of KISS1 leading to increased invasiveness in breast cancer cells Oncogenee-pub ahead of print January 14 2013doi: 10.1038/onc.2012.565 [DOI] [PMC free article] [PubMed]
- Timmer MR, Beuers U, Fockens P, Ponsioen CY, Rauws EA, Wang KK, Krishnadath KK. Genetic and epigenetic abnormalities in primary sclerosing cholangitis-associated cholangiocarcinoma. Inflamm Bowel Dis. 2013;19 (8:1789–1797. doi: 10.1097/MIB.0b013e318281f49a. [DOI] [PubMed] [Google Scholar]
- Vandewalle C, Van Roy F, Berx G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci. 2009;66 (5:773–787. doi: 10.1007/s00018-008-8465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8 (10:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- Wang M, Xiao J, Jiang J, Qin R. CD133 and ALDH may be the molecular markers of cholangiocarcinoma stem cells. Int J Cancer. 2011;128 (8:1996–1997. doi: 10.1002/ijc.25519. [DOI] [PubMed] [Google Scholar]
- Wang M, Xiao J, Shen M, Yahong Y, Tian R, Zhu F, Jiang J, Du Z, Hu J, Liu W, Qin R. Isolation and characterization of tumorigenic extrahepatic cholangiocarcinoma cells with stem cell-like properties. Int J Cancer. 2011;128 (1:72–81. doi: 10.1002/ijc.25317. [DOI] [PubMed] [Google Scholar]
- Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, Brunton VG, Morton J, Sansom O, Schuler J, Stemmler MP, Herzberger C, Hopt U, Keck T, Brabletz S, Brabletz T. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11 (12:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131 (6:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yang F, Yuan JH, Yuan SX, Zhou WP, Huo XS, Xu D, Bi HS, Wang F, Sun SH. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis. 2013;34 (3:577–586. doi: 10.1093/carcin/bgs381. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.