Abstract
Background:
Glutathione S-transferase mu 3 (GSTM3) has been proven to be downregulated in renal cell carcinoma (RCC). We aimed to characterise the role of GSTM3 and its genetic predisposition on the occurrence and postoperative prognosis of RCC.
Methods:
The effect of GSTM3 on RCC aggressiveness was examined using transfection and silencing methods. Glutathione S-transferase mu 3 expression in renal tissues was examined by immunohistochemistry. The associations of rs1332018 (A-63C) and rs7483 (V224I) polymorphisms with RCC risk were examined using 400 RCC patients and 802 healthy controls. The factors contributing to postoperative disease-specific survival of RCC patients were evaluated using the Cox proportional hazard model.
Results:
Glutathione S-transferase mu 3 silencing increased the invasion and anchorage-independent growth of RCC cell lines. rs1332018 (AC+CC vs AA), which correlated with low expression of GSTM3 in kidney, was associated with RCC risk (odds ratio, 1.446; 95% confidence interval (CI), 1.111–1.882). rs1332018 variants and low GSTM3 expression significantly predicted unfavourable postoperative survivals of RCC patients (P<0.05). rs1332018 variants independently predicted a poor prognosis (hazard ratio, 2.119; 95% CI, 1.043–4.307).
Conclusion:
Glutathione S-transferase mu 3 may function as a tumour suppressor in RCC. rs1332018 genetic variants predispose the host to downregulating GSTM3 expression in kidney, facilitate carcinogenesis, and predict an unfavourable postoperative prognosis of RCC.
Keywords: renal cell carcinoma, GSTM3, polymorphism, risk, prognosis
Renal cell carcinoma (RCC) is a highly heterogeneous tumour, accounting for 90–95% of total kidney tumours. Clear cell RCC (ccRCC) is the major histological subtype (approximately 80%). Although the RCC incidence rate varies worldwide, it has continued to rise over the past two decades (Chow et al, 2010). Renal cell carcinoma incidence is two-fold higher in men than in women and its prognosis differs by race (Chow et al, 2010, 2013; Motzer et al, 2013). Surgical resection remains the mainstay of curative treatment. Renal cell carcinoma does not respond to radiotherapy and chemotherapy, but partially responds to targeted therapies or immunotherapy. Although the use of targeted therapies for advanced RCC is promising, there are many challenges for the application of these new approaches (Angevin et al, 2013; Figlin et al, 2013; Motzer et al, 2013). Little progress has been made with regard to the development of specific risk or prognostic biomarkers for RCC.
Compelling epidemiological evidence suggest that occupational exposure to trichloroethylene and pesticide, hypertension, smoking, obesity, urolithiasis, nitrite from animal food, and diabetes are risk factors, whereas green tea intake, cruciferous and dark green vegetables, increased physical activity, the use of statins, alcohol consumption, and occupational ultraviolet exposure are protective factors of ccRCC (Chow et al, 2010; Lew et al, 2011; Wang et al, 2012; Dellavalle et al, 2013). Polymorphisms of xenobiotic metabolism-related genes such as glutathione S-transferase mu (GSTM), cytochrome P450 mono-oxygenases, and folate metabolism genes and their interactions with environmental exposure are related to RCC risk (Semenza et al, 2001; Karami et al, 2008; Moore et al, 2008; Simic et al, 2009). Recent genome-wide association studies (GWAS) revealed that specific loci mapped on 2p21 (rs11894252), 2q22.3 (rs12105918), 11q13.3 (rs7105934), 12p11.23 (rs718314, rs1049380), and 12q24.31 (rs4765623) were significantly associated with RCC susceptibility (Purdue et al, 2011; Schödel et al, 2012; Wu et al, 2012; Henrion et al, 2013). However, the genetic risk factors found in European population are sometimes unable to be replicated in Asians (Su et al, 2013). Recent studies have shown that some molecules in tumours, genetic variations, and the neutrophil–lymphocyte ratio predict poor prognosis of RCC (Hirata et al, 2007; Kleinrath et al, 2007; Lin et al, 2010; Doberstein et al, 2011; Li et al, 2011; Beuselinck et al, 2013; Pichler et al, 2013). However, the causal factors for this malignancy in Asians remain obscure.
In our previous study, we identified a group of deregulated genes in ccRCC using cDNA microarray and GSTM3, a GSTM-class subunit, was found to be not only downregulated in metastatic ccRCC cells compared with primary ccRCC cells, but also downregulated in primary ccRCC tissues compared the adjacent normal renal tissues (Tan et al, 2008). The role of GSTM3 in the occurrence and progression of RCC is not clear. Here we report that GSTM3 can function as a tumour suppressor in RCC and a specific polymorphism predisposing to downregulating GSTM3 in kidney is significantly associated with the occurrence and postoperative prognosis of RCC.
Materials and methods
Cell culture, transfection, and knockdown of GSTM3
NRCC, a ccRCC cell line isolated from a patient without distant metastasis in our laboratory (Tan et al, 2013), and 786-O and ACHN ccRCC cell lines purchased from American Tissue Culture Collection (ATCC) (Manassas, VA, USA) within 2 months before our in vitro study were maintained in DMEM or RPMI 1640 (Hyclone, Thermo Fisher Scientific, Pittsburgh, PA, USA) supplemented with 10% fetal calf serum (GIBCO, Invitrogen, Grand Island, NY, USA), 100 U ml–1 penicillin, and 100 μg ml–1 streptomycin. A full-length of human GSTM3 cDNA clone (MHS4771-99611057) was purchased from Thermo Fisher Scientific (Pittsburgh, PA, USA), released by BamHI and XhoI digestion, and inserted into mammalian expression vector pcDNA3.1/V5-His TOPO (Invitrogen, Carlsbad, CA, USA). After identified with direct DNA sequencing, pcDNA3.1/V5-His TOPO-GSTM3 construct was transfected into NRCC cells using lipofectamine 2000 Reagent (Invitrogen, Grand Island, NY, USA). Stable GSTM3-expressing cells were selected in the presence of 1400 μg ml–1 G418 (Sigma-Aldrich, St Louis, MO, USA) for 14 days. Transient transfection of pcDNA3.1/V5-His TOPO-GSTM3 construct into 786-O and ACHN cells was performed using FuGENE HD Transfection Reagent (Promega, Madison, WI, USA) and lipofectamine LTX and Plus Reagent (Invitrogen, Grand Island, NY, USA) without antibiotics selection, respectively.
The retrovirus packing vector Pegpam 3e, RDF vector and pSuper-retro vector were kindly provided by Dr Yang (Sun et al, 2010). The sequences of small hairpin RNA (shRNA) targeting to GSTM3 were 5′-AATCGCTGCCTACTTACAGTC-3′ (shGSTM3-1); 5′-AACAACAAGATGCCCAGTGG-3′ (shGSTM3-2); 5′-CTGGCATCGGTGTGGATGA-3′ (shsiscramble: a pSUPER-vector construct containing a non-gene targeting DNA sequence, served as a negative control). Retroviral supernatants were generated as previously described (Sun et al, 2010). NRCC cells were incubated with virus-containing medium supplemented with 4 μg ml–1 polybrene (Sigma-Aldrich). Stable GSTM3-knockdown cells were selected in the presence of 3.5 μg ml–1 puromycin (Sigma-Aldrich) for 7 days. Transient transfection of shRNA constructs targeted to GSTM3 was performed in 786-O and ACHN cells without antibiotics selection.
Glutathione S-transferase mu 3 expression was examined by quantitative reverse transcription PCR (qRT-PCR) and western blot as previously described (Sun et al, 2010; Tan et al, 2013). The primers for the amplification of GSTM3 cDNA were 5′-TTGGAAGAGCTACCTGGACAA-3′ (forward) and 5′-CCTTCAGGTTTGG GAACTCA-3′ (reverse), for GAPDH (an internal control) were 5′-GACCCCTTCATTGACCTCAAC-3′ (forward) and 5′-CTTCTCCATGGTGGTGAAGA-3′ (reverse).
Anchorage-independent growth, invasion, and cell cycle analysis
Anchorage-independent growth potential was evaluated by double-layered soft agarose culture system. Cell invasion assay was performed using 8-μm pore size 24-well tissue culture Transwell plates (Corning, NY, USA). Cell cycle of NRCC-shsiscramble and NRCC-shGSTM3-1 cells was examined with flow cytometry (FACSCalibur, BD Biosciences, San Jose, CA, USA). These assays were carried out as previously described (Tan et al, 2013). All examinations were performed in triplicate.
Study population
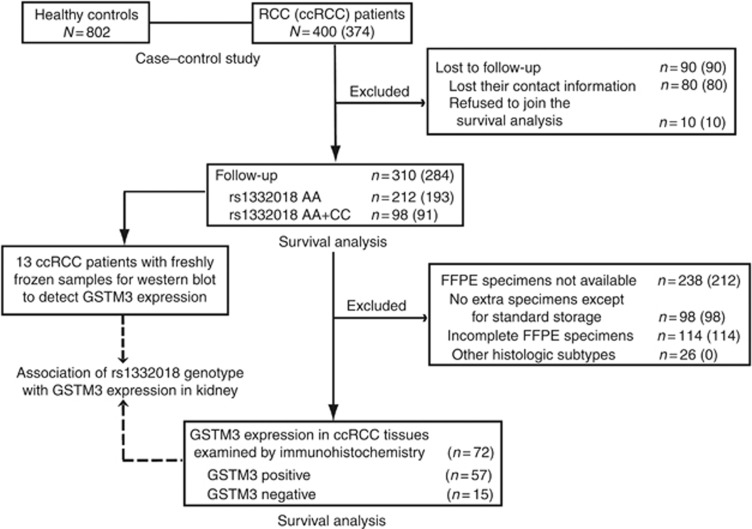
We enrolled a total of 400 newly diagnosed RCC patients and 802 healthy controls (free of diagnosed cancers of any histotype and other clinical diseases). The RCC patients received curative nephrectomy at Department of Urology in Changhai and Changzheng hospitals affiliated to Second Military Medical University from May 1998 to November 2011. Surgical specimens were pathologically identified by a pathologist (YY). None of the enrolled patients received other treatments before or after surgery. Healthy controls were randomly selected from those who received annual physical examination at Physical Examination Center of Changhai hospital from May 2006 to November 2011. The characteristics of RCC patients and healthy controls in the case–control study are included in Supplementary Table 1. Cases and controls were matched on gender while healthy controls were older than RCC patients (P<0.001) according to frequency matching. Of the 400 RCC patients, 374 were pathogenically diagnosed as ccRCC and 43 were at advanced stage (AJCC stage III, 26; stage IV, 17). We also acquired freshly frozen tumour tissues and their paired adjacent renal tissues from 13 ccRCC patients who received surgery at Department of Urology in Changhai hospital for comparative analysis of GSTM3 expression. Flow diagram of study subjects enrolled onto a case–control study and postoperative survival analyses is presented in Figure 1. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki, and was approved by the Institutional Ethical Review Board of Second Military Medical University. A written informed consent was obtained from each participant.
Figure 1.
Flow diagram of study subjects enrolled onto case–control study and survival analysis. Number in parentheses referred to the number of ccRCC patients.
Selection of single-nucleotide polymorphism (SNP) and genotyping
We searched International HapMap Project (www.hapmap.org) for GSTM3-related SNPs with the minor allele frequency of>20% in Chinese Han population. rs1332018 (in the promoter, A-63C) is in a transcription factor-binding site and splicing site, with predicted regulatory potential (http://snpinfo.niehs.nih.gov/cgi-bin/snpinfo/snpfunc.cgi). Thus, rs1332018 was predicted as a functional SNP. rs7483 is in the coding region, resulting in an amino acid change V224I. Both SNPs were selected because they were also the representatives of two haplotype blocks as determined using online Haploview 4.2 software (http://hapmap.ncbi.nlm.nih.gov).
Peripheral blood samples were collected from the patients just before nephrectomy and healthy controls during physical examination and preserved in freezers. Genomic DNA was extracted as previously described (Wang et al, 2012). rs1332018 and rs7483 were genotyped using fluorescent-probe real-time quantitative PCR in LightCycler480 (Roche, Basel, Switzerland) and PCR-confronting two-pair primers (PCR-CTPP; Hamajima et al, 2000), respectively. Primers and probes were designed and synthesised by GeneCore BioTechnologies (Shanghai, China). Sequences of the primers for rs1332018 were 5′-GGAGGCAAGGGACGGAGA-3′ (forward) and 5′-TTCCGAGCCTTCGAGGACTAG-3′ (reverse). The primers for rs7483 were 5′-CCACCCTGCCATCCTCAAGT-3′ (forward) and 5′-CCCAAGAGAAACTCAGCTGGACA-3′ (reverse). The probes for rs1332018 were FAM-CGCCCCCTTATGTAGGGTATAAAGCCC-TAMRA and HEX-CGCCCCCTTATGTCGGGTATAAAGC-TAMRA. The sequences of allele-specific primers for rs7483 were 5′-CAGTGGGGCAACAAGCCTA-3′ (forward) and 5′-TGCCTCCTGCTCAGCATAC-3′ (reverse). The PCR reaction for genotyping rs1332018 was 95 °C for 10 s, 40 cycles of 95 °C for 10 s, 60 °C for 30 s, and 40 °C for 1 s; for genotyping rs7483 was 95 °C for 3 min, 40 cycles of 95 °C for 30 s, 58 °C for 30 s, 72 °C for 50 s. Allele-specific PCR products were visualised on a 1.5% agarose gel with ethidium bromide staining. The genotyping was performed with two blank controls in each 96-well format of PCR. More than 15% of samples were randomly selected to repeat, yielding a 100% concordance. The success rates of genotyping for the two SNPs were >99%.
Western blot
Protein was extracted from RCC cells and freshly frozen adjacent renal tissues of 13 ccRCC patients, quantified, and subjected to western blot with rabbit polyclonal antibodies against GSTM3 (1 : 1000; ProteinTech Group, Chicago, IL, USA) and mouse monoclonal antibody to β-actin (1 : 1000; Cell Signal, Danvers, MA, USA) as previously described (Tan et al, 2013). Signal intensity of each band was quantified using Genetools software (version 4.02, Cambridge, UK). Relative expression was calculated as that signal intensity of GSTM3 divided by signal intensity of β-actin in each lane. A value of ⩾0.8 was defined as a high expression.
Immunohistochemistry
Formalin-fixed paraffin-embedded (FFPE) specimens of tumours and their paired adjacent pathologically normal renal tissues from 72 ccRCC patients were examined immunohistochemically for GSTM3 expression. Polyclonal antibodies against GSTM3 (1 : 100; ProteinTech Group) were used for immunostaining. The sections were processed as previously described (Tan et al, 2008). Immunostains were independently assessed by four researchers (XT, TS, YH, and YY) who were blinded of clinical information. Glutathione S-transferase mu 3 expression in glomeruli, proximal convoluted tubule, and distal convoluted tubule plus collecting ducts as three independent sites were separately evaluated in each section. For each visual field, the percentage of positive epithelial and endothelial cells was graded as 0 (0–4%), 1 (5–24%), 2 (25–49%), 3 (50–74%), and 4 (>75%); the intensity of immunostaining was rated as 0 (negative), 1 (weak), 2 (moderate), and 3 (intense). Values of the intensity and the extent were multiplied as an immunoreactive score. We selected three visual fields in each site and scored each visual field independently. A mean score of the three fields was given to each site. The scores of the three sites were sum up and ranked the total score of each sample by −/+ (score 0–3), ++ (score 3–12), and +++ (score >12). Disagreements on immunoreactive scores for GSTM3 expression in eight (11.11%) patients among the researchers were resolved by consensus.
Follow-up after curative nephrectomy
Clinicopathological variables before surgery were documented. All the RCC patients were followed up by phone or face-to-face interview every 6 months according to our standard epidemiologic procedure. We excluded patients who lost contact information and those refused to be enrolled in the survival analysis. Death from RCC relapse was defined as an event. Patients alive at the last follow-up and deaths caused by conditions unrelated to RCC were censored. Time scale of the follow-up was from 3 to 176.4 months after surgery. Median follow-up time was 43.5 months (interquartile range: 18.3–67.3 months). The last follow-up date was 27 January 2013. Disease-specific survival (DSS) was measured in months from surgery to death from RCC relapse or censored.
Statistical analysis
Hardy–Weinberg equilibrium (HWE) was examined by using online analytical tools (http://ihg.gsf.de). The association of rs1332018 genotypes with GSTM3 expression or with AJCC stage was evaluated by Pearson correlation analysis. For the main effect of SNPs, unconditional logistic regression model was conducted to calculate odds ratios (ORs) and their 95% confidence intervals (95% CIs), adjusting for age and gender, in case–control study. For postoperative survival analysis, DSSs and their 95% CIs were estimated by the Kaplan–Meier method. The log-rank test was used to compare DSS between groups. A test of the proportional hazards, a required assumption of Cox regression, was performed for each covariate and globally using the Breslow method in R (http://www.r-project.org/). Multivariate stage-stratified Cox proportional hazards model was applied to estimate the hazard ratios (HRs) with 95% CIs for DSS. Age (continuous variable), gender (male vs female), and rs1332018 genotype (AC+CC vs AA) as predictor variables and AJCC stage (II+IV vs I+II) as a stratification variable were introduced into this model. All statistical tests were two-sided and conducted using Statistical Program for Social Sciences (SPSS 16.0, Chicago, IL, USA) and R. A P-value of<0.05 was considered as statistically significant.
Results
GSTM3 expression inversely related to the aggressiveness of RCC cells
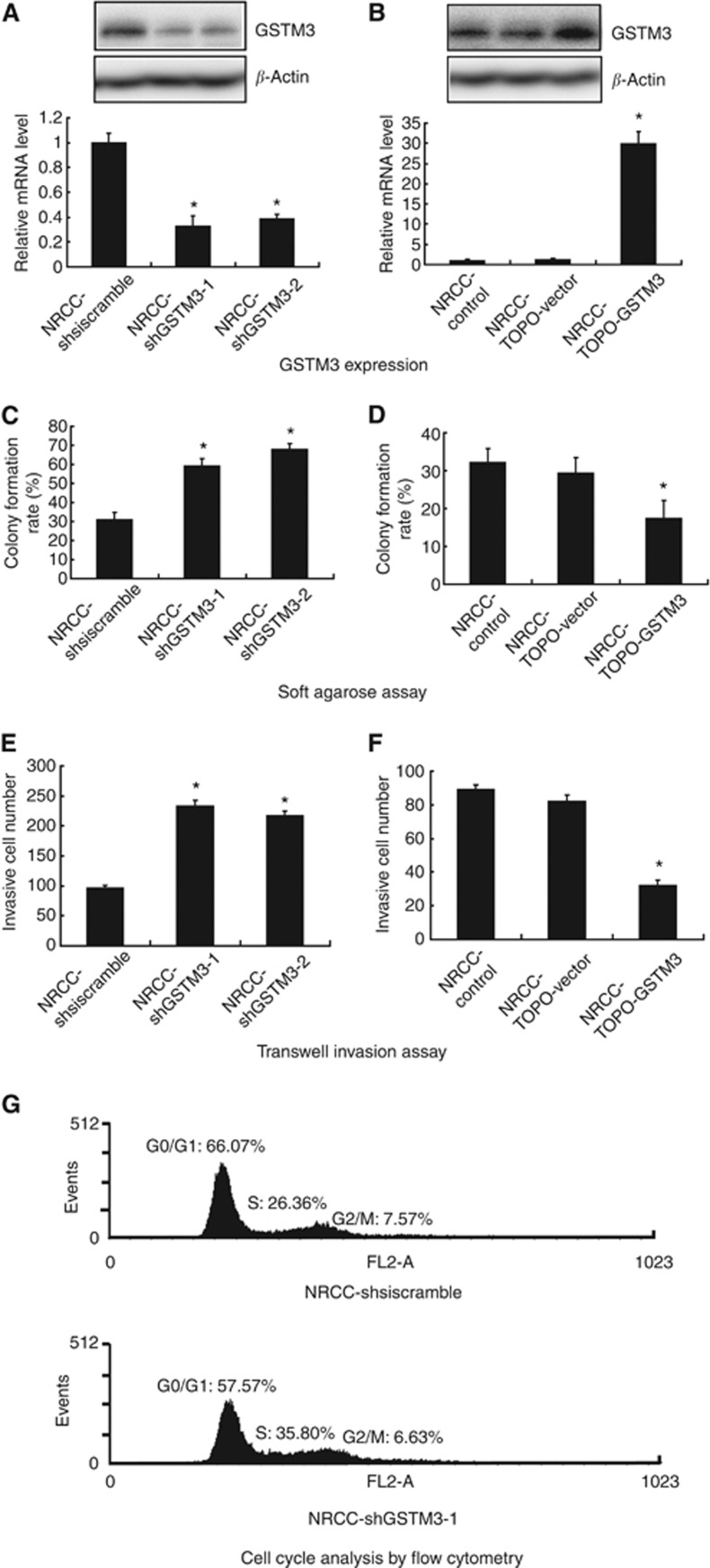
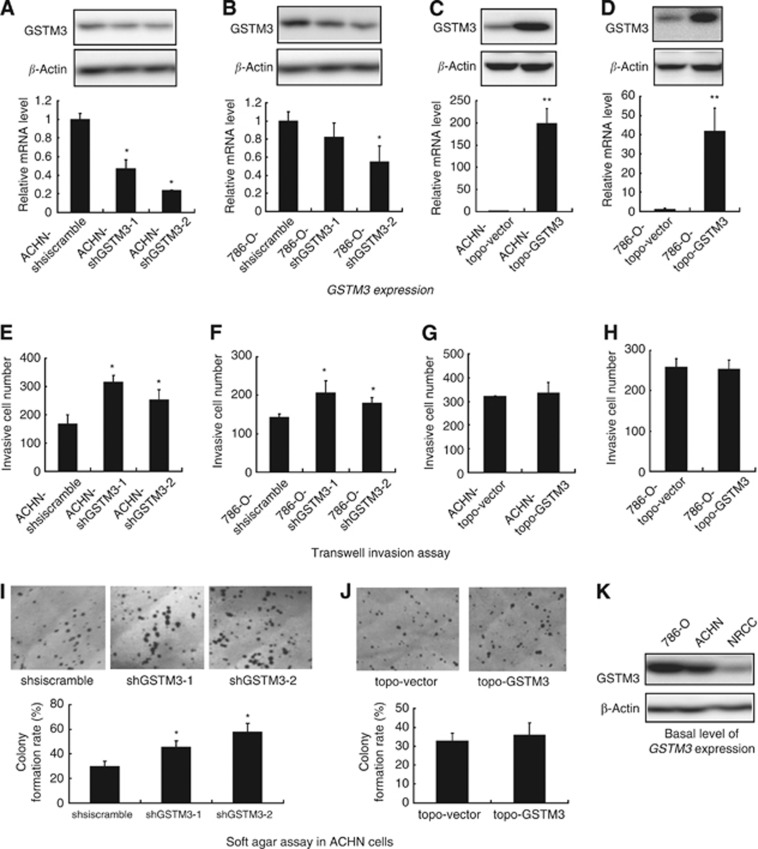
We initially generated stable GSTM3-knockdown and GSTM3-overexpression NRCC cells. Glutathione S-transferase mu 3 expression was significantly decreased in NRCC-shGSTM3-1 or NRCC-shGSTM3-2 cells compared with shsiscramble control cells as examined by western blot and qRT-PCR (Figure 2A). Notably, knockdown of GSTM3 in NRCC cells enhanced anchorage-independent growth in agarose and invasiveness in Transwell plates as compared with the control cells (Figure 2C and E). As expected, GSTM3 expression was significantly upregulated in NRCC cells after GSTM3 transfection (Figure 2B). Ectopic overexpression of GSTM3 significantly reduced anchorage-independent growth and invasive potential of ccRCC cells (Figure 2D and F). Cytometry analysis indicated that NRCC-shGSTM-1 cells had a higher S phase fraction than did the control cells (Figure 2G). Transient transfection of shRNA-GSTM3 significantly decreased GSTM3 expression while transient transfection of TOPO-GSTM3 significantly increased GSTM3 expression in 786-O and ACHN cells. Furthermore, downregulation of GSTM3 expression significantly increased the invasive abilities in 786-O and ACHN cells and anchorage-independent growth in ACHN cells (Figure 3).
Figure 2.
Anchorage-independent growth and invasive potential of ccRCC cell line NRCC with altered expression of GSTM3 by stable transfection. (A) Knockdown of GSTM3 expression by shRNA. (B) Ectopic overexpression of GSTM3 by the transfection. (C) Knockdown of GSTM3 increased anchorage-independent growth. (D) Ectopic overexpression of GSTM3 decreased anchorage-independent growth. (E) Knockdown of GSTM3 increased invasive potential. (F) Ectopic overexpression of GSTM3 decreased invasive potential. (G) Cell cycle analysis by flow cytometry. *P<0.05.
Figure 3.
Anchorage-independent growth and invasive potential of ccRCC cell lines 786-O and ACHN with altered expression GSTM3 by transient transfection. *P<0.05, **P<0.01. (A–D) Expression levels of GSTM3 in ACHN and 786-O cells after transient transfection with shRNA constructs and TOPO-GSTM3 construct, respectively, as detected by qRT-PCR and western blot. (E–H) Invasive potential ACHN and 786-O cells after the transient transfection, as detected by Transwell assay. (I–J) Anchorage-independent growth in ACHN cells after transient transfection with shRNA constructs and TOPO-GSTM3 construct, as detected by the double-layered soft agarose culture system. (K) Background level of GSTM3 expression in ACHN, 786-O and NRCC cells, as detected by western blot.
Association of SNPs with RCC risk
rs1332018 and rs7483 were conformed to HWE in healthy controls, with P-values of 0.947 and 0.388, respectively. Of the two SNPs, rs1332018 variant genotypes were significantly associated with an increased risk of RCC (Table 1). Compared with rs1332018 AA genotype, the AC and AC+CC genotypes were significantly associated with increased risks of RCC, with ORs of 1.442 (P=0.008) and 1.446 (P=0.006) adjusting for age and gender, respectively. Compared with the A allele of rs1332018, the C allele was significantly associated with an increased risk of RCC. As ccRCC patients accounted for 93.5% of the patients enrolled, we also analysed the proportions of rs1332018 genotypes in 374 ccRCC patients and 802 healthy controls. The C allele of rs1332018 was significantly associated with an increased risk of ccRCC, with an adjusted OR of 1.393 (P=0.006). rs7483 genotype was not significantly associated with the risk of RCC or ccRCC.
Table 1. The associations of GSTM3 polymorphisms with the risk of RCC and ccRCC.
| Genotype | RCC patients (%) | ccRCC cases (%) | Controls (%) | Adjusted OR (95% CI) a | P-valuea | Adjusted OR (95% CI) b | P-valueb |
|---|---|---|---|---|---|---|---|
|
rs1332018 | |||||||
| AA | 265 (66.3) | 246 (65.8) | 596 (74.0) | 1.000 (Reference) | — | — | — |
| AC | 125 (31.2) | 118 (31.5) | 191 (23.8) | 1.442 (1.100–1.890) | 0.008 | 1.463 (1.111–1.928) | 0.007 |
| CC | 10 (2.5) | 10 (2.7) | 15 ( 1.9) | 1.524 (0.670–3.466) | 0.315 | 1.639 (0.721–3.726) | 0.238 |
| AC+CC | 135 (33.7) | 128 (34.2) | 206 (25.7) | 1.446 (1.111–1.882) | 0.006 | 1.473 (1.126–1.926) | 0.005 |
| A allele | 655 (81.9) | 610 (81.6) | 1383 (86.2) | 1.000 (Reference) | — | — | — |
| C allele |
145 (18.1) |
138 (18.4) |
221 (13.8) |
1.365 (1.083–1.721) |
0.008 |
1.393 (1.101–1.762) |
0.006 |
|
rs7483 | |||||||
| AA | 224 (56.0) | 206 (55.1) | 452 (56.4) | 1.000 (Reference) | — | ||
| GA | 149 (37.3) | 142 (38.0) | 294 (36.7) | 1.040 (0.804–1.345) | 0.766 | 1.071 (0.824–1.393) | 0.608 |
| GG | 27 (6.7) | 26 (6.9) | 56 ( 7.0) | 0.950 (0.582–1.552) | 0.837 | 0.993 (0.604–1.633) | 0.978 |
| GA+GG | 176 (44.0) | 168 (44.9) | 350 (43.6) | 1.024 (0.801–1.307) | 0.852 | 1.057 (0.823–1.356) | 0.665 |
| A allele | 597 (85.1) | 554 (74.1) | 1198 (74.7) | 1.000 (Reference) | — | — | — |
| G allele | 203 (14.9) | 194 (25.9) | 406 (25.3) | 1.004 (0.824–1.222) | 0.970 | 1.030 (0.843–1.259) | 0.772 |
Abbreviations: CI=confidence interval; ccRCC=clear cell renal cell carcinoma; GSTM3=glutathione S-transferase mu 3; OR=odds ratio; RCC=renal cell carcinoma.
RCC patients vs healthy controls.
ccRCC patients vs healthy controls.
Association of rs1332018 genotypes with GSTM3 expression in kidney
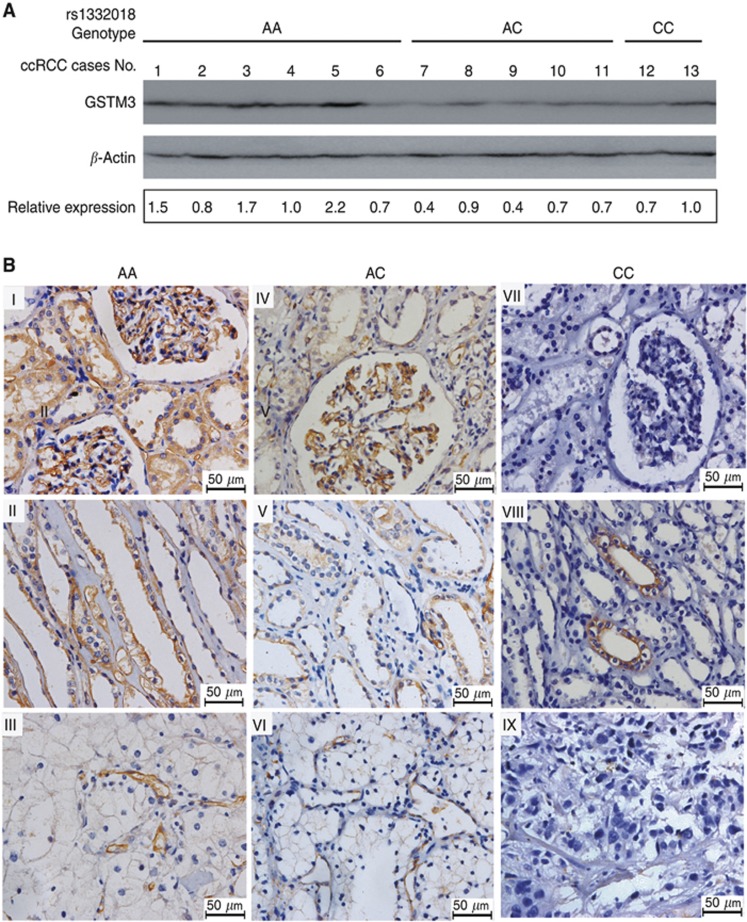
To determine whether rs1332018 genotype was related to the level of GSTM3 expression in kidney, we initially demonstrated GSTM3 expression in freshly frozen adjacent normal renal samples from 13 available ccRCC patients with different rs1332018 genotypes by western blot. We found that GSTM3 expression was higher in five of six samples with the AA genotype and lower in five of seven samples with the CC+AC genotypes (Figure 4A). We then examined GSTM3 expression in the FFPE specimens of adjacent renal specimens of 72 ccRCC patients using immunohistochemistry. In the patients with rs1332018 AA genotype, GSTM3 staining was mainly positive in the membrane and cytoplasm of vascular endothelial cells of glomeruli and epithelial cells of proximal convoluted tubules and distal convoluted tubules and collecting ducts. Interestingly, GSTM3 was selectively positive in a fragment of distal convoluted tubules or collecting ducts but negative in glomeruli and proximal convoluted tubules in those with rs1332018 CC genotype. In RCC tissues with rs1332018 AA or AC genotype, GSTM3 expression was detected in endothelial cells but not detected in cancer epithelial cells; whereas in RCC tissues with rs1332018 CC genotype, GSTM3 expression was seldom detected in both of endothelial cells and cancer epithelial cells. These data are shown in Figure 4B. The total immunoreactive scores of GSTM3 expression in adjacent normal renal tissues correlated with rs1332018 AA genotype, in contrast to those with the AC+CC genotypes (Pearson r=−0.326, P=0.005); however, the scores were not significantly correlated to AJCC stages (Pearson r=−0.093, P=0.436; Table 2).
Figure 4.
The level and pattern of GSTM3 expression in freshly frozen adjacent renal tissues and formalin-fixed paraffin-embedded specimens of ccRCC patients with different rs1332018 genotypes. (A) Expression level of GSTM3 protein in freshly frozen adjacent renal tissues examined by western blot. (B) The immunostaining pattern of GSTM3 expression in glomeruli and proximal convoluted tubules (I, IV, and VII), distal convoluted tubules and collecting ducts (II, V, and VIII) of adjacent renal tissues and the paired RCC tissues (III, VI, and IX) from three patients with rs1332018 genotype of AA, AC, and CC, respectively.
Table 2. The association of rs1332018 genotypes and AJCC stage with GSTM3 expression in the adjacent normal renal tissues of 72 ccRCC patients.
|
Total immunoreactive scores of GSTM3 (no.) |
|
||||
|---|---|---|---|---|---|
| −/+ | ++ | +++ | P-value | r | |
|
rs1332018 | |||||
| AA | 13 | 14 | 22 | 0.005 | −0.326 |
| AC+CC |
10 |
11 |
2 |
|
|
|
AJCC stage | |||||
| I | 13 | 16 | 17 | 0.436 | −0.093 |
| II | 4 | 2 | 1 | ||
| III | 3 | 7 | 4 | ||
| IV | 3 | 0 | 2 | ||
Abbreviations: AJCC=American Joint Committee on Cancer; ccRCC=clear cell renal cell carcinoma; GSTM3=glutathione S-transferase mu 3.
Associations of rs1332018 genotypes and GSTM3 expression in adjacent renal tissues with the survival of RCC patients after surgery
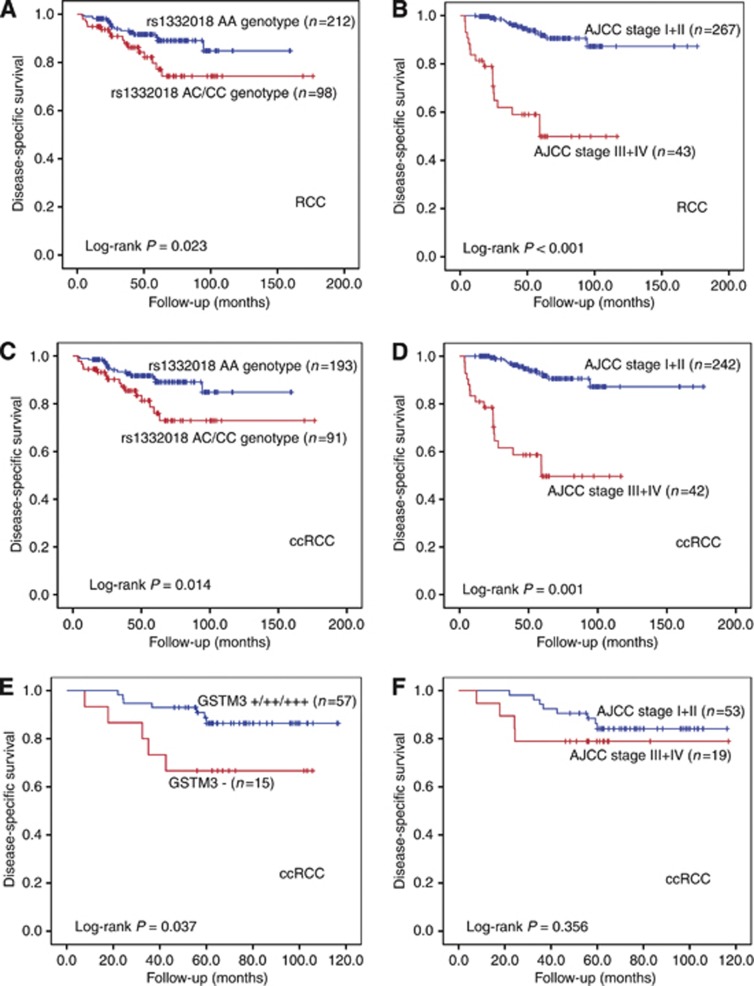
In all, 310 of the 400 RCC patients enrolled in the case–control study were successfully followed up after surgery. Ninety patients lost to follow-up. Major reason of the failure to follow-up was contact information lost (88.9%). These patients were mostly from other cities and had only temporary local phone number in their medical record. Of the 310 RCC patients, 32 died of RCC relapse. Kaplan–Meier analysis showed that the RCC patients with rs1332018 AA genotype had a favourable postoperative DSS compared with those with the C allele (AC+CC genotypes; log-rank P=0.023, Figure 5A). Compared with the patients at AJCC stages III+IV, those at the stages I+II had a good prognosis (log-rank P<0.001, Figure 5B). In the ccRCC patients, rs1332018 AC+CC genotypes and late AJCC stages also significantly predicted an unfavourable prognosis, respectively (Figure 5C and D). Of the 310 RCC patients, 72 ccRCC patients were analysed for GSTM3 expression by using immunohistochemical analysis of their FFPE specimens. Of the 72 patients, 12 died of ccRCC relapse. Positive GSTM3 expression in adjacent renal tissues (positive vs negative) significantly predicted an unfavourable DSS (log-rank P=0.037; Figure 5E), whereas advanced AJCC stages did not significantly predict an unfavourable DSS (Figure 5F) in the 72 ccRCC patients. Multivariate stage-stratified Cox proportional hazards analysis of the 310 patients demonstrated that rs1332018 AC+CC genotypes independently predict an unfavourable postoperative prognosis of RCC or ccRCC (Table 3).
Figure 5.
Kaplan–Meier analysis of DSS of RCC patients with different rs1332018 genotypes or with different GSTM3 expression levels in adjacent renal tissues. (A) RCC patients with rs1332018 AA genotype vs those with the variant (AC+CC) genotype. (B) RCC patients at AJCC stages I+II vs those at AJCC stages III+IV. (C) ccRCC patients with rs1332018 AA genotype vs those with the variant (AC+CC) genotype. (D) ccRCC patients at AJCC stages I+II vs those at AJCC stages III+IV. (E) Positive GSTM3 expression vs negative GSTM3 expression in the adjacent renal tissues of the 72 ccRCC patients. (F) AJCC stages I+II vs those at AJCC stages III+IV of the 72 ccRCC patients.
Table 3. Factors significantly predicted disease-specific survival in multivariate stage-stratified Cox proportional hazards model.
| Variables | HR (95% CI) a | P-valuea | HR (95% CI) b | P-value b |
|---|---|---|---|---|
| Age |
1.005 (0.978, 1.033) |
0.720 |
1.005 (0.977, 1.034) |
0.740 |
| Gender (male vs female) |
1.102 (0.532, 2.283) |
0.790 |
1.127 (0.538, 2.362) |
0.750 |
| rs1332018 genotype (AC+CC vs AA) | 2.119 (1.043, 4.307) | 0.038 | 2.248 (1.091, 4.631) | 0.028 |
Abbreviations: CI=confidence interval; ccRCC=clear cell renal cell carcinoma; HR=hazard ratio; RCC=renal cell carcinoma.
For RCC.
For ccRCC.
Discussion
In this study, we found that downregulation of GSTM3 expression by shRNA significantly increased the invasive potential and anchorage-independent growth of the three ccRCC cell lines; whereas ectopic overexpression of GSTM3 in NRCC cells decreased the invasive potential and anchorage-independent growth (Figure 2D and F). We did not observe the effect of GSTM3 overexpression by transient transfection in 786-O and ACHN cells possibly because the expression background of GSTM3 was high in the two cell lines (Figure 3M). Cytometry analysis indicated that ccRCC cells tended to proliferate faster when GSTM3 was downregulated (Figure 2G). These data, together with our previous findings (Tan et al, 2008), suggest that GSTM3 may function as a tumour suppressor in ccRCC. Glutathione S-transferase mu 3 has been shown to be downregulated in ovarian cancer by using proteomics analysis (Lim et al, 2011). Epigenetic inactivation (promoter hypermethylation) of GSTM3 has been found in Barrett's adenocarcinoma (Peng et al, 2009). A significant reduction of GSTM3 expression has been observed in chemical-induced hepatocarcinogenesis (Quiles-Perez et al, 2010). These data indicate that GSTM3 expression is downregulated in these cancer types. However, it has been shown that GSTM3 expression correlates with lymph node metastasis and advanced stage of colon cancer and low GSTM3 expression is associated with better survival in patients with bladder cancer (Mitra et al, 2009; Meding et al, 2012). Thus, GSTM3 function appears to be context dependent and may vary with different cancer types.
We found that a polymorphism (rs1332018) in the promoter region influenced GSTM3 expression in adjacent pathologically normal renal tissues. rs1332018 AA genotype was correlated to high GSTM3 expression while the variant genotype (CC+AC) were correlated to low GSTM3 expression (Table 2). This is consistent with the previous observations that the CC genotype of rs1332018 is associated with lower GSTM3 expression in brain tumours, normal breast tissues, lymphoblast cells, and glioma cell lines by means of altering binding of RNA polymerase II (Liu et al, 2005; Yu et al, 2010; Searles Nielsen et al, 2011). This SNP was shown to be functional in previous studies (Liu et al, 2005). Interestingly, we found that GSTM3 was predominantly expressed in vascular endothelial cells of glomeruli and epithelial cells of proximal convoluted tubules in tissues with rs1332018 AA genotype; whereas GSTM3 was exclusively expressed in a fragment of distal convoluted tubules or collecting ducts in those with rs1332018 CC genotype (Figure 4). The underlying mechanism for the rs1332018 genotype-specific GSTM3 expression patterns in renal units remains unknown, but we believe that GSTM3 and its functional genetic predisposition have important roles in renal function and diseases. Although there is some evidence supporting the concept that ccRCC might origin from collecting ducts and distal tubules (Straube et al, 2011), it is generally believed that ccRCC originates from proximal tubular epithelial cells. Lack of GSTM3 in proximal tubular epithelial cells with rs1332018 variants may facilitate renal carcinogenesis if exposed to carcinogen and by-products of oxidative stress, as GSTM3 is basically a cytosolic enzyme involved in prostaglandin and leukotriene synthesis and in the metabolism of toxic compounds (Hayes et al, 2005).
Our case–control study suggested that the C allele of rs1332018 was significantly associated with an increased risk of RCC (Table 1). However, this SNP was not found to be a risk locus of RCC in recent GWAS, possibly because of the race of study population, coverage of the probes, and correction of multiple comparisons in GWAS (Purdue et al, 2011; Wu et al, 2012; Henrion et al, 2013). Importantly, the Bonferroni correction in GWAS does not take into account SNP function. In this regard, a key finding of this study is that the C allele of rs1332018 independently predicted an unfavourable postoperative DSS of RCC (Table 3), indicating the importance of this functional SNP in RCC. It has been shown that the variant genotype of rs1332018 is significantly associated with increased risks of brain tumour and breast cancer (Yu et al, 2010; Searles Nielsen et al, 2011). Although the activity of GSTM3 promoter with different rs1332018 genotypes should be examined in the RCC cells, we believe that the C allele of rs1332018 predisposes the host to decreasing GSTM3 expression in kidney. We also demonstrated that negative GSTM3 expression in adjacent renal tissues was significantly associated with an unfavourable DSS (Figure 5). Silencing of GSTM3 expression in renal tissues might facilitate renal carcinogenesis upon carcinogen exposure and also promote disease relapse and metastasis. These data support that GSTM3 is a putative tumour suppressor in these cancer types including RCC. To our knowledge, this is the first study to report that rs1332018 variant genotype was not only a risk factor and but also a prognostic factor of RCC.
Our study had several limitations. First, 90 of the 400 genotyped RCC patients were lost to follow-up. This limitation might not bias the associations of rs1332018 genotypes with RCC risk and postoperative prognosis because contact information lost was the main reason of the lost to follow-up. However, a selection bias might be introduced, at least theoretically, by this percentage (22.5%) of the patients lost to follow-up. Second, RCC-free patients with diseases that increase RCC risk, such as hypertension, should be also considered as controls in this case–control study. Third, the mechanism by which rs1332018 genotypes regulate GSTM3 expression was not fully examined in this study. The effect of rs1332018 variants on the promoter activity of GSTM3 in renal cells and the role of GSTM3 expression pattern in renal subunits of the individuals with distinct rs1332018 genotypes on renal function warrant further investigation.
In summary, we demonstrated, for the first time, that GSTM3 may function as a tumour suppressor in RCC. The C allele of rs1332018 predisposes host to downregulating GSTM3 in their renal tissues, especially in the membrane and cytoplasm of vascular endothelial cells of glomeruli and epithelial cells of proximal convoluted tubules. rs1332018 variant genotype was not only a significant genetic risk factor and but also a significant prognostic factor for RCC. These findings will be helpful in the surveillance and prognosis prediction of RCC, as well as in the development of targeted therapy for this malignancy. Our report also points to the need for further studies of regulation of GSTM3 expression in RCC.
Acknowledgments
This study was supported by National Natural Science Foundation of China (30873041, 81025015, and 81101928), Shanghai Health Bureau (2010Y099) and, in part, by the United States National Institutes of Health supported MD Anderson Cancer Center Support Grant (CA16672).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Angevin E, Lopez-Martin JA, Lin CC, Gschwend JE, Harzstark A, Castellano D, Soria JC, Sen P, Chang J, Shi M, Kay A, Escudier B. Phase I study of dovitinib (TKI258), an oral FGFR, VEGFR, and PDGFR inhibitor, in advanced or metastatic renal cell carcinoma. Clin Cancer Res. 2013;19:1257–1268. doi: 10.1158/1078-0432.CCR-12-2885. [DOI] [PubMed] [Google Scholar]
- Beuselinck B, Karadimou A, Lambrechts D, Claes B, Wolter P, Couchy G, Berkers J, Paridaens R, Schöffski P, Méjean A, Verkarre V, Lerut E, de la Taille A, Tourani JM, Bigot P, Linassier C, Négrier S, Berger J, Patard JJ, Zucman-Rossi J, Oudard S. Single-nucleotide polymorphisms associated with outcome in metastatic renal cell carcinoma treated with sunitinib. Br J Cancer. 2013;108:887–900. doi: 10.1038/bjc.2012.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow WH, Dong LM, Devesa SS. Epidemiology and risk factors for kidney cancer. Nat Rev Urol. 2010;7:245–257. doi: 10.1038/nrurol.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow WH, Shuch B, Linehan WM, Devesa SS. Racial disparity in renal cell carcinoma patient survival according to demographic and clinical characteristics. Cancer. 2013;119:388–394. doi: 10.1002/cncr.27690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellavalle CT, Daniel CR, Aschebrook-Kilfoy B, Hollenbeck AR, Cross AJ, Sinha R, Ward MH. Dietary intake of nitrate and nitrite and risk of renal cell carcinoma in the NIH-AARP Diet and Health Study. Br J Cancer. 2013;108:205–212. doi: 10.1038/bjc.2012.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doberstein K, Wieland A, Lee SB, Blaheta RA, Wedel S, Moch H, Schraml P, Pfeilschifter J, Kristiansen G, Gutwein P. L1-CAM expression in ccRCC correlates with shorter patients survival times and confers chemoresistance in renal cell carcinoma cells. Carcinogenesis. 2011;32:262–270. doi: 10.1093/carcin/bgq249. [DOI] [PubMed] [Google Scholar]
- Figlin RA, Kaufmann I, Brechbiel J. Targeting PI3K and mTORC2 in metastatic renal cell carcinoma: new strategies for overcoming resistance to VEGFR and mTORC1 inhibitors. Int J Cancer. 2013;133:788–796. doi: 10.1002/ijc.28023. [DOI] [PubMed] [Google Scholar]
- Hamajima N, Saito T, Matsuo K, Kozaki K, Takahashi T, Tajima K. Polymerase chain reaction with confronting two-pair primers for polymorphism genotyping. Jpn J Cancer Res. 2000;91:865–868. doi: 10.1111/j.1349-7006.2000.tb01026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- Henrion M, Frampton M, Scelo G, Purdue M, Ye Y, Broderick P, Ritchie A, Kaplan R, Meade A, McKay J, Johansson M, Lathrop M, Larkin J, Rothman N, Wang Z, Chow WH, Stevens VL, Ryan Diver W, Gapstur SM, Albanes D, Virtamo J, Wu X, Brennan P, Chanock S, Eisen T, Houlston RS. Common variation at 2q22.3 (ZEB2) influences the risk of renal cancer. Hum Mol Genet. 2013;22:825–831. doi: 10.1093/hmg/dds489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H, Hinoda Y, Kikuno N, Kawamoto K, Suehiro Y, Tanaka Y, Dahiya R. MDM2 SNP309 polymorphism as risk factor for susceptibility and poor prognosis in renal cell carcinoma. Clin Cancer Res. 2007;13:4123–4129. doi: 10.1158/1078-0432.CCR-07-0609. [DOI] [PubMed] [Google Scholar]
- Karami S, Boffetta P, Rothman N, Hung RJ, Stewart T, Zaridze D, Navritalova M, Mates D, Janout V, Kollarova H, Bencko V, Szeszenia-Dabrowska N, Holcatova I, Mukeria A, Gromiec J, Chanock SJ, Brennan P, Chow WH, Moore LE. Renal cell carcinoma, occupational pesticide exposure and modification by glutathione S-transferase polymorphisms. Carcinogenesis. 2008;29:1567–1571. doi: 10.1093/carcin/bgn153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinrath T, Gassner C, Lackner P, Thurnher M, Ramoner R. Interleukin-4 promoter polymorphisms: a genetic prognostic factor for survival in metastatic renal cell carcinoma. J Clin Oncol. 2007;25:845–851. doi: 10.1200/JCO.2006.07.8154. [DOI] [PubMed] [Google Scholar]
- Lew JQ, Chow WH, Hollenbeck AR, Schatzkin A, Park Y. Alcohol consumption and risk of renal cell cancer: the NIH-AARP diet and health study. Br J Cancer. 2011;104:537–541. doi: 10.1038/sj.bjc.6606089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Tan X, Yu Y, Chen H, Chang W, Hou J, Xu D, Ma L, Cao G. D9S168 microsatellite alteration predicts a poor prognosis in patients with clear cell renal cell carcinoma and correlates with the down-regulation of protein tyrosine phosphatase receptor delta. Cancer. 2011;117:4201–4211. doi: 10.1002/cncr.26028. [DOI] [PubMed] [Google Scholar]
- Lim R, Lappas M, Ahmed N, Permezel M, Quinn MA, Rice GE. 2D-PAGE of ovarian cancer: analysis of soluble and insoluble fractions using medium-range immobilized pH gradients. Biochem Biophys Res Commun. 2011;406:408–413. doi: 10.1016/j.bbrc.2011.02.056. [DOI] [PubMed] [Google Scholar]
- Lin J, Horikawa Y, Tamboli P, Clague J, Wood CG, Wu X. Genetic variations in microRNA-related genes are associated with survival and recurrence in patients with renal cell carcinoma. Carcinogenesis. 2010;31:1805–1812. doi: 10.1093/carcin/bgq168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Campbell MR, Pittman GS, Faulkner EC, Watson MA, Bell DA. Expression-based discovery of variation in the human glutathione S-transferase M3 promoter and functional analysis in a glioma cell line using allele-specific chromatin immunoprecipitation. Cancer Res. 2005;65:99–104. [PubMed] [Google Scholar]
- Meding S, Balluff B, Elsner M, Schöne C, Rauser S, Nitsche U, Maak M, Schäfer A, Hauck SM, Ueffing M, Langer R, Höfler H, Friess H, Rosenberg R, Walch A. Tissue-based proteomics reveals FXYD3, S100A11 and GSTM3 as novel markers for regional lymph node metastasis in colon cancer. J Pathol. 2012;228:459–470. doi: 10.1002/path.4021. [DOI] [PubMed] [Google Scholar]
- Mitra AP, Pagliarulo V, Yang D, Waldman FM, Datar RH, Skinner DG, Groshen S, Cote RJ. Generation of a concise gene panel for outcome prediction in urinary bladder cancer. J Clin Oncol. 2009;27:3929–3937. doi: 10.1200/JCO.2008.18.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LE, Hung R, Karami S, Boffetta P, Berndt S, Hsu CC, Zaridze D, Janout V, Kollarova H, Bencko V, Navratilova M, Szeszenia-Dabrowska N, Mates D, Mukeria A, Holcatova I, Yeager M, Chanock S, Garcia-Closas M, Rothman N, Chow WH, Brennan P. Folate metabolism genes, vegetable intake and renal cancer risk in central Europe. Int J Cancer. 2008;122:1710–1715. doi: 10.1002/ijc.23318. [DOI] [PubMed] [Google Scholar]
- Motzer RJ, Escudier B, Bukowski R, Rini BI, Hutson TE, Barrios CH, Lin X, Fly K, Matczak E, Gore ME. Prognostic factors for survival in 1059 patients treated with sunitinib for metastatic renal cell carcinoma. Br J Cancer. 2013;108:2470–2477. doi: 10.1038/bjc.2013.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng DF, Razvi M, Chen H, Washington K, Roessner A, Schneider-Stock R, El-Rifai W. DNA hypermethylation regulates the expression of members of the Mu-class glutathione S-transferases and glutathione peroxidases in Barrett's adenocarcinoma. Gut. 2009;58:5–15. doi: 10.1136/gut.2007.146290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler M, Hutterer GC, Stoeckigt C, Chromecki TF, Stojakovic T, Golbeck S, Eberhard K, Gerger A, Mannweiler S, Pummer K, Zigeuner R. Validation of the pre-treatment neutrophil-lymphocyte ratio as a prognostic factor in a large European cohort of renal cell carcinoma patients. Br J Cancer. 2013;108:901–907. doi: 10.1038/bjc.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdue MP, Johansson M, Zelenika D, Toro JR, Scelo G, Moore LE, Prokhortchouk E, Wu X, Kiemeney LA, Gaborieau V, Jacobs KB, Chow WH, Zaridze D, Matveev V, Lubinski J, Trubicka J, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Bucur A, Bencko V, Foretova L, Janout V, Boffetta P, Colt JS, Davis FG, Schwartz KL, Banks RE, Selby PJ, Harnden P, Berg CD, Hsing AW, Grubb RL, 3rd, Boeing H, Vineis P, Clavel-Chapelon F, Palli D, Tumino R, Krogh V, Panico S, Duell EJ, Quirós JR, Sanchez MJ, Navarro C, Ardanaz E, Dorronsoro M, Khaw KT, Allen NE, Bueno-de-Mesquita HB, Peeters PH, Trichopoulos D, Linseisen J, Ljungberg B, Overvad K, Tjønneland A, Romieu I, Riboli E, Mukeria A, Shangina O, Stevens VL, Thun MJ, Diver WR, Gapstur SM, Pharoah PD, Easton DF, Albanes D, Weinstein SJ, Virtamo J, Vatten L, Hveem K, Njølstad I, Tell GS, Stoltenberg C, Kumar R, Koppova K, Cussenot O, Benhamou S, Oosterwijk E, Vermeulen SH, Aben KK, van der Marel SL, Ye Y, Wood CG, Pu X, Mazur AM, Boulygina ES, Chekanov NN, Foglio M, Lechner D, Gut I, Heath S, Blanche H, Hutchinson A, Thomas G, Wang Z, Yeager M, Fraumeni JF, Jr, Skryabin KG, McKay JD, Rothman N, Chanock SJ, Lathrop M, Brennan P. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13.3. Nat Genet. 2011;43:60–65. doi: 10.1038/ng.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiles-Perez R, Muñoz-Gámez JA, Ruiz-Extremera A, O'Valle F, Sanjuán-Nuñez L, Martín-Alvarez AB, Martín-Oliva D, Caballero T, Muñoz de Rueda P, León J, Gonzalez R, Muntané J, Oliver FJ, Salmerón J. Inhibition of poly adenosine diphosphate-ribose polymerase decreases hepatocellular carcinoma growth by modulation of tumor-related gene expression. Hepatology. 2010;51:255–266. doi: 10.1002/hep.23249. [DOI] [PubMed] [Google Scholar]
- Schödel J, Bardella C, Sciesielski LK, Brown JM, Pugh CW, Buckle V, Tomlinson IP, Ratcliffe PJ, Mole DR. Common genetic variants at the 11q13.3 renal cancer susceptibility locus influence binding of HIF to an enhancer of cyclin D1 expression. Nat Genet. 2012;44:420–425. doi: 10.1038/ng.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searles Nielsen S, Mueller BA, Preston-Martin S, Farin FM, Holly EA, McKean-Cowdin R. Childhood brain tumors and maternal cured meat consumption in pregnancy: differential effect by glutathione S-transferases. Cancer Epidemiol Biomarkers Prev. 2011;20:2413–2419. doi: 10.1158/1055-9965.EPI-11-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza JC, Ziogas A, Largent J, Peel D, Anton-Culver H. Gene-environment interactions in renal cell carcinoma. Am J Epidemiol. 2001;153:851–859. doi: 10.1093/aje/153.9.851. [DOI] [PubMed] [Google Scholar]
- Simic T, Savic-Radojevic A, Pljesa-Ercegovac M, Matic M, Mimic-Oka J. Glutathione S-transferases in kidney and urinary bladder tumors. Nat Rev Urol. 2009;6:281–289. doi: 10.1038/nrurol.2009.49. [DOI] [PubMed] [Google Scholar]
- Straube T, Elli AF, Greb C, Hegele A, Elsässer HP, Delacour D, Jacob R. Changes in the expression and subcellular distribution of galectin-3 in clear cell renal cell carcinoma. J Exp Clin Cancer Res. 2011;30:89. doi: 10.1186/1756-9966-30-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T, Han Y, Yu Y, Tan X, Li X, Hou J, Du Y, Shen J, Wang G, Ma L, Jiang S, Zhang H, Cao G. A GWAS-identified susceptibility locus on chromosome 11q13.3 and its putative molecular target for prediction of postoperative prognosis of human renal cell carcinoma. Oncol Lett. 2013;6:421–426. doi: 10.3892/ol.2013.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Tan X, Shi Y, Xu G, Mao R, Gu X, Fan Y, Yu Y, Burlingame S, Zhang H, Rednam SP, Lu X, Zhang T, Fu S, Cao G, Qin J, Yang J. USP11 negatively regulates TNFalpha-induced NF-kappaB activation by targeting on IkappaBalpha. Cell Signal. 2010;22:386–394. doi: 10.1016/j.cellsig.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, He S, Han Y, Yu Y, Xiao J, Xu D, Wang G, Du Y, Chang W, Yin J, Su T, Hou J, Cao G. Establishment and characterization of clear cell renal cell carcinoma cell lines with different metastatic potential from Chinese patients. Cancer Cell Int. 2013;13:20. doi: 10.1186/1475-2867-13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Zhai Y, Chang W, Hou J, He S, Lin L, Yu Y, Xu D, Xiao J, Ma L, Wang G, Cao T, Cao G. Global analysis of metastasis-associated gene expression in primary cultures from clinical specimens of clear-cell renal-cell carcinoma. Int J Cancer. 2008;123:1080–1088. doi: 10.1002/ijc.23637. [DOI] [PubMed] [Google Scholar]
- Wang G, Hou J, Ma L, Xie J, Yin J, Xu D, Chang W, Tan X, Su T, Zhang H, Cao G. Risk factor for clear cell renal cell carcinoma in Chinese population: a case-control study. Cancer Epidemiol. 2012;36:177–182. doi: 10.1016/j.canep.2011.09.006. [DOI] [PubMed] [Google Scholar]
- Wu X, Scelo G, Purdue MP, Rothman N, Johansson M, Ye Y, Wang Z, Zelenika D, Moore LE, Wood CG, Prokhortchouk E, Gaborieau V, Jacobs KB, Chow WH, Toro JR, Zaridze D, Lin J, Lubinski J, Trubicka J, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, Jinga V, Bencko V, Slamova A, Holcatova I, Navratilova M, Janout V, Boffetta P, Colt JS, Davis FG, Schwartz KL, Banks RE, Selby PJ, Harnden P, Berg CD, Hsing AW, Grubb RL, 3rd, Boeing H, Vineis P, Clavel-Chapelon F, Palli D, Tumino R, Krogh V, Panico S, Duell EJ, Quirós JR, Sanchez MJ, Navarro C, Ardanaz E, Dorronsoro M, Khaw KT, Allen NE, Bueno-de-Mesquita HB, Peeters PH, Trichopoulos D, Linseisen J, Ljungberg B, Overvad K, Tjønneland A, Romieu I, Riboli E, Stevens VL, Thun MJ, Diver WR, Gapstur SM, Pharoah PD, Easton DF, Albanes D, Virtamo J, Vatten L, Hveem K, Fletcher T, Koppova K, Cussenot O, Cancel-Tassin G, Benhamou S, Hildebrandt MA, Pu X, Foglio M, Lechner D, Hutchinson A, Yeager M, Fraumeni JF, Jr, Lathrop M, Skryabin KG, McKay JD, Gu J, Brennan P, Chanock SJ. A genome-wide association study identifies a novel susceptibility locus for renal cell carcinoma on 12p11.23. Hum Mol Genet. 2012;21:456–462. doi: 10.1093/hmg/ddr479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu KD, Fan L, Di GH, Yuan WT, Zheng Y, Huang W, Chen AX, Yang C, Wu J, Shen ZZ, Shao ZM. Genetic variants in GSTM3 gene within GSTM4-GSTM2- GSTM1-GSTM5-GSTM3 cluster influence breast cancer susceptibility depending on GSTM1. Breast Cancer Res Treat. 2010;121:485–496. doi: 10.1007/s10549-009-0585-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.