Abstract
Species inhabit complex environments and respond to selection imposed by numerous abiotic and biotic conditions that vary in both space and time. Environmental heterogeneity strongly influences trait evolution and patterns of adaptive population differentiation. For example, heterogeneity can favor local adaptation, or can promote the evolution of plastic genotypes that alter their phenotypes based on the conditions they encounter. Different abiotic and biotic agents of selection can act synergistically to either accelerate or constrain trait evolution. The environmental context has profound effects on quantitative genetic parameters. For instance, heritabilities measured in controlled conditions often exceed those measured in the field; thus, laboratory experiments could overestimate the potential for a population to respond to selection. Nevertheless, most studies of the genetic basis of ecologically relevant traits are conducted in simplified laboratory environments, which do not reflect the complexity of nature. Here, we advocate for manipulative field experiments in the native ranges of plant species that differ in mating system, life-history strategy and growth form. Field studies are vital to evaluate the roles of disparate agents of selection, to elucidate the targets of selection and to develop a nuanced perspective on the evolution of quantitative traits. Quantitative genetics field studies will also shed light on the potential for natural populations to adapt to novel climates in highly fragmented landscapes. Drawing from our experience with the ecological model system Boechera (Brassicaceae), we discuss advancements possible through dedicated field studies, highlight future research directions and examine the challenges associated with field studies.
Keywords: agents of selection, conservation genetics, mating system, phenotypic plasticity, quantitative trait loci, targets of selection
Introduction
Species evolve in complex environments where conditions vary in space and time. To appreciate the effects of environmental heterogeneity on the evolution of quantitative traits, we must link classical quantitative genetics to ecology using manipulative experiments in realistic natural settings. Field studies can uncover novel functions of previously characterized genes (Brock et al., 2009), examine how divergent selection contributes to local adaptation at the organismal level (Hall and Willis, 2006), address the genetic basis of adaptive population differentiation (Leinonen et al., 2013), expose genetic correlations among traits that could constrain adaptation (Brock et al., 2009) and reveal whether genetic variation is sufficient to enable evolutionary responses to selection imposed by anthropogenic disturbance (Etterson and Shaw, 2001).
The importance of field experiments for quantitative genetics can be illustrated by work on flowering phenology. The Arabidopsis thaliana flowering time gene network has been extensively investigated under controlled laboratory conditions, and consists of several pathways: photoperiod (Putterill et al., 1995; Valverde et al., 2004), circadian clock (Brachi et al., 2010), vernalization (exposure to winter temperatures, Kim et al., 2009), autonomous (Blazquez et al., 2003), temperature (Kumar et al., 2012) and gibberellin (a plant hormone) (Mutasa-Göttgens and Hedden, 2009). However, even in Arabidopsis, there could be additional genetic pathways regulated by other environmental variables. In nature, A. thaliana and other species flower in response to water availability (Eckhart et al., 2004), seasonal fluctuations in temperature (Aikawa et al., 2011), atmospheric CO2 concentration (Springer and Ward, 2007), soil nutrient levels (Stanton et al., 2000), foliar herbivory (Strauss et al., 1996; Poveda et al., 2003; Kawagoe and Kudoh, 2010; Brys et al., 2011) and seed predation (Parachnowitsch and Caruso, 2008), among other conditions (Forrest and Miller-Rushing, 2010). Yet, we know very little about how plants sense and respond to these abiotic and biotic conditions at the molecular level. Quantitative genetic field experiments can illuminate the genetic basis of flowering phenology in nature (Weinig et al., 2002; Brock et al., 2009; Brachi et al., 2010).
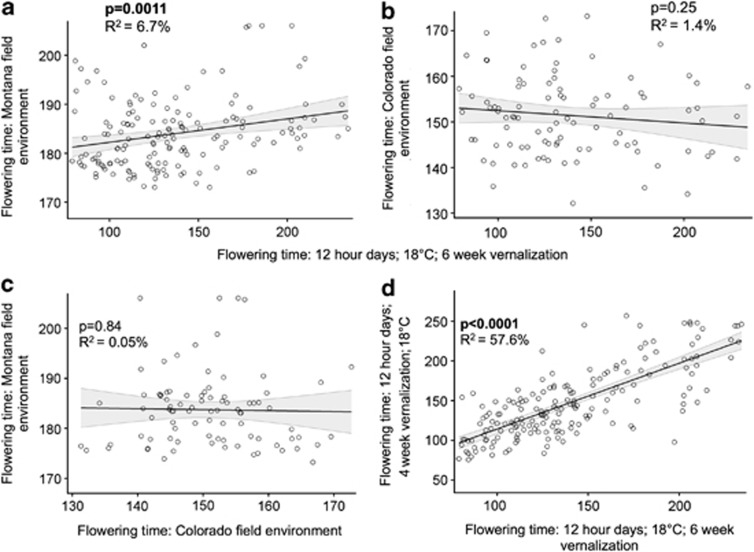
Flowering phenology has clearly evolved in response to complex natural environments that are difficult to simulate in the laboratory (Weinig et al., 2002; Brachi et al., 2010; Anderson et al., 2011; Hancock et al., 2011). Field studies of A. thaliana have detected quantitative trait loci (QTL) for flowering time that do not colocalize with flowering time genes known from laboratory studies (Brachi et al., 2010). Conversely, several QTL that control the timing of flowering in the lab were not detected in the field, suggesting that their importance might be exaggerated under simplified laboratory conditions (Brachi et al., 2010). Similarly, in the closely related mustard Boechera stricta, some QTL mapped for the timing and developmental stage at first flowering under laboratory conditions did not correspond with QTL mapped for these traits in a field experiment in native habitats (Anderson et al., 2011). The same recombinant inbred lines of B. stricta were exposed to two field and six growth chamber environments, yet the genetic correlation of flowering time in the field and the laboratory was low (Anderson et al., 2011), suggesting that controlled conditions did not effectively replicate natural environments (Figure 1). Future manipulative studies in the field could uncover QTL and quantitative trait nucleotides (QTN) that influence flowering time response to environments that would be nearly impossible to model in the laboratory, such as pollinator or herbivore communities.
Figure 1.
Genetic correlations of flowering times in the same B. stricta recombinant inbred lines exposed to six laboratory and two field environments. If laboratory conditions reasonably simulated natural environments, we would expect a tight correlation between flowering phenology in lab and field. Instead, our data indicate that family-mean flowering times are significantly correlated in laboratory and Montana field conditions (a), but little of the genotypic variation in flowering time in the field is explained by conditions in the growth chamber, reflected in the low R2 value. We found no significant correlation between family-mean flowering times in our Colorado garden and any of our six growth chamber conditions (one representative growth chamber treatment is displayed in (b)). Furthermore, flowering time values are uncorrelated between our disparate field sites (c), highlighting the importance of investigating life-history transitions and ecologically relevant traits under multiple natural environments. The genetic correlation of flowering time is much tighter in comparisons of different growth chamber treatments (d). The data presented in these panels come from Anderson et al. (2011, 2012a), and are available in the associated Dryad files.
In this review, we highlight the importance of field studies in advancing our understanding of evolutionary genetics. Field experiments can measure components of fitness in native habitats, enabling tests of evolutionary hypotheses in historically relevant environments. Researchers rarely have a detailed a priori understanding of the entire complement of biotic and abiotic agents of selection that influence complex trait evolution. Interdisciplinary approaches combined with field studies allow researchers to investigate quantitative trait evolution across levels of biological organization, and to test whether results from laboratory studies of short-lived annual species apply to species with different life histories in nature. Building off our own work in the ecological model system Boechera (Rushworth et al., 2011), we emphasize advances that have been made because of large-scale field studies replicated in space and time, discuss limitations to current data and suggest future directions. We focus on long-standing conceptual issues in quantitative genetics. First, we review the evolutionary responses of natural populations to temporal and spatial environmental heterogeneity. We then discuss how field studies can shed light on the complex suite of abiotic and biotic conditions that impose selection on natural populations, and the phenotypic and genetic targets of that selection, with a particular focus on gene expression (eQTL) and small-effect QTL. We address the effects of inbreeding on quantitative traits and fitness in nature. Finally, we consider the practical relevance of these concepts to emerging global problems.
Genetic and plastic responses to environmental heterogeneity
Abiotic and biotic conditions vary spatially and temporally, thereby altering the strength and direction of natural selection across the landscape and over time. In turn, selection can lead to the evolution of: (1) specialization, where ecotypes are adapted to a narrow range of conditions; (2) generalization, where an intermediate fixed phenotype persists in all environments; and (3) adaptive phenotypic plasticity, where individuals adjust their phenotypes to suit specific conditions (van Tienderen, 1997). In a heterogeneous landscape, when should natural selection favor specialization, generalization or plasticity?
Fundamentally, the scale at which individuals interact with the local environment may determine the answer. Empirical and theoretical evidence indicates that strongly divergent natural selection in contrasting habitats promotes specialization, particularly when environmental conditions change slowly relative to the lifespan of an individual or the dispersal distances of progeny (coarse-grained spatial variation) (Hedrick, 1986; Alpert and Simms, 2002; Kawecki and Ebert, 2004; Hereford, 2009). In contrast, generalization or phenotypic plasticity may evolve when individuals experience multiple abiotic and/or biotic conditions during their lifetime (fine-grained temporal variation) (Moran, 1992; Stratton and Bennington, 1998; Baythavong, 2011). Gene flow between habitats can introduce maladapted alleles into local populations, restricting the evolution of specialization (Lenormand, 2002). However, models suggest that interhabitat gene flow may actually enhance selection for phenotypic plasticity (van Tienderen, 1997; Sultan and Spencer, 2002). Even if parents have only ever experienced one set of environmental conditions, phenotypic plasticity can be advantageous if propagules (seeds or pollen) establish in non-parental habitat types (Alpert and Simms, 2002). Therefore, plasticity could theoretically evolve if the spatial extent of gene flow exceeds the spatial scale of habitat variation, such that a species experiences patchy environments where habitat boundaries occur over small spatial scales (fine-grained spatial variation) (Via and Lande, 1985; van Tienderen, 1997).
Phenotypic plasticity affects a huge variety of phenotypes in response to many stimuli in numerous organisms (e.g., Dudley and Schmitt, 1996; Donohue and Schmitt, 1999; Dorn et al., 2000; Sultan, 2000; Givnish, 2002; Casal et al., 2004; Callahan et al., 2005; Mommer and Visser, 2005; Nussey et al., 2005; Caruso et al., 2006; Levin, 2009; Baythavong and Stanton, 2010; Holeski et al., 2010; Maherali et al., 2010; Moczek et al., 2011); however, it remains challenging to test whether plasticity confers a fitness advantage. Neither the genetic basis nor the adaptive value of phenotypic plasticity is clear, and both have been subjects of much debate (Via et al., 1995). It is certain, however, that in most cases plasticity can be studied through the lens of quantitative genetics—whether viewed as a polygenic trait in its own right (Schlichting and Levin, 1986; Scheiner and Lyman, 1991) or as a by-product of differential selection on polygenic traits in multiple environments (Via and Lande, 1985; Gomulkiewicz and Kirkpatrick, 1992; Via, 1993). Surprisingly, few studies have explicitly tested whether selection favors plasticity (but see Dudley and Schmitt, 1996; Bell and Galloway, 2007; Galloway and Etterson, 2007; Baythavong, 2011). A quantitative genetics perspective is essential for testing whether plasticity is an adaptive response to spatial and temporal variation in native field environments (for example, Baythavong and Stanton, 2010), or is simply an effect of stressful conditions, such as resource limitation (for example, Bell and Galloway, 2007). Laboratory studies may fail to capture the appropriate axes of environmental variation to which species have responded over their evolutionary history. For example, a trait that appears to change as a function of one environmental factor in the field may, in fact, be responding to an unmeasured, correlated factor. In this case, attempts to replicate the causal gradient in the lab are likely to be unsuccessful. Furthermore, plastic responses may require complex combinations of environmental factors that are only found in nature.
Field experiments hold great promise for elucidating the role of plasticity in adaptive evolution because they expose lineages to the spatial and temporal variation in the native environments in which they originally evolved (Pigliucci, 2005; Baythavong and Stanton, 2010). For example, families may exhibit coarse-grained plasticity when replicated over multiple field sites in geographically separated habitats, as well as fine-grained plasticity across multiple blocks within each site. Field experiments can address long-standing questions, such as: Is phenotypic plasticity costly? What is the genetic architecture underlying plastic responses and how do epigenetic modifications contribute to heritable plasticity? Does plasticity constrain or accelerate adaptive evolution (Ghalambor et al., 2007; Paenke et al., 2007; Moczek et al., 2011)? Answers to these questions have practical implications for agriculture and conservation biology in an era of rapidly changing climate and increased abundance of invasive species (for example, Richards et al., 2006; Chown et al., 2007).
Reciprocal transplant and common garden experiments can disentangle local adaptation from phenotypic plasticity and assess how selection on traits changes along environmental gradients. Multiyear field studies, in particular, have the distinct advantage of detecting general patterns of adaptation and/or plasticity at multiple life-history stages even when there is interannual variation in conditions (Kittelson and Maron, 2001; Mojica and Kelly, 2010; Ågren and Schemske, 2012). The extent and directionality of natural selection can change through ontogeny, and selection detected at one life-history stage may misrepresent selection integrated across the lifetime.
Field experiments can also examine the constraining effects of gene flow on adaptive population differentiation (Kawecki, 2008; Holt and Barfield, 2011). For example, all else being equal, local adaptation is expected to be more pronounced in remote populations that experience limited interhabitat gene flow than in populations at ecotones where interhabitat gene flow could be high. Phenotypic variation among wild populations reflects both genetic and environmental differences, as well as genotype × environment interactions—manipulative field experiments are required to infer the relative importance of these factors.
Agents of selection: the importance of ecology for quantitative genetics
To understand the interface between ecology and evolution, we must investigate biotic and abiotic agents of selection. Various abiotic and biotic pressures could interact to intensify or alleviate selection imposed by only a single agent. A quantitative trait that affects an organism's interactions with multiple species in the community, or increases fitness under specific abiotic conditions, could cause evolutionary tradeoffs that maintain genetic variation through antagonistic pleiotropy (Mitchell-Olds et al., 2007). The true effect of community-level interactions on the heritability of polygenic traits and selection on these traits can only be inferred through dedicated experiments that manipulate the putative agent(s) of selection.
Quantitative genetic studies have mostly neglected to isolate the effects of specific agents of selection, perhaps because of the difficulty of maintaining the large sample sizes needed to estimate quantitative genetic parameters, while manipulating environmental conditions. Nevertheless, manipulative quantitative genetic experiments have successfully revealed the magnitude and direction of selection on floral traits imposed by biotic agents of selection such as pollinators and seed predators (for example, Campbell et al., 1994; Parachnowitsch and Caruso, 2008; Sletvold et al., 2010). Indeed, by preferentially visiting flowers of only one color, pollinators can even cause prezygotic reproductive isolation between congeneric plant species with overlapping ranges (Hopkins and Rausher, 2012), a conclusion that could only be drawn from a field study in native habitats. Interestingly, polymorphism in flower color can be maintained not only by pollinator preferences but also by herbivores, pathogens, temperature and UV light, indicating a role for both biotic and abiotic agents of selection in the evolution of a key floral trait (Dick et al., 2011). A recent meta-analysis found, however, that traits under biotic selection are more likely than those under abiotic selection to be controlled by QTL of large effect, suggesting that biotic environments are more complex and offer a more rugged adaptive landscape than abiotic environments (Louthan and Kay, 2011).
In some cases, we understand the functional and adaptive significance of intermediate-level phenotypes—and even their molecular basis—without understanding the mechanisms of selection at the ecological level. For example, glucosinolates are a family of secondary chemicals (Halkier and Gershenzon, 2006) that protect against insect herbivory in the model plant A. thaliana and other Brassicaceous plants such as B. stricta (Schranz et al., 2009; Manzaneda et al., 2010; Prasad et al., 2012). The genetic and biochemical basis of glucosinolate production is well established (Halkier and Gershenzon, 2006). Glucosinolate diversity has functional significance (Manzaneda et al., 2010) and has been evolving non-neutrally (Benderoth et al., 2006; Prasad et al., 2012). The exact agents and mechanisms of selection remain unknown, however, which raises more questions about the ecological role of this quantitative trait. Why are different types of glucosinolates more effective in different habitats (Prasad et al., 2012)? In the absence of herbivory, do the selective advantages of various plant defenses persist or disappear? How do antiherbivore defenses such as glucosinolates respond to selective pressures imposed by other agents, such as pathogens (Sanchez-Vallet et al., 2010) and microbial mutualists (Bressan et al., 2009)? More generally, what role do different abiotic and biotic agents of selection play in the evolution of complex traits? These questions reflect quantitative genetic issues relevant to any ecological system that can be addressed using carefully designed field experiments manipulating biotic communities.
Targets of selection: Dissecting the genetic basis of quantitative traits and fitness components
Most species maintain considerable intra- and interpopulation variation in morphology, development, physiology and susceptibility to abiotic and biotic stresses. The polygenic nature of these complex phenotypes is the primary reason why very few genes underlying ecologically relevant traits have been identified (Ingvarsson and Street, 2011). In traits with simple Mendelian inheritance, the strong association between genotype and phenotype means that experiments can easily quantify effects of chromosomal regions <1 cM. However, it is much more difficult to resolve the genetic basis of complex polygenic traits owing to locus heterogeneity (mutations in different genes explain variation in one trait), epistasis, variation in the extent to which a given allele produces a given phenotype (variable expressivity), pleiotropy, genes with small effects and limited statistical power.
Despite these difficulties, QTL mapping directly links phenotypes and/or fitness to the genotype of a specific region of the genome and can be a first step in dissecting the genetic basis of complex traits (Mackay et al., 2009). Artificial laboratory environments are unlikely to reflect the selective pressures experienced by a species throughout its evolutionary history, and different QTL have been detected in field and laboratory settings (Weinig et al., 2002; Brachi et al., 2010; Anderson et al., 2011). Therefore, to understand the evolution of complex traits in natural environments, researchers must conduct QTL mapping and genome-wide association studies in natural environments in which the parental lines originally evolved. Ideally, these field studies should be replicated in space and time to account for microhabitat and interannual environmental heterogeneity. Identifying QTL and quantitative trait nucleotides for complex traits will enable researchers to investigate whether allelic variation at these loci is associated with environmental conditions that vary across the range of a species, and to illuminate the roles of divergent selection, population history, gene flow and genetic drift in maintaining genetic variation.
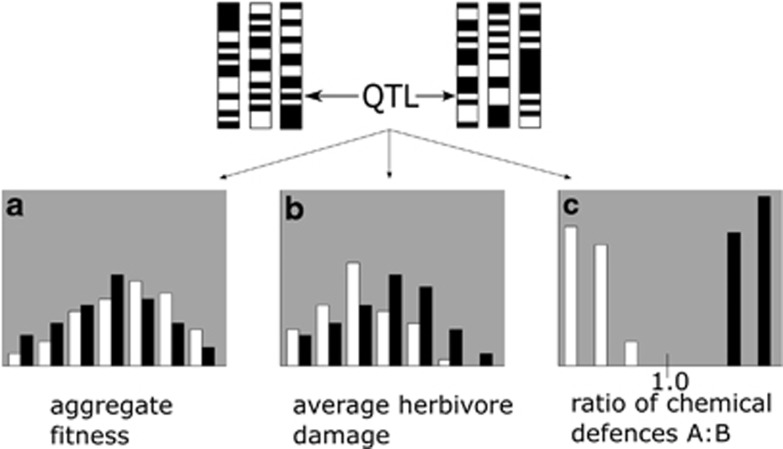
As evolutionary ecologists, we are interested in the genetic basis of traits that affect fitness, because molecular properties such as network position and epistatic interactions determine how readily a gene will respond to selection (Olson-Manning et al., 2012). Although fitness can be considered a quantitative trait—one that is under strong positive selection—in practice, mapping QTL for fitness and ultimately identifying causal genes continues to be difficult (Barrett and Hoekstra, 2011). For one, fitness data do not generally follow a normal distribution, but rather binomial (for example, survivorship) or Poisson (for example, number of offspring), and lifetime fitness is often zero-inflated (over-representation of zero values). These distribution patterns can increase the difficulty of detecting QTL for components of fitness. In addition, like other high-level phenotypes that are aggregates of simpler traits, fitness is ultimately influenced by every locus with an effect on one of the underlying phenotypes. Consequently, the influence of each QTL on fitness might be so diluted that it barely registers even in mapping studies with high statistical power (Figure 2; Rockman, 2012). This limitation is especially true when measuring fitness in the field, where noise introduced by environmental stochasticity could mask even large-effect QTL. Therefore, in many cases a combination of field and laboratory work is likely to be necessary to identify systematically QTL that affect fitness in nature. Below, we describe a recent successful example of such an approach.
Figure 2.
Distributions of fitness and underlying phenotypes in a mapping population for individuals with two alleles (‘white' and ‘black') at a putatively adaptive QTL controlling the ratio of two types of chemical defenses. (a) The fitness distribution has a large variance and very little differentiation between the groups, indicating that this QTL has a small effect on fitness. (b) The QTL has a more defined effect on herbivory; variance is smaller and individuals with the black allele suffer significantly more herbivore damage than those with the white allele when grown together in a common garden. (c) The phenotype A:B has low variance and strong contrast, making it easy to map to the same QTL that has more diluted effects on herbivory and fitness. Herbivore resistance is controlled by fewer genes than fitness (a) but more genes than chemical defense ratio (c).
The genetic basis of adaptation: a case study in herbivory and secondary chemistry
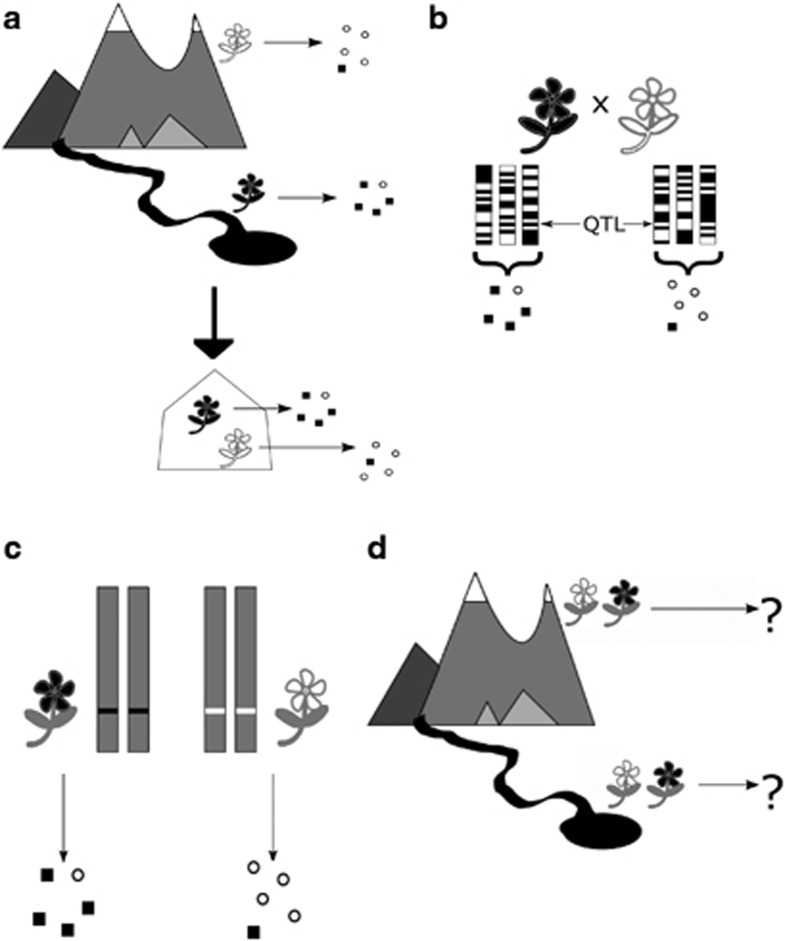
To demonstrate the adaptive significance of loci involved in complex traits, it may be necessary first to show that a locus influences an intermediate-level phenotype that in turn affects fitness (Figure 3). The underlying developmental, biochemical or physiological traits may be controlled by fewer genes than more complex, composite phenotypes; hence, information on functional traits may reveal strong QTL (Figure 2). In cases where a strong QTL for a putatively adaptive trait is known, but cannot immediately be shown to influence fitness or other higher-level polygenic traits, a stepwise experimental approach can validate the QTL's adaptive importance.
Figure 3.
Stepwise approach to discovery and verification of adaptive QTL in natural environments. (a) Genetic control of a putatively adaptive phenotypic difference observed between two geographically separated ecotypes must be confirmed by growing them in a common location. (b) Family-based genetic mapping can be used to identify genomic regions associated with the phenotype of interest. (c) To pinpoint the effects of the candidate adaptive QTL, confounding genetic variation can be eliminated via repeated backcrossing of heterozygotes, so that the segregating alleles of the QTL are preserved. (d) The resulting near-isogenic lines, genetically identical except for the QTL of interest, can be grown together in the parental environments. Phenotypic and fitness differences between the genotypes reflect variation at the candidate QTL.
We used precisely this strategy to establish that allelic variation at the BCMA (Branched Chain Methionine Allocation) locus has important fitness consequences for B. stricta growing in its native habitats (Prasad et al., 2012). BCMA was first identified in a mapping experiment as a QTL controlling both insect damage and glucosinolate profiles in the laboratory (Schranz et al., 2009). That study, which used F2 and F3 generations, did not measure reproductive fitness (Schranz et al., 2009). Although BCMA showed a significant QTL for both glucosinolates and foliar damage, its effects on fitness in the field might have been too small to stand out from those of the many other loci influencing fitness (Figure 2). Furthermore, QTL controlling fitness under lab conditions (as in Schranz et al., 2009) may be unrelated to QTL controlling fitness in the field. Nevertheless, subsequent field experiments focusing on this locus detected a fitness effect of BCMA (Figure 3; Prasad et al., 2012). The original identification of a large effect QTL controlling >20% of variation in glucosinolate content (Figure 3b; Schranz et al., 2009) allowed us to breed near-isogenic lines segregating only for BCMA and a small flanking genomic region (Figure 3c). By eliminating confounding genetic variation, we demonstrated that BCMA is under strong selection in the field (Prasad et al., 2012).
Approaching small-effect QTL and eQTL
The BCMA story illustrates one of the biggest challenges for evolutionary ecologists studying polygenic traits: characterizing QTL of small effect (Rockman, 2012). In the original experiments BCMA explained only 17.9% of variation in herbivore damage, but 80.4% of the variation in a simpler underlying phenotype, glucosinolate biochemistry (Schranz et al., 2009). Although herbivore damage is a much more complex phenotype than glucosinolate biochemistry, it can also be considered a relatively simple trait compared with reproductive fitness; therefore, BCMA would have explained an even smaller fraction of the variation in fitness (Figure 2). Only through methodical breeding and field experimentation were we able to demonstrate that this QTL influences fitness in the field.
Clearly, the effect size of any given locus depends on the phenotype under consideration and the environmental context. For example, thousands of QTL influencing steady-state gene transcript levels are routinely detected in eQTL studies (Gilad et al., 2008). Undoubtedly, most of these loci would have vanishingly small impacts on ecologically salient phenotypes, and therefore would not be detected in mapping experiments at an ecological level. Although the influence of transcript abundance for organismal fitness is unclear, it is likely that eQTL are among the numerous minor QTL that, together, determine evolutionarily important higher-level phenotypes (Hansen et al., 2008).
Phenotypic variation can result from differences in protein coding sequence in the causal gene(s), or small changes in transcriptional regulation (Cubillos et al., 2012). It is now straightforward to map eQTL to detect genomic regions that regulate differential gene expression (Kliebenstein, 2009). Furthermore, mapping eQTLs can clarify the relationship between gene expression polymorphism and phenotypic variation for a given trait (Hansen et al., 2008). Gene expression is often heritable and has a complex underlying genetic architecture (for example, Stam and Laurie, 1996; Kliebenstein, 2009; Cubillos et al., 2012). As prices for next generation sequencing decline, studies of complex variation in gene expression may identify functionally important polymorphisms that contribute to components of fitness in nature. Combining QTL mapping with eQTL analysis could strengthen our ability to investigate the genetic basis of phenotypic variation and adaptation (Hansen et al., 2008; Kliebenstein, 2009 and references therein).
Despite recent progress in understanding transcriptomics in the laboratory, very few studies have investigated gene expression in complex field environments. Aikawa et al. (2011) quantified expression of the FLC flowering locus in field populations of perennial Arabidopsis halleri, and found that FLC expression levels integrate temperature signals over the 6 weeks preceding flowering. Recently, Richards et al. (2012) determined that the expression of many A. thaliana genes changed through ontogeny under natural conditions. Furthermore, these gene expression changes varied with accession, indicating that the genetic background contributes to expression polymorphism (Richards et al., 2012). Finally, genes regulated by abiotic and biotic stresses were significantly over-represented in many coexpressed gene clusters (Richards et al., 2012). Investigating molecular networks and gene expression profiles in the wild will allow us to link gene function to ecologically relevant plant responses, illuminating the molecular basis of adaptation to natural environments. Although the transcriptional response to a single stimulus may be most easily studied in the lab, understanding its role among the myriad other stimuli and responses experienced in nature can only be addressed through field experimentation.
Linking the immediate molecular effects of genetic variants to their impact (if any) on an organism's ecology and evolution is a major undertaking. Nevertheless, starting with low-level molecular processes, such as physiological differences or changing gene expression, may be a good way to discover the majority of loci that control ecologically important polygenic traits. The functional importance of any given QTL may only become apparent after extensive experimentation. This demanding process may be more feasible, and more rewarding, in some systems than others; but applied efficiently and judiciously, this bottom-up approach could eventually reveal causal genes with unforeseen links to fitness (Gilad et al., 2008).
The effects of reproductive system on fitness and quantitative traits in nature
The evolutionary dynamics of quantitative traits are inextricably linked with the breeding system of the study organism, which may itself be dependent on environment. The predominant breeding systems in plants are outcrossing, self-fertilization and apomixis (asexual reproduction via seed). Evolutionary theory states that most plants should reproduce either via complete self-fertilization or complete outcrossing, with mixed mating existing only ephemerally (Lande and Schemske, 1985). However, recent work suggests that outcrossing and intermediate selfing are both more common than highly selfing taxa, and that a mixed mating strategy might be more stable than previously thought (Goodwillie et al., 2005; Johnston et al., 2009). The ratio of selfing to outcrossing in mixed mating system plants may vary among or within populations, depending on biotic factors including pollinator limitation and pollen discounting (Goodwillie et al., 2005), as well as abiotic environmental factors such as light and water availability (for example, Waller, 1980). A recent review suggests that selfing rates change as a result of anthropogenic disturbance, whether through biotic or abiotic agents (Eckert et al., 2010). Thus, it is imperative that experiments addressing mating system variation take place in the field, while considering the quality of the environment as well as its similarity to the study organism's ancestral habitat.
Breeding system affects evolutionary parameters such as genetic diversity, patterns of gene flow and sex ratios, which subsequently influence selection, genetic response to selection and adaptation (Charlesworth, 2003). To our knowledge, no comparative experiment has definitively shown adaptive deficiencies in selfers, leaving an outstanding gap in our understanding of comparative patterns of adaptation in selfing and outcrossing lineages. Nevertheless, carefully designed field experiments focusing on the native agents of natural selection, combined with extensive biological and genetic characterization of a system, may offer a solution.
Theory predicts that inbreeding dramatically increases homozygosity, decreases the effective population size and the effective recombination rate of a given species, diminishes genetic diversity (Charlesworth, 2003) and increases linkage disequilibrium in comparison with outcrossing (Nordborg, 2000). These factors result in the accumulation of deleterious alleles in inbred lineages (Lynch et al., 1995), and a reduced efficacy of natural selection compared with outcrossing species (Glémin et al., 2006; Wright et al., 2008). However, one recent comparative study found no evidence of elevated nonsynonymous mutations in selfing species of Triticaceae, suggesting that selection may not be relaxed in selfing species (Escobar et al., 2010). Several empirical studies have demonstrated reduced genetic diversity in self-fertilizing species (Glémin et al., 2006). However, average genome-wide linkage disequilibrium decays surprisingly quickly (within 10 kb) in the selfing mustards A. thaliana (Kim et al., 2007) and B. stricta (Song et al., 2009).
Inbreeding depression (ID) reduces survival, flower number, fruit number, plant height, seed weight and seed mass, all of which are important and polygenic components of fitness (Charlesworth and Willis, 2009). It is commonly accepted that the magnitude of ID can vary across environments. One classic study of three environments (natural, greenhouse and common garden) uncovered a 50% reduction in fitness in selfed vs outcrossed progeny of the self-compatible Sabatia angularis—but only in natural conditions (Dudash, 1990). Since this groundbreaking work, it has become clear that the effects of ID on fitness should be studied in the field, and in multiple environments (Dudash, 1990; Cheptou and Donohue, 2011).
ID may increase in stressful environmental conditions (Fox and Reed, 2011, but see Armbruster and Reed, 2005), which could reduce the potential for adaptive responses to anthropogenic disturbance or rapidly changing climate. To address the fitness implications of climate change, habitat fragmentation and environmental degradation for inbred populations, future research must focus on quantifying ID in both ancestral and changing environments.
Finally, as natural selection changes throughout ontogeny, so does the magnitude of ID, with variable results; some studies find strong ID early in life, while others find stronger ID in later life stages (Husband and Schemske, 1996). More recent work has not resolved this complication, highlighting a strong need for more field studies incorporating fitness and ID estimates at multiple life stages.
Emerging global problems
A quantitative genetics framework can provide much needed insight into conservation biology by exploring whether anthropogenic disturbance alters natural or sexual selection and whether populations harbor sufficient genetic variation to respond via adaptation (for example, Hoffmann and Sgrò, 2011; Shaw and Etterson, 2012). For example, Franks et al. (2007) demonstrated rapid evolution of drought avoidance in Brassica rapa by comparing ancestral lineages (derived from seeds collected before a multiyear drought) with descendent and hybrid lineages. Extremely strong selection for earlier flowering, along with high heritabilities and short generation time, resulted in rapid adaptive evolution in response to climate change (Franks et al., 2007). Climate change has also imposed selection on body coloration in animals (Karell et al., 2011). Historically, for tawny owls in Europe, viability selection during snowy winters favored individuals with pale (gray) plumage over brown morphs (Karell et al., 2011). However, recent warming has resulted in milder winters, and relaxation of selection against owls with brown plumage; concomitantly, the frequency of the brown morph has increased in this population (Karell et al., 2011). Thus, evolution can proceed rapidly in response to anthropogenic disturbance (see also, Bradshaw and Holzapfel, 2001; Réale et al., 2003; Nussey et al., 2005; Franks et al., 2007; Gienapp et al., 2008). As heritabilities and genetic (co)variances are often inflated under controlled laboratory conditions, field studies are critical for estimating the evolutionary response to novel selection imposed by anthropogenic disturbance (Geber and Griffen, 2003).
We have begun to investigate potential adaptive responses to climate change in B. stricta, with a focus on flowering phenology because this life-history trait is highly responsive to complex and interacting climatic variables (for example, Eckhart et al., 2004; Sherry et al., 2007; Kim et al., 2009; Wilczek et al., 2009, 2010; Lambert et al., 2010; Blackman et al., 2011; Hancock et al., 2011; Chew et al., 2012). Current projections indicate that temperatures will continue to rise at an increasing rate (Williams et al., 2007) and growing seasons could begin earlier because of advancing snowmelt (Anderson et al., 2012a), but other conditions that influence the optimal timing of flowering, such as photoperiod, are unlikely to change. Decoupling of previously reliable environmental variables could depress fitness in species that depend on multiple signals to trigger seasonal life-history transitions (Wilczek et al., 2010). As flowering phenology is well-characterized at the genetic level (Wilczek et al., 2010), it offers excellent future opportunities for tracking temporal changes in allele frequencies at candidate genes associated with vernalization and temperature (Kumar et al., 2012).
Our fieldwork with B. stricta detected strong directional selection for earlier flowering, along with moderate heritability for the timing of first flowering (Anderson et al., 2012a). Consistent with contemporary directional selection, flowering phenology has advanced by roughly 3.4 days per decade in B. stricta since the mid 1970s (Anderson et al., 2012a), a figure that is similar to long-term phenological changes in a diverse array of taxa, including plants, birds, butterflies and other insects, and amphibians (Parmesan and Yohe, 2003; Jonzén et al., 2006; Beaubien and Hamann, 2011). Our field and laboratory studies suggest that the majority of the observed long-term advancement in flowering observed from 1973 to 2011 is due to plasticity, not adaptive evolution (Anderson et al., 2011, 2012a). However, we estimated a potential evolutionary response to contemporary levels of selection of 0.2–0.5 days acceleration in flowering per generation using the breeder's equation and Robertson–Price identity, respectively (Anderson et al., 2012a), which would result in 1–2.5 days acceleration per decade, assuming generation time is 2 years. Thus, it seems possible that B. stricta could adapt its flowering phenology to a warming climate.
It remains to be seen whether adaptive evolution will keep pace with rapid anthropogenic climate change and habitat degradation (Hoffmann and Sgrò, 2011). Reduction in effective population size due to habitat loss will decrease the likelihood of adaptive evolution (Anderson et al., 2012b). Species with short generation times may be capable of adapting to novel selection, whereas for long-lived species, strategies such as phenotypic plasticity and distributional shifts may be likelier methods of surviving anthropogenic disturbance.
Conclusions and future directions
Recently, there has been a major shift in ecology to trait-based approaches that investigate how phenotypes influence community structure (Cornwell and Ackerly, 2010). Quantitative geneticists have long appreciated the importance of traits in the evolutionary dynamics of natural and crop species. Perhaps, it is now time for evolutionary geneticists to borrow the manipulative experimental approaches of field ecologists. Via complementary field and laboratory studies, researchers can examine evolutionary processes that constrain adaptive population differentiation, identify traits that are subject to selection, investigate the effects of breeding system on important complex traits and perhaps uncover novel QTL or causal genes (Lowry et al., 2009). Quantitative genetic studies rarely manipulate abiotic and biotic conditions in the field, but such investigations can tease apart the effects of different agents of selection on ecologically relevant traits (Parachnowitsch and Caruso, 2008) and could potentially identify intriguing tradeoffs that would otherwise go unnoticed. For example, exclosures or enclosures could be used to investigate herbivore-mediated selection on traits, and to map QTL for herbivore resistance and tolerance in response to the entire herbivore community or specific herbivores. Experiments that manipulate temperature, drought and other climatic variables (for example, snowpack) could advance our understanding of fitness consequences of global change while providing valuable insight about climate-mediated selection on phenotypes. Such experiments are increasingly important in a rapidly changing world.
Acknowledgments
We acknowledge support from the NIH (R01 GM086496 to TM-O, Training Grant 5T32GM007754-32 to Duke University Program in Genetics and Genomics), NSF (EF-0723447 to TM-O) and the University of South Carolina. We apologize to the numerous authors whose excellent studies we were unable to cite here. We thank Bruce Walsh and two anonymous reviewers for helpful comments on a previous draft.
The authors declare no conflicts of interest.
References
- Ågren J, Schemske DW. Reciprocal transplants demonstrate strong adaptive differentiation of the model organism Arabidopsis thaliana in its native range. New Phytologist. 2012;194:1112–1122. doi: 10.1111/j.1469-8137.2012.04112.x. [DOI] [PubMed] [Google Scholar]
- Aikawa S, Kobayashi MJ, Satake A, Shimizu KK, Kudoh H. Robust control of the seasonal expression of the Arabidopsis FLC gene in a fluctuating environment. Proc Natl Acad Sci USA. 2011;107:11632–11637. doi: 10.1073/pnas.0914293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpert P, Simms EL. The relative advantages of plasticity and fixity in different environments: when is it good for a plant to adjust. Evol Ecol. 2002;16:285–297. [Google Scholar]
- Anderson JT, Inouye D, McKinney A, Colautti R, Mitchell-Olds T. Phenotypic plasticity and adaptive evolution contribute to advancing flowering phenology in response to climate change. Proc R Soc Se B. 2012a;279:3843–3852. doi: 10.1098/rspb.2012.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JT, Lee CR, Mitchell-Olds T. Life-history QTLs and natural selection on flowering time in Boechera stricta, a perennial relative of Arabidopsis. Evolution. 2011;65:771–787. doi: 10.1111/j.1558-5646.2010.01175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JT, Panetta AM, Mitchell-Olds T. Evolutionary and ecological responses to anthropogenic climate change. Plant Physiol. 2012b;160:1728–1740. doi: 10.1104/pp.112.206219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster P, Reed DH. Inbreeding depression in benign and stressful environments. Heredity. 2005;95:235–242. doi: 10.1038/sj.hdy.6800721. [DOI] [PubMed] [Google Scholar]
- Barrett RDH, Hoekstra H. Molecular spandrels: tests of adaptation at the genetic level. Nat Rev Genet. 2011;12:767–780. doi: 10.1038/nrg3015. [DOI] [PubMed] [Google Scholar]
- Baythavong BS. Linking the spatial scale of environmental variation and the evolution of phenotypic plasticity: selection favors adaptive plasticity in fine-grained environments. Am Nat. 2011;178:75–87. doi: 10.1086/660281. [DOI] [PubMed] [Google Scholar]
- Baythavong BS, Stanton ML. Characterizing selection on phenotypic plasticity in response to natural environmental heterogeneity. Evolution. 2010;64:2904–2920. doi: 10.1111/j.1558-5646.2010.01057.x. [DOI] [PubMed] [Google Scholar]
- Beaubien E, Hamann A. Spring flowering response to climate change between 1936 and 2006 in Alberta, Canada. Bioscience. 2011;61:514–524. [Google Scholar]
- Bell DL, Galloway LF. Plasticity to neighbour shade: fitness consequences and allometry. Funct Ecol. 2007;21:1146–1153. [Google Scholar]
- Benderoth M, Textor S, Windsor A, Mitchell-Olds T, Gershenzon J, Kroymann J. Positive selection driving diversification in plant secondary metabolism. Proc Natl Acad Sci. 2006;103:9118–9123. doi: 10.1073/pnas.0601738103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman BK, Michaels SD, Rieseberg LH. Connecting the sun to flowering in sunflower adaptation. Mol Ecol. 2011;20:3503–3512. doi: 10.1111/j.1365-294X.2011.05166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez MA, Ahn JH, Weigel D. A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat Genet. 2003;33:168. doi: 10.1038/ng1085. [DOI] [PubMed] [Google Scholar]
- Brachi B, Faure N, Horton M, Flahauw E, Vazquez A, Nordborg M, et al. Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet. 2010;6:e1000940. doi: 10.1371/journal.pgen.1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw WE, Holzapfel C. Genetic shift in photoperiodic response correlated with global warming. Proc Natl Acad Sci. 2001;98:14509–14511. doi: 10.1073/pnas.241391498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan M, Roncato MA, Bellvert F, Comte G, el Zahar Haichar F, Achouak W, et al. Exogenous glucosinolate produced by Arabidopsis thaliana has an impact on microbes in the rhizosphere and plant roots. ISME J. 2009;3:1243–1257. doi: 10.1038/ismej.2009.68. [DOI] [PubMed] [Google Scholar]
- Brock MT, Stinchcombe JR, Weinig C. Indirect effects of FRIGIDA: floral trait (co)variances are altered by seasonally variable abiotic factors associated with flowering time. J Evol Biol. 2009;22:1826–1838. doi: 10.1111/j.1420-9101.2009.01794.x. [DOI] [PubMed] [Google Scholar]
- Brys R, Shefferson RP, Jacquemyn H. Impact of herbivory on flowering behaviour and life history trade-offs in a polycarpic herb: a 10-year experiment. Oecologia. 2011;166:293–303. doi: 10.1007/s00442-010-1842-7. [DOI] [PubMed] [Google Scholar]
- Callahan HS, Dhanoolal N, Ungerer MC. Plasticity genes and plasticity costs: a new approach using an Arabidopsis recombinant inbred population. New Phytol. 2005;166:129–139. doi: 10.1111/j.1469-8137.2005.01368.x. [DOI] [PubMed] [Google Scholar]
- Campbell DR, Waser NM, Price MV. Indirect selection of stigma position in Ipomopsis aggregata via a genetically correlated trait. Evolution. 1994;48:55–68. doi: 10.1111/j.1558-5646.1994.tb01294.x. [DOI] [PubMed] [Google Scholar]
- Caruso CM, Maherali H, Sherrard M. Plasticity of physiology in Lobelia: testing for adaptation and constraint. Evolution. 2006;60:980–990. [PubMed] [Google Scholar]
- Casal JJ, Fankhauser C, Coupland G, Blazquez MA. Signalling for developmental plasticity. Trends Plant Sci. 2004;9:309–314. doi: 10.1016/j.tplants.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Charlesworth D. Effects of inbreeding on the genetic diversity of populations. Philos Trans R Soc Lond Ser B. 2003;358:1051–1070. doi: 10.1098/rstb.2003.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Willis J. Fundamental concepts in genetics: the genetics of inbreeding depression. Nat Rev Genet. 2009;10:783–796. doi: 10.1038/nrg2664. [DOI] [PubMed] [Google Scholar]
- Cheptou P, Donohue K. Environment-dependent inbreeding depression: its ecological and evolutionary significance. New Phytol. 2011;189:395–407. doi: 10.1111/j.1469-8137.2010.03541.x. [DOI] [PubMed] [Google Scholar]
- Chew YH, Wilczek AM, Williams M, Welch SM, Schmitt J, Halliday KJ. An augmented Arabidopsis phenology model reveals seasonal temperature control of flowering time. New Phytol. 2012;194:654–665. doi: 10.1111/j.1469-8137.2012.04069.x. [DOI] [PubMed] [Google Scholar]
- Chown SL, Slabber S, McGeoch MA, Janion C, Leinaas HP. Phenotypic plasticity mediates climate change responses among invasive and indigenous arthropods. Proc R Soc Ser B. 2007;274:2531–2537. doi: 10.1098/rspb.2007.0772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornwell WK, Ackerly DD. A link between plant traits and abundance: evidence from coastal California woody plants. J Ecol. 2010;98:814–821. [Google Scholar]
- Cubillos F, Yansouni J, Khalili H, Balzergue S, Elftieh S, Martin-Magniette M-L, et al. Expression variation in connected recombinant populations of Arabidopsis thaliana highlights distinct transcriptome architectures. BMC Genom. 2012;13:117. doi: 10.1186/1471-2164-13-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick CA, Buenrostro J, Butler T, Carlson ML, Kliebenstein DJ, Whittall JB. Arctic mustard flower color polymorphism controlled by petal-specific downregulation at the threshold of the anthocyanin biosynthetic pathway. PLoS One. 2011;6:e18230. doi: 10.1371/journal.pone.0018230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue K, Schmitt J. The genetic architecture of plasticity to density in Impatiens capensis. Evolution. 1999;53:1377–1386. doi: 10.1111/j.1558-5646.1999.tb05402.x. [DOI] [PubMed] [Google Scholar]
- Dorn LA, Hammond Pyle E, Schmitt J. Plasticity to light cues and resources in Arabidopsis thaliana: testing for adaptive value and costs. Evolution. 2000;54:1982–1994. doi: 10.1111/j.0014-3820.2000.tb01242.x. [DOI] [PubMed] [Google Scholar]
- Dudash MR. Relative fitness of selfed and outcrossed progeny in a self-compatible, protandrous species, Sabatia angularis L. (Gentianaceae): a comparison in three environments. Evolution. 1990;44:1129–1139. doi: 10.1111/j.1558-5646.1990.tb05220.x. [DOI] [PubMed] [Google Scholar]
- Dudley SA, Schmitt J. Testing the adaptive plasticity hypothesis: density-dependent selection on manipulated stem length in Impatiens capensis. Am Nat. 1996;147:445–465. [Google Scholar]
- Eckert CG, Kalisz S, Geber MA, Sargent R, Elle E, Cheptou PO, et al. Plant mating systems in a changing world. Trends Ecol Evol. 2010;25:35–43. doi: 10.1016/j.tree.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Eckhart V, Geber M, McGuire CM. Experimental studies of adaptation in Clarkia xantiana. I. Sources of trait variation across a subspecies border. Evolution. 2004;58:59–70. doi: 10.1111/j.0014-3820.2004.tb01573.x. [DOI] [PubMed] [Google Scholar]
- Escobar J, Cenci A, Bolognini J, Haudry A, Laurent S, David J, et al. An integrative test of the dead-end hypothesis of selfing evolution in Triticeae (Poaceae) Evolution. 2010;64:2855–2872. doi: 10.1111/j.1558-5646.2010.01045.x. [DOI] [PubMed] [Google Scholar]
- Etterson JR, Shaw RG. Constraint to adaptive evolution in response to global warming. Science. 2001;294:151–154. doi: 10.1126/science.1063656. [DOI] [PubMed] [Google Scholar]
- Forrest J, Miller-Rushing AJ. Toward a synthetic understanding of the role of phenology in ecology and evolution. Philos Trans R Soc Lond Ser B. 2010;365:3101–3112. doi: 10.1098/rstb.2010.0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox C, Reed D. Inbreeding depression increases with environmental stress: an experimental study and meta-analysis. Evolution. 2011;65:246–258. doi: 10.1111/j.1558-5646.2010.01108.x. [DOI] [PubMed] [Google Scholar]
- Franks S, Sim S, Weis A. Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proc Natl Acad Sci. 2007;104:1278–1282. doi: 10.1073/pnas.0608379104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway LF, Etterson JR. Transgenerational plasticity is adaptive in the wild. Science. 2007;318:1134–1136. doi: 10.1126/science.1148766. [DOI] [PubMed] [Google Scholar]
- Geber M, Griffen LR. Inheritance and natural selection on functional traits. Int J Plant Sci. 2003;164:S21–S42. [Google Scholar]
- Ghalambor CK, McKay JK, Carroll SP, Reznick DN. Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments. Funct Ecol. 2007;21:394–407. [Google Scholar]
- Gienapp P, Teplitsky C, Alho J, Mills J, Merila J. Climate change and evolution: disentangling environmental and genetic responses. Mol Ecol. 2008;17:167–178. doi: 10.1111/j.1365-294X.2007.03413.x. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Rifkin SA, Pritchard JK. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 2008;24:408–415. doi: 10.1016/j.tig.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givnish TJ. Ecological constraints on the evolution of plasticity in plants. Evol Ecol. 2002;16:213–242. [Google Scholar]
- Glémin S, Bazin E, Charlesworth D. Impact of mating systems on patterns of sequence polymorphism in flowering plants. Proc R Soc Ser B. 2006;273:3011–3019. doi: 10.1098/rspb.2006.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomulkiewicz R, Kirkpatrick M. Quantitative genetics and the evolution of reaction norms. Evolution. 1992;46:390–411. doi: 10.1111/j.1558-5646.1992.tb02047.x. [DOI] [PubMed] [Google Scholar]
- Goodwillie C, Kalisz S, Eckert CG. The evolutionary enigma of mixed mating systems in plants: occurrence, theoretical explanations, and empirical evidence. Annu Rev Ecol Evol Syst. 2005;36:47–79. [Google Scholar]
- Halkier BA, Gershenzon J. Biology and biochemistry of glucosinolates. Annu Rev Plant Biol. 2006;57:303–333. doi: 10.1146/annurev.arplant.57.032905.105228. [DOI] [PubMed] [Google Scholar]
- Hall MC, Willis JH. Divergent selection on flowering time contributes to local adaptation in Mimulus guttatus populations. Evolution. 2006;60:2466–2477. [PubMed] [Google Scholar]
- Hancock AM, Brachi B, FAure N, Horton MW, Jarymowycz LB, Sperone FG, et al. Adaptation to climate across the Arabidopsis thaliana genome. Science. 2011;334:83–86. doi: 10.1126/science.1209244. [DOI] [PubMed] [Google Scholar]
- Hansen BG, Halkier BA, Kliebenstein DJ. Identifying the molecular basis of QTLs: eQTLs add a new dimension. Trends Plant Sci. 2008;13:72–77. doi: 10.1016/j.tplants.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. Genetic polymorphism in heterogeneous environments: a decade later. Annu Rev Ecol Syst. 1986;17:535–566. [Google Scholar]
- Hereford J. A quantitative survey of local adaptation and fitness trade-offs. Am Nat. 2009;173:579–588. doi: 10.1086/597611. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Sgrò C. Climate change and evolutionary adaptation. Nature. 2011;470:479–485. doi: 10.1038/nature09670. [DOI] [PubMed] [Google Scholar]
- Holeski LM, Chase-Alone R, Kelly JK. The genetics of phenotypic plasticity in plant defense: trichome production in Mimulus guttatus. Am Nat. 2010;175:391–400. doi: 10.1086/651300. [DOI] [PubMed] [Google Scholar]
- Holt RD, Barfield M. Theoretical perspectives on the statics and dynamics of species' borders in patchy environments. Am Nat. 2011;178:S6–S25. doi: 10.1086/661784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins R, Rausher MD. Pollinator-mediated selecton on flower color allele drives reinforcement. Science. 2012;335:1090–1092. doi: 10.1126/science.1215198. [DOI] [PubMed] [Google Scholar]
- Husband BC, Schemske DW. Evoltuion of the magnitude and timing of inbreeding depression in plants. Evolution. 1996;50:54–70. doi: 10.1111/j.1558-5646.1996.tb04472.x. [DOI] [PubMed] [Google Scholar]
- Ingvarsson PK, Street NR. Association genetics of complex traits in plants. New Phytol. 2011;189:909–922. doi: 10.1111/j.1469-8137.2010.03593.x. [DOI] [PubMed] [Google Scholar]
- Johnston MO, Porcher E, Cheptou P-O, Eckert CG, Elle E, Geber M, et al. Correlations among fertility components can maintain mixed mating in plants. Am Nat. 2009;173:1–11. doi: 10.1086/593705. [DOI] [PubMed] [Google Scholar]
- Jonzén N, Lindén A, Ergon T, Knudsen E, Vik JO, Rubolini D, et al. Rapid advance of spring arrival dates in long-distance migratory birds. Science. 2006;312:1959–1961. doi: 10.1126/science.1126119. [DOI] [PubMed] [Google Scholar]
- Karell P, Ahola K, Karstinen T, Valkama J, Brommer JE. Climate change drives microevolution in a wild bird. Nat Commun. 2011;2:208. doi: 10.1038/ncomms1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagoe T, Kudoh H. Escape from floral herbivory by early flowering in Arabidopsis halleri subsp. gemmifera. Oecologia. 2010;164:713–720. doi: 10.1007/s00442-010-1709-y. [DOI] [PubMed] [Google Scholar]
- Kawecki TJ. Adaptation to marginal habitats. Annu Rev Ecol Evol Syst. 2008;39:321–342. [Google Scholar]
- Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecol Lett. 2004;7:1225–1241. [Google Scholar]
- Kim D, Doyle M, Sung S, Amasino R. Vernalization: winter and the timing of flowering in plants. Annu Rev Cell Dev Biol. 2009;25:277–299. doi: 10.1146/annurev.cellbio.042308.113411. [DOI] [PubMed] [Google Scholar]
- Kim S, Plagnol V, Hu TT, Toomajian C, Clark RM, Ossowski S, et al. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet. 2007;39:1151–1155. doi: 10.1038/ng2115. [DOI] [PubMed] [Google Scholar]
- Kittelson PM, Maron JL. Fine-scale genetically based differentiation of life-history traits in the perennial shrub Lupinus arboreus. Evolution. 2001;55:2429–2438. doi: 10.1111/j.0014-3820.2001.tb00758.x. [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ. Quantitative genomics: analyzing intraspecific variation using global gene expression polymorphisms or eQTLs. Annu Rev Plant Biol. 2009;60:93–114. doi: 10.1146/annurev.arplant.043008.092114. [DOI] [PubMed] [Google Scholar]
- Kumar SV, Lucyshyn D, Jaeger KE, Alos E, Alvey E, Harberd NP, et al. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature. 2012;484:242–245. doi: 10.1038/nature10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert AM, Miller-Rushing AJ, Inouye DW. Changes in snowmelt date and summer precipitation affect the flowering phenology of Erythronium grandiflorum (glacier lily; Liliaceae) Am J Bot. 2010;97:1431–1437. doi: 10.3732/ajb.1000095. [DOI] [PubMed] [Google Scholar]
- Lande R, Schemske DW. The evolution of self-fertilization and inbreeding depression in plants. I. Genetic models. Evolution. 1985;39:24–40. doi: 10.1111/j.1558-5646.1985.tb04077.x. [DOI] [PubMed] [Google Scholar]
- Leinonen PH, Remington DL, Leppala J, Savolainen O. Genetic basis of local adaptation and flowering time variation in Arabidopsis lyrata. Mol Ecol. 2013;22:709–723. doi: 10.1111/j.1365-294X.2012.05678.x. [DOI] [PubMed] [Google Scholar]
- Lenormand T. Gene flow and the limits to natural selection. Trends Ecol Evol. 2002;17:183–189. [Google Scholar]
- Levin DA. Flowering-time plasticity facilitates niche shifts in adjacent populations. New Phytol. 2009;183:661–666. doi: 10.1111/j.1469-8137.2009.02889.x. [DOI] [PubMed] [Google Scholar]
- Louthan AM, Kay KM. Comparing the adaptive landscape across trait types: larger QTL effect size in traits under biotic selection. BMC Evol Biol. 2011;11:60. doi: 10.1186/1471-2148-11-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry DB, Hall MC, Salt DE, Willis J. Genetic and physiological basis of adaptive salt tolerance divergence between coastal and inland Mimulus guttatus. New Phytol. 2009;183:776–788. doi: 10.1111/j.1469-8137.2009.02901.x. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery J, Burger R. Mutation accumulation and the extinction of small populations. Am Nat. 1995;146:489–518. [Google Scholar]
- Mackay TF, Stone EA, Ayroles JF. The genetics of quantitative traits: challenges and prospects. Nat Rev Genet. 2009;10:565–577. doi: 10.1038/nrg2612. [DOI] [PubMed] [Google Scholar]
- Maherali H, Caruso CM, Sherrard ME, Latta RG. Adaptive value and costs of physiological plasticity to soil moisture limitation in recombinant inbred lines of Avena barbata. Am Nat. 2010;175:211–224. doi: 10.1086/649598. [DOI] [PubMed] [Google Scholar]
- Manzaneda AJ, Prasad K, Mitchell-Olds T. Variation and fitness costs for tolerance to different types of herbivore damage in Boechera stricta genotypes with contrasting glucosinolate structures. New Phytol. 2010;188:464–477. doi: 10.1111/j.1469-8137.2010.03385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T, Willis JH, Goldstein DB. Which evolutionary processes influence natural genetic variation for phenotypic traits. Nat Rev Genet. 2007;8:845–856. doi: 10.1038/nrg2207. [DOI] [PubMed] [Google Scholar]
- Moczek AP, Sultan SE, Foster S, Ledon-Rettig C, Dworkin I, Nijhout HF, et al. The role of developmental plasticity in evolutionary innovation. Proc R Soc Ser B. 2011;278:2705–2713. doi: 10.1098/rspb.2011.0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica JP, Kelly JK. Viability selection prior to trait expression is an essential component of natural selection. Proc R Soc Ser B. 2010;277:2945–2950. doi: 10.1098/rspb.2010.0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mommer L, Visser EJW. Underwater photosynthesis in flooded terrestrial plants: a matter of leaf plasticity. Ann Bot. 2005;96:581–589. doi: 10.1093/aob/mci212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. The evolutionary maintenance of alternative phenotypes. Evolution. 1992;139:971–989. [Google Scholar]
- Mutasa-Göttgens E, Hedden P. Gibberellin as a factor in floral regulatory networks. J Exp Biol. 2009;60:1979–1989. doi: 10.1093/jxb/erp040. [DOI] [PubMed] [Google Scholar]
- Nordborg M. Linkage disequilibrium, gene trees and selfing: an ancestral recombination graph with partial self-fertilization. Genetics. 2000;154:923–929. doi: 10.1093/genetics/154.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussey DH, Postma E, Gienapp P, Visser ME. Selection on heritable phenotypic plasticity in a wild bird population. Science. 2005;310:304–306. doi: 10.1126/science.1117004. [DOI] [PubMed] [Google Scholar]
- Olson-Manning C, Wagner MR, Mitchell-Olds T. Adaptive evolution: evaluating empirical support for theoretical predictions. Nat Rev Genet. 2012;13:867–877. doi: 10.1038/nrg3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paenke I, Sendhoff B, Kawecki TJ. Influence of plasticity and learning on evolution under directional selection. Am Nat. 2007;170:E47–E58. doi: 10.1086/518952. [DOI] [PubMed] [Google Scholar]
- Parachnowitsch AL, Caruso CM. Predispersal seed herbivores, not pollinators, exert selection on floral traits via female fitness. Ecology. 2008;89:1802–1810. doi: 10.1890/07-0555.1. [DOI] [PubMed] [Google Scholar]
- Parmesan C, Yohe G. A globally coherent fingerprint of climate change impacts across natural systems. Nature. 2003;421:37–42. doi: 10.1038/nature01286. [DOI] [PubMed] [Google Scholar]
- Pigliucci M. Evolution of phenotypic plasticity: where are we going now. Trends Ecol Evol. 2005;20:481–486. doi: 10.1016/j.tree.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Poveda K, Steffan-Dewenter I, Scheu S, Tscharntke T. Effects of below- and above-ground herbivores on plant growth, flower visitation and seed set. Oecologia. 2003;135:601–605. doi: 10.1007/s00442-003-1228-1. [DOI] [PubMed] [Google Scholar]
- Prasad K, Song B-H, Olson-Manning C, Anderson JT, Lee C-R, Schranz ME, et al. A gain of function polymorphism controlling complex traits and fitness in nature. Science. 2012;336:1081–1084. doi: 10.1126/science.1221636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- Réale D, McAdam A, Boutin S, Berteaux D. Genetic and plastic responses of a northern mammal to climate change. Proc R Soc Ser B. 2003;270:591–596. doi: 10.1098/rspb.2002.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards CL, Bossdorf O, Muth NZ, Gurevitch J, Pigliucci M. Jack of all trades, master of some? On the role of phenotypic plasticity in plant invasions. Ecol Lett. 2006;9:981–993. doi: 10.1111/j.1461-0248.2006.00950.x. [DOI] [PubMed] [Google Scholar]
- Richards CL, Rosas U, Banta JA, Bhambhra N, Purugganan M. Genome-wide patterns of Arabidopsis gene expression in nature. PLoS Genet. 2012;8:e1002662. doi: 10.1371/journal.pgen.1002662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV. The QTN program and the alleles that matter for evolution: all that's gold does not glitter. Evolution. 2012;66:1–17. doi: 10.1111/j.1558-5646.2011.01486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushworth C, Song B-H, Lee C-R, Mitchell-Olds T. Boechera, a model system for ecological genomics. Mol Ecol. 2011;20:4843–4857. doi: 10.1111/j.1365-294X.2011.05340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Vallet A, Ramos B, Bednarek P, Lopez G, Pislewska-Bednarek M, Schulze-Lefert P, et al. Tryptophan-derived secondary metabolites in Arabidopsis thaliana confer non-host resistance to necrotrophic Plectosphaerella cucumerina fungi. Plant J. 2010;63:115–127. doi: 10.1111/j.1365-313X.2010.04224.x. [DOI] [PubMed] [Google Scholar]
- Scheiner SM, Lyman RF. The genetics of phenotypic plasticity. II. Response to selection. J Evol Biol. 1991;3:23–50. [Google Scholar]
- Schlichting CD, Levin DA. Phenotypic plasticity: an evolving plant character. Biol J Linn Soc. 1986;29:37–47. [Google Scholar]
- Schranz ME, Manzaneda AJ, Windsor AJ, Clauss MJ, Mitchell-Olds T. Ecological genomics of Boechera stricta: identification of a QTL controlling the allocation of methionine- vs branched-chain amino acid-derived glucosinolates and levels of insect herbivory. Heredity. 2009;102:465–474. doi: 10.1038/hdy.2009.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RG, Etterson JR. Rapid climate change and the rate of adaptation: insight from experimental quantitative genetics. New Phytol. 2012;195:752–765. doi: 10.1111/j.1469-8137.2012.04230.x. [DOI] [PubMed] [Google Scholar]
- Sherry R, Zhou X, Gu S, Arnone J, III, Schimel D, Verburg P, et al. Divergence of reproductive phenology under climate warming. Proc Natl Acad Sci. 2007;104:198–202. doi: 10.1073/pnas.0605642104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sletvold N, Grindeland JM, Ågren J. Pollinator-mediated selection on floral display, spur length and flowering phenology in the deceptive orchid Dactylorhiza lapponica. New Phytol. 2010;188:385–392. doi: 10.1111/j.1469-8137.2010.03296.x. [DOI] [PubMed] [Google Scholar]
- Song B, Windsor AJ, Schmid KJ, Ramos-Onsins S, Schranz ME, Heidel AJ, et al. Multilocus patterns of nucleotide diversity, population structure and linkage disequilibrium in Boechera stricta, a wild relative of Arabidopsis. Genetics. 2009;181:1021–1033. doi: 10.1534/genetics.108.095364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer C, Ward J. Flowering time and elevated atmospheric CO2. New Phytol. 2007;176:243–255. doi: 10.1111/j.1469-8137.2007.02196.x. [DOI] [PubMed] [Google Scholar]
- Stam LF, Laurie CC. Molecular dissection of a major gene effect on a quantitative trait: the level of alcohol dehydrogenase expression in Drosophila melanogaster. Genetics. 1996;144:1559–1564. doi: 10.1093/genetics/144.4.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton ML, Roy BA, Thiede D. Evolution in stressful environments. I. Phenotypic variability, phenotypic selection and response to selection in five distinct environmental stresses. Evolution. 2000;54:93–111. doi: 10.1111/j.0014-3820.2000.tb00011.x. [DOI] [PubMed] [Google Scholar]
- Stratton DA, Bennington CC. Fine-grained spatial and temporal variation in selection does not maintain genetic variation in Erigeron annuus. Evolution. 1998;52:678–691. doi: 10.1111/j.1558-5646.1998.tb03693.x. [DOI] [PubMed] [Google Scholar]
- Strauss SH, Conner J, Rush SL. Foliar herbivory affects floral characters and plant attractiveness to pollinators: implications for male and female plant fitness. Am Nat. 1996;147:1098–1107. [Google Scholar]
- Sultan SE. Phenotypic plasticity for plant development, function and life-history. Trends Plant Sci. 2000;5:537–542. doi: 10.1016/s1360-1385(00)01797-0. [DOI] [PubMed] [Google Scholar]
- Sultan SE, Spencer HG. Metapopulation structure favors plasticity over local adaptation. Am Nat. 2002;160:271–283. doi: 10.1086/341015. [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- van Tienderen PH. Generalists, specialists and the evolution of phenotypic plasticity in sympatric species of distinct species. Evolution. 1997;51:1372–1380. doi: 10.1111/j.1558-5646.1997.tb01460.x. [DOI] [PubMed] [Google Scholar]
- Via S. Phenotypic plasticity: target or by-product of selection in a variable environment. Am Nat. 1993;142:352–365. doi: 10.1086/285542. [DOI] [PubMed] [Google Scholar]
- Via S, Gomulkiewicz R, De Jong G, Scheiner SM, Schlichting CD, Van Tienderen PH. Adaptive phenotypic plasticity: consensus and controversy. Trends Ecol Evol. 1995;10:212–217. doi: 10.1016/s0169-5347(00)89061-8. [DOI] [PubMed] [Google Scholar]
- Via S, Lande R. Genotype–environment interaction and the evolution of phenotypic plasticity. Evolution. 1985;39:505–522. doi: 10.1111/j.1558-5646.1985.tb00391.x. [DOI] [PubMed] [Google Scholar]
- Waller DM. Environmental determinants of outcrossing in Impatiens capensis (Balsaminaceae) Evolution. 1980;34:747–761. doi: 10.1111/j.1558-5646.1980.tb04014.x. [DOI] [PubMed] [Google Scholar]
- Weinig C, Ungerer MC, Dorn LA, Kane NC, Toyonaga Y, Halldorsdottir SS, et al. Novel loci control variation in reproductive timing in Arabidopsis thaliana in natural environments. Genetics. 2002;162:1875–1884. doi: 10.1093/genetics/162.4.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczek AM, Burghardt LT, Cobb AR, Cooper MD, Welch SM, Schmitt J. Genetic and physiological bases for phenological response to current and predicted climates. Philos Trans R Soc Lond Ser B. 2010;365:3129–3147. doi: 10.1098/rstb.2010.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczek AM, Roe JL, Knapp MC, Cooper MD, Lopez-Gallego C, Martin LJ, et al. Effects of genetic perturbation on seasonal life history plasticity. Science. 2009;323:930–934. doi: 10.1126/science.1165826. [DOI] [PubMed] [Google Scholar]
- Williams J, Jackson S, Kutzbach J. Projected distributions of novel and disappearing climates by 2100 AD. Proc Natl Acad Sci. 2007;104:5738–5742. doi: 10.1073/pnas.0606292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Ness RW, Foxe JP, Barrett SCH. Genomic consequences of outcrossing and selfing in plants. Int J Plant Sci. 2008;169:105–118. [Google Scholar]