Abstract
The additive genetic variance with respect to absolute fitness, VA(W), divided by mean absolute fitness, 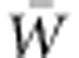 , sets the rate of ongoing adaptation. Fisher's key insight yielding this quantitative prediction of adaptive evolution, known as the Fundamental Theorem of Natural Selection, is well appreciated by evolutionists. Nevertheless, extremely scant information about VA(W) is available for natural populations. Consequently, the capacity for fitness increase via natural selection is unknown. Particularly in the current context of rapid environmental change, which is likely to reduce fitness directly and, consequently, the size and persistence of populations, the urgency of advancing understanding of immediate adaptive capacity is extreme. We here explore reasons for the dearth of empirical information about VA(W), despite its theoretical renown and critical evolutionary role. Of these reasons, we suggest that expectations that VA(W) is negligible, in general, together with severe statistical challenges of estimating it, may largely account for the limited empirical emphasis on it. To develop insight into the dynamics of VA(W) in a changing environment, we have conducted individual-based genetically explicit simulations. We show that, as optimizing selection on a trait changes steadily over generations, VA(W) can grow considerably, supporting more rapid adaptation than would the VA(W) of the base population. We call for direct evaluation of VA(W) and
, sets the rate of ongoing adaptation. Fisher's key insight yielding this quantitative prediction of adaptive evolution, known as the Fundamental Theorem of Natural Selection, is well appreciated by evolutionists. Nevertheless, extremely scant information about VA(W) is available for natural populations. Consequently, the capacity for fitness increase via natural selection is unknown. Particularly in the current context of rapid environmental change, which is likely to reduce fitness directly and, consequently, the size and persistence of populations, the urgency of advancing understanding of immediate adaptive capacity is extreme. We here explore reasons for the dearth of empirical information about VA(W), despite its theoretical renown and critical evolutionary role. Of these reasons, we suggest that expectations that VA(W) is negligible, in general, together with severe statistical challenges of estimating it, may largely account for the limited empirical emphasis on it. To develop insight into the dynamics of VA(W) in a changing environment, we have conducted individual-based genetically explicit simulations. We show that, as optimizing selection on a trait changes steadily over generations, VA(W) can grow considerably, supporting more rapid adaptation than would the VA(W) of the base population. We call for direct evaluation of VA(W) and 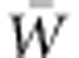 in support of prediction of rates adaptive evolution, and we advocate for the use of aster modeling as a rigorous basis for achieving this goal.
in support of prediction of rates adaptive evolution, and we advocate for the use of aster modeling as a rigorous basis for achieving this goal.
Keywords: adaptation, aster models, breeding value, fitness, quantitative genetics, stabilizing selection
‘the question was never really, How much genetic variation is there...? but rather, What is the nature of genetic variation for fitness in a population?' RC Lewontin, 1974.
Introduction
Adaptation proceeds through the differential genetic contributions among individuals. This idea is formalized in the Fundamental Theorem of Natural Selection (FTNS), which quantitatively predicts the increase in a population's mean fitness as the simple ratio of its additive genetic variance in absolute fitness, VA(W), to its mean absolute fitness, 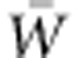 (Fisher, 1930; Bürger, 2000, Section 6.5 and Ewens, 2004, Section 7.4.5 explain the breadth of applicability of the FTNS). This ratio, thus, indicates a population's capacity to adapt to current conditions, given its present genetic composition. Potential for adaptation over an extended period must also take into account eventual contributions of new mutations to VA(W), but the initial rarity of new mutations makes their role in immediate adaptation negligible.
(Fisher, 1930; Bürger, 2000, Section 6.5 and Ewens, 2004, Section 7.4.5 explain the breadth of applicability of the FTNS). This ratio, thus, indicates a population's capacity to adapt to current conditions, given its present genetic composition. Potential for adaptation over an extended period must also take into account eventual contributions of new mutations to VA(W), but the initial rarity of new mutations makes their role in immediate adaptation negligible.
The FTNS has served as an important conceptual foundation of evolutionary biology. However, although it has also motivated some empirical research, its full predictive power has yet to be realized because it has rarely been applied quantitatively to predict a population's adaptive capacity. The effort required to obtain full fitness records for many individuals in a pedigreed population makes estimation of the key parameters for applying the FTNS, VA(W) and 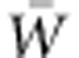 , a daunting challenge. Yet even more influential reasons that the FTNS has not been applied empirically may be the expectation that VA(W) is negligible in many populations, as well as statistical challenges to rigorous estimation of VA(W). Here, we examine the grounds for expecting negligible VA(W) and show that change in selection regime can lead to considerable increase in VA(W). We underscore that the magnitude of VA(W) is an empirical question and that insight into the capacity for adaptive evolution in response to changing environment requires estimation of VA(W), which recently developed aster modeling renders tractable.
, a daunting challenge. Yet even more influential reasons that the FTNS has not been applied empirically may be the expectation that VA(W) is negligible in many populations, as well as statistical challenges to rigorous estimation of VA(W). Here, we examine the grounds for expecting negligible VA(W) and show that change in selection regime can lead to considerable increase in VA(W). We underscore that the magnitude of VA(W) is an empirical question and that insight into the capacity for adaptive evolution in response to changing environment requires estimation of VA(W), which recently developed aster modeling renders tractable.
Conceptual impediments to direct study of the adaptive process
In formulating the theory of evolution by natural selection, Darwin (1859) emphasized his view of adaptation as an extremely slow process, spanning many generations, a view that discourages direct study of ongoing adaptation. As the theory of population and quantitative genetics has developed, it has implicitly reinforced this perspective by focusing on solutions at evolutionary equilibrium under static conditions (Charlesworth, 1987). It is now clear that various processes, including mutation, migration and spatial or temporal variation in selection, can maintain VA(W) at equilibrium; nevertheless, for parameter values considered realistic, the general conclusion has been that VA(W) is expected to be slight at equilibrium (reviewed in Charlesworth, 1987). Yet environments in nature change, and populations occupying them may rarely be subject to conditions that are sufficiently stable (that is, either static or changing with regularity) for attainment of evolutionary equilibrium. Thus, although, following Darwin, some early evolutionists considered rates of adaptation too slow, relative to human lifetime, for direct study (and many cases are), others forged ahead (for example, the classic studies of selection favoring melanic morphs of Biston betularia in areas of Britain subject to heavy industrial pollution (Kettlewell, 1955, 1956)).
Now a large and growing body of evidence documents adaptation over tens of generations (Antonovics and Bradshaw, 1970; Hairston et al., 1999) or fewer (Franks et al., 2007). Many are cases of evolutionary response to abrupt, drastic environmental alteration. They confirm that adaptation is often quite rapid and that the process of adaptation is, in many cases, amenable to direct study. Major environmental changes under way have prompted questions about the capacity of the biota to adapt to them: both what rates of adaptation can be expected (Shaw and Etterson, 2012) and the likelihood of evolutionary rescue, that is, adaptation via natural selection of an isolated population from negative to positive population growth rate (Gomulkiewicz et al., 2010). It is often assumed that differences among populations in the amount of their molecular variation reflect their differences in adaptive capacity. However, Reed and Frankham (2001) showed that the correlation between molecular genetic variation and genetic variation with respect to quantitative traits is slight. Thus, the goal of addressing questions about adaptive capacity necessitates direct study of the adaptive process, and this entails evaluation of populations' additive genetic variance for absolute fitness, VA(W), and their mean absolute fitness, 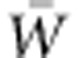 , under conditions currently prevailing and conditions expected in the immediate future.
, under conditions currently prevailing and conditions expected in the immediate future.
As a central concept in evolutionary biology, fitness is well understood, yet its precise definition is elusive (Beatty, 1994; Keller, 1994; Paul, 1994). A convenient definition of an individual's fitness is the number of offspring it leaves to the next generation. This definition simultaneously captures an individual's demographic contribution to its population and its genetic representation, via its descendants. Overlap of generations complicates the definition of fitness (Charlesworth, 1980), but here, we set this complexity aside and consider fitness as the expected number of offspring over a defined period of time.
Using a classic quantitative genetic model to study VA(W) and its dynamics
Tachida and Cockerham (1988) derived expressions for the genetic components of variance for fitness for the classic model of optimizing (quadratic) selection on a quantitative trait. Their results, although informative, are not directly amenable to use in empirical research; the complicated expressions cannot be evaluated without knowledge of the distribution of effects of individual alleles at all loci that contribute to variation in the traits under consideration, including fourth moments. Turelli and Barton (1994) have since shown that, even under strong selection (including truncation and disruptive selection), the distribution of breeding values for the trait is very well approximated by the normal distribution. This justifies a simpler approach to investigate the distribution of genetic effects on fitness, one in which we can treat in aggregate the contributions of the many loci throughout the genome to variation in the trait and fitness.
Fitness under Gaussian selection
For simplicity, we here take the trait values as arising from strictly additive gene action, together with environmental effects, z=a+e, where the additive genetic effects, a, are normally distributed with mean, μz, and variance, 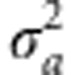 , and the environmental effects are normally distributed with mean zero and variance,
, and the environmental effects are normally distributed with mean zero and variance, 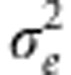 . Under these assumptions, we seek the distribution of the genotypic effects on absolute fitness, E(W | a), under optimizing selection on a trait, z, using a Gaussian function for the relationship between absolute fitness and the trait,
. Under these assumptions, we seek the distribution of the genotypic effects on absolute fitness, E(W | a), under optimizing selection on a trait, z, using a Gaussian function for the relationship between absolute fitness and the trait,
with the width of the fitness function given by ω2, which is inversely related to the strength of selection. This function, ranging from 0 to 1, is routinely used to model Gaussian selection on a quantitative trait (for example, Turelli and Barton, 1994).
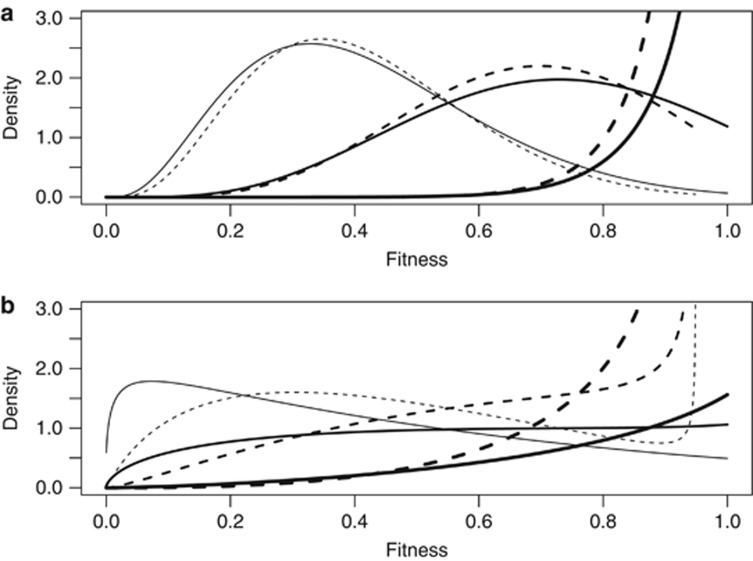
We have derived expressions for the probability density functions for the genotypic fitness effects in the appendix. As illustrated in Figure 1, the genotypic distribution for fitness is concentrated toward high values when the trait mean coincides with the optimum, reflecting the generally high-degree of adaptation. However, for populations that are less well-adapted, with the trait mean deviating from the optimum, the distribution can be approximately symmetric, with mode well below the maximum expressed fitness values. If the additive genetic variance for the trait, 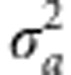 , is similar in magnitude to the width of the fitness function, ω2, and the trait mean is far from the optimum, the distribution can be bimodal with modes toward opposite extremes of the range. The inclusion in the model of environmental variance as a contributor to variation in the trait causes the genotypic distribution for fitness to be shifted toward intermediate values. In that case, no genotype has an expected fitness that coincides with the maximal fitness, and genotypes with low-fitness values in the absence of environmental variance have higher expected fitness when it is included (Figure 1).
, is similar in magnitude to the width of the fitness function, ω2, and the trait mean is far from the optimum, the distribution can be bimodal with modes toward opposite extremes of the range. The inclusion in the model of environmental variance as a contributor to variation in the trait causes the genotypic distribution for fitness to be shifted toward intermediate values. In that case, no genotype has an expected fitness that coincides with the maximal fitness, and genotypes with low-fitness values in the absence of environmental variance have higher expected fitness when it is included (Figure 1).
Figure 1.
Probability density functions for fitness f(W) (Equation 3) when fitness is a Gaussian function of trait value with width ω=3 and z is normally distributed with mean μz and s.d. σa. In both panels, μz=0 thick lines; μz=3 medium lines; μz=4.5 thin lines. Continuous lines, h2=1; corresponding dashed lines, h2=0.5, that is, when 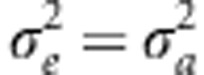 . (a) σa=1. (b) σa=2.
. (a) σa=1. (b) σa=2.
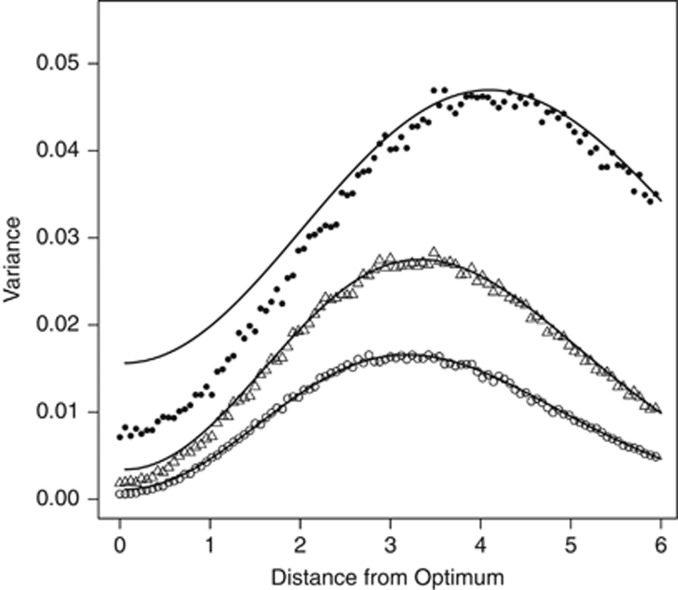
Although the distributions of genotypic effects on fitness provide insight into the nature of selection at the genotypic level, in a sexual population, the response to natural selection, in terms of the rate of change in fitness, depends on the distribution of the additive genetic effects on fitness (that is, fitness breeding values). In principle, the nonlinearity of the fitness function induces nonadditive genetic effects on fitness, even when the trait under selection is subject to strictly additive genetic effects (Wright, 1935). Consequently, the additive genetic variance for fitness, VA(W), can be substantially less than the variance of the genotypic fitness distribution (for example, those shown in Figure 1). Nevertheless, unless 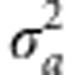 approaches ω2 in magnitude, the genotypic variance of fitness, VG(W), is largely attributable to VA(W) over a wide range of difference of the trait mean from the optimum, as Figure 2 illustrates. VG(W) is maximized when μz is close to the value of ω, where the fitness function is steepest, and this is also true for VA(W), at least for modest values of the trait variance.
approaches ω2 in magnitude, the genotypic variance of fitness, VG(W), is largely attributable to VA(W) over a wide range of difference of the trait mean from the optimum, as Figure 2 illustrates. VG(W) is maximized when μz is close to the value of ω, where the fitness function is steepest, and this is also true for VA(W), at least for modest values of the trait variance.
Figure 2.
Variance of f(W), denoted VG(W) as a function of μz for σa=0.5 (curve through open circles), σa=1 (curve through open triangles); σa=2 (curve through filled circles). Points denote means over 100 REML estimates of VA(W), the additive genetic variance for fitness, for simulated data sets having the given μz and σa and 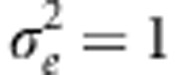 . Througout, ω=3.
. Througout, ω=3.
Dynamics of fitness variation in a changing environment
To gain insight into the evolutionary dynamics of adaptive capacity, we examine the dynamics of genetic variance for fitness for a population subjected to Gaussian selection, with the fitness optimum changing over generations (Pease et al., 1989). Bürger and Lynch (1995) used this model as the basis for examining persistence of populations in the face of environmental change. They presented the dynamics of genetic variation for the selected trait, along with expected time of extinction for populations subject to changing environment such that the trait optimum changes over time. Our independent simulation study presents the dynamics of both VG(W) and VA(W) while replicating their findings for the dynamics of the trait mean and its variance.
The simulation approach has been previously described in detail (Ronce et al., 2009). In brief, each individual's genotypic trait value is the sum of allelic effects over L unlinked loci. An independent environmental effect also contributes to the phenotypic value. During gametogenesis, mutations are generated at a rate U per diploid genome. For a mutation occurring in a particular allele, its effect is modeled as that of the original allele, plus a deviation drawn from a normal distribution with zero mean and variance 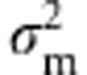 . The genetic variance of the trait is thus free to evolve. At the fertilization stage, the total number of juveniles generated is taken as a Poisson random variable, with mean equal to f times the number of reproducing individuals. Genetic variation in fitness is expressed through variation in survival only, imposed via a Gaussian fitness function with trait optimum θ and strength of selection ω2, as above. If the population remains above its carrying capacity K after this selection, then it is reduced to size K by elimination of individuals at random. Here, we set L=50,
. The genetic variance of the trait is thus free to evolve. At the fertilization stage, the total number of juveniles generated is taken as a Poisson random variable, with mean equal to f times the number of reproducing individuals. Genetic variation in fitness is expressed through variation in survival only, imposed via a Gaussian fitness function with trait optimum θ and strength of selection ω2, as above. If the population remains above its carrying capacity K after this selection, then it is reduced to size K by elimination of individuals at random. Here, we set L=50, 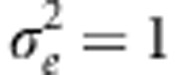 , U=0.01,
, U=0.01, 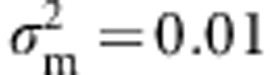 , f=5, ω=3 and K=400, with exceptions noted below. At the outset, the population was allowed to evolve for 1000 generations to reach mutation-selection-drift balance (that is, we detected no change in the genetic variance averaged over replicates after generation 500; not shown). At this point, a steady, directional change in the fitness function was imposed via a change in the trait optimum, θ, at a constant rate (θ=kt, where k is the rate and t is in generations). We chose values of k and f for which the population persists; at higher rates of change in the trait optimum, extinction is common (Pease et al., 1989; Bürger and Lynch, 1995).
, f=5, ω=3 and K=400, with exceptions noted below. At the outset, the population was allowed to evolve for 1000 generations to reach mutation-selection-drift balance (that is, we detected no change in the genetic variance averaged over replicates after generation 500; not shown). At this point, a steady, directional change in the fitness function was imposed via a change in the trait optimum, θ, at a constant rate (θ=kt, where k is the rate and t is in generations). We chose values of k and f for which the population persists; at higher rates of change in the trait optimum, extinction is common (Pease et al., 1989; Bürger and Lynch, 1995).
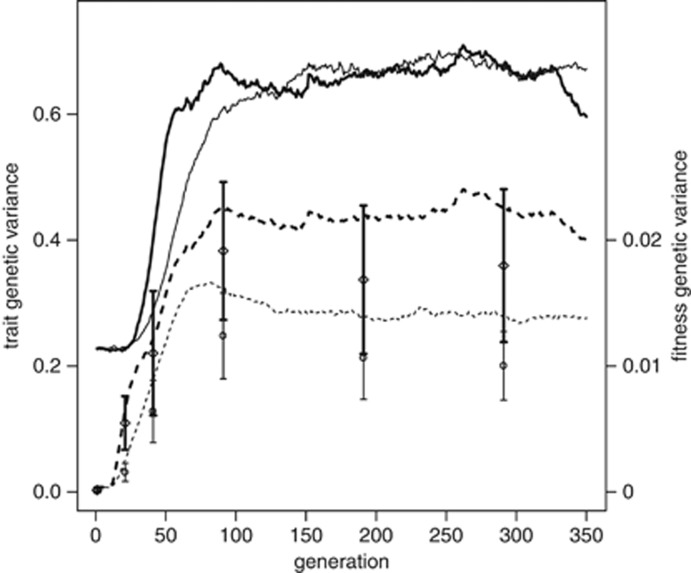
Under this regime, the population evolves such that the trait mean follows the optimum, with some lag. In Figure 3, the additive genetic variance of the trait, 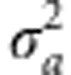 , and the genotypic variance of absolute fitness, VG(W), are shown for two different rates of change in the optimum, θ. When selection begins to change, VG(W) increases rapidly, with increase in
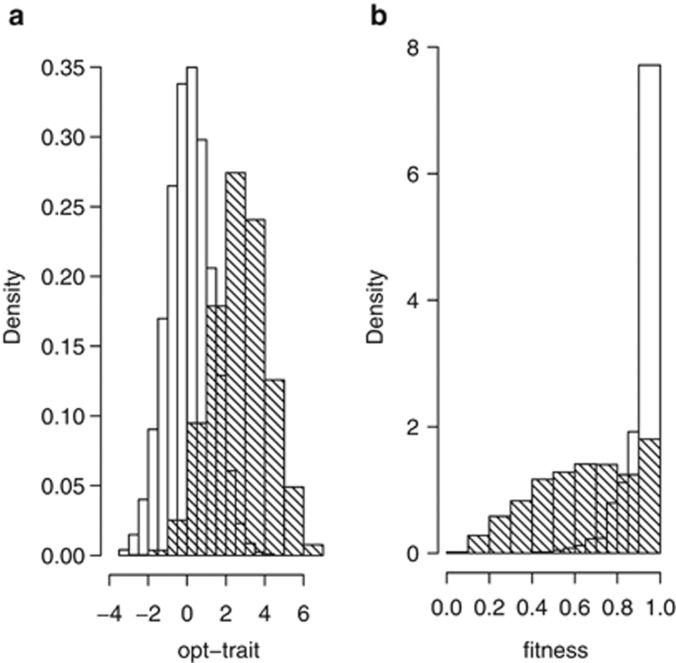
, and the genotypic variance of absolute fitness, VG(W), are shown for two different rates of change in the optimum, θ. When selection begins to change, VG(W) increases rapidly, with increase in 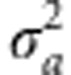 following. The genetic variances eventually stabilize at values substantially higher than when selection is constant; for the chosen parameters, it takes over 50 generations for these variances to be reached. For the slower rate of change, k=0.06=0.02ω per generation, the lag of the trait mean behind the optimum, θ, settles down to 1.03 (s.d. 0.032 over 50 replicate runs) after 150 generations. For the larger rate of change, k=0.2=0.067ω, the lag is at 3.74 (s.d. 0.14), close to the value of ω. Consequently, μz is close to the point on the fitness surface that yields maximal VG(W). Concordant with the analytical results (Appendix and Figure 1), the distribution of genetic effects on fitness changes with the offset or lag of the trait mean from the optimum. This agreement is found, even though the analytical treatment assumes a Gaussian distribution for both the phenotypic and breeding values for the trait, whereas the simulation need not satisfy this assumption. In further agreement with the analytical results, the simulations show a sharply skewed distribution of the breeding values for fitness when the trait mean is near the optimum, and broad distribution when the trait mean is lagging approximately ω units behind the optimum (Figure 4).
following. The genetic variances eventually stabilize at values substantially higher than when selection is constant; for the chosen parameters, it takes over 50 generations for these variances to be reached. For the slower rate of change, k=0.06=0.02ω per generation, the lag of the trait mean behind the optimum, θ, settles down to 1.03 (s.d. 0.032 over 50 replicate runs) after 150 generations. For the larger rate of change, k=0.2=0.067ω, the lag is at 3.74 (s.d. 0.14), close to the value of ω. Consequently, μz is close to the point on the fitness surface that yields maximal VG(W). Concordant with the analytical results (Appendix and Figure 1), the distribution of genetic effects on fitness changes with the offset or lag of the trait mean from the optimum. This agreement is found, even though the analytical treatment assumes a Gaussian distribution for both the phenotypic and breeding values for the trait, whereas the simulation need not satisfy this assumption. In further agreement with the analytical results, the simulations show a sharply skewed distribution of the breeding values for fitness when the trait mean is near the optimum, and broad distribution when the trait mean is lagging approximately ω units behind the optimum (Figure 4).
Figure 3.
Time series for genetic variance of trait and fitness as trait optimum, θ, changes, for two rates of change (0.1 per generation, thin lines; 0.2 per generation, thick lines). The optimum begins to move at generation 10. Trait (additive) genetic variance (with scale on left); fitness genetic variance, VG(W) (dashed, with scale on right). Points and confidence regions give results of 50 estimates of VA(W) plus and minus 1 s.d. for generations 1, 21, 31, 81, 181 and 281.
Figure 4.
Histograms of phenotypic values for trait and fitness when the trait optimum, θ changes at the rate of 0.2 per generation. (a) θ−μz, generation 1 (unshaded) and generation 181 (shaded). (b) Phenotypic fitness values before selection, generation 1 (unshaded) and generation 181 (shaded).
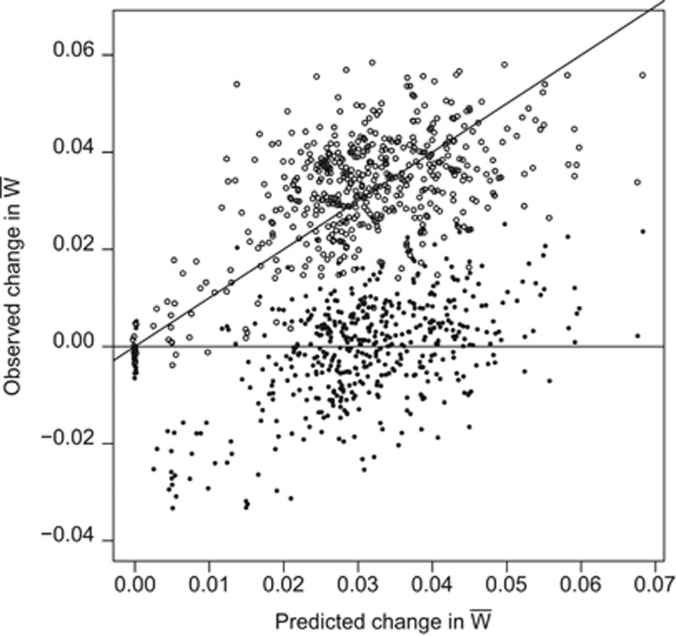
We evaluated the accuracy, within the simulation, of the prediction from the FTNS (Figure 5), estimating VA(W) as follows. From the individuals remaining after selection and producing progeny for the next generation, a subset was chosen at random to produce a separate set of 1200 progeny, via a mating scheme of full sibs nested within half sibs, with inheritance as described above to obtain trait values and corresponding fitness measures. Despite considerable sampling variance, the change in mean fitness is accurately predicted by 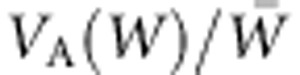 , with VA(W) estimated by REML (Patterson and Thompson, 1971) (Figure 5). When the population is evaluated in the environment in which the parental generation underwent selection, the observed change in mean fitness is similar to the predicted increase. The strong, linear relationship between observed and predicted change in mean fitness with slope=1 demonstrates the accuracy of the prediction, despite expected scatter (Figure 5). When the population is evaluated in the current environment, the slope of this relationship between observed and predicted adaptation is the same. However, in this case, the fitness increment is reduced by a constant amount due to the direct effect of change in environment on mean fitness, the Red Queen effect (Van Valen, 1973).
, with VA(W) estimated by REML (Patterson and Thompson, 1971) (Figure 5). When the population is evaluated in the environment in which the parental generation underwent selection, the observed change in mean fitness is similar to the predicted increase. The strong, linear relationship between observed and predicted change in mean fitness with slope=1 demonstrates the accuracy of the prediction, despite expected scatter (Figure 5). When the population is evaluated in the current environment, the slope of this relationship between observed and predicted adaptation is the same. However, in this case, the fitness increment is reduced by a constant amount due to the direct effect of change in environment on mean fitness, the Red Queen effect (Van Valen, 1973).
Figure 5.
Scatterplot of change in fitness versus predicted change in fitness (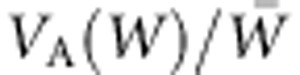 ), lines y=x, y=0. Points are from 20 replicate runs, every 10th generation, for 350 generations after optimum movement of 0.2 per generation begins. Values for change in fitness are calculated as the difference between the fitness of the population before selection in generation k and that at generation k+1 using the optimum for generation k (open) or k+1 (solid). Values for VA(W) estimated using REML from data generated from the simulations (see text).
), lines y=x, y=0. Points are from 20 replicate runs, every 10th generation, for 350 generations after optimum movement of 0.2 per generation begins. Values for change in fitness are calculated as the difference between the fitness of the population before selection in generation k and that at generation k+1 using the optimum for generation k (open) or k+1 (solid). Values for VA(W) estimated using REML from data generated from the simulations (see text).
Empirical study of the quantitative genetics of fitness and attendant challenges
These simulations show that, when the selection regime begins to change, VA(W) can increase steadily over an extended period and, with continued change in selection, stabilize at values considerably greater than at the equilibrium under constant selection. Zhang (2012) has obtained expressions for equilibrium levels of VG(W) for several scenarios of change in optimizing selection, showing the conditions under which it can be greatly increased relative to its value in a stable environment. Because VA(W) bears directly on the rate of ongoing adaptation, estimates of 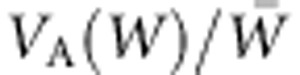 indicate the immediate adaptive capacity of populations, as the simulations illustrate (Figure 5). Although individual fitness is readily modeled mathematically, it is a challenging organismal property to measure, particularly for long-lived species. Nevertheless, it is clear that an individual's fitness, its contribution to the next generation, depends on traits of the life history, including longevity and components of reproduction.
indicate the immediate adaptive capacity of populations, as the simulations illustrate (Figure 5). Although individual fitness is readily modeled mathematically, it is a challenging organismal property to measure, particularly for long-lived species. Nevertheless, it is clear that an individual's fitness, its contribution to the next generation, depends on traits of the life history, including longevity and components of reproduction.
Estimates of genetic variance and heritability (h2) for life-history traits have long been available for numerous populations (Mousseau and Roff, 1987; Roff and Mousseau, 1987; Burt, 1995). They have tended to show relatively little genetic variation for the individual components of fitness, compared with morphological traits, and this has reinforced the expectation that genetic variance for fitness itself is slight. However, genetic variances of components of fitness, many estimated in lab studies, are often found to be sizable in absolute terms, with h2 averaging 0.26 in the survey by Mousseau and Roff (1987). Even so, as these authors and others have noted, genetically based tradeoffs among fitness components could constrain VA(W) to a negligible magnitude, despite substantial h2 of the components of fitness (see also, Charlesworth, 1987). Conversely, though it is less often suggested, VA(W) could be considerably larger than that of its components in cases where genetic differences in fitness expressed early in life are compounded over the life span.
Thus, estimates of genetic variance for fitness components do not resolve the question of whether genetic variance of fitness itself is typically negligible. Recently, substantial efforts have been devoted to evaluating quantitative genetic variation in lifetime fitness and its components for wild vertebrate populations whose pedigrees were partially known from long-term observations and were otherwise inferred from behavior or molecular markers (for example, Kruuk et al., 2000; Merila and Sheldon, 2000; Kruuk, 2004; McCleery et al., 2004; Morrissey and Ferguson, 2010). In several of these studies, VA in the measure of lifetime fitness has not been detectable (Kruuk et al., 2000; McCleery et al., 2004), despite significant VA for components of fitness. For example, from observations of nearly 4000 individuals of Parus major from 1960 to 1998, genetic variation was not detected for lifetime reproductive success, although genetic variation was substantial for traits deemed likely to bear on fitness, such as clutch size (McCleery et al., 2004). However, in a population of Ficedula albicollis, records of ∼3000 birds monitored over 17 years (1980–1997) yielded a sizable estimate of narrow-sense heritability (h2=0.2, CVa=29; P<0.01) for lifetime reproductive success of females (Merila and Sheldon, 2000). Although they are few, these studies demonstrate the possibility of assessing adaptive capacity in natural populations. Nevertheless, such strictly observational studies are subject to two important concerns. First, despite advances in estimating quantitative genetic parameters with relationships inferred from marker data (Thomas and Hill, 2000; Thomas et al., 2000), uncertainties in the inferred relationships erode the precision and accuracy of quantitative genetic estimation (Thomas and Hill, 2000). Second, inferences about the genetic variance for any trait may be biased because the resemblances between relatives, from which these estimates are obtained, can result from shared environmental effects, as well as gene sharing. Individual-based statistical modeling of quantitative traits (the ‘animal model') can account for known shared environments, such as nest boxes or territories. However, this does not eliminate biases due to shared environment primarily because the recorded indicators of environment may not fully account for environment-sharing relevant to expression of the trait. This issue has famously plagued research on quantitative traits of humans for many decades (Lewontin, 1975; Keller, 2010). On the other hand, if genetic and environmental effects covary, techniques to account for effects of environment may ‘overcorrect' and produce underestimates of genetic variance. Consequently, it is problematic to predict evolutionary capacity from heritability estimates obtained in observational studies (see for example, Stopher et al., 2012).
In contrast to study of variation in fitness itself, the form and magnitude of selection on traits in natural populations has been much more extensively evaluated, stimulated by two influential works (Lande and Arnold, 1983; Endler, 1986). These studies have largely documented phenotypic relationships between traits and components of fitness, but studies to determine the genetic relationship between them, and hence to evaluate genetic selection, have also appeared, especially since Rausher (1992). They have also suggested temporal and spatial variation in selection on specific traits (for example, Kalisz, 1986; Siepielski et al., 2009; but see Morrissey and Hadfield, 2012). Few clear generalizations about the form and strength of selection have emerged from consideration of these studies collectively (Kingsolver et al., 2001; Hereford, 2009; Kingsolver and Diamond, 2011). In view of the heterogeneity of traits, taxa, measures of fitness and, presumably, conditions in which the studied populations were undergoing selection, it would, perhaps, have been surprising if strong commonalities in selection had been evident.
When it has been possible to identify traits that are strongly associated with fitness (for example, bill depth in Geospiza fortis, Grant and Grant, 1995; stigma exsertion in Ipomopsis aggregata, Campbell, 1991; Campbell et al., 1994), this approach has informed understanding of natural selection on those traits. Yet it can mislead, for example, when differential mortality with respect to focal traits occurs before the traits are expressed (Bennington and McGraw, 1995; Mojica and Kelly, 2010) or when variation in inbreeding has not been taken into account (Willis, 1996). Moreover, statistical power to identify the traits under selection may be inadequate, even in studies of substantial scale (Shaw and Geyer, 2010). Beyond this, analysis of selection on traits does not reveal the overall extent of selection (whether phenotypic or genotypic). Lande and Arnold (1983) noted, Mitchell-Olds and Shaw (1987) emphasized and many authors have since acknowledged that phenotypic selection analysis can evaluate selection only on the traits that are included in the analysis. A related limitation that is not often explicitly recognized is that no empirically manageable set of traits can be expected to account fully (or even nearly) for the overall variation in fitness. Thus, there may be far more genetic variation in fitness, and hence, capacity for adaptation, than is manifested by selection on the traits under consideration in a given study. Fortunately, it is not necessary to know or discover which traits are under selection in order to study adaptation.
Direct study of VA(W)
In view of the central importance of the genetic variation with respect to fitness, VA(W), in a population's adaptive potential (Fisher, 1930; Bürger, 2000; Ewens, 2004), the scarcity of evidence about the magnitude of VA(W) compromises prediction of the capacity of natural populations for ongoing adaptation and the rates at which adaptation could proceed. As for any quantitative trait, VA(W) is subject to direct evaluation via experimental approaches, but this has rarely been accomplished. Gardner et al. (2005) have demonstrated considerable VA(W) in the Dahomey strain of Drosophila melanogaster by following the dynamics of non-recombining balancer chromosomes in replicate population cages over 300 days (see also Fowler et al., 1997). The authors argue convincingly against laboratory artifacts inflating the fitness variation. Fry (2008) has pointed out that the few observed cases of transient changes in frequency are more likely due to evolution of modifiers mitigating the deleterious effects of the balancers than G × E interaction, as suggested by the original authors, but this does not undermine the conclusion that, even for a population maintained in a relatively stable laboratory environment for 30 years, VA(W) was found to be sizable and persistent. Long et al. (2009) have also documented substantial VA(W) for lifetime fitness of female D. melanogaster.
These laboratory studies of quantitative genetics of fitness reveal the capacity of a population to adapt to conditions imposed by the experimentalist. However, given the sensitivity of fitness to environmental conditions (Antonovics et al., 1988), it would be presumptuous to extrapolate from lab-based estimates of VA(W) to the adaptive capacity of natural populations to conditions that impinge on them in nature. Accurate prediction of rates of adaptation in nature will require quantitative genetic experiments on wild populations under natural conditions. Such studies offer the realism of fitness expression in quasi-natural conditions for populations evolving under the vagaries of environmental change, while avoiding the problems of observational studies of confounded environmental effects and uncertainty of pedigrees. Unfortunately, although there are examples of this approach (for example, Mitchell-Olds, 1986; Schmitt and Antonovics, 1986; Shaw, 1986; Schwaegerle and Levin, 1991; Campbell 1996), this body of work remains relatively small, and estimates of VA(W) in nature are few.
Two recent experimental studies in nature evaluated quantitative genetics of lifetime fitness. In an investigation of an annual plant Dicerandra linearifolia (Lamiaceae), Winn (2004) used a paternal halfsib crossing design and planted seedling progeny into the field site from which the parents had been sampled. For two key components of the life history, survival and number of flowers, very low estimates of h2 (<0.01, ns) were obtained. Etterson (2004) likewise carried out a paternal halfsib crossing design within three populations of the annual legume Chamaecrista fasciculata and planted the resulting seedling progeny into sites near to each of the source sites, employing the reciprocal transplant approach. In this case, additive genetic variation for overall fitness, assessed as total number of seeds per individual, was reported as significant for one population (from Oklahoma, USA) in all three sites, with h2 ranging from 0.1 to 0.27. For this population in its home site, the estimate of h2(W) was 0.22, indicating considerable further capacity for adaptation to local conditions. The other two populations showed significant (or marginally so) VA(W) in at least one site, with h2(W) about 0.08, evidencing the populations' capacity for ongoing adaptation.
Aster modeling to support study of VA(W)
A long-standing and vexing impediment to the study of lifetime fitness has been that the complexity of fitness expression throughout the life cycle, which contrasts with the simplicity of its representation in mathematical models, results in an empirical distribution that is not well approximated by any conventional statistical distribution. Lifetime fitness is the composite of survival over successive intervals and multiple components of reproduction, within reproductive episodes, as well as over possibly many episodes in a lifetime. Accordingly, fitness has a compound distribution that is far from Gaussian or any other standard sampling distribution (see for example, Figure 4 in Wagenius et al., 2010). Consequently, assumptions of standard methods, such as linear models, are routinely violated, necessitating caveats about statistical inferences and their implications (for example, Shaw, 1986) and sometimes multiple approaches to analysing the data at hand (Antonovics and Ellstrand, 1984). The unconventional phenotypic distribution of fitness is especially problematic when interest focuses on estimating a population's additive genetic variance for fitness, as required for evolutionary prediction. Statistical inference for random effects models, which are the basis of variance partitioning, relies heavily on the Normal distribution (Fisher, 1918).
Aster modeling has been developed to fill this methodological gap (Geyer et al., 2007; Shaw et al., 2008). Using a likelihood framework, aster explicitly models the components of fitness, including the dependence of later expressed components on earlier ones, for example, the number of offspring produced in a particular year on survival up to that year, on reproductive status and on the number of mates. For each component, it employs a suitable sampling distribution, for example, Bernoulli for survival and reproductive status, and Poisson for number of mates. Given these specifications and an appropriate definition of lifetime fitness, for example, the total number of offspring produced over a specified period of time, aster analysis yields inferences of interest, including comparisons of mean fitness and estimates of the form of the fitness function (see examples in Shaw et al., 2008; Shaw and Geyer, 2010).
Aster has now been extended to accommodate random effects (Geyer et al., 2012). This development allows estimation of components of variance for fitness, while explicitly accounting for the compound nature of fitness expression and for its resulting distribution. Application of random effects models in aster to reanalyze the data of Etterson (2004) revealed significant variance in fitness among paternal halfsib groups (with fitness defined as expected number of fruits produced per individual) in all three populations grown in the Kansas and Oklahoma locations. This result implies the availability of VA(W) that would support further adaptation of the local population, as well as the foreign ones, to those two sites.
Further analysis gave no indication of substantial deviation from a Gaussian distribution of breeding values for fitness (Geyer et al., 2012, Section 8.5). We caution against overinterpretation of this finding. As others have noted (Hadfield et al., 2010), extreme uncertainty in individual estimates of breeding values renders problematic inference from estimates of breeding values per se. Nevertheless, it is intriguing that the inferred distributions of the breeding values are approximately symmetric, rather than negatively skewed as expected in a population subjected to consistent selection over a long period (Figure 4b, unshaded).
Conclusions
Theoretical arguments have led to an expectation that natural selection rapidly exhausts additive genetic variance for fitness, VA(W), as adaptation proceeds, and, therefore, that populations are likely to harbor little VA(W). In conjunction with the considerable logistical and statistical challenges of evaluating VA(W), this view has tended to discourage direct study of it, particularly in nature. For components of fitness, genetic variance has often been found to be appreciable; such findings have been reconciled with the expectation of negligible VA(W) by invoking tradeoffs among fitness components, which have in some cases been demonstrated (for example, Rose and Charlesoworth, 1981a, 1981b). More recently, however, evidence of genetic variance in lifetime fitness has begun to accumulate, and this demands reconsideration of the view that standing additive genetic variation for fitness is generally slight.
Our simulations show that VA(W) can rise over tens of generations at the outset of a period of change in selection regime. The initial increase in VA(W) results from the change in the fitness function alone, and thus, begins before the increase in genetic variance for the trait under selection shown by Bürger and Lynch (1995). Thereafter, changes in allele frequencies also contribute to the increase in VA(W). Both the increase in VA(W) and the decline in mean fitness, 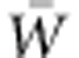 , due to the direct effects of the changed environment jointly imply augmentation of the population's adaptive capacity, as represented by the ratio of VA(W) to
, due to the direct effects of the changed environment jointly imply augmentation of the population's adaptive capacity, as represented by the ratio of VA(W) to 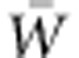 . Nevertheless, the adaptation that proceeds from one generation to the next due to VA(W) may be entirely counteracted by further change in the direct environmental effects on fitness, such that
. Nevertheless, the adaptation that proceeds from one generation to the next due to VA(W) may be entirely counteracted by further change in the direct environmental effects on fitness, such that 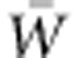 remains the same or, in fact, declines, as Fisher (1930) and many since have noted and as shown in Figure 5 (filled circles).
remains the same or, in fact, declines, as Fisher (1930) and many since have noted and as shown in Figure 5 (filled circles).
The rise in VA(W) can continue over dozens of generations, given our choice of parameters, before VA(W) reaches a new steady-state considerably greater than in the constant environment. These findings about the dynamics of VA(W) as selection begins to change complement the results of Zhang (2012), showing that equilibrium levels of VG(W) under various scenarios of environmental change can be far greater than the values expected in a constant environment. In addition to addressing the case of a linearly changing optimum, Zhang (2012) also considered randomly varying change in optimum, as well as autocorrelated change, finding that VG(W) may attain dramatically higher values than in a constant environment, depending on the nature of the change in the trait optimum and number of traits under selection.
Environmental conditions always vary over generations to some degree, yet it is unclear to what extent and on what time scale selective environment, in the sense of Antonovics et al. (1988), has changed in nature. To evaluate this empirically would require experiments in which replicate genetic populations are grown at different times; we are unaware of such experiments in natural populations. If change in selective environment has frequently accompanied typical environmental change over recent past generations, then it may be that populations currently harbor substantial VA(W), as in the steady-states of the cases considered by Zhang (2012). However, to the extent that current temporal change in selective environment is a novel consequence of the newly dramatic and rapid changes in environment, then our simulations suggest that populations may be expressing somewhat greater VA(W) in conjunction with recent change in conditions, but they may remain well below their potential adaptive capacity for several dozens of generations.
In the context of rapid change in environmental conditions globally, the urgent practical need to assess the immediate adaptive capacity of the biota augments the importance of calibrating this property as the essential basis for evolutionary explanation of rates of ongoing adaptation. These considerations argue for renewal of efforts to evaluate the additive genetic variance for absolute fitness, VA(W), and mean absolute fitness, 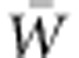 , so that per generation change in mean absolute fitness can be predicted for natural populations under realistic conditions. The long established experimental methodology of quantitative genetics is indispensable in achieving this goal, and it is reinforced by the new statistical methodology of aster modeling for fitness. Conducting quantitative genetic experiments in nature will always be an arduous task, especially when the objective is evaluation of additive genetic variance in lifetime fitness. Nevertheless, the motivations for assessing rates of ongoing adaptation increasingly demand it.
, so that per generation change in mean absolute fitness can be predicted for natural populations under realistic conditions. The long established experimental methodology of quantitative genetics is indispensable in achieving this goal, and it is reinforced by the new statistical methodology of aster modeling for fitness. Conducting quantitative genetic experiments in nature will always be an arduous task, especially when the objective is evaluation of additive genetic variance in lifetime fitness. Nevertheless, the motivations for assessing rates of ongoing adaptation increasingly demand it.
Data archiving
There were no data to deposit.
Acknowledgments
We gratefully acknowledge the help of Thomas Shaw, and discussions with many insightful colleagues, especially Charles Geyer. J Stanton-Geddes, A Eule-Nashoba, S Flint and N Goldsmith commented helpfully on the manuscript. We particularly thank Michael Morrissey for his thorough, constructive review.
Appendix
We begin our derivation of expressions for the distribution of additive genetic effects on fitness by assuming that there is no environmental variance so that 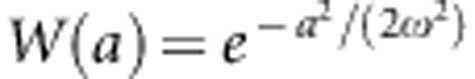 . We will later include environmental variation. To find the cumulative distribution function for the genotypic fitness effects, we must calculate, for each W∈(0,1), the probability that W(a)<W. Solving this inequalty for a yields a2<−2ω2 ln(W) so that, for
. We will later include environmental variation. To find the cumulative distribution function for the genotypic fitness effects, we must calculate, for each W∈(0,1), the probability that W(a)<W. Solving this inequalty for a yields a2<−2ω2 ln(W) so that, for 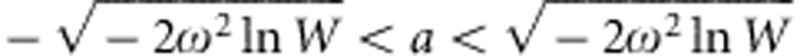 we have W(a)>W. Integrating the probability density function for a between these two limits and subtracting the result from one gives the probability that W(a)<W. Thus, we find the cumulative distribution function for the genotypic fitness effects to be
we have W(a)>W. Integrating the probability density function for a between these two limits and subtracting the result from one gives the probability that W(a)<W. Thus, we find the cumulative distribution function for the genotypic fitness effects to be
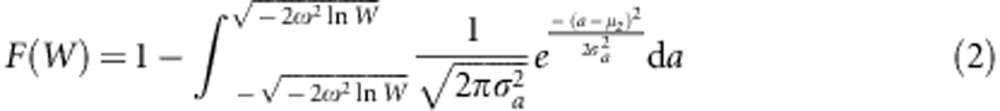 |
and its probability density function to be
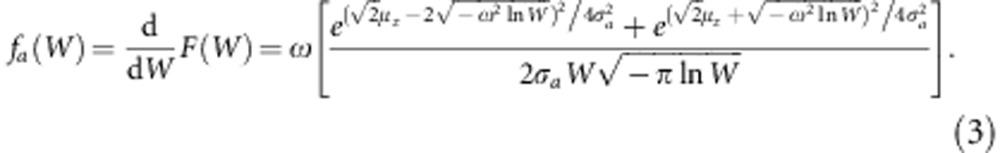 |
The shape of this probability density function depends on the mean of the trait relative to the optimum of the fitness function, the additive genetic variance for the trait, and the strength of stabilizing selection on it, ω. We take the mean of the trait to be identical to the mean of the breeding values, μz. To introduce environmental effects on the trait, we now consider each breeding value a as the mean of a normal distribution with variance 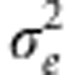 . We find the distribution of the fitness effects for a particular a. By analogy to Equation 3, this is
. We find the distribution of the fitness effects for a particular a. By analogy to Equation 3, this is
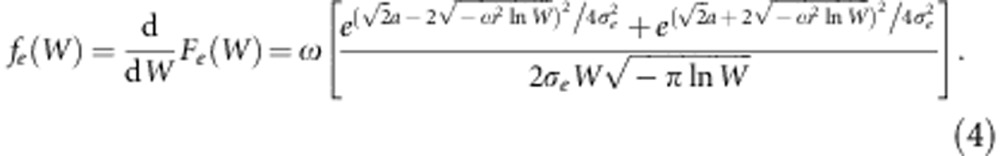 |
The mean of this distribution, for the breeding value a, is
 |
where 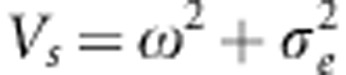 . We note that We(a)=W(a) when Vs=ω2, that is, when there is no environmental variance. Substituting We(a) for W(a) in the derivation of fa(W) yields
. We note that We(a)=W(a) when Vs=ω2, that is, when there is no environmental variance. Substituting We(a) for W(a) in the derivation of fa(W) yields
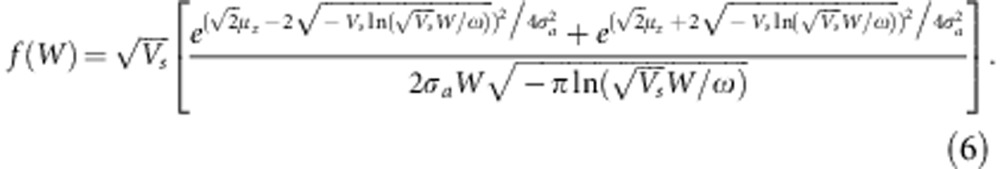 |
The authors declare no conflict of interest.
References
- Antonovics J, Ellstrand NC. Experimental studies of the evolutionary significance of sexual reproduction. I. A test of the frequency-dependent selection hypothesis. Evolution. 1984;38:103–115. doi: 10.1111/j.1558-5646.1984.tb00263.x. [DOI] [PubMed] [Google Scholar]
- Antonovics J, Ellstrand NC, Brandon RN.1988Environmental variation and genetic variation: expectations and experimentsIn: Gottlieb LD, Jain SK, (eds).Plant Evolutionary Biology Chapman and Hall: N.Y.275–303. [Google Scholar]
- Antonovics J, Bradshaw AD. Evolution in closely adjacent plant populations. VIII. Clinal patterns at a mine boundary. Heredity. 1970;25:349–362. doi: 10.1038/sj.hdy.6800835. [DOI] [PubMed] [Google Scholar]
- Beatty J.1994Fitness: theoretical contextsIn: Keller EF, Lloyd EA, (eds)Keywords in Evolutionary Biology Harvard University Press: Cambridge; 115–119. [Google Scholar]
- Bennington CC, McGraw JB. Phenotypic selection in an artificial population of Impatiens pallida: the importance of the invisible fraction. Evolution. 1995;49:317–324. doi: 10.1111/j.1558-5646.1995.tb02244.x. [DOI] [PubMed] [Google Scholar]
- Bürger R, Lynch M. Evolution and extinction in a changing environment: a quantitative-genetic analysis. Evolution. 1995;49:151–163. doi: 10.1111/j.1558-5646.1995.tb05967.x. [DOI] [PubMed] [Google Scholar]
- Bürger R. The Mathematical Theory of Selection, Recombination, and Mutation. John Wiley & Sons: Chichester; 2000. [Google Scholar]
- Burt A. Perspective: the evolution of fitness. Evolution. 1995;49:1–8. doi: 10.1111/j.1558-5646.1995.tb05954.x. [DOI] [PubMed] [Google Scholar]
- Campbell DR. Effects of floral traits on sequential components of fitness in Ipomopsis aggregata. Am Nat. 1991;137:713–737. [Google Scholar]
- Campbell DR, Waser NM, Price MV. Indirect selection of stigma position in Ipomopsis aggregata via a genetically correlated trait. Evolution. 1994;48:55–68. doi: 10.1111/j.1558-5646.1994.tb01294.x. [DOI] [PubMed] [Google Scholar]
- Campbell DR. Evolution of floral traits in a hermaphroditic plant: field measurements of heritabilities and genetic correlations. Evolution. 1996;50:1442–1453. doi: 10.1111/j.1558-5646.1996.tb03918.x. [DOI] [PubMed] [Google Scholar]
- Charlesworth B.1987The heritability of fitnessIn: Bradbury JW, Andersson MB, (eds).Sexual Selection: Testing the Alternatives Wiley: Chichester; 21–40. [Google Scholar]
- Charlesworth B. Evolution in Age-Structured Populations. Cambridge University Press: Cambridge; 1980. [Google Scholar]
- Darwin D.1859The Origin of Species6th edn.Appleton: New York, NY, USA [Google Scholar]
- Endler JA. Natural Selection in the Wild. Princeton University Press: Princeton; 1986. [Google Scholar]
- Etterson JR. Evolutionary potential of Chamaecrista fasciculata in relation to climate change. II. Genetic architecture of three populations reciprocally planted along an environmental gradient in the Great Plains. Evolution. 2004;58:1459–1471. doi: 10.1111/j.0014-3820.2004.tb01727.x. [DOI] [PubMed] [Google Scholar]
- Ewens WJ. Mathematical Population Genetics. I. Theoretical Introduction. Springer: New York; 2004. [Google Scholar]
- Fisher RA. The correlations between relatives on the supposition of Mendelian inheritance. Proc Roy Soc. 1918;52:399–433. [Google Scholar]
- Fisher RA. The Genetical Theory of Natural Selection. Clarendon Press: Oxford; 1930. [Google Scholar]
- Fowler K, Semple C, Barton NH, Partridge L. Genetic variation for total fitness in Drosophila melanogaster. Proc Biol Sci. 1997;264:191–199. doi: 10.1098/rspb.1997.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks S, Sim S, Weis AE. Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proc Natl Acad Sci USA. 2007;104:1278–1282. doi: 10.1073/pnas.0608379104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry JD. Genotype-environment interaction for total fitness in Drosophila. J Genet. 2008;87:355–362. doi: 10.1007/s12041-008-0058-7. [DOI] [PubMed] [Google Scholar]
- Gardner MP, Fowler K, Barton NH, Partridge L. Genetic variation for total fitness in Drosophila melanogaster: complex yet replicable patterns. Genetics. 2005;169:1553–1571. doi: 10.1534/genetics.104.032367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer CJ, Wagenius S, Shaw RG. Aster models for life history analysis. Biometrika. 2007;94:415–426. [Google Scholar]
- Geyer CJ, Ridley CE, Latta RG, Etterson JR, Shaw RG. Aster Models With Random Effects Via Penalized Likelihood. Technical Report No. 692. University of Minnesota School of Statistics: Minneapolis, MN, USA; 2012. [Google Scholar]
- Gomulkiewicz R, Holt RD, Barfield M, Nuismer SL. Genetics, adaptation, and invasion in harsh environments. Evol Appl. 2010;3:97–108. doi: 10.1111/j.1752-4571.2009.00117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PR, Grant BR. Predicting microevolutionary responses to directional selection on heritable variation. Evolution. 1995;49:241–251. doi: 10.1111/j.1558-5646.1995.tb02236.x. [DOI] [PubMed] [Google Scholar]
- Hadfield JD, Wilson AJ, Garant D, Sheldon BC, Kruuk LE. The misuse of BLUP in ecology and evolution. Am Nat. 2010;175:116–125. doi: 10.1086/648604. [DOI] [PubMed] [Google Scholar]
- Hairston NG, Jr, Lampert W, Caceres CE, Holtmeier CL, Weider LJ, Gaedke U, et al. Lake ecosystems—rapid evolution revealed by dormant eggs. Nature. 1999;401:446–446. [Google Scholar]
- Hereford J. A quantitative survey of local adaptation and fitness trade-Offs. Am Nat. 2009;173:579–588. doi: 10.1086/597611. [DOI] [PubMed] [Google Scholar]
- Kalisz S. Variable selection on the timing of germination in Collinsia verna (Scrophulariaceae) Evolution. 1986;40:479–491. doi: 10.1111/j.1558-5646.1986.tb00501.x. [DOI] [PubMed] [Google Scholar]
- Keller EF.1994Fitness: reproductive ambiguitiesIn: Keller EF, Lloyd EA, (eds).Keywords in Evolutionary Biology Harvard University Press: Cambridge; 115–121. [Google Scholar]
- Keller EF. The Mirage of a Space between Nature and Nurture. Duke University Press: Durham, NC; 2010. [Google Scholar]
- Kettlewell HBD. Selection experiments on industrial melanism in the Lepidoptera. Heredity. 1955;9:323–342. [Google Scholar]
- Kettlewell HBD. Further selection experiments on industrial melanism in the Lepidoptera. Heredity. 1956;10:287–301. [Google Scholar]
- Kingsolver JG, Diamond SE. Phenotypic selection in natural populations: what limits directional selection. Am Nat. 2011;177:346–357. doi: 10.1086/658341. [DOI] [PubMed] [Google Scholar]
- Kingsolver JG, Hoekstra HE, Hoekstra JM, Berrigan D, Vignieri SN, Hill CE, et al. The strength of phenotypic selection in natural populations. Am Nat. 2001;157:245–261. doi: 10.1086/319193. [DOI] [PubMed] [Google Scholar]
- Kruuk LEB. Estimating genetic parameters in natural populations using the ‘animal model'. Phil Trans R Soc Lond B Biol Sci. 2004;359:873–890. doi: 10.1098/rstb.2003.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruuk LEB, Clutton-Brock TH, Slate J, Pemberton JM, Brotherstone S, Guinness FE. Heritability of fitness in a wild mammal population. Proc Natl Acad Sci USA. 2000;97:698–703. doi: 10.1073/pnas.97.2.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R, Arnold SJ. The measurement of selection on correlated characters. Evolution. 1983;37:1210–1226. doi: 10.1111/j.1558-5646.1983.tb00236.x. [DOI] [PubMed] [Google Scholar]
- Lewontin RC. Genetic Basis of Evolutionary Change. Columbia University Press: New York; 1974. [Google Scholar]
- Lewontin RC. Genetic aspects of intelligence. Annu Rev Genet. 1975;9:387–405. doi: 10.1146/annurev.ge.09.120175.002131. [DOI] [PubMed] [Google Scholar]
- Long TAF, Miller PM, Stewart AD, Rice WR. Estimating the heritability of female lifetime fecundity in a locally adapted Drosophila melanogaster population. J Evol Biol. 2009;22:637–643. doi: 10.1111/j.1420-9101.2008.01676.x. [DOI] [PubMed] [Google Scholar]
- McCleery RH, Pettifor RA, Armbruster P, Meyer K, Sheldon BC, Perrins CM. Components of variance underlying fitness in a natural population of the great tit parus major. Am Nat. 2004;164:E62–E72. doi: 10.1086/422660. [DOI] [PubMed] [Google Scholar]
- Merila J, Sheldon BC. Lifetime reproductive success and heritability in nature. Am Nat. 2000;155:301–310. doi: 10.1086/303330. [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T. Quantitative genetics of survival and growth in Impatiens capensis. Evolution. 1986;40:107–116. doi: 10.1111/j.1558-5646.1986.tb05722.x. [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T, Shaw RG. Regression analysis of natural selection: statistical inference and biological interpretation. Evolution. 1987;41:1149–1161. doi: 10.1111/j.1558-5646.1987.tb02457.x. [DOI] [PubMed] [Google Scholar]
- Mojica JP, Kelly JK. Viability selection prior to trait expression is an essential component of natural selection. Proc Biol Sci. 2010;277:2945–2950. doi: 10.1098/rspb.2010.0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey MG, Ferguson MM. A test for the genetic basis of natural selection: an individual-based longitudinal study in a stream-dwelling fish. Evolution. 2010;65:1037–1047. doi: 10.1111/j.1558-5646.2010.01200.x. [DOI] [PubMed] [Google Scholar]
- Morrissey MB, Hadfield JD. Directional selection in temporally replicated studies is remarkably constant. Evolution. 2012;66:1037–1047. doi: 10.1111/j.1558-5646.2011.01444.x. [DOI] [PubMed] [Google Scholar]
- Mousseau TA, Roff DA. Natural selection and the heritability of fitness components. Heredity. 1987;59:181–197. doi: 10.1038/hdy.1987.113. [DOI] [PubMed] [Google Scholar]
- Patterson HD, Thompson R. Recovery of inter-block information when block sizes are unequal. Biometrika. 1971;58:545–554. [Google Scholar]
- Paul D.1994Fitness: historical perspectivesIn: Keller EF, Lloyd EA, (eds).Keywords in Evolutionary Biology Harvard University Press: Cambridge; 115–119. [Google Scholar]
- Pease CM, Lande R, Bull JJ. A model of population growth, dispersal and evolution in a changing environment. Ecology. 1989;70:1657–1664. [Google Scholar]
- Rausher MD. The measurement of selection on quantitative traits: biases due to environmental covariances between traits and fitness. Evolution. 1992;46:616–626. doi: 10.1111/j.1558-5646.1992.tb02070.x. [DOI] [PubMed] [Google Scholar]
- Reed DH, Frankham R. How closely correlated are molecular and quantitative measures of genetic variation? A meta-analysis. Evolution. 2001;55:1095–1103. doi: 10.1111/j.0014-3820.2001.tb00629.x. [DOI] [PubMed] [Google Scholar]
- Roff DA, Mousseau TA. Quantitative genetics and fitness: lessons from Drosophila. Heredity. 1987;58:103–118. doi: 10.1038/hdy.1987.15. [DOI] [PubMed] [Google Scholar]
- Ronce O, Shaw FH, Rousset F, Shaw RG. Is inbreeding depression lower in maladapted populations? A quantitative genetic model. Evolution. 2009;63:1807–1819. doi: 10.1111/j.1558-5646.2009.00678.x. [DOI] [PubMed] [Google Scholar]
- Rose MR, Charlesoworth B. Genetics of life history in Drosophila melanogaster. I. Sib analysis of adult females. Genetics. 1981a;9:175–186. doi: 10.1093/genetics/97.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MR, Charlesoworth B. Genetics of life history in Drosophila melanogaster. II. Exploratory selection experiments. Genetics. 1981b;97:187–196. doi: 10.1093/genetics/97.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt J, Antonovics J. Experimental studies of the evolutionary significance of sexual reproduction. III. Maternal and paternal effects during seedling establishment. Evolution. 1986;40:817–829. doi: 10.1111/j.1558-5646.1986.tb00541.x. [DOI] [PubMed] [Google Scholar]
- Schwaegerle KE, Levin DA. Quantitative genetics of fitness traits in a wild population of Phlox. Evolution. 1991;45:169–177. doi: 10.1111/j.1558-5646.1991.tb05275.x. [DOI] [PubMed] [Google Scholar]
- Shaw RG. Response to density in a wild population of the perennial herb Salvia lyrata: variation among families. Evolution. 1986;40:492–505. doi: 10.1111/j.1558-5646.1986.tb00502.x. [DOI] [PubMed] [Google Scholar]
- Shaw RG, Etterson JR. Rapid climate change and the rate of adaptation: insight from experimental quantitative genetics. New Phytologist. 2012;195:752–765. doi: 10.1111/j.1469-8137.2012.04230.x. [DOI] [PubMed] [Google Scholar]
- Shaw RG, Geyer CJ, Wagenius S, Hangelbroek HH, Etterson JR. Unifying life history analyses for inference of fitness and population growth. Am Nat. 2008;172:E35–E47. doi: 10.1086/588063. [DOI] [PubMed] [Google Scholar]
- Shaw RG, Geyer CJ. Inferring fitness landscapes. Evolution. 2010;64:2510–2520. doi: 10.1111/j.1558-5646.2010.01010.x. [DOI] [PubMed] [Google Scholar]
- Siepielski AM, DiBattista J, Carlson S. It's about time: the temporal dynamics of phenotypic selection in the wild. Ecol Lett. 2009;11:1261–1276. doi: 10.1111/j.1461-0248.2009.01381.x. [DOI] [PubMed] [Google Scholar]
- Stopher KV, Walling CA, Morris A, Guinness FE, Clutton-Brock TH, Pemberton JM, et al. Shared spatial effects on quantitative genetic parameters: accounting for spatial autocorrelation and home range overlap reduces estimates of heritability in red deer. Evolution. 2012;66:2411–2426. doi: 10.1111/j.1558-5646.2012.01620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachida H, Cockerham CC. Variance components of fitness under stabilizing selection. Genet Res. 1988;51:47–53. doi: 10.1017/s0016672300023934. [DOI] [PubMed] [Google Scholar]
- Thomas SC, Hill WG. Estimating quantitative genetic parameters using sibships reconstructed from marker data. Genetics. 2000;155:1961–1972. doi: 10.1093/genetics/155.4.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SC, Pemberton JM, Hill WG. Estimating variance components in natural populations using inferred relationships. Heredity. 2000;84:427–436. doi: 10.1046/j.1365-2540.2000.00681.x. [DOI] [PubMed] [Google Scholar]
- Turelli M, Barton NH. Genetic and statistical analyses of strong selection on polygenic traits: what, me normal. Genetics. 1994;138:913–941. doi: 10.1093/genetics/138.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valen L. A new evolutionary theory. Evol Theory. 1973;1:1–30. [Google Scholar]
- Wagenius S, Hangelbroek HH, Ridley CE, Shaw RG. Biparental inbreeding and inter-remnant mating in a perennial prairie plant: fitness consequences for progeny in their first eight years. Evolution. 2010;64:761–771. doi: 10.1111/j.1558-5646.2009.00860.x. [DOI] [PubMed] [Google Scholar]
- Willis JH. Measures of phenotypic selection are biased by partial inbreeding. Evolution. 1996;50:1501–1511. doi: 10.1111/j.1558-5646.1996.tb03923.x. [DOI] [PubMed] [Google Scholar]
- Winn AA. Natural selection, evolvability and bias due to environmental covariance in the field in an annual plant. J Evol Biol. 2004;17:1073–1083. doi: 10.1111/j.1420-9101.2004.00740.x. [DOI] [PubMed] [Google Scholar]
- Wright S. The analysis of variance and the correlations between relatives with respect to deviations from an optimum. Genetics. 1935;30:243–256. [Google Scholar]
- Zhang X-S. Fisher's geometrical model of fitness landscape and variance in fitness within a changing environment. Evolution. 2012;66:2350–2368. doi: 10.1111/j.1558-5646.2012.01610.x. [DOI] [PubMed] [Google Scholar]