Abstract
Maize is the most widely grown cereal in the world. In addition to its role in global agriculture, it has also long served as a model organism for genetic research. Maize stands at a genetic crossroads, as it has access to all the tools available for plant genetics but exhibits a genetic architecture more similar to other outcrossing organisms than to self-pollinating crops and model plants. In this review, we summarize recent advances in maize genetics, including the development of powerful populations for genetic mapping and genome-wide association studies (GWAS), and the insights these studies yield on the mechanisms underlying complex maize traits. Most maize traits are controlled by a large number of genes, and linkage analysis of several traits implicates a ‘common gene, rare allele' model of genetic variation where some genes have many individually rare alleles contributing. Most natural alleles exhibit small effect sizes with little-to-no detectable pleiotropy or epistasis. Additionally, many of these genes are locked away in low-recombination regions that encourage the formation of multi-gene blocks that may underlie maize's strong heterotic effect. Domestication left strong marks on the maize genome, and some of the differences in trait architectures may be due to different selective pressures over time. Overall, maize's advantages as a model system make it highly desirable for studying the genetics of outcrossing species, and results from it can provide insight into other such species, including humans.
Keywords: maize, nested association mapping, quantitative traits, genome-wide association, heterosis
Introduction
Every year, more maize is harvested than any other crop on Earth; global output for 2010 exceeded 844 million tonnes (FAOSTAT, http://faostat.fao.org/). Apart from its impact as feed and fuel, maize also has a long history as a model genetic system. Both Charles Darwin and Gregor Mendel, for example, made early observations regarding maize inheritance (Darwin, 1876; Coe, 2001), and in the Twentieth Century maize was often at the forefront of genetics research. Some of its more notable contributions include the discovery of and/or significant insights into hybrid vigor (Shull, 1908), quantitative genetics (Emerson and East, 1913), chromosome translocation (McClintock, 1930), cytoplasmic inheritance (Rhodes, 1931), transposons (McClintock, 1950) and epigenetic inheritance (Kermicle, 1970) (see also Coe, 2001 for a review).
As a model system, maize has many advantages. It has more phenotypic diversity than any other model system, and the separate male (tassel) and female (ear) inflorescences make controlled crosses simple. Though naturally outcrossing, maize can be easily self-pollinated, and a single plant can yield hundreds of progeny. Diverse germplasm collections are readily accessible from repositories around the world, with the most notable ones in Mexico (CIMMYT), the United States (USDA-ARS) and France (INRA).
All these benefits make maize an important, if unlikely, bridge between human and plant genetics. On the one hand, maize encompasses many tools prized by plant geneticists: large-scale replication, ease of creating controlled crosses and inbred lines, the ability to create doubled haploids and the like. On the other hand, its outcrossing nature makes its genetics more similar to other outcrossing species, including humans, mice and Drosophila, than to self-pollinating plants like rice, wheat and Arabidopsis (for example, Rafalski and Morgante 2004).
If one takes Shull's observation of heterosis as a (somewhat arbitrary) beginning for maize quantitative genetics (Shull, 1908), we are now just past the century mark of such research. Moving forward into the second century of maize quantitative genetics promises a plethora of new insights and findings. In this review, we highlight several of these insights that have been gleaned over the past few years. They address pressing questions about maize quantitative genetics, including the general architecture of maize traits, the prevalence of epistasis and pleiotropy, the influence of recombination on traits in general and heterosis specifically, the types of polymorphisms underlying known maize variation and the influence of selection and domestication on the maize genome. We also briefly speculate on the role population history may have had in determining trait architecture. Finally, we conclude with a prospective on maize genetics, including several key questions yet to be answered.
The blossoming of maize genetics
Maize has a rich history of genetics studies, with insights and mechanisms stretching back through most of the past century (Coe, 2001). The study of maize quantitative traits stretches back nearly as long (for example, Shull, 1908; Emerson and East, 1913), but in most cases the insights were gleaned from specific experimental populations that did not always apply to the larger maize population or species. Even as recently as five years ago the majority of studies occurred in unique biparental populations that, although powerful in themselves, often arrived at conflicting results or ones that failed to hold up in different backgrounds.
The advent of inexpensive, high-throughput genotyping has changed maize genetics just as it has many other fields. Genotyping is no longer an experimental bottleneck, and large, diverse populations have now been assembled to take full advantage of the increased capacity.
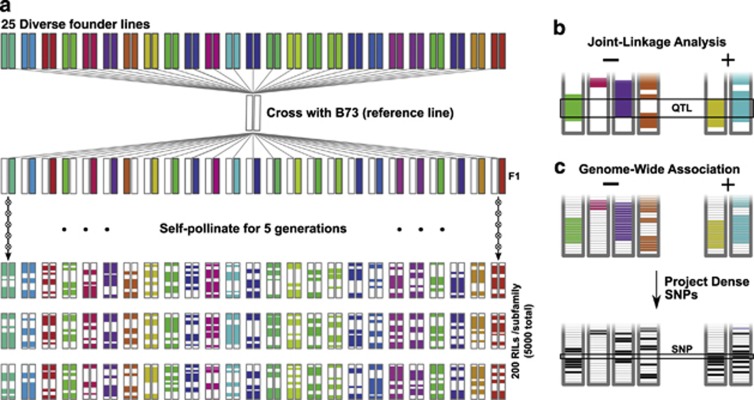
The largest of these new populations is the maize nested association mapping (NAM) population, a collection of 5000 recombinant inbred lines made by crossing 25 diverse parental inbred lines with the reference line, B73 (Figure 1a; McMullen et al., 2009). Using a common parent is slightly suboptimal from a statistical perspective (Stich, 2009) but provides several agronomic and physiological benefits for studying the population (Yu et al., 2008). As NAM is essentially a series of 25 parallel biparental populations linked by a common parent, it captures the advantages of both linkage mapping (high linkage disequilibrium within families, high allele frequencies) and association mapping (low linkage disequilibrium among families, diverse array of haplotypes). One of the great strengths of the NAM design is that detailed genotypes can be created for the whole population by genotyping the founders deeply and then projecting these genotypes onto progeny based on low-density genetic markers. This allows genome-wide association studies (GWAS) to take full advantage of ancient recombination histories to identify associations between traits and genetic loci at a very fine scale (Figure 1c). Due to the success of the original NAM population, additional populations are currently being prepared in China and Europe; similar designs are also being implemented in cotton, soybean and sorghum. Aside from their statistical power, these populations help unite the research community by providing a permanent resource for phenotyping and analysis. In this way they parallel the large, standardized resources of other organisms, such as the International HapMap Project (The International HapMap Consortium, 2003) and the Arabidopsis Multiparent Advanced Generation Inter-Cross (MAGIC) population (Kover et al., 2009).
Figure 1.
Nested association mapping (NAM) design. (a) The maize NAM population was created by crossing 25 diverse founder lines by the reference line B73. Single-seed descent and self-pollination for five generations were then used to generate 200 recombinant inbred lines (RILs) for each subfamily. Figure based on Yu et al. (2008). (b, c) Identifying significant associations in NAM proceeds through two routes. Joint linkage mapping (b) across the subfamilies can identify quantitative trait loci (QTL) for specific traits at moderate resolution by taking advantage of the shared B73 line in all the subfamilies (Li et al., 2011). Genome-wide association (c) instead uses the information of which chromosomal segments were inherited from which parent (top) to project dense genotyping from the founder lines onto the progeny for improved resolution (bottom). Colored bars show which parent the chromosomal segment originated from. Single-nucleotide polymorphisms (SNPs) in panel c are shown as either matching the allele present in B73 (white) or as the alternative allele (black).
In addition to NAM, many dedicated association populations have been and continue to be assembled for GWAS analysis (for example, Flint-Garcia et al., 2005; Camus-Kulandaivelu et al., 2006; Beló et al., 2008; Yang et al., 2011). As these populations sample extant variation, they are faster and simpler to set up than a controlled population like NAM. The greatest challenge with association populations, however, is the need for very dense genotyping. Due to low linkage disequilibrium in maize, it is estimated that at least 10 million single-nucleotide polymorphisms (SNPs) are needed to saturate the genome, nearly 100 times the number needed for Arabidopsis (Myles et al., 2009). This problem could be alleviated through projecting SNPs from a dense haplotype map, such as the ∼55 million SNPs in Maize HapMap2 (Chia et al., 2012). Eighty percent of these SNPs are in high linkage disequilibrium (r2>0.8) with at least one neighbor, allowing for relatively accurate projections. However, the same study showed that read-depth variants (a surrogate for moderate (2–10 kb) insertions, duplications, and deletions) were enriched for significant associations with phenotypic differences, indicating that SNPs do not capture the entirety of genetic variation. Thus, how effective this sort of projection will prove in practice remains to be seen.
General architecture of maize traits
As high-throughput genotyping and GWAS studies became possible, two major questions in the field of quantitative genetics were, first, how many genes influenced each trait, and second, what was the allele distribution within those genes? This debate was most visible for human genetic diseases, but it applies to maize and other organisms equally well. The initial view in both human genetic diseases and maize genetics was that quantitative variation is mostly due to large-effect alleles at relatively high frequencies. Subsequent GWAS studies consistently eroded this hypothesis, however, as even the best studies could account for only a small portion of total variability (Gibson, 2012).
The question of where all the ‘missing heritability' lies is still quite pressing in human genetic diseases, with rare alleles, epistasis, epigenetics, genotype-by-environment interaction and other mechanisms vying for evidence in their support (Manolio et al., 2009; Gibson, 2012). In maize, though, most of the missing heritability appears to have been found. In powerful populations such as NAM, for example, genetic models can often account for ⩾80% of the genetic variance (for example, Buckler et al., 2009; Kump et al., 2011; Tian et al., 2011). With several traits of varying complexity thus analyzed, we can begin to answer the two key questions raised earlier.
First, in terms of gene number, the majority of maize traits appear to be controlled by a large number of small-effect genes. The largest-effect sizes typically explain <5% of total variation, with total identifiable genetic variation spread across up to several dozen genes (for example, Laurie et al., 2004; Buckler et al., 2009; Kump et al., 2011; Tian et al., 2011; Cook et al., 2012). A prominent exception is kernel carotenoid content, which seems to be primarily controlled by variation in just three genes: y1 (Buckner et al., 1990), lcyE (Harjes et al., 2008) and crtB1 (Yan et al., 2010). The general pattern of many small-effect genes is also present in maize's wild ancestor, teosinte (Weber et al., 2008).
Additional evidence for a large number of genes affecting traits comes from an analysis of segregation distortion in the NAM population (McMullen et al., 2009). Segregation distortion occurs when a chromosomal segment from one parent is disproportionately inherited by the recombinant offspring. Such distortion is usually due to selection against the alternative allele. Tests of 1106 low-density SNP markers in NAM showed fully 54% of them to have consistent segregation distortion across subfamilies. These effects were significant but small, with most still within 10% of the hypothetical (1:1) segregation ratio. This result implies that most of the maize genome has a significant (albeit minor) effect on fitness. Of the five sites with extreme distortion, four of them could be identified with known genetic features, such as the su1 locus in sweet corn that enhances flavor but negatively affects germination. Conclusions about a general trait such as fitness may not be directly applicable to other, more specific traits, yet the fact that over half of the maize genome showed a detectable fitness effect implies that a large number of genes are not only involved in fitness but have segregating alleles with significant effects.
Second, though the allelic distribution within genes is not completely proven, it appears to follow a ‘common gene, rare variant' distribution where different lines often have distinct polymorphisms in the same genetic region. The strongest evidence for this again comes from NAM, where multiple studies consistently find quantitative trait loci (QTL) with different (and sometimes opposite) effect sizes mapping to the same locations in different subfamilies. This pattern holds for flowering time (Buckler et al., 2009), leaf architecture (Tian et al., 2011) and disease resistance (Kump et al., 2011; Poland et al., 2011), along with kernel carotenoid content mapped in different populations (Harjes et al., 2008; Yan et al., 2010). Given this variety of traits, this same pattern will probably hold for many—though not necessarily all—other maize traits.
Interestingly, although the QTL identified in these studies are sometimes enriched for candidate genes, many known candidate genes are not detected and most QTL do not contain any such genes at all (for example, Kump et al., 2011; Tian et al., 2011; Weng et al., 2011; Cook et al., 2012; Hung et al., 2012). With only 26 founder lines, NAM cannot capture all of maize diversity and so it is not surprising if some candidate genes are not rediscovered. Many of these candidate genes were also discovered by mutagenesis experiments, where mutants are generally selected based on their large phenotypic effect. If the effect size in nature is similar to that seen in the lab, variation in genes identified this way may be less relevant to natural populations because large, deleterious effects should be quickly purged by natural selection.
Large-scale genetic dissection is still in its infancy, so one can only make tentative comparisons between the genetic architecture of maize and that of other species. Some human studies, for example, also favor the ‘common gene, rare variant' hypothesis, though the support behind it is far from proven (Gibson, 2012). Maize also appears to have a much more dispersed genetic architecture than other model plants, such as rice or Arabidopsis. Often in these plants only a few QTL exert the majority of control on traits that, in maize, are controlled by dozens of QTL. For example, flowering time in maize is influenced by over 30 small-effect QTL (Buckler et al., 2009), while in rice (Huang et al., 2012), sorghum (Lin et al., 1995) and Arabidopsis (Salomé et al., 2011) most variation is explained by less than a dozen QTL (and in the case of sorghum, only one). Maize genetics thus appears, at least in some respects, to act more like human genetics than those of selfing plants.
Recombination rate
The difference in genetic architecture between maize and sorghum, rice and Arabidopsis is probably due to the different effective recombination rate each species experiences. Maize is an outcrossing species, so genotypes get shuffled and recombined every generation. Selfing species like rice and Arabidopsis, however, are generally homozygous at all loci, so any recombination that does occur does not change the genotype (Rafalski and Morgante, 2004) and leads to a low effective recombination rate. This fixed genotype in selfing species allows mutations to build genetic networks in relative isolation, whereas in maize any such networks get shuffled around with every mating. Selection may thus favor smaller effect sizes in outcrossing species, because small effects have a lower chance of disrupting critical networks. In the specific case of flowering time, reproduction itself is probably also exerting pressure as a maize plant must flower both at the proper environmental time and also at the same time as its neighbors. This situation would intrinsically disfavor large deviations in flowering time and favor many independent, small-effect genes because such an arrangement helps keep an individual's response near the local optimum (Buckler et al., 2009).
This argument does not apply equally to all maize genes, however. The degree to which recombination isolates specific genes depends on the local recombination rate, which varies across the genome and is especially low around the centromeres (Tenaillon et al., 2001). Before the maize genome was sequenced, it was thought that these pericentromeric regions would contain relatively few genes. Subsequent analysis, however, found that the pericentromeric regions in maize cover 60–100 megabases each and include fully 21% of its genes (Gore et al., 2009; Schnable et al., 2009).
The low recombination rate in these regions has several important implications for maize genetics. As they only rarely recombine, these regions tend to be inherited as large gene blocks (haplotypes), more akin to what occurs in a selfing species. Allele-interaction networks and epistatic relationships should be easier to create and maintain within these blocks than they are across the genome. The effect would still be less than in selfing species, however, as most such blocks would be heterozygous in any given organism. Also, the difficulty of breaking such networks apart would make them hard to identify in the lab.
Another consequence of low recombination in these regions is that any deleterious alleles are strongly linked to their neighboring genes. Many such deleterious alleles are probably present in maize germplasm, and the low recombination rates mean that substituting them with a beneficial allele has only rarely occurred during recent maize breeding. Using genotyping to select for specific recombinants in these regions could potentially introduce powerful new allelic combinations into maize germplasm.
One potential way to accelerate targeted recombination is to select for alleles that increase global recombination rates, thereby increasing the chances of a favorable crossover event. Such controllers have been found in other species, including humans (Kong et al., 2008). Although recombination rates vary among the NAM subfamilies, no shared, global controllers of recombination rates were found (McMullen et al., 2009). Instead, only local controllers such as small inversions or transposon insertions were found, most of which repress recombination. Given the high power and molecular diversity of NAM, the absence of obvious modifiers of global recombination implies that they may not exist in natural maize variation. Such a situation would be unfortunate from the breeding perspective, though it could probably be overcome via transgenic or mutagenesis methods.
Heterosis
Heterosis is the emergent property wherein the progeny of a cross perform better than either parent. Heterosis in maize was first recorded by Darwin (1876) and then formally defined by Shull over a century ago (Shull, 1908). Since then, maize breeders have turned a simple field observation into the basis of almost all modern maize breeding (Duvick, 2001). Despite decades of work, however, the actual molecular mechanisms behind heterosis are still debated (Duvick, 2001; Garcia et al., 2008; Larièpe et al., 2012).
Three major (and not necessarily incompatible) mechanisms have been proposed to explain heterosis: dominance, where superior alleles in one parent complement poor alleles from the other parent; epistasis, where interactions with genes encoded elsewhere in the genome leads to superior performance; and overdominance, where having two different alleles is superior to either one alone. A variant mechanism known as ‘pseudo-overdominance' occurs when a series of linked dominant and recessive genes appears to act like a single overdominant locus due to low local recombination (Larièpe et al., 2012). Evidence that pseudo-overdominance is the major contributor to heterosis in maize has been around for nearly a century (for example, Jones, 1917, Moll et al., 1964), and modern analyses tend to support this conclusion (reviewed in Larièpe et al., 2012). Another consequence of having these linked gene blocks is that they limit the gains available to breeders. This is known as the Hill–Robertson effect (Hill and Robertson, 1966), which states that selection cannot act perfectly on loci in a population if they are linked to loci with an opposed fitness effect (that is, a beneficial allele linked to a harmful one, or vice-versa).
Blocks of linked genes are exactly what results from the low recombination rate around maize centromeres, so these blocks could be responsible for heterosis in maize via pseudo-overdominance. In support of this model, heterosis QTL frequently overlap the centromeric regions (for example, Schön et al., 2010; Larièpe et al., 2012). Additionally, the NAM population shows significant excess heterozygosity around every centromere (McMullen et al., 2009). This pattern implies strong selection to maintain these regions in heterozygous states, probably due to their containing alleles that are highly deleterious when homozygous.
This again contrasts with the situation in rice, where epistasis seems to have the stronger role in heterosis (Garcia et al., 2008). The differing reproductive modes between the species probably cause the difference in heterosis mechanisms. Not only does self-pollination help maintain epistatic networks, but it also quickly exposes recessive, deleterious alleles to selection pressure. Outcrossing, however, allows such alleles to persist in the population by hiding in the heterozygous state. Inbred lines used in modern breeding inherit this genetic load, which results in pronounced inbreeding depression and then a correspondingly large heterotic effect when complementary lines are mated.
Epistasis
Epistasis has been put forth as the explanation for a wide variety of genetic conundrums, including both missing heritability and heterosis (Gibson, 2012; Larièpe et al., 2012). The degree to which epistasis has a role in these is unknown, however, and in some cases appears to differ markedly by organism.
Maize studies to date indicate only a small role for epistasis in general gene regulation (for example, Stuber et al., 1992; Laurie et al., 2004; Buckler et al., 2009; Zhang et al., 2010a; Tian et al., 2011; Larièpe et al., 2012). Little evidence of two-gene epistasis was found during the examination of NAM, for example (McMullen et al., 2009), though in this and other cases it may be due to the effects being too small to detect after correcting for multiple testing. Epistatic interactions in heterosis have been noted (for example, Larièpe et al., 2012), but they account for only a minor portion of the total heterotic effect.
As mentioned earlier, the low recombination rate in pericentromeric regions should allow local epistatic networks to be created and maintained. The lack of evidence for widespread epistasis appears to contradict this assertion. Current studies may not have the power to detect these interactions, either due to statistical limitations or lack of sufficient recombination to isolate the individual network components. Alternatively, such epistatic interactions may exist but still only minimally contribute to the overall trait effects. The latter is supported by several studies that have used QTL identified in NAM to ‘predict' the phenotype of the 26 founder lines (Brown et al., 2011; Kump et al., 2011; Poland et al., 2011; Tian et al., 2011). These predictions are generally highly accurate, which is not what one would expect if these lines contained significant epistatic networks (which were presumably broken apart in the recombinant inbred lines).
An important caveat for these studies is that most of them are not designed to consistently detect epistasis on a genome-wide level (Gibson, 2012). As such, many biologically relevant epistatic effects may still be hidden by our inability to detect them. This view is supported by work in animals, where highly controlled experiments in mice and Drosophila reveal extensive epistasis, whereas GWAS in humans, which generally has lower power than in these controlled populations, has shown almost none (Flint and Mackay, 2009). The situation in maize is similar, in that experiments testing specific genes can find significant epistasis (for example, Weber et al., 2008; Studer and Doebley, 2011), whereas genome-wide studies generally do not. Biological epistasis (meaning different genes interacting and affecting each other) is thus probably widespread, but the effect on total variance is so small that statistical epistasis (meaning the interaction of two terms (genes) in a model) cannot be detected.
Pleiotropy
Pleiotropy—one gene affecting multiple traits—appears to be minimal in maize, with pleiotropic effects confined to closely related traits. Flowering time traits, for example, exhibit some pleiotropy among themselves (Buckler et al., 2009) and with inflorescence architecture (Brown et al., 2011). Traits related to central carbon and nitrogen metabolism also show pleiotropy among themselves, though this is probably due to common regulation and pathways (Zhang et al., 2010a).
The majority of QTL are not pleiotropic, however, and several maize traits seem to exhibit no pleiotropy at all. For example, a presumed correlation between flowering time and resistance to several diseases appears to be simply due to population structure (Kump et al., 2011; Poland et al., 2011). Almost no pleiotropy was found among several leaf architecture traits either (Tian et al., 2011), even though one might expect these to be closely correlated. Selection may favor independence of traits, as combinations that are advantageous in one environment may be detrimental in a different environment. Being able to select on different traits independently would have helped wild teosinte adapt to different conditions across its native habitat; later, it facilitated adaptation of maize to many different local conditions as farmers spread it across the Americas (Figure 2).
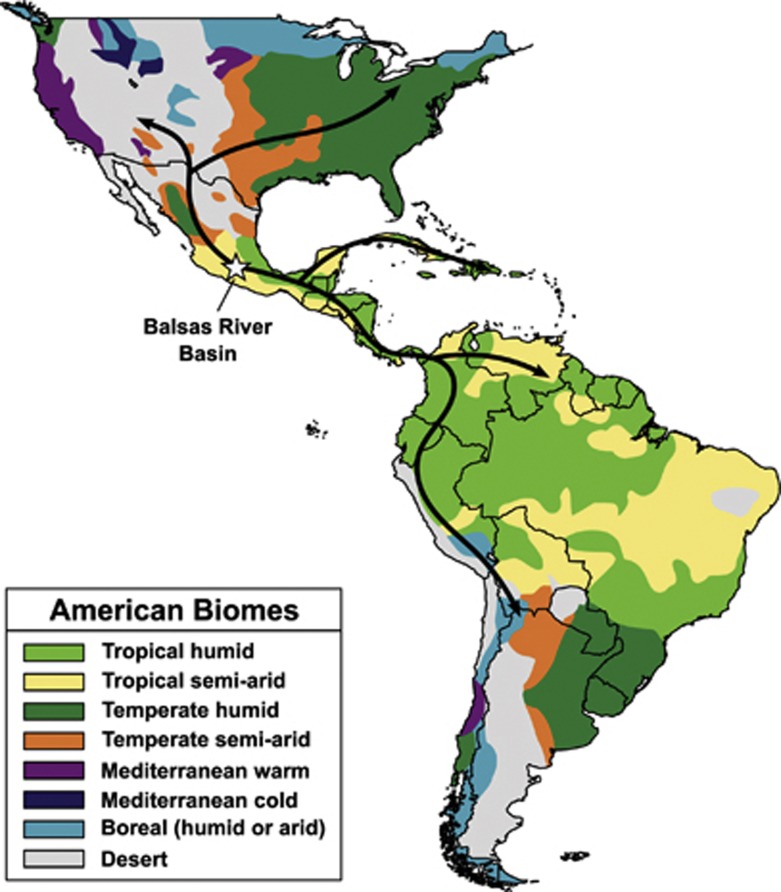
Figure 2.
Maize dispersion and diversity. Maize originated in the Balsas river basin of southwestern Mexico approximately 9000 years ago (Matsuoka et al., 2002). Over the next several thousand years, it spread through the Americas, crossing (and adapting to) deserts, mountains, tropics and almost every other environment present across both continents before contact with European explorers spread it throughout the world. Arrows show probable routes of pre-Columbian dispersion based on phylogenetic reconstruction (Matsuoka et al., 2002); biome data are from the USDA National Resource Conservation Service (http://www.nrcs.usda.gov).
One question that remains unanswered is whether the pleiotropic alleles mentioned above are truly pleiotropic or whether they are simply a series of tightly linked genes where each affects different traits. Resolving this question will require fine-mapping of the QTL to see if the effects can be separated or not, although the small effect sizes will make this difficult in most cases.
Molecular polymorphisms
Known trait variation in maize has been shown to be due to SNPs (Wang et al., 2005; Salvi et al., 2007; Harjes et al., 2008; Zheng et al., 2008; Zhang et al., 2010b), transposon insertions (Palaisa et al., 2003; Salvi et al., 2007; Yan et al., 2010; Studer et al., 2011) and other insertion/deletion variants (indels) (Palaisa et al., 2003; Salvi et al., 2007; Harjes et al., 2008; Yan et al., 2010), but the relative importance of each is hard to determine. A recent resequencing of >100 maize and teosinte lines (Chia et al., 2012) found extensive variation at both the single-nucleotide level and for read-depth variants (indicators of mid-sized (2–10 kb) insertions, duplications and deletions). GWAS analysis of five traits found that read-depth variants were strongly enriched relative to SNPs for significant associations, especially in intergenic regions. Read-depth variants still formed only a minority (15–27%) of associated loci, though this could be an underestimate due to the relatively crude methods of identifying them. On a site-per-site basis, then, indels may have a stronger propensity to cause phenotypic variation, likely because of their larger mutational effect. The maize genome has an exceptional diversity of indels and related variation, such as copy-number variation and presence–absence variation (Wang and Dooner, 2006; Swanson-Wagner et al., 2010), so these variants could be a rich source of maize phenotypic diversity.
Several instances are known of allelic series where different polymorphisms accomplish similar genetic effects (for example, Salvi et al., 2007; Harjes et al., 2008; Yan et al., 2010). This interchangeability implies that most variation in maize focuses around disruptive mutations, as many different polymorphisms can disrupt a given genetic feature. Disruption does not automatically mean a loss of function, however, as disruption of regulatory elements can cause a gain of function for the regulated gene. This situation is exactly the case for the phytoene synthase y1 gene, where two insertions in the promoter region appear to cause ectopic expression in the endosperm (Palaisa et al., 2003). Similarly, a pair of transposon insertions ∼60 kb upstream of teosinte branched1 (tb1) increase expression by ∼twofold in reporter constructs (Studer et al., 2011), implying that the insertion disrupts some sort of repressor element.
Although several known polymorphisms affect protein sequence (for example, Wang et al., 2005; Zheng et al., 2008; Zhang et al., 2010b), differences in gene expression appear more pervasive. For example, expression-level differences underlie variation in kernel carotenoid content (Palaisa et al., 2003; Harjes et al., 2008; Yan et al., 2010), flowering time (Salvi et al., 2007) and plant architecture (Studer et al., 2011). These all reflect polymorphisms in cis, where the causative polymorphism is located on the same DNA strand and is usually close to or within the gene it influences. A linkage analysis of enzyme activity, however, implies that most QTL act in trans (Zhang et al., 2010a). Of the 73 QTL identified for 10 different enzyme activities, only 3 actually overlapped genes with the specified activity; all others apparently work in trans.
Given the prevalence of trans-acting QTL, it is not surprising that transcription factors are frequently identified as important drivers of variation. Well-studied examples of important maize transcription factors include tb1 (Studer et al., 2011), which controls the dominance of a single stalk over secondary branching; vegetative to generative transition (vtg1) (Salvi et al., 2007), an important gene for flowering time; and teosinte glume architecture1 (tga1) (Wang et al., 2005), which controls the kernel casing and was a major target during maize domestication. GWAS, meanwhile, has identified transcription factors associated with leaf architecture (Tian et al., 2011), disease resistance (Kump et al., 2011; Poland et al., 2011), kernel composition (Cook et al., 2012) and abscisic acid content (Setter et al., 2011). Transcription factors have also been strongly implicated in maize domestication (Swanson-Wagner et al., 2012; Hufford et al., 2012a). Transcriptional control thus seems to be the primary driver of variation in maize, with structural mutations having an important secondary role.
Epigenetics
Despite its active epigenetics research community (for example, Arteaga-Vazquez and Chandler, 2010; Eichten et al., 2011; Miclaus et al., 2011) and the fact that much of the initial research on epigenetics occurred in maize (for example, Kermicle, 1970), evidence for epigenetic influence on maize quantitative traits is lacking. This is primarily due to lack of data, though we expect that studies specifically testing for epigenetics in maize inheritance will be published soon. In the meantime, the fact that several studies have been able to account for the majority of genetic variance without invoking epigenetic mechanisms (for example, Buckler et al., 2009; Kump et al., 2011; Poland et al., 2011; Larièpe et al., 2012) implies that such mechanisms are either of minor importance to QTL or that they are stably associated with underlying genetic polymorphisms.
The effects of domestication and selection
Maize has been shaped by domestication more than any other crop. Domestication selected for genes controlling many of its most important agronomic traits (kernel architecture, plant architecture, harvestable yield and so on), so determining which parts of its genome were most affected by domestication can indicate genes that control these traits. Searching for these genes in maize is facilitated by its being an outcrossing species, as the high effective recombination efficiently breaks up haplotypes over time. The weak bottleneck during maize domestication (Wright et al., 2005) also helps, as it allowed fully 83% of the standing variation in teosinte to pass into maize unscathed (Hufford et al., 2012a). Thus when comparing maize to teosinte, domestication regions appear as small areas of low maize diversity surrounded by much larger swaths of near-equivalent diversity.
Early experiments indicated that only five or six major loci were involved in the transition from teosinte to maize (Beadle, 1980). Subsequent fine-mapping experiments, however, indicate that these loci are probably just linked collections of genes with the largest effects (Quijada et al., 2009). Genome-wide analysis now indicates several thousand genes that show evidence of selection during domestication (Wright et al., 2005; Hufford et al., 2012a). These domestication genes suffered a bottleneck 10 times as strong as the rest of the maize genome, though an excess of rare alleles indicates that their diversity may be starting to recover (Hufford et al., 2012a).
Although specific domestication genes are known, such as tb1 (Studer et al., 2011) and tga1 (Wang et al., 2005), it is still unknown to what degree various protein classes had a role in domestication. Transcription factors and other regulatory genes are obvious candidates, as changes in one result in downstream effects in all its targets. A test of 72 regulatory genes found minimal enrichment for domestication genes, however, except in the case of kinase/phosphatase genes (Zhao et al., 2008). On the other hand, a specific analysis of MADS-box transcription factors found them to be significantly enriched for genes involved in both maize domestication and recent improvement (Zhao et al., 2011). Genome-wide analysis of domestication loci also reveals a slight enrichment for transcription factors or genes with DNA-binding properties (Swanson-Wagner et al., 2012; Hufford et al., 2012a). Stronger evidence for the importance of transcription during domestication comes from RNA expression analysis. Compared with expression in teosinte, genes in maize show significantly altered expression levels, and many of the transcriptional correlations present in teosinte have been disrupted in maize. (Hufford et al., 2012a; Swanson-Wagner et al., 2012). Many selected genes have higher, more consistent expression in maize than in teosinte (Hufford et al., 2012a), a possible result of early farmers selecting for plants with robust, consistent phenotypes.
In addition to the diversity that passed through domestication, maize has also been enriched by subsequent introgression from wild teosinte (van Heerwaarden et al., 2011; Hufford et al., 2012a, 2012b). Although most such introgressions were probably purged owing to unfavorable effects on the crop, some have been maintained due to their presumed adaptive potential (Hufford et al., 2012a, 2012b). Maize and teosinte still grow together in some regions of Mexico, and it thus seems likely that gene flow between the two is still occurring. Such flow is a double-edged sword, as it not only creates the possibility of novel variation coming into maize landraces but also allows for teosinte's natural variation to be eroded by introgression from improved maize lines. Preservation of existing variation in genebanks is thus of high priority to preserve this pool of natural variation for future use.
Phenotype prediction
Maize is arguably the most important crop in the world, and the ability to predict phenotypes from genotype alone would be an enormous boon to breeders. The primary method for doing so is through genomic selection (GS) models (Lorenz et al., 2011), a strategy that is well established in animal breeding but still in its infancy among plant breeders.
GS uses a training population to create a statistical model and assigns breeding values to all markers across the genome (instead of just the significant ones) using both phenotypes and genotypes. This model is then used to assign breeding values to new individuals based solely on their genotype. Simulation studies and early field studies indicate strong potential for GS in maize and other crop species (Bernardo and Yu, 2007; Bernardo, 2009; Albrecht et al., 2011; Heslot et al., 2012), while other studies have shown promise by predicting maize phenotypes from transcriptomes (Fu et al., 2012), metabolites (Riedelsheimer et al., 2012a, 2012b) and spectral reflectance (Weber et al., 2012). The ultimate goal of all these techniques is to shorten the time per breeding cycle and/or reduce the need for phenotyping in order to make breeding both faster and more efficient.
The biggest current challenge is that models based on one training population lose a significant amount of accuracy when transferred to divergent populations (Albrecht et al., 2011). Presumably this is due to different allele frequencies and novel haplotypes in the divergent populations that make them a poor fit for the statistical model. This pattern even holds among elite maize lines with relatively narrow gene pools (Massman et al., 2012), severely limiting the utility of any single model.
Almost all existing reports on GS in maize are prospective, estimating how well it should work based on simulation data or existing populations. The true test of its potential will come with the publication of multi-year breeding programs that rely heavily or completely on GS and compare the results with those based solely on phenotypic selection. If successful, such a program could potentially breed maize many times faster than has been historically possible (Lorenz et al., 2011). Such programs are already operating in the private sector, and they are currently being implemented in the public sector.
Determinants of trait architecture
Across all the different maize traits, an important question is what determines the relative complexity of one trait versus another. As mentioned earlier, most maize traits are controlled by many genes of small effect, but there are a few exceptions to this general pattern. Carotenoids, for example, have only a few genes controlling most of their variation (Buckner et al., 1990; Harjes et al., 2008; Yan et al., 2010), whereas ear architecture has many genes of (relatively) large effect (Brown et al., 2011).
When comparing traits, it is helpful to remember that maize has gone through several different population phases during its history (Figure 3). Up until domestication, it was a wild grass (teosinte) that likely had a large effective population size and plenty of time to adapt to natural changes in the environment. Domestication, however, mildly bottlenecked the whole genome and severely reduced diversity in domestication-related genes (Hufford et al., 2012a). The subsequent spread of maize through the Americas by native farmers then exposed it to strong new selection pressures as it adapted to drastically different environments (Figure 2). Most recently, modern maize breeding placed strong selection pressure on existing varieties, not just due to pressure to increase yield potential but also by relying heavily on self-pollinated inbred lines, a situation that maize never faced in nature. These different phases of maize history have each left their mark on the maize genome, and one would expect that traits that were selected for in each phase would have genetic architectures reflecting their history.
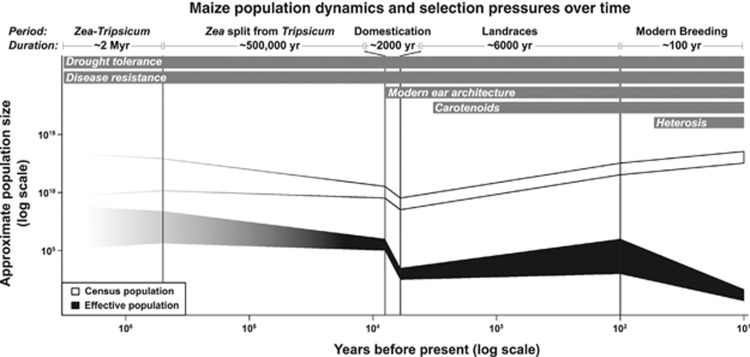
Figure 3.
Maize population dynamics and trait architecture. The genetic pool leading to modern maize has fluctuated greatly over its history, both in terms of absolute (census) population and effective population size, and also the selective pressures operating on it. Major divisions in maize history are indicated, with approximate durations noted. As many of the population values are unknown, the indicated population sizes are a range of likely estimates, with relative differences more important than absolute ones. Differences in the genetic architectures of certain traits may be due to the different lengths of time and population regimes that the traits have evolved under. (Note that although heterosis is not truly a trait, breeding for distinct heterotic groups can be seen as diversifying selection with heterosis as the final goal (Duvick, 2005)).
Traits that have been important throughout maize and teosinte's history, such as disease resistance and drought tolerance, encompass millions of years of accumulated variation. These genes should have very diffuse genetic architectures, with a very large number of genes each contributing a small amount to the phenotype as any large-effect alleles for these traits would presumably have been fixed rapidly (Orr, 1998). On the other hand, traits that came under selection during and after domestication, such as kernel carotenoids and maize-specific ear morphology, should have architectures with fewer genes and/or larger effect sizes because of the shorter amount of time for variation to accumulate. Finally, traits that have only become important during the past century of intense maize breeding should have the simplest architectures of all. The best example of this latter class is heterosis. Even though heterosis is more of an emergent property than a trait per se, selection to form distinct groups with good combining ability can be seen as a type of diversifying selection (Duvick, 2005). This selection has resulted in only two to three major heterotic groups that underlie essentially all modern hybrid maize production.
The way forward
Maize genetics is at a powerful point in its history, with large experimental populations combining with inexpensive genotyping to enable rigorous investigation of its genetic mechanisms. Although we have made great strides in just the past few years, many more questions remain to be answered. Among these, some of the most pressing are:
How can we best find rare alleles?
Most maize alleles seem to be rare, which makes them difficult to find. Part of the power of NAM is that it artificially boosts the allele frequency such that even the rarest allele will occur in enough progeny to be detected, so long as it is present in one of the founders. Those founders contain only part of total maize diversity, however, and such an approach is impractical for panels consisting of hundreds or thousands of unique maize lines. How, then, can we best locate rare alleles in these large populations? Better statistical and experimental methods are needed to identify these rare alleles, and development of such should be a high priority in the coming years.
How can recombination be further harnessed to accelerate breeding?
The point of breeding is to make better combinations of alleles than are currently available. Though breeders have always taken advantage of recombination to form new lines, the question now is how to use modern molecular and genetic knowledge to further accelerate the breeding process. Part of this will be determining factors that increase the rate of recombination. Another aspect is creating methods to enhance and/or select for recombination in specific regions to generate novel diversity while minimizing genetic load. Targeted homologous recombination around centromeres, for example, could create novel haplotypes and/or replace deleterious alleles in these normally low-recombination regions. One could even apply the same process to specific genes to get a desired haplotype that is not found in existing populations.
What is the molecular basis of heterosis?
Although many studies indicate dominance and pseudo-overdominance as the primary mechanisms of heterosis in maize, the degree to which each contributes is still unclear. Heterosis underlies much of modern maize breeding, so an understanding of its mechanisms has the potential to greatly improve maize production. Studies of heterosis using more diverse lines, such as from NAM or a diversity panel, could be especially insightful due to the different genotypes involved.
To what extent does epistasis have a role in maize traits?
Epistasis occurs in maize but, so far, appears to have only a minor role in trait architecture. Is this due to a particular facet of maize biology, or is it that current studies simply lack the power to identify epistatic interactions? Are extensive epistatic networks hidden in low-recombination regions where they can avoid being broken apart by recombination? Also, when epistasis is noticed it appears more prevalent with some traits than others (Larièpe et al., 2012). Knowing the reason for this would provide valuable insight into the role of epistasis in maize genetics.
How much of maize quantitative variation is due to epigenetic inheritance?
Although current studies loosely imply that epigenetics has little role in maize inheritance, confirmation of this hypothesis is still needed. Given the strong history of epigenetic studies in maize, its importance to global agriculture and the numerous epigenetic tools now available, we expect that studies specifically addressing this question will be quickly forthcoming.
How can we best predict phenotype from genotype?
Genotyping costs have fallen so much in the past decade that they are now frequently among the least-expensive parts of an experiment. There is still room for improvement, of course, especially in intergenic regions; nonetheless, current genotyping quality is good enough that many researchers' focus is on shifting from phenotypes to genotypes as the basis for selection because of the promise of both reduced costs and faster breeding gains. Early results of such GS are promising, but there is still much debate in the field as to the best methodology to use (Heslot et al., 2012). Ideally, one would also want side-by-side comparisons of breeding programs using GS versus standard phenotypic selection to evaluate their relative performances and determine how well the predicted accuracies hold up in practice. Other major questions regard what size of training set is necessary to get accurate predictions and how closely it needs to be related to the target population. Will 1000 maize lines be enough, or will it require 100 000? How many environmental replicates are sufficient? Will training on a diverse maize population allow accurate prediction of any subpopulation, or must the makeup of the training population closely match the target? These and other considerations are informed by simulations but will require field trials to answer definitively.
Ultimately, maize quantitative genetics aims to improve maize yield for agriculture. The insights gained in the process can have broader impacts, however. As maize has inherited its genetic architecture from a history of outcrossing, its genetics bear at least some resemblance to other outcrossing species, including humans. Combining this with the advantages of doing genetics in maize—ease of replication, large number of progeny, controlled crosses and the like—makes maize a strong platform for informing broader genetic investigations. Human genetics will have its own quirks and insights, of course, and ultimately a plant is no substitute for a human no matter how much you know about its genetics. Such comparisons can form a starting point, however. Maize is much more than just kernels on a plate, and the discoveries flowing from it promise to keep it at the forefront of genetics research for many years to come.
Data archiving
There were no data to deposit.
Acknowledgments
We thank the members of the Buckler lab for helpful discussions during the writing of this manuscript, and Peter Bradbury, Denise Costich and Sara Miller for help in editing it for publication. This work was supported by NSF grant 0820619.
The authors declare no conflict of interest.
References
- Albrecht T, Wimmer V, Auinger H-J, Erbe M, Knaak C, Ouzunova M, et al. Genome-based prediction of testcross values in maize. Theor Appl Genet. 2011;123:339–350. doi: 10.1007/s00122-011-1587-7. [DOI] [PubMed] [Google Scholar]
- Arteaga-Vazquez MA, Chandler VL. Paramutation in maize: RNA mediated trans-generational gene silencing. Curr Opin Genet Dev. 2010;20:156–163. doi: 10.1016/j.gde.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle G. The ancestry of corn. Scientific American. 1980;242:112–119. [Google Scholar]
- Beló A, Zheng P, Luck S, Shen B, Meyer DJ, Li B, et al. Whole genome scan detects an allelic variant of fad2 associated with increased oleic acid levels in maize. Mol Genet Genomics. 2008;279:1–10. doi: 10.1007/s00438-007-0289-y. [DOI] [PubMed] [Google Scholar]
- Bernardo R. Genomewide selection for rapid introgression of exotic germplasm in maize. Crop Sci. 2009;49:419–425. [Google Scholar]
- Bernardo R, Yu J. Prospects for genomewide selection for quantitative traits in maize. Crop Sci. 2007;47:1082–1090. [Google Scholar]
- Brown PJ, Upadyayula N, Mahone GS, Tian F, Bradbury PJ, Myles S, et al. Distinct genetic architectures for male and female inflorescence traits of maize. PLoS Genet. 2011;7:e1002383. doi: 10.1371/journal.pgen.1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, et al. The genetic architecture of maize flowering time. Science. 2009;325:714–718. doi: 10.1126/science.1174276. [DOI] [PubMed] [Google Scholar]
- Buckner B, Kelson TL, Robertson DS. Cloning of the yl locus of maize, a gene involved in the biosynthesis of carotenoids. Plant Cell. 1990;2:897–876. doi: 10.1105/tpc.2.9.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus-Kulandaivelu L, Veyrieras J-B, Madur D, Combes V, Fourmann M, Barraud S, et al. Maize adaptation to temperate climate: relationship between population structure and polymorphism in the Dwarf8 gene. Genetics. 2006;172:2449–2463. doi: 10.1534/genetics.105.048603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia J-M, Song C, Bradbury PJ, Costich D, de Leon N, Doebley J, et al. Maize HapMap2 identifies extant variation from a genome in flux. Nat Genet. 2012;44:803–807. doi: 10.1038/ng.2313. [DOI] [PubMed] [Google Scholar]
- Coe EH. The origins of maize genetics. Nat Rev Genet. 2001;2:898–905. doi: 10.1038/35098524. [DOI] [PubMed] [Google Scholar]
- Cook JP, McMullen MD, Holland JB, Tian F, Bradbury P, Ross-Ibarra J, et al. Genetic architecture of maize kernel composition in the nested association mapping and inbred association panels. Plant Physiol. 2012;158:824–834. doi: 10.1104/pp.111.185033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C.1876The Effects of Cross and Self Fertilization in the Vegetable Kingdom John Murray: London, UK; p.482 [Google Scholar]
- Duvick DN. Biotechnology in the 1930s: the development of hybrid maize. Nat Rev Genet. 2001;2:69–74. doi: 10.1038/35047587. [DOI] [PubMed] [Google Scholar]
- Duvick DN. The contribution of breeding to yield advances in maize (Zea mays L.) Adv Agron. 2005;86:83–145. [Google Scholar]
- Eichten SR, Swanson-Wagner RA, Schnable JC, Waters AJ, Hermanson PJ, Liu S, et al. Heritable epigenetic variation among maize inbreds. PLoS Genet. 2011;7:e1002372. doi: 10.1371/journal.pgen.1002372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson RA, East EM.1913The inheritance of quantitative characters in maize Bulletin of the Agricultural Experiment Station of Nebraska Lincoln, Nebraska; vol. 2,pp1–120. [Google Scholar]
- Flint J, Mackay TFC. Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res. 2009;19:723–733. doi: 10.1101/gr.086660.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint-Garcia S, Thuillet A-C, Yu J, Pressoir G, Romero SM, Mitchell SE, et al. Maize association population: a high-resolution platform for quantitative trait locus dissection. Plant J. 2005;44:1054–1064. doi: 10.1111/j.1365-313X.2005.02591.x. [DOI] [PubMed] [Google Scholar]
- Fu J, Falke KF, Thiemann A, Schrag TA, Melchinger AE, Scholten S, et al. Partial least squares regression, support vector machine regression, and transcriptome-based distances for prediction of maize hybrid performance with gene expression data. Theor Appl Genet. 2012;124:825–833. doi: 10.1007/s00122-011-1747-9. [DOI] [PubMed] [Google Scholar]
- Garcia AAF, Wang S, Melchinger AE, Zeng Z-B. Quantitative trait loci mapping and the genetic basis of heterosis in maize and rice. Genetics. 2008;180:1707–1724. doi: 10.1534/genetics.107.082867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 2012;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore MA, Chia J-M, Elshire RJ, Sun Q, Ersoz ES, Hurwitz BL, et al. A first-generation haplotype map of maize. Science. 2009;326:1115–1117. doi: 10.1126/science.1177837. [DOI] [PubMed] [Google Scholar]
- Harjes CE, Rocheford TR, Bai L, Brutnell TP, Kandianis CB, Sowinski SG, et al. Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science. 2008;319:330–333. doi: 10.1126/science.1150255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslot N, Yang H-P, Sorrells ME, Jannink J-L. Genomic selection in plant breeding: a comparison of models. Crop Sci. 2012;52:146–160. [Google Scholar]
- Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966;8:269–294. [PubMed] [Google Scholar]
- Huang X, Zhao Y, Wei X, Li C, Wang A, Zhao Q, et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat Genet. 2012;44:32–39. doi: 10.1038/ng.1018. [DOI] [PubMed] [Google Scholar]
- Hufford MB, Xu X, van Heerwaarden J, Pyhäjärvi T, Chia J-M, Cartwright RA, et al. Comparative population genomics of maize domestication and improvement. Nat Genet. 2012a;44:808–811. doi: 10.1038/ng.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufford MD, Lubinsky P, Pyhäjärvi P, Devengenzo M, Ellstrand NC, Ross-Ibarra J.2012bThe genomic signature of crop-wild introgression in maize arXiv 12083894v2[q-bio.PE]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung H-Y, Shannon LM, Tian F, Bradbury PJ, Chen C, Flint-Garcia SA, et al. ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc Natl Acad Sci USA. 2012;109:E1913–E1921. doi: 10.1073/pnas.1203189109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DF. Dominance of linked factors as a means of accounting for heterosis. Genetics. 1917;2:466–479. doi: 10.1093/genetics/2.5.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle JL. Dependence of the R-mottled aleurone phenotype in maize on mode of sexual transmission. Genetics. 1970;66:69–85. doi: 10.1093/genetics/66.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Stefansson H, Masson G, Helgason A, Gudbjartsson DF, et al. Sequence variants in the RNF212 gene associate with genome-wide recombination rate. Science. 2008;319:1398–1401. doi: 10.1126/science.1152422. [DOI] [PubMed] [Google Scholar]
- Kover PX, Valdar W, Trakalo J, Scarcelli N, Ehrenreich IM, Purugganan MD, et al. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 2009;5:e1000551. doi: 10.1371/journal.pgen.1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kump KL, Bradbury PJ, Wisser RJ, Buckler ES, Belcher AR, Oropeza-Rosas MA, et al. Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat Genet. 2011;43:163–168. doi: 10.1038/ng.747. [DOI] [PubMed] [Google Scholar]
- Larièpe A, Mangin B, Jasson S, Combes V, Dumas F, Jamin P, et al. The genetic basis of heterosis: multiparental quantitative trait loci mapping reveals contrasted levels of apparent overdominance among traits of agronomical interest in maize (Zea mays L.) Genetics. 2012;190:795–811. doi: 10.1534/genetics.111.133447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie CC, Chasalow SD, LeDeaux JR, McCarroll R, Bush D, Hauge B, et al. The genetic architecture of response to long-term artificial selection for oil concentration in the maize kernel. Genetics. 2004;168:2141–2155. doi: 10.1534/genetics.104.029686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Bradbury P, Ersoz E, Buckler ES, Wang J. Joint QTL linkage mapping for multiple-cross mating design sharing one common parent. PLoS One. 2011;6:e17573. doi: 10.1371/journal.pone.0017573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Schertz K, Paterson AH. Comparative analysis of QTLs affecting plant height and maturity across the Poaceae, in reference to an interspecific sorghum population. Genetics. 1995;141:391–411. doi: 10.1093/genetics/141.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A, Chao S, Asoro F, Heffner EL, Hayashi T, Iwata H, et al. Genomic selection in plant breeding: knowledge and prospects. Adv Agron. 2011;110:77–123. [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massman JM, Gordillo A, Lorenzana RE, Bernardo R. Genomewide predictions from maize single-cross data. Theor Appl Genet. 2012. pp. 13–22. [DOI] [PubMed]
- Matsuoka Y, Vigouroux Y, Goodman MM, Sanchez GJ, Buckler E, Doebley J. A single domestication for maize shown by multilocus microsatellite genotyping. Proc Natl Acad Sci USA. 2002;99:6080–6084. doi: 10.1073/pnas.052125199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. A cytological demonstration of the location of an interchange between two non-homologous chromosomes of Zea mays. Proc Natl Acad Sci USA. 1930;16:791–796. doi: 10.1073/pnas.16.12.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci USA. 1950;36:344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen MD, Kresovich S, Villeda HS, Bradbury P, Li H, Sun Q, et al. Genetic properties of the maize nested association mapping population. Science. 2009;325:737–740. doi: 10.1126/science.1174320. [DOI] [PubMed] [Google Scholar]
- Miclaus M, Xu J-H, Messing J. Differential gene expression and epiregulation of alpha zein gene copies in maize haplotypes. PLoS Genet. 2011;7:e1002131. doi: 10.1371/journal.pgen.1002131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll RH, Lindsey MF, Robinson HF. Estimates of genetic variances and level of dominance in maize. Genetics. 1964;49:411–423. doi: 10.1093/genetics/49.3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles S, Peiffer J, Brown PJ, Ersoz ES, Zhang Z, Costich DE, et al. Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell. 2009;21:2194–2202. doi: 10.1105/tpc.109.068437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. The population genetics of adaptation: the distribution of factors fixed during adaptive evolution. Evolution. 1998;52:935–949. doi: 10.1111/j.1558-5646.1998.tb01823.x. [DOI] [PubMed] [Google Scholar]
- Palaisa KA, Morgante M, Williams M, Rafalski A. Contrasting effects of selection on sequence diversity and linkage disequilibrium at two phytoene synthase loci. Plant Cell. 2003;15:1795–1806. doi: 10.1105/tpc.012526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland JA, Bradbury PJ, Buckler ES, Nelson RJ. Genome-wide nested association mapping of quantitative resistance to northern leaf blight in maize. Proc Natl Acad Sci USA. 2011;108:6893–6898. doi: 10.1073/pnas.1010894108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quijada P, Shannon LM, Glaubitz JC, Studer AJ, Doebley J. Characterization of a major maize domestication QTL on the short arm of chromosome 1. Maydica. 2009;54:401–408. [Google Scholar]
- Rafalski R, Morgante M. Corn and humans: recombination and linkage disequilibrium in two genomes of similar size. Trends Genet. 2004;20:103–111. doi: 10.1016/j.tig.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Rhodes MM. Cytoplasmic inheritance of male sterility in Zea mays. Science. 1931;73:340–341. doi: 10.1126/science.73.1891.340. [DOI] [PubMed] [Google Scholar]
- Riedelsheimer C, Czedik-Eysenberg A, Grieder C, Lisec J, Technow F, Sulpice R, et al. Genomic and metabolic prediction of complex heterotic traits in hybrid maize. Nat Genet. 2012b;44:217–220. doi: 10.1038/ng.1033. [DOI] [PubMed] [Google Scholar]
- Riedelsheimer C, Lisec J, Czedik-Eysenberg A, Sulpice R, Flis A, Grieder C, et al. Genome-wide association mapping of leaf metabolic profiles for dissecting complex traits in maize. Proc Natl Acad Sci USA. 2012a;109:8872–8877. doi: 10.1073/pnas.1120813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé AP, Bomblies K, Laitinen RAE, Yant L, Mott R, Weigel D. Genetic architecture of flowering-time variation in Arabidopsis thaliana. Genetics. 2011;188:421–433. doi: 10.1534/genetics.111.126607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi S, Sponza G, Morgante M, Tomes D, Niu X, Fengler KA, et al. Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc Natl Acad Sci USA. 2007;104:11376–11381. doi: 10.1073/pnas.0704145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- Schön CC, Dhillon BS, Utz HF, Melchinger AE. High congruency of QTL positions for heterosis of grain yield in three crosses of maize. Theor Appl Genet. 2010;120:321–332. doi: 10.1007/s00122-009-1209-9. [DOI] [PubMed] [Google Scholar]
- Setter TL, Yan J, Warburton M, Ribaut J-M, Xu Y, Sawkins M, et al. Genetic association mapping identifies single nucleotide polymorphisms in genes that affect abscisic acid levels in maize floral tissues during drought. J Exp Bot. 2011;62:701–716. doi: 10.1093/jxb/erq308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull G. The composition of a field of maize. Rep Am Breeders Assoc. 1908;4:296–301. [Google Scholar]
- Stich B. Comparison of mating designs for establishing nested association mapping populations in maize and Arabidopsis thaliana. Genetics. 2009;183:1525–1534. doi: 10.1534/genetics.109.108449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuber CW, Lincoln SE, Wolff DW, Helentjaris T, Lander ES. Identification of genetic factors contributing to heterosis in a hybrid from two elite maize inbred lines using molecular markers. Genetics. 1992;132:823–839. doi: 10.1093/genetics/132.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer A, Zhao Q, Ross-Ibarra J, Doebley J. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet. 2011;43:1160–1163. doi: 10.1038/ng.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer AJ, Doebley JF. Do large effect QTL fractionate? AcCase study at the maize domestication QTL teosinte branched1. Genetics. 2011;188:673–681. doi: 10.1534/genetics.111.126508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner R, Briskine R, Schaefer R, Hufford MB, Ross-Ibarra J, Myers CL, et al. Reshaping of the maize transcriptome by domestication. Proc Natl Acad Sci USA. 2012;109:11878–11883. doi: 10.1073/pnas.1201961109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner RA, Eichten SR, Kumari S, Tiffin P, Stein JC, Ware D, et al. Pervasive gene content variation and copy number variation in maize and its undomesticated progenitor. Genome Res. 2010;20:1689–1699. doi: 10.1101/gr.109165.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.) Proc Natl Acad Sci USA. 2001;98:9161–9166. doi: 10.1073/pnas.151244298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Tian F, Bradbury PJ, Brown PJ, Hung H, Sun Q, Flint-Garcia S, et al. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat Genet. 2011;43:159–162. doi: 10.1038/ng.746. [DOI] [PubMed] [Google Scholar]
- van Heerwaarden J, Doebley J, Briggs WH, Glaubitz JC, Goodman MM, Sanchez-Gonzales J, et al. Genetic signals of origin, spread, and introgression in a large sample of maize landraces. Proc Natl Acad Sci USA. 2011;108:1088–1092. doi: 10.1073/pnas.1013011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wagler TN, Li B, Zhao Q, Vigouroux Y, Faller M, et al. The origin of the naked grains of maize. Nature. 2005;436:714–719. doi: 10.1038/nature03863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Dooner HK. Remarkable variation in maize genome structure inferred from haplotype diversity at the bz locus. Proc Natl Acad Sci USA. 2006;103:17644–17649. doi: 10.1073/pnas.0603080103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber AL, Briggs WH, Rucker J, Baltazar BM, Sanchez-Gonzales J, Feng P, et al. The genetic architecture of complex traits in teosinte (Zea mays ssp. parviglumis): new evidence from association mapping. Genetics. 2008;180:1221–1232. doi: 10.1534/genetics.108.090134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber VS, Araus JL, Cairns JE, Sanchez C, Melchinger AE, Orsini E, et al. Prediction of grain yield using reflectance spectra of canopy and leaves in maize plants grown under different water regimes. Field Crops Res. 2012;128:82–90. [Google Scholar]
- Weng J, Xie C, Hao Z, Wang J, Liu C, Li M, et al. Genome-wide association study identifies candidate genes that affect plant height in Chinese elite maize (Zea mays L.) inbred lines. PLoS One. 2011;6:e29229. doi: 10.1371/journal.pone.0029229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S, Bi IV, Schroedr SG, Yamasaki M, Doebley JF, McMullen MD, et al. The effects of artificial selection on the maize genome. Science. 2005;308:1310–1314. doi: 10.1126/science.1107891. [DOI] [PubMed] [Google Scholar]
- Yan J, Kandianis CB, Harjes CE, Bai L, Kim E-H, Yang X, et al. Rare genetic variation at Zea mays crtRB1 increases beta-carotene in maize grain. Nat Genet. 2010;42:322–327. doi: 10.1038/ng.551. [DOI] [PubMed] [Google Scholar]
- Yang X, Gao S, Xu S, Zhang Z, Prasanna BM, Li L, et al. Characterization of a global germplasm collection and its potential utilization for analysis of complex quantitative traits in maize. Mol Breed. 2011;28:511–526. [Google Scholar]
- Yu J, Holland JB, McMullen MD, Buckler ES. Genetic design and statistical power of nested association mapping in maize. Genetics. 2008;178:539–551. doi: 10.1534/genetics.107.074245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Gur A, Gibon Y, Sulpice R, Flint-Garcia S, McMulled D, et al. Genetic analysis of central carbon metabolism unveils an amino acid substitution that alters maize NAD-dependent isocitrate dehydrogenase activity. PLoS One. 2010b;5:e9991. doi: 10.1371/journal.pone.0009991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Gibon Y, Gur A, Chen C, Lepak N, Hohne M, et al. Fine quantitative trait loci mapping of carbon and nitrogen metabolism enzyme activities and seedling biomass in the maize IBM mapping population. Plant Phys. 2010a;154:1753–1765. doi: 10.1104/pp.110.165787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Thuillet A-C, Uhlmann NK, Weber A, Rafalski JA, Allen SM, et al. The role of regulatory genes during maize domestication: evidence from nucleotide polymorphism and gene expression. Genetics. 2008;178:213–2143. doi: 10.1534/genetics.107.085506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Weber AL, McMullen MD, Guill K, Doebley J. MADS-box genes of maize: frequent targets of selection during domestication. Genet Res Camb. 2011;93:65–75. doi: 10.1017/S0016672310000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Allen WB, Roesler K, Williams M, Zhang S, Li J, et al. A phenylalanine in DGAT is a key determinant of oil content and composition in maize. Nat Genet. 2008;40:367–372. doi: 10.1038/ng.85. [DOI] [PubMed] [Google Scholar]